Abstract
Aim:
We explore HER3 expression in lung adenocarcinoma (adeno-NSCLC) and identify potential mechanisms of HER3 expression.
Materials & methods:
Tumor samples from 45 patients with adeno-NSCLC were analyzed. HER3 and HER2 expression were identified using immunohistochemistry and bioinformatic interrogation of The Cancer Genome Atlas (TCGA).
Results:
HER3 was highly expressed in 42.2% of cases. ERBB3 copy number did not account for HER3 overexpression. Bioinformatic analysis of TCGA demonstrated that MEK activity score (a surrogate of functional signaling) did not correlate with HER3 ligands. ERBB3 RNA expression levels were significantly correlated with MEK activity after adjusting for EGFR expression.
Conclusion:
HER3 expression is common and is a potential therapeutic target by virtue of frequent overexpression and functional downstream signaling.
Keywords: : ERBB3, HER3, MEK, NSCLC
Tweetable abstract
HER3 expression is common in adeno-NSCLC and is a potential therapeutic target by virtue of frequent overexpression and functional downstream signaling.
Background
Non-small-cell lung carcinoma
Lung cancer is the leading cause of cancer mortality worldwide. There is a poor 5-year survival rate in patients with regional and distant disease in non-small-cell lung carcinoma (NSCLC), of 33 and 6%, respectively [1]. Systemic treatment with chemotherapy and immunotherapy is standard of care, while a small subset of cancers demonstrates oncogene-addiction with effective targeted treatments. These include EGFR mutated, ALK rearranged and newer targets such as ROS, RET and NTRK.
HER3
HER3 is a member of the EGFR family of receptors. Investigation of HER3 as a therapeutic target has not been previously prioritized due to its impaired kinase activity [2]. HER3 acts as a heterodimeric partner for other members of the EGFR family, namely EGFR/HER-1 and ERB2/HER2/Neu [3–6]. HER3 possesses weak tyrosine kinase activity and so preferentially forms heterodimers with kinase-proficient receptor tyrosine kinases in the presence of its ligands neuregulin (NRG 1 and NRG 2) [2]. It has also been demonstrated that HER3 can form heterodimers with MET, AXL and IGF1R [7–9]. EGFR has a number of ligands, including amphiregulin, epidermal growth factor and transforming growth factor, while HER2 has no known ligand and remains in an ‘open’, active conformation [10–12]. However, HER2/HER3 dimerization can occur independently of ligand binding [13]. More recently it has been appreciated that HER3 can also homodimerize on binding to the heregulin ligands, NRG1 and NRG2, so having intrinsic, non-EGFR or HER2 mediated activity [14,15]. Downstream signaling of HER3 is mainly via the PI3K/AKT pathway, with MAPK/ERK and JAK/STAT pathways also activated [16,17].
ERBB3, the HER3 gene, is under the regulation of several miRNAs including miR125a, miR125b and miR205 [18–20]. Studies have shown that the ubiquitin-proteasome pathway plays a significant role in cancer initiation and progression by regulating protein levels [9,21–25]. E3 ubiquitin ligases regulate EGFR family receptors. NRDP1 mediates HER3 degradation [9,22]. NRG1 stabilizes USP8 which in turn stabilizes NRDP1 [21] NEDD4 is a novel interaction partner and ubiquitin E3 ligase of HER3 [23].
Evidence for the role of HER3 in lung cancer pathogenesis is largely drawn from retrospective data sets correlating HER3 expression with poor prognosis and metastatic development [26–29]. Accumulating evidence supports the role of HER3 in the development of resistance to EGFR targeted therapies [30–35]. From preclinical data, dual targeting of EGFR and HER3 is capable of overcoming acquired resistance to EGFR inhibition [32,35]. MET amplification or overexpression is a mechanism of resistance to EGFR-TKI occurring in 5% of cases [36]. HER3 phosphorylation is observed in EGFR mutant tumors with MET amplification, indicating that MET activates PI3K/AKT pathway via HER3 phosphorylation [37]. MEK inhibition causes a transcriptional upregulation of both HER2 and HER3 and the formation of heterodimeric complexes in KRAS mutant NSCLC and colon cancer and combined MEK and dual EGFR-HER2 inhibition has a synergistic effect [34]. Mutations of HER3 have been described in the extra-cellular domain or tyrosine kinase domain as a rare event in lung cancer and can influence the activity of HER3 receptor as well as the dimerization affinity with HER2 and EGFR [38–40].
Here, we explore HER3 expression levels in adenocarcinoma of the lung (adeno-NSCLC) patients and correlate it with clinical parameters. We also interrogate large genomic datasets to identify the potential mechanism of HER3 expression and propose that HER3 could be an alternate pathway for MEK pathway activation.
Materials & methods
Patients & tissue samples
The Drug Development Unit at the Royal Marsden recruits patients with advanced solid malignancies to early phase clinical trials. Patients with adeno-NSCLC attending consultations in our Phase I clinic provided informed written consent. The archival or fresh tissue from biopsies were used in the study. Approval was gained from the local research and ethics committee for the use of the clinical data. Archival formalin fixed paraffin embedded tumor blocks were analyzed for HER2 and HER3 protein expression and genomic analysis for HER3 copy number was performed. Patient clinical data were retrospectively collected from the Royal Marsden hospital electronic patient record system.
Immunohistochemistry & scoring system
Tissue blocks were freshly sectioned and only considered for immunohistochemistry (IHC) analyses of HER2 and HER3 if adequate material was present. Protein expression on 3 μm thick formalin-fixed paraffin-embedded (FFPE) sections was assessed by a pathologist blinded to the clinical data.
HER3 and HER2 antibody validation by siRNA and immunohistochemical assays were performed as previously described. Briefly, HER3 immunoreactivity was investigated using the rabbit monoclonal anti-HER3 antibody clone D22C5 (#12708, Cell Signaling Technology, London, UK). HER2 immunoreactivity was investigated using the mouse monoclonal anti-HER2 antibody clone CB11 (#NCL-L-CB11, Leica Biosystems, Newcastle, UK). Rabbit and Mouse IgGs (#I-1000 and I-2000, Vector, CA, USA) were used as negative controls. For HER3 IHC, antigen retrieval was conducted by heating slides in high pH buffer using a microwave, staining was subsequently performed using BioGenex i6000 autostainer (Launch Diagnostics, Kent, UK). HER3 immunoreactivity was detected using Dako EnVision Flex high pH kit (#K800021-2, Agilent, Glostrup, Denmark). For HER2 IHC automated antigen retrieval and detection were performed using Leica Bond RX (Leica Biosystems). HER2 immunoreactivity was detected using the Bond Polymer Refine Detection system (#DS9800, Leica Biosystems).
For HER2, membranous expression was scored according to College of American Pathologists HER2 scoring guidelines [41]. No staining or membrane staining that is incomplete and is faint/barely perceptible and in ≤10% of tumor cells (score 0); incomplete membrane staining that is faint/barely perceptible and in >10% of tumor cells (score 1+); weak-to-moderate complete membrane staining observed in >10% of tumor cells (score 2+) and circumferential membrane staining that is complete, intense and in >10% of tumor cells (score 3+). For HER3 staining, the same guidelines were used to generate an ad hoc HER2-like score for membranous staining. Additionally, HER2 and HER3 cytoplasmic staining was visually assessed, percentage of cells staining weakly, moderately or strongly were noted and H score calculated.
Digital PCR
Digital PCR was performed on a Q×100 droplet digital PCR system (Bio-Rad, CA, USA). Copy number assays were performed with a FAM-ERBB3 validated Bio-Rad assay, reference ID: dHsaCP2500395 (67 bp amplicon) and a recommended reference copy number Hex-assay for AP3B1, assay ID: dHsaCP2500348 (60 bp amplicon). Digital PCR analysis was performed using QuantaSoft v1.3.2.0 software from Bio-Rad to assess the number of positive droplets. At least two negative control wells with no DNA were included in every run and each assay was run at least in duplicate (see Supplementary methods).
Statistical methods
Statistical analysis of IHC and clinical data was performed on Graphpad Prism v7.0d. The association between HER3 expression and other binary data was analyzed using two sided Fisher’s exact test. For survival Kaplan–Meier curves were constructed and Gehan–Breslow–Wilcoxon test compared populations. Survival was taken from diagnosis to death and those alive at follow-up were censored.
Bioinformatic methods
Genomic data and gene expression (RNA-Seq by expectation maximization) for 507 adeno-NSCLC was downloaded from cBioportal for analysis [42]. The RNA-Seq by expectation maximization value represents the normalized expression level of each gene in each tumor. It was calculated by: estimated number of reads that aligned to a gene and; the scaled version of the raw counts using a standard 75th-percentile normalization approach [43]. This approach followed the The Cancer Genome Atlas (TCGA) Research Network recommendations and TCGA lung adenocarcinoma publication [43]. We interrogated the TCGA to determine the relationship between ERBB3 expression and particular oncogenes of interest, namely EGFR and KRAS mutations.
The MEK activity score is an accumulation measurement of MEK pathway activity based on a previously validated 18-gene transcript signature of MEK signaling [44]. Linear regression was used to assess the associations between gene expression levels (and with MEK signaling). All statistical test was performed using R (v1.1.43).
Results
Clinical characteristics
From December 2010 to December 2018, 45 patients with adeno-NSCLC were referred to the Drug Development Unit, consented to tissue analysis and had available archival tumor specimens. The majority of samples were from biopsies (41), the rest being from surgical resection (4). There were relatively fewer males (19) than females (26) in the cohort. The median age at diagnosis was 58 (range: 38–72) and the median overall survival (OS) was 38 months. The median number of lines of treatment for advanced disease was 3 (range: 0–7): 43 patients received chemotherapy (median 2: range 1–4), 22 patients received EGFR tyrosine kinase inhibitor (TKI) (median 1: range 1–3) and nine patients received immunotherapy. All patients who had an EGFR mutation received an EGFR TKI and 25 patients were enrolled in a Phase I trial and received at least one Phase I trial drug.
HER2 & HER3 protein expression
HER3 protein expression showed a membranous distribution with no nuclear expression (Figure 1A). High expression of HER3 (2+ or 3+) was seen in 42.2% of cases (Table 1). HER2 expression was positive in 31.1% of cases, while HER2 and HER3 co-expression was seen in 11.1% (5/45). No correlation between HER2 and HER3 expression was demonstrated (Fisher’s exact test; p > 0.9999). Patient characteristics according to HER3 status are described (Table 2).
Figure 1. . Adeno-non-small-cell lung carcinoma patient samples.
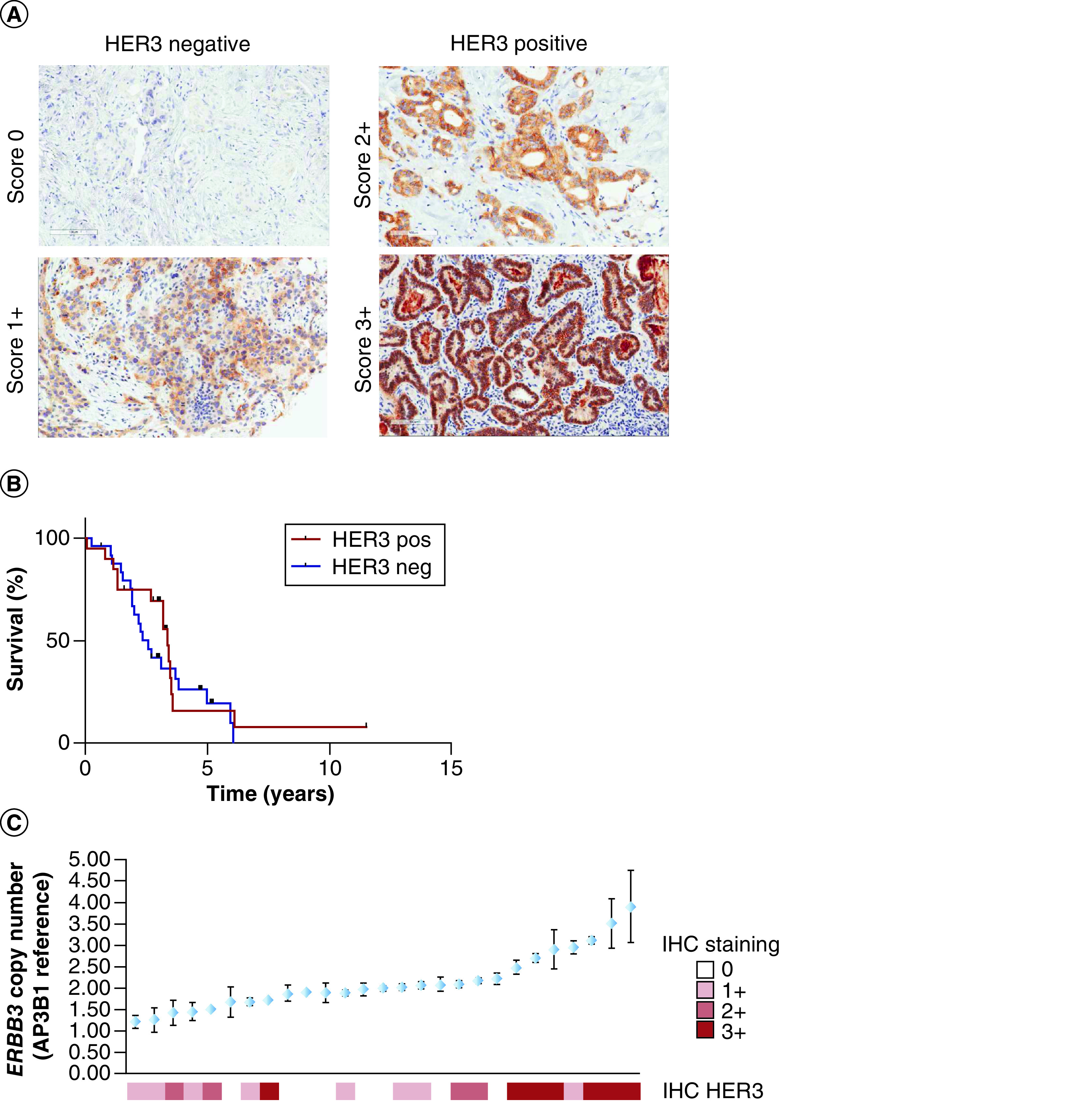
(A) Panel of HER3 negative cases (0–1+) and HER3 positive cases (2–3+). For HER3 staining, the guidelines from CAP HER2 scoring guidelines were used to generate an ad-hoc HER2 like score. (B) Kaplan–Meier curve of HER3 positive versus HER3 negative cases. (C) ERBB3 gene copy number and HER3 immunohistochemistry.
Table 1. . Membranous HER3 and HER2 protein expression by immunohistochemistry.
| HER3 staining | % (n) | |
|---|---|---|
| 0 | 35.5% (16) | 57.8% (26) |
| 1 | 22.2% (10) | |
| 2 | 15.5% (7) | 42.2% (19) |
| 3 | 26.7% (12) |
| HER2 staining | % (n) | |
|---|---|---|
| 0 | 33.3% (15) | 68.9% (31) |
| 1 | 35.5% (16) | |
| 2 | 22.2% (10) | 31.1% (14) |
| 3 | 8.8% (4) |
Table 2. . Clinical characteristics for HER3 positive and HER3 negative patients.
| Clinical characteristics | HER3 positive, % (n) | HER3 negative, % (n) |
|---|---|---|
| Male | 26% (5) | 74% (14) |
| Female | 54% (14) | 46% (12) |
| Median age (years) | 58 | 57.5 |
| Smoking status | ||
| Non smoker | 16% (3) | 12% (3) |
| Current smoker | 5% (1) | 12% (3) |
| Ex-smoker | 47% (9) | 35% (9) |
| Unknown | 32% (6) | 42% (11) |
| Prior therapy | ||
| No prior treatment | 2% (1) | 2% (1) |
| Prior chemotherapy | 35% (17) | 43% (25) |
| Prior immunotherapy | 10% (5) | 7% (4) |
| Prior TKI | 23% (11) | 22% (13) |
| Phase I clinical trial | 17% (10) | 26% (15) |
TKI: Tyrosine kinase inhibitor.
Correlation of HER3 protein expression by IHC with clinical characteristics
The median OS from diagnosis for HER3 positive patients was 40.5 months, and for HER3 negative patients 30.8 months. There was no statistical difference in survival between HER3 positive and HER3 negative cases (Gehan–Breslow–Wilcoxon; p = 0.4596) (Figure 1B). There was no difference in median survival between HER3 positive and negative cases in those patients with EGFR mutations (median OS: 42.6 months HER3 positive, median OS: 43.9 months HER3 negative; Gehan–Breslow–Wilcoxon; p = 0.4927).
HER3 gene amplification
Digital PCR of ERBB3 was performed in 27 cases. There was low frequency of small copy number gain in HER3 with 11% (3/27) having copy number variant of 3–4 (Figure 1C). Six out of the seven samples with this highest copy number variant scored 3+ on HER3 IHC.
EGFR & KRAS mutations & correlation with HER3
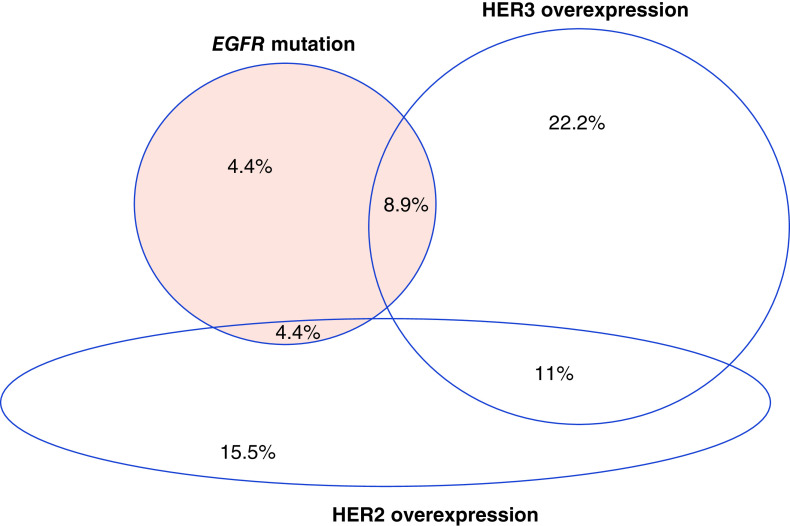
As standard of care, EGFR mutational analysis had been performed in all 45 patients and KRAS mutational analysis in 28 patients. EGFR mutation was present in 17.8% (eight cases). HER3 protein overexpression was present in 8.9% (four cases) of EGFR mutation and in 14.2% (four cases) of KRAS mutation. There was no correlation between HER3 overexpression and EGFR positivity (Fisher’s exact test; p = 0.7043) or HER3 and KRAS positivity (Fisher’s exact test; p = 0.6908) (Figure 2).
Figure 2. . Adeno-non-small-cell lung carcinoma patient samples.
Frequency of aberrations in adeno-non-small-cell lung carcinoma as percentage of total population.
Baseline genomic data in TCGA
Interrogation of the PanCancer TCGA of 507 patients with adeno-NSCLC showed a genetic alteration rate of 31% in the ERBB family with the most common genetic alteration seen in EGFR and ERBB4, which was evident in 16% (79 cases) and 10% (50 cases). In ERBB2 and ERBB3, genetic alterations were less frequent, seen in 4% (19 cases) and 5% (25 cases), respectively (Figure 3A). Among the different types of genetic alteration, amplification was the most common alteration in ERBB3, observed in 48% out of all alterations (12 cases). ERBB3 copy number change demonstrated a positive association with ERBB3 RNA expression (r = 0.26; p = 8 × 10-6) (Figure 3B); amplification cases had 1.7-times higher expression of ERBB3 RNA than diploid cases (p = 0.004).
Figure 3. . The Cancer Genome Atlas adeno-non-small-cell lung carcinoma.
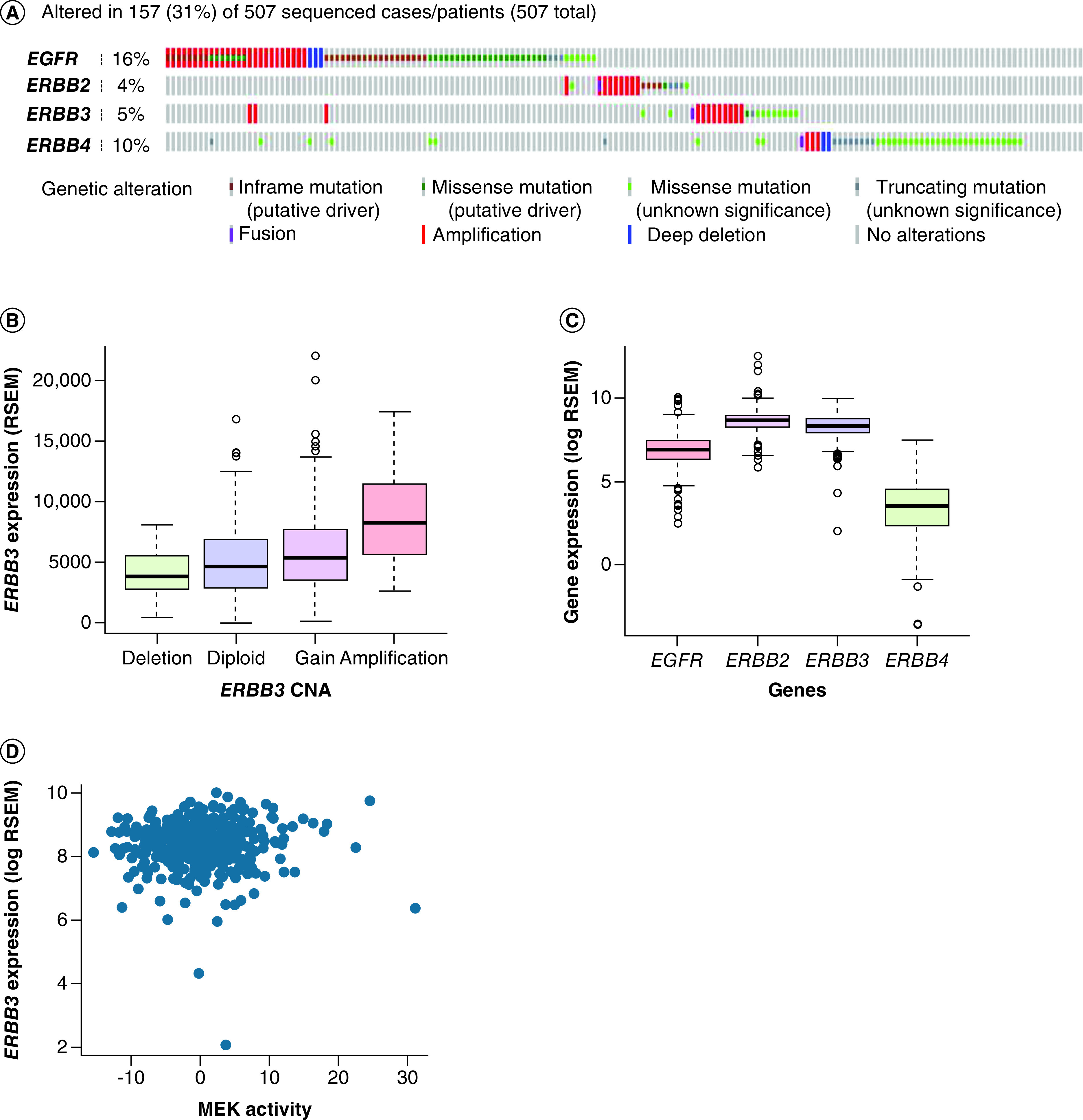
(A) Frequency of HER family genetic alterations. (B) ERBB3 RNA expression with ERBB3 CNA. (C) Gene expression of EGFR, ERBB2, ERBB3 and ERBB4. (D) Correlation of ERBB3 expression level with MEK activity.
RSEM: RNA-Seq by expectation maximization.
In the TCGA dataset ERBB3 amplification co-occurred with EGFR amplification; the amplification of ERBB3 was four-times more likely to co-exist with EGFR amplification than not, though this did not reach statistical significance (p = 0.1). There was no co-amplification between ERBB3 and ERBB2, but the expression of ERBB3 was highly correlated with ERBB2 expression levels (r = 0.09; p = 2.5 × 10-23) (Supplementary Figure 1A). ERBB3 expression also positively correlated with ERBB4 expression (r = 0.1; p = 0.001) but there was no correlation with the expression levels of EGFR (r = -0.12; p = 0.5). EGFR and ERBB2 gene copy number also increased with gene expression (EGFR: r = 0.4; p < 2 × 10-16; and ERBB3: r = 0.35; p = 4 × 10-16) (Supplementary Figure 1B). ERBB3 gene copy number decreased with ERBB2 gene expression (r = -0.1; p = 0.09) (Supplementary Figure 1C).
MEK activity score correlation with key nodes in ERBB pathway in TCGA data
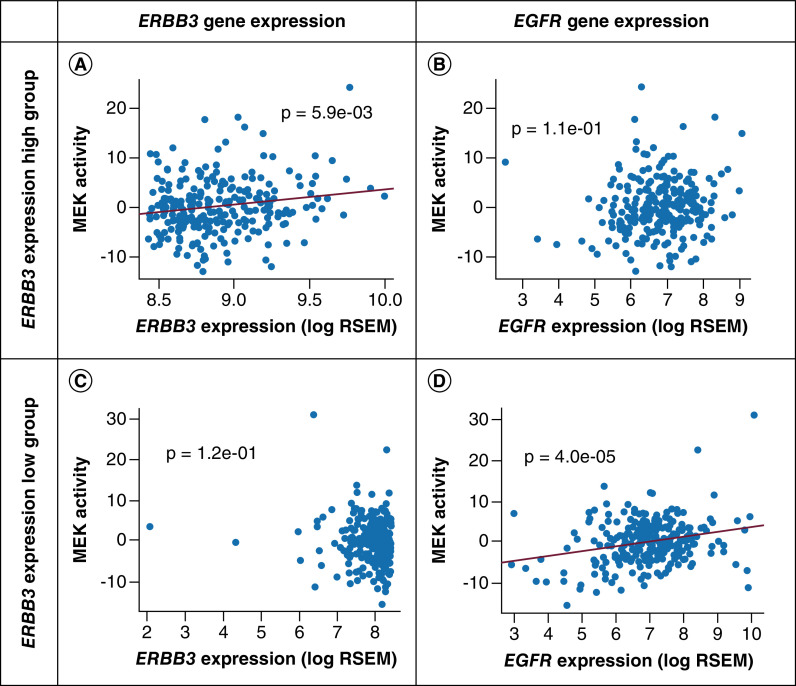
We performed further analyses of RNA sequencing data from TCGA. We observed ERBB2 and ERBB3 RNA expression levels were higher than EGFR and ERBB4 in adeno-NSCLC (Figure 3C). We measured absolute gene expression level of ERBB family genes and derived MEK pathway activity using a validated 18-gene MEK signature [44]. EGFR RNA expression levels correlated with MEK activity (r = 0.16; p = 3.0 × 10-5), ERBB3 RNA expression did not correlate with MEK activity (r = 0.09; p = 0.78) (Figure 3D). We further characterized MEK activation score with gene expression levels of ERBB ligand and proteins of ubiquitination (Supplementary Table 1). AREG (EGFR ligand) and RNF41 (an ubiquitin ligase) expression had a significant positive correlation with MEK activity score.
In order to stratify ERBB3 RNA expression effects on MEK activity, we grouped patients into ERBB3 ‘expression high’ (higher 50% of ERBB3 RNA expression cases) and ERBB3 ‘expression low’ (lower 50% of ERBB3 RNA expression cases). In the ERBB3 expression high group, ERBB3 RNA expression levels were significant and positively correlated with MEK activity score (r = 0.19; p = 0.006), whereas no correlation was observed in the ERBB3 expression low group (p = 0.12) (Figure 4). The expression of EGFR did not correlate with MEK activity score in the ERBB3 expression high group (p = 0.11), though did correlate with MEK activity score in the ERBB3 expression low group (r = 0.2; p = 4 × 10-5). In the ERBB3 expression high group, ERBB3 expression levels still highly positively correlated with MEK activity even after adjusting for EGFR expression effects (p = 0.007).
Figure 4. . Correlation of gene expression with MEK activity in The Cancer Genome Atlas adeno-non-small-cell lung carcinoma.
(A) Correlation of ERBB3 expression level with MEK activity in ERBB3 expression high cancers. (B) Correlation of EGFR expression with MEK activity in ERBB3 expression high cancers. (C) Correlation of ERBB3 expression level with MEK activity in ERBB3 expression low cancers. (D) Correlation of EGFR expression with MEK activity in ERBB3 expression low cancers.
RSEM: RNA-Seq by expectation maximization.
Discussion
HER3 protein levels in adeno-NSCLC & mechanism of overexpression
Reports of the incidence of HER3 protein overexpression in NSCLC vary greatly according to the cohort. Previous groups have reported rates ranging from 6.5 to 86.1% [28,45–49]. IHC methods and scoring have not been previously standardized. Our data use a validated and robust scoring system based on optical density and H-score. Although our patient cohort is small, we demonstrate that overexpression of HER3 protein is common in advanced adeno-NSCLC, occurring in over 40% of patients.
The rate of ERBB3 gene amplification in NSCLC is low. The Cancer Genome Atlas Research Group sequenced 230 cases of lung adenocarcinoma and did not identify any ERBB3 amplification [43]. The cBioportal and GENIE data sets report amplification in just over 1% frequency [42]. It was not possible to perform ERBB3 PCR on all cases in our cohort as tumor tissue was limited, but a sample of 27 cases had sufficient tumor sample to pass quality control. There appeared to be clustering of HER3 overexpressing cases in those with ERBB3 copy number gain but this does not account for the majority of HER3 overexpression. We interrogated the PanCancer TCGA dataset of adeno-NSCLC samples. This dataset includes transcriptome (RNA) and genomic (DNA) data. The genomic data support our finding of low levels of ERBB3 amplification. It also provided the confirmation that ERBB3 copy number alteration highly associates with its gene and HER3 protein expression.
HER3 expression as a prognostic marker
Yi et al. demonstrated that HER3 overexpression is associated with a poor prognosis in a cohort of 443 patients with advanced NSCLC [28]. The median overall survival in our study was not significantly different between the HER3 negative and positive patient group. This Phase I patient cohort may not be truly reflective of the broader population of NSCLC patients, as these patients tend to be fitter with fewer or no comorbidities.
HER3 in relation to oncogenic drivers in adeno-NSCLC
HER2 protein overexpression is reported in 6–30% of patients with lung cancer, which in keeping with our results [50–52]. HER2 expression is less frequently observed in NSCLC than HER3 expression. There was some overlap, though no statistically significant association between, HER3 expression with HER2 expression or KRAS and EGFR mutations though this is somewhat limited by small sample size. TCGA data demonstrated correlation of ERBB3 and ERBB2 expression. Previous data have demonstrated that HER3 plays a role in EGFR TKI resistance. Half of the cases in this cohort with EGFR mutation also co-expressed HER3. No difference in overall survival was seen between HER3 expressing and nonexpressing groups in the EGFR-mutant cases in this cohort though patient numbers were small.
HER3 & functional activity in adeno-NSCLC
The MEK activity score is an 18 gene signature derived from an experiment to test a mixed-tumor response to the MEK inhibitor, selumetinib [44]. The MEK activity score is a surrogate marker of MEK pathway activation though not specific for activation through the HER3 channel as the MEK pathway is also initiated upon other receptor activation. As HER3 activity is predominantly via HER2/HER3 and EGFR/HER3 hetero-dimerization, we inferred that the MEK activity score could be used as a surrogate of, but not specific to, HER3 protein expression. If a key node in the HER3 pathway is found to correlate with MEK activity score, it can be concluded that, in that cohort, the node may play a key part in pathway. The lack of correlation with ERBB3 ligands (NRG 1 and 2) expression is an important negative finding (i.e., HER3 protein expression does not appear to be ligand dependent).
In the TCGA dataset MEK activity score was associated with EGFR and AREG (EGFR ligand) gene expression. This suggests that signaling via EGFR is key to the oncogenic behavior in lung cancer. EGFR has been previously reported as a key driver of oncogenesis in lung (and other) cancers [53]. Although our initial analysis did not show correlation of ERBB3 with MEK signature, we stratified ERBB3 expression into two groups; ‘expression low’, where ERRB3 expression was less than 50% of the median expression and ‘expression high’, where ERBB3 expression was 50% or more than the median expression levels; and analyzed this along with EGFR gene expression. In the ERBB3 expression high group, ERBB3 RNA expression levels positively correlated with MEK activity score but not in the expression low group. The expression of EGFR correlated with MEK activity score in the ERBB3 expression low group. HER3 may be an alternative pathway for MEK activation in the HER3 expression high group. This analysis implies a degree of functional downstream pathway signaling via HER3. In effect, this suggests that when EGFR drives the cancer, the effect of HER3 is minimal as EGFR drives the downstream signaling. However, as HER3 expression increases this becomes the main driver of pathway activation.
Conclusion
We conclude that HER3 is commonly over-expressed in NSCLC and co-expresses with other HER family receptors. HER3 over-expression was not fully attributable to gene copy-number change. In the search for nongenomic causes of HER3 protein overexpression, the MEK activity score was used as a surrogate of HER3 overexpression; however no correlation with ligands of HER3 were seen. Further investigation, therefore, is needed to continue to elucidate the mechanism of HER3 overexpression in adeno-NSCLC. We have a simple tool in the form of IHC that can be used to ‘prescreen’ and stratify patients to receive HER3 targeting compounds. HER3 targeting compounds are in development such as the HER3 antibody drug conjugate patritumab deruxtecan [54], and the monoclonal antibodies lumretuzumab [55] and seribantumab [56]. There are some patients where HER3 expression occurs concurrently with driver mutations (EGFR). The role of HER3 in these groups, including in drug resistance, is the subject of ongoing investigation. Additionally, we propose that HER3 may signal via the MEK pathway (oncogenic role, for further functional validation). The ‘clinical reality’ of targeting HER3 may be more complex with heterogeneity of response depending on other genomic and as yet, undefined, features.
Future perspective
We foresee remarkable change in lung cancer care, with increasingly more targeted therapies, stratifying patients according to novel targetable aberrations, and to overcome resistance. Antibody drug conjugates as well as bispecific antibodies use will be routine practice and enable precise targeting and delivery of cytotoxics.
Summary points.
In a cohort of 45 patients with adeno-non-small-cell lung carcinoma (NSCLC), HER3 protein overexpression occurred in 42.2% of cases and was more frequently seen than HER2 overexpression. There was no statistically significant association between HER3 expression and HER2 expression, EGFR or KRAS mutations.
The medial overall survival in our study was not significantly different between the HER3 negative and positive patient groups.
On The Cancer Genome Atlas analysis, the mechanism of HER3 overexpression was not fully attributable to ERBB3 copy number nor was ERBB3 ligand dependent. Further investigation is required to elucidate the mechanism of HER3 overexpression.
MEK activity score was used as a functional surrogate marker for HER3 overexpression. In the ERBB3 expression high group, ERBB3 RNA expression levels positively correlated with MEK activity score but not in the expression low group. The expression of EGFR correlated with MEK activity score in the ERBB3 expression low group. This supports the hypothesis that HER3 is an alternative pathway for MEK activation in the HER3 expression high group.
HER3 overexpression is common, and has functional downstream signaling, particularly in non-EGFR driven adeno-NSCLC.
Supplementary Material
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/lmt-2020-0031
Author contributions
Interpretation of data has been contributed by T Manickavasagar and A Minchom. Preparation of manuscript has been contributed by A Minchom, T Manickavasagar and W Yuan. Data analysis has been contributed by A Minchom, W Yuan, S Carreira, B Gurel, S Miranda, A Ferreira, M Crespo, R Riisnaes and C Baker. Review of manuscript has been contributed by S Carreira, B Gurel, S Miranda, A Ferreira, M Crespo, R Riisnaes, C Baker, M O’Brien, J Bhosle, S Popat, U Banerji, J Lopez and JD Bono.
Financial & competing interests disclosure
M O'Brien sits on advisory boards for MSD, Abbvie, BMS, BI and Pierre Fabre. S Popat has received honoraria from BMS, Roche, Takeda, AstraZeneca, Pfizer, MSD, EMD Serono, Guardant Health, AbbVie, Boehringer Ingelheim, Medscape, Tesaro, Paradox, Incyte, OncLive and receives direct funding from Elsevier. U Banerji has received honoraria fron Astellas, Novartis, Karus Therapuetics, Pheonix Solutions, Eli Lilly, Astex, Vernalis and Boehringer Ingelheim. U Banerji is a recipient of an NIHR Research Professorship Award and has received CRUK funding: Cancer Research UK Scientific Executive Board, Cancer Research UK Centre Award. Cancer Research UK Drug Discovery Committee – Programme Award. J Lopez has received research grant funding from Roche, Basilea and Genmab unrelated to this work, and is on the Editorial Board for British Journal of Cancer. J de Bono has served on advisory boards and received fees from many companies including AstraZeneca, Astellas, Bayer, Boehringer Ingelheim, Cellcentric, Daiichi, Genentech/Roche, Genmab, GSK, Janssen, Merck Serono, Merck Sharp & Dohme, Menarini/Silicon Biosystems, Orion, Pfizer, Qiagen, Sanofi Aventis, Sierra Oncology, Taiho and Vertex Pharmaceuticals. He is an employee of The ICR, which has received funding or other support for his research work from AZ, Astellas, Bayer, Cellcentric, Daiichi, Genentech, Genmab, GSK, Janssen, Merck Serono, MSD, Menarini/Silicon Biosystems, Orion, Sanofi Aventis, Sierra Oncology, Taiho, Pfizer, Vertex and which has a commercial interest in abiraterone, PARP inhibition in DNA repair defective cancers and PI3K/AKT pathway inhibitors (no personal income). He was named as an inventor, with no financial interest, for patent 8,822,438. He has been the CI/PI of many industry sponsored clinical trials. He is a National Institute for Health Research (NIHR) Senior Investigator. A Minchom has served on advisory boards and received fees from Merck, FARON, Novartis, Bayer and Janssen. The authors also acknowledge funding from Cancer Research UK and the Experimental Cancer Centre Initiative. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Disclaimer
This study represents independent research supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at the Royal Marsden NHS Foundation Trust and the Institute of Cancer Research. The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care.
Ethical conduct of research
Informed written consent was gained from patients to use the archival of fresh tumor samples. Approval was gained from the local research and ethics committee for the use of the clinical data.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.American Cancer Society. Lung cancer survival rates. (2019). https://www.cancer.org/cancer/lung-cancer/detection-diagnosis-staging/survival-rates.html
- 2.Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc. Natl Acad. Sci. USA 107(17), 7692–7697 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallasch C, Weiss FU, Niederfellner G, Jallal B, Issing W, Ullrich A. Heregulin-dependent regulation of HER2/neu oncogenic signaling by heterodimerization with HER3. EMBO J. 14(17), 4267–4275 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinkas-Kramarski R, Soussan L, Waterman H et al. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 15(10), 2452–2467 (1996). [PMC free article] [PubMed] [Google Scholar]
- 5.Sergina NV, Rausch M, Wang D et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature 445(7126), 437–441 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelman JA, Janne PA, Mermel C et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc. Natl Acad. Sci. USA 102(10), 3788–3793 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanizaki J, Okamoto I, Sakai K, Nakagawa K. Differential roles of trans-phosphorylated EGFR, HER2, HER3, and RET as heterodimerisation partners of MET in lung cancer with MET amplification. Br. J. Cancer 105(6), 807–813 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Revach OY, Sandler O, Samuels Y, Geiger B. Cross-Talk between receptor tyrosine kinases AXL and ERBB3 regulates invadopodia formation in melanoma cells. Cancer Res. 79(10), 2634–2648 (2019). [DOI] [PubMed] [Google Scholar]; • HER3 has weak tyrosine kinase activity, preferentially forming heterodimers with a variety of kinase-proficient receptor tyrosine kinases, including EGFR, HER2, MET and AXL.
- 9.Chen L, Siddiqui S, Bose S et al. Nrdp1-mediated regulation of ErbB3 expression by the androgen receptor in androgen-dependent but not castrate-resistant prostate cancer cells. Cancer Res. 70(14), 5994–6003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho HS, Mason K, Ramyar KX et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 421(6924), 756–760 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Garrett TP, Mckern NM, Lou M et al. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol. Cell 11(2), 495–505 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Burgess AW, Cho HS, Eigenbrot C et al. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol. Cell 12(3), 541–552 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Lee-Hoeflich ST, Crocker L, Yao E et al. A central role for HER3 in HER2-amplified breast cancer: implications for targeted therapy. Cancer Res. 68(14), 5878–5887 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Steinkamp MP, Low-Nam ST, Yang S, Lidke KA, Lidke DS, Wilson BS. erbB3 is an active tyrosine kinase capable of homo- and heterointeractions. Mol. Cell. Biol. 34(6), 965–977 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waterman H, Sabanai I, Geiger B, Yarden Y. Alternative intracellular routing of ErbB receptors may determine signaling potency. J. Biol. Chem. 273(22), 13819–13827 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Hellyer NJ, Cheng K, Koland JG. ErbB3 (HER3) interaction with the p85 regulatory subunit of phosphoinositide 3-kinase. Biochem. J. 333(Pt 3), 757–763 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vijapurkar U, Cheng K, Koland JG. Mutation of a Shc binding site tyrosine residue in ErbB3/HER3 blocks heregulin-dependent activation of mitogen-activated protein kinase. J. Biol. Chem. 273(33), 20996–21002 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Liao H, Deng Z et al. miRNA-205 affects infiltration and metastasis of breast cancer. Biochem. Biophys. Res. Commun. 441(1), 139–143 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Iorio MV, Casalini P, Piovan C et al. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 69(6), 2195–2200 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J. Biol. Chem. 282(2), 1479–1486 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Cao Z, Wu X, Yen L, Sweeney C, Carraway KL 3rd. Neuregulin-induced ErbB3 downregulation is mediated by a protein stability cascade involving the E3 ubiquitin ligase Nrdp1. Mol. Cell. Biol. 27(6), 2180–2188 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carraway KL 3rd. E3 ubiquitin ligases in ErbB receptor quantity control. Semin. Cell Dev. Biol. 21(9), 936–943 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Z, Choi BK, Mujoo K et al. The E3 ubiquitin ligase NEDD4 negatively regulates HER3/ErbB3 level and signaling. Oncogene 34(9), 1105–1115 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Mujoo K, Choi BK, Huang Z, Zhang N, An Z. Regulation of ERBB3/HER3 signaling in cancer. Oncotarget 5(21), 10222–10236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingalla EQ, Miller JK, Wald JH et al. Post-transcriptional mechanisms contribute to the suppression of the ErbB3 negative regulator protein Nrdp1 in mammary tumors. J. Biol. Chem. 285(37), 28691–28697 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poller DN, Spendlove I, Baker C et al. Production and characterization of a polyclonal antibody to the c-erbB-3 protein: examination of c-erbB-3 protein expression in adenocarcinomas. J. Pathol. 168(3), 275–280 (1992). [DOI] [PubMed] [Google Scholar]
- 27.Muller-Tidow C, Diederichs S, Bulk E et al. Identification of metastasis-associated receptor tyrosine kinases in non-small cell lung cancer. Cancer Res. 65(5), 1778–1782 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Yi ES, Harclerode D, Gondo M et al. High c-erbB-3 protein expression is associated with shorter survival in advanced non-small cell lung carcinomas. Mod. Pathol. 10(2), 142–148 (1997). [PubMed] [Google Scholar]; •• This article showed that in a cohort of 443 patients with NSCLC, HER3 overexpression is associated with poor prognosis.
- 29.Sun M, Behrens C, Feng L et al. HER family receptor abnormalities in lung cancer brain metastases and corresponding primary tumors. Clin. Cancer Res. 15(15), 4829–4837 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cappuzzo F, Toschi L, Domenichini I et al. HER3 genomic gain and sensitivity to gefitinib in advanced non-small-cell lung cancer patients. Br. J. Cancer 93(12), 1334–1340 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yonesaka K, Hirotani K, Kawakami H et al. Anti-HER3 monoclonal antibody patritumab sensitizes refractory non-small cell lung cancer to the epidermal growth factor receptor inhibitor erlotinib. Oncogene 35(7), 878–886 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Huang S, Li C, Armstrong EA et al. Dual targeting of EGFR and HER3 with MEHD7945A overcomes acquired resistance to EGFR inhibitors and radiation. Cancer Res. 73(2), 824–833 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• HER3 has a role in the development of resistance to EGFR inhibition, which can be overcome with dual targeting of EGFR and HER3.
- 33.Fujimoto N, Wislez M, Zhang J et al. High expression of ErbB family members and their ligands in lung adenocarcinomas that are sensitive to inhibition of epidermal growth factor receptor. Cancer Res. 65(24), 11478–11485 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Sun C, Hobor S, Bertotti A et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 7(1), 86–93 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Noto A, De Vitis C, Roscilli G et al. Combination therapy with anti-ErbB3 monoclonal antibodies and EGFR TKIs potently inhibits non-small cell lung cancer. Oncotarget 4(8), 1253–1265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sequist LV, Waltman BA, Dias-Santagata D et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 3(75), 75ra26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engelman JA, Zejnullahu K, Mitsudomi T et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316(5827), 1039–1043 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Jaiswal BS, Kljavin NM, Stawiski EW et al. Oncogenic ERBB3 mutations in human cancers. Cancer Cell 23(5), 603–617 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Kiavue N, Cabel L, Melaabi S et al. ERBB3 mutations in cancer: biological aspects, prevalence and therapeutics. Oncogene 39(3), 487–502 (2020). [DOI] [PubMed] [Google Scholar]; • ERBB3 mutations in lung cancer is a rare event and can influence the activity of HER3 receptor.
- 40.Littlefield P, Liu L, Mysore V, Shan Y, Shaw DE, Jura N. Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations. Sci.Signal 7(354), ra114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 41.Wolff AC, Hammond MEH, Allison KH et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline focused update. J. Clin. Oncol. 36(20), 2105–2122 (2018). [DOI] [PubMed] [Google Scholar]
- 42.cBioPortal for Cancer Genomics. (2019). https://www.cbioportal.org
- 43.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511(7511), 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dry JR, Pavey S, Pratilas CA et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244). Cancer Res. 70(6), 2264–2273 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The MEK activity score is a functional surrogate marker of MEK pathway activation. It is an 18 gene signature derived from tumor response to Selumetinib, an MEK inhibitor.
- 45.Koutsopoulos AV, Mavroudis D, Dambaki KI et al. Simultaneous expression of c-erbB-1, c-erbB-2, c-erbB-3 and c-erbB-4 receptors in non-small-cell lung carcinomas: correlation with clinical outcome. Lung Cancer 57(2), 193–200 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Cstoth I, Anthoine G, Berghmans T et al. C-erbB-3 expression in non-small cell lung cancer (NSCLC) patients treated by Erlotinib. Anticancer Res. 31(1), 281–285 (2011). [PubMed] [Google Scholar]
- 47.Lai WW, Chen FF, Wu MH et al. Immunohistochemical analysis of epidermal growth factor receptor family members in stage I non-small cell lung cancer. Ann. Thorac. Surg. 72(6), 1868–1876 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Fontanini G, De Laurentiis M, Vignati S et al. Evaluation of epidermal growth factor-related growth factors and receptors and of neoangiogenesis in completely resected stage I–IIIA non-small-cell lung cancer: amphiregulin and microvessel count are independent prognostic indicators of survival. Clin. Cancer Res. 4(1), 241–249 (1998). [PubMed] [Google Scholar]
- 49.Kumagai T, Tomita Y, Nakatsuka SI et al. HER3 expression is enhanced during progression of lung adenocarcinoma without EGFR mutation from stage 0 to IA1. Thorac. Cancer 9(4), 466–471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rouquette I, Lauwers-Cances V, Allera C et al. Characteristics of lung cancer in women: importance of hormonal and growth factors. Lung Cancer 76(3), 280–285 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Pellegrini C, Falleni M, Marchetti A et al. HER-2/Neu alterations in non-small cell lung cancer: a comprehensive evaluation by real time reverse transcription-PCR, fluorescence in situ hybridization, and immunohistochemistry. Clin. Cancer Res. 9(10 Pt 1), 3645–3652 (2003). [PubMed] [Google Scholar]
- 52.Langer CJ, Stephenson P, Thor A, Vangel M, Johnson DH. Eastern Cooperative Oncology Group S. Trastuzumab in the treatment of advanced non-small-cell lung cancer: is there a role? Focus on Eastern Cooperative Oncology Group study 2598. J. Clin. Oncol. 22(7), 1180–1187 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Sigismund S, Avanzato D, Lanzetti L. Emerging functions of the EGFR in cancer. Molecul. Oncol. 12(1), 3–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu HA, Baik CS, Gold K et al. LBA62 Efficacy and safety of patritumab deruxtecan (U3-1402), a novel HER3 directed antibody drug conjugate, in patients (pts) with EGFR-mutated (EGFRm) NSCLC. Ann. Oncol. 31, S1189–S1190 (2020). [Google Scholar]; • Drugs targeting HER3 for NSCLC are in development and early phase trials show promising results.
- 55.Meulendijks D, Jacob W, Martinez-Garcia M et al. First-in-human Phase I study of lumretuzumab, a glycoengineered humanized anti-HER3 monoclonal antibody, in patients with metastatic or advanced HER3-positive solid tumors. Clin. Cancer Res. 22(4), 877–885 (2016). [DOI] [PubMed] [Google Scholar]; • Drugs targeting HER3 for NSCLC are in development and early phase trials show promising results.
- 56.Sequist LV, Gray JE, Harb WA et al. Randomized Phase II trial of seribantumab in combination with erlotinib in patients with EGFR wild-type non-small cell lung cancer. Oncologist 24(8), 1095–1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Drugs targeting HER3 for NSCLC are in development and early phase trials show promising results.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.