Abstract
Background
Viral RNA amplification by real-time RT-PCR still represents the gold standard for the detection of SARS-CoV-2, but the development of rapid, reliable and easy-to-perform diagnostic methods is crucial for public health, because of the need of shortening the time of result-reporting with a cost-efficient approach.
Objectives
The aim of our research was to assess the performance of FREND™ COVID-19 Ag assay (NanoEntek, South Korea) as a ultra-rapid frontline test for SARS-CoV-2 identification, in comparison with RT-PCR and another COVID-19 antigen fluorescence immunoassay (FIA).
Study design
The qualitative FIA FREND™ test, designed to detect within 3 min the Nucleocapsid protein of SARS-CoV-2, was evaluated using nasopharyngeal swabs in Universal Transport Medium (UTM™, Copan Diagnostics Inc, US) from suspected COVID-19 cases who accessed the Emergency Room of the Ospedale Policlinico San Martino, Genoa, Liguria, Northwest Italy. Diagnostic accuracy was determined in comparison with SARS-CoV-2 RT-PCR and STANDARD F™ COVID-19 Ag FIA test (SD BIOSENSOR Inc., Republic of Korea).
Results
In November 2020, 110 nasopharyngeal samples were collected consecutively; 60 resulted RT-PCR positive. With respect to RT-PCR results, sensitivity and specificity of FREND™ COVID-19 Ag test were 93.3 % (95 % CI: 83.8−98.2) and 100 % (95 % CI: 92.9−100), respectively.
FREND™and STANDARD F™ COVID-19 Ag FIA assays showed a concordance of 96.4 % (Cohen’s k = 0.93, 95 % CI: 0.86−0.99).
Conclusions
FREND™ FIA test showed high sensitivity and specificity in nasopharyngeal swabs. The assay has the potential to become an important tool for an ultra-rapid identification of SARS-CoV-2 infection, particularly in situations with limited access to molecular diagnostics.
Keywords: SARS-CoV-2 infection, COVID-19 identification, Rapid antigen diagnostic test, Fluorescence immunoassay (FIA)
1. Background
According to the World Health Organization (WHO), Real Time reverse transcription polymerase chain reaction (RT-qPCR) on respiratory samples (nasopharyngeal swabs) is the current recommended laboratory method to diagnose SARS-CoV-2 acute infection, the cause of 2019 coronavirus disease (COVID-19) (Corman et al., 2020; OMS, 2020).
In 2020, the increasing molecular analysis demand due to the COVID-19 pandemic raised several critical issues such as requirement of special equipment, laboratory reagents and skilled staff.
Therefore, looking for alternative diagnostic solutions to implement a molecular screening strategy extended to a large number of subjects and to counter the SARS-CoV-2 spread has become a priority for the health care systems (Mak et al., 2020).
In the last months, several easy to perform rapid antigen detection tests were developed and recommended in some countries as first line laboratory strategy for COVID-19 diagnostic (Scohy et al., 2020; Porte et al., 2020).
2. Objectives
The aim of the present research was to assess the performance of FRENDTM™COVID-19 Ag assay (NanoEntek, South Korea) as a frontline test for SARS-CoV-2 identification in comparison to the available molecular techniques and to the STANDARD F COVID-19 Ag FIA test (SD BIOSENSOR Inc., Republic of Korea), a rapid fluorescent immunoassay test currently used in clinical practice (Cerutti et al., 2020).
3. Study design
In November 2020, at the regional reference laboratory for COVID-19 diagnostic located into Ospedale Policlinico San Martino, Hygiene Unit, Genoa, Liguria, Northwest Italy, 110 nasopharyngeal samples from 110 patients who accessed the Emergency Room of the hospital with symptoms attributable to SARS-CoV-2 infection were consecutively collected. Swabs were carried out using a flocked probe and eluted in Universal Transport Medium (UTM™, Copan Diagnostics Inc, US). RT-qPCR, FREND™ COVID-19 Ag and STANDARD F COVID-19 Ag FIA tests were conducted within 8 h from the swabs’ arrival at the laboratory.
Each respiratory swab was set up for PCR using the extraction-free method on Nimbus IVD, (Seegene, South Korea) using the Allplex™ SARS-CoV-2 Assay kit (Seegene, South Korea), according to the manufacturer’s instructions. The obtained material was tested for the identification of SARS-CoV-2 through a one-step multiplex RT-qPCR on Biorad CFX96™ thermal cycler. It targeted the nucleoprotein region (N), the RNA-dependent RNA-polymerase region (RdRp), the Spike protein (S) and the Envelop region (E), respectively. Samples showing a cycle threshold (Ct) value <35 for all the target genes were considered positive.
Positive swabs were tested in parallel with FREND™ COVID-19 Ag rapid diagnostic test for SARS-CoV-2 infection. It was a qualitative fluorescence immunoassay (FIA) designed to detect in nasopharyngeal swabs within 3 min the Nucleocapsid protein (N) of SARS-CoV-2 and intended for use with FREND™ system (NanoEntek, South Korea). Results were displayed as positive or negative for COVID-19 antigen and a numeric value was provided (≥1.00 for positive samples).
As comparison, we performed the STANDARD F COVID-19 Ag FIA test (SD BIOSENSOR Inc., Republic of Korea). It was a fluorescent immunoassay capable of detecting SARS-CoV-2 viral nucleoprotein antigens in human nasopharyngeal UTM swab specimens. Results were provided within 30 min and the observed values were expressed as a CutOff Index (COI) value, with COI ≥ 1 to be considered as sample positive for SARS-CoV-2 antigen.
IBM SPSS Statistics version 25 (manufactured by IBM in Armonk, NY, USA) was used to calculate 95 % confidence interval for sensitivity, specificity, negative and positive predictive values, and Cohen’s kappa coefficient.
4. Results
Fiftyfive percent (60/110) of swabs tested positive in RT-qPCR while in the remaining 45 % (50/110) no virus was detected. Within the 60 positive samples, 23 had a Ct value for the N gene lower than 26, 32 a Ct ranging from 26 to 30 and the remaining 5 a Ct ranging from 31 and 35.
In Table 1 , FREND™ COVID-19 Ag test results are shown in comparison with STANDARD F COVID-19 Ag FIA test and RT-qPCR results, for positive and negative samples, according Ct values for N gene and time from the onset of symptoms to sample obtention. With respect to RT-qPCR results, sensitivity, specificity, negative and positive predictive values of FREND™ COVID-19 Ag test were 93.3 % (95 % CI: 83.8−98.2), 100 % (95 % CI: 92.9−100), 92.5 % (95 % CI: 82.6–96.9) and 100 % (95 % CI: n.a.), respectively. Concordance between the two techniques was 96.3 % (Cohen’s k = 0.93, 95 % CI: 0.86−0.99). As far as STANDARD F COVID-19 Ag FIA results, sensitivity, specificity, negative and positive predictive values were 86.7 % (95 % CI: 75.4−94.1), 100 % (95 % CI: 92.9−100), 86.0 % (95 % CI: 76.3–92.1) and 100 % (95 % CI: n.a.), respectively, compared with RT-PCR. FREND™ COVID-19 Ag test showed higher sensitivity both for samples with N gene Ct values from 26 to 35 and for samples collected up to 14 days after the onset of symptoms. Concordance between the two techniques was 92.7 % (Cohen’s k = 0.86, 95 % CI: 0.76−0.95). The two Ag FIA assays showed a concordance of 96.4 % (Cohen’s k = 0.93, 95 % CI: 0.86−0.99). In particular, for 4 samples (2 with a Ct value for N gene <30 and 2 with a Ct values from 31 to 35) there were discordant results between the two FIA methods. For these swabs, the mean value for FREND™ COVID-19 Ag test was 1.78, whereas for STANDARD F COVID-19 Ag FIA test was 0.88.
Table 1.
Sensitivity and specificity of FREND™ COVID-19 Ag test (NanoEntek, South Korea) and STANDARD F COVID-19 Ag FIA test (SD BIOSENSOR Inc., Republic of Korea), compared with RT-qPCR results. Swabs were divided into 3 groups based on RT-qPCR Ct values for N gene (<26, 26-30, 31-35) and into 2 groups according time from the onset of symptoms to sample obtention (<7 days, 7-14 days).
| Swabs | FRENDTM COVID-19 Ag |
STANDARD F COVID-19 Ag FIA |
||
|---|---|---|---|---|
| Sensitivity(%, 95 % C.I.) | Specificity(%, 95 % C.I.) | Sensitivity(%, 95 % C.I.) | Specificity(%, 95 % C.I.) | |
| RT-qPCR positive (n = 60) | 93.3 (83.8−98.2) | 86.7 (75.4−94.1) | ||
| RT-qPCR negative (n = 50) | 100 (92.9−100) | 100 (92.9−100) | ||
| N gene Ct values | ||||
| Ct <26 (n = 23) | 100 (85.2−100) | 100 (85.2−100) | ||
| Ct 26−30 (n = 32) | 96.9 (83.8−99.9) | 90.6 (75−98) | ||
| Ct 31−35 (n = 5) | 40 (5.3−85.3) | 0 (0−52.1) | ||
| Time from the onset of symptoms | ||||
| <7 days (n = 45) | 95.6 (84.9−99.5) | 88.9 (76−96.3) | ||
| 7−14 days (n = 15) | 86.7 (59.5−98.3) | 80 (51.9−95.7) | ||
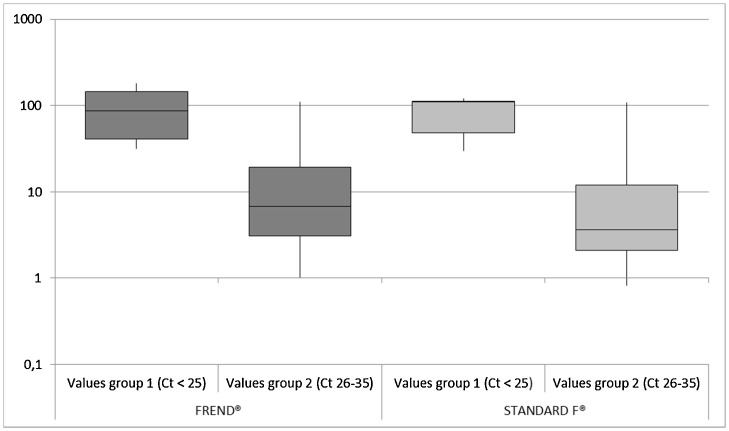
Fig. 1 compares numeric values provided by FREND™ COVID-19 Ag test and STANDARD F COVID-19 Ag FIA test for positive SARS-CoV-2 swabs in RT-qPCR. For positive samples with a RT-qPCR Ct value lower than 25, median value provided by FREND™was 87.1 (interquartile range 40.8–146.0), whereas median COI provided by STANDARD F was 110.6 (interquartile range 48.2-112.6); as far as positive samples with a RT-qPCR Ct value ranging from 26 to 35, median value provided by FREND™was 5.9 (interquartile range 2.1–18.3), whereas median COI provided by STANDARD F was 3.3 (interquartile range 1.7–11.6). With respect to time from the onset of symptoms to sample obtention, for positive samples collected up to 7 days after onset of symptoms median value provided by FREND™was 30.2 (interquartile range 5.2–71.3), whereas median COI provided by STANDARD F was 15.2 (interquartile range 3.1–50.2); as far as positive samples collected from 7 to 14 days after onset of symptoms, median value provided by FREND™was 7.6 (interquartile range 2.4–58.5), whereas median COI provided by STANDARD F was 6.2 (interquartile range 1.9–71.4).
Fig. 1.
Box plot of numeric values provided by FREND™ COVID-19 Ag test (NanoEntek, South Korea) and STANDARD F COVID-19 Ag FIA test (SD BIOSENSOR Inc., Republic of Korea) for positive SARS-CoV-2 swabs in RT-qPCR. A value ≥1.00 is considered positive for both assay. Group 1: positive samples with a RT-qPCR Ct value for N gene lower than 25. Group 2: positive samples with a RT-qPCR Ct value for N gene ranging from 26 to 35.
5. Discussion
In the ongoing COVID-19 pandemic context, diagnostic testing for SARS-CoV-2 is crucial in order to limit the spread of the virus as well as appropriately manage infected patients. Different diagnostic test manufacturers have developed rapid tests based on SARS-CoV-2 proteins detection in respiratory samples. However, the analytical performances of these rapid antigenic tests depend on different factors including the viral load, the quality of the specimen and how it is processed.
In this study, we determined the performance characteristics of the FREND™ COVID-19 Ag test for detecting SARS-CoV-2 virus in respiratory samples and compared the results with RT-qPCR as the gold standard and another FIA antigen test. Our data showed that FREND™ COVID-19 Ag proved to be more sensitive and had several advantages such as the rapid answer in 3 min and the non-requirement of special equipment or personnel skills compared with molecular techniques.
The number of swabs collected and tested in this study could be a limit for our research; moreover, the Ct values of positive samples were never higher than 35. However, the study was performed in a “real world” clinical setting and within a few days, in order to evaluate the possible use of FREND™ COVID-19 Ag as a screening tool in a large reference hospital.
Although negative results cannot rule out SARS-CoV-2 infection and made this test of little help when it is necessary to evaluate the progress of the disease and therefore, the state of recovery of the patient, our data suggest that FREND™ system could be a useful device particularly in situations with limited access to molecular diagnostics and the need of rapid results in order to appropriately manage people and resources, i.e. Emergency Rooms or outpatient facilities.
In conclusion, FREND™ COVID-19 Ag assay represents a valid frontline test for COVID-19 screening and it could ease the burden on the laboratories, reducing the time spent for diagnosis and the use of RT-qPCR.
Funding
We thank Arrow Diagnostics Srl for the donation of the FREND™COVID-19 Ag kits to pursue the study. No other specific grant from public funding agencies was received.
CRediT authorship contribution statement
Andrea Orsi: Writing - original draft, Formal analysis, Conceptualization. Beatrice Marina Pennati: Writing - original draft, Formal analysis, Conceptualization. Bianca Bruzzone: Conceptualization, Writing - review & editing, Supervision. Valentina Ricucci: Validation, Investigation, Data curation. Diego Ferone: Validation, Investigation. Paolo Barbera: Validation, Investigation. Eleonora Arboscello: Validation, Investigation. Chiara Dentone: Validation, Investigation. Giancarlo Icardi: Conceptualization, Writing - review & editing, Supervision.
Declaration of Competing Interest
None.
Acknowledgements
We thank all staff of the Hygiene Unit, Regional Reference Laboratory for COVID-19 diagnostic, for the technical assistance offered for the study conduction. We special thank Simona Boccotti, Patrizia Caligiuri, Rita Cerruti, Valerio Chessa, Vanessa De Pace, Gaetano De Rosa, Monica Ferraris, Barbara Galano, Giulia Guarona, Nicola Nigro, Rexhina Qosja, Nadia Randazzo, Matteo Theimer and Serena Varesano for their precious and indispensable work during the current SARS-CoV-2 pandemic.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2021.114201.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Cerutti F., Burdino E., Milia M.G., Allice T., Gregori G., Bruzzone B., et al. Urgent need of rapid tests for SARS CoV-2 antigen detection: evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J. Clin. Virol. 2020;132 doi: 10.1016/j.jcv.2020.104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C.S., Lam E.T., et al. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;129:104500. doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OMS Laboratory testing strategy recommendations for COVID-19. Interim Guidance. 2020;21(March) [Google Scholar]
- Porte L., Legarraga P., Vollrath V., Aguilera X., Munita J.M., Araos R., et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int. J. Infect. Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scohy A., Anantharajah A., Bodéus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.