Abstract
The introduction of immune checkpoint inhibitor (ICI)-based therapy for non-oncogene addicted non-small cell lung cancer (NSCLC) has significantly transformed the treatment landscape of the disease. Inhibitors of the programmed cell death protein 1/programmed death-ligand 1 (PD-1/PD-L1) immune checkpoint axis, which were initially considered as a late-line treatment option, gradually became the standard of care as first-line treatment for subgroups of NSCLC patients. However, a significant fraction of patients either fails to respond or progresses after a partial response to ICI treatment. Thus, the identification of mechanisms responsible for innate and acquired resistance to immunotherapy within a rapidly evolving tumor microenvironment (TME) is urgently required, as is the identification of reliable predictive biomarkers beyond PD-L1 expression. The deregulation of the epigenome is a key driver of cancer initiation and progression, and it has also been shown to drive therapeutic resistance. Tumor education of infiltrating myeloid cells towards an immuno-suppressive phenotype as well as induction of T-cell dysfunction in the TME is also driven by epigenome reprogramming. As it stands and, given their dynamic nature, epigenetic changes in cancer and non-cancer cells represent an attractive target to increase immunotherapy activity in NSCLC. Accordingly, clinical trials of combinatorial immuno-epigenetic drug regimens have been associated with tumor response in previously immunotherapy-resistant NSCLC patients irrespective of their PD-L1 status. Moreover, epigenetic signatures might represent valuable theragnostic biomarkers as they can be assayed easily in liquid biopsy and provide multiple layers of information. In this review, we discuss the current knowledge regarding the dysregulated epigenetic mechanisms contributing to immunotherapy resistance in NSCLC. Although the clinical data are still maturing, we highlight the attractive perspective that the synergistic model of immuno-epigenetic strategies might overcome the current limitations of immunotherapy alone and will be translated into durable clinical benefit for a broader NSCLC population.
Keywords: anti-PD-1, combination immunotherapy, epigenetics, immunotherapy, non-small cell lung cancer (NSCLC)
Introduction
Lung cancer continues to be the primary cause of cancer-related deaths for both genders.1 Non-small cell lung cancer (NSCLC), accounting approximately for 85% of newly diagnosed lung cancer cases, is subclassified into two main histologies: lung adenocarcinoma (LUAD) and squamous cell lung carcinoma (SQLC).2 Following histopathological diagnosis, NSCLC tumors and, especially, LUAD are tested routinely for identifying druggable molecular alterations in genes encoding primarily receptor tyrosine kinases. Epidermal growth factor receptor (EGFR)-activating mutations are the most frequent alterations in NSCLC; their prevalence ranges from 10% to 30%, increasing up to 60% in East Asian individuals. The second most common alteration is the anaplastic lymphoma kinase (ALK) gene translocation that can be found in 2–5% of NSCLC. Other relatively common molecular alterations that have been identified in 0.5–3% of NSCLC include ROS1 and RET gene rearrangements, NRTK gene fusions, BRAF V600E, and MET exon 14 skipping mutations. However, these kinase inhibitors (KIs)-sensitizing alterations are found predominantly in LUAD.3 Other therapeutic options, such as radiation, chemotherapy, or immunotherapy, either as single agents or in combination are available for LUAD patients testing negative for kinase alterations as well as for SQLC, for which approved targeted therapies are still lacking.4
The identification of inhibitory immune checkpoint molecules represented a groundbreaking discovery that provided new targets for anticancer treatment.5 Immune checkpoint inhibitors (ICIs) such as monoclonal antibodies against two immune checkpoint molecules, cytotoxic T-lymphocyte antigen 4 (CTLA-4) and programmed cell death protein 1 (PD-1), were introduced rapidly in clinical practice for the treatment of several solid tumors.6 PD-1 is normally expressed on mature and active immune cells, such as T and B lymphocytes and natural killers in peripheral tissues, while its ligand programmed death-ligand 1 (PD-L1) is expressed mainly by macrophages and dendritic cells.7,8 CTLA-4 is expressed primarily by T-cells including T regulatory cells, while B7 ligands (including CD80) are expressed by antigen-presenting cells (APC).9 Both the PD-1/PD-L1 and CTLA-4 axes act as immunological breaks for halting immune response at the appropriate time by controlling T-cell tolerance and preventing autoimmunity.10 Among other mechanisms, cancer cells also highjack this immune checkpoint mechanism, for example, by expressing PD-L1 to prevent anti-tumor immunity and evade immunological surveillance (Figure 1). Interfering with this PD-1/PD-L1 checkpoint using ICIs proved effective in extending the survival of subsets of lung cancer patients, thus reconfiguring its treatment management.11
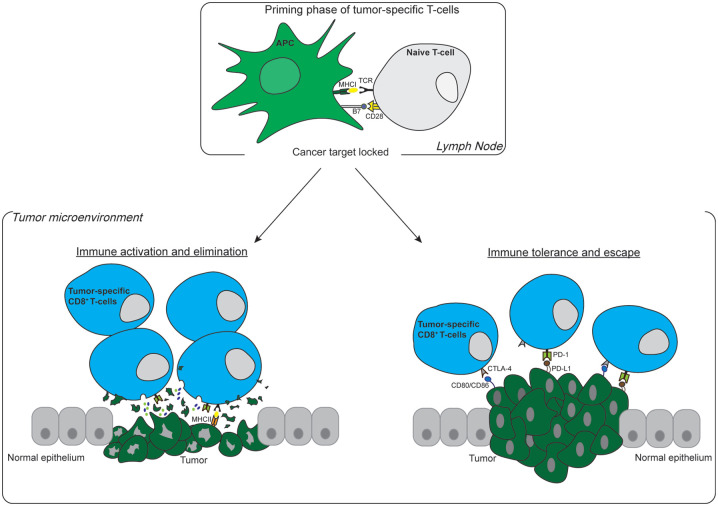
Figure 1.
Fates of CD8+ T-cells upon recognition of tumor antigen. The priming phase of tumor-specific T-cells requires antigen presentation by professional APCs such as activated dendritic cells to the naive T-cells. This procedure takes place in lymph nodes and requires two activation signals. First, the TCR of the CD8+ T-cells binds to the antigen captured in MHCI on the surface of APCs (signal I). Second, B7 ligands located on APCs interact with the CD28 receptor on CD8+ T-cells (signal II). After activation, CD8+ T-cells head to the TME to act against tumor cells. Left panel: CD8+ T-cells recognize tumor antigens and release chemokines and small molecules such as perforin/granzyme resulting in tumor elimination. Right panel: tumor cells express T-cell exhaustion-inducing ligands (PD-L1 and CD80/CD86), which interact with their receptors on the surface of T-cells (PD-1 and CTLA-4) and allow tumor cells to escape immune surveillance.
APC, antigen-presenting cell; CTLA-4, cytotoxic T-lymphocyte-associated protein 4; MHCI, major histocompatibility complex class I; PD-1, programmed cell death protein 1; PD-L1, programmed death-ligand 1; TCR, T-cell receptor; TME, tumor microenvironment.
NSCLC is among the tumor types most responsive to anti-PD-1/PD-L1 therapies.12 Results of the KEYNOTE-024 study enrolling NSCLC patients harboring greater than 50% tumoral PD-L1 expression demonstrated that the use of the anti-PD-1 pembrolizumab as first-line therapy significantly extended the survival of patients with NSCLC, including those with SQLC.13 However, a considerable fraction of patients with NSCLC either does not respond or even experiences rapid enlargement of tumor during immunotherapy – the latter termed hyperprogressive disease.14–16 Furthermore, it is still unclear whether patients with oncogene-driven NSCLC, eligible for KI treatment, benefit from immune checkpoint blockade.17 Currently, assessment of PD-L1 tumor expression in formalin-fixed paraffin-embedded (FFPE) tissue samples by immunohistochemistry (IHC) is the only clinically validated biomarker to identify patients likely to respond to immunotherapy.18 Different clinical studies have reported that high PD-L1 protein expression is associated with improved survival in patients with NSCLC treated with immunotherapy.19,20 While useful for enriching responses to anti-PD-1/PD-L1 therapies, the predictive value of PD-L1 IHC is not perfectly accurate and several other biomarkers are currently under investigation. Indeed, latest data suggest that the assessment of tumor mutational burden (TMB), tumor neoantigens, and human leukocyte antigen (HLA) expression in this specific context might add value to the evaluation of PD-L1 expression.21 Interestingly, all the events described can be controlled directly or indirectly by epigenetic mechanisms, including DNA methylation, post-translational histone modifications, and non-coding RNAs.22 In this review, we describe how epigenetics control immune mechanisms in NSCLC and specifically highlight different candidate mechanisms that govern PD-1/PD-L1 regulation. Moreover, we present available clinical findings that involve predominantly NSCLC patients with no actionable genetic alterations. In light of the necessity of maximizing immunotherapy benefit for NSCLC patients, epigenetic biomarkers and targets might be useful to the efficient stratification of patients as well as to the design of innovative synergistic treatment modalities.
Predictive and prognostic role of circulating and tissue non-coding RNAs in ICI-treated NSCLC
Non-coding ribonucleic acids (ncRNAs) are functional RNA molecules that are not translated into proteins. ncRNAs are divided into two main groups: (i) short ncRNAs, which usually do not exceed a length of 200 nucleotides; (ii) and long ncRNAs, which are transcripts containing more than 200 nucleotides.23 MicroRNAs (miRNAs) are a class of short ncRNAs, around 22 nucleotides in length, that participate in the post-transcriptional regulation of protein-coding genes. In general, miRNAs bind through their seed region on complementary sequences located at the 3′ untranslated region (UTR) of target mRNAs. Depending on the degree of miRNA/mRNA complementarity, mRNA is either repressed or degraded, leading to translation inhibition.24 Numerous miRNAs have been found deregulated in NSCLC, and their aberrant levels have been associated with resistance to chemotherapy or KIs as well as with poor survival.25,26 Moreover, miRNAs and circular RNAs (circRNAs), due to their abundance in biofluids such as blood, saliva, and urine, have been studied extensively and characterized as non-invasive predictive and prognostic biomarkers in NSCLC.27 Specifically, a plethora of ncRNAs has been suggested as determinants of immunotherapy responses in NSCLC patients (Table 1).Serum miRNA profile was assessed by next-generation sequencing in a training set of 20 NSCLC patients before treatment initiation with nivolumab, a PD-1 blocking antibody.
Table 1.
Possible determinants of immunotherapy responses in NSCLC patients.
| Candidate biomarker | Type of biomarker | Expression/presence | Material origin | Correlation | Reference |
|---|---|---|---|---|---|
| 7 miRNA-based signature | Prognostic | 6 upregulated miRNAs/1 downregulated miRNA | Serum | OS >6 months after nivolumab treatment | Halvorsen et al.28 |
| MSC (24 miRNAs) | Prognostic | High/intermediate/low-risk MSC | Plasma | High-risk MSC and worse prognosis after ICI treatment | Boeri et al.29 |
| miRNA-320b miRNA-320c miRNA-320d | Predictive | High expression | Plasma | Poor response to PD-1 blockade/disease progression | Peng et al.30 |
| miRNA-320b miRNA-375 | Predictive | Downregulated before nivolumab initiation | Plasma | Clinical benefit | Costantini et al.31 |
| miRNA-33a | Prognostic | High | Tissue | Downregulation of immune checkpoint molecules | Boldrini et al.34 |
| miRNA-140 | − | Downregulated | Tissue | Higher tumor size and PD-L1 upregulation | Xie et al.35 |
| Methylation status (CpG sites) | Predictive | High | Tissue | High TMB | Cai et al.36 |
| EPIMMUNE signature/Unmethylated FOXP1 | Prognostic | Presence | Tissue | Response to anti-PD-1 Abs/Tumor-infiltrating immune cells | Duruisseaux et al.37 |
| Methylation status PDCD1/CTLA-4 | - | Low | Tissue | Higher expression of PDCD1/CTLA-4 transcripts | Marwitz et al.38 |
| PD-L1 promoter polymorphism/PD-L1 CNG | - | CC genotype in rs822335/presence | Tissue | Higher expression of PD-L1 | Krawczyk et al.39 |
ICI, immune checkpoint inhibitor; miRNA, microRNA; MSC, miRNA-signature classifier; NSCLC, non-small cell lung cancer; OS, overall survival; PD-1, programmed cell death protein 1; PD-L1, programmed death-ligand 1; TMB, tumor mutational burden.
A miRNA-based signature of six upregulated (miRNA-215-5p, miRNA-411-3p, miRNA-493-5p, miRNA-494-3p, miRNA-495-3p, miRNA-548j-5p) and one downregulated (miRNA-93-3p) miRNAs was identified as a prognostic factor and associated with overall survival (OS) >6 months. The findings were confirmed in a cohort of 31 NSCLC patients by a quantitative PCR (qPCR) assay, which demonstrated 71% sensitivity and 90% specificity. However, this potentially prognostic signature remains to be validated in a larger and independent cohort.28 Plasma is another rich source of circulating molecules. Indeed, a plasma miRNA-signature classifier (MSC) involving reciprocal ratios of 24 miRNAs categorized NSCLC patients into three risk levels (low, intermediate, and high) regarding the likelihood of experiencing benefit from ICIs. Application of this algorithm to a prospective study enrolling 140 immunotherapy-naïve NSCLC patients demonstrated that patients belonging to MSC high-risk have worse prognosis as opposed to MSC intermediate and low-risk patients. Interestingly, all patients with high-risk MSC failed to respond to immunotherapy, while longitudinal collection of plasma samples and assessment of MSC risk level mirrored disease evolution during ICI treatment, suggesting its potential use as a disease-monitoring tool.29 In another study, plasma samples from 30 EGFR/ALK wild-type NSCLC patients were collected prior to anti-PD-1/PD-L1 therapy. Detection of high levels of three exosomal miRNAs (miRNA-320b, miRNA-320c, miRNA-320d) correlated with poor response to PD-1 blockade and disease progression.30 The authors also reported the miRNA-125b-5p as the only miRNA to be differentially expressed (downregulated) comparing the pre- and post-treatment plasma miRNA profile of patients with partial responses to anti-PD-1.30 In keeping with this, miRNA-125b-5p was suggested as a suppressor of T-cell activity and inducer of immunotherapy resistance by abrogating the secretion of interferon gamma (IFN-γ) and tumor necrosis factor alpha (TNF-α) from T-cells, which are known to trigger powerful anti-cancer effects.30 Another study analyzing miRNA profile in plasma collected at baseline of nivolumab treatment from NSCLC patients, showed that elevated levels of miRNA-320b and miRNA-375 were associated with progression of disease (n = 9) as opposed to the clinical benefit observed in patients with low levels of the two miRNAs.31 Interestingly, these miRNAs interact with genes that are associated with proliferation (MYC, Hippo pathway) and immune-related pathways (Wnt pathway, JAK2, TGF-β2), and have also been associated with ICI resistance and modulation of CD8+-mediated immune response in other tumor types, such as melanoma and colon cancer.31–33 Interestingly, miRNA-33a was found upregulated in young female patients with early stage and low-grade LUAD who experienced better survival. MiRNA-33a levels were inversely correlated with the expression of its direct targets PD-1, PD-L1, and CTLA-4 while LUAD patients with miRNA-33a/PD-1 high ratio had a better prognosis than those with miRNA-33a/PD-1 low ratio.34 Another miRNA with a potential link to immunity is miRNA-140, which was reported downregulated in NSCLC samples and was associated with increased tumor size. NSCLC patient-derived cells exhibited a negative regulation between miRNA-140 and PD-L1 expression, while PD-L1 was associated positively with the cyclin E – a factor that has been associated with invasive NSCLC and poor prognosis. Functional studies suggested PD-L1 as a direct target of miRNA-140 and, after overexpression of miRNA-140, NSCLC cell proliferation was decreased. Interestingly, genetic perturbation of PD-L1 resulted in a significant decrease of cyclin E, suggesting a potential signaling cascade of miRNA-140/PD-L1/cyclin E.35
Genetic and epigenetic regulators orchestrate PD-L1 expression in NSCLC
Different patterns of genetic and epigenetic alterations could modulate the expression of PD-L1 gene at different levels, establishing an intricate PD-L1 signaling network that permits tumor cells to activate various resistance mechanisms, potentially escaping immune surveillance.40 The p53 protein is a dominant tumor suppressor and more than half of cancers are expected to have inactivating mutations in the TP53 gene. In addition to well-established pro-apoptotic functions, p53 is also implicated in the regulation of immune response.41 Members of the miRNA-34 family are transcriptionally activated by p53 and exert several anti-tumor activities.42 Interestingly, in the absence of wild-type p53, NSCLC cell lines and patient samples expressed lower levels of miRNA-34a and higher levels of PD-L1. In contrast, wild-type p53 cell models and NSCLC samples expressed reduced levels of PD-L1, suggesting an inverse correlation between them, confirmed also by The Cancer Genome Atlas (TCGA) data analysis. Interestingly, a miRNA-34 binding site is located in the 3′ UTR region of the PD-L1 mRNA and miRNA-34 overexpression downregulated PD-L1 protein levels. Collectively, these indications describe a p53/miRNA-34/PD-L1 axis as another mechanism of tumor immune surveillance.43 Exogenous administration of MRX34 – a miRNA-34a mimic-loaded nanoparticle – into a genetically and immunologically compatible mouse model decreased PD-L1 mRNA and protein levels while simultaneously promoted tumor-infiltrating CD8+ cells and IFN-γ, resulting in tumor growth arrest. Intriguingly, the combination of MRX34 with conventional radiotherapy produced the most profound results and restricted even more tumor growth, thus suggesting an innovative immunotherapy approach combined with existing treatments.43
CircRNAs are covalently linked single-stranded RNAs derived from a particular splicing mechanism, called back-splicing. Due to this loop structure, circRNAs are highly resistant to RNA-degrading enzymes.44 After their biogenesis, circRNAs are translocated from nucleus to cytoplasm, where they participate in the regulation of different cellular processes through various mechanisms. Specifically, circRNAs contain multiple miRNA binding sites and act as miRNA sponges, protecting target mRNAs from miRNA-mediated degradation. Moreover, circRNAs can also interact with RNA binding proteins and act as protein sponges, regulating their function.45 The role of circRNAs in disease development has gained attention lately, and their expression has been associated with cancer pathogenesis and development.46 In this regard, recent evidence suggests a potential role for circRNA-002178 in promoting PD-1/PD-L1 axis in LUAD tissues and cell lines by neutralizing miRNA-34a – a PD-L1 suppressor.47 Exosomal circRNA-002178 is also able to shape the tumor microenvironment (TME) by promoting PD-1 expression on CD8+ T-cells. Specifically, circRNA-002178 delivered to cytotoxic T-cells is able to block the activity of the miRNA-28-5p that targets PD-1 by binding to its 3′ UTR.47
Tumor cell transdifferentiation (e.g., epithelial to mesenchymal transition, EMT) is a complex and well-orchestrated process that relies on reprogramming of the cancer epigenome. Usually, the EMT phenotype is associated with more aggressive behavior.48 EMT also plays a pivotal role in immune suppression and it has been proposed that bidirectional regulation between EMT and PD-L1 signaling might contribute to therapy resistance. As an example, a two-step model of triggering PD-L1 expression was reported in A549 LUAD cells undergoing the EMT process. TNF-α/transforming growth factor beta 1 (TGF-β1) downregulated DNA methyl transferases (DNMTs) in A549 LUAD cells, thus promoting demethylation of the PD-L1 promoter. Then, in presence of TNF-α, NF-κB recruitment to the PD-L1 promoter is enhanced, leading to PD-L1 gene induction. Interestingly, NSCLC tissues positive for classical EMT marker vimentin also overexpressed PD-L1.49
Candidate biomarkers of immunotherapy efficacy and epigenetic profile of NSCLC
Lately, TMB has received increased attention as potential predictive biomarker of immunotherapy, thus creating very high expectations. However, the latest clinical trials have reported that TMB status, as assessed by whole exome sequencing (WES), was not significantly associated with pembrolizumab efficacy in patients with NSCLC.50,51 Nevertheless, the potential clinical utility of TMB is still under close investigation because numerous key challenges such as standardization of TMB assessment methodology and accurate definition of tumor type-specific TMB cutoffs need to be addressed.52,53 Recently, emerging insights from the epigenome of NSCLC shed light on the potential utility of TMB in NSCLC treatment management. Integration of DNA methylation and TMB data from WES has been explored recently in 89 Chinese patients with NSCLC. Based on the number of nonsynonymous somatic mutations, patients were divided into low (1.1–2.5 mutations/Mb), medium (2.5–4.1 mutations/Mb), and high (4.2–13.9 mutations/Mb) TMB groups. Patients with high TMB showed elevated levels of methylated CpG sites as compared with patients with low TMB.36 Methylated cytosines are prone to spontaneous deamination, which might ultimately result in a G–T mismatch, thus potentially explaining the link between high level of methylation and increased mutational burden.54,55 The potential association of methylation profile and response to immunotherapy was evaluated also in a multicenter study including stage IV NSCLC patients that have been treated with anti-PD-1 mAbs (nivolumab or pembrolizumab).37 An epigenetic signature of 301 differentially methylated CpG sites (mCpGs), named EPIMMUNE, was identified comparing the levels of mCpGs between responders and non-responders to PD-1 blockade.37 EPIMMUNE positive (responders) patients had improved clinical benefit after PD-1 blockade, in terms of both progression-free survival (PFS) and OS. Intriguingly, this potential biomarker was not correlated with other validated or candidate biomarkers such as PD-L1 expression or TMB. Moreover, tissues from EPIMMUNE positive patients were characterized by the presence of a considerable quantity of tumor-infiltrating lymphocytes (CD4+, CD8+, NK cells), while tissues from EPIMMUNE negative patients were dominated by tumor-associated macrophages, tumor-associated neutrophils, and cancer-associated fibroblasts (CAFs). Of the mCpGs composing the signature, 63% (191/301) were associated with known genes. Among them, a regulatory region of forkhead box P1 (FOXP1) gene presented the highest CpG methylation difference between responders and non-responders. Patients harboring an unmethylated regulatory region of FOXP1 had prolonged PFS. Interestingly, validation of the methylation state of this potential marker was performed by pyrosequencing-based PCR assay, thus representing another cost-effective laboratory option.37
In another cohort of NSCLC patients, hypomethylation of PDCD1 (encoding for PD-1) and CTLA-4 was found associated with increased expression of the corresponding transcripts.38 Regulation of PD-L1 expression in NSCLC has been instead associated with gene copy number changes or promoter polymorphism (rs822335).39 Collectively, these genetic and epigenetic changes could serve as potential predictive factors of immunotherapy effectiveness.38,39
Modulation of TME favors response to immunotherapy
TME is a dynamic mixture of numerous cellular and acellular components, including CAFs, microvesicles, chemokines, extracellular matrix, and immune cells, all interacting with tumor cells to define disease trajectories.56 Polarization of stromal cells into pro-tumorigenic populations is also associated with dramatic changes in their epigenome,57,58 which has long suggested the possibility of using epigenetic modulators [e.g., DNMT inhibitors and histone deacetylase (HDAC) inhibitors] to rewire immune cells into anti-tumor cells. In keeping with this, HDAC inhibitors were shown to improve CD4+ and CD8+ activity in breast and lung cancer.59,60 Interestingly, different immune cell populations could migrate in response to chemokines inducing an inflamed TME, which has been associated with improved prognosis and higher response to immunotherapy.61 Two different HDAC inhibitors, romidepsin and vorinostat, showed the ability to induce expression of Th1 chemokines Ccl5, Cxcl9, and Cxcl10 in human and murine LUAD models. Similarly, in vivo experiments confirmed the enhanced romidepsin-induced T-cell chemokines expression, which in turn increased CD8+ T-cell infiltration and was associated with stable disease. Interestingly, further in vivo experiments evaluating the efficacy of romidepsin or anti-PD-1 treatment reported that, despite an initial response, tumor progression was observed following monotherapy. In contrast, combination of these two agents induced tumor regression or complete rejection of the tumor without reappearance across different lung cancer models. The synergistic value of the combination was highlighted also by the fact that T-cells produced increased IFN-γ in the presence of romidepsin. Since poor T-cell infiltration seems to sabotage response to anti-PD-1 therapy, a combination of HDAC inhibitor and ICIs could represent a promising treatment approach.62
Immuno-epigenetic drug combinations in NSCLC preclinical and clinical studies
The dynamic nature of the epigenome represents an attractive target for the development of therapeutic compounds. As of today, the few epigenetic drugs that have reached the clinic are used in hematological malignancies, yet numerous epigenetic drugs are under extensive clinical investigation for numerous solid tumor types.63 A treatment rationale that involves combination of ICI and epigenetic agents, as presented in Figure 2, has been already applied in patients with immunotherapy resistant and refractory NSCLC and although the reported response rates are currently moderate, the perspectives of this novel immunomodulatory strategy are promising.
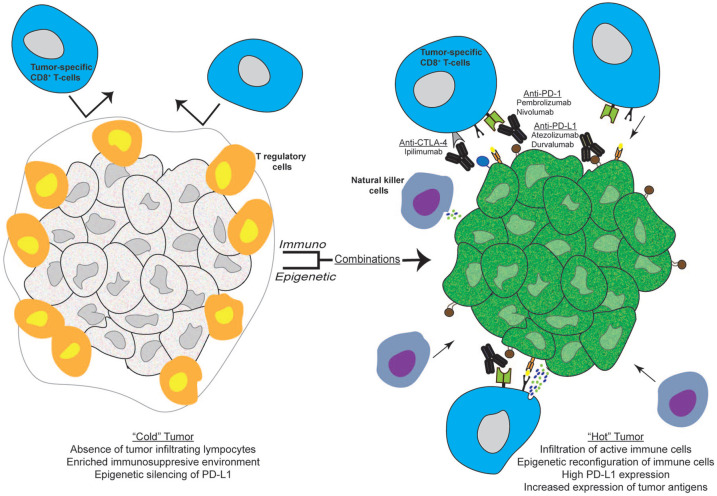
Figure 2.
Immuno-epigenetic drug combinations reverse immunologically “cold” tumors into “hot” ones. Left panel: an immunologically “cold” tumor is characterized by poor infiltration of tumor-specific T-cells, professional APCs and NK cells. Moreover, the strong immunosuppresive TME is reinforced by the abundance of Tregs as well as low expression of tumor antigens and PD-L1 by tumor cells, eventually inducing resistance to immunotherapy. Right panel: combination of ICIs with epigenetic modulating agents such as DNMT inhibitors (decitabine, azacytidine) and HDAC inhibitors (vorinostat, entinostat, mocetinostat) might increase the expression of tumor antigens, induce reprogramming of immune cells in TME and restore the expression of PD-L1 by tumor cells rendering tumor more susceptible to ICIs, resulting in the formation of a more inflamed tumor infiltrated by tumor-fighting immune cells.
APC, antigen-presenting cell; CTLA-4, cytotoxic T-lymphocyte-associated protein 4; DNMT, DNA methyl tranferase; HDAC, histone deacetylase; ICIs, immune-checkpoint inhibitors; NK, natural killer; PD-1, programmed cell death protein 1; PD-L1, programmed death-ligand 1; TME, tumor microenvironment; Tregs, T regulatory cells.
In a phase I/II clinical trial involving a combinatorial treatment in recurrent metastatic NSCLC, patients received repetitive low doses of azacytidine and entinostat, a DNMT inhibitor and a HDAC inhibitor, respectively. Study conclusions reported that only 4% of treated patients showed marked responses to the epigenetic combination.64 Interestingly, after completion of the combinatorial epigenetic treatment, a subset of patients received subsequent systematic anticancer therapies, and 21% (4/19) reported major responses (including one patient treated with anti-PD-1 mAb), indicating that epigenetic therapy might sensitize patients for higher response to ICI.64 These clinical findings prompted a preclinical analysis in NSCLC cell lines for generation of potential immuno-epigenetic-related biomarkers. The treatment of NSCLC cell lines, in the majority LUAD, with the DNMT inhibitor azacytidine, resulted in the modulation of a broad range of immune-related signaling pathways. Specifically, azacytidine promoted the expression of PD-L1, of genes involved in antigen presentation, and increased type I and II IFN-γ signaling. Among other targets, azacytidine treatment reversed the heavily methylated promoter of interferon regulatory factor 7 (IRF7) – a transcription factor that has been associated with the activation of interferon pathway – in the NSCLC cell lines. Projection of the preclinical findings to TCGA-derived NSCLC samples confirmed that tumor profiles match the NSCLC cell lines pre-azacytidine treatment profile, for example, IRF7 methylation.65 The potential biomarker utility of these azacytidine-induced immune-related expression signatures is currently being evaluated in a phase II clinical trial [ClinicalTrials.gov identifier: NCT01928576] where pretreated NSCLC patients receive an epigenetic combination (azacytidine plus entinostat) and afterwards nivolumab.65
A phase II study explored the efficacy of pembrolizumab plus CC-486 (oral version of azacytidine) combination versus pembrolizumab plus placebo as second-line therapy in previously chemotherapy-treated advanced NSCLC patients. DNA methylation analysis in the group of pembrolizumab plus CC-486 revealed that methylation levels of subset of patients were decreased >10%. However, addition of CC-486 resulted in worse PFS and OS compared with the pembrolizumab-treated arm, although results were not statistically significant. Severe side-effects were detected in the pembrolizumab plus CC-486 arm leading to treatment interruptions and likely explaining the poor outcomes. Considering the described effect of CC-486 on methylation status, optimization of dosing scheme of this epigenetic drug may present a combination with meaningful clinical benefit.66
Another study evaluated pembrolizumab plus vorinostat in chemotherapy-treated, immunotherapy-pretreated or naïve NSCLC patients. Interestingly, 3 of 24 patients from the immunotherapy-pretreated group presented partial responses. Moreover, the reactivation of stromal CD8+ T-cells and their mitigation towards tumor in patients who benefited might be associated with the treatment combination. Although the combination was generally well tolerated by patients, around half of them needed a dosing readjustment due to emerging adverse events.67
In the ENCORE-601 study, a phase II trial of entinostat plus pembrolizumab in previously anti-PD-1/PD-L1-treated and progressed NSCLC patients, it was observed that 11% (6/57) responded to the combinatorial treatment. Interestingly, four out of six patients who responded were PD-L1 negative at enrollment.68 More details of completed or ongoing clinical trials are presented in Table 2.
Table 2.
Completed or ongoing clinical trials.
| ClinicalTrials.gov identifier: | Study status | Eligible patients | Regimen | Conclusions | Safety profile | Reference |
|---|---|---|---|---|---|---|
| NCT02546986 | Phase II | Chemotherapy-pretreated aNSCLC | Pembrolizumab + CC-486 versus Pembrolizumab + placebo | No difference in PFS between arms | GI-related adverse events | Levy et al.66 |
| NCT02638090 | Phase I/Ib | Chemotherapy-pretreated - ICI- pretreated/naïve aNSCLC | Pembrolizumab + Vorinostat | PR in 3/24 ICI-pretreated patients | 50% patients required adjustment of vorinostat dosing | Gray et al.67 |
| NCT02437136 | Phase II (ENCORE-601) | Chemotherapy and ICI-pretreated aNSCLC | Pembrolizumab + entinostat | 6/57 PR | Well tolerated | Hellmann et al.68 |
| NCT02805660 | Phase I/II | ICI-pretreated NSCLC | Durvalumab + Mocetinostat | Completed | NR | − |
| NCT03233724 | Phase I/II | Pretreated aNSCLC | DAC-THU + Pembrolizumab | Recruiting | NR | − |
| NCT01928576 | Phase II | Pretreated aNSCLC | Azacytidine + Entinostat, then Nivolumab | Recruiting | NR | − |
| NCT02635061 | Phase Ib | Pretreated unresectable NSCLC | Nivolumab + ACY 241 | Active | NR | − |
DAC, decitabine; GI, gastrointestinal; ICI, immune checkpoint inhibitor; aNSCLC, advanced non-small cell lung cancer; OS, overall survival; NR, not reported; PFS, progression-free survival; THU, tetrahydrouridine.
Conclusions
Introduction of immunotherapy in the NSCLC treatment landscape was a breakthrough, especially for specific subtypes such as non-oncogene driven LUAD and SQLC, which lack any type of targeted therapy options. However, tumor intrinsic and acquired resistance mechanisms eventually emerge, overcoming its efficacy, and a substantial percentage of NSCLC patients fail to experience clinical benefit. The emerging toxicity in non-responders and the high financial cost of immunotherapy has rendered vital the efficient stratification of eligible patients for immunotherapy. In this review, we explored whether two rapidly progressing cancer research fields, immunotherapy and epigenetics, can work in concert towards the development of more efficacious preventive strategies. PD-L1 expression is the mothership of evading mechanisms from immune surveillance but its regulatory mechanisms in NSCLC are not well understood. Here, we report that PD-L1 expression is associated with epigenetic characteristics and use of artificial miRNA technology, and epigenetic modulating agents to control PD-L1 gene expression represent a promising research perspective that might be translated into effective clinical treatments. As reviewed here, different epigenetic patterns such as methylation profiles and circulating miRNA signatures could distinguish patients likely to benefit from immunotherapy. Further studies will need to explore whether epigenetic biomarkers can actually add accuracy or be superior to PD-L1 expression in identifying patients likely to respond to immunotherapy. Although preliminary clinical findings hold promise for improving immunotherapy efficacy after incorporating epigenetic modulating agents in NSCLC, we strongly encourage efforts to identify and verify epigenetic patterns on immunomodulatory genes with potential predictive value in larger cohorts of NSCLC patients. Moreover, we suggest including in these cohorts more oncogene-addicted NSCLC exploring the aspect that specific KI-induced epigenetic phenotypes are responsible for the generally poor response to immunotherapy in this subgroup of patients. Further, based on the plethora of available epigenetic modulators, we suggest the implementation of further clinical studies applying immuno-epigenetic protocols. Of note, it is becoming evident from completed studies that emerging toxicity should be taken into serious consideration. Adverse events were generally manageable in most cases, but adjustment of the dosage schedule might lead to improved safety profile and enhanced benefit. Collectively, immunotherapy has revolutionized the treatment landscape of NSCLC but its (long-lasting) efficacy is not certain. In our opinion, a deeper understanding of the NSCLC epigenome and appropriate pharmacological interference might serve as a valuable “wingman” in order to turn an immunologically “cold” tumor into a “hot” and immunotherapy-favorable tumor in virtually all NSCLC patients.
Footnotes
Conflict of interest statement: E.B. received speaker’s and travel fees from MSD, AstraZeneca, Celgene, Pfizer, Helsinn, Eli-Lilly, BMS, Novartis, and Roche. E.B received consultant’s fee from Roche and Pfizer. E.B. received institutional research grants from AstraZeneca and Roche.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: E.B. is supported by Institutional funds of Università Cattolica del Sacro Cuore (UCSC-project D1-2019-2020). E.B. is currently supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) under Investigator Grant (IG) No. IG20583. VC is supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) under Start Up Grant No. 18718. V.C. and P.D. are supported by Fondazione Nadia Valsecchi.
Contributor Information
Anastasios Gkountakos, ARC-NET Applied Research on Cancer Center, University of Verona, P.le L.A. Scuro 10, Verona, 37134, Italy.
Pietro Delfino, Department of Diagnostics and Public Health, University of Verona, Verona, Italy.
Rita T. Lawlor, ARC-NET Applied Research on Cancer Center, University of Verona, Verona, Italy
Aldo Scarpa, ARC-NET Applied Research on Cancer Center, University of Verona, Verona, Italy; Department of Diagnostics and Public Health, University of Verona, Verona, Italy.
Vincenzo Corbo, ARC-NET Applied Research on Cancer Center, University of Verona, Verona, Italy; Department of Diagnostics and Public Health, University of Verona, Verona, Italy.
Emilio Bria, Comprehensive Cancer Center, Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Rome, Italy; Department of Medical Oncology, Università Cattolica Del Sacro Cuore, Rome, Italy.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020; 70: 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 2015; 10: 1243–1260. [DOI] [PubMed] [Google Scholar]
- 3. Yuan M, Huang LL, Chen JH, et al. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct Target Ther 2019; 4: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alexander M, Kim SY, Cheng H. Update 2020: management of non-small cell lung cancer. Lung 2020; 198: 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karachaliou N, Cao MG, Teixido C, et al. Understanding the function and dysfunction of the immune system in lung cancer: the role of immune checkpoints. Cancer Biol Med 2015; 12: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gravbrot N, Gilbert-Gard K, Mehta P, et al. Therapeutic monoclonal antibodies targeting immune checkpoints for the treatment of solid tumors. Antibodies (Basel) 2019; 8: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Han Y, Liu D, Li L. PD-1/PD-L1 pathway: current researches in cancer. Am J Cancer Res 2020; 10: 727–742. [PMC free article] [PubMed] [Google Scholar]
- 8. Oh SA, Wu D-C, Cheung J, et al. PD-L1 expression by dendritic cells is a key regulator of T-cell immunity in cancer. Nat Cancer 2020; 1: 681–691. [DOI] [PubMed] [Google Scholar]
- 9. Rowshanravan B, Halliday N, Sansom DM. CTLA-4: a moving target in immunotherapy. Blood 2018; 131: 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buchbinder EI, Desai A. CTLA-4 and PD-1 pathways: similarities, differences, and implications of their inhibition. Am J Clin Oncol 2016; 39: 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pacheco JM, Camidge DR, Doebele RC, et al. A changing of the guard: immune checkpoint inhibitors with and without chemotherapy as first line treatment for metastatic non-small cell lung cancer. Front Oncol 2019; 9: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pham T, Roth S, Kong J, et al. An update on immunotherapy for solid tumors: a review. Ann Surg Oncol 2018; 25: 3404–3412. [DOI] [PubMed] [Google Scholar]
- 13. Reck M, Rodriguez-Abreu D, Robinson AG, et al. Updated analysis of KEYNOTE-024: pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. J Clin Oncol 2019; 37: 537–546. [DOI] [PubMed] [Google Scholar]
- 14. Ferrara R, Mezquita L, Texier M, et al. Hyperprogressive disease in patients with advanced non-small cell lung cancer treated with PD-1/PD-L1 inhibitors or with single-agent chemotherapy. JAMA Oncol 2018; 4: 1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim CG, Kim KH, Pyo KH, et al. Hyperprogressive disease during PD-1/PD-L1 blockade in patients with non-small-cell lung cancer. Ann Oncol 2019; 30: 1104–1113. [DOI] [PubMed] [Google Scholar]
- 16. Adashek JJ, Subbiah IM, Matos I, et al. Hyperprogression and immunotherapy: fact, fiction, or alternative fact? Trends Cancer 2020; 6: 181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karachaliou N, Gonzalez-Cao M, Sosa A, et al. The combination of checkpoint immunotherapy and targeted therapy in cancer. Ann Transl Med 2017; 5: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Incorvaia L, Fanale D, Badalamenti G, et al. Programmed Death Ligand 1 (PD-L1) as a predictive biomarker for pembrolizumab therapy in patients with advanced Non-Small-Cell Lung Cancer (NSCLC). Adv Ther 2019; 36: 2600–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016; 375: 1823–1833. [DOI] [PubMed] [Google Scholar]
- 20. Aguilar EJ, Ricciuti B, Gainor JF, et al. Outcomes to first-line pembrolizumab in patients with non-small-cell lung cancer and very high PD-L1 expression. Ann Oncol 2019; 30: 1653–1659. [DOI] [PubMed] [Google Scholar]
- 21. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019; 19: 133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duruisseaux M, Esteller M. Lung cancer epigenetics: from knowledge to applications. Semin Cancer Biol 2018; 51: 116–128. [DOI] [PubMed] [Google Scholar]
- 23. Diamantopoulos MA, Tsiakanikas P, Scorilas A. Non-coding RNAs: the riddle of the transcriptome and their perspectives in cancer. Ann Transl Med 2018; 6: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther 2016; 1: 15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang N, Zhu S, Lv X, et al. MicroRNAs: pleiotropic regulators in the tumor microenvironment. Front Immunol 2018; 9: 2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu J, Zhan Y, Feng J, et al. MicroRNAs associated with therapy of non-small cell lung cancer. Int J Biol Sci 2018; 14: 390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Inamura K, Ishikawa Y. MicroRNA in lung cancer: novel biomarkers and potential tools for treatment. J Clin Med 2016; 5: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Halvorsen AR, Sandhu V, Sprauten M, et al. Circulating microRNAs associated with prolonged overall survival in lung cancer patients treated with nivolumab. Acta Oncol 2018; 57: 1225–1231. [DOI] [PubMed] [Google Scholar]
- 29. Boeri M, Milione M, Proto C, et al. Circulating miRNAs and PD-L1 tumor expression are associated with survival in advanced NSCLC patients treated with immunotherapy: a prospective study. Clin Cancer Res 2019; 25: 2166–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peng X-X, Yu R, Wu X, et al. Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR/ALK wild-type advanced non-small cell lung cancer. J Immunother Cancer 2020; 8: e000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Costantini A, Julie C, Dumenil C, et al. Predictive role of plasmatic biomarkers in advanced non-small cell lung cancer treated by nivolumab. Oncoimmunology 2018; 7: e1452581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramos RN, Piaggio E, Romano E. Mechanisms of resistance to immune checkpoint antibodies. Handb Exp Pharmacol 2018; 249: 109–128. [DOI] [PubMed] [Google Scholar]
- 33. Pai SG, Carneiro BA, Mota JM, et al. Wnt/beta-catenin pathway: modulating anticancer immune response. J Hematol Oncol 2017; 10: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boldrini L, Giordano M, Niccoli C, et al. Role of microRNA-33a in regulating the expression of PD-1 in lung adenocarcinoma. Cancer Cell Int 2017; 17: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xie WB, Liang LH, Wu KG, et al. MiR-140 expression regulates cell proliferation and targets PD-L1 in NSCLC. Cell Physiol Biochem 2018; 46: 654–663. [DOI] [PubMed] [Google Scholar]
- 36. Cai L, Bai H, Duan J, et al. Epigenetic alterations are associated with tumor mutation burden in non-small cell lung cancer. J Immunother Cancer 2019; 7: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duruisseaux M, Martinez-Cardus A, Calleja-Cervantes ME, et al. Epigenetic prediction of response to anti-PD-1 treatment in non-small-cell lung cancer: a multicentre, retrospective analysis. Lancet Respir Med 2018; 6: 771–781. [DOI] [PubMed] [Google Scholar]
- 38. Marwitz S, Scheufele S, Perner S, et al. Epigenetic modifications of the immune-checkpoint genes CTLA4 and PDCD1 in non-small cell lung cancer results in increased expression. Clin Epigenetics 2017; 9: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krawczyk P, Grenda A, Wojas-Krawczyk K, et al. PD-L1 gene copy number and promoter polymorphisms regulate PD-L1 expression in tumor cells of non-small cell lung cancer patients. Cancer Genet 2019; 237: 10–18. [DOI] [PubMed] [Google Scholar]
- 40. Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity 2018; 48: 434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Blagih J, Buck MD, Vousden KH. p53, cancer and the immune response. J Cell Sci 2020; 133: jcs237453. [DOI] [PubMed] [Google Scholar]
- 42. Rokavec M, Li H, Jiang L, et al. The p53/miR-34 axis in development and disease. J Mol Cell Biol 2014; 6: 214–230. [DOI] [PubMed] [Google Scholar]
- 43. Cortez MA, Ivan C, Valdecanas D, et al. PDL1 regulation by p53 via miR-34. J Natl Cancer Inst 2016; 108: djv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci 2019; 26: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019; 20: 675–691. [DOI] [PubMed] [Google Scholar]
- 46. Zhang C, Ma L, Niu Y, et al. Circular RNA in lung cancer research: biogenesis, functions, and roles. Int J Biol Sci 2020; 16: 803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang J, Zhao X, Wang Y, et al. CircRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis 2020; 11: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chockley PJ, Keshamouni VG. Immunological consequences of epithelial-mesenchymal transition in tumor progression. J Immunol 2016; 197: 691–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Asgarova A, Asgarov K, Godet Y, et al. PD-L1 expression is regulated by both DNA methylation and NF-kB during EMT signaling in non-small cell lung carcinoma. Oncoimmunology 2018; 7: e1423170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Garassino M, Rodriguez-Abreu D, Gadgeel S, et al. OA04.06 Evaluation of TMB in KEYNOTE-189: pembrolizumab plus chemotherapy vs placebo plus chemotherapy for nonsquamous NSCLC. In: World conference on lung cancer, Barcelona, Spain, 7–10 September 2019. Winnipeg, Canada: WCLC. [Google Scholar]
- 51. Langer C, Gadgeel S, Borghaei H, et al. OA04.05 KEYNOTE-021: TMB and outcomes for carboplatin and pemetrexed with or without pembrolizumab for nonsquamous NSCLC. In: World conference on lung cancer, Barcelona, Spain, 7–10 September 2019. Winnipeg, Canada: WCLC. [Google Scholar]
- 52. Wu Y, Xu J, Xu J, et al. The predictive value of tumor mutation burden for immune checkpoint inhibitors therapy in non-small cell lung cancer is affected by patients’ age. Biomark Res 2020; 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sholl LM, Hirsch FR, Hwang D, et al. The promises and challenges of tumor mutation burden as an immunotherapy biomarker: a perspective from the International Association for the Study of Lung Cancer Pathology Committee. J Thorac Oncol 2020; 15: 1409–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ehrlich M, Zhang XY, Inamdar NM. Spontaneous deamination of cytosine and 5-methylcytosine residues in DNA and replacement of 5-methylcytosine residues with cytosine residues. Mutat Res 1990; 238: 277–286. [DOI] [PubMed] [Google Scholar]
- 55. Cooper DN, Mort M, Stenson PD, et al. Methylation-mediated deamination of 5-methylcytosine appears to give rise to mutations causing human inherited disease in CpNpG trinucleotides, as well as in CpG dinucleotides. Hum Genomics 2010; 4: 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Baghban R, Roshangar L, Jahanban-Esfahlan R, et al. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun Signal 2020; 18: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ghoneim HE, Fan Y, Moustaki A, et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 2017; 170: 142–157.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Youngblood B, Oestreich KJ, Ha S-J, et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8+ T cells. Immunity 2011; 35: 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McCaw TR, Li M, Starenki D, et al. Histone deacetylase inhibition promotes intratumoral CD8+ T-cell responses, sensitizing murine breast tumors to anti-PD1. Cancer Immunol Immunother 2019; 68: 2081–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Adeegbe DO, Liu Y, Lizotte PH, et al. Synergistic immunostimulatory effects and therapeutic benefit of combined histone deacetylase and bromodomain inhibition in non-small cell lung cancer. Cancer Discov 2017; 7: 852–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vilgelm AE, Richmond A. Chemokines modulate immune surveillance in tumorigenesis, metastasis, and response to immunotherapy. Front Immunol 2019; 10: 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zheng H, Zhao W, Yan C, et al. HDAC inhibitors enhance T-cell chemokine expression and augment response to PD-1 immunotherapy in lung adenocarcinoma. Clin Cancer Res 2016; 22: 4119–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cheng Y, He C, Wang M, et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther 2019; 4: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 2011; 1: 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wrangle J, Wang W, Koch A, et al. Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget 2013; 4: 2067–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Levy BP, Giaccone G, Besse B, et al. Randomised phase 2 study of pembrolizumab plus CC-486 versus pembrolizumab plus placebo in patients with previously treated advanced non-small cell lung cancer. Eur J Cancer 2019; 108: 120–128. [DOI] [PubMed] [Google Scholar]
- 67. Gray JE, Saltos A, Tanvetyanon T, et al. Phase I/Ib study of pembrolizumab plus vorinostat in advanced/metastatic non-small cell lung cancer. Clin Cancer Res 2019; 25: 6623–6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hellmann M, Jänne P, Opyrchal M, et al. OA05.01 Efficacy/Safety of Entinostat (ENT) and Pembrolizumab (PEMBRO) in NSCLC patients previously treated with anti-PD-(L)1 therapy. J Thorac Oncol 2018; 13: S330. [Google Scholar]