Supplemental Digital Content is available in the text.
Keywords: Antibody, Case ascertainment ratio, COVID-19, Cumulative incidence, Infection fatality ratio, SARS-CoV-2, Seroprevalence, Waning antibody
Background:
Serology tests can identify previous infections and facilitate estimation of the number of total infections. However, immunoglobulins targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been reported to wane below the detectable level of serologic assays (which is not necessarily equivalent to the duration of protective immunity). We estimate the cumulative incidence of SARS-CoV-2 infection from serology studies, accounting for expected levels of antibody acquisition (seroconversion) and waning (seroreversion), and apply this framework using data from New York City and Connecticut.
Methods:
We estimated time from seroconversion to seroreversion and infection fatality ratio (IFR) using mortality data from March to October 2020 and population-level cross-sectional seroprevalence data from April to August 2020 in New York City and Connecticut. We then estimated the daily seroprevalence and cumulative incidence of SARS-CoV-2 infection.
Results:
The estimated average time from seroconversion to seroreversion was 3–4 months. The estimated IFR was 1.1% (95% credible interval, 1.0%, 1.2%) in New York City and 1.4% (1.1, 1.7%) in Connecticut. The estimated daily seroprevalence declined after a peak in the spring. The estimated cumulative incidence reached 26.8% (24.2%, 29.7%) at the end of September in New York City and 8.8% (7.1%, 11.3%) in Connecticut, higher than maximum seroprevalence measures (22.1% and 6.1%), respectively.
Conclusions:
The cumulative incidence of SARS-CoV-2 infection is underestimated using cross-sectional serology data without adjustment for waning antibodies. Our approach can help quantify the magnitude of underestimation and adjust estimates for waning antibodies.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019 (COVID-19), rapidly spread across the world in 2020.1 Globally, there have been 36 million documented cases and more than 1 million COVID-19 associated fatalities as of 15 October 2020, with case counts continuing to increase.2 Reliable measurement of infection history in a population is a critical epidemiologic outcome, and is needed to derive several key epidemiologic indices such as the infection fatality ratio (IFR). However, the number of SARS-CoV-2 infections reported through public health surveillance mechanisms is underestimated because of the limited capacity of testing and surveillance systems, an overwhelmed healthcare system, low healthcare-seeking behavior among those with mild disease, imperfect sensitivity of diagnostic tests (especially rapid antigen tests), and a large fraction of infections that are asymptomatic.3 Imperfect specificity could in theory overestimate the number of cases, but this may be less of a concern because the specificity of the relevant assays is generally high (e.g., 99.3% as described in the Methods). Assays that detect viral antigen or genomic material cannot identify individuals who were previously infected once viral material is no longer present. In contrast, serology tests measuring the level of immunoglobulins have the potential to identify those previously infected. Serology results can be used to generate an estimate of the cumulative incidence, but this requires the relatively strong assumption that antibodies persist permanently after infection. Because antibody levels for SARS-CoV-2 wane over time and can become undetectable, some previously infected individuals could have already returned to a seronegative status at the time of testing (i.e., seroreversion). Hence, new methods are needed to account for waning antibodies when using cross-sectional serology data to estimate cumulative incidence of SARS-CoV-2 infection.
Population-level serosurveys for SARS-CoV-2 have been conducted across the world, with variation in geographic scale, sample demographics, sampling mechanisms, and testing methods.4,5 Although the reported incidence of COVID-19 varies widely by location, a consistent finding in all settings is that total estimated infections vastly outnumber confirmed cases. For example, the Centers for Disease Control and Prevention (CDC) and commercial laboratories conducted large-scale geographic longitudinal serosurveys in 10 sites in the United States in the spring and summer of 2020.6 Seroprevalence ranged from 1% [95% confidence interval (CI) = 0.3%, 2.4%] to 6.9% (95% CI = 5.0%, 8.9%) across sites. That study estimated that the number of total infections was 6–24 times higher than that of documented cases, while acknowledging that these ratios varied widely depending on the timing of sampling or the stage of the epidemic in each location. These early population-level serosurveys provided critical insights on the true burden of COVID-19; however, as we enter the second year of the US epidemic, the ability to estimate the cumulative incidence directly from serosurveys is increasingly limited because antibody levels continue to wane and the corresponding serologic “record” of historical infection is lost. Indeed, Ibarrondo et al. estimated that the half-life of anti-SARS-CoV-2 spike receptor-binding domain IgG was 36 days.7 Patel et al. reported that over half (11/19) of health care personnel who tested seropositive in early April became seronegative at the second visit in June (approximately 60 days after the baseline).8 Taken together, these findings suggest that serosurveys have likely failed to recognize previous infection in those whose antibody levels have already waned below the detectable limit at the time of sampling. An adjustment for waning antibody kinetics must account for the interacting timescales of antibody kinetics, case incidence and the period over which a serosurvey is conducted.
Here, we describe a framework for estimating the cumulative incidence and IFR of SARS-CoV-2 from population-level cross-sectional serology data and mortality data, by adjusting for the timeline of seroconversion (acquisition of the detectable level of antibodies) and seroreversion (loss of detectable antibodies). We apply this framework to data from New York City and Connecticut because these two sites observed a large wave of COVID-19 cases that lasted for a relatively short period in spring 2020, followed by low case counts in the summer and early fall (eFigure 1; http://links.lww.com/EDE/B804). We note that it is critical to distinguish the ability to detect antibodies from immunity to reinfection, which may persist longer than antibodies are detectable.9 We make no claim that the time from seroconversion to seroreversion estimated from this framework reflects the duration of immunity protection against reinfection.
METHODS
Population-level Cross-sectional Seroprevalence Data
Details of the serosurvey conducted by the CDC and commercial laboratories can be found elsewhere.6 Briefly, the survey collected convenience samples of deidentified residual patient sera in 10 US sites (Connecticut, Louisiana, Minneapolis–St Paul–St Cloud metro area, Missouri, New York City metro area, Philadelphia metro area, San Francisco Bay area, South Florida, Utah, and Western Washington State) from March to July 2020, and expanded the serosurvey to all 50 states in August. Blood specimens were originally collected for reasons unrelated to COVID-19, such as for routine medical care or sick visits, but information on the reason for specimen collection was not available. Most of the samples were from outpatients. Multiple rounds of surveys have been conducted at each site, approximately every 3–4 weeks (eTable 1; http://links.lww.com/EDE/B804). Each round tested approximately 1,800 samples from each site.10 Samples were tested by an enzyme-linked immunosorbent assay (ELISA) against the SARS-CoV-2 spike protein that detects the total immunoglobulin response (IgA, IgM, and IgG) with sensitivity 96% [95% confidence interval (CI) = 98.3%, 99.9%] and specificity 99.3% (95% CI = 98.3%, 99.9%).6 We downloaded publicly available seroprevalence data adjusted for age and sex distributions on October 8, 2020.11 We focused our analysis on New York City and Connecticut because the single, short wave of infection in the spring allowed us to evaluate how the antibodies acquired over a short period subsequently waned during a time when minimal new infections were occurring in these locations (eFigure 1; http://links.lww.com/EDE/B804). More details can be found in eAppendix 1.
Mortality Data and Case Data
Mortality data are less subject to changes in testing capacity and guidelines over time than case data, so we used mortality data to estimate parameters. We analyzed daily time series data on COVID-19 associated deaths from March to September 2020 in New York City and Connecticut. We downloaded New York City data 2 October 2020 and Connecticut data on 6 October 2020 from their government websites.12,13 To account for undercounting of COVID-19 deaths, especially at the beginning of the pandemic, we used the total (i.e., probable and confirmed) deaths for our mortality time series (eAppendix 2; http://links.lww.com/EDE/B804). To account for a delay distribution between symptom onset and death, we used data on the date of symptom onset and date of death for 6,999 COVID-19 associated deaths in Georgia, USA from February to October 2020.
To estimate case ascertainment ratios (see the “Model” section for detail), we also utilized daily time series data for the number of documented cases in New York City and Connecticut from their government websites (eAppendix 2; http://links.lww.com/EDE/B804).12,13
Model
Our Bayesian model aims to estimate the cumulative incidence and IFR, accounting for the time of positivity between seroconversion and seroreversion. The model first determines the timing of symptom onset from the COVID-19 mortality time series based on empirical data on the distribution of time from symptom onset to death from Georgia. The model then estimates two parameters, the IFR and the average duration of seropositivity (time from seroconversion to serorerversion), by comparing the simulated seroprevalence with the observed data (CDC longitudinal seroprevalence) using binomial likelihood calculation. These two parameters were then used to calculate corresponding daily seroprevalence, cumulative incidence, and the case ascertainment ratio.
We estimated the IFR and time from seroconversion to seroreversion using the reported number of deaths and CDC serosurvey data (Figure 1). Using a Markov chain Monte Carlo (MCMC) analysis, we estimated the number of seropositive individuals on day t (St) as follows:
FIGURE 1.
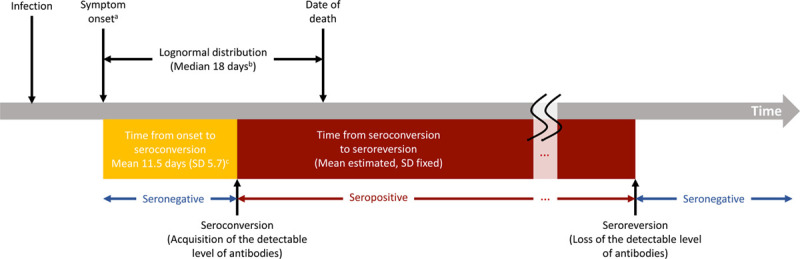
Structure of the analytic framework. aOnset of infectiousness for asymptomatic cases; bData from Georgia Department of Public Health; cIyer et al. medRxiv 2020.
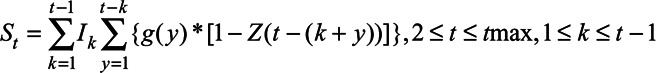 |
(1) |
where 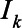 is the total number of SARS-CoV-2 infections on day k, which is the number of reported deaths divided by the estimated IFR, after accounting for the delay between symptom onset and death (eAppendix 3; http://links.lww.com/EDE/B804). Therefore, Equation 1 calculates, of those who were infected on day k, how many of them have acquired the detectable level of antibodies but have not lost them by day t. We estimated the IFR, assuming that the IFR was constant over time in the main analysis and relaxed this assumption in a sensitivity analysis. Additional parameters include tmax, the total number of days in the daily time series data for COVID-19 deaths, and g, the probability density function of the Weibull distribution for time from symptom onset to seroconversion with the mean 11.5 days and SD 5.7 days,14 which is consistent with other reports.15–18 For asymptomatic cases, g represents the time from onset of infectiousness to seroconversion (eAppendix 3; http://links.lww.com/EDE/B804). Z is the cumulative density function of the Weibull distribution for time from seroconversion to seroreversion. Therefore,
is the total number of SARS-CoV-2 infections on day k, which is the number of reported deaths divided by the estimated IFR, after accounting for the delay between symptom onset and death (eAppendix 3; http://links.lww.com/EDE/B804). Therefore, Equation 1 calculates, of those who were infected on day k, how many of them have acquired the detectable level of antibodies but have not lost them by day t. We estimated the IFR, assuming that the IFR was constant over time in the main analysis and relaxed this assumption in a sensitivity analysis. Additional parameters include tmax, the total number of days in the daily time series data for COVID-19 deaths, and g, the probability density function of the Weibull distribution for time from symptom onset to seroconversion with the mean 11.5 days and SD 5.7 days,14 which is consistent with other reports.15–18 For asymptomatic cases, g represents the time from onset of infectiousness to seroconversion (eAppendix 3; http://links.lww.com/EDE/B804). Z is the cumulative density function of the Weibull distribution for time from seroconversion to seroreversion. Therefore, 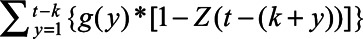
in Equation 1 represents the probability that an individual seroconverted before day t and seroreverted after day t, which in other words means the probability that an individual remains seropositive on day t. We estimated the mean of the Weibull distribution for time from seroconversion to seroreversion, while fixing SD at 50 days. We calculated the daily seroprevalence (Pt) by dividing 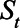 by the population (8.3 million for New York City and 3.7 million for Connecticut). We compared the estimated
by the population (8.3 million for New York City and 3.7 million for Connecticut). We compared the estimated 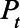 with the reported seroprevalence in each round of the CDC commercial laboratory serosurvey and calculated the log-likelihood assuming the binomial distribution, which we then used in the MCMC analysis for parameter estimation (eAppendix 4; http://links.lww.com/EDE/B804). We used the random-walk Metropolis–Hastings algorithm to sample new candidate values centered at current values for the IFR and the average duration of seropositivity. The uncertainty of a model parameter is quantified and captured by its posterior distribution which is being estimated by our MCMC algorithm.
with the reported seroprevalence in each round of the CDC commercial laboratory serosurvey and calculated the log-likelihood assuming the binomial distribution, which we then used in the MCMC analysis for parameter estimation (eAppendix 4; http://links.lww.com/EDE/B804). We used the random-walk Metropolis–Hastings algorithm to sample new candidate values centered at current values for the IFR and the average duration of seropositivity. The uncertainty of a model parameter is quantified and captured by its posterior distribution which is being estimated by our MCMC algorithm.
Given an estimated model from above, the number of individuals who seroconverted on day t (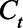 ) may be calculated as follows:
) may be calculated as follows:
| (2) |
where G is the cumulative density function of the aforementioned Weibull distribution for time from symptom onset to seroconversion. 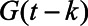 represents the probability that infected individuals on day
represents the probability that infected individuals on day 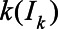 had seroconverted by day t. We calculated the cumulative incidence on day t by dividing the cumulative sum of
had seroconverted by day t. We calculated the cumulative incidence on day t by dividing the cumulative sum of 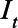 by the population. We also calculated a cumulative case ascertainment ratio
by the population. We also calculated a cumulative case ascertainment ratio 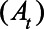 as follows:
as follows:
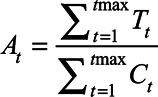 |
(3) |
where 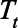 is the number of documented cases on day t (eAppendix 2; http://links.lww.com/EDE/B804). The ascertainment bias was calculated as 1/At.
is the number of documented cases on day t (eAppendix 2; http://links.lww.com/EDE/B804). The ascertainment bias was calculated as 1/At.
The median of the posterior samples was reported as a point estimate, and 2.5th and 97.5th percentiles were reported as 95% credible intervals (CrIs). We performed all analyses with R (Vienna, Austria). The code can be found in the following github repository: https://github.com/lopmanlab/SARS-CoV-2_CumInc_WaningAntibodies.
Sensitivity Analysis
We performed sensitivity analyses to evaluate how sensitive our results were to different assumptions. We changed the fixed value of the SD for time from seroconversion to seroreversion from 50 days to 20 and 70 days. We selected these values based on previous findings.19 We also relaxed the assumption of the constant IFR. It has been reported that the IFR and case fatality ratio for COVID-19 declined over time in New York City20 and in other countries.21 Therefore, we assumed that the IFR decreased by 5% per week from mid-March to the end of July 2020, reflecting findings reported by the previous studies,20 and the model estimated the IFR in mid-March (before the decline started). Lastly, we used data from Wuhan, China to inform delay between symptom onset and death,22 instead of the Georgia data to evaluate the impact of variation in this distribution on parameter inference.23
Ethical Considerations
Seroprevalence data, mortality data, and case data in New York City and Connecticut were publicly available, deidentified and aggregated. The Georgia Department of Public Health Institutional Review Board (IRB) has determined that the project is exempt from the requirement for IRB review and approval.
RESULTS
Estimated Timeline of Seroreversion, IFR, and Case Ascertainment Ratio
We estimated the average time from seroconversion to seroreversion to be 4.0 months (95% CrI = 3.6, 4.6 months) using the New York City data and 3.0 months (95% CrI = 2.3, 4.1 months) using the Connecticut data with the SD fixed at 50 days (Figure 2). More than 85% of the infected individuals were estimated to become seronegative due to waning antibodies within 6 months after seroconversion (Table).
FIGURE 2.
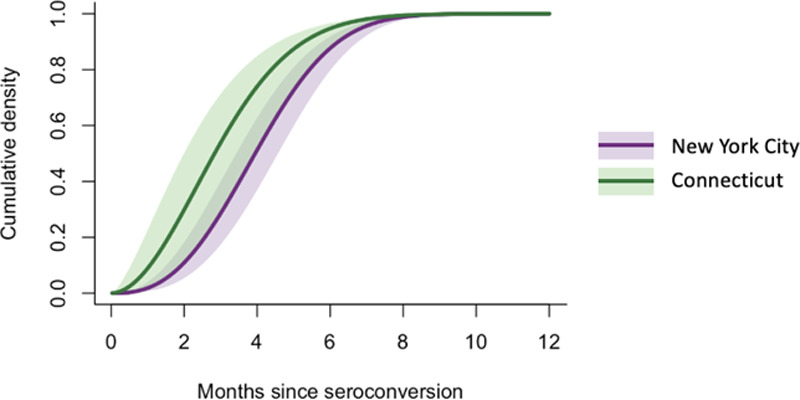
Cumulative density function for the estimated Weibull distribution for time from seroconversion to seroreversion (New York City and Connecticut). Lines and shaded areas represent the posterior median and 95% credible intervals, respectively.
TABLE.
Percentage of Infected Individuals Whose Antibody Level has Declined and Become Undetectable by Serologic Assays Within t Months Since Seroconversion
| Months Since Seroconversion (months) | New York City (95% CrI) | Connecticut (95% CrI) |
|---|---|---|
| 1 | 1.8% (0.6, 3.9) | 8.7% (1.7, 22.2) |
| 2 | 10.5% (5.2, 17.7) | 29.2% (10.0, 49.4) |
| 3 | 28.4% (17.3, 39.8) | 53.7% (27.5, 71.2) |
| 4 | 50.8% (36.5, 62.3) | 73.6% (49.7, 84.7) |
| 5 | 72.5% (59.8, 80.8) | 87.3% (71.7, 92.6) |
| 6 | 87.5% (79.4, 91.7) | 94.6% (87.0, 96.6) |
| 9 | 99.8% (99.6, 99.8) | 99.8% (99.8, 99.8) |
| 12 | 100% (100, 100) | 100% (100, 100) |
The mean of the Weibull distribution for time from seroconversion to seroreversion was estimated while fixing the SD at 50 days.
CrI, credible interval.
We estimated the IFR at 1.1% (95% CrI = 1.0%, 1.2%) for New York City and 1.4% (95% CrI = 1.1%, 1.7%) for Connecticut. The estimated case ascertainment ratio increased rapidly in the early phase of the pandemic and continued to gradually increase from May to October in both sites (Figure 3). The ascertainment ratio was estimated to reach 13% (95% CrI = 12%, 14%) in New York City and 18% (95% CrI = 14%, 23%) in Connecticut at the end of September 2020, suggesting that the number of estimated infections was 7.7 times greater than the number of documented cases in New York City and 5.6 times greater in Connecticut.
FIGURE 3.
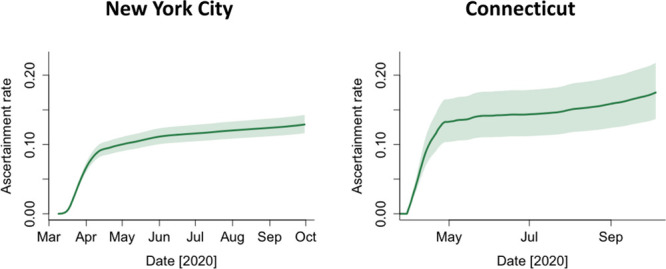
Estimated case ascertainment ratio in New York City and Connecticut in 2020. The case ascertainment ratio was calculated in Equation 3. Lines represent the 50th percentile of the posterior distributions, and shaded areas represent 95% credible intervals.
Estimated Daily Seroprevalence and Cumulative Incidence
In both New York City and Connecticut, the estimated daily seroprevalence decreased over time after the first wave of COVID-19, diverging from the estimated cumulative incidence adjusted for waning antibodies (Figure 4). The cumulative incidence was estimated to reach 26.8% (95% CrI = 24.2%, 29.7%) in New York City and 8.8% (95% CrI = 7.1%, 11.3%) in Connecticut at the end of September 2020. In contrast, the estimated daily seroprevalence peaked at 22.1% (95% CrI = 20.5%, 23.6%) in mid-May and declined to 4.9% (95% CrI = 3.9%, 6.9%) by the end of September in New York City. In Connecticut, the estimated daily seroprevalence peaked at 6.1% (95% CrI = 5.3%, 7.1%) at the end of May and early June and declined to1.3% (95% CrI = 0.9%, 2.0%) by the end of September.
FIGURE 4.
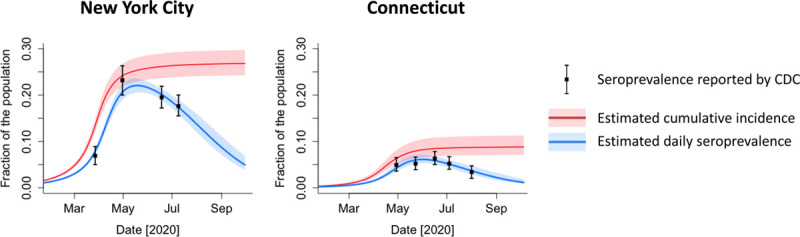
Estimated daily seroprevalence and cumulative incidence of SARS-CoV-2 infection in New York City and Connecticut in 2020. Lines and shaded areas represent the posterior median and 95% credible intervals, respectively. CDC, Centers for Disease Control and Prevention; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Sensitivity Analysis
We ran the analysis with different values of the SD for time from seroconversion to seroreversion (20, 50, and 70 days). The estimated IFR and average time from seroconversion to seroreversion were robust to this variation for Connecticut, but the estimated mean time increased as the SD increased for the New York City data (eTable 2; http://links.lww.com/EDE/B804). Although changes in the SD had small effects on the estimated cumulative incidence, the speed of decline in the daily seroprevalence after the peak varied by SD (eFigure 2; http://links.lww.com/EDE/B804). When the SD was 20 days, the daily seroprevalence declined rapidly after the peak and became 1.2% (95% CrI = 0.9%, 2.1%) at the end of September, while it declined more slowly with the SD 70 days [7.4% (95% CrI = 6.1%, 8.7%) at the end of September].
Under the assumption of the decreasing IFR (eFigure 3; http://links.lww.com/EDE/B804), the average time from seroconversion to seroreversion was estimated to be 4.0 months (95% CrI = 3.5, 4.8 months) with the SD 50 days in New York City, which was consistent with the result using the constant IFR (eTable 2; http://links.lww.com/EDE/B804). The estimated IFR in mid-March was 2.9% (95% CrI = 2.6%, 3.3%), which automatically decreased by 5% per week and reached 1.1% (95% CrI = 0.9%, 1.2%) at the end of July (eFigure 3; http://links.lww.com/EDE/B804). The average IFR from March to September was 1.4% (95% CrI = 1.2%, 1.6%). The daily seroprevalence and cumulative incidence did not appreciably change between the models using the constant IFR vs. decreasing IFR (eTable 2; http://links.lww.com/EDE/B804). We also used data from Wuhan, China to account for variation in the delay between symptom onset and death (eFigure 4; http://links.lww.com/EDE/B804), and found that results were robust to this change (eTable 2; http://links.lww.com/EDE/B804).
DISCUSSION
A reliable estimate of the cumulative number of people who have been infected with SARS-CoV-2 is critical for understanding and, ultimately, controlling the COVID-19 pandemic. Measuring severity (in the form of the IFR), monitoring progress towards a herd immunity threshold, and predicting the impact of vaccination all depend on a robust estimate of cumulative incidence of infection. Given evidence that anti-SARS-CoV-2 antibodies wane below the detection limit among a substantial portion of the population, we developed an analytical framework to estimate cumulative incidence of infection from cross-sectional serologic surveys. First, our model was able to capture the observed declines in seroprevalence after the first wave of COVID-19 in the spring of 2020 in New York City and Connecticut. Second, we were able to estimate that the cumulative incidence was 26.8% (95% CrI = 24.2%, 29.7%) in New York City and 8.8% (95% CrI = 7.1%, 11.3%) in Connecticut by the end of September 2020, which was greater than the peak daily seroprevalence of 22.1% (95% CrI = 20.5%, 23.6%) in mid-May in New York City and 6.1% (95% CrI = 5.3%, 7.1%) at the end of May in Connecticut. Cumulative incidence could be underestimated by cross-sectional serosurveys only a few months after the first cases of SARS-CoV-2 in a population because individuals no longer have a detectable level of antibodies 3–4 months after seroconversion on average. Taken together, our findings suggest that cumulative incidence of SARS-CoV-2 should not be estimated directly from cross-sectional serology data, especially at later stages of an outbreak. Rather, cumulative incidence should be adjusted given impacts of waning of detectable antibodies in serologic assays.
The timeline of seroconversion and seroreversion should ideally be determined by individual-level longitudinal studies that follow up each infected individual multiple times, as frequently as possible, for a long enough period to observe seroreversion. The timeline for seroreversion after SARS-CoV-2 infection reported by the longitudinal studies published to date varies. Some studies have reported rapid waning of IgG, with substantial attrition of the seropositive population in as little as 60 days,7,24 while others reported that antibodies remained above the detectable threshold for at least 82 days after symptom onset25 or 120 days after qPCR diagnosis of SARS-CoV-2.26 These differences are likely due to limitations or heterogeneity of the studies reported to date, including short follow-up times, small sample sizes, and different serology testing methods and analytic sensitivities.27 Most of the longitudinal studies published to date have followed participants for 14–150 days after symptom onset or baseline visits, which is not long enough to understand the complete timeline of seroreversion for all participants, particularly for IgG.7,8,18,19,24,25 The variation in clinical and demographic characteristics and severity of infection of participants in each study has also likely influenced these different findings.15,16,19,24 Infected individuals with mild or no symptoms who may exhibit lower titer and shorter time to seroreversion are often not included in these longitudinal studies. Therefore, assessing the timeline using population-level data, such as cross-sectional serology data and mortality data, is a viable alternative approach. Using our framework, infected individuals were estimated to remain seropositive for about 3–4 months on average. The average duration estimated by the New York City data and Connecticut data were mostly in agreement, with overlapping CrIs. Differences in these average durations could be attributable to differences in demographic and clinical characteristics of infected individuals and differences in the testing and reporting practices. It is also important to note that the timeline of seroconversion and seroreversion is dependent on serologic assays and targeted immunoglobulins that may have different thresholds to define seropositivity.
We estimated that the IFR was 1.1% (95% CrI = 1.0%, 1.2%) in New York City, which was consistent with estimates from the routine care group at Mount Sinai Hospital in New York City.28 Yang et al. estimated the time-varying IFR for New York City (1.39%; 95% CrI = 1.04, 1.77),20 matching with our estimate under the assumption of decreasing IFR [1.4% (95% CrI = 1.2%, 1.6%) on average between March and September 2020]. Although the sensitivity analyses did not change our central conclusions, we note that different values of the SD for time from seroconversion to seroreversion changed the estimated seroprevalence in the later phase of the study period (August and September 2020; Figure 4 and eFigure 2; http://links.lww.com/EDE/B804). We ran the model with SD 20, 50, and 70 days, because the estimated SD for time from seroconversion to seroreversion for IgA and IgM was approximately 50 and 25 days, respectively.19 Our SD could be larger than those estimated for small study populations, as we used population-level data that likely have greater variation in demographic and clinical characteristics. Analyzing seroprevalence data in August and after could provide critical information on how population-level seroprevalence declines over time and would enable better estimates of the timeline of antibody waning.
The data used in this study have certain limitations. The samples collected for the CDC seroprevalence data may not be representative of the general population, as Havers et al. discussed.6 The data on the date of symptom onset among cases who died in Georgia may be incorrectly recorded and subject to recall bias upon case identification, as onset dates are largely self-reported. Also, because the model adjusts for the delay between the date of symptom onset and date of death, estimated cumulative incidence towards the end of the time series could be underestimated. This is not a concern for this particular study because the daily number of COVID-19 deaths was very small in the last few months; however, researchers need to be careful when using this model during the growing phase of the epidemic. Changes in testing and reporting practice over time likely affected the number of reported deaths, although mortality data are less sensitive to these changes compared to case data. We conducted the sensitivity analysis with a different source of data on time from onset to death and considered time-varying IFR instead of constant IFR, and found that results were robust to these changes (eTable 2; http://links.lww.com/EDE/B804).
In addition to limitations on timing, it is critical to note that detection of antibodies by serologic assays may not correlate to protection from reinfection or disease, and thus, time from seroconversion to seroreversion estimated in our study is not necessarily equivalent to the duration of protective immunity. Several studies have noted strong correlation between results from assays based on antibody binding (such as ELISA) and neutralization testing,15,19,25 which may allow protective immunity to be inferred from simpler serologic tests once more comprehensive longitudinal data sets are available. Moreover, T cell immunity may make an important contribution to protective immunity but is not assessed by serologic methods.29 Therefore, additional studies are needed to define immunologic determinants of SARS-CoV-2 protection against reinfection and severe disease.
Our findings suggest that the cumulative incidence estimated from serology data needs to be adjusted for seroreversion. We intend to apply this framework to other seroprevalence studies30 and we suggest others conducting serologic surveys consider doing so as well. Our framework could readily be applied to other data to estimate the duration of seroreversion, IFR, ascertainment ratios, daily seroprevalence, and cumulative incidence.
ACKNOWLEDGMENTS
The authors thank Dr. Laura Edison from the Georgia Department of Public Health for sharing the data. We also thank Dr. Manish Patel from the Centers for Disease Control and Prevention, Carly Adams, Avnika B. Amin, and Dr. Julia Baker from Emory University for providing feedback on this study.
Supplementary Material
Footnotes
This study was supported by the US National Science Foundation (grant 2032082 to J.S.W. and grant 2032084 to B.A.L.) and the US National Institute of Allergy and Infectious Diseases (3R01AI143875-02S1 to P.S.S. and A.J.S.).
B.A.L. reports personal fees from Takeda Pharmaceutical, personal fees from CDC Foundation, and personal fees from Hall Booth Smith, P.C., outside the submitted work. The other authors have no conflicts to report.
Description of the process by which someone else could obtain the data and computing code required to replicate the results reported in our submission: COVID-19 associated mortality data and longitudinal seroprevalence data are available on government websites that are cited in the article. The code can be found in the following github repository: https://github.com/lopmanlab/SARS-CoV-2_CumInc_WaningAntibodies.
Supplemental digital content is available through direct URL citations in the HTML and PDF versions of this article (www.epidem.com).
REFERENCES
- 1.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johns Hopkins Coronavirus Resource Center. COVID-19 Map. Available at: https://coronavirus.jhu.edu/map.html. Accessed 8 October 2020.
- 3.Wu SL, Mertens AN, Crider YS, et al. Substantial underestimation of SARS-CoV-2 infection in the United States. Nat Commun. 2020;11:4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckerle I, Meyer B. SARS-CoV-2 seroprevalence in COVID-19 hotspots. Lancet. 2020;396:514–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arora RK, Joseph A, Wyk JV, et al. SeroTracker: a global SARS-CoV-2 seroprevalence dashboard. Lancet Infect Dis. 2021;21:e75–e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Havers FP, Reed C, Lim T, et al. Seroprevalence of antibodies to SARS-CoV-2 in 10 Sites in the United States, March 23-May 12, 2020. JAMA Intern Med. 180:1576–1586.Published online July 21, 2020; [DOI] [PubMed] [Google Scholar]
- 7.Ibarrondo FJ, Fulcher JA, Goodman-Meza D, et al. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild Covid-19. New Engl J Med. 2020;383:1085–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel MM, Thornburg NJ, Stubblefield WB, et al. Change in antibodies to SARS-CoV-2 over 60 days among health care personnel in Nashville, Tennessee. JAMA. 2020;324:1781–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poland GA, Ovsyannikova IG, Kennedy RB. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396:1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. Coronavirus Disease 2019 (COVID-19) Commercial Laboratory Seroprevalence Surveys. Published February 11, 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/commercial-lab-surveys.html. Accessed 8 October 2020.
- 11.Centers for Disease Control and Prevention. CDC COVID Data Tracker. Published March 28, 2020. Available at: https://covid.cdc.gov/covid-data-tracker. Accessed 8 October 2020.
- 12.Connecticut Data. COVID-19 Tests, Cases, Hospitalizations, and Deaths (Statewide). Available at: https://data.ct.gov/Health-and-Human-Services/COVID-19-Tests-Cases-Hospitalizations-and-Deaths-S/rf3k-f8fg. Accessed 15 October 2020.
- 13.NYC Health. COVID-19: Data Details on Deaths. Available at: https://www1.nyc.gov/site/doh/covid/covid-19-data-deaths.page. Accessed 15 October 2020.
- 14.Iyer AS, Jones FK, Nodoushania A, et al. Dynamics and significance of the antibody response to SARS-CoV-2 infection. medRxiv. Published online July 20, 2020. doi: 10.1101/2020.07.18.20155374 [Google Scholar]
- 15.Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2−specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26:1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin Infect Dis. 2020;71:2027–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang AT, Garcia-Carreras B, Hitchings MDT, et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun. 2020;11:4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.To KK-W, Tsang OT-Y, Leung W-S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iyer AS, Jones FK, Nodoushani A, et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol. 2020;5:eabe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang W, Kandula S, Huynh M, et al. Estimating the infection-fatality risk of SARS-CoV-2 in New York City during the spring 2020 pandemic wave: a model-based analysis. Lancet Infect Dis. 2021;21:203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Our World in Data. Mortality risk of COVID-19 – statistics and research. Available at: https://ourworldindata.org/mortality-risk-covid. Accessed October 11, 2020.
- 22.Wu JT, Leung K, Bushman M, et al. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med. 2020;26:506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Driscoll M, Dos Santos GR, Wang L, et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590:140–145. [DOI] [PubMed] [Google Scholar]
- 24.Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26:1200–1204. [DOI] [PubMed] [Google Scholar]
- 25.Wajnberg A, Amanat F, Firpo A, et al. SARS-CoV-2 infection induces robust, neutralizing antibody responses that are stable for at least three months. medRxiv. Published online July 17, 2020:2020.07.14.20151126. doi: 10.1101/2020.07.14.20151126 [Google Scholar]
- 26.Gudbjartsson DF, Norddahl GL, Melsted P, et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383:1724–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi S, Greenhouse B, Rodríguez-Barraquer I. Are SARS-CoV-2 seroprevalence estimates biased? J Infect Dis. 2020222:1772–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stadlbauer D, Tan J, Jiang K, et al. Repeated cross-sectional sero-monitoring of SARS-CoV-2 in New York City. Nature. 2021;590:146–150. [DOI] [PubMed] [Google Scholar]
- 29.Cox RJ, Brokstad KA. Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nat Rev Immunol. 2020;20:581–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegler AJ, Sullivan PS, Sanchez T, et al. Protocol for a national probability survey using home specimen collection methods to assess prevalence and incidence of SARS-CoV-2 infection and antibody response. Ann Epidemiol. 2020;49:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


