Supplemental Digital Content is available in the text.
Keywords: biomarker, immune response, infection and inflammation, leukocytes activation, Raman spectroscopy, sepsis diagnosis
Abstract
Objectives:
Leukocytes are first responders to infection. Their activation state can reveal information about specific host immune response and identify dysregulation in sepsis. This study aims to use the Raman spectroscopic fingerprints of blood-derived leukocytes to differentiate inflammation, infection, and sepsis in hospitalized patients. Diagnostic sensitivity and specificity shall demonstrate the added value of the direct characterization of leukocyte’s phenotype.
Design:
Prospective nonrandomized, single-center, observational phase-II study (DRKS00006265).
Setting:
Jena University Hospital, Germany.
Patients:
Sixty-one hospitalized patients (19 with sterile inflammation, 23 with infection without organ dysfunction, 18 with sepsis according to Sepsis-3 definition).
Interventions:
None (blood withdrawal).
Measurements AND MAIN RESULTS:
Individual peripheral blood leukocytes were characterized by Raman spectroscopy. Reference diagnostics included established clinical scores, blood count, and biomarkers (C-reactive protein, procalcitonin and interleukin-6). Binary classification models using Raman data were able to distinguish patients with infection from patients without infection, as well as sepsis patients from patients without sepsis, with accuracies achieved with established biomarkers. Compared with biomarker information alone, an increase of 10% (to 93%) accuracy for the detection of infection and an increase of 18% (to 92%) for detection of sepsis were reached by adding the Raman information. Leukocytes from sepsis patients showed different Raman spectral features in comparison to the patients with infection that point to the special immune phenotype of sepsis patients.
Conclusions:
Raman spectroscopy can extract information on leukocyte’s activation state in a nondestructive, label-free manner to differentiate sterile inflammation, infection, and sepsis.
Sepsis is a life-threatening multiple organ dysfunction due to infection resulting in high mortality (1). The progression from infection to sepsis is characterized by a complex dysregulated host response (2–4). This complexity cannot be fully mapped by available biomarkers. Diagnostic accuracy of biomarkers to differentiate sepsis from the immune response to severe, noninfectious stimuli or from localized infection with a regulated host response is therefore limited (5). New diagnostic strategies are needed to yield detailed information on the immunological host response of the patient.
Leukocytes are an interesting research target as they orchestrate the response of the immune system against infection and, thus, can provide insights into the host response (6). The immune system of individuals determines tolerance or resistance to infection, as well as a special predisposition to progress to sepsis (7–9). Activation of innate and adaptive immunity during infection and sepsis leads to molecular and functional changes of leukocytes (10, 11). The importance of leukocytes for assessment of the host response has been recognized by several studies: for example, a sepsis-induced immunosuppression a few days after sepsis onset was shown in neutrophils (12), leading to a higher presence of immature neutrophils during systemic inflammation (13). The volume of immune cells was found to change during severe infection and sepsis (14, 15), and the altered response of immune cells in sepsis can be determined by changes in the phenotype of leukocytes (16).
Raman spectroscopy is a label-free, nondestructive method and can characterize the biochemical composition of leukocytes by analyzing the inelastically scattered light from the cells after laser illumination. The Raman spectrum can be regarded as a molecular fingerprint of the whole cell (17) (Fig. 1). This fingerprint allows to distinguish different leukocyte subpopulations (18–22) and can reveal their specific activation state, as was shown for in vitro stimulated monocytes (23), neutrophils (24), T-lymphocytes (25), and macrophages (26) and also for T-lymphocytes after in vivo activation in an endotoxemia mouse model (27). The translation to patient diagnostics and patient stratification has not yet been shown, and the HemoSpec study is the first to do so.
Figure 1.
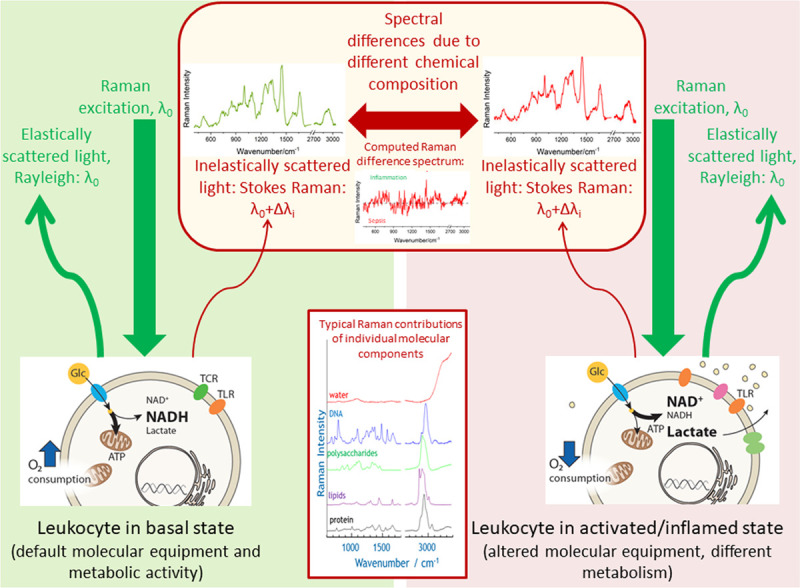
Schematics visualizing the Raman effect and highlighting the molecular origin of different Raman spectra of leukocytes in different disease states. The Raman spectrum plots the wavelength shift Δλi as Raman intensity over wavenumber νi = 1/Δλi. Origin of that wavelength shift is the inelastic scattering of the monochromatic excitation light with wavelength λ0 on the different molecular vibrations of the chemical components in the leukocytes. Upon infection or exposure to pathogens, most immune cells undergo dramatic rewiring of their cellular energy metabolism which also affects immune cells’ phenotype and function (e.g., expression of different receptors [e.g., TCR = T cell receptor, TLR = toll-Like receptor). In basal state glucose (Glc) is used for efficient energy production (ATP = adenosine triphosphate) in the mitochondria. In activated state decoupling is observed with increased lactate production and increased levels of oxidized nicotinamide adenine dinucleotide (NAD+) (NADH = reduced form of NAD, H for hydrogen). As Raman spectroscopy probes intrinsic properties of the molecules (molecular vibrational states), it is a label-free method. Laser light intensities in the visible are low to keep Raman spectroscopy a nondestructive method. Spectral differences in complex biological systems, such as immune cells, are usually difficult to see by naked eye and require the use of statistical data analysis methods. The simplest way to visualize the variations between the groups is by computing a difference spectrum between the group means. In this work, also supervised statistical models are used to extract the spectral features characteristic for group differentiation as well as to assign class membership in an automated manner.
The goal of this study was to assess the potential of Raman spectroscopy for leukocyte phenotyping and as a diagnostic tool in sepsis and acute systemic inflammation. For this, we characterized leukocytes of hospitalized patients using Raman spectroscopy in a diagnostic phase-II trial with the aim to detect sepsis and differentiate patients with infection.
MATERIALS AND METHODS
Study Design and Patients
The study “Spectroscopic differentiation of leukocytes and detection of their activation patterns for rapid differentiation of systemic inflammatory reactions in the early diagnosis of sepsis” (HemoSpec) was a prospective, single-center, diagnostic phase-II study, registered at the German clinical trials register (DRKS00006265) and approved by the local ethics committee (approval number: 4004-02/14, ethic commission of Jena University Hospital). Patients were included if they were above or equal to 18 years old and had 1) systemic inflammation with at least one systemic inflammatory response syndrome (SIRS) criterion without infection, 2) infection without organ dysfunction, and 3) sepsis as defined according to Sepsis-3 (28), and written informed consent was obtained. All patients showed least one SIRS criterion: 1) either fever greater than 38.0°C or hypothermia less than 36.0°C, 2) tachycardia greater than 90 beats/min, 3) tachypnea greater than 20 breaths/min, or 4) leukocytosis greater than 12 × 109/L or leukopenia less than 4 × 109/L.
Excluded were patients with HIV infection, immunosuppressive medications, malignancies, neutropenia less than 1,000/mm3 of noninfectious genesis, or immunodeficiency of noninfectious genesis.
All patients were identified by experienced study personal, and the classification was confirmed by a physician. In preparation of the article, all cases were reviewed by another physician certified in critical care medicine and with extensive research experience in sepsis and infection research, including chart review. All cases of infection or sepsis were classified as microbiologically or clinically proven (Table 1). Patients with SIRS needed to have a causing inflammatory stimulus (e.g., cardiac surgery) and at least one SIRS criterion.
TABLE 1.
Clinical Characterization of the HemoSpec Patients
| Characteristics | Inflammation | Infection | Sepsisa |
|---|---|---|---|
| Number of patients | 24 | 19 | 18 |
| Sex, male, n (%) | 15 (63) | 13 (68) | 15 (83) |
| Age, median (interquartile range) | 70 (63–77) | 73 (70–83) | 64 (59–75) |
| SIRS after cardiac surgery, n (%) | 22 (92) | ||
| SIRS after traumatic brain injury, n (%) | 1 (4) | ||
| SIRS after liver resection, n (%) | 1 (4) | ||
| Infection microbiologically proven, n (%) | 9 (47) | 13 (72) | |
| Respiratory tract infection, n (%) | 13 (68) | 6 (33) | |
| Urinary tract infection, n (%) | 3 (16) | 2 (11) | |
| Abdominal infection, n (%) | 4 (22) | ||
| Wounds or soft-tissue infection, n (%) | 4 (22) | ||
| Other or unknown infection focus, n (%) | 3 (16) | 2 (11) | |
| Hypo- or hyperthermia, n (%) | 5 (21) | 5 (26) | 7 (39) |
| Tachycardia, n (%) | 10 (42) | 10 (53) | 17 (94) |
| Tachypnea or ventilation, n (%) | 22 (92) | 6 (32) | 17 (94) |
| Leukocytosis, leukopenia, or left shift, n (%) | 10 (42) | 9 (47) | 15 (83) |
| Acute Physiology And Chronic Health Evaluation II, median (interquartile range) | 21 (11–23) | 16 (15–23)11,b | 21 (16–32) |
| Sequential Organ Failure Assessment, median (interquartile range) | 7 (4–8) | 6 (3–9)11,b | 8 (7–12) |
| C-reactive protein, mg/L, median (interquartile range) | 3.9 (2–10) | 156 (92–187) | 216 (85–327) |
| Interleukin-6, pg/mL, median (interquartile range) | 228 (143–450) | 43 (16–92) | 617 (148–3,000) |
| Procalcitonin in ng/mL, median (interquartile range) | 0.1 (0.1–0.2) | 0.4 (0.16–1.8) | 3.4 (1.2–13.5) |
| Soluble urokinase-type plasminogen activator receptor, ng/mL, median (interquartile range) | 2.5 (1.8–2.9)7 | 4.4 (3.0–8.8) | 10.3 (6.1–14.7)2 |
| Leukocytes Giga particles per litre, median (interquartile range) | 10 (8.9–14.5) | 10.5 (6.9–12.0)1 | 18.5 (9.6–23.0) |
| Relative neutrophil count, %, median (interquartile range) | 82 (76–83)5 | 80 (73–87)2 | 86 (83–88)2 |
| Relative lymphocyte count, %, median (interquartile range) | 13 (10–17)5 | 9 (6–14)2 | 5 (2–9)2 |
| Relative monocyte count, %, median (interquartile range) | 6 (5–8)5 | 8 (7–9)2 | 6 (5–7)2 |
SIRS = systemic inflammatory response syndrome.
aSepsis: sepsis or septic shock according to Sepsis-3 definition.
bScores are missing for 11 ward patients never treated in the ICU in the infection group.
n indicates the number of missing values in each group.
Clinical information (blood count, biomarkers, demographic information) were entered into a web-based clinical trial database system (OpenClinica; LLC, Waltham, MA).
Blood Sampling
Blood samples were obtained following standard protocols within 24 hours from patients after being diagnosed with sepsis and from patients with inflammation within 24 hours after admission to the hospital or after causative surgery. An aliquot of 500 µL blood from EDTA blood was used for Raman spectroscopy. Further blood samples were used for measuring automated blood cell counts and biomarker analysis (procalcitonin, C-reactive protein [CRP], interleukin [IL]–6). More details are presented in Supplemental Digital Content (http://links.lww.com/CCX/A583).
Leukocytes Isolation
Blood collection protocols and leukocyte isolation procedure have been published previously (18). Briefly, it involves RBC lysis using NH4Cl solution, centrifugation-based collection of leukocytes, chemical fixation with formaldehyde, and resuspension in phosphate buffered saline. For the Raman measurements, leukocytes were spin-coated onto CaF2 slides using a cell suspension of 1 × 106 leukocytes/100 µL 0.9% NaCl solution. For more details, see Supplemental Digital Content (http://links.lww.com/CCX/A583).
Raman Spectroscopic Investigation of Peripheral Blood Leukocytes
Raman spectroscopic analysis was performed on single-cell level as described previously (18) with a commercial upright micro-Raman setup (CRM 300; WITec GmbH, Ulm, Germany) using a 785 nm diode laser (75 mW in the object plane; Toptica GmbH, Munich, Germany) for excitation. Raman images were recorded through a 100× objective (NA 0.9; Zeiss, Göttingen, Germany) in the scanning mode with a step size of 0.3 µm and an integration time of 1 s/spectrum, yielding ~ 500 Raman spectra/cell. On average, 20–30 cells were measured per patient, yielding in total 1,816 cells from 61 patients. Raman spectroscopic analysis was completed within 8 hours after blood withdrawal.
Data Analysis
GNU R (29) was used for data analysis with in-house written code. The Raman data preprocessing involved cosmic ray removal (30), wavenumber calibration using 4-acetamidophenol, sorting-out the spectra with low signal-to-noise ratio, and extended multiplicative signal correction model-based preprocessing (31). Further details are given in the Supplemental Digital Content (http://links.lww.com/CCX/A583).
In order to separate the patient classes using a small number of components with well-interpretable loadings, we used canonical-powered partial least squares (CPPLS) discriminant analysis (32, 33) to build supervised models. One model was built for each pair of target classes, that is, to detect infection (including sepsis) versus sterile inflammation and to discriminate sepsis versus no-sepsis (inflammation and infection without organ damage). The target responses were converted into a dummy response matrix (two columns with 0 and 1, specifying the class membership). Data were balanced by providing weights to account for different numbers of cells/patients per class. Model bias (e.g., due to laboratory relocation, age, and sex of patients) was suppressed by including additional responses, which are used during model training for generating so-called temporary loadings (32). Those addition responses were not considered for prediction (Figs. S1−S3, http://links.lww.com/CCX/A583). Leave-one-patient-out cross-validation (LPOCV) was used to generate predicted Raman scores on single-cell level. The threshold for assigning class membership (i.e., infection vs no-infection and sepsis vs no-sepsis) was defined within the cross-validation loop, so the data from the predicted patient could not influence it. To obtain predictions per patient from multiple leukocytes per sample, the median of the predicted Raman scores was calculated and the respective threshold used. The median value was chosen because it is less sensitive to outliers than the mean value and better represents the central tendency of skewed distributions.
To demonstrate added diagnostic value of Raman spectroscopy, the predicted Raman scores (median value per patient) were combined with biomarker values (IL–6, CRP, procalcitonin) in subsequent combined CPPLS models to yield improved predictions of patient status (Fig. S2, http://links.lww.com/CCX/A583). Biomarker values were scaled to their sds, and scaling coefficients were stored to be available for new test datasets. In the CPPLS algorithm, balanced datasets and additional responses to suppress demographic bias were used as described above.
The combined models were validated by LPOCV. A threshold for the predicted value to assign the class label was estimated inside the cross-validation loop. All prediction metrics and demonstrated receiver operating characteristic (ROC) curves are generated from the cross-validated predictions. Balanced accuracy was calculated by averaging the sensitivity and specificity values of the two-class models. The values for balanced accuracy of sepsis and infection detection are presented along with the upper and lower margins of the 95% credible intervals (CIs). Details on calculation of CI are given in the Supplemental Digital Content (http://links.lww.com/CCX/A583).
RESULTS
Patient Characteristics
The clinical trial included 24 patients with sterile inflammation (mostly after cardiac surgery), 19 with infection (mostly respiratory infections), and 18 with sepsis (different foci of infection) as shown in Figure 2A. Clinical characteristics of the patients are shown in Table 1 and the detected pathogens in Table S1 (http://links.lww.com/CCX/A583). Patient groups are similar regarding age and gender distribution and with no ethnic diversity. Sepsis patients show higher biomarkers of inflammation such as procalcitonin, IL-6, and soluble urokinase-type plasminogen activator receptor compared with the other groups.
Figure 2.
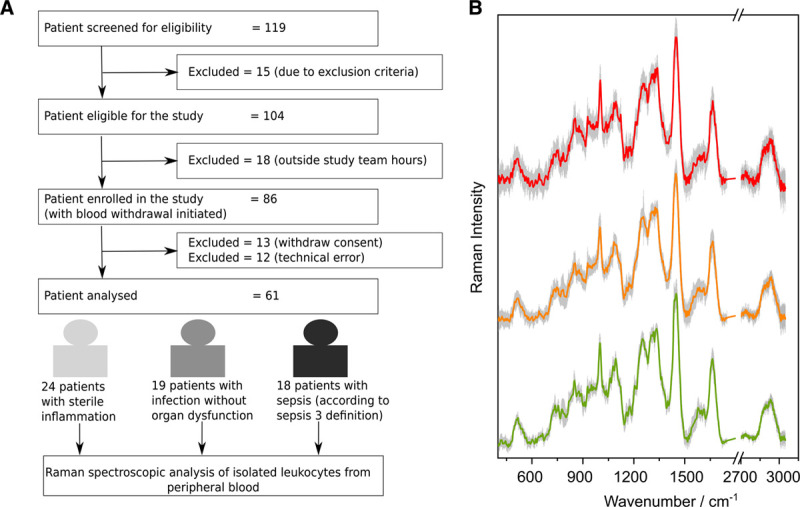
Patient flow chart and Raman spectra per patient group. A, Flow chart for patient recruitment during HemoSpec study. Technical error: not possible to withdraw the required volume of blood or nonavailability of the device. B, Mean preprocessed Raman spectra are shown together with sd (gray shadows) for sepsis (red, top), infection without organ failure (orange, center), and sterile inflammation (green, bottom). The spectra are shifted on the y-axis to avoid overlap.
Raman Spectra of Peripheral Leukocytes
Mean preprocessed Raman spectra of leukocytes per patient group are depicted in Figure 2B. Number of cells contributing to the mean spectrum is given in Table S2 (http://links.lww.com/CCX/A583). The Raman spectral features of leukocytes isolated from the patients having inflammation, infection, and sepsis show similar spectral profile with typical spectral features of leukocytes. Subtle variations in the spectra reflect an overall different chemical composition. Logistic regression analysis was used for a first data overview and to visualize the spectral separation of the groups (Fig. S4, http://links.lww.com/CCX/A583). Here, a single score value was used to map the disease condition: 0: sterile inflammation, 0.5: infection without organ dysfunction, and 1: sepsis. The cross-validated predicted values in Figure S4 (http://links.lww.com/CCX/A583) enable a separation of patients with inflammation, infection, and sepsis. The spectral features responsible for this separation were extracted from the regression coefficients. The extracted features can be assigned mainly to Raman bands of nucleic acids (662, 778, 1,074, 1,580 cm–1) and proteins (934, 1,676, 2,926 cm–1) (18, 25–27). Negative Raman bands in the regression coefficient for nucleic acids indicate that leukocytes from patients with inflammation of noninfectious origin have slightly higher overall nucleic acid content than leukocytes from sepsis patients. The protein composition differs in all three patient cohorts as seen from positive and negative Raman bands for proteins.
Raman Spectroscopy as a Diagnostic Marker Using CPPLS Analysis
In order to use the leukocyte’s spectral characteristics in the different disease groups for diagnostic purposes, CPPLS classification models were trained. The resulting ROC curves after LPOCV are depicted in Figure 3A for differentiating patients with infection (including sepsis patients) from patients with sterile inflammation and in Figure 3B for differentiating patients with sepsis from all other patients. With area under the curve (AUC) of 0.84 (balanced accuracy 76%; 95% CI, 65–85%) and 0.82 (balanced accuracy 72%; 95% CI, 49–82%) for infection and sepsis, respectively, these accuracies are comparable to those obtained with (combined) established biomarkers, such as IL-6, CRP, and procalcitonin (34) as can be seen from the ROC curves in Figure 3.
Figure 3.
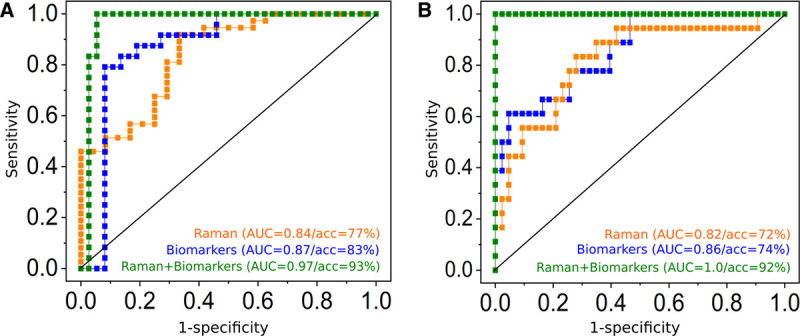
Added value of Raman leukocyte analysis to detect infections (A) and to identify sepsis (B). Receiver operating characteristic (ROC) curves obtained from canonical-powered partial least squares (CPPLS) analysis. All models were validated using leave-one-patient-out cross-validation. Predictions using Raman spectroscopic data were obtained on a single-cell level and were aggregated to obtain a single value (median of all leukocytes) per patient (orange curves). A, ROC curves for infection detection (against sterile inflammation) using predictions based on Raman spectroscopic data (orange curve), biomarker scores (C-reactive protein, procalcitonin, and interleukin-6, blue curve), and on a combined model using Raman scores and biomarkers scores (green curve). B, ROC curves for sepsis detection (against sterile inflammation and infection without organ failure) using predictions from Raman spectroscopic data (orange curve), from biomarkers (blue curve) and using a combined CPPLS models using Raman scores and biomarker values (green curve). Scatterplots of the data used for sepsis detection based on Raman spectroscopic scores and biomarkers are shown in Figure S6 (http://links.lww.com/CCX/A583); the model loadings and the fitted model in Figure S7 (http://links.lww.com/CCX/A583). The combined models (green curves in A and B) show superior diagnostic power. Please note that balanced accuracies (acc) can be different from 1, also if area under the curve (AUC) is 1, as there is only a small margin between the patient groups. Thus, when a patient is excluded from the training data within the cross-validation loop, the model and the estimation of the optimal threshold change slightly. This might lead to a misprediction of some patients when the threshold is set for the prediction within the cross-validation loop, even if the cross-validated predicted values perfectly separate the groups.
The CPPLS loadings provide insights which Raman spectral features are responsible for the differentiation. The first two CPPLS loadings for the model to predict infection are displayed in Figure 4A, the ones to detect sepsis in Figure 4B (CPPLS scatter plots in Fig. S5, http://links.lww.com/CCX/A583). More prominent nucleic acid bands (1,582 cm–1, 778 cm–1) are visible in the leukocytes of patients with sterile inflammation compared with the leukocytes of patients with infection/sepsis. Raman bands around 1,664 cm–1 and 1,434 cm–1 indicate altered protein and lipid composition.
Figure 4.
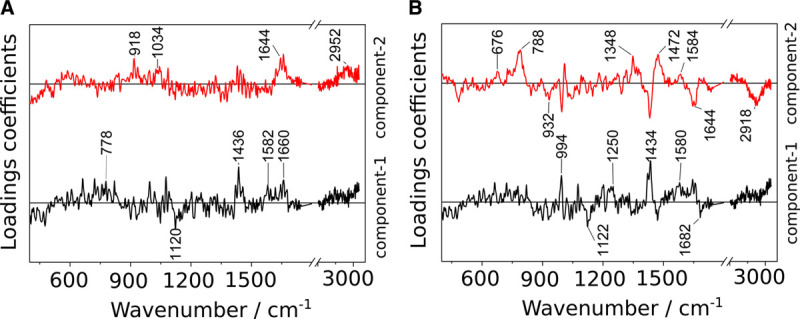
Canonical-powered partial least squares coefficients for the first two latent variables in the Raman models for the detection of (A) infection and (B) sepsis. Respective scatter plots are depicted in Figure S5 (http://links.lww.com/CCX/A583).
In order to improve diagnostic accuracy, the predicted values obtained from cross-validation of Raman-based model discussed above were combined with the molecular biomarkers CRP, PCT, and IL-6 in a combined CPPLS model (Fig. 3) (Table S3, http://links.lww.com/CCX/A583). The balanced accuracy and AUC could significantly be improved and reached up to 93% (95% CI, 84–97%) and AUC equals to 0.97, respectively, for infection (Fig. 3A). A patient with sepsis could be identified with a balanced accuracy of 92% (95% CI, 80–97%; AUC = 1) when clinical biomarker and Raman data were evaluated together (Fig. 3B).
Also, among the patients with infections n = 37), Raman spectroscopy could contribute significant added value to detect sepsis patients (Table S3, http://links.lww.com/CCX/A583; Fig. S8, http://links.lww.com/CCX/A583). Together with the biomarker scores, the Raman data could achieve a perfect identification in our study cohort (AUC = 1.0; balanced accuracy 100%; 95% CI, 91–100%).
DISCUSSION
HemoSpec trial was designed as a pilot diagnostic trial to assess the translational clinical use of Raman spectroscopic leukocyte phenotyping for differentiating systemic inflammatory reactions under sterile (e.g., surgical intervention) and infectious (infection vs sepsis) conditions. We could show that the characteristic spectroscopic fingerprint of patients’ leukocytes allows discrimination between sepsis, infection, and sterile inflammation. The AUCs from the ROC curves for identifying patients with sepsis (AUC = 0.82) and infection (AUC = 0.84) based on the Raman spectroscopic fingerprint of leukocytes are comparable with AUCs obtained from molecular biomarkers reported in the literature (5, 35, 36).
In order to better reflect the complex host response during sepsis, the use of a combinative index of different biomarkers has been shown to provide an improved sepsis patient stratification (5, 37). In line with these studies, a model combining the score values from Raman spectra of leukocytes with scores of inflammatory mediators (procalcitonin, CRP, and IL-6) is best in detecting sepsis with an AUC equals to 1.0 (balanced accuracy of 92%) in our dataset. This indicates that the Raman spectroscopic fingerprint of leukocytes holds high potential to be used as a marker for patient stratification by itself and to provide added diagnostic value when combined with conventional biomarkers.
Raman spectroscopic analysis captures the overall biochemical composition. Although it is rather difficult (or impossible without specific molecular tags) to identify individual molecules that are affected and changed, a meaningful and specific fingerprint-like spectral profile is obtained from each leukocyte. Differences between the spectral fingerprints of different patient groups can be analyzed to extract overall molecular changes. For an interpretation of the Raman profiles and to use them for prediction of patient characteristics, CPPLS classification models were trained. The first loading coefficients of both CPPLS models, that is, the model for detection infection and the model for sepsis, are similar (Fig. 4) and show typical features also observed in in vitro infection models where isolated human leukocyte subpopulations are challenged with defined fungal and bacterial pathogens (38). In particular, features relevant for the differentiation of infected versus noninfected neutrophils and a few Raman bands from the lymphocyte infection model (38) are present in loading coefficient 1 of the CPPLS models of the HemoSpec trial. This supports the presumption that spectral features responsible for distinguishing leukocytes from patients with infectious versus sterile inflammation are indeed due to infection-induced stimulation. The particular presence of neutrophil and lymphocyte signal can be explained with the fact that due to their relative abundance in full blood; mainly neutrophils (48–74%) and lymphocytes (11–13%) were characterized in the HemoSpec study, whereas only between 5.5% and 7.5% of the measured cells were monocytes (Table S3, http://links.lww.com/CCX/A583).
The second loading coefficient in the statistical model for sepsis detection (Fig. 4) shows some similarities to the coefficient of the logistic regression model used for the data overview (Fig. S4, http://links.lww.com/CCX/A583), where the progression of infection to sepsis is included in a single score value. The distinct spectral features of loading coefficient 2 cannot be found in the in vitro infection model indicating that simple pathogen-induced changes are not enough to simulate the complex spectral signatures of leukocyte’s “sepsis phenotype.” This is in agreement with current understanding of sepsis immunology where sepsis-associated alterations in the functions and phenotype of all leukocytes subtypes are described (4, 39). Gene expression profiling studies indicate characteristic transcriptional changes in leukocytes during sepsis (40–42). Those infection- and sepsis-induced changes in the overall biochemical composition of the immune cells are reflected in the Raman signal.
We see high potential for the label-free Raman technology. From only 500 µL of blood, the overall chemical composition of all leukocyte subpopulations can be measured. Sample preparation is simple and involves standard clinical laboratory routines. Furthermore, as Raman spectroscopy is based on fast (instantaneous) light-matter interactions, the method has the potential to provide the result within the recommended “Golden Hour” for the diagnosis of sepsis. Recently, Raman technology has advanced to high-throughput devices which enable the analysis of greater than 1,000 cells within only 1 hour (19, 43) paving the way for larger, multicenter clinical trials to validate spectroscopic immunophenotyping. This will enable to overcome the limitation of the current study and to include more patients, so that the impact of timing of sampling, type of pathogen as well as age, sex, ethnicity, or race of the patient can be fully assessed.
CONCLUSIONS
Label-free, nondestructive Raman spectroscopy was shown in the HemoSpec study to be a powerful tool to differentiate the activation profile of circulating leukocytes from peripheral blood. The overall molecular information captured in the Raman spectra reflects the different chemical profiles of the leukocytes from patients with inflammation, infection, and sepsis. Those results merit further investigation as they could potentially be used for patient stratification. They add value to conventional biomarkers for the discrimination of sterile inflammation, uncomplicated infection, and sepsis. Future studies analyzing a larger number of subjects and cells per subject shall help to confirm our findings and also enable analysis of leukocyte subpopulation-resolved host response.
ACKNOWLEDGMENT
Drs. E. Shahangi, S. Weis, and T. Bruns are highly acknowledged for their support and their participation in patient recruitment. We thank Prof. C. Kroegel and team (Internal Medicine I, Pneumology) for support for blood withdrawal from healthy volunteers, and the Center for Clinical Studies (Prof. F. M. Brunkhorst and team), the study nurses as well as C. Richert and J. Köhler for their support.
Supplementary Material
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
Drs. Ramoji, Thomas-Rüddel, and Ryabchykov contributed equally. Drs. Bauer, Giamarellos-Bourboulis, Popp, and Neugebauer designed the study. Drs. Thomas-Rüddel, Bauer, and Bloos supervised clinical trial, recruited patients, and collected and approved clinical data. Drs. Ramoji, Thomas-Rüddel, Arend, Eugen-Olsen, and Kiehntopf performed measurements and recorded data. Drs. Ramoji, Thomas-Rüddel, Ryabchykov, and Bocklitz analyzed data. Drs. Rybachkov and Bocklitz developed Raman analysis algorithms. All authors discussed and interpreted results. All authors wrote, corrected, and approved the article.
Supported, in part, by the German Federal Ministry of Education and Research via the Integrated Research and Treatment Center “Center for Sepsis Control and Care” (FKZ 01EO1502) and by the European Union via the project “HemoSpec” (Seventh Framework Programme, CN 611682). The Jena Biophotonic, Imaging Laboratory (PO 563/29-1, BA 1601/10-1), Research Campus InfectoGnostics (FKZ 13GW0096F), the Leibniz Science Campus InfectoOptics (W8/2018) are gratefully acknowledged for providing the infrastructure.
The authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.Rudd KE, Johnson SC, Agesa KM, et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: Analysis for the global burden of disease study. Lancet. 2020; 395:200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venet F, Chung C-S, Huang X, et al. Lymphocytes in the development of lung inflammation: A role for regulatory CD4(+) T cells in indirect pulmonary lung injury. J Immunol. 2009; 183:3472–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer M, Coldewey SM, Leitner M, et al. Deterioration of organ function as a hallmark in sepsis: The cellular perspective. Front Immunol. 2018; 9:1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubio I, Osuchowski MF, Shankar-Hari M, et al. Current gaps in sepsis immunology: New opportunities for translational research. Lancet Infect Dis. 2019; 19:e422–e436 [DOI] [PubMed] [Google Scholar]
- 5.Pierrakos C, Velissaris D, Bisdorff M, et al. Biomarkers of sepsis: Time for a reappraisal. Crit Care. 2020; 24:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodin P, Davis MM. Human immune system variation. Nat Rev Immunol. 2017; 17:21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weis S, Carlos AR, Moita MR, et al. Metabolic adaptation establishes disease tolerance to sepsis. Cell. 2017; 169:1263–1275.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chousterman BG, Swirski FK, Weber GF. Cytokine storm and sepsis disease pathogenesis. Semin Immunopathol. 2017; 39:517–528 [DOI] [PubMed] [Google Scholar]
- 9.van der Poll T, van de Veerdonk FL, Scicluna BP, et al. The immunopathology of sepsis and potential therapeutic targets. Nat Rev Immunol. 2017; 17:407–420 [DOI] [PubMed] [Google Scholar]
- 10.Kumar V. Immunometabolism: Another road to sepsis and its therapeutic targeting. Inflammation. 2019; 42:765–788 [DOI] [PubMed] [Google Scholar]
- 11.Bermejo-Martin JF, Andaluz-Ojeda D, Almansa R, et al. Defining immunological dysfunction in sepsis: A requisite tool for precision medicine. J Infect. 2016; 72:525–536 [DOI] [PubMed] [Google Scholar]
- 12.Demaret J, Venet F, Friggeri A, et al. Marked alterations of neutrophil functions during sepsis-induced immunosuppression. J Leukoc Biol. 2015; 98:1081–1090 [DOI] [PubMed] [Google Scholar]
- 13.Mare TA, Treacher DF, Shankar-Hari M, et al. The diagnostic and prognostic significance of monitoring blood levels of immature neutrophils in patients with systemic inflammation. Crit Care. 2015; 19:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crouser ED, Parrillo JE, Seymour CW, et al. Monocyte distribution width: A novel indicator of sepsis-2 and sepsis-3 in high-risk emergency department patients. Crit Care Med. 2019; 47:1018–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crouser ED, Parrillo JE, Seymour C, et al. Improved early detection of sepsis in the ED with a novel monocyte distribution width biomarker. Chest. 2017; 152:518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rimmelé T, Payen D, Cantaluppi V, et al. ; ADQI XIV Workgroup. Immune cell phenotype and function in sepsis. Shock. 2016; 45:282–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler HJ, Ashton L, Bird B, et al. Using Raman spectroscopy to characterize biological materials. Nat Protoc. 2016; 11:664–687 [DOI] [PubMed] [Google Scholar]
- 18.Ramoji A, Neugebauer U, Bocklitz T, et al. Toward a spectroscopic hemogram: Raman spectroscopic differentiation of the two most abundant leukocytes from peripheral blood. Anal Chem. 2012; 84:5335–5342 [DOI] [PubMed] [Google Scholar]
- 19.Schie IW, Rüger J, Mondol AS, et al. High-throughput screening raman spectroscopy platform for label-free cellomics. Anal Chem. 2018; 90:2023–2030 [DOI] [PubMed] [Google Scholar]
- 20.Managò S, Mirabelli P, Napolitano M, et al. Raman detection and identification of normal and leukemic hematopoietic cells. J Biophotonics. 2018; 11:e201700265. [DOI] [PubMed] [Google Scholar]
- 21.Hobro AJ, Kumagai Y, Akira S, et al. Raman spectroscopy as a tool for label-free lymphocyte cell line discrimination. Analyst. 2016; 141:3756–3764 [DOI] [PubMed] [Google Scholar]
- 22.Chen MZ, Hudson CA, Vincent EE, et al. Bariatric surgery in morbidly obese insulin resistant humans normalises insulin signalling but not insulin-stimulated glucose disposal. PLoS One. 2015; 10:e0120084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Topfer N, Muller MM, Dahms M, et al. Raman spectroscopy reveals LPS-induced changes of biomolecular composition in monocytic THP-1 cells in a label-free manner [published online ahead of print May 13. 2019]. Integr Biol (Camb). 2019; 11:87–98 [DOI] [PubMed] [Google Scholar]
- 24.Arend N, Pittner A, Ramoji A, et al. Detection and differentiation of bacterial and fungal infection of neutrophils from peripheral blood using raman spectroscopy. Anal Chem. 2020; 92:10560–10568 [DOI] [PubMed] [Google Scholar]
- 25.Ichimura T, Chiu LD, Fujita K, et al. Non-label immune cell state prediction using Raman spectroscopy. Sci Rep. 2016; 6:37562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavillon N, Hobro AJ, Akira S, et al. Noninvasive detection of macrophage activation with single-cell resolution through machine learning. Proc Natl Acad Sci U S A. 2018; 115:E2676–E2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramoji A, Ryabchykov O, Galler K, et al. Raman spectroscopy follows time-dependent changes in T lymphocytes isolated from spleen of endotoxemic mice. Immunohorizons. 2019; 3:45–60 [DOI] [PubMed] [Google Scholar]
- 28.Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA. 2016; 315:801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Team: R: A language and environment for statistical computing. Vienna, Austria, R Foundation for Statistical Computing. 2018. Available at: https://www.R-project.org/ [Google Scholar]
- 30.Ryabchykov O, Bocklitz T, Ramoji A, et al. Automatization of spike correction in Raman spectra of biological samples. Chemometr Intell Lab Syst. 2016; 155:1–6 [Google Scholar]
- 31.Martens H, Stark E. Extended multiplicative signal correction and spectral interference subtraction: New preprocessing methods for near infrared spectroscopy. J Pharm Biomed Anal. 1991; 9:625–635 [DOI] [PubMed] [Google Scholar]
- 32.Indahl UG, Liland KH, Næs T. Canonical partial least squares—a unified PLS approach to classification and regression problems. J Chemom. 2009; 23:495–504 [Google Scholar]
- 33.Mevik B-H, Wehrens R, Liland K: R Package: ‘pls’: Partial Least Squares and Principal Component regression, J Stat Softw. 2007; 18 [Google Scholar]
- 34.Ríos-Toro JJ, Márquez-Coello M, García-Álvarez JM, et al. Soluble membrane receptors, interleukin 6, procalcitonin and C reactive protein as prognostic markers in patients with severe sepsis and septic shock. PLoS One. 2017; 12:e0175254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen J, Vincent JL, Adhikari NK, et al. Sepsis: a roadmap for future research. Lancet Infect Dis. 2015; 15:581–614 [DOI] [PubMed] [Google Scholar]
- 36.Sinha M, Jupe J, Mack H, et al. Emerging technologies for molecular diagnosis of sepsis. Clin Microbiol Rev. 2018; 31:e00089-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolin HH, Papadimos TJ, Stepkowski S, et al. A novel combination of biomarkers to herald the onset of sepsis prior to the manifestation of symptoms. Shock. 2018; 49:364–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pistiki A, Ramoji A, Ryabchykov O, et al. Biochemical analysis of in-vitro and in-vivo activated leukocytes by bacterial and fungal pathogens using Raman spectroscopy. under submission, 2021 [DOI] [PMC free article] [PubMed]
- 39.Monneret G, Gossez M, Aghaeepour N, et al. How clinical flow cytometry rebooted sepsis immunology. Cytometry A. 2019; 95:431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang BM, McLean AS, Dawes IW, et al. Gene-expression profiling of peripheral blood mononuclear cells in sepsis. Crit Care Med. 2009; 37:882–888 [DOI] [PubMed] [Google Scholar]
- 41.Tang BM, McLean AS, Dawes IW, et al. Gene-expression profiling of gram-positive and gram-negative sepsis in critically ill patients. Crit Care Med. 2008; 36:1125–1128 [DOI] [PubMed] [Google Scholar]
- 42.Tang BM, McLean AS, Dawes IW, et al. The use of gene-expression profiling to identify candidate genes in human sepsis. Am J Respir Crit Care Med. 2007; 176:676–684 [DOI] [PubMed] [Google Scholar]
- 43.Mondol AS, Töpfer N, Rüger J, et al. New perspectives for viability studies with high-content analysis Raman spectroscopy (HCA-RS). Sci Rep. 2019; 9:12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


