Figure S4.
In bovine zygotes, unclustered chromosomes are more likely to missegregate, related to Figure 3
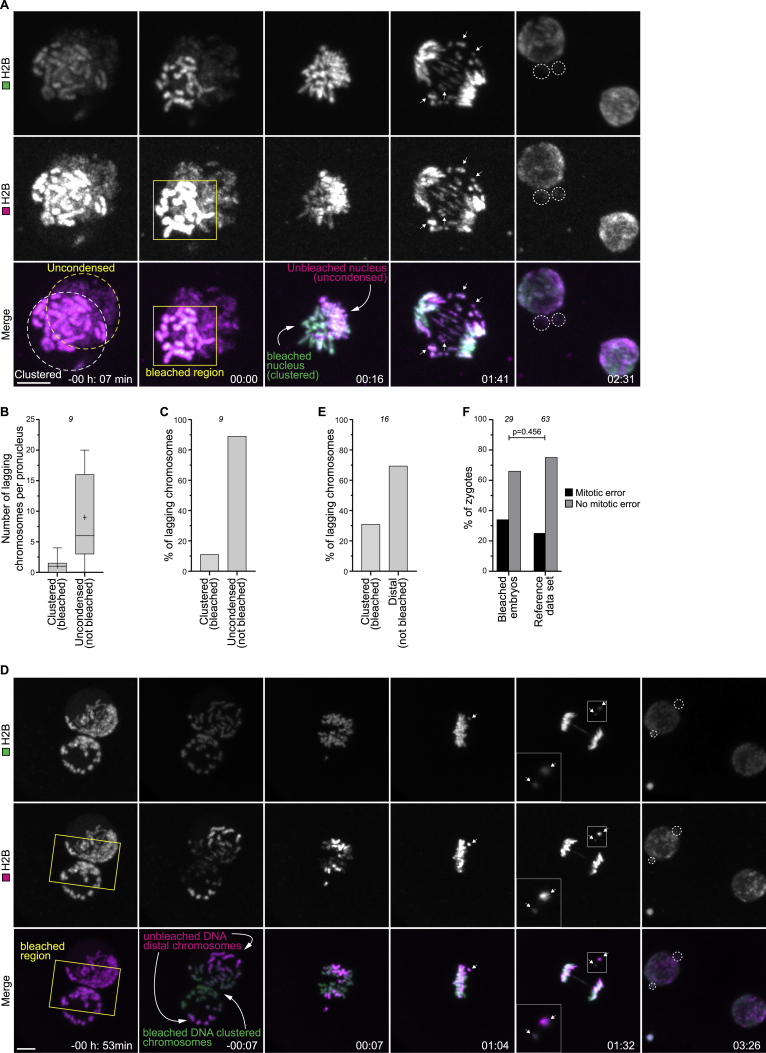
(A) Representative stills from time-lapse movies of a bovine zygote with one pronucleus having uncondensed chromatin (yellow dashed line). The H2B-mScarlet chromatin signal in the pronucleus with clustered chromatin (left) was bleached upon NEBD in the region indicated by the yellow rectangle. The bleached DNA has a green signal, while the unbleached DNA is visible both in green and magenta. Lagging chromosomes are magenta and green (arrows), indicating that they originated from the uncondensed pronucleus. Green, DNA (H2B-mClover3). Magenta, DNA (H2B-mScarlet). Dashed lines indicate micronuclei. Time, h:min, 00:00 is NEBD. Z-projections, 9 sections every 1.76 μm.
(B) Number of lagging chromosomes that originated from pronuclei with clustered DNA (bleached) or uncondensed DNA (not bleached).
(C) Percentage of total lagging chromosomes that originated from pronuclei with clustered DNA (bleached) or uncondensed DNA (not bleached).
(D) Representative stills from time-lapse movies of a zygote with unclustered chromosomes. The H2B-mScarlet chromatin signal between pronuclei was bleached before NEBD in the region indicated by the yellow rectangle. Lagging chromosomes are magenta and green (arrows), indicating that they were not bleached and were peripheral chromosomes. Green, DNA (H2B-mClover3). Magenta, DNA (H2B-mScarlet). Dashed lines indicate micronuclei. Time, h:min, 00:00 is NEBD. Z-projections, 18 sections every 1.76 μm.
(E) Percentage of total lagging chromosomes that originated from clustered DNA (bleached) or distal DNA (not bleached).
(F) Zygotes displaying mitotic errors after bleaching and in reference dataset (Figure 2J, merge of clustered and unclustered groups).
Data are from four (B, C, E, F-bleached embryos) and eleven (F-reference dataset) independent experiments. The number of analyzed zygotes is specified in italics. p values were calculated using Fisher’s exact test. Scale bars, 10 μm.