Abstract
We rationally designed and developed caged siRNA nanoparticles (Multi-Chol-siRNA) self-assembled with cholesterol-modified multimerized caged siRNAs for photomodulation of siRNA gene silencing activity. Strong resistance to serum nuclease and RNase A was observed for these cholesterol-modified caged siRNA nanoparticles due to the formation of nanostructures with high intensity of siRNA. These caged Multi-Chol-siRNA self-assembled nanoparticles were successfully used to achieve photochemical regulation of both exogenous GFP and endogenous Eg5 gene expressions with a GFP/RFP transient transfection system and Eg5-associated assays, respectively. Further, Two-in-One caged Multi-Chol-siGFP/siEg5 self-assembled nanoparticles simultaneously targeting GFP and Eg5 genes were also developed. The caged Multi-Chol-siRNA self-assembled nanoparticles have demonstrated the effectiveness of enhancing photomodulation of multiple RNAi-induced gene silencing activities in cells.
Upon light irradiation, multimerized self-assembled caged Two-in-One siRNA nanoparticles (Multi-Chol-siRNA) were collapsed to release trapped siRNAs for multiple RNAi-induced gene silencing activity.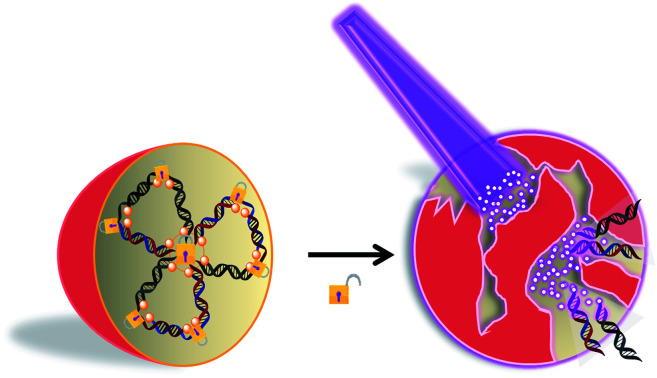
Introduction
Currently, RNA interference (RNAi)-based gene therapy has four major challenges, including delivery systems, stability, the immune response, and off-target effects.1–3 It is difficult for small interfering RNA (siRNA) drugs to penetrate the cell membrane by themselves due to their size and negative charges. A siRNA delivery process includes the accumulation of target tissues/organs, the target cell uptake and the effective endosomal escape. siRNA is prone to vascular degradation and rapid renal clearance, with a short half-life and poor serum stability. Although siRNA is sequence-specific for silencing target mRNA, off-target effects still exist and are concentration-dependent. Therefore, chemical modifications of siRNA drugs and their delivery materials have been shown to be more successful and effective methods to deal with several unsolved issues for siRNA drugs.4 Strategies involving chemically modified siRNA with 3′ terminal cholesterol conjugation with a long linker are one of the most promising and popular methods to improve siRNA PK/PD properties and therapeutic gene silencing activity.5–7 ARC-520, BMT101 and OLX10010 are the typical cholesterol-modified siRNA drugs in clinical trials.8–10
There are several classes of synthetic siRNAs, such as short hairpin RNA (shRNA), synthetic shRNA (sshRNA), small segmented siRNA (sisiRNA), siRibonucleic neutral (siRNN) and single-stranded siRNA (ss-siRNA) etc.3 In addition to chemical synthesis of novel siRNAs, self-assembled siRNA nanoparticles are currently a hotspot in the field of nucleic acid drug research. Nucleic acid self-assembled nanomaterials with a three-dimensional structure have potential application prospects in in vivo imaging and gene delivery, such as tetrahedral oligonucleotide nanoparticles (ONP) and RNAi microsponges.11,12 Recently, Ke et al. reported self-assembly of cholesterol-conjugated DNA nanorods and spherical micelles under acidic conditions.13 Multivalent Comb-aptamer-siRNA conjugates and multimeric siRNAs using combinatorial RNAi technology (co-RNAi) with the disulfide bonds have also been developed for gene delivery and gene therapy, which are responsible for endogenous reduced glutathione (GSH).14,15
Light is a clear, simple and rapid, non-invasive trigger. A photolabile linker was used as a promising photochemical switch, such as DMNPE, NPE, NPP, NPOM and CD-DMNPE.16 Photocaging strategies have been demonstrated to be successful and effective for temporal and spatial control of the gene expression in vitro and in vivo, including caged plasmids, caged antisense oligonucleotides, caged sgRNAs,17–22 caged miRNAs23–25, caged siRNAs etc.26,27 These caging siRNA strategies usually apply one or more photolabile moieties to randomly or precisely insert into the sequences of siRNA and sterically block the association of siRNA with the RNA-induced silencing complex (RISC), or prevent mRNA cleavage.28,29 RNAi-induced gene silencing activity of caged siRNAs was temporarily masked until light irradiation. Light-activated RNAi was first reported by Friedman et al. in 2005; they applied Diazo-DMNPE to randomly couple to the backbone of siRNA.30 Deiters and colleagues developed a caged siRNA strategy using NPP-modified caged dG/dT or NPOM-modified caged rG/rU to precisely position the seed region of siRNA.31,32 The 5′ terminal phosphorylation of antisense strand of siRNA is one of the most important binding sites for RICS formation and RNAi exertion of gene silencing according to the fixed-end model.2,33 Caged siRNA with photolabile terminal modifications has been reported to be more effective for light-activated RNAi, where terminal modification moieties include biotin,34 CD-DMNPE,35 vitamin E,36 cholesterol,37 cRGD, MUC1/AS1411 aptamers,38 or natural hydrophilic polymers,39 or circular siRNAs with 5′ and 3′ terminal crosslinking.27,40,41
In this work, we developed cholesterol-modified caged siRNA self-assembled nanoparticles, which included Multi-Chol-siGFP and Multi-Chol-siEg5 nanoparticles as well as Two-in-One Multi-Chol-siGFP/siEg5 nanoparticles simultaneously targeting exogenous GFP and endogenous Eg5 genes. These caged Multi-Chol-siRNA nanoparticles were self-assembled through hybridization between the complementary sequences of single-stranded caged Dimer-Chol-antisense and Dimer-Chol-Sense. Even without the addition of cationic condensing species (such as Lipo 2000, and PEI), caged Multi-Chol-siRNA self-assembled nanoparticles showed high resistance to 20% FBS and RNase A. More importantly, these as-prepared caged siRNA nanoparticles could be a novel caging strategy to temporally inhibit RNAi-induced gene silencing activity. Upon light irradiation, the caged Multi-Chol-siRNA self-assembled nanoparticles collapsed and quickly released the trapped siRNAs to recover their siRNA activities. In GFP/RFP and Eg5-associated assays, we demonstrated that Two-in-One caged Multi-Chol-siRNA self-assembled nanoparticles could be successfully applied to achieve photoregulation of both exogenous GFP and endogenous Eg5 gene expressions simultaneously.
Results and discussion
Rational design and syntheses of multimerized caged siRNAs with cholesterol modification
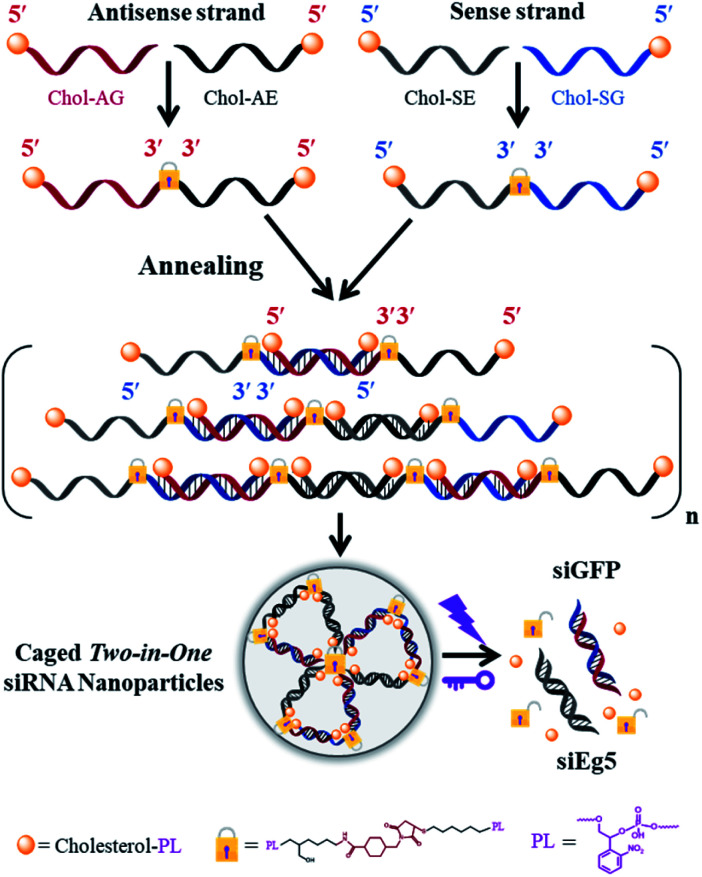
The phosphoramidites of thiol-modified C6 S-S (SS), a photolabile linker (p) and cholesterol (Chol) were sequentially coupled to RNA oligonucleotides using an ABI394 DNA/RNA solid-phase synthesizer (Fig. S1†). Terminal 5′-cholesterol- and 3′-thiol-modified caged RNA oligonucleotides (Chol-p-oligos-p-SS) targeting GFP and Eg5 were synthesized (Table S1†). All RNA oligonucleotides were then purified using reversed-phase HPLC and further characterized using ESI-MS (see the ESI†). We then constructed the single-stranded caged sense dimer (Dimer-Chol-Sense) and caged antisense dimer (Dimer-Chol-Antisense) through the conjugation of bis(maleimido)hexane (BMH, a bismaleimide crosslinker) and corresponding 3′ end thiol-modified Chol-p-sense-p-HS or Chol-p-antisense-p-HS oligonucleotides respectively with 5′-3′-3′-5′ sequence directions, as shown in Fig. S2 and S3.† The synthetic products of single-stranded caged Dimer-Chol-Sense or Dimer-Chol-Antisense were purified using reversed-phase HPLC (Fig. S2†) and were further characterized using ESI-MS (see the ESI†). Chemically cross-linked multimerized caged siRNAs with terminal cholesterol modification (Multi-Chol-siRNA) were then prepared through hybridization of single-stranded caged Dimer-Chol-Sense and Dimer-Chol-Antisense via base pairing between the two complementary sequences of siRNA, which resulted in the formation of caged siRNA nanoparticles (Fig. S3† and 1).
Fig. 1. Scheme of multimerized self-assembled caged Two-in-One siRNA nanoparticles (Multi-Chol-siRNA). Chol, cholesterol; PL (or p), photolinker; A, antisense strand RNA; S, sense strand RNA; G, GFP; E, Eg5.
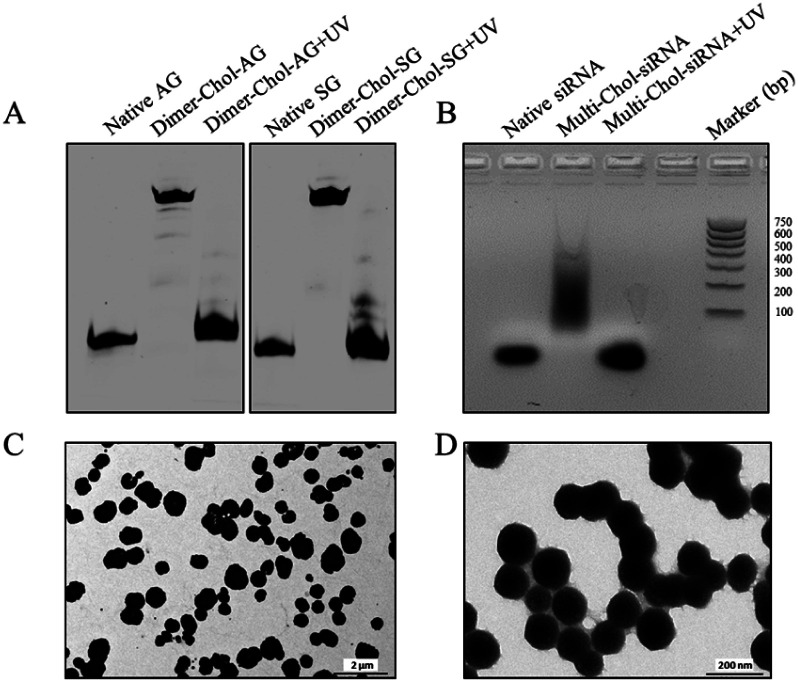
Fig. 2A shows the dimerization and photo-uncaging ability of caged Dimer-Chol-Sense and Dimer-Chol-Antisense analyzed with 20% PAGE gels. The single-stranded caged Dimer-Chol-Sense or Dimer-Chol-Antisense showed slower migration in PAGE gels than that observed for the native control oligonucleotides. Upon 3 min of light irradiation (365 nm, 7 mW cm−2), the photolinkers of caged Dimer-Chol-Sense and Dimer-Chol-Antisense were nearly photocleaved, which led to the release of terminal phosphorylated oligonucleotides with a similar shift in PAGE gels in comparison to the corresponding native oligonucleotides, and these uncaged oligonucleotides with terminal phosphate groups were further characterized using ESI-MS (see the ESI†). By mixing caged Dimer-Chol-Sense and Dimer-Chol-Antisense, the formation of caged Multi-Chol-siRNA nanoparticles with cholesterol modification was verified using a transmission electron microscope (TEM) and 1% agarose gels (Fig. 2). The average diameter of caged Multi-Chol-siRNA self-assembled nanoparticles was determined to be ∼500 nm that could be further reduced to ∼130 nm in the presence of a condensing agent (10 pmol Multi-Chol-siRNA/0.5 μL Lipo 2000) and a high density of siRNAs in nanoparticles was achieved. The photo-uncaging properties of caged Multi-Chol-siRNA nanoparticles in solution was also analyzed. Light irradiation broke most of the caged Multi-Chol-siRNA nanoparticles as indicated in the TEM image (Fig. S4†) which triggered the release of naked siRNA that moved to a similar position in agarose gel electrophoresis as demonstrated for the corresponding control siRNA (Fig. 2B). The photosensitivity of caged Multi-Chol-siRNA nanoparticles was evaluated at different irradiation times of 1–4 min and different light intensities (7–28 mW cm−2) (Fig. S5†). And 3 min of light irradiation (7 mW cm−2) was enough for photo-cleavage of caged Multi-Chol-siRNA nanoparticles in solution.
Fig. 2. Characterization of caged Multi-Chol-siRNA self-assembled nanoparticles. (A) Dimerization and photo-uncaging properties of caged Dimer-Chol-Sense and Dimer-Chol-Antisense analyzed with 20% PAGE gels. (B) Photo-uncaging ability of caged Multi-Chol-siRNA nanoparticles analyzed with 1% agarose gels. (C) TEM image of caged Multi-Chol-siRNA nanoparticles. (D) TEM image of caged Multi-Chol-siRNA nanoparticles after condensation with Lipo 2000.
Analysis of serum and RNase A stability of caged Multi-Chol-siRNA nanoparticles
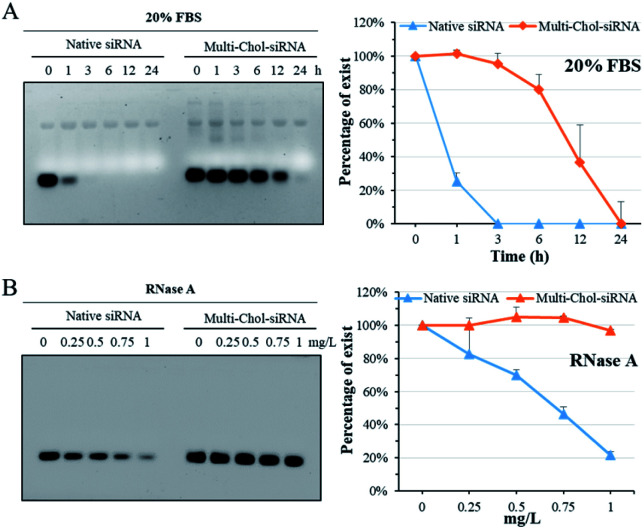
The stability of caged Multi-Chol-siRNA self-assembled nanoparticles against fetal bovine serum (FBS) was evaluated without any cationic condensing agent. The native siRNA and caged Multi-Chol-siRNA self-assembled nanoparticles were incubated with 20% FBS solution at 37 °C for 1, 3, 6, 12, and 24 h, respectively, and each sample was then irradiated to release siRNA for gel characterization. As shown in Fig. 3A, the native linear siRNA quickly and completely degraded after 3 h of incubation in 1% agarose gels. However, the serum stability of caged Multi-Chol-siRNA self-assembled nanoparticles was significantly improved in comparison to that of control linear siRNA with around 95% siRNA intact for 3 h. Prolonged incubation led to the gradual degradation of caged Multi-Chol-siRNA by FBS. RNase A, another most important nuclease for siRNA, was also chosen for evaluating the stability of caged Multi-Chol-siRNA self-assembled nanoparticles without the addition of cationic condensing species. The native siRNA and caged Multi-Chol-siRNA self-assembled nanoparticles were incubated in RNase A solution at 37 °C with RNase A concentrations of 0, 0.25, 0.5, 0.75, and 1 mg L−1, respectively. Each sample was then irradiated to release siRNA for gel characterization. Fig. 3B shows that the band of native siRNA gradually degraded and up to 80% siRNA degradation was observed in an RNase A concentration of 1 mg L−1 in 0.5% agarose gels. The caged Multi-Chol-siRNA self-assembled nanoparticles showed much higher stability against RNase A in our predetermined concentrations (0–1 mg L−1), and no obvious degradation of siRNA in caged Multi-Chol-siRNA self-assembled nanoparticles was observed even at 1 mg L−1 RNase A concentration. These results demonstrated that caged Multi-Chol-siRNA self-assembled nanoparticles showed a dramatic increase in stability in the presence of either FBS or RNase A even without condensation.
Fig. 3. Fetal bovine serum (FBS) and RNase A stability of native siRNA and caged Multi-Chol-siRNA self-assembled nanoparticles. The native siRNA and caged Multi-Chol-siRNA self-assembled nanoparticles were incubated with 20% FBS solution at 37 °C for predetermined time intervals (A) and with 0–1 mg L−1 RNase A (B). Percentages of siRNA were evaluated by normalizing the relative intensity of each band over the control band on the same gel using Image J software. Each sample was irradiated to release siRNA for gel electrophoresis. Data are presented as mean ± S.E.M. (n = 3).
Photochemical regulation of GFP gene silencing with caged Multi-Chol-siGFP self-assembled nanoparticles
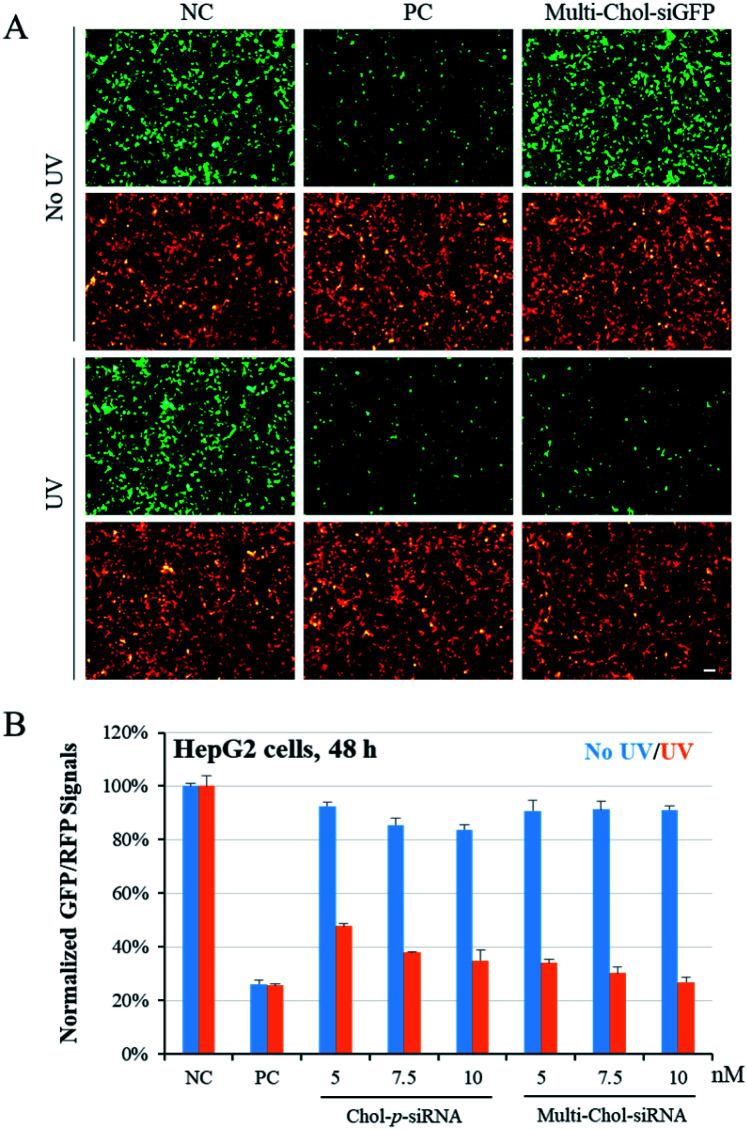
To confirm the photochemical regulation of GFP gene silencing with caged Multi-Chol-siGFP self-assembled nanoparticles in cells, we used a GFP/RFP transient transfection system, where the GFP gene was chosen as the target gene and the RFP gene as an internal control. HepG2 cells were cotransfected with pEGFP-N1 and pDsReds-N2 plasmids as well as the corresponding native siGFP (AG/SG) or Multi-Chol-siGFP self-assembled nanoparticles for 5 h, and the transfected cells were further incubated for 48 h. As shown in Fig. 4A, GFP fluorescence of HepG2 cells with siGFP (AG/SG) transfection (positive control, PC) significantly decreased in comparison to that of the only plasmid transfection groups (negative control, NC), while there was no obvious difference for the gene expression of RFP in HepG2 cells as displayed in cell fluorescence images. The gene expressions of GFP and RFP in caged Multi-Chol-siGFP nanoparticle-treated groups without light irradiation were similar to those of the negative control groups. This observation indicated that caged Multi-Chol-siRNA self-assembled nanoparticles showed high ability to temporally trap siRNA and inhibit further RISC complex formation and siRNA processing until light activation. However, the GFP gene expression was significantly downregulated in cells treated with caged Multi-Chol-siGFP self-assembled nanoparticles upon 3 min light irradiation, indicating that light collapsed caged siRNA nanoparticles and released siGFP for gene silencing.
Fig. 4. Photoregulation of GFP gene silencing with caged Multi-Chol-siGFP self-assembled nanoparticles. (A) Photochemical regulation of GFP gene silencing was observed from fluorescence images. (B) Comparison of the differences in the photomodulation of gene silencing between single cholesterol-modified caged siRNA (Chol-p-siRNA) and the caged Multi-Chol-siGFP self-assembled nanoparticles. The concentration of the native siGFP (AG/SG, PC) is 5 nM. Scale bar = 25 μm. Data are presented as mean ± S.E.M. (n = 3).
We further quantified the dose effect of photochemical regulation of the GFP gene expression using flow cytometry with caged Multi-Chol-siGFP self-assembled nanoparticles in both HepG2 and HeLa cells (Fig. S6†). Without light activation, the normalized mean GFP fluorescence intensity in cells treated with caged Multi-Chol-siGFP self-assembled nanoparticles was similar to that of the negative control group with almost no decease of relative GFP fluorescence intensity, which indicated that caged Multi-Chol-siGFP nanoparticles lost their RNAi-induced gene silencing activity even with the concentration increase of caged Multi-Chol-siGFP nanoparticles. After light irradiation, caged Multi-Chol-siGFP self-assembled nanoparticles were light-triggered to dose-dependently decrease the GFP fluorescence signal in HepG2 cells. The GFP intensity of caged Multi-Chol-siGFP self-assembled nanoparticles at a concentration of 5 nM after 48 h of incubation was downregulated to 34% in comparison to that of the negative control, indicating that caged Multi-Chol-siGFP nanoparticles almost recovered RNAi-induced gene silencing activity upon 3 min light irradiation.
We also compared the differences in the photomodulation of gene silencing activity between single cholesterol-modified siRNA (Chol-p-siRNA) and caged Multi-Chol-siGFP self-assembled nanoparticles. As shown in Fig. 4B, we demonstrated that the photoregulation of the target gene expression could be achieved with the single cholesterol-modified caged siRNA. However, the leaking RNAi activity increased with the increase of the Chol-p-siRNA concentration in 48 h. For caged Multi-Chol-siGFP self-assembled nanoparticles with a higher density of siRNA, almost no decrease of the GFP expression level was observed with the increase of the caged siRNA concentration before light activation. Upon 3 min of light irradiation, native siRNA was fully released and greater enhancement of photomodulation of RNAi-induced gene silencing activity was successfully realized than the single cholesterol-modified caged siRNA. Thus, caged Multi-Chol-siGFP self-assembled nanoparticles were successfully used to achieve photoregulation of RNAi-induced GFP gene silencing activity in cells with low leaking activity.
Photochemical regulation of the endogenous Eg5 gene expression with caged Multi-Chol-siEg5 self-assembled nanoparticles
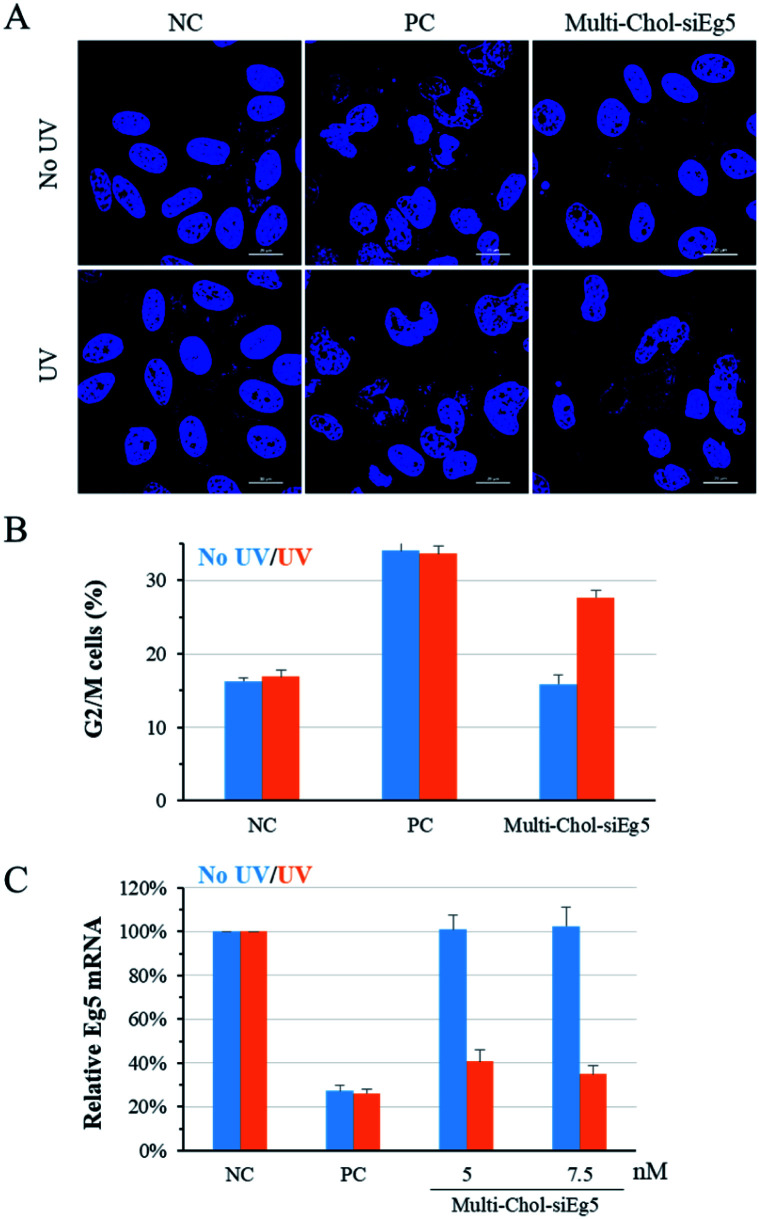
To confirm the generality of gene silencing using caged Multi-Chol-siRNA self-assembled nanoparticles, we also investigated the photoregulation of the endogenous Eg5 gene expression in cells. Eg5 gene silencing by siEg5 (positive control, PC) inhibited the expression of mitotic motor protein, resulting in the formation of irregular nuclei phenotypes and arresting in prometaphases of the cell cycle. As shown in Fig. 5A, the nuclei phenotypes of HepG2 cells treated with caged Multi-Chol-siEg5 self-assembled nanoparticles without light irradiation were similar to those of the negative control groups with the normal shape of cell nuclei. This observation indicated that caged Multi-Chol-siEg5 self-assembled nanoparticles effectively blocked RNAi-induced Eg5 gene silencing. After 3 min of light irradiation, caged Multi-Chol-siEg5 self-assembled nanoparticles collapsed and the trapped siEg5 was released from the nanoparticles to exert Eg5 gene silencing activity in cells. Obvious irregular nucleic phenotypes were observed for HepG2 cells treated with light-activated caged Multi-Chol-siEg5 nanoparticles, as displayed in the cell image in Fig. 5A using a confocal laser scanning microscope.
Fig. 5. Photoregulation of Eg5 gene silencing with the caged Multi-Chol-siRNA self-assembled nanoparticles. (A) Nuclei phenotypes of HepG2 cells using a confocal laser scanning microscope. Scale bar = 20 μm. (B) Percentage of cells in G2/M phases of the cell cycle quantified via a cycle analysis kit. (C) Photoregulation of Eg5 mRNA levels determined by real-time qPCR. The concentration of PC (AE/SE) is 5 nM. Data are presented as mean ± S.E.M. (n = 3).
Further quantification of cell arresting in G2/M phases was used to further examine the photomodulation of Eg5 gene silencing with caged Multi-Chol-siRNA self-assembled nanoparticles (Fig. 5B). Without light irradiation, the G2/M phase percentage of cells treated with caged Multi-Chol-siRNA self-assembled nanoparticles was around 15.9%, similar to that of the negative control group (16.3%). Light activation triggered the release of trapped siEg5 from caged Multi-Chol-siEg5 self-assembled nanoparticles, which successfully arrested cells in mitotic prometaphases. The percentage of cells in G2/M phases increased to 27.7% under Multi-Chol-siRNA transfection following 3 min of light irradiation. Under the same experimental conditions, photochemical regulation of Eg5 mRNA levels using caged Multi-Chol-siEg5 self-assembled nanoparticles was also examined using real-time qPCR analysis. As shown in Fig. 5C and S7,† Eg5 mRNA level in cells treated with caged Multi-Chol-siRNA self-assembled nanoparticles were similar to those of the negative control group with no decrease at both 5 nM and 7.5 nM. However, light activation successfully decreased the Eg5 mRNA level of cells treated with caged Multi-Chol-siRNA self-assembled nanoparticles in a dose-dependent manner. All these results showed that caged Multi-Chol-siRNA self-assembled nanoparticles were successfully applied to achieve photochemical regulation of the endogenous gene expression in cells.
Two-in-One caged Multi-Chol-siGFP/siEg5 self-assembled nanoparticles
To further extend the application of silencing multiple target genes simultaneously using caged Multi-Chol-siRNA self-assembled nanoparticles, we rationally developed Two-in-One caged Multi-Chol-siGFP/siEg5 self-assembled nanoparticles. These caged Multi-Chol-siGFP/siEg5 self-assembled nanoparticles were designed based on combinatorial RNAi technology that can simultaneously silence both GFP and Eg5 genes. Both RNA strands of siGFP were first synthesized with 5′ end photolabile linker/cholesterol and 3′ end photolabile linker/amine modifications. By conjugation with an SMCC crosslinker, maleimide moieties were introduced to 3′ ends of both RNA strands of siGFP. At the same time, both RNA strands of siEg5 were synthesized with 5′ end photolabile linker/cholesterol and 3′ end photolabile linker/thiol modifications. As shown in Fig. S8,† single-stranded caged sense Dimer-Chol-SG/SE (sense strand RNAs of siGFP and siEg5) and caged antisense Dimer-Chol-AG/AE (antisense strand RNAs of siGFP and siEg5) with 5′-3′-3′-5′ sequence directions were respectively synthesized using the maleimide-thiol based bioconjugation strategy at 3′ ends. Dimer-Chol-SG/SE and Dimer-Chol-AG/AE were further purified through reverse phase HPLC and characterized using ESI-MS (see the ESI†). The dimerization and photo-uncaging ability of caged Dimer-Chol-SG/SE and Dimer-Chol-AG/AE were then evaluated using PAGE gels (Fig. S8†). In the PAGE gels, caged Dimer-Chol-SG/SE and Dimer-Chol-AG/AE showed slow migration. 3 min of light irradiation nearly photocleaved the photolinker between the two sense RNA strands of caged Dimer-Chol-SG/SE or two antisense RNA strands of Dimer-Chol-AG/AE, and released single sense or antisense RNA oligonucleotides. Chemically cross-linked Two-in-One caged Multi-Chol-siGFP/siEg5 self-assembled nanoparticles were formed after the hybridization of caged sense Dimer-Chol-SG/SE and caged antisense Dimer-Chol-AG/AE through corresponding complementary sense and antisense sequences of siGFP and siEg5.
Further simultaneous gene silencing activities for both GFP and Eg5 genes using Two-in-One caged Multi-Chol-siGFP/siEg5 self-assembled nanoparticles were evaluated in cells. As shown in Fig. S9 and S10,† fluorescence images and the quantitative results of the normalized mean GFP fluorescence intensity showed that Two-in-One caged Multi-Chol-siGFP/siEg5 nanoparticles without light irradiation did not affect the gene expression of GFP and RFP, as well as Eg5. Upon light irradiation, the GFP gene expression significantly decreased in a dose-dependent manner in cells treated with Two-in-One caged Multi-Chol-siGFP/siEg5 self-assembled nanoparticles, but no effect on the RFP expression. Meanwhile, caged Multi-Chol-siGFP/siEg5 nanoparticles also demonstrated the effective photochemical regulation of Eg5 mRNA levels determined by real-time qPCR (Fig. S10†). Upon 3 min of light activation, these caged Multi-Chol-siGFP/siEg5 self-assembled nanoparticles successfully arrested cells in mitotic prometaphases. HepG2 cells treated with Two-in-One caged Multi-Chol-siGFP/siEg5 nanoparticles showed obvious irregular nucleic phenotypes after 48 h of incubation (Fig. S9†).
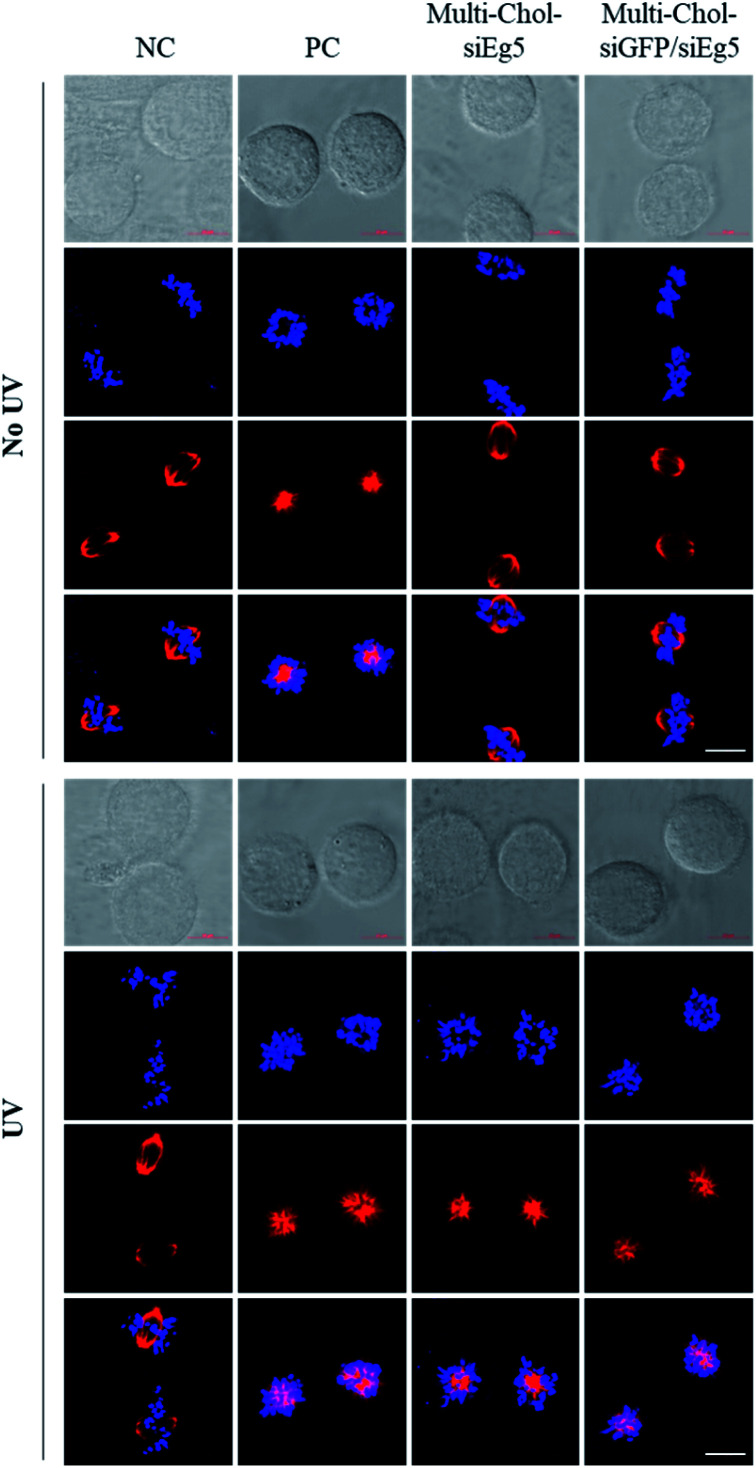
Importantly, as shown in Fig. 6, the mitotic spindle phenotypes of HepG2 cells treated with 7.5 nM Multi-Chol-siEg5 and 15 nM Two-in-One Multi-Chol-siGFP/siEg5 self-assembled nanoparticles (7.5 nM siEg5 and 7.5 nM siGFP in nanoparticles) without light irradiation were similar to those of the negative control groups with the normal mitotic spindle bipolarity and chromosomes near the spindle equator. After irradiation, HepG2 cells treated with light-activated caged Multi-Chol-siEg5 and Multi-Chol-siGFP/siEg5 nanoparticles were arrested in prometaphases with rosette-like monoastral spindle phenotypes surrounded by chromosomes due to the inhibition of the kinesin Eg5 gene in mitotic HepG2 cells (Fig. 6).
Fig. 6. Mitotic spindle and chromosome analysis of HepG2 cells using a confocal laser scanning microscope. Cells were treated with AE/SE (PC, 5 nM), 7.5 nM Multi-Chol-siEg5 (equivalent to siEg5 in nanoparticles), and 15 nM Two-in-One Multi-Chol-siGFP/siEg5 self-assembled nanoparticles (7.5 nM siEg5 and 7.5 nM siGFP in nanoparticles). Scale bar = 10 μm. Hoechst (blue), tubulin (red).
All these results confirmed that the Two-in-One caged Multi-Chol-siGFP/siEg5 self-assembled nanoparticles have great potential for photoregulation of RNAi-induced gene silencing of both GFP and Eg5 genes simultaneously. Self-assembled caged Two-in-One siRNA nanoparticles will provide a novel caging strategy for simultaneous optical control of two pathogenic gene expressions with spatial and temporal resolutions, especially in their application in stimuli-responsive combinatorial RNAi therapy.
Experimental
Synthesis and purification of chemically modified caged RNA oligonucleotides with terminal cholesterol modification
The phosphoramidites of β-[bis(4-methoxyphenyl)-phenylmethoxy]-2-nitro-benzene ethanol (photolabile linker, p) and cholesterol (Chol) were synthesized according to our previous study,37,39 and characterized by NMR (see the ESI†). RNA oligonucleotides were then synthesized according to standard phosphoramidite chemistry using an ABI394 DNA/RNA solid-phase synthesizer. Chemically modified Chol-p-oligos-p-SS oligonucleotides with a 3′-thiol group were synthesized through sequentially coupling with the phosphoramidites of the thiol-modified C6 S-S, different nucleotides, a photolabile linker and cholesterol, respectively. Chol-p-oligos-p-NH2 oligonucleotides with a 3′-amino group were synthesized using 3′-amino-modifier C7 CPGs through sequentially coupling with the phosphoramidites of different nucleotides, a photolabile linker and cholesterol. These synthetic RNA oligonucleotides were cleaved from solid CPGs using 1 mL AMA solution (ammonium hydroxide/methylamine) at 25 °C for 16 h, followed by the removal of TBDMS groups using the Et3N·3HF reagent at 55 °C for 2 h. RNA oligonucleotides were precipitated using a 3 M sodium acetate/ethanol method and were collected via centrifugation (14 000g for 15 min) at 4 °C. RNA oligonucleotides were further purified using reversed-phase HPLC with the elution gradient of ACN/TEAB buffer (ACN: 5 → 65% in 30 min, 1 mL min−1), and finally characterized using ESI-MS (see the ESI†).
Synthesis of chemically cross-linked multimerized caged siRNAs with cholesterol modification
These as-prepared Chol-p-oligos-p-SS oligonucleotides with disulfide bonds were reduced by 5 mM TCEP at 25 °C for 1 h, and then purified using a centrifugal filter (3 kD, Millipore) at 14 000g by washing several times using DEPC-treated water. The purified 3′-thiol-modified Chol-p-oligos-p-SH oligonucleotides were then conjugated with a BMH crosslinker in 100 mM PBS buffer (pH = 7.2, 1 M NaCl) overnight to obtain single-stranded caged Dimer-Chol-Sense and Dimer-Chol-Antisense with 5′-3′-3′-5′ sequence directions, respectively. Chol-p-oligos-p-NH2 oligonucleotides (5 nmol) directly reacted with 0.6 μmol SMCC crosslinker (50 μL DMSO) for 1.5 h in 150 μL PBS buffer (100 mM, pH = 7.2), and were then purified with a centrifugal filter (3 kD, Millipore) at 14 000g by washing for several times to obtain maleimide-modified Chol-p-oligos-p-Mal oligonucleotides. Single-stranded Two-in-One caged Dimer-Chol-AG/AE and Dimer-Chol-SG/SE with 5′-3′-3′-5′ sequence directions were then prepared using maleimide-thiol based bioconjugation chemistry, where Chol-p-oligos-p-Mal oligonucleotides were conjugated with Chol-p-oligos-p-HS oligonucleotides at a 1 : 1 ratio in PBS buffer (100 mM, pH = 7.2). The as-prepared single-stranded caged Dimer-Chol-oligos were further purified using reverse phase HPLC with the elution gradient of ACN/TEAB buffer (ACN: 10 → 40% in 10 min, 40 → 55% in 20 min). Chemically cross-linked multimerized caged siRNAs (Multi-Chol-siGFP, Multi-Chol-siEg5 or Two-in-One Multi Chol-siGFP/siEg5) were then respectively prepared by hybridization of single-stranded caged Dimer-Chol-Sense (sense strand) and Dimer-Chol-Antisense (antisense strand) at a 1 : 1 ratio through base paring between the two corresponding complementary sequences of siRNA to form caged siRNA self-assembled nanoparticles.
Electrophoretic mobility gel-shift assay and serum stability assay
The dimerization and photo-uncaging ability of single-stranded caged Dimer-Chol-oligos were examined using 20% native PAGE gels in our study. The PAGE gels were stained with SYBR Gold nucleic acid gel stain (Invitrogen, S11494) after running for 1 h at 200 V in RNase-free 1× TBE buffer. The photocleavage of caged Multi-Chol-siRNA nanoparticles in solution was performed under 1, 2, 3, and 4 min of UV light irradiation (7 mW cm−2), and at 7, 14, 21, and 28 mW cm−2 light intensities (1 min) (LED lamp, Lamplic, No. UVDRV-8-A-0150-30). The polymerization and photo-uncaging properties as well as the serum stability of Multi-Chol-siRNA self-assembled nanoparticles were determined using 1% agarose gels which were pre-stained with Gel-red stain and run for 20 min at 150 V in RNase-free 1× TAE buffer. The PAGE and agarose gels were imaged using gel imaging systems (ChemiDoc XRS). In the serum stability study, the native siRNA and caged Multi-Chol-siRNA were incubated with 20% FBS solution at 37 °C for 1, 3, 6, 12, and 24 h, respectively. In RNase A stability assay, the native siRNA and caged Multi-Chol-siRNA were incubated with RNase A at 37 °C in an incubator for 10 min with RNase A concentrations of 0, 0.25, 0.5, 0.75, and 1 mg L−1, respectively. Each sample was irradiated to release siRNA for gel electrophoresis.
Transmission electron microscopy
10 μL of caged Multi-Chol-siRNA self-assembled nanoparticles (1 μM) with or without UV light irradiation (3 min, 7 mW cm−2) was dropped onto the surface of a carbon-coated copper grid and dried for TEM analysis. The TEM images of the caged Multi-Chol-siRNA self-assembled nanoparticles were obtained using a JEM-1200EX instrument (120 kV).
Cell culture and GFP/RFP transfection assay
HepG2 cells and HeLa cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37 °C in a 5% CO2 atmosphere. In GFP/RFP transfection assay, cells were plated into 24-well plates at a density of 10 × 104 cells per well for 24 h incubation, and then cotransfected with 300 ng pEGFP-N1 and 450 ng pDsReds-N2 plasmids as well as the corresponding native siRNA (5 nM) or caged Multi-Chol-siGFP self-assembled nanoparticles (2.5–10 nM, equivalent to siRNA) using 1 μL Lipofectamine 2000 transfection reagent (Invitrogen). After 5 h of incubation, cells were treated with 3 min of light irradiation (UV = 365 nm, 7 mW cm−2) or kept under dark conditions (no light), followed by the replacement of fresh DMEM for 48 h incubation. Under the same imaging conditions, the GFP and RFP fluorescence intensities of cells were monitored and imaged using an automatic inverted fluorescence microscope (Olymups IX83). The excitation wavelengths of GFP and RFP protein are 488 and 560 nm. The normalized mean GFP fluorescence intensity of cells was further quantitated and analyzed by flow cytometry (CytoFLEX, Beckman).
Polymerase chain reaction
HepG2 cells were transfected with positive siRNA (5 nM) or Multi-Chol-siEg5 self-assembled nanoparticles (5 and 7.5 nM, equivalent to siRNA) using the Lipofectamine 2000 transfection reagent, respectively. After 5 h of incubation, HepG2 cells were irradiated with UV light (3 min, 7 mW cm−2) or kept under dark conditions (no light), followed by the replacement of fresh DMEM for 48 h incubation. Total RNA was extracted from HepG2 cells using the BioZol reagent (Biodee). RNA was reverse-transcribed to the complementary DNA which was used for RT-PCR and real-time qPCR using the GoTaq qPCR master mix reagent according to the manufacturer's protocol. GAPDH was used as an internal control in our study. Sequences of GAPDH and Eg5 primers are listed as follows:
GAPDH forward primer: 5′-TGCACCACCAACTGCTTAGC-3′ and reverse primer: 5′-GGCATGGACTGTGGTCATGAG-3′; Eg5 forward primer: 5′-CAGCTGAAAAGGAAACAGCC-3′ and reverse primer: 5′-ATGAACAATCCACACCAGCA-3′.
Cell cycle analysis
HepG2 cells were transfected with 5 nM positive siEG5 or 7.5 nM Multi-Chol-siEg5 self-assembled nanoparticles using the Lipofectamine 2000 reagent. After 6 h of incubation, cells were treated with light irradiation (UV = 365 nm for 3 min) or kept under dark conditions (no light). HepG2 cells were trypsinized using trypsin and washed with ice-cold 1× PBS, and then fixed in ice-cold 70% ethanol at 4 °C for 6 h. These as-prepared HepG2 cells were further mixed with 50 μg mL−1 propidium iodide (PI) and 20 μg mL−1 RNase A for 30 min at 37 °C in an incubator. Finally, HepG2 cells were examined using flow cytometry (CytoFLEX, Beckman) and the percentages of cells in the G2/M phases were quantitated and analyzed using ModFit LT software.
Confocal laser scanning microscope
HepG2 cells were transfected with positive siEg5 (AE/SE, 5 nM) or 7.5 nM Multi-Chol-siEg5, or 15 nM Two-in-One Multi-Chol-siGFP/siEg5 self-assembled nanoparticles (7.5 nM siEg5 and 7.5 nM siGFP in nanoparticles) using the Lipofectamine 2000 transfection reagent. After 5 h of incubation, HepG2 cells were irradiated with UV light or kept under dark conditions, followed by the replacement of fresh DMEM for 36 h incubation. Hoechst 33342 and tubulin-tracker red were added to stain the chromosomes and mitotic spindle phenotypes of mitotic HepG2 cells, respectively. Cell imaging was performed using a confocal laser scanning microscope (Zeiss, LSM880).
Conclusions
We have rationally designed and developed cholesterol-modified caged Multi-Chol-siGFP and Multi-Chol-siEg5 self-assembled nanoparticles as well as Two-in-One caged Multi-Chol-siGFP/siEg5 self-assembled nanoparticles for simultaneous gene silencing of both exogenous GFP and endogenous Eg5 genes. These caged Multi-Chol-siRNA self-assembled nanoparticles were formed by simple hybridization of caged Dimer-Chol-antisense and Dimer-Chol-Sense through base-paring between the corresponding complementary sense and antisense sequences of siRNAs. Great enhancement in nuclease resistance to FBS and RNase A was observed. Temporal trapping of siRNAs and efficient inhibition of RNAi processing were successfully achieved with these caged Multi-Chol-siRNA self-assembled nanoparticles with low leaking activity in comparison to non-assembled caged siRNA. Upon short light irradiation, these caged Multi-Chol-siRNA self-assembled nanoparticles collapsed to release the trapped siRNAs for the RNAi-induced gene silencing activity, and both exogenous GFP and endogenous Eg5 were simultaneously photomodulated with Two-in-One caged Multi-Chol-siGFP/siEg5. These results indicated that caged Multi-Chol-siRNA self-assembled nanoparticles as a novel caging strategy showed great promise for photochemical regulation of multiple target genes in cells simultaneously.
Conflicts of interest
The authors declare no conflict of competitive interest.
Supplementary Material
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (Grant No. 81821004, 21877001, and 22077005), National Major Scientific and Technological Special Project for “Significant New Drugs Development” (Grant No. 2017ZX09303013), and Fujian Medical University (Grant No. XRCZX2019027).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc03562a
Notes and references
- Wu S. Y. Lopez-Berestein G. Calin G. A. Sood A. K. Sci. Transl. Med. 2014;6:240ps247. doi: 10.1126/scitranslmed.3008362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M. Doudna J. A. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- Setten R. L. Rossi J. J. Han S. P. Nat. Rev. Drug Discovery. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- Chen C. Yang Z. Tang X. Med. Res. Rev. 2018;38:829–869. doi: 10.1002/med.21479. [DOI] [PubMed] [Google Scholar]
- DiFiglia M. Sena-Esteves M. Chase K. Sapp E. Pfister E. Sass M. Yoder J. Reeves P. Pandey R. K. Rajeev K. G. Manoharan M. Sah D. W. Zamore P. D. Aronin N. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raouane M. Desmaele D. Urbinati G. Massaad-Massade L. Couvreur P. Bioconjugate Chem. 2012;23:1091–1104. doi: 10.1021/bc200422w. [DOI] [PubMed] [Google Scholar]
- Bruck J. Pascolo S. Fuchs K. Kellerer C. Glocova I. Geisel J. Dengler K. Yazdi A. S. Rocken M. Ghoreschi K. J. Immunol. 2015;195:2216–2223. doi: 10.4049/jimmunol.1402989. [DOI] [PubMed] [Google Scholar]
- Rozema D. B. Lewis D. L. Wakefield D. H. Wong S. C. Klein J. J. Roesch P. L. Bertin S. L. Reppen T. W. Chu Q. Blokhin A. V. Hagstrom J. E. Wolff J. A. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluep T. Lickliter J. Hamilton J. Lewis D. L. Lai C. L. Lau J. Y. Locarnini S. A. Gish R. G. Given B. D. Clin. Pharmacol. Drug Dev. 2017;6:350–362. doi: 10.1002/cpdd.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B. Weng Y. Xia X. Liang X. Huang Y. J. Gene Med. 2019;21:e3097. doi: 10.1002/jgm.3097. [DOI] [PubMed] [Google Scholar]
- Lee H. Lytton-Jean A. K. R. Chen Y. Love K. T. Park A. I. Karagiannis E. D. Sehgal A. Querbes W. Zurenko C. S. Jayaraman M. Peng C. G. Charisse K. Borodovsky A. Manoharan M. Donahoe J. S. Truelove J. Nahrendorf M. Langer R. Anderson D. G. Nat. Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh Y. H. Deng J. Z. Dreaden E. C. Park J. H. Yun D. S. Shopsowitz K. E. Hammond P. T. Angew. Chem., Int. Ed. 2016;55:3347–3351. doi: 10.1002/anie.201508978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Peng R. Xu F. Ke Y. Bioconjugate Chem. 2019;30:1845–1849. doi: 10.1021/acs.bioconjchem.9b00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. Jung H. Kim S. A. Mok H. Chem. Commun. 2014;50:6765–6767. doi: 10.1039/C4CC01620C. [DOI] [PubMed] [Google Scholar]
- Lee S. H. Mok H. Jo S. Hong C. A. Park T. G. Biomaterials. 2011;32:2359–2368. doi: 10.1016/j.biomaterials.2010.11.062. [DOI] [PubMed] [Google Scholar]
- Ankenbruck N. Courtney T. Naro Y. Deiters A. Angew. Chem., Int. Ed. 2018;57:2768–2798. doi: 10.1002/anie.201700171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. Brown W. Bardhan A. Delaney M. Ilk A. S. Rauen R. R. Kahn S. I. Tsang M. Deiters A. Angew. Chem., Int. Ed. Engl. 2020;59:8998–9003. doi: 10.1002/anie.201914575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Ling X. Su X. Zhang S. Wang J. Zhang P. Feng W. Zhu Y. Liu T. Tang X. Angew. Chem., Int. Ed. Engl. 2020 doi: 10.1002/anie.202009890. [DOI] [PubMed] [Google Scholar]
- Wang S. Wei L. Wang J. Ji H. Xiong W. Liu J. Yin P. Tian T. Zhou X. ACS Chem. Biol. 2020;15:1455–1463. doi: 10.1021/acschembio.0c00105. [DOI] [PubMed] [Google Scholar]
- Moroz-Omori E. V. Satyapertiwi D. Ramel M. C. Hogset H. Sunyovszki I. K. Liu Z. Wojciechowski J. P. Zhang Y. Grigsby C. L. Brito L. Bugeon L. Dallman M. J. Stevens M. M. ACS Cent. Sci. 2020;6:695–703. doi: 10.1021/acscentsci.9b01093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Zou R. S. He S. Nihongaki Y. Li X. Razavi S. Wu B. Ha T. Science. 2020;368:1265–1269. doi: 10.1126/science.aay8204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill J. Borchardt E. K. Brown K. Asokan A. Deiters A. J. Am. Chem. Soc. 2015;137:5642–5645. doi: 10.1021/ja512664v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Sun Y. Li J. Zhang Y. Chem. Commun. 2020;56:627–630. doi: 10.1039/C9CC08277H. [DOI] [PubMed] [Google Scholar]
- Zhao J. Chu H. Zhao Y. Lu Y. Li L. J. Am. Chem. Soc. 2019;141:7056–7062. doi: 10.1021/jacs.9b01931. [DOI] [PubMed] [Google Scholar]
- Lucas T. Schafer F. Muller P. Eming S. A. Heckel A. Dimmeler S. Nat. Commun. 2017;8:15162. doi: 10.1038/ncomms15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruble B. K. Yeldell S. B. Dmochowski I. J. J. Inorg. Biochem. 2015;150:182–188. doi: 10.1016/j.jinorgbio.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y. Shu Z. Ito M. Abe N. Nakamoto K. Tomoike F. Shuto S. Ito Y. Abe H. Chem. Commun. 2020;56:466–469. doi: 10.1039/C9CC04872C. [DOI] [PubMed] [Google Scholar]
- Brieke C. Rohrbach F. Gottschalk A. Mayer G. Heckel A. Angew. Chem., Int. Ed. 2012;51:8446–8476. doi: 10.1002/anie.201202134. [DOI] [PubMed] [Google Scholar]
- Matsushita-Ishiodori Y. Ohtsuki T. Acc. Chem. Res. 2012;45:1039–1047. doi: 10.1021/ar200227n. [DOI] [PubMed] [Google Scholar]
- Shah S. Rangarajan S. Friedman S. H. Angew. Chem., Int. Ed. Engl. 2005;44:1328–1332. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]
- Govan J. M. Young D. D. Lusic H. Liu Q. Y. Lively M. O. Deiters A. Nucleic Acids Res. 2013;41:10518–10528. doi: 10.1093/nar/gkt806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikat V. Heckel A. RNA. 2007;13:2341–2347. doi: 10.1261/rna.753407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas M. M. Schlegel M. K. Harbison C. E. Yilmaz V. O. Jiang Y. Parmar R. Zlatev I. Castoreno A. Xu H. Shulga-Morskaya S. Rajeev K. G. Manoharan M. Keirstead N. D. Maier M. A. Jadhav V. Nat. Commun. 2018;9:723. doi: 10.1038/s41467-018-02989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Q. N. Chavli R. V. Marques J. T. Conrad, Jr. P. G. Wang D. He W. Belisle B. E. Zhang A. Pastor L. M. Witney F. R. Morris M. Heitz F. Divita G. Williams B. R. McMaster G. K. Biochim. Biophys. Acta. 2006;1758:394–403. doi: 10.1016/j.bbamem.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Jain P. K. Shah S. Friedman S. H. J. Am. Chem. Soc. 2011;133:440–446. doi: 10.1021/ja107226e. [DOI] [PubMed] [Google Scholar]
- Ji Y. Yang J. Wu L. Yu L. Tang X. Angew. Chem., Int. Ed. Engl. 2016;55:2152–2156. doi: 10.1002/anie.201510921. [DOI] [PubMed] [Google Scholar]
- Yang J. Chen C. Tang X. Bioconjugate Chem. 2018;29:1010–1015. doi: 10.1021/acs.bioconjchem.8b00080. [DOI] [PubMed] [Google Scholar]
- Zhang L. Chen C. Fan X. Tang X. Chembiochem. 2018;19:1259–1263. doi: 10.1002/cbic.201700623. [DOI] [PubMed] [Google Scholar]
- Chen C. Wang Z. Zhang J. Fan X. Xu L. Tang X. Bioconjugate Chem. 2019;30:1459–1465. doi: 10.1021/acs.bioconjchem.9b00204. [DOI] [PubMed] [Google Scholar]
- Zhang L. Liang D. Wang Y. Li D. Zhang J. Wu L. Feng M. Yi F. Xu L. Lei L. Du Q. Tang X. Chem. Sci. 2018;9:44–51. doi: 10.1039/C7SC03842A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Liang D. Chen C. Wang Y. Amu G. Yang J. Yu L. Dmochowski I. J. Tang X. Mol. Ther.--Nucleic Acids. 2018;10:237–244. doi: 10.1016/j.omtn.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.