Abstract
Background
Mortality due to COVID-19 caused by SARS-CoV-2 infection varies among populations. Functional relevance of genetic variations in Angiotensin-converting enzyme 2 (ACE2) and Transmembrane serine protease 2 (TMPRSS2), two crucial host factors for viral entry, might explain some of this variation.
Methods
In this comparative study in Indian subjects, we recruited 510 COVID-19 patients and retrieved DNA from 520 controls from a repository. Associations between variants in ACE2 and TMPRSS2 with disease severity were identified by whole exome sequencing (WES, n = 20) and targeted genotyping (n = 1010). Molecular dynamic simulations (MDS) were performed to explore functional relevance of the variants. Cleavage of spike glycoprotein by wild and variant TMPRSS2 was determined in HEK293T cells. Potential effects of confounders on the association between genotype and disease severity were tested (Mantel-Haenszel test).
Results
WES identified deleterious variant in TMPRSS2 (rs12329760, G > A, p. V160M). The minor allele frequency (MAF) was 0·27 in controls, 0·31 in asymptomatic, 0·21 in mild-to-moderately affected and 0·19 in severely affected COVID-19 patients. Risk of severity increased with decreasing MAF: Asymptomatic: Odds ratio-0·69 (95% CI–0·52–0·93; p = 0·01); mild-to-moderate: Odds ratio-1·89 (95% CI–1·22–2.92;p = 0·004) and severe: Odds ratio-1·79 (95% CI–1·11–2.88;p = 0·01). No confounding effect of diabetes and hypertension were observed on the risk of developing severe COVID-19 disease with respect to genotype. MDS revealed decreased stability of TMPRSS2 with 160 M variant. Spike glycoprotein cleavage by TMPRSS2 reduced ~2·4-fold in cells expressing 160 M variant.
Conclusion
We demonstrate association of TMPRSS2 variant rs12329760 with decreased disease severity in COVID-19 patients from India.
Keywords: COVID-19, SARS-CoV-2, TMPRSS2, ACE2, Genotyping, Disease severity
1. Introduction
SARS-CoV-2 infection exhibits varied infectivity and mortality rates across geographical locations. The United States and European countries have documented a large number of infections associated with higher mortality in the initial months of the pandemic, compared to South Asian countries. Although rates of infection have increased in South Asian countries, mortality rates remain low during the evolution of the pandemic (Yamamoto and Bauer, 2020). These variations could be due to differences in virulence of viral strains (Islam et al., 2020; Eaaswarkhanth et al., 2020) and host factors (Chen et al., 2020) including genetic makeup (Carter-Timofte et al., 2020). Few studies are available regarding disease-modifying genetic variations in the host (Ellinghaus et al., 2020). Based on the known importance of ACE2 and TMPRSS2 in SARS-CoV-2 infection (Zhou et al., 2020; Hoffmann et al., 2020), two previous studies of large, publicly available genome databases (Asselta et al., 2020; Hou et al., 2020) identified deleterious variants that might predict differences in COVID-19 disease severity among populations. These studies emphasized the urgent need for these genetic associations to be tested in COVID-19 positive patients.
Increased mortality rates in COVID-19 are associated with age of the population, male sex and comorbidities such as diabetes, hypertension and obesity (Ioannidis et al., 2020; Williamson et al., 2020). In India, COVID-19 related mortality rates are lower (1·6%) than those in the United States (3·01%), Brazil (3·06%) and United Kingdom (12·0%) (Worldometers, n.d.). Despite high prevalence of diabetes (State-Level Disease Burden Initiative Diabetes Collaborators, 2018) and metabolic syndrome (Deedwania et al., 2014) in India, the observed differences in the rates of mortality among countries raises an intriguing question whether functionally relevant variants in ACE2 and TMPRSS2 contribute to differences in infection rates and mortality.
The aim of our study was to i) identify functionally relevant variants in ACE2 and TMPRSS2 in COVID-19 positive patients compared to controls, ii) demonstrate the clinical relevance of the variant, and iii) explore the association of the variants with severity of COVID-19. To identify the variants, we sequenced complete exonic regions of healthy individuals. We report a variant rs12329760 in the TMPRSS2 gene that is associated with mild symptoms of COVID-19.
2. Methods
2.1. Participants
In this comparative study, 510 consecutive COVID-19 patients, who tested positive for SARS-CoV-2 by qRT-PCR during 5th May 2020 and 30th August 2020 were recruited at AIG Hospitals, a tertiary care hospital in Hyderabad, India. All the participants provided written informed consent. Patients with flu-like symptoms who tested negative for SARS-CoV-2 by qRT-PCR and had a normal CT study were excluded. Whole blood (3 ml) was collected from all COVID-19 patients for genotyping. The diagnosis and classification of COVID-19 patients was established based on the diagnostic criteria of the Diagnostic and Therapeutic Program of Novel Coronavirus Pneumonia (6th Version for Trial Implementation) (National Health Commission of the People's Republic of China, 2020). Patients with a positive qRT-PCR for SARS-CoV-2 were classified into the following categories of COVID-19: i) asymptomatic: no symptoms; ii) mild: headache, dry cough, myalgia with or without ground glass opacities; iii) moderate: fever, breathlessness, and ground glass opacities on imaging; iv) severe: respiratory distress (respiratory rate ≥ 30 breaths/min), oxygen requirement more than 6 L/min or requirement of non-invasive/invasive ventilator support, with lesions significantly progressing >50% within 24–48 h on pulmonary imaging (Verity et al., 2020). In addition, DNA from healthy individuals (n = 520),retrieved from AIG Hospital's DNA repository (collected during 2016–2018 before COVID-19 for other research purposes) was considered as control group to generate allelic frequency for the identified variants.
2.1.1. Ethics
The study participants were recruited after the protocol was approved by the Institutional Ethics Committee of AIG Hospitals. All the participants had provided informed consent to collect clinical data and blood samples. Confidentiality of all the samples was maintained.
2.1.2. Whole exome sequencing and targeted genotyping
Key genotyping methods are summarized here, with additional details provided as Supplementary Material. DNA was extracted from whole blood using a commercial kit (Bioserve Biotechnologies, India). Complete exonic regions (n = 20 DNA samples considered as controls) were amplified employing Ion Ampliseq Exome RDY kit and sequenced on the Next generation sequencer (NGS-Ion Proton; Life technologies, USA). Generated sequences were aligned to reference human genome (hg19) and annotated using Ion reporter. Functionally relevant variants were identified using Polyphen and SIFT scores. Primers were designed using Primer-Z software (Tsai et al., 2007)(Supplementary Table S1) to amplify the flanking regions of variants in ACE2 and TMPRSS2.All the amplicons were sequenced on Beckman GeXP system. Genotypes were interpreted using Genome Lab GeXP software (v10·2). Minor allele frequency (MAF) for the Indian ethnicity was calculated based on the genotype data from this study. Insilico analysis was performed using I-mutant (Capriotti et al., 2005) and PhyreRisksoftware (Ofoegbu et al., 2019). All the protocols conformed to standard kit instructions. Quality datasets generated and analyzed for Whole exome and sequencing data are available at the Mendeley data repository accessible at.
https://data.mendeley.com/datasets/kn9jx7mgzd/draft?a=3d3903f0-c68d-4cb9-994f-fbf640a351de.
2.1.3. Correlation of TMPRSS2-MAF with mortality
The MAF for rs12329760 variant in TMPRSS2 for various ethnicities were retrieved from ENSEMBL genome browser and the MAF generated in this study was used for the Indian ethnicity. Corresponding mortality rates for the ethnicities were retrieved from Worldometer (https://www.worldometers.info/coronavirus/). Pearson's correlation coefficient (r) was used to measure the strength of association between MAF and mortality per million.
2.1.4. Genotype-based TMPRSS2 mRNA and protein expression in human lung tissue
To assess the genotype-based differences in the mRNA and protein expression of TMPRSS2, paraffin embedded human normal lung tissue blocks were identified and retrieved (n = 10) from AIG Hospital's pathology repository. RNA was isolated, converted to cDNA and relative gene expression was evaluated (Sybr chemistry). Samples were analyzed in duplicates and data were normalized against human GAPDH. Relative gene quantification was performed using the 2-ΔΔCT method (Pfaffl, 2001) and expressed as Log2 fold change. Genotyping for TMPRSS2 variant was performed following genotyping protocol.
Tissue sections (~0·5 μm) of paraffin embedded blocks were immunostained with anti-human rabbit TMPRSS2 antibodies (Invitrogen USA), followed by anti-rabbit HRP conjugated secondary antibodies and stained with DiaminoBenzidine (DAB) after antigen retrieval and counter stain. Images were captured using light microscope (Olympus Tokyo Japan).
2.1.5. Functional relevance of the variant V160M in spike protein cleavage
The role of TMPRSS2 variant V160M in spike protein cleavage was studied in a cell culture system employing HEK-293 T cells which do not endogenously express TMPRSS2. HEK293T cells were procured from ATCC and maintained routinely in DMEM supplemented with 10% fetal bovine serum at 37 °C with 5% CO2. HEK 293 T cells were transfected using polyethylenimine (PEI; Polysciences). Approximately 4 million cells were seeded in a 60 mm dish and were transfected the following day with 4 μg each of CoV-2 spike (pCMV14-3×-Flag-SARS-CoV-2 S was a gift from Zhaohui Qian (Addgene plasmid # 145780; http://n2t.net/addgene:145780; RRID: Addgene_145780) and TMPRSS2 wild type or variant constructs (Wild type and Val160Met TMPRSS2 variant plasmids were obtained from Genscript) using 40 μl of PEI (1 mg/ml) for 24 h. In experiments comparing the protein levels of wild type and variant TMPRSS2, cells were transfected with 4 μg of each of the plasmid. Total DNA concentration in all the transfections was made up with pCDNA3.1 plasmid. Approximately 24 h post transfection, cells were washed with ice cold PBS prior to lysis in TENNS lysis buffer as described earlier (Behera et al., 2018). Cellular lysates were resolved on SDS PAGE, transferred to PVDF membrane and probed overnight with Flag tag antibody (M2 clone of Spike protein; Sigma-Aldrich, USA) at 1:1000 dilution. On the third day, membranes were washed 3 times with TBST and probed with anti-rabbit and anti-mouse secondary antibodies (Cell Signaling, USA) for 90 min. B-actin and GAPDH were used to normalize. Membranes were washed 3 times with TBST and the signals were developed with ECL (Hyper HRP, Takara).
2.2. Statistical analysis
The study was powered based on existing literature (0·22 MAF for GIH-retrieved from ENSEMBL) (Ensembl Genome Browser, n.d.). A sample size of 313 is adequately powered with confidence level at 95% to generate MAF in controls. Continuous variables were expressed as mean and standard deviation, categorical variables as proportions. Patient characteristics were compared using ANOVA for continuous variables and Chi-square or Fisher's exact test for categorical variables. To obtain Odds ratio, we compared the genotypes between Controls Vs Asymptomatic; Asymptomatic Vs Mild to moderate and Asymptomatic Vs Severe categories under Dominant and Recessive genetic models. “A” allele was considered as protective. Pearson's correlation coefficient (r) was used to measure the strength of association between MAF and mortality per million reported in various ethnicities. Chi-square goodness-of-fit was used to confirm the agreement of the observed genotype frequencies with those of expected (Hardy-Weinberg equilibrium) for all the variants. Multivariate logistic regression was used to identify significant independent variables associated with disease severity. Confounding effect of the variables was explored using Mantel-Haenszel Test. The data was analyzed using Statistical package for Social Sciences (SPSS Version 25). A two-tailed ‘p’ value ≤0·05 was considered statistically significant.
3. Results
3.1. Demographics and clinical characteristics of study participants
The mean age was 32·46 ± 9·65 years for controls and 44·42 ± 17·0 for patients with COVID-19, with both groups comprising predominantly males of Indian ethnicity. The clinical characteristics of patients with COVID-19 are given in Table 1 and those of the control group in supplementary Table S2.
Table 1.
Clinical characteristics of the study participants.
| Parameter | Asymptomatic (n = 299) | Mild to Moderate# (n = 119) | Severe (n = 92) |
|---|---|---|---|
| Age in years (Mean ± SD) | 37·25 ± 15·09 | 52·0 ± 14·2 | 57·9 ± 13·6* |
| Age Range in Years - | 1·5–80 | 24–90 | 26–92 |
| Gender (Male) | 189 (63·2%) | 83 (69·8%) | 69 (75·0%) |
| BMI (kg/m2) | 22·04 ± 1·27 | 24·03 ± 4·17 | 27·16 ± 4·66* |
| Comorbidities | |||
| Normal | 285 (95·3%) | 55 (46·2%) | 41 (44·6%) |
| Only diabetes | 3 (1·0%) | 15 (12·6%) | 14 (15·2%)** |
| Only Hypertension | 2 (0·7%) | 20 (16·8%) | 11 (11·9%)** |
| Diabetes/Hypertension | 9 (3·0%) | 29 (24·4%) | 26 (28·3%)** |
| Blood Counts | |||
| Haemoglobin (g/dL) | 13·4 ± 2·0 | 13·9 ± 9·5 | 12·6 ± 2·7* |
| RBC (cells/mcL) | 4·9 ± 0·6 | 4·5 ± 0·7 | 4·4 ± 0·9* |
| WBC cells/μl | 7823·4 ± 2594·6 | 7301·7 ± 3629·0 | 11,277·2 ± 6409·4* |
| Neutrophils (%) | 54·3 ± 11·1 | 68·0 ± 13·3 | 82·6 ± 9·3* |
| Lymphocytes (%) | 37·0 ± 9·9 | 24·8 ± 11·5 | 11·7 ± 8·2* |
| Eosinophils (%) | 3·2 ± 2·8 | 1·8 ± 1·5 | 1·3 ± 0·7* |
| Monocytes (%) | 5·2 ± 2·2 | 5·2 ± 2·1 | 4·1 ± 2·2* |
| Platelets (mcL) | 3·0 ± 1·3 | 2·4 ± 1·0 | 2·4 ± 1·1* |
| Oxygen Delivery | |||
| On room air | None | 92 (77·3%) | 5 (5·4%) |
| On face Mask | None | 2 (1·7%) | 3 (3·3%) |
| Nasal Prongs | None | 24 (20·2%) | 10 (10·9%) |
| On NIV | None | 1 (0·8%) | 22 (23·9%) |
| On HFNC | None | 0 | 2 (2·2%) |
| NRBM Mask | None | 0 | 3 (3·2%) |
| Ventilator | None | 0 | 47 (51·1%)** |
| FiO2 | None | 22·7 ± 5·5 | 74·2 ± 24·8* |
| Serum Ferritin Level (ng/ml) | 127·4 ± 71·2 | 568·6 ± 638·9 | 1178·6 ± 930·4* |
| IL-6 Levels (pg/ml) | 9·5 ± 5·2 | 41·0 ± 97·2 | 230·9 ± 211·2* |
| D Dimer (ng/ml) | < 200 | 441·0 ± 560·8 | 1217·1 ± 1615·1* |
| Chest X-ray | |||
| Normal | 239 (79·9%) | 37 (31·1%) | 0 (0%) |
| Unilateral infiltration | 60 (2·0%) | 31 (26·1%) | 1 (1·1%) |
| Bilateral infiltration | 0 | 51 (42·8%) | 91 (98·9%)** |
| CT Scan | |||
| CT CO-RAD Score | NA | 4·5 ± 0·9 | 4·9 ± 0·5* |
#Among the 119 patients in the mild-to-moderate group, 36 (30·2%) had moderate severity. Baseline age (p = 0·17), gender (p = 0·41) and proportion with diabetes (p = 0·27), hypertension (p = 0·77) and both (p = 0·91) were similar for the patients with mild (n = 83) vs moderate (n = 36) severity; therefore, these groups were combined in the analysis.
*One-way ANOVA (P < 0·05) **Fishers exact (P < 0·05).
SD – standard deviation; g – gram, dL – deci liter; mcl/μl – micro liter; ng – nanogram; pg – pico gram; ml – milli liter; CT – Computerized Tomography; % - percent; NIV – Non-invasive ventilation; HFNC – High flow nasal cannula; NRBM – Non-rebreather masks; FiO2 –Fraction of inspired oxygen; NA- Not applicable; ND – Not done. CORAD - COVID-19 Reporting and Data System
3.1.1. Exome Data identifies functionally relevant variants in ACE2 and TMPRSS2
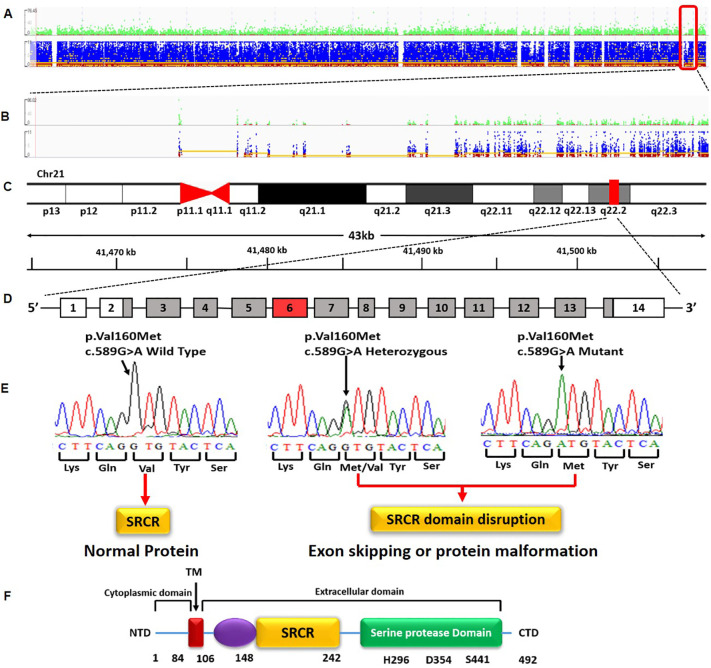
Whole exome sequencing on NGS for 20 healthy controls yielded 6·5–7·5 GB data and ~ 56,000 variants per sample across the genome. We identified 3 variants in ACE2 and 9 in TMPRSS2 applying relevant filters (Fig. S2). Of these 12 variants, two variants-rs971249 inACE2 and rs12329760 in TMPRSS2 were selected and replicated in an independent control sample to generate MAF. The selected variant in ACE2 (rs971249) is an eQTL (expression Quantitative trait loci) and the variant in TMPRSS2 (rs12329760; a valine to methionine substitution at position 160;V160M) is present in an exonic splicing enhancer site (Srp40),associated with an increased chance of exon skipping or malformation of the protein (Fig. 1 .) (FitzGerald et al., 2008; Cartegni et al., 2003). In addition, V160M is a residue overlap splice site variant and is predicted to be deleterious and damaging by SIFT and Polyphen respectively.
Fig. 1.
Representative image depicting Whole exome sequencing data and the identification of a variant in TMPRSS2. (A) Variant representation at the whole exome level extracted from Integrative Genomics Viewer (IGV) (B), Variant representation at the chromosome level (chr. 21) extracted from Integrative Genomics Viewer (IGV) (C), Localization of the TMPRSS2 gene to q 22·2 loci (D) the variant c.589G> A is located in the 6th exon of TMPRSS2 gene that has 14 exons (E) representative images of the Wild type, Heterozygous and Mutant genotypes with the variant mapped to SRCR domain (Scavenger Receptor Cysteine-Rich protein domain. While the wild type produces a normal protein, the variant is located in an exonic splicing enhancer site (Srp40), that is associated with an increased chance of exon skipping or protein malformation that is due to disruption of potential exonic splicing enhancer site. (F) domains of TMPRSS2. NTD – N terminal domain; CTD – C terminal domain; T M transmembrane domain; Chr – chromosome, kb – kilo bases.
3.1.2. Risk for severe disease increases with decrease in MAF of TMPRSS2 variant
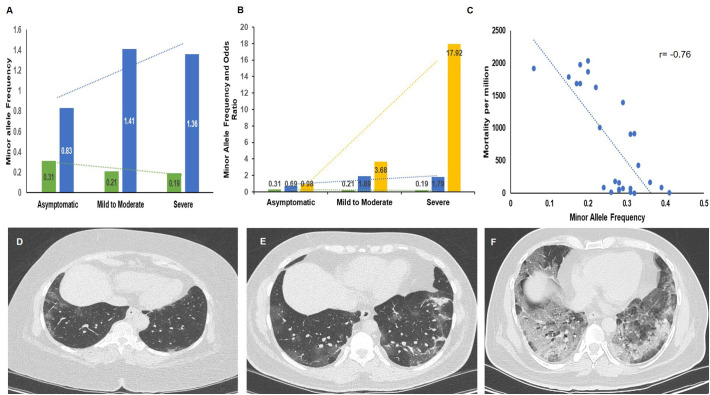
While there were 277 individuals with the GG genotype, 180 with GA and 43 with the AA genotype in the Controls, there were 139 with GG, 134 with GA and 26 with AA in the Asymptomatic, 74 with GG, 42 with GA and 3 with AA genotypes in the Mild to Moderate and 56 with GG, 36 with GA and none with the AA genotypes in the Severe group. The allelic, genotype frequencies and associated Odds ratio for the variant are presented in Table 2 . There was a significant difference in the genotype frequencies for the TMPRSS2 variant under different genetic models (Dominant and Recessive), however there was no significant difference in the genotypes with the Recessive model for the Controls Vs asymptomatic group. The MAF for rs12329760 in TMPRSS2 was 0·27 in controls, 0·31 in the asymptomatic, 0·21 in the mild-to-moderate and 0·19 in the severe COVID-19 patients (Fig. 2A). The risk for severity increased with decrease in MAF (Fig. 2B). There was a negative correlation between MAF and mortality rates (Pearson's correlation coefficient; r = −0·76; p = 0.001; Fig. 2C, Supplementary Table S4). Genotype based CT severity score is given in Fig. 2D,E,F). MAF of the ACE2 variant (rs971249-T) was 0·24 in Indians based on data obtained from our study [143 wild type (CC); 18 heterozygous (CT) and 40 homozygous (TT)], which was similar to other ethnicities and therefore was not replicated in the patients.
Table 2.
Association between TMPRSS2 variant and severity using the dominant and recessive genetic models.
| Type of Comparison |
Minor Allele (A) Frequency |
Model |
Genotype |
Controls |
Patients |
P value |
Odds Ratio |
95% CI |
P value |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||||
| Controls Vs Asymptomatic | 0.27 Vs 0.31 | Dominant | GG Vs (GA + AA) | 277 and 223 | 139 and 160 | 0.01 | 0.69 | 0.52 | 0.93 | 0.01 |
| Recessive | (GG + GA) Vs AA | 457 and 43 | 273 and 26 | 0.96 | 0.98 | 0.59 | 1.64 | 0.96 | ||
| Asymptomatic Vs Mild to Moderate | 0.31 Vs 0.21 | Dominant | GG Vs (GA + AA) | 139 and 160 | 74 and 45 | 0.003 | 1.89 | 1.22 | 2.92 | 0.004 |
| Recessive | (GG + GA) Vs AA | 273 and 26 | 116 and 3 | 0.02 | 3.68 | 1.09 | 12.40 | 0.03 | ||
| Asymptomatic Vs Severe | 0.31 Vs 0.19 | Dominant | GG Vs (GA + AA) | 139 and 160 | 56 and 36 | 0.01 | 1.79 | 1.11 | 2.88 | 0.01 |
| Recessive | (GG + GA) Vs AA | 273 and 26 | 92 and 0 | 0.003 | 17.92 | 1.08 | 297.07 | 0.04 | ||
Genotype GG is wild type, GA is heterozygous and AA homozygous mutant. “A” allele was considered as protective. To obtain Odds ratio, we compared the genotypes between Controls Vs Asymptomatic; Asymptomatic Vs Mild to moderate and Asymptomatic Vs Severe categories under Dominant and Recessive genetic models.
Fig. 2.
Bar graphs depicting Minor allele frequency, Odds ratio, correlation and CT images in the study group (A) Minor allele Frequency (MAF) in Controls, asymptomatic, mild-to-moderate and severe patients. Decreasing trend was noted in MAF with increasing severity (B) MAF and Odds ratio (Dominant and Recessive Models) in Asymptomatic, Mild-to-moderate and Severe COVID19 patients. An inverse trend was seen between risk for severity and MAF. Green bars depict Minor allele frequency, Blue bars depict Odds ratio (Dominant Model) and Yellow bars depict Odds ratio (Recessive Model). Green colour dotted line depicts Linear for MAF, blue colour dotted line depicts Linear for Odds ratio (Dominant Model) and Yellow colour dotted line depicts Linear for Odds ratio (Recessive model) (C) A negative correlation (Pearson's correlation coefficient; r = −0·76) was seen between MAF and mortality rates (D and E) Representative CT images of a patient with mild symptoms with genotype GA and AA(variant), (F) severe disease with GG genotype (wild).
3.1.3. Multivariate logistic regression analysis for confounding effect of variables
Multivariate analysis identified diabetes, hypertension and TMPRSS2 genotype to be independently associated with risk of developing severe disease. Details are provided in Supplementary Table S3. The Mantel-Haenszel test for identifying the confounding effect of the variables revealed no significant difference (P = 0.43) in the unadjusted and adjusted relative risks (Relative risk −1·39 corrected for diabetes in the mild-to-moderate group and RR = 1·28 in the severe group; Relative risk of 1·28 corrected for hypertension in the mild-to-moderate group and RR = 1·76 in the severe group).
3.1.4. V160M decreases the stability of TMPRSS2
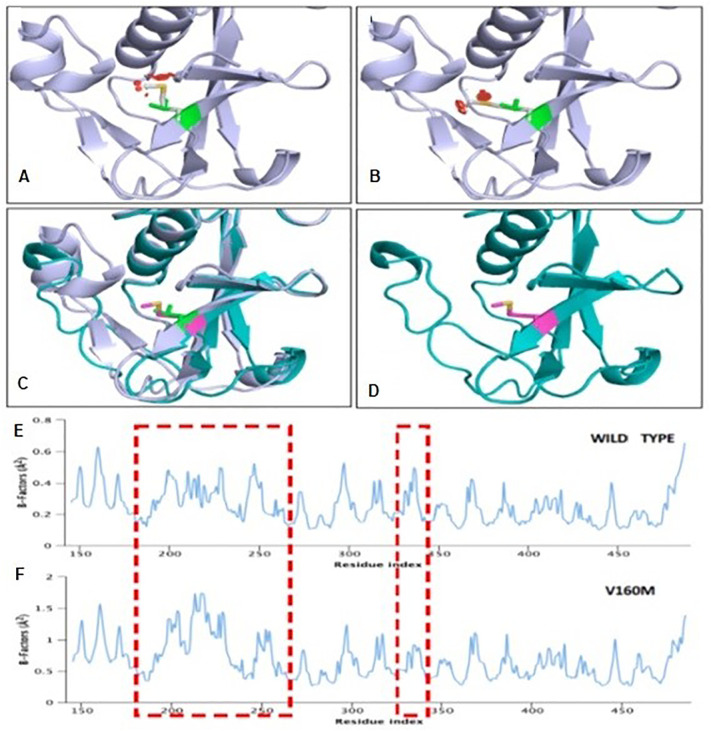
It is known that variant rs12329760 in TMPRSS2 results in substitution of valine with methionine (V160M). Structurally the V160 was found to be stable with several polar residues creating hydrophobic pockets. Replacement of 160 M shows a steric hindrance clash with the surrounding residues and does not accommodate Methionine due to the topology and charge limit (Fig. 3 ). In order to understand the influence of V160M on the overall structural stability of TMPRSS2, we performed MDS studies. We observed that the longer methionine residue substitution V/M160 induces a significant increase in the stability factor of TMPRSS2 decreasing the stability of the protein.
Fig. 3.
Molecular dynamics and simulation analysis of WT and V160M mutant TMPRSS2. Cartoon representation showing the N-terminal domain TMPRSS2 (A) Wild-Type TMPRSS2, (B) V160M mutant TMPRSS2 and (C and D) Superimposing of Wild-Type and V160M structures. The position of amino-acid Valine and Methionine are shown in pink sticks and change in the TMPRSS2 secondary structure is noticeable. (E and F) B-factor profiles of wild type and V160M mutant structure of TMPRSS2. The difference in the domain oscillation or B-factor resulting in structural change are marked in red dashed box.
3.1.5. Genotype-based expression revealed increased TMPRSS2 levels in variant carriers
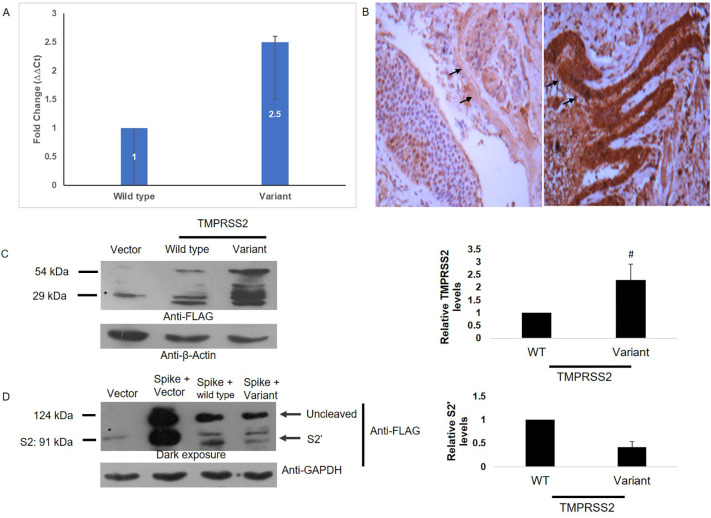
Genotyping of TMPRSS2 in normal lung tissues (n = 10) revealed 6 tissues to be wild type and 4 to be heterozygous. A ~ 2·5-fold increase in mRNA (Fig. 4A) and protein levels (Fig. 4B) were observed in variant carriers. In consistent with this, ectopic expression of wild and variant TMPRSS2 also revealed ~2·3-fold increase in HEK 293 T cells (Fig. 4C).
Fig. 4.
TMPRSS2 Expression. (A) Representative image showing higher mRNA levels of TMPRSS2 in variant carriers as compared to wild type (B)Representative IHC image (10×) showing higher TMPRSS2 protein expression in variant carriers (right panel) as compared to wild type (left panel). Arrows indicate the epithelial lining (C) Left Panel indicating Immunoblot analysis of HEK 293 T cells transfected with indicated constructs using Flag tag antibody. Membranes were stripped and re-probed with β-actin antibody to ensure uniform loading. *, non-specific and Right Panel depicts Densitometry quantification of unprocessed band (~54 kDa form) intensity using ImageJ. Data represents mean + SD of 4 independent experiments. Statistical analysis was performed by Student's ‘t’ test. #, p < 0·05. (D) Left panel representing Immunoblot analysis of HEK 293 T cells transfected with the indicated constructs using Flag tag antibody. Membranes were stripped and re-probed with GAPDH antibody to ensure uniform loading. Uncleaved, S2 and S2’ fragments highlighted. *, non-specific and Right Panel depicting Densitometry quantification of S2’ band intensity using ImageJ. Data represents mean + SD of 4 independent experiments. Statistical analysis was performed by Student's ‘t’ test. ##, p < 0·01.
3.1.6. Reduced spike cleavage in V160M TMPRSS2 over-expressing cells
To address the importance of Val160Met variant in the cleavage of spike, HEK 293 T cells were transfected with spike alone or along with wild type or variant TMPRSS2 plasmids and were analyzed for cellular lysates by western blotting 24 h post transfection (Fig. 4D).Expression of spike protein alone in HEK 293 T cells resulted in two bands: an unprocessed band >124 kDa and a processed band at ~91 kDa corresponding to S2 fragment of spike (Fig. 4D).Over-expression of TMPRSS2 resulted in marked disappearance of S2 band, while a slightly lower migrating S2’ band was evident at ~80 kDa (Fig. 4D) indicating the processing of spike protein by TMPRSS2 at S2’ region. However, in Val160Met TMPRSS2 over-expressing cells, the processing of spike protein was reduced by ~2·4 fold.
4. Discussion
Although the SARS-CoV-2 virus emerged from China (Mackenzie and Smith, 2020) and spread to various regions of the world, significant differences in the infectivity and mortality rates cannot be completely explained by the various quasi-sub species of the virus. This suggests that the host factors ACE2 receptor and TMPRSS2, may play a role in the varied infection and mortality due to severe disease. This is the first study to report clinical significance of a variant in TMPRSS2 in COVID-19 patients.
At present, India is reported to have the second largest infections in terms of number (22.66 million), second only to the USA (33.47 million), however, the recovery rate is high (~83%) and mortality is low (India:177/1 million, USA:1791/million)0.13 It is interesting to note that the MAF of the identified variant in TMPRSS2is 0·27 in India and 0·15 in USA (Ensembl Genome Browser, n.d.). Further the MAF was relatively lower in the European countries (0·22) as compared to South Asian countries (0·39–0·41) (Ensembl Genome Browser, n.d.). We found a negative correlation (r = −0·76) between MAF and mortality in COVID-19. Despite, strong negative correlation, a few ethnicities with a higher MAF recorded higher mortality as in South Africans, which could be due to risk variants identified in other loci (Ellinghaus et al., 2020). Although significant difference in the genotype frequencies was noted between controls and asymptomatic by dominant model, there was no difference in the genotype frequencies in the Recessive model. This is probably because only SARS-CoV-2 positive patients were recruited and it is highly likely that individuals with the homozygous mutant genotype are largely protected from infection and may test negative. This is also reiterated from the fact that the homozygous mutant genotype (AA) is significantly decreasing in the patient group with increasing level of severity. The MAF of 0·31 in asymptomatic patients, 0·21 in mild-to-moderate patients and 0·19 in severe COVID-19 patients that we determined further reiterates the significant association of the variant with milder disease. Although there was a significant higher mean age and BMI in the symptomatic (mild-to-moderate and severe groups) as compared to asymptomatic group (Table 1), multivariate logistic regression revealed no confounding effect of age (Regression coefficient-0·5; Standard error-0·3; OR-1·62; 95%CI-0·88–2·97; p = 0·11) and BMI (Regression coefficient-0·5; Standard error-0·3; OR-1·68; 95% CI-0·82–3.18; p = 0·14) with respect to variant. No confounding effect of diabetes and hypertension on the relative risk of developing severe disease was identified.
Most of the studies published in literature used bioinformatics approaches on large datasets from the public domain and have consistently identified rs12329760 in TMPRSS2 as the functionally relevant variant in COVID-19 (Asselta et al., 2020; Hou et al., 2020; Senapati et al., 2020). Recently it was reported that higher nasal expression of TMPRSS2 may contribute to the higher burden of COVID-19 in black individuals (Bunyavanich et al., 2020)however, this study has not performed genotype-based expression. More studies are needed to clarify the clinical relevance of the variant beyond extrapolation of expression data to estimate functional relevance. Our study extends previous findings by predicting decreased stability of the TMPRSS2 protein in variant carriers, demonstrating reduced cleavage of spike protein in vitro and association of the variant with decreased disease severity in a large cohort of COVID-19 patients. The results generated in this study suggest that development of small molecules with a potential to decrease the stability of the TMPRSS2 protein could be explored as a prophylactic option to minimize severity of COVID-19.
Having observed higher expression levels of TMPRSS2 in lung tissue at mRNA and protein levels in variant carriers, we compared the levels of ectopically expressed wild type and variant TMPRSS2 and the spike protein cleavage efficiency. It is interesting to note that similar increase in variant TMPRSS2 protein was observed in over expressing HEK 293 T cells. Further, expression data retrieved from public data bases also reveal increase in expression of TMPRSS2 in the variant carriers as compared to the wild type (https://gtexportal.org/home/, n.d.). However, MDS demonstrated increased B factor (stability factor) in TMPRSS2 variant carriers indicating decreased stability of the protein, likely affecting its ability to cleave the spike protein. Consistent with this finding, spike cleavage decreased by 2·4 fold in variant TMPRSS2 expressing cells, indicating decreased viral entry, although its expression was increased. Defective functional protein might elicit constant trigger resulting in increased expression.
Our findings show that spike cleavage function of TMPRSS2 is decreased in the presence of variant, leading to possible reduced viral entry into epithelial cells, causing milder disease. In clinical practice, milder disease associates with reduced mortality. There may be other host factors responsible for less severe disease in South East Asian countries and India including host immunity. These will have to be studied further.
The prevailing pandemic has enforced certain limitations in conducting this study. We could not conduct lung biopsies to demonstrate variant-based viral loads in human lung epithelium because of safety concerns for the COVID-19 patients. Our study demonstrates the potential for targeting TMPRSS2 in high risk groups. Nasal spray targeting TMPRSS2 might have greater efficacy in reducing viral entry and decreasing disease severity. Further studies in large, multi-ethnic cohorts would enable the inclusion of the variant in screening algorithm for prognosis of COVID-19.
While the present work was being reviewed, there is a manuscript in the pre-print server [medRxiv; doi: https://doi.org/10.1101/2021.03.04.21252931] that independently replicated the variant identified in this study and reported protection against severe COVID-19. We were one of the earliest groups to identify this association [doi: https://doi.org/10.1101/2020.06.30.179663 ] and successful replication of the variant in multiple ethnicities confirms its protective role.
In conclusion, our study demonstrates significant association of the variant rs12329760 in TMPRSS2 with decreased disease severity in COVID-19 patients.
Credit author statement
VR: Data curation (Whole exome Sequenicng and Genotyping); Formal analysis; Project administration; Software; Supervision; original draft; Writing - review & editing.
MS: Project administration; Supervision; original draft; Writing - review & editing.
VN: Software; Formal analysis (Molecular dynamic simulations); Writing - review & editing.
SSL: Data curation (functional validation); Formal analysis.
KVLP: Data curation (functional validation); Formal analysis.
KV: Software; Formal analysis (Molecular dynamic simulations).
RA: Writing - review & editing.
SA: and BG Data curation (Whole exome Sequenicng and Genotyping); Software.
KR, DSK, BS: Data curation (Patient recruitment).
GVR: Data curation (Patient recruitment); Writing - review & editing.
DNR: Data curation (Patient recruitment); Project administration; Supervision; Writing - review & editing.
Declaration of Competing Interest
We declare no competing interests.
Acknowledgements
The authors acknowledge the funding received from Asian Healthcare Foundation. The authors acknowledge Dr. HVV Murthy, Statistician, Asian Healthcare Foundation for his help with statistical analyses. We thank the Monash University Software Platform for license and access to the concerned software. SSL acknowledges post-doctoral fellowship from Dr. Reddy's Institute of Life Sciences. KVLP and MS acknowledge the financial support from Scientific and Engineering Research Board (SERB; CRG2019002570).
We are grateful to Dr. Kanneganti Thirumala Devi, Vice Chair of the St. Jude Children's Research Hospital, Department of Immunology, for her suggestions in revising the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mgene.2021.100930.
Appendix A. Supplementary data
Supplementary material 1
References
- Asselta R., Paraboschi E.M., Mantovani A., Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging (Albany NY) 2020;12:10087–10098. doi: 10.18632/aging.103415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera S., Kapadia B., Kain V., et al. ERK1/2 activated PHLPP1 induces skeletal muscle ER stress through the inhibition of a novel substrate AMPK. BiochimBiophysActaMol Basis Dis. 2018;1864(5 Pt A):1702–1716. doi: 10.1016/j.bbadis.2018.02.019. [DOI] [PubMed] [Google Scholar]
- Bunyavanich S., Grant C., Vicencio A. Racial/ethnic variation in nasal gene expression of Transmembrane serine protease 2 (TMPRSS2) JAMA. 2020;324:1–2. doi: 10.1001/jama.2020.17386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Fariselli P., Casadio R., et al. Nucleic Acids Res. 2005;33(Web Server issue):W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-Timofte M.E., Jørgensen S.E., Freytag M.R., et al. Deciphering the role of host genetics in susceptibility to severe COVID-19. Front. Immunol. 2020;11:1606. doi: 10.3389/fimmu.2020.01606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Yu J., He W., et al. Risk factors for death in 1859 subjects with COVID-19. Leukemia. 2020;34:2173–2183. doi: 10.1038/s41375-020-0911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deedwania P.C., Gupta R., Sharma K.K., et al. High prevalence of metabolic syndrome among urban subjects in India: a multisite study. Diabetes MetabSyndr. 2014;8:156–161. doi: 10.1016/j.dsx.2014.04.033. [DOI] [PubMed] [Google Scholar]
- Eaaswarkhanth M., Al Madhoun A., Al-Mulla F. Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int. J. Infect. Dis. 2020;96:459–460. doi: 10.1016/j.ijid.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Degenhardt F., Bujanda L., et al. Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensembl Genome Browser www.ensembl.org accessed at. (Accessed 15th September 2020)
- FitzGerald L.M., Agalliu I., Johnson K., et al. Association of TMPRSS2-ERG gene fusion with clinical characteristics and outcomes: results from a population-based study of prostate cancer. BMC Cancer. 2008;8:230. doi: 10.1186/1471-2407-8-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Zhao J., Martin W., Kallianpur A., Chung M.K., Jehi L., Sharifi N., Erzurum S., Eng C., Cheng F. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18:216. doi: 10.1186/s12916-020-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://gtexportal.org/home/ (accessed 14th September 2020)
- Ioannidis J.P.A., Axfors C., Contopoulos-Ioannidis D.G. Population-level COVID-19 mortality risk for non-elderly individuals overall and for non-elderly individuals without underlying diseases in pandemic epicenters. Environ. Res. 2020;188:109890. doi: 10.1016/j.envres.2020.109890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.R., Hoque M.N., Rahman M.S., et al. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020;10:14004. doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J.S., Smith D.W. COVID-19: a novel zoonotic disease caused by a coronavirus from China: what we know and what we don’t. Microbiol Aust. 2020:MA20013. doi: 10.1071/MA20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Health Commission of the People's Republic of China The Notification of Printing and Distributing New Coronavirus Pneumonia Management (Trial Version 6) 2020. http://www.nhc.gov.cn/yzygj/s7653p/202002/8334a8326dd94d329df351d7da8aefc2.sht in Chinese. (accessed 24 February 2020)
- Ofoegbu T.C., David A., Kelley L.A., et al. PhyreRisk: a dynamic web application to bridge genomics, proteomics and 3D structural data to guide interpretation of human genetic variants. J. Mol. Biol. 2019;431:2460–2466. doi: 10.1016/j.jmb.2019.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapati S., Kumar S., Singh A.K., et al. Assessment of risk conferred by coding and regulatory variations of TMPRSS2 and CD26 in susceptibility to SARS-CoV-2 infection in human. J. Genet. 2020;99:53. doi: 10.1007/s12041-020-01217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- India State-Level Disease Burden Initiative Diabetes Collaborators The increasing burden of diabetes and variations among the states of India: the Global Burden of Disease Study 1990–2016. Lancet Glob. Health. 2018:e1352–e1362. doi: 10.1016/S2214-109X(18)30387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.F., Lin Y.J., Cheng Y.C., et al. PrimerZ: streamlined primer design for promoters, exons and human SNPs. Nucleic Acids Res. 2007;35(Web Server issue) doi: 10.1093/nar/gkm383. (W63–5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verity R., Okell L.C., Dorigatti I., et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson E.J., Walker A.J., Bhaskaran K., et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worldometers https://www.worldometers.info/coronavirus/#countries Available from: (Accessed 10th May 2021)
- Yamamoto N., Bauer G. Apparent difference in fatalities between Central Europe and East Asia due to SARS-COV-2 and COVID-19: four hypotheses for possible explanation. Med. Hypotheses. 2020;144:110160. doi: 10.1016/j.mehy.2020.110160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1