Abstract
Purpose:
MicroRNAs play an important role in osteosarcoma (OS) development and progress. Although miR-1253 was considered as a tumor-inhibitor in some cancers, it’s function in the OS is not clear.
Methods:
In our study, we examined the expression of miR-1253 in OS cells and osteoblast cells using quantitative real-time PCR. The proliferation of OS cells was measured by BrdU assay, and we performed transwell to detect migration and invasion of OS cells. Meanwhile, EMT proteins were tested by western blot. We used Bioinformatics to predict the target genes of miR-1253 and found out Matrix metalloproteinases9 (MMP9) was one of that. The direct combination between miR-1253 and MMP9 was verified by double luciferase reporting experiment. Quantitative real-time PCR and western blot were used to detect the expression of MMP9.
Results:
We found that the expression level of miR-1253 in OS cells was significantly lower than that in osteoblast cells. Overexpression of miR-1253 could significantly inhibit OS cell proliferation, migration, invasion and EMT. And then, MMP9 was predicted as a downstream target of miR-1253 by Bioinformatics analysis. Further experiments showed that miR-1253 could reduce the protein level of MMP9 by directly binding to the 3’-UTR of MMP9. Afterward, we performed a rescue experiment, in which both MMP9 and miR-1253 were overexpressed. Compared with the groups overexpressed miR-1253 alone, cell proliferation, migration and invasion in co-overexpression groups were improved.
Conclusions:
In summary, these results suggested that miR-1253 down-regulated in OS cells, and could suppress the proliferation, migration and invasion of OS cells. Its molecular regulatory mechanism was that inhibits the expression of the downstream target gene MMP9 by directly binding, thus affect OS cell functions. Therefore, miR-1253 has the potential to become a biomarker and therapeutic target for OS therapy.
Keywords: miR-1253, MMP9, proliferation, migration, invasion
Introduction
Osteosarcoma (OS) is a common malignant bone tumor in adolescents or children under 20 years of age.1 Before the 1970s, the treatment of the OS was relatively simple. Surgical resection of the tumor segment was the only standard treatment method and the long-term survival rate was less than 20%.2 In recent years, the application of multi-agent chemotherapy regimens significantly improved the prognosis for un-metastasized OS patients and improved the long-term survival rate to 65–70%.3 Approximately 15–20% of patients have clinical metastasis at presentation, and their 5-year survival rate was less than 30%.4 To improve the prognosis and survival rates of OS patients, it is very important to find novel biomarkers for early diagnosis and therapeutic targets for therapy in OS.
MicroRNAs (miRNAs), consisting of 21 bases, are a type of non-coding small RNAs, which inhibit gene expression by binding to the 3’ -untranslated region (3’ -UTR) of the target mRNAs. As a negative regulator of gene expression, miRNAs widely participate in the regulation of cell proliferation, differentiation, apoptosis, migration and transformation.5
In recent years, more and more evidence has shown that abnormal expression and dysfunction of miRNAs have a vital impact on OS cells.6 Liao et al reported that decreased miR-374b expression increases the expression and secretion of vascular endothelial growth factor-A (VEGF-A) and promotes angiogenesis in OS cell lines.7 The expression level of miR-329 was decreased in human OS tissues, and it was negatively correlated with cancer stage.8 Similarly, the miRNAs which were down-regulated in OS and inhibited the tumor cell proliferation and metastasis also include miR-384 and miR-335.9,10
MiR-1253 has been reported to be associated with the progression of several cancers repeatedly. MiR-1253 suppressed cell proliferation and invasion of non-small cell lung carcinoma (NSCLC) by targeting WNT5A.11 In prostate cancer, miR-1253 and its target EZH2 were regulated by FOXC2-AS1 and that pathway participated in tumor growth.12 In human pancreatic ductal adenocarcinoma, miR-1253, as a downstream miRNA of circ_0030235, participated in cell growth promotion.13 But in OS, little research has been reported on the function of miR-1253, only Huang et al found that miR-1253 was absorbed by circNASP in OS cells. MiR-1253 and its target gene FOXF1 regulated the OS cell proliferation and invasion.14
In this study, we examined the expression of miR-1253 in OS cells and its effects on the proliferation, migration and invasion of OS cells were also investigated. Because we found that Matrix metalloproteinases9 (MMP9) was a downstream target gene of miR-1253 by Bioinformatics analysis, so the regulation between miR-1253 and MMP9 in OS was also researched.
Materials and Methods
Cell Lines
The osteoblast cell line hFOB1.19, OS cell lines 143B and MG63 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). hFOB1.19 and MG63 cell were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Life Technologies, Gaithersburg, MD, USA) and 143B cell was cultured in Roswell Park Memorial Institute-1640 (RPMI-1640, Gibco, Life Technologies, Gaithersburg, MD, USA) at 37°C with 5% CO2. All cell lines supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO, USA), 100U/ml penicillin and 100μg/ml streptomycin.
RT-qPCR
Total RNA from cell lines was extracted using the RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA). Used One Step Prime-Script cDNA Synthesis Kit (Takara, Dalian, China) to perform the reverse transcription. Quantitative real-time PCR (RT-qPCR) was performed on the ABI7500 Real-time PCR system (Applied Biosystems, Foster City, CA, USA) using the SYBR® Premix Ex Taq II Kit (Takara, Dalian, China). Used GAPDH and β-actin as internal controls. We applied 2-ΔΔCT to calculate the relative expression quantity. The primers were listed as follows: miR-1253 forward 5’-ACACTCCAGCTGGGAGAGAAGAAGATCAG-3’ and reverse 5’-CTCAACTGGTGTCGTGGA-3’; β-actin forward 5’- TAAAGACCTCTATGCCAACACAGT-3’ and reverse 5’- CACGATGGAGGGGCCGGACTCATC-3’; MMP9 forward 5’- TCTGCCTGCACCACCGACG -3’ and reverse 5’-CTGGGTGTAGAGTCTCTCG-3’; GAPDH forward 5’-GTCTCCTCTGACTTCAACAGCG-3’ and reverse 5’-ACCACCCTGTTGCTGTAGCCAA-3’.
Transfection
MiR-1253 mimics, inhibitor and negative control (NC) were synthesized by Sangon and pcDNA3.1-MMP9 was purchased from Ribobio. The transfection of miRNAs and pcDNA3.1-MMP9 were performed by lipofection using Lipofectamine RNAiMAX transfection reagent (Life Technologies).
BrdU Proliferation Assay
The cells were seeded into 96-well plates. 48 hours after transfection, BrdU (10 μg/mL) incubated for 2 h. Fixed with 4% paraformaldehyde for 30 min. The sections were first denatured at 2mol/L HCl at 37°C for 30 min, and then neutralized in 0.1 M boric acid buffer (pH = 8.5) for 10 min. Then 1 ml of 0.2% Tritonx-100 was added and incubated for 10 min. Added 1 ml 3% BSA and blocked at room temperature for 1 h. BrdU monoclonal antibody (Abcam, ab8152) was treated overnight at 4°C. The sections were then incubated with Alexa Fluor®647-labeled secondary antibodies (Abcam, ab150115). Finally, DAPI (Millipore, USA) was used to stain the nucleus. The number of proliferating cells (BrdU positive) was calculated in 3 random regions/slides.
Transwell Assay
Transwell for migration, Boyden chambers with uncoated polycarbonate filters (8 µm pore size, Corning) were used, while invasion assays used polycarbonate filters (8 µm pore size, Corning) pre-coated with Matrigel Matrix (BD Biosciences). 300µl medium of different groups (1 × 105 cells) and 600µl blank medium (10% FBS) were added in the upper and lower chamber respectively. The migrated cells on the lower chamber were fixed in 4% paraformaldehyde for 20 min, then the cells were stained by hematoxylin. The number of migrated cells was counted under an upright microscope (6 fields per chamber). The invaded cells were undergoing the same operations after removing the matrigel. Each migration and invasion assay was repeated in 3 independent experiments.
Dual-Luciferase Reporter Gene Assay
Two databases: TargetScan(http://www.targetscan.org/Release7.1) and microRNA.org(http://www.microrna.org/microrna/home.do) were used to predict the target genes of miR-1253. We amplified the wide-type (WT) and mutant 3’-UTR of MMP9 and cloned them into the psi-CHECK2 vector to construct the double luciferase reporter vectors. The WT or mutant plasmid (30 ng) was transfected into OS cells couple with miR-1253 mimics, inhibitor or negative control (50nM). Renilla and firefly luciferase activity were measured by the Dual-Luciferase Assay kit (Promega).
Western Blot Analysis
100µl lysate containing RIPA (Beyotime, Shanghai, China) was added to each tube of cells which lysed on the ice for 30 minutes. Centrifuged at 14000 rpm at 4°C for 10mins. Protein concentrations were determined by using the bicinchoninic acid protein assay kit (BCA1-1KT, Sigma-Aldrich). Mixed the total protein (30μg/lane) and 5× loading buffer in 4:1 ratio, and boiled for 10 min. Added the mixture to sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel and run electrophoresis. Then, the isolated protein was transferred from gel to polyvinylidene fluoride (PVDF) membrane. Then, rinsed it with TBST for 5 min and blocked for 2 hours with 5% skim milk powder at room temperature. Membranes were incubated with anti-MMP9, anti-E-cadherin, anti- N-cadherin, anti-vimentin or anti-GAPDH (Abcam) overnight at 4°C. The secondary antibody (Invitrogen) conjugated horseradish peroxidase (HRP) was used to combine with primary antibodys. Finally, bands were visualized by Enhanced Chemiluminescence reagent (PerkinElmer Life Sciences, MA, USA). The density was scanned by Image J software.
Statistical Analysis
We used SPSS 17.0 (SPSS Inc., Chicago, IL, USA) to analyze all experimental data. A student’s t-test was performed for statistical analysis of the differences between 2 groups. P < 0.05 was considered as statistical significance.
Results
miR-1253 Was Downregulated in OS Cell Lines
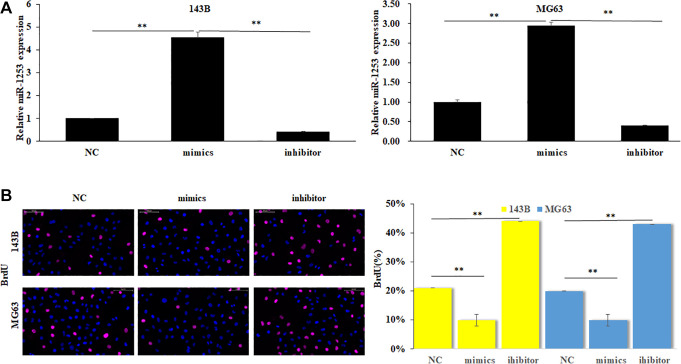
In order to determine whether miR-1253 differentially expressed in OS cells, we used RT-qPCR to analyze the expression of miR-1253. The result showed that the miR-1253 expression was significantly decreased in OS cell lines 143B and MG63 comparing with normal cell line hFOB1.19 (Figure 1).
Figure 1.
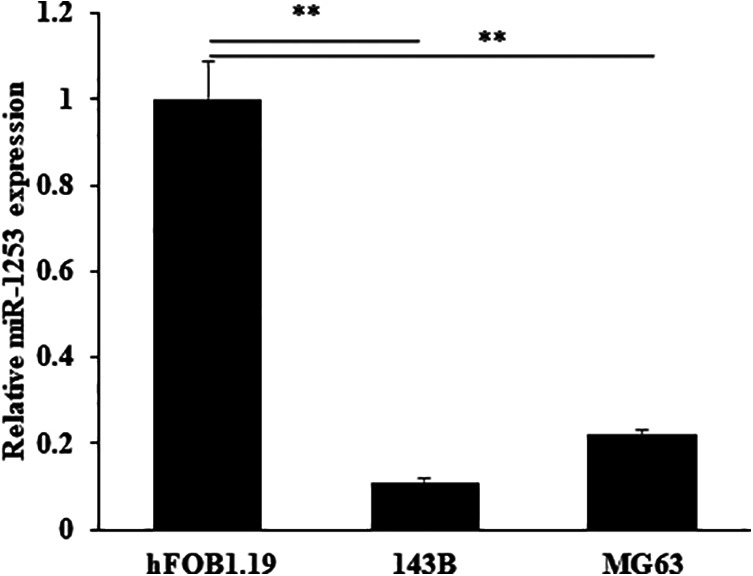
MiR-1253 was down-regulated in OS cells. The expression of miR-1253 was detected by RT-qPCR in OS cells and normal cells.
**P < 0.01.
miR-1253 Suppressed the Proliferation of OS Cells
To evaluate the effect of miR-1253 on OS cell proliferation, we transfected miR-1253 mimics, inhibitor and NC respectively into both 143B and MG63 cells to change the expression of miR-1253 (Figure 2A) (P < 0.01). Then, cell proliferation was determined by BrdU assay. The results show that the BrdU positive number of OS cells was significantly decreased when miR-1253 was overexpressed; on the contrary, when the expression of miR-1253 was inhibited, the BrdU positive number of OS cells was greatly increased (Figure 2B) (P < 0.01). These results revealed that miR-1253 could suppress the proliferation of OS cells.
Figure 2.
miR-1253 suppressed the proliferation of OS cells. (A) miR-1253 expression was detected by RT-PCR when transfecting miR-1253 mimics, inhibitor and negative control into both 143B and MG63 cells. (B) The cell proliferation of all groups was determined by BrdU assay. **P < 0.01.
miR-1253 Suppressed the Migration and Invasion of OS Cells
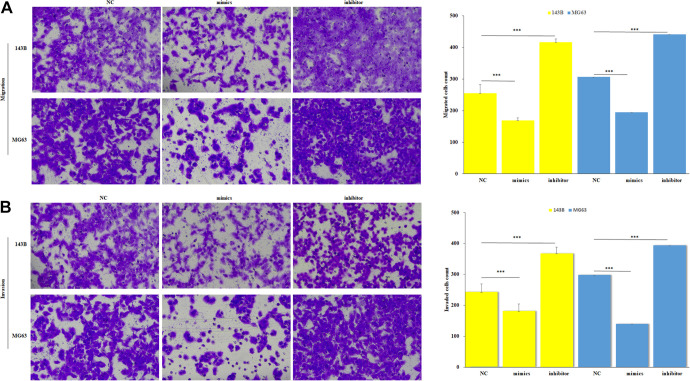
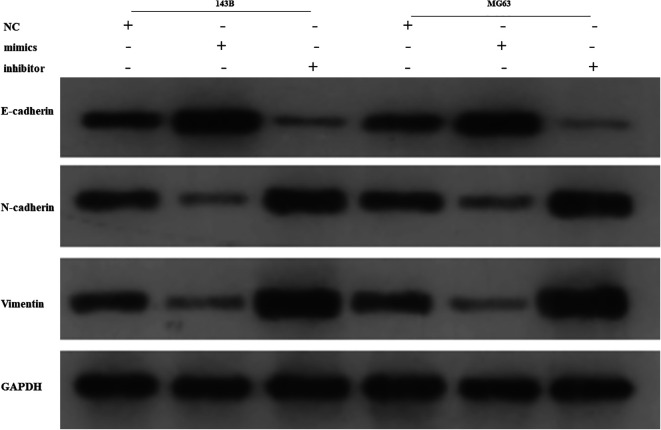
Transwell assay was used to investigate the function of miR-1253 on migration and invasion. The migration and invasion ability of OS cells were significantly decreased in the group of miR-1253 overexpression compared to the negative control (Figure 3A, 3B) (p < 0.001), while the results were completely reversed in the inhibition group. Epithelial-mesenchymal transition (EMT) proteins detection showed that E-cadherin expression increased while N-cadherin and vimentin expression decreased in the miR-1253 overexpression group. The inhibition group did the opposite (Figure 4). These results suggested that miR-1253 can suppress the metastasis of OS cells.
Figure 3.
MiR-1253 suppressed the migration and invasion of OS cells. The migration (A) and invasion (B) of all groups were detected by transwell assay after transfecting. ***P < 0.001.
Figure 4.
MiR-1253 suppressed the EMT of OS cells. The proteins level of E-cadherin, N-cadherin and vimentin were detected by western blot.
MMP9 Was the Direct Target Gene of miR-1253
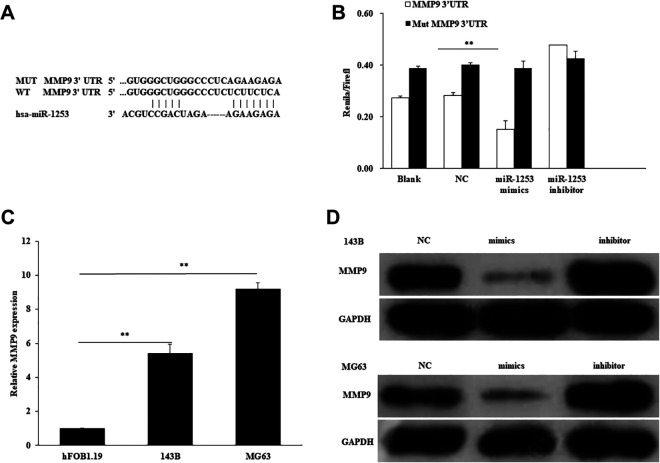
To clarify the molecular mechanisms of miR-1253 in the OS cells function, the downstream targets of miR-1253 were predicted by the TargetScan and microRNA.org databases. We recognized that MMP9 has a binding site of miR-1253 at the 3′-UTR (Figure 5A). Then, luciferase reporter assay was performed to detect whether miR-1253 directly binds to MMP9. WT or mutant psi-CHECK2-MMP9-3′UTR was transfected into different groups of miR-1253 mimics, inhibitor or NC. Their results showed that the luciferase activity of WT psi-CHECK2-MMP9-3′UTR, but not the mutant, was significantly decreased by miR-1253 mimics (Figure 5B). These results indicated MMP9 was the direct target of miR-1253.
Figure 5.
MMP9 was the direct target gene of miR-1253. (A) The binding site of miR-1253 and it’s mutant at the 3′-UTR of MMP9. (B) The Luciferase reporter assay was performed by co-transfecting WT or mutant psi-CHECK2-MMP9-3′UTR with miR-1253 mimics, inhibitor or negative control, then luciferase activity/renilla luciferase activity was measured. **P < 0.01. (C) The expression of MMP9 in OS cells and normal cells was detected. (D) 143B and MG63 cells were transfected with miR-1253 mimics, inhibitor or its control, the protein level of MMP9 was detected by western blot.
miR-1253 Negatively Regulated the Expression of MMP9 in OS Cells
To verify the relationship between miR-1253 and MMP9 in OS cells, we first examined the expression of MMP9 in OS cells, then the protein level of MMP9 was detected by western blot assay when transfecting miR-1253 mimics, inhibitor or NC. The results showed that the expression of MMP9 in OS cells was significantly higher than that in the normal cells (Figure 5C). Overexpression of miR-1253 significantly decreased the protein level of MMP9 as compared to the negative control, while the inhibitory group increased the MMP9 expression (Figure 5D). These results suggested that miR-1253 negatively regulated MMP9 in OS cells.
miR-1253 Suppressed Proliferation, Migration and Invasion of OS Cells by Targeting MMP9
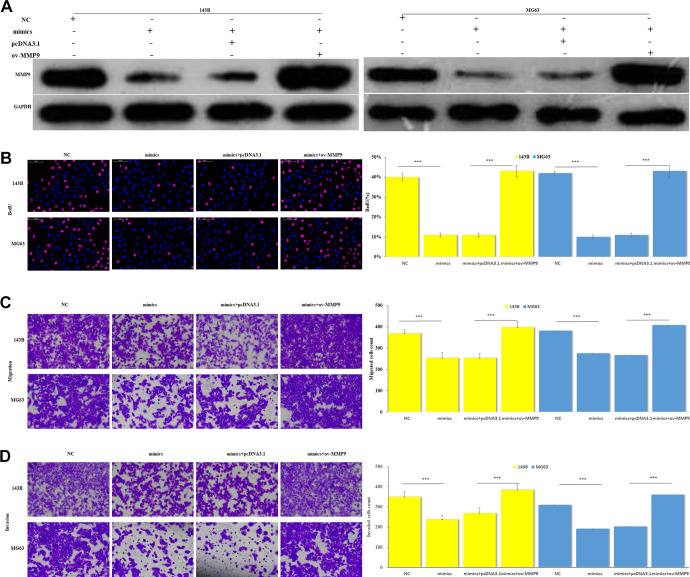
To further verify the effect of this negative correlation between MMP9 and miR-1253 on OS cells, we co-transfected miR-1253 mimics with pcDNA3.1-MMP9 or its NC control into 143B and MG63 cells. Western blot was used to detect the protein level of MMP9. BrdU assay was used to detect the cell proliferation of each group, transwell experiments were used to detect cell migration and invasion abilities. The results showed that after co-transfecting miR-1253 mimics and pcDNA3.1-MMP9, the inhibitory effect of miR-1253 on the MMP9 protein level was weakened (Figure 6A). While the proliferation (Figure 6B), migration (Figure 6C) and invasion (Figure 6D) abilities of co-overexpression groups were enhanced compared with the mimics group. These results suggested that the overexpression of MMP9 in OS cells can offset the reduction of proliferation, migration and invasion abilities induced by miR-1253, which means miR-1253 suppressed proliferation, migration and invasion of OS cells by targeting MMP9.
Figure 6.
Restoration of the MMP9 expression could attenuate the effects of miR-1253 on OS cells. (A) Western blot assay was performed to detect the MMP9 restoration transfecting miR-1253 mimics into pcDNA3.1-MMP9 and its NC control cells on 143B and MG63 cells, using GAPDH as the internal reference. (B) The cell proliferation of different groups was determined by BrdU assay. (C) The migration of different groups was detected by transwell assay. (D) The invasion of different groups was detected by transwell assay. **P < 0.001.
Discussion
In recent years, numerous studies have confirmed that miRNAs are indeed involved in the development and progression of many cancers. In addition, certain miRNAs in the blood of patients with several cancers showed significant specific abnormal expression. Therefore, miRNAs are likely to have potential as diagnostic markers for cancer. On the other hand, in vitro studies have found that changing the expression of miRNAs can change the proliferation or metastasis ability of tumor cells. Therefore, the potential of miRNAs as drug targets cannot be ignored.
In our study, to elucidate the function of miR-1253 in OS, we detected the expression of miR-1253, showed a downward trend in the OS cells, it’s similar to the researches of miR-1253 in other cancers.11-13 Then we focus on its functions on OS cells, miR-1253 significantly reduced the proliferation, migration and invasion of OS cells. These results revealed that miR-1253 plays a tumor-suppressive role in OS.
It is well known that the function of miRNA depends on the regulation of the downstream target gene expression. 23 types of MMPs have been reported in humans.15 Of these enzymes, MMP9 has been reported to be involved in the migration and infiltration of OS cells. Cho et al confirmed disulfiram regulated the invasion of human OS cells by inhibiting expression of MMP2 and MMP9.16 Another research suggested that MMP9 as one of the target genes of miRNA-218 participated in OS cell migration and invasion.17 It has also been reported that MMP9 cooperating with serum alkaline phosphatase could predict the metastasis and poor prognosis in patients with early OS.18 In our study, the bioinformatics database predicted that MMP9 was a target gene of miR-1253, so we furthermore investigated whether the suppression of miR-1253 on OS cell proliferation, migration and invasion is realized by targeting MMP9.
Our study indicated that miR-1253 directly bound to the 3’-UTR of MMP9 and negatively regulated the protein level of MMP9 in OS cells. Then, we restored the expression of MMP9 in groups that had overexpressed miR-1253 and we found the inhibitory effect of miR-1253 on OS cell proliferation, migration and invasion were attenuated.
This study suggested that miR-1253 maybe has the potential to be a novel biomarker because of the aberrant expression in OS. Our studies also mean that miR-1253 might be a potential therapeutic target for OS because of its inhibition on cell growth and metastasis of OS cells. Meanwhile, drugs that target the dysregulated expression of miR-1253 are also valuable. Some researchers have already been trying to verify the therapeutic effect of abnormal miRNAs on OS. Osaki used atelocollagen as a carrier to intravenous inject miR-143 to a spontaneous OS lung metastasis model, results showed the metastasis of the primary lesion was inhibited.19 Furthermore, Xin et al found that injection of miR-22 inhibited OS tumor growth and metastasis in an animal model.20 Of course, whether miR-1253 can be used as a biomarker or a drug target still has a long way to go, and more and deeper experimental studies are needed, which is also our next direction.
Conclusion
This research enriched the role of miR-1253 in the OS process. Down-regulated miR-1253 was found in OS cells and was related to the proliferation, migration and invasion of OS. Excessive miR-1253 induces inhibition of metastasis of cancer cells by negatively regulating MMP9. Though more experiments from vivo are needed to verify the deeply mechanism, this study already indicated that the miR-1253/MMP9 axis maybe is a new direction of OS research. MiR-1253 may have the potential to be a biomarker and therapeutic target for OS.
Abbreviations
- BrdU
Bromodeoxyuridine
- EMT
epithelial-mesenchymal transition
- HRP
horseradish peroxidase
- MMP9
Matrix metalloproteinases9
- OS
Osteosarcoma
- PVDF
polyvinylidene fluoride
- RT-PCR
Quantitative real-time PCR
- UTR
3′-untranslated region
Footnotes
Authors’ Contributions: Jianwen Mo, Tiansheng Zheng performed the experiments, wrote the manuscript and prepared the figures making an equal contribution to this study. Li Lei, Peng Dai, Jun Liu, Huabin He, Jin Shi, Xi Chen, Tianting Guo and Bin Yuan researched the literatures and collected the experiments data. Guanglin Ji and Jianwen Mo designed the experiments and reviewed the manuscript and revised the manuscript according to the comments of the reviewers.
All coauthors have reviewed and approved of the manuscript prior to submission.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by the science and technology research project of education department of Jiangxi provincial (Project No. GJJ180811) and science and technology plan of health and family planning commission of Jiangxi provincial (Project No. 20195375).
ORCID iD: Jianwen Mo, MD  https://orcid.org/0000-0003-0502-0716
https://orcid.org/0000-0003-0502-0716
References
- 1. Tabone MD, Terrier P, Pacquement H, et al. Outcome of radiation-related osteosarcoma after treatment of childhood and adolescent cancer: a study of 23 cases. J Clin Oncol. 1999;17(9):2789–2795. [DOI] [PubMed] [Google Scholar]
- 2. Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33(27):3029–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. [DOI] [PubMed] [Google Scholar]
- 4. Gill M, McCarthy M, Murrells T, Silcocks P. Chemotherapy for the primary treatment of osteosarcoma: population effectiveness over 20 years. Lancet. 1988;1(8587):689–692. [DOI] [PubMed] [Google Scholar]
- 5. Worku T, Rehman ZU, Talpur HS, et al. MicroRNAs: new insight in modulating follicular atresia: a review. Int J Mol Sci. 2017;18 (2):2789–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sasaki R, Osaki M, Okada F. MicroRNA-based diagnosis and treatment of metastatic human osteosarcoma. Cancers. 2019;11(4):553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liao YY, Tsai HC, Chou PY, et al. CCL3 promotes angiogenesis by dysregulation of miR-374b/VEGF-A axis in human osteosarcoma cells. Oncotarget. 2016;7(4):4310–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiang W, Liu J, Xu T, Yu X. MiR-329 suppresses osteosarcoma development by downregulating Rab10. FEBS Lett. 2016;590(17):2973–2981. [DOI] [PubMed] [Google Scholar]
- 9. Wang Y, Huang H, Li Y. Knocking down miR-384 promotes growth and metastasis of osteosarcoma MG63 cells by targeting SLBP. Artif Cells Nanomed Biotechnol. 2019;47(1):1458–1465. [DOI] [PubMed] [Google Scholar]
- 10. Xie Y, Deng H, Wei R, et al. Overexpression of miR-335 inhibits the migration and invasion of osteosarcoma by targeting SNIP1. Int J Biol Macromol. 2019;133:137–147. [DOI] [PubMed] [Google Scholar]
- 11. Liu M, Zhang Y, Zhang J, et al. MicroRNA-1253 suppresses cell proliferation and invasion of non-small-cell lung carcinoma by targeting WNT5A. Cell Death Dis. 2018;9(2):189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y, Gu M, Liu C, et al. Long noncoding RNA FOXC2-AS1 facilitates the proliferation and progression of prostate cancer via targeting miR-1253/EZH2. Gene. 2019;686(undefined):37–42. [DOI] [PubMed] [Google Scholar]
- 13. Xu Y, Yao Y, Gao P, Cui Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun. 2019;509(1):138–142. [DOI] [PubMed] [Google Scholar]
- 14. Huang L, Chen M, Pan J, Yu W. Circular RNA circNAS P modulates the malignant behaviors in osteosarcoma via miR-1253/FOXF1 pathway. Biochem Biophys Res Commun. 2018;500(2):511–517. [DOI] [PubMed] [Google Scholar]
- 15. Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2(3):161–174. [DOI] [PubMed] [Google Scholar]
- 16. Cho HJ, Lee TS, Park JB, et al. Disulfiram suppresses invasive ability of osteosarcoma cells via the inhibition of MMP-2 and MMP-9 expression. J Biochem Mol Biol. 2007;40(6):1069–1076. [DOI] [PubMed] [Google Scholar]
- 17. Jin J, Cai L, Liu ZM, Zhou XS. miRNA-218 inhibits osteosarcoma cell migration and invasion by down-regulating of TIAM1, MMP2 and MMP9. Asian Pac J Cancer Prev. 2013;14(6):3681–3684. [DOI] [PubMed] [Google Scholar]
- 18. Han J, Yong B, Luo C, Tan P, Peng T, Shen J. High serum alkaline phosphatase cooperating with MMP-9 predicts metastasis and poor prognosis in patients with primary osteosarcoma in Southern China. World J Surg Oncol. 2012;10(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Osaki M, Takeshita F, Sugimoto Y, et al. MicroRNA-143 regulates human osteosarcoma metastasis by regulating matrix metalloprotease-13 expression. Mol Ther. 2011;19(6):1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xin M, Qiao Z, Li J, et al. miR-22 inhibits tumor growth and metastasis by targeting ATP citrate lyase: evidence in osteosarcoma, prostate cancer, cervical cancer and lung cancer. Oncotarget. 2016;7(28):44252–44265. [DOI] [PMC free article] [PubMed] [Google Scholar]