Abstract
De novo germline variants of the casein kinase 2α subunit (CK2α) gene (CSNK2A1) have been reported in individuals with the congenital neuropsychiatric disorder Okur–Chung neurodevelopmental syndrome (OCNS). Here, we report on two unrelated children with OCNS and review the literature to explore the genotype–phenotype relationship in OCNS. Both children showed facial dysmorphism, growth retardation, and neuropsychiatric disorders. Using whole-exome sequencing, we identified two novel de novo CSNK2A1 variants: c.479A>G p.(H160R) and c.238C>T p.(R80C). A search of the literature identified 12 studies that provided information on 35 CSNK2A1 variants in various protein-coding regions of CK2α. By quantitatively analyzing data related to these CSNK2A1 variants and their corresponding phenotypes, we showed for the first time that mutations in protein-coding CK2α regions appear to influence the phenotypic spectrum of OCNS. Mutations altering the ATP/GTP-binding loop were more likely to cause the widest range of phenotypes. Therefore, any assessment of clinical spectra for this disorder should be extremely thorough. This study not only expands the mutational spectrum of OCNS, but also provides a comprehensive overview to improve our understanding of the genotype–phenotype relationship in OCNS.
Keywords: Casein kinase 2α subunit, CSNK2A1, Okur–Chung neurodevelopmental syndrome, genotype–phenotype relationship, mutational spectrum, whole-exome sequencing
Introduction
Neurodevelopmental disorders (NDDs) are a group of neuropsychiatric disorders (NPDs) that are diagnosed during childhood and encompass a wide range of severities.1 They include global developmental delay (GDD), intellectual disability (ID), autism spectrum disorder (ASD), attention deficit hyperactivity disorder (ADHD), motor disorder (MD), and many behavioral disorders such as sleep problems and temper tantrums. They affect approximately 2% to 5% of children. Identifying the etiology of NDDs has been challenging because of the diversity of non-genetic and genetic causes. Trio-based whole exome sequencing (trio-WES) is emerging as an effective diagnostic tool for identifying children with phenotypically similar and etiologically diverse NDDs, and for discovering new genes and conditions. Many of these conditions result from de novo variants of genes with important roles in neuronal development or function.2
In 2016, de novo germline variants of the α subunit of casein kinase 2 (CK2α) gene (CSNK2A1) were shown to be associated with a novel NDD with multisystemic involvement: Okur–Chung neurodevelopmental syndrome (OCNS, OMIM#617062).3 CK2 is a pleiotropic ubiquitous serine/threonine kinase that participates in a variety of cellular processes such as DNA repair, cell cycle regulation, cell differentiation, and cell signaling.4 It is a heterodimeric complex consisting of two catalytic subunits (CK2α and/or CK2α′) and two regulatory subunits (CK2β/CK2β).5 Disruption of CSNK2A1 in animal embryos can induce neural tube defects and other neurodevelopmental abnormalities.6
Clinical features of OCNS include GDD/ID, ASD, ADHD, MD, behavioral problems, and various dysmorphic facial features.4 Up to October 2020, 33 individuals with de novo germline CSNK2A1 variants had been reported in various countries with detailed phenotypes and genotypes.3–5,7–14 However, the genotype–phenotype relationship in OCNS has not been fully elucidated.
Here, we describe two unrelated individuals from two non-consanguineous Chinese families who exhibited NDDs with growth retardation and distinct dysmorphic facial features. In each child, we identified a heterozygous CSNK2A1 mutation in a protein-coding region of CK2α using trio-WES. Furthermore, by systematically reviewing the literature and comparing the clinical features of all identified patients with OCNS (including the two patients in this report), we determined the phenotypic spectra of organ anomalies and NPDs associated with each CSNK2A1 variant, providing information on the genotype–phenotype relationship in OCNS.
Subjects and Methods
Patients and ethical compliance
Two children with facial dysmorphism, growth retardation, and NPDs were investigated in this study. Informed written consent for genetic testing and publication was obtained from their parents. Genomic DNA was extracted from the whole blood of patients and their parents. Detailed clinical information was obtained from the Children’s Neuroendocrinology Department at Sun Yat-sen Memorial Hospital (Sun Yat-sen University) after written permission was obtained from their parents. This study was approved by the Institutional Review Board of Sun Yat-sen Memorial Hospital, Sun Yat-sen University (approval no. SYSEC-KY-KS-2019-160).
Genetic investigations
Routine G-banded karyotyping and fragile X chromosome analysis were performed according to standard protocols,13 and the International System for Human Cytogenetic Nomenclature was applied to analyze chromosome abnormalities. Array comparative genomic hybridization (aCGH) was employed for the two patients and their parents using the Affymetrix Cytoscan HD suite (Affymetrix, Santa Clara, CA, USA). Labeling and hybridization procedures were conducted following the manufacturer’s instructions. Raw data from the chromosomal microarray were analyzed by Affymetrix Chromosome Analysis Suite Software (Affymetrix). Trio-WES was performed in both families using a trio-design with exon targets isolated by capture using the SureSelectXT Human All Exon V4, V5, or V6 (Agilent Technologies, Santa Clara, CA, USA). Captured libraries were sequenced using the Illumina HiSeq 2000 or 2500 system (Illumina, San Diego, CA, USA). Sequencing methodology and variant annotation were performed according to the manufacturer’s protocols. Final variants were interpretated using ANNOtate VARiation software to determine the predictive value of the functional impact of coding variants, and to assess the allele frequency using the 1000 Genome Project (https://www.genome.gov/27528684/1000-genomes-project), Exome Aggregation Consortium (ExAC) (https://exac.broadinstitute.org), and Genome Aggregation Database (gnomAD) (https://gnomad-sg.org/). All candidate variants were confirmed by Sanger sequencing using an ABI 3130xl Genetic Analyzer (Applied Biosystems, Waltham, MA, USA) according to the manufacturer’s instructions.
In silico analysis
The pathogenicity of candidate variants was predicted by a variety of in silico tools including MutationTaster (https://www.mutationtaster.org), SIFT/PROVEAN (https://provean.jcvi.org/index.php), PolyPhen-2 (https://genetics.bwh.harvard.edu/pph2), and CADD (https://cadd.gs.washington.edu). Nucleotide conservation prediction was perfomed using MEGA tools (https://www.megasoftware.net).
Literature search and selection
The systematic review aimed to identify the phenotypic spectra of OCNS reported in the medical literature and to provide a comprehensive overview of the genotype–phenotype relationship of CSNK2A1 variants in various protein-coding regions of CK2α. The review was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.15 PubMed was thoroughly searched up to October 2020 using the search terms: ‘Okur–Chung syndrome’ OR ‘Okur–Chung neurodevelopmental syndrome’ OR ‘CSNK2A1’.
The following types of studies were included: cohort studies, case–control studies, case series, and case reports. We only included studies with available full texts and selected those that involved patients with OCNS and CSNK2A1 germline variants with full details of phenotypes and genotypes. We excluded articles that described the function and/or mechanism of CSNK2A1 in neoplastic diseases or non-NDDs. Finally, we excluded papers with insufficient phenotype and genotype information.
Data extraction and analysis
The titles and abstracts were screened by two independent reviewers who then read the full texts of selected articles that appeared to meet the eligibility criteria. The accuracy of the extracted data was ensured by discussion and consensus. For papers that met the eligibility criteria, we collected information related to CSNK2A1 mutations, associated clinical features and phenotypes of OCNS, and corresponding references. To further analyze the genotype–phenotype relationship in OCNS, we classified phenotypic spectra into two types: (1) congenital anomalies including brain, craniofacial, eye/ear, skeletal, limb, skin, gastroesophageal, endocrine system, and immune system abnormalities to determine the spectrum of affected organs in OCNS, and (2) NPDs including seizures, dystonia, ASD, ADHD, GDD/ID, MD, sleep problems, and temper tantrums/socially inappropriate behavior to determine the spectrum of neuropsychiatric disorders in OCNS. We then quantitively analyzed the associations between these phenotypic spectra and CSNK2A1 variants located in different regions of CK2α using cumulative scores.
The cumulative scores adopted in the study were designed as a scoring system, with one score given to each congenital anomaly phenotype and another to each NPD phenotype. The total congenital involvement and NPDs were then calculated for each patient. If both the cumulative anomaly score and the cumulative NPD score were zero in the same individual, this was considered to be within the normal range of phenotypic spectra.
Statistical analysis
The cumulative organ anomaly score and NPD score among CSNK2A1 variants in different regions of CK2α were presented as means ± SD, and differences between two cumulative scores were compared using Mann–Whitney U tests. P values <0.05 were considered significant. All statistical analyses were carried out using IBM SPSS software version 22 (IBM Inc., Armonk, NY, USA).
Results
Clinical features
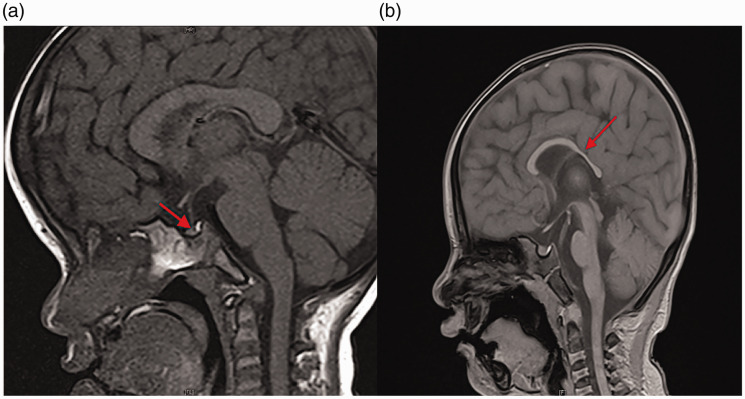
Patient 1 was a 3-year-old girl who presented with severe growth retardation, febrile convulsions, and distinct facial features including a prominent forehead, almond-shaped eyes, and low-set ears. Her parents were non-consanguineous healthy Chinese individuals, and she had a healthy twin brother. She was delivered after an uncomplicated 38-week twin pregnancy, and had no pathological features at the time of birth. Her body weight, length, and head circumference at birth were 2750 g (−1.26 SD), 49 cm (−0.41 SD), and 33 cm (−0.83 SD), respectively. All birth measurements were within normal ranges. Her motor development was slightly delayed but her speech development was normal. Head control was achieved at 6 months, sitting unsupported at 8 months, standing independently at 1 year, and walking unaided at 1 year and 6 months. She spoke meaningful words at 1 year and showed no autistic features or other behavioral problems. Since the age of 9 months, she has experienced five bouts of febrile convulsions. Almost all electroencephalograms (EEGs) showed no obvious abnormal findings, except for one urgent EEG which revealed a small amount of focal left temporal spike-wave discharges. All convulsions were self-terminating and she had not required any anti-epileptic treatments. During her initial consultation at the age of 3 years, physical examination revealed short stature (89 cm, −2.26 SD) with a mild delayed bone age (Tanner–Whitehouse 3 [TW3]-radius, ulna, and short-bone [RUS]: −4 months, TW3-Carpal: −5 months) compared with her twin brother, underweight (11.0 kg, −2.25 SD), and low-normal head circumference (47.9 cm, −0.69 SD). Administering levodopa (10 mg/kg) plus pyridostigmine (1 mg/kg) in a growth hormone (GH) secretion test revealed that the peak GH level was only 7.8 µg/L, indicating partial GH deficiency. Levels of other anterior pituitary gland hormones including thyroid-stimulating hormone, adrenocorticotropic hormone, and gonadotropins were all within normal ranges. Cranial magnetic resonance imaging (MRI) showed a Rathke’s cleft cyst and a mildly reduced anterior pituitary gland (Figure 1a). Using the Chinese version of the Denver Developmental Screening Test (DDST), a questionnaire developed for children aged 0 to 6 years, 13 her developmental quotient (DQ) was tested at the age of 42 months. T-values of adaptive, gross motor, fine motor, and personal–social DQ were 57, 55, 63, and 68, respectively, indicating only mild developmental delay. Routine immunological and biochemical testing of peripheral blood revealed mild-to-moderate hypogammaglobulinemia (serum IgG: 4.271 g/L) and severe IgA deficiency (serum IgA: 0.159 g/L). Other laboratory testing, including organic acids, lactic acids, amino acids, and lymphocyte subset measures, were all unremarkable.
Figure 1.
MRI imaging characteristics of two patients with de novo CSNK2A1 variants. (a) Sagittal T1-weighted pituitary MRI of patient 1 at 3 years of age showing Rathke’s cysts and a reduced anterior pituitary gland (red arrow). (b) Sagittal T1-weighted brain MRI of patient 2 at 2 years of age showing dysplasia of the corpus callosum (red arrow).
MRI, magnetic resonance imaging; CSNK2A1, casein kinase 2α subunit gene.
Patient 2 was a 2-year-old girl who presented with moderate-to-severe growth retardation, GDD, and dysmorphic facial features including arched eyebrows, epicanthic folds, a broad nasal bridge, and micrognathia. Her parents were non-consanguineous healthy Chinese individuals. She was delivered after an uneventful 40-week pregnancy. Her body weight, length, and head circumference at birth were 2850 g (−1.00 SD), 47.5 cm (−1.29 SD), and 33 cm (−0.83 SD), respectively. All birth measurements were within normal ranges. All developmental milestones were severely delayed. She started gaining head control at 8 months and cannot yet sit or stand without support. She also cannot speak any meaningful words. During her initial consultation at the age of 2 years, physical examination showed microcephaly (43.4 cm, −2.97 SD), low-normal height (83.1 cm, −1.80 SD), an approximately normal bone age, and low-normal weight (10.3 kg, −1.55 SD). Considering the presence of growth retardation, her parents requested a GH secretion test involving levodopa (10 mg/kg) plus pyridostigmine (1 mg/kg). The peak GH level was 13.2 µg/L, indicating normal GH stimulation. However, cranial MRI revealed dysplasia of the corpus callosum with a normal-sized pituitary gland (Figure 1b). DDST (Chinese version) testing of her DQ at 25 months of age revealed t-values of adaptive, gross motor, fine motor, and personal–social DQ of 37, 30, 25, and 20, respectively, indicating severe developmental delay. Routine immunological and biochemical testing of peripheral blood and other laboratory testing, including organic acids, lactic acids, amino acids, and lymphocyte subset measures, were all unremarkable.
Identification of de novo CSNK2A1 variants
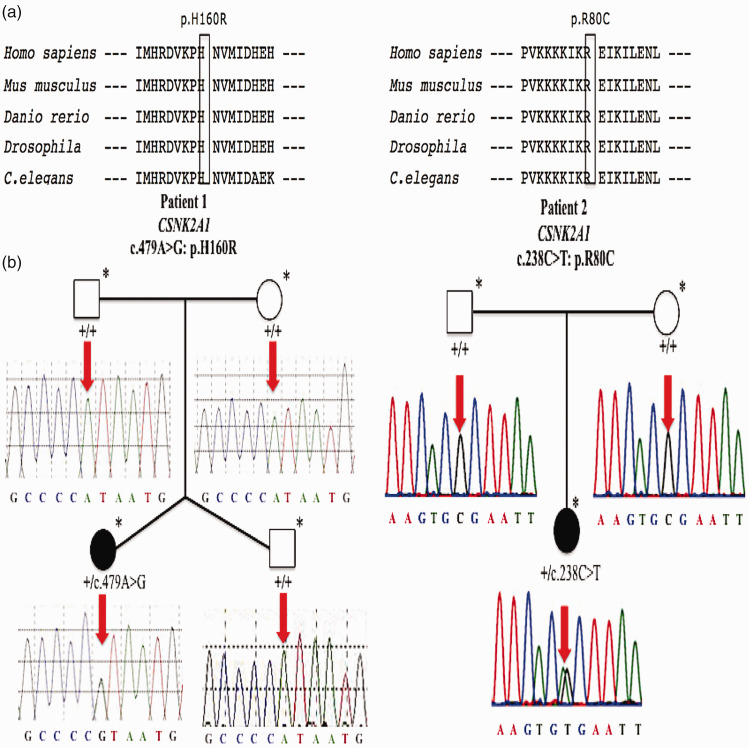
Routine genetic investigations including karyotype, fragile X-chromosome analysis, and aCGH testing were all normal in both patients. Trio-WES was conducted on genomic DNA extracted from whole blood samples from both subjects and their parents to identify variants consistent with autosomal/X-linked dominant or recessive models (including those that were de novo). We found a novel heterozygous CSNK2A1 variant (NM_177559.2: c.479A>G, p.H160R) in patient 1, and another heterozygous CSNK2A1 variant (NM_177559.2: c.238C>T, p.R80C) in patient 2. We classified the two de novo CSNK2A1 variants according to American College of Medical Genetics and Genomics variant classification guidelines.16 First, the variants were both predicted to be deleterious based on multiple lines of computational evidence (PP3) (Table 1). They were also absent from major allele frequency databases, including the 1000 Genomes Project, gnomAD, and ExAC databases (PM2). Additionally, the mutational locus p.R80C was previously reported in a patient with OCNS (PS1).5 His 160 (H160) and Arg 80 (R80) in CK2α are highly evolutionarily conserved and the corresponding missense variants are located in the evolutionarily conserved activation segment of CK2α (Figure 2a), which is critical for protein function, with no benign variations (as shown by the red boxes in Figure 4a) (PM1). Furthermore, using Sanger sequencing, we confirmed that the two variants occurred de novo in the respective families (PS2) (Figure 2b). Considering these findings, including the in silico results, we concluded that the de novo missense CSNK2A1 variants, c.479A>G (p.H160R) and c.238C>T (p.R80C), could be classified as likely pathogenic and pathogenic, respectively. Consequently, they were the most plausible causative variants in our two patients, and the genetic diagnosis of OCNS was made for each of the two subjects.
Table 1.
In silico pathogenicity prediction of detected de novo CSNK2A1 variants.
| Refseq | Variant | chr20 co-ordinates (GRCH37/hg19) | Origin | Mutation Taster | SIFT | PROVEAN | PolyPhen-2 HumVar | CADD phred |
|---|---|---|---|---|---|---|---|---|
| NM_177559.2 | c.479A>G, p.(H160R) | 476394 | De novo | Disease causing (0.999) | Damaging (0) | Deleterious (–7.84) | Damaging (0.900) | 25.6 |
| NM_177559.2 | c.238C>T, p.(R80C) | 480554 | De novo | Disease causing (0.999) | Damaging (0) | Deleterious (–7.63) | Damaging (0.999) | 32.0 |
SIFT, Sorting Intolerant From Tolerant; PROVEAN, Protein Variation Effect Analyzer; PolyPhen-2 HumVar, Polymorphism Phenotyping v2 human variation; CADD phred, Combined Annotation Dependent Depletion Phil’s read editor.
Figure 2.
Comparative sequence alignments and Sanger sequencing electropherogram of two patients with de novo CSNK2A1 variants. (a) Comparative sequence alignments revealing that the two missense variants, H160R (right) and R80C (left), occur at evolutionarily conserved amino acids of the CK2α protein (black boxes). (b) Familial pedigrees. Affected individuals including patient 1 (right) and patient 2 (left) are represented by black circles; other unaffected family members are represented by white circles/squares. Family members who underwent whole-exome sequencing are marked by an asterisk (*). Sanger sequencing electropherograms and genotypes are shown for each family member examined. Wild-type alleles are represented by + and mutational locus is indicated by a red arrow.
CSNK2A1, casein kinase 2α subunit gene; CK2α, casein kinase 2α subunit.
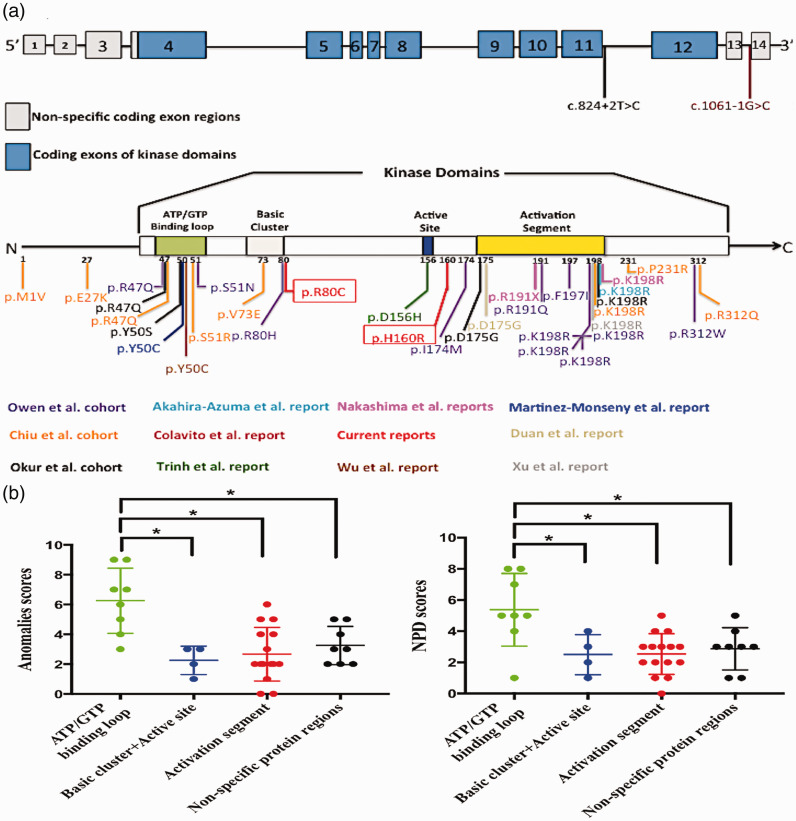
Figure 4.
Comparisons of genotypic and phenotypic spectra among reported variants located in different regions of CK2α or CSNK2A1. (a) Distributions of genotypic spectra of all reported variants, including current ones, with schematics of the CSNK2A1 exon structure (above) and CK2α protein (below). (b) Scatter plots showing scoring differences for phenotypic spectra of congenital anomalies (right) and neuropsychiatric disorders (left) among variants residing in the ATP/GTP binding loop, activation segment, and other protein regions. *Significant differences between the two groups were set at P < 0.05.
CSNK2A1, casein kinase 2α subunit gene; CK2α, casein kinase 2α subunit; NPD, neuropsychiatric disorders.
Systematic review of the genotype–phenotype relationship in identified CSNK2A1 variants
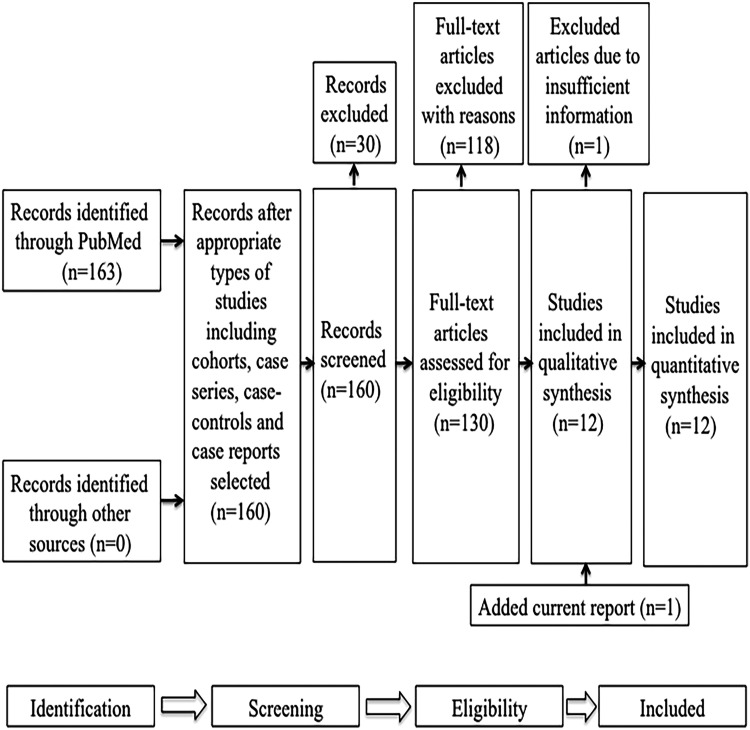
The initial literature search yielded 163 articles. After selecting full-text reports on cohort studies, case–control studies, case series, and case reports in humans, 130 remained. Of these, 118 did not meet eligibility criteria, but 12 did. However, 1 of the 12 lacked detailed information on CSNK2A1 mutations and associated clinical features, so was excluded.2 We added our current study to give a total of 12 that met eligibility criteria (Figure 3).3–5,7–14 Thirty-five CSNK2A1 germline variants/patients with OCNS with details regarding genotypes and phenotypes were included in the final analysis.
Figure 3.
PRISMA chart. Search results for studies describing patients with OCNS carrying CSNK2A1 variants.
PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-Analyses.
CSNK2A1 was previously shown to have a large protein kinase domain (encoded by exons 4–12), including an ATP/GTP-binding loop, a basic cluster, an active site, an activation segment, and non-specific kinase domains.3After summarizing the genotypic features of the 35 patients with de novo CSNK2A1 variants (Figure 4a), we found that only three variants (p.M1V, p.E27K, and c.1061-1G>C) were located outside of the protein kinase domain/exons 4 to 12, whereas the remainder were located within this domain (32/35, 91.43%). Among these 32 variants, 15 involved the activation segment (15/32, 46.86%), 8 involved the ATP/GTP-binding loop (8/32, 25.00%), 5 involved non-specific kinase domains (5/32, 15.63%), 3 involved the basic cluster (3/32, 9.38%), and 1 involved the active site (1/32, 3.13%). We speculate that both the activation segment and ATP/GTP-binding loop are mutation hotspots.
We next reviewed phenotypic information on previously described patients with OCNS and the two current patients, and found that individuals with de novo CSNK2A1 variants had various congenital anomalies and NPDs with different ranges of phenotypic spectra. The most commonly reported phenotypes of congenital anomalies were facial dysmorphism (19/35, 54.29%), gastroesophageal reflux disorder (17/35, 48.57%), short stature (17/35, 48.57%), and brain abnormalities (16/35, 45.71%) (Table 2), and the most frequently reported NPDs were GDD/ID (28/35, 80.00%) and dystonia (21/35, 60.00%) (Table 3). To compare the phenotypic spectra of congenital organ anomalies and NPDs among variants located in different protein-coding regions of CK2α (the non-specific kinase domains and the non-kinase domains), we used cumulative scores to quantify them. No individual was found to have a normal range of phenotypic spectra. As shown in the scatter plots (Figure 4b), the cumulative organ anomaly score was significantly higher for variants in the ATP/GTP-binding loop than for those in the activation segment (6.25 ± 2.19 vs 2.67 ± 1.80, p = 0.001), basic cluster and active site domains (6.25 ± 2.19 vs 2.25 ± 0.96, p = 0.008), and non-specific protein-coding regions (6.25 ± 2.19 vs 3.25 ± 1.28, p = 0.007). Additionally, the cumulative NPD score was significantly higher for variants in the ATP/GTP-binding loop than for those in the activation segment (5.38 ± 2.33 vs 2.53 ± 1.30, p = 0.003), basic cluster and active site domains (5.38 ± 2.33 vs 2.50 ± 1.29, p = 0.039), and non-specific protein-coding regions (5.38 ± 2.33 vs 2.88 ± 1.36, p = 0.018). We therefore speculate that variants in different domains of CK2α play important roles in the range of OCNS phenotypic spectra. Moreover, variants in the ATP/GTP-binding loop are more likely to lead to the widest range of phenotypic spectra of organ anomalies and NPDs compared with variants in the activation segment or other protein-coding regions.
Table 2.
Congenital anomalies of reported variants residing in different regions of the CK2α protein
| Individual |
Variant | Reference | Age (y) | Sex | Anomalies |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Structural domain | Brain | Facial | Eyes/Ears | Skeletal | Limbs | Skin | Cardiac | Renal | Imm | GHD | SS | Micro | GR | Score | ||||||
| 1 | ATP/GTP binding loop | 1 | p.R47Q | 5 | 6.1 | M | + | + | + | + | 4 | ||||||||||
| 2 | 2 | p.R47Q | 3 | 6 | F | + | + | + | + | + | 5 | ||||||||||
| 3 | 3 | p.R47Q | 11 | 2 | F | + | + | + | + | + | + | + | + | + | 9 | ||||||
| 4 | 4 | p.Y50S | 3 | 2 | F | + | + | + | + | + | + | + | + | + | 9 | ||||||
| 5 | 5 | p.Y50C | 13 | 8 | M | + | + | + | + | + | + | + | 7 | ||||||||
| 6 | 6 | p.Y50C | 12 | 5 | F | + | + | + | + | + | + | + | 7 | ||||||||
| 7 | 7 | p.S51R | 11 | 4 | M | + | + | + | + | + | + | 6 | |||||||||
| 8 | 8 | p.S51Y | 5 | 11.1 | F | + | + | + | 3 | ||||||||||||
| 9 | Basic cluster | 1 | p.V73E | 11 | 8 | M | + | + | 2 | ||||||||||||
| 10 | 2 | p.R80H | 5 | 10.7 | M | + | 1 | ||||||||||||||
| 11 | 3 | p.R80C | Current report | 2 | M | + | + | + | 3 | ||||||||||||
| 12 | Active site | 1 | p.D156H | 9 | 7 | M | + | + | + | 3 | |||||||||||
| 13 | Non-specific kinase domain | 1 | c.824 + 2T>C | 3 | 13 | F | + | + | + | 3 | |||||||||||
| 14 | 2 | p.P231R | 11 | 4 | M | + | + | + | 3 | ||||||||||||
| 15 | 3 | p.R312W | 5 | 6 | F | + | + | + | + | + | 5 | ||||||||||
| 16 | 4 | p.R312Q | 11 | 14 | M | + | + | 2 | |||||||||||||
| 17 | 5 | p.H160R | Current report | 3 | F | + | + | + | + | + | 5 | ||||||||||
| 18 | Non-kinase domain | 1 | p.M1V | 11 | 2.5 | F | + | + | 2 | ||||||||||||
| 19 | 2 | p.E27K | 11 | 14 | M | + | + | + | + | 4 | |||||||||||
| 20 | 3 | c.1061-1G>C | 7 | 1 | M | + | + | 2 | |||||||||||||
| 21 | Activation segment | 1 | p.I174M | 5 | 10.9 | M | + | 1 | |||||||||||||
| 22 | 2 | p.R191Q | 5 | 10 | F | + | + | 2 | |||||||||||||
| 23 | 3 | p.D175G | 8 | 1.7 | M | + | + | + | + | 4 | |||||||||||
| 24 | 4 | p.R191* | 4 | 1.7 | M | 0 | |||||||||||||||
| 25 | 5 | p.K198R | 10 | 8 | M | + | + | + | 3 | ||||||||||||
| 26 | 6 | p.K198R | 4 | 15 | F | + | + | + | + | + | 5 | ||||||||||
| 27 | 7 | p.F197I | 5 | 8 | F | + | + | 2 | |||||||||||||
| 28 | 8 | p.K198R | 5 | 7.3 | F | + | + | 2 | |||||||||||||
| 29 | 9 | p.K198R | 5 | 10.9 | M | 0 | |||||||||||||||
| 30 | 10 | p.K198R | 5 | 18.3 | M | + | + | 2 | |||||||||||||
| 31 | 11 | p.K198R | 5 | 18.7 | F | + | + | 2 | |||||||||||||
| 32 | 12 | p.K198R | 11 | 5 | M | + | + | 2 | |||||||||||||
| 33 | 13 | p.K198R | 3 | 4.5 | F | + | + | + | + | + | + | 6 | |||||||||
| 34 | 14 | p.K198R | 14 | 6.8 | M | + | + | + | + | 4 | |||||||||||
| 35 | 15 | p.D175G | 3 | 4 | F | + | + | + | + | + | 5 | ||||||||||
Y, years; M, male; F, female;
SS, Short stature; Micro, Microcephaly; Imm, Immunological abnormalities; GR, Gastroesophageal reflux disorder; GHD, Growth hormone deficiency
Table 3.
Neuropsychiatric disorders of reported variants in different regions of the CK2α protein
| Individual |
Mutation | Reference | Age (y) | Sex | NPD |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Structural domain | Seizure | Dystonia | ASD | ADHD | Sleep | Temper | MD | ID/GDD | Score | |||||
| 1 | ATP/GTP binding loop | 1 | p.R47Q | 5 | 6.1 | M | + | + | + | + | 4 | ||||
| 2 | 2 | p.R47Q | 3 | 6 | F | + | + | + | + | + | 5 | ||||
| 3 | 3 | p.R47Q | 11 | 2 | F | + | + | + | + | + | 5 | ||||
| 4 | 4 | p.Y50S | 3 | 2 | F | + | + | + | + | + | 5 | ||||
| 5 | 5 | p.Y50C | 13 | 8 | M | + | + | + | + | + | + | + | + | 8 | |
| 6 | 6 | p.Y50C | 12 | 5 | F | + | + | + | + | + | + | + | 7 | ||
| 7 | 7 | p.S51R | 11 | 4 | M | + | + | + | + | + | + | + | + | 8 | |
| 8 | 8 | p.S51Y | 5 | 11.1 | F | + | 1 | ||||||||
| 9 | Basic cluster | 1 | p.V73E | 11 | 8 | M | + | + | 2 | ||||||
| 10 | 2 | p.R80H | 5 | 10.7 | M | + | + | + | + | 4 | |||||
| 11 | 3 | p.R80C | Current report | 2 | M | + | 1 | ||||||||
| 12 | Active site | 1 | p.D156H | 9 | 7 | M | + | + | + | 3 | |||||
| 13 | Non-specific kinase domain | 1 | c.824 + 2T>C | 3 | 13 | F | + | + | + | 3 | |||||
| 14 | 2 | p.P231R | 11 | 4 | M | + | + | + | + | 4 | |||||
| 15 | 3 | p.R312W | 5 | 6 | F | + | + | + | 3 | ||||||
| 16 | 4 | p.R312Q | 11 | 14 | M | + | 1 | ||||||||
| 17 | 5 | p.H160R | Current report | 3 | F | + | 1 | ||||||||
| 18 | Non-kinase domain | 1 | p.M1V | 11 | 2.5 | F | + | + | + | 3 | |||||
| 19 | 2 | p.E27K | 11 | 14 | M | + | + | + | + | + | 5 | ||||
| 20 | 3 | c.1061-1G>C | 7 | 1 | M | + | + | + | 3 | ||||||
| 21 | Activation segment | 1 | p.I174M | 5 | 10.9 | M | + | + | + | 3 | |||||
| 22 | 2 | p.R191Q | 5 | 10 | F | + | + | 2 | |||||||
| 23 | 3 | p.D175G | 8 | 1.7 | M | + | + | 2 | |||||||
| 24 | 4 | p.R191* | 4 | 1.7 | M | + | + | 2 | |||||||
| 25 | 5 | p.K198R | 10 | 8 | M | + | + | + | + | 4 | |||||
| 26 | 6 | p.K198R | 4 | 15 | F | + | + | + | 3 | ||||||
| 27 | 7 | p.F197I | 5 | 8 | F | + | 1 | ||||||||
| 28 | 8 | p.K198R | 5 | 7.3 | F | + | + | + | + | + | 5 | ||||
| 29 | 9 | p.K198R | 5 | 10.9 | M | + | 1 | ||||||||
| 30 | 10 | p.K198R | 5 | 18.3 | M | + | + | + | 3 | ||||||
| 31 | 11 | p.K198R | 5 | 18.7 | F | + | + | 2 | |||||||
| 32 | 12 | p.K198R | 11 | 5 | M | 0 | |||||||||
| 33 | 13 | p.K198R | 3 | 4.5 | F | + | + | + | 3 | ||||||
| 34 | 14 | p.K198R | 14 | 6.8 | M | + | + | + | 3 | ||||||
| 35 | 15 | p.D175G | 3 | 4 | F | + | + | + | + | 4 | |||||
Y, years; M, male; F, female; NPD: Neuropsychiatric disorders; GDD: Global developmental delay; ID: Intellectual disability; ASD: Suspected/diagnostic autism spectrum disorders; ADHD: Suspected/diagnostic attention deficit hyperactivity disorder; MD: Motor disorders; Sleep: Sleep problems; Temper: Temper tantrums/socially inappropriate behavior
Discussion
In the present study, we observed a characteristic clinical feature of OCNS (abnormality of the pituitary gland) in patient 1. Five other recently reported patients (individual 5,13 individual 6,12 individual 7,11 individual 25,10 and individual 34,14) also presented with a reduced anterior pituitary gland/pituitary gland duplication and/or GH deficiency (Table 2). These findings suggest that pituitary gland abnormalities are common in patients with OCNS, and that they may occur more often than previously thought. The abnormality also indicates the possibility of pituitary dysplasia in patients with OCNS.
The CK2 protein has been shown to colocalize and interacts with the LHX3 protein, a transcription-related phosphoprotein that is essential in pituitary development.17 CK2 mediates the phosphorylation of specific amino acid residues in the LHX3 epitope, promoting pituitary development at an early stage and pituitary hormone secretion. The dysregulation of LHX3 phosphorylation reduces its capacity to upregulate the transcription of pituitary developmental genes, resulting in pituitary gland abnormalities and dysfunctional pituitary hormone secretion.18 This implies that loss of the CK2 kinase function may be involved in the pathogenesis underlying pituitary gland abnormalities and/or pituitary hormone deficiency. However, it is still unclear how CK2α dysfunction influences pituitary development and hormone secretion, so further research is required.
Missense variants have previously been shown to be the primary CSNK2A1 variant type in OCNS,11 and both variants identified in our current patients were missense. However, the lack of clinical features specific to OCNS makes the attribution of OCNS to CSNK2A1 variants, especially missense variants, particularly challenging. Nevertheless, elucidating the genotype–phenotype relationship of CSNK2A1 variants is required to better understand the mechanisms by which germline CSNK2A1 variants cause OCNS. Previous studies have mainly focused on either genotypic or phenotypic features of patients with germline CSNK2A1 variants. They reported that CSNK2A1 variants were mostly missense variants in the protein kinase domains and that congenital anomalies and NPDs were the most common phenotypes of CK2α-related disorders.4,5,11 No analysis of the genotype–phenotype relationship in CK2α-related disorders has yet been reported.
By thoroughly reviewing current and previously reported CSNK2A1 variants and analyzing their associations with phenotypic spectra, this study shows for the first time that variants in different regions of CK2α result in varied phenotypic spectra. In particular, variants in the ATP/GTP-binding loop appear more likely to lead to the widest range of phenotypic spectra compared with variants in other protein-coding regions of CK2α. It is conceivable that the ATP/GTP-binding loop has a central role in maintaining the substrate recognition and ATP/GTP-dependent catalysis of CK2α, and in stabilizing its normal conformational shifts. As a key element of the ATP/GTP-dependent catalytic subunit, destruction of the N-terminal ATP/GTP-binding loop leads to total loss of catalytic activity and its disruption likely has the most deleterious effects on the brain and other organs, compared with other protein-coding regions of CK2α.19 We propose that systematic examinations should be conducted of patients with OCNA and variants in the ATP/GTP-binding loop to assess the clinical spectra of their affected organs and NPDs.
Our present study was limited in that we could not obtain all clinical information for previously described patients with OCNS, especially details of the degree of each phenotype. It was therefore difficult to evaluate the clinical severity with respect to variants in different protein-coding regions of CK2α and to discriminate severe from less severe phenotypes by cumulative score analysis. We were also unable to ascertain the association between phenotypic spectrum and phenotypic severity in patients with OCNS because we lacked information about phenotype severity. Additionally, giving the same score to each phenotype may have caused unavoidable ascertainment bias when assessing the range of congenital organ anomaly and NPD clinical spectra in patients with OCNS. Therefore, the finding that those patients with variants in the ATP/GTP-binding loop of CK2α have the widest range of phenotypic spectra requires further confirmation. Finally, because few cases involving CSNK2A1 variants are available for analysis, the associations between phenotype and genotype clinical spectra in OCNS are not fully understood. However, we believe that with the increasing accessibility of trio-WES analysis, more patients with CSNK2A1 variants will be identified, allowing an expansion of our knowledge regarding OCNS.
Acknowledgements
We acknowledge the patients and their families for providing relevant medical information and clinical images, as well as their time and patience to take part in the research project. We also thank all clinicians and technicians involved in this study.
Footnotes
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
ORCID iD: Zhan-wen He https://orcid.org/0000-0001-8408-6398
References
- 1.Harris JC. New classification for neurodevelopmental disorders in DSM-5. Curr Opin Psychiatry 2014; 27: 95–97. DOI: 10.1097/YCO.0000000000000042. [DOI] [PubMed] [Google Scholar]
- 2.Deciphering Developmental Disorders Study . Prevalence and architecture of de novo mutations in developmental disorders. Nature 2017; 542: 433–438. DOI: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okur V, Cho MT, Henderson L, et al. De novo mutations in CSNK2A1 are associated with neurodevelopmental abnormalities and dysmorphic features. Hum Genet 2016; 135: 699–705. DOI: 10.1007/s00439-016-1661-y. [DOI] [PubMed] [Google Scholar]
- 4.Nakashima M, Tohyama J, Nakagawa E, et al. Identification of de novo CSNK2A1 and CSNK2B variants in cases of global developmental delay with seizures. J Hum Genet 2019; 64: 313–322. DOI: 10.1038/s10038-018-0559-z. [DOI] [PubMed] [Google Scholar]
- 5.Owen CI, Bowden R, Parker MJ, et al. Extending the phenotype associated with the CSNK2A1-related Okur-Chung syndrome-A clinical study of 11 individuals. Am J Med Genet A 2018; 176: 1108–1114. DOI: 10.1002/ajmg.a.38610. [DOI] [PubMed] [Google Scholar]
- 6.Dominguez I, Degano IR, Chea K, et al. CK2alpha is essential for embryonic morphogenesis. Mol Cell Biochem 2011; 356: 209–216. DOI: 10.1007/s11010-011-0961-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colavito D, Del Giudice E, Ceccato C, et al. Are CSNK2A1 gene mutations associated with retinal dystrophy? Report of a patient carrier of a novel de novo splice site mutation. J Hum Genet 2018; 63: 779–781. DOI: 10.1038/s10038-018-0434-y. [DOI] [PubMed] [Google Scholar]
- 8.Duan HL, Peng J, Pang N, et al. A case of Okur-Chung syndrome caused by CSNK2A1 gene variation and review of literature. Zhonghua Er Ke Za Zhi 2019; 57: 368–372. DOI: 10.3760/cma.j.issn.0578-1310.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Trinh J, Huning I, Budler N, et al. A novel de novo mutation in CSNK2A1: reinforcing the link to neurodevelopmental abnormalities and dysmorphic features. J Hum Genet 2017; 62: 1005–1006. DOI: 10.1038/jhg.2017.73. [DOI] [PubMed] [Google Scholar]
- 10.Akahira-Azuma M, Tsurusaki Y, Enomoto Y, et al. Refining the clinical phenotype of Okur-Chung neurodevelopmental syndrome. Hum Genome Var 2018; 5: 18011. DOI: 10.1038/hgv.2018.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiu ATG, Pei SLC, Mak CCY, et al. Okur-Chung neurodevelopmental syndrome: Eight additional cases with implications on phenotype and genotype expansion. Clin Genet 2018; 93: 880–890. DOI: 10.1111/cge.13196. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Monseny AF, Casas-Alba D, Arjona C, et al. Okur-Chung neurodevelopmental syndrome in a patient from Spain. Am J Med Genet A 2020; 182: 20–24. DOI: 10.1002/ajmg.a.61405. [DOI] [PubMed] [Google Scholar]
- 13.Wu R, Tang W, Liang L, et al. Identification of a novel de novo variant of CSNK2A1 gene in a boy with Okur-Chung neurodevelopmental syndrome. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2020; 37: 641–644. DOI: 10.3760/cma.j.issn.1003-9406.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Xu S, Lian Q, Wu J, et al. Dual molecular diagnosis of tricho-rhino-phalangeal syndrome type I and Okur-Chung neurodevelopmental syndrome in one Chinese patient: a case report. BMC Med Genet 2020; 21: 158. DOI: 10.1186/s12881-020-01096-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009; 6: e1000097. DOI: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424. DOI: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter CS, Malik RE, Witzmann FA, et al. LHX3 interacts with inhibitor of histone acetyltransferase complex subunits LANP and TAF-1beta to modulate pituitary gene regulation. PLoS One 2013; 8: e68898. DOI: 10.1371/journal.pone.0068898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellsworth BS, Butts DL, Camper SA. Mechanisms underlying pituitary hypoplasia and failed cell specification in Lhx3-deficient mice. Dev Biol 2008; 313: 118–129. DOI: 10.1016/j.ydbio.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niefind K, Raaf J, Issinger OG. Protein kinase CK2 in health and disease: Protein kinase CK2: from structures to insights. Cell Mol Life Sci 2009; 66: 1800–1816. DOI: 10.1007/s00018-009-9149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]