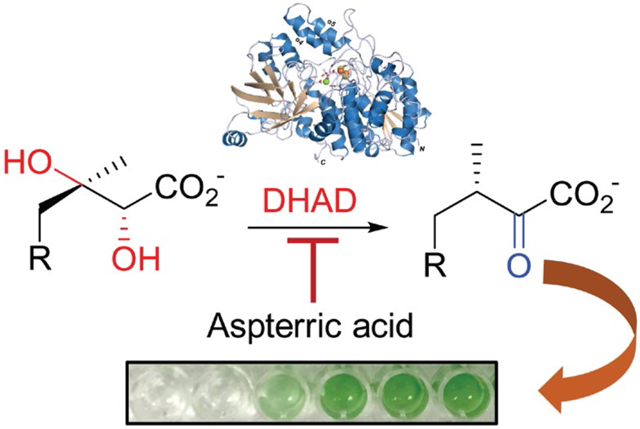
Abstract
Microbes are essential to the global ecosystem but undesirable microbial growth causes issues ranging from food spoilage and infectious diseases to harmful cyanobacterial blooms. The use of chemicals to control microbial growth has achieved significant successes, while specific roles for a majority of essential genes in growth control remain unexplored. Here, we show the growth inhibition of cyanobacterial species by targeting an essential enzyme for the biosynthesis of branched-chain amino acids. Specifically, we report the biochemical, genetic, and structural characterization of dihydroxyacid dehydratase from the model cyanobacterium Synechocystis sp. PCC 6803 (SnDHAD). Our studies suggest that SnDHAD is an oxygen-stable enzyme containing a [2Fe-2S] cluster. Furthermore, we demonstrate that SnDHAD is selectively inhibited in vitro and in vivo by the natural product aspterric acid, which also inhibits the growth of representative bloom-forming Microcystis and Anabaena strains but has minimal effects on microbial pathogens with [4Fe-4S] containing DHADs. This study suggests DHADs as a promising target for the precise growth control of microbes and highlights the exploration of other untargeted essential genes for microbial management.
Graphical Abstract
INTRODUCTION
Microbes occupy almost all known environmental niches and are essential players in the global ecosystem. For example, they are key nutrient suppliers to living organisms by fixating elements (e.g., carbon, oxygen and nitrogen) and recycling nutrients from the environment.1, 2 Throughout evolution, microbes have established themselves in a variety of environments and deviations in these associations can be linked to the health and well-being of humans, animals, plants, and the overall ecosystem.3, 4 Indeed, in the United States, microbial pathogen infections cause millions of office visits every year, while exuberant growth of cyanobacteria and algae have increasingly resulted in harmful blooms that adversely affect the quality of water bodies and cause enormous economic losses every year.5 Discovery and development of capable approaches to control and manipulate microbial growth under various situations are thus critically needed.
Microbes require many essential gene products to execute life processes, indicated by the minimal bacterial genome.6 By targeting a subset of these proteins, primarily those involved in the biosynthesis of proteins, RNA, DNA, cell wall components, and folic acid, a number of antimicrobials have been developed to treat human infectious diseases with tremendous success.7 In this regard, targeting essential but unexplored microbial gene products can lead to the discovery and development of new promising strategies for managing undesirable microbial growth.8
The branched-chain amino acids (BCAAs, L-Val, L-Leu and L-lle) are among the most abundant essential amino acids. In addition building blocks of proteins and peptides, BCAAs are precursors to branched fatty acids and other essential amino acids.9 However, humans and animals are unable to synthesize BCAAs de novo and have to acquire them from the diet. In contrast, the majority of microbes and plants biosynthesize BCAAs, as detailed in Fig. 1.10, 11 In the biosynthesis of L-Ile, the threonine dehydratase IlvA first produces 2-ketobutyrate from L-Thr. The next four steps are sequentially catalyzed by acetohydroxyacid synthase (AHAS) IlvBN, which requires pyruvate as a co-substrate, ketoacid reductoisomerase IlvC, dihydroxyacid dehydratase (DHAD) IlvD, and branched-chain aminotransferase IlvE. The same enzyme set is shared for the biosynthesis of L-Val, where IlvBN forms 2(S)-acetolactate from two molecules of pyruvate (Fig. 1). The biosynthesis of L-Leu requires three additional enzymes (LeuA, LeuC/D, and LeuB) that together divert 2-ketoisovalerate, the immediate precursor of L-Val, to 2-ketoisocaproate, which is then converted to L-Leu by IlvE.10 Given the indispensability of BCAAs, their biosynthetic enzymes have been centered in a number of studies.10 Of note, AHAS is the proven target of one main category of herbicides.12 Importantly, the constrained availability of BCAAs has led to the reduced survival and virulence of multiple microbial pathogens,13, 14 indicating that the biosynthetic pathway to BCAAs offers opportunities for controlling microbial growth.
Figure 1.
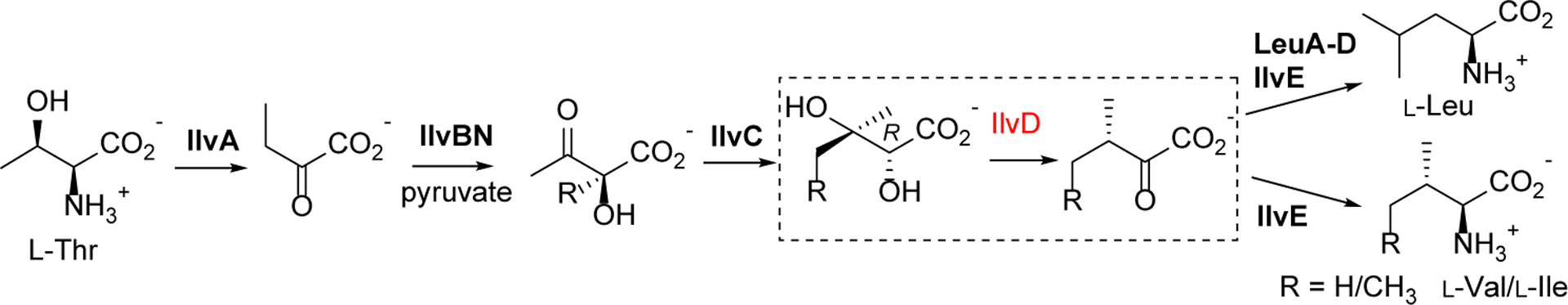
The biosynthetic pathway to branched-chain amino acids. The reaction of IlvD (DHAD shown in red) is depicted in the dashed box. LeuA, isopropylmalate synthase; LeuB, isopropylmalate dehydrogenase; LeuC/D, isopropylmalate isomerase.
DHADs of diverse origins form a large enzyme family of close to 20,000 members with 90% or less of amino acid identities in the UniProt database,15 indicating notable sequence variations. These enzymes produce 2-ketoisovalerate (KIV) and (3R)-2-ketomethylvalerate from (2R)-2,3-dihydroxyisovalerate (DHIV) and (2R,3R)-2,3-dihydroxymethylvalerate, respectively (Fig. 1). KIV is the precursor of both L-Leu and pantothenate, an intermediate to the essential cofactor coenzyme A.10 To date, nine DHADs has been biochemically or genetically characterized,13, 16–25 e.g., those from Escherichia coli (EcDHAD), Mycobacterium tuberculosis (MbDHAD), Sulfolobus solfataricus (SsDHAD), and Arabidopsis thaliana (AtDHAD). Together, these studies establish DHAD as a member of the IlvD/EDD protein family with a [2Fe-2S] or [4Fe-4S] cluster, which generally leads to the enzyme being oxygen sensitive.17 In addition, the structural basis of DHAD catalysis was revealed only recently in the studies of AtDHAD25 and MbDHAD.18 Notably, AtDHAD and MbDHAD, which both contain a [2Fe-2S] cluster, are inhibited to varying degrees by the natural product aspterric acid (AA).
Cyanobacteria are among the most ancient organisms on Earth and have an enormous impact on the evolution of other species and the formation of an aerobic ecology. In recent years, cyanobacteria have received significant attention for the engineered production of a wide variety of chemicals using CO2 as a carbon source.26 Conversely, eutrophication and climate changes have benefited the dominant growth of cyanobacteria (e.g., Microcystis sp. and Anabaena sp.) in aquatic ecosystems, resulting in the increasing frequency and intensity of harmful cyanobacterial blooms (HCBs) worldwide.27–29 Multiple strategies have been developed to mitigate HCBs, mainly based on physical, chemical and biological manipulations along with nutrient input reductions.30 However, new strategies are continuously needed to ensure success in dynamic changing environments and promote long-term sustainability.
Here, we describe the biochemical, genetic, and structural characterization of an oxygen-stable DHAD from the cyanobacterium Synechocystis sp. PCC 6803 (SnDHAD), a species that has long served as a model system for cyanobacterial and biotechnological applications.31 Our studies detail its biochemical properties and suggest new structural features in comparison with AtDHAD,25 the oxygen-sensitive MbDHAD,18 and two sugar dehydratases of the IlvD/EDD protein family, l-arabinonate dehydratase (ArDHT) and d-xylonate dehydratase (XyDHT).32, 33 We further show that SnDHAD is potently inhibited by aspterric acid (AA) in vitro. In addition, this compound inhibits the growth of bloom-forming cyanobacterial species but shows no activity toward [4Fe-4S]-containing DHADs and associated microbes. This work provides the first comprehensive study of enzymes for the biosynthesis of essential BCAAs in cyanobacteria34, 35 and suggests the BCAA biosynthesis as a promising target for selectively controlling microbial growth.
RESULTS AND DISCUSSION
SnDHAD is an oxygen-stable enzyme carrying a [2Fe-2S] cluster.
We obtained recombinant SnDHAD and EcDHAD from E. coli after affinity purification. Denaturing SDS-PAGE analysis showed the expected molecular weights of highly pure SnDHAD and EcDHAD at 62 kD and 67 kD, respectively (Fig. 2A). Native gel analysis revealed EcDHAD as a homodimer (Fig. 2B), agreeing with a previous report16 and the same oligomeric state as SsDHAD20 along with the spinach enzyme.23 Interestingly, SnDHAD appeared monomeric on gel analysis (Fig. 2B).
Figure 2.
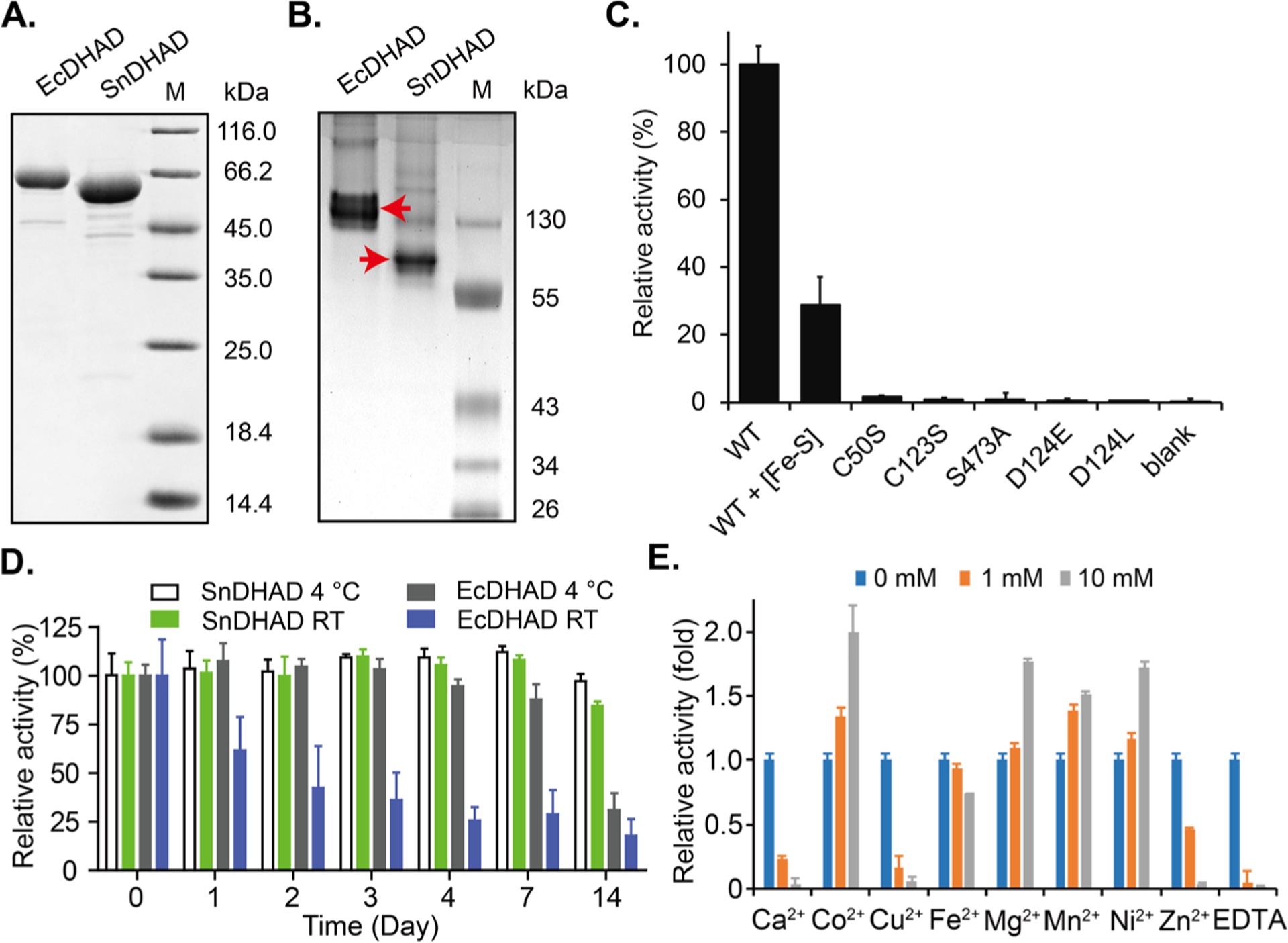
Biochemical characterization of SnDHAD. A: Recombinant EcDHAD and SnDHAD showed expected molecular weights in SDS-PAGE analysis and (B:) different oligomeric states in native gel analysis. C: Relative catalytic activities of SnDHAD and its mutants. The amount of KIV in the wild type (WT) enzyme reaction was set as 100% for normalizing the relative activities of others. The blank had no enzyme. D: Relative catalytic activities of SnDHAD and EcDHAD after exposed to air at room temperature (RT) and 4 °C. Their residual activities at indicated days were determined. E: Metal dependence of the SnDHAD reaction. The enzyme activity in the reaction without any divalent metal was set as 1 for normalizing its relative activity when supplemented with serial metal ions and EDTA.
As-purified SnDHAD is brown in color, suggesting the presence of a bound [Fe-S] cluster. To characterize the cluster nature, we first performed bioinformatics prediction by aligning the sequence of SnDHAD against 34 selected prokaryotic and eukaryotic DHADs, highlighting the three highly conserved cysteine residues Cys50, Cys123 and Cys195, a characteristic feature of DHADs with a [2Fe-2S] cluster (Fig. S1, Table S1). By contrast, DHADs with a [4Fe-4S] cluster (e.g., EcDHAD) have only two conserved cysteine residues.24 All DHADs with the [2Fe-2S] cluster also form one major clade, distinct from [4Fe-4S] containing DHADs in a constructed phylogenetic tree (Fig. S2). Consistent with the bioinformatics analysis, the UV-visible spectrum of as-purified recombinant SnDHAD revealed diagnostic absorption peaks of a [2Fe-2S] cluster at 320–330, 420 and 460 nm (shoulder) (Fig. S3).36 However, as-purified SnDHAD carried 0.62 ± 0.14 Fe per monomer as determined by the ferrozine assay.37 We then reconstituted the [Fe-S] cluster by incubating as-purified SnDHAD with excess amounts of Na2S and (NH4)2Fe(SO4)2 under anaerobic conditions.38 After purification, the reconstituted SnDHAD contained 2.29 ± 0.17 Fe per monomer but showed a lower intensity and broader peaks in spectral analysis (Fig. S3). The similar spectral changes were observed with some other reconstituted [2Fe-2S] containing enzymes, e.g., ArDHT,32 and presumably caused by the formation of [Fe-S] clusters other than the [2Fe-2S]. Indeed, the spectrum of the reconstituted SnDHAD demonstrated partial feature of the [4Fe-4S] cluster whose UV-visible spectrum is known with a broad shoulder centered at 420 nm (Fig. S3).39 Collectively, bioinformatics and spectral analysis indicated that SnDHAD carries a [2Fe-2S] cluster and future studies could elucidate more details of the cluster nature in as-purified and reconstituted enzymes.
As-purified SnDHAD was catalytically competent in aerobic conditions as it converted ~ 43% of (2R)-DHIV (1 mM) into KIV in 30 min (0.25 μM enzyme, Fig. S4), though it carried a suboptimal amount of Fe/monomer. KIV was quantitated after derivatization with 2,4-dinitrophenylhydrazine and confirmed by LC-MS analysis (Fig. S5A–B). Under the same reaction conditions, the reconstituted SnDHAD retained 30% of the relative activity of the as-purified enzyme, presumably suggesting the detrimental effect of other types of [Fe-S] cluster on its catalysis (Fig. S3, Fig. 2C). A similar result was observed with SsDHAD showing a 78% loss of activity after in vitro activation.20 In the same assay, both SnDHAD C50S and C123S mutants completely lost the catalytic activity (Fig. 2C, Figs. S6), further supporting that SnDHAD is a [2Fe-2S] containing enzyme.
We next assessed the oxygen sensitivity of as-purified SnDHAD using EcDHAD as control. EcDHAD lost ~80% of activity after aerobic incubation for four days at room temperature or after two weeks at 4 °C (Fig. 2D), likely caused by oxidative degradation of the [4Fe-4S] cluster.16 By contrast, as-purified SnDHAD retained 95% of activity at both temperatures after two weeks (Fig. 2D). This result demonstrated the subpopulation of as-purified SnDHAD carrying the intact [2Fe-2S] cluster is likely oxygen stable, a feature shared with archaean SsDHAD20 and plant enzymes.23, 25
We further probed the biochemical properties of as-purified SnDHAD under aerobic conditions. The enzyme favored optimal conditions of pH 8.0 and 50 °C for the chemistry (Fig. S7A–B). The thermal treatment at 37 °C for 2 h had no effect on the enzyme activity. However, the enzyme lost ~60 % of activity after pre-incubation at 50 °C for 2 h, and 65 % and 100 % at 65 °C and 80 °C for 15 min, respectively (Fig. S7C), indicating a mild thermostability. We observed that divalent cations Co2+, Mg2+, Mn2+ and Ni2+ at 1 and 10 mM improved the enzyme activity from 1.2 to 2.0 folds, while Ca2+, Cu2+, and Zn2+ significantly inhibited the activity at 10 mM (Fig. 2E). The enzyme divalent metal dependence was further supported by the complete inhibition of its activity by 1 mM EDTA. Under the above-determined optimal conditions, the reaction with 0.05 μM of as-purified SnDHAD followed Michaelis-Menten kinetics with Km and Vmax values of 136.3 ± 9.0 μM and 1.4 ± 0.02 μM⋅min−1, respectively, giving a turnover value (kcat) of about 28.0 min−1 (Fig. S8). Given only the portion of the as-purified enzyme catalytically active, the actual catalytic efficiency of SnDHAD can be higher.
SnDHAD supports the growth of the engineered E. coli mutant.
We next sought to investigate in vivo function of SnDHAD as part of BCAA biosynthesis in Synechocystis sp. PCC 6803 (hereafter Synechocystis). As-purified SnDHAD was potently inhibited in vitro by AA (Ki = 9 nM), the recently discovered inhibitor of AtDHAD (Ki = 0.3 μM) and MbDHAD (Ki = 10.1 μM) (Fig. S9A–C). The drastically different in vitro inhibition potency of AA toward the three DHADs with the same [2Fe-2S] cluster suggests that the DHAD family members can obtain different biochemical and functional properties in evolution. Indeed, SnDHAD, AtDHAD and MbDHAD share only about 50% of the amino acid identity, and their separation into three groups in a sequence similarity network analysis demonstrates that SnDHAD can represent an unstudied subfamily (Fig. S10).40Furthermore, AA inhibited the growth of Synechocystis with a minimal inhibitory concentration (MIC) value at around 63 nM (Fig. 3A). Exogenous BCAAs at 0.05 mM significantly alleviated the AA-induced growth inhibition (Fig. 3A, Fig. S11), although BCAAs exhibited a dose-dependent inhibition of cyanobacterial growth (MIC = 2.5 mM, Fig. S12).35, 41 Tartronic acid (TA) is a known inhibitor of [4Fe-4S] containing EcDHAD (Ki = 30 μM)42 but even at 0.5 mM, it reduced only around 35% of SnDHAD activity in vitro (Fig. S9D). The MIC of TA toward Synechocystis was 0.25 mM (Fig. 3A, Fig. S11), ~4,000 times higher than AA. BCAAs partially rescued TA-related growth inhibition, but similar to other organic acids, TA at 0.25 mM may exhibit a general toxicity.43 Overall, these results reveal that DHADs with a [2Fe-2S] and [4Fe-4S] cluster can be inhibited separately by chemicals, and further state that SnDHAD carries a [2Fe-2S] cluster.
Figure 3.
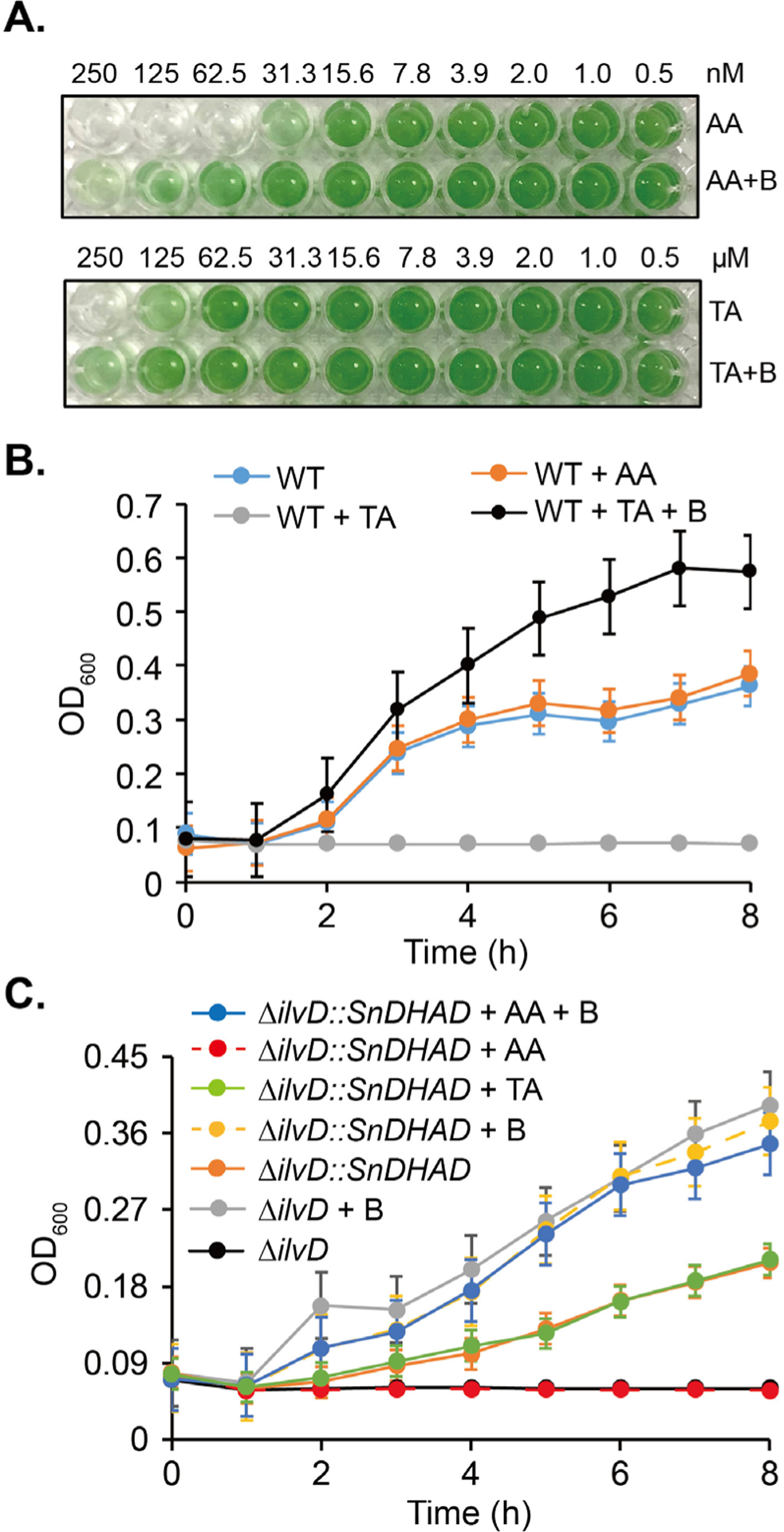
In vivo functional characterization of DHADs. A: Growth inhibition of TA and AA toward Synechocystis. BCAAs (B) at 0.05 mM were supplemented to BG-11 medium. B-C: Growth inhibition of TA (50 mM) and AA (10 μM) toward wild type (WT) E. coli, ΔilvD and ΔilvD::SnDHAD mutants in M9 medium. BCAAs at 1 mM were supplemented. Cell optical density at 600 nm was measured at indicated time points.
We further probed the separate inhibition of DHADs by AA and TA when expressed in E. coli. In the M9 minimal medium, E. coli was completely insensitive to 10 μM AA, while its growth was inhibited partially by TA at 25 mM and completely at 50 mM (Fig. 3B). BCAAs (1 mM) enhanced the growth of TA-treated E. coli by 1.6 times after 8 h, compared with the untreated control. This result demonstrated that EcDHAD is selectively sensitive to TA. Next, we deleted the ilvD gene of E. coli BL21(DE3) and showed that growth of the resultant mutant, E. coli ΔilvD, was dependent on supplemented BCAAs (Fig. 3C). When transformed with the SnDHAD gene, the 8-h growth of E. coli ΔilvD::SnDHAD was restored to 60% of the WT and further improved by supplemented BCAAs (Fig. 3C). Importantly, the growth of E. coli ΔilvD::SnDHAD became insensitive to 50 mM TA but completely inhibited by 1 μM AA. The growth inhibition of the mutant was rescued by 1 mM BCAAs. Collectively, these results validated the in vivo function of SnDHAD and suggested the in vivo function exchangeability of DHADs with a [2Fe-2S] and [4Fe-4S] cluster. Importantly, this study reiterated that the two types of DHADs can be separately inhibited by chemicals, offering the new, promising target for precise growth control.
The overall structure of SnDHAD
To provide a molecular basis of the observed function of SnDHAD, the X-ray structure of the as-purified enzyme was solved to 2.3 Å resolution (Fig. 4A). The SnDHAD monomer has two domains, similar to AtDHAD (PDB: 5ZE4)25 and MbDHAD (PDB: 6OVT).18 The N-terminal domain contains a four-stranded β-sheet surrounded by two α-helices on each side, forming an αβα sandwich, while the C-terminal domain contains an eight-stranded mixed β-sheet folded onto itself due to a β-bulge at Pro407. The C-terminal β-sheet also has two α-helices on its interior face that are in proximity to the active site.
Figure 4.
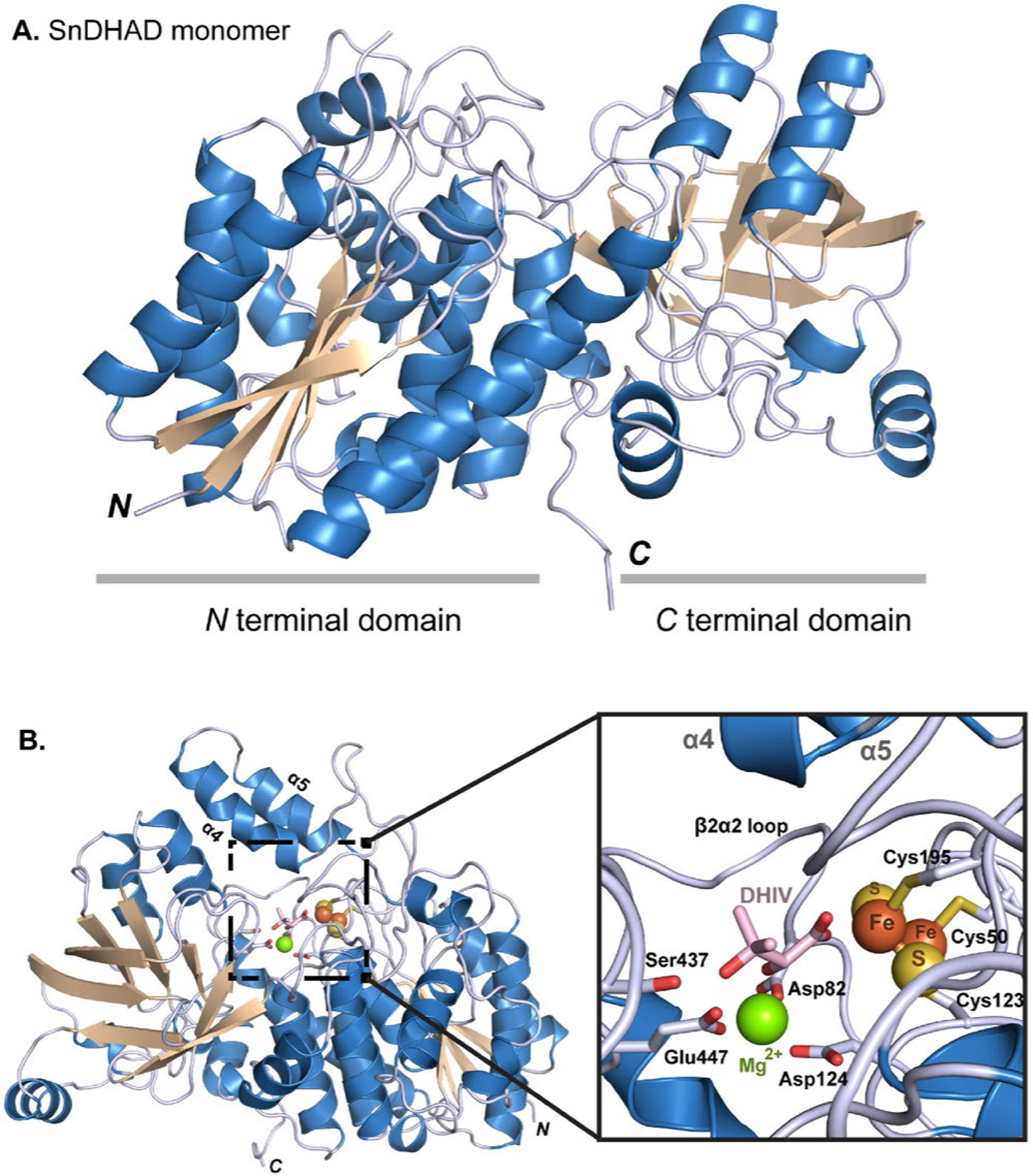
The crystal structure of SnDHAD. A: Ribbon diagram of the monomer with the two subdomains highlights. B: Ribbon representation of the SnDHAD monomer with modeled active site and substrate DHIV. Expanded view of the active site of SnDHAD showing modeled DHIV coordinated to the [2Fe-2S] cluster and Mg2+.
Unlike other IlvD/EDD family members that are dimeric, SnDHAD was monomeric in solution (Fig. 2B) and as opposed to AtDHAD and MbDHAD, the active site of SnDHAD is formed within the single monomer. In the structures of XyDHT33 and ArDHT,32 a key recognition loop at the N-terminus of one monomer extends into the active site of the other (Fig. S13), forming a catalytically active dimer. The corresponding N-terminal recognition loop (1 to 54) in SnDHAD was missing in the solved structure. Instead, the β2α2 loop of SnDHAD extends further over the active site, which perturbs the region that the recognition loop should bind, presumably forcing the loop to move up and away from the active site and to become unresolved in the structure. As comparison, AtDHAD has the β2α2 loop in a similar orientation to SnDHAD, and its N-terminal recognition loop is away from the active site (Fig. S13).
The active site of SnDHAD
Our experimentally solved structure of SnDHAD was missing three loops (residues 44–54, 80–88, and 155–195) predicted to be involved in metal binding (e.g., the conserved Cys50) and catalysis. We therefore modeled the key loops with RosettaCM45 along with an Fe-S cluster and Mg2+ into the active site with RosettaScripts (Fig. 4B).46 The active site of SnDHAD is flanked by the β2α2 loop region and the α4 and α5 helices acting as a lid (Fig. 4B). The lid is significantly flexible in our modelling result and has no significant contacts with the active-site motif. Indeed, homologous helices in all available DHAD homolog structures have a higher than average B-factor. Substrate binding and oxygen insensitive catalysis could be facilitated and mediated by opening and closing of the lid (Fig. 4B), similar to the helix-loop-helix structure in XyDHT33 and ArDHT.32
The conserved Cys50, Cys123, and Cys195 all interact with the [2Fe-2S] cluster (Fig. 4B, Fig. S1), leaving one coordinate site on one Fe atom free for a potential oxygenic ligand.23 The modeled Mg2+ ion is coordinated with three acidic residues Asp82, Asp124, and Glu447. Based on structural homology, the substrate binding site of SnDHAD is between the Fe-S cluster and Ser473 of the β9β10 loop, and adjacent to the coordinated Mg2+ ion (Fig. 4B). In all top ten scored substrate binding models, the C2-OH of DHIV is in close proximity to (2.3 Å) and coordinates with the active-site Mg2+, while the carboxylate of the ligand provides one coordination to the partially occupied Fe atom of the [2Fe-2S] cluster (Fig. 4B, Fig. S14). This binding mode of SnDHAD differs from models suggested with other DHADs,18, 23, 24, 42 where the C3-OH of the substrate binds to the cluster. Interestingly, EPR data of previous studies support both types of substrate binding modes.23, 24 Furthermore, the binding of S-malate and fumarate in the active site of [4Fe-4S]-containing fumarate hydratase from Leishmania major involves a similar Fe-carboxylate oxygen coordination.47 However, further studies are needed to investigate the newly suggested binding mode of DHIV. Nonetheless, the SnDHAD structure indicates that substrate 2R-configuration is required to properly interact with the active-site Mg2+. To initiate the catalysis, the α-proton is removed by a general base. In the new substrate binding mode proposed here, the carboxylate side-chain of Asp124, which also coordinates the binding of the Mg2+, probably mediates the removal of the α-proton likely by coordinating one water molecule (Fig. S15). Asp124 is absolutely conserved in DHADs (Fig. S1), and mutating Asp124 into Glu or Leu completely inactivated the enzyme (Fig. 2C, Fig. S6). The next step is the elimination of 3-OH which can be facilitated by the conserved Ser473 as a general acid, while the released water could partially interact with the serine alkoxide and the Mg2+ (Fig. 4B, Fig. S15). The SnDHAD S473A mutant was completely inactive (Fig. 2C), while retaining a similar level of Fe content (0.44 ± 0.07 Fe per monomer) as the WT, suggesting its catalytic role.
The modeled active site of SnDHAD is highly similar to AtDHAD and MbDHAD, both of which are ligand free,18, 25 although neither was used as a modelling template. For example, the catalytic activity of as-purified MbDHAD (0.25 μM) was about 50% of SnDHAD and dropped by over 60% after air exposure at room temperature for 2 h, indicating rapid oxygen inactivation (Fig. S16). However, the structures of SnDHAD and MbDHAD are very similar in the coordination with the cluster and Mg2+ and their key catalytic residues are in similar positions, supporting our modelling results. On the other hand, the loop containing the active site serine residue extends further into the active site of MbDHAD (Fig. S17A). As such, MbDHAD and SnDHAD could represent the “closed” and “open” states of enzyme, respectively. For MbDHAD, only when the substrate is present, would the active-site loop move to accommodate the substrate, showing as the “closed” state, which would also preclude oxygen access to the cluster. Furthermore, structural comparison of MbDHAD and SnDHAD indicated that residues at position 477 (495 for MbDHAD) sitting over the active-site could play a role in regulating the enzyme function (Fig. S17B). Interestingly, histidine or tyrosine is conserved in the equivalent position of many cyanobacterial and plant DHADs (e.g., AtDHAD),25 while threonine or serine is shared by oxygen-sensitive [4Fe-4S] containing DHADs (Fig. S1). To probe the potential role of this position, we attempted to create SnDHAD Y477H and Y477T and MbDHAD T495H and T495Y., Despite many attempts, we were unable to generate the recombinant SnDHAD Y477T, while the other three were readily available after their expression in E. coli (Figs. S6 and S16). The conversion rate of SnDHAD Y477H was 1.6 times higher than the WT under kinetic reaction conditions (Fig. S18). Kinetic analysis further revealed that this mutation enhanced the turnover value (kcat) from 28.0 min−1 to 38.0 min−1, while slightly affecting the Km (108.9 ± 10.7 μM vs. 136.3 ± 9.0 μM) (Fig. S8). SnDHAD Y477H showed the same level of Fe content and the similar UV-visible spectrum as the WT. On the other hand, as-purified MbDHAD T495H and T495Y retained about 60% and 6% of the catalytic activity of the WT, respectively, while were similarly sensitive to oxygen (Fig. S16). Together, these results supported the functional role of the position 477, and suggested that the X-ray structure of as-purified SnDHAD provides useful information for understanding the structure-function-relationship of this enzyme family.
AA selectively inhibits the growth of cyanobacteria.
We next examined AA as a selective antimicrobial toward microbes with a [2Fe-2S]-containing DHAD. The growth of Microcystis aeruginosa NIES 298 and Anabaena sp. PCC 7120, representing two top-abundant genera of HCBs, was nearly completely inhibited by AA at 0.1 μM, which was recovered by 0.05 mM BCAAs to various degrees (Fig. 5A). The importance of BCAA biosynthesis to cyanobacterial growth was further confirmed by nicosulforon, a known herbicide inhibiting plant AHASs (Fig. 1). However, the inhibition potency of nicosulforon was hundreds of times weaker than AA (Fig. S19). As expected, AA even at 0.1 mM showed no growth inhibition to E. coli, Pseudomonas aeruginosa, and Staphylococcus aureus, whose DHADs carry the [4Fe-4S] cluster (Fig. 5B, Fig. S20). On the other hand, the growth of the fungal pathogen Candida albicans, containing a [2Fe-2S] DHAD, was reduced to 32.2 ± 1.2% by AA, which was recovered to about 47% by 1 mM BCAAs. These results suggested that targeting the BCAA biosynthesis could be a promising strategy to control HCBs and microbial growth. Importantly, screening campaigns and medicinal chemistry efforts are likely to discover and develop new selective potent inhibitors toward DHADs with a [2Fe-2S] and [4Fe-4S] cluster for the precise growth control.
Figure 5.
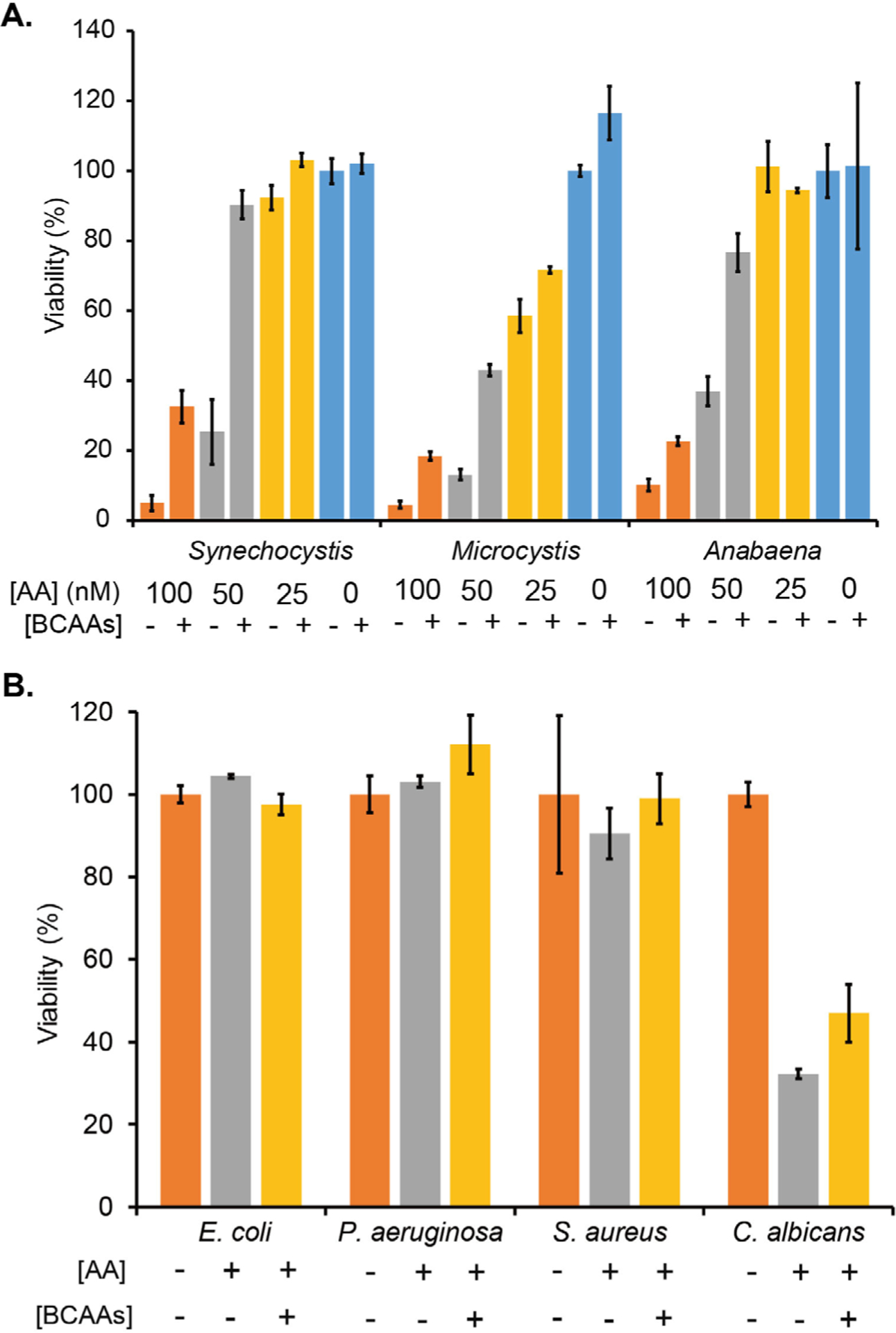
DHADs are a promising target for selective growth control. A: The growth of Synechocystis, Microcystis aeruginosa NIES 298 and Anabaena sp. PCC 7120 was inhibited by AA in a dose-dependent manner. The optical density (730 nm) of each cyanobacterium in BG-11 was determined and set as 100% for normalizing the strain growth in the presence of AA and/or 0.05 mM BCAAs. B: Microbial pathogens showed varied responses to AA (0.1 mM). The optical density (600 nm) of each strain in minimal medium was determined and set as 100% for normalizing the strain growth in the presence of AA and/or 1 mM BCAAs.
Conclusions:
The essentiality of BCAAs to cyanobacterial growth remains largely unstudied. In this work, we provide detailed biochemical, functional and structural characterization of an essential biosynthetic enzyme of BCAAs, DHAD, in the model cyanobacterial strain Synechocystis. Although as-purified SnDHAD contains the suboptimal amount of Fe, bioinformatics, UV-visible spectral, mutagenesis, biochemical, genetic, and chemical inhibition studies provide multiple lines of evidence to support SnDHAD as a highly oxygen-stable [2Fe-2S]-containing enzyme (Figs. 2 and 3). Furthermore, the structure of SnDHAD offers useful insights into the enzyme function (Fig. 4). Despite years of extensive efforts with both as-purified and reconstituted enzymes, the final structure of SnDHAD lacks three loops which contain some residues predicted to participate in the coordination of the Fe-S cluster and Mg2+. Nonetheless, comparison with the two available DHAD structures helps advance the understanding of the enzyme structure-function-relationship.18, 25 Importantly, we demonstrate the successful control of cyanobacterial growth by inhibiting de novo BCAA biosynthesis (Fig. 5). Owing to global climate changes (e.g., increased CO2 level and warming) and excessive nutrient inputs into aquatic systems, HCBs have increasingly occurred worldwide.28–30 Effective management of cyanobacterial growth is vital to sustain and restore ecosystem function. This study suggests that the BCAA biosynthetic pathway can be targeted to control HCBs (e.g., AA at the nM range), particularly in relatively closed water bodies (e.g., fishing ponds). Recently, one report revealed that AA inhibits the plant growth at the μM level (e.g., 50 to 250 μM).25 The thousands of times higher inhibitory concentrations suggest that even AA itself can be useful to selectively control cyanobacterial growth while having minimal to no effects on aquatic plants. This work further demonstrates exploration of untapped essential microbial proteins for a wide range of applications. Future studies are needed to evaluate the effects of low levels of AA on other aquatic organisms including plants, and importantly, to discover additional selective inhibitors in medicinal chemistry research. Furthermore, the current work highlights the BCAA biosynthetic pathway of bacterial and fungal pathogens as a promising new target of novel families of antimicrobials,18 which can find synergistic actions with existing drugs.
Methods
Reconstitution of [Fe-S] cluster
Aerobically purified SnDHAD was incubated with 30-fold molar excess of Na2S and (NH4)2Fe(SO4)2 under anaerobic condition (Whitley DG250 Workstation) for 3 h. The resultant protein solution, dark in color, was anaerobically purified twice using a PD-10 column with the storage buffer (50 mM Tris-Cl, pH 8, 1 mM DTT) to remove nonspecific bound iron and sulfide. The reconstituted SnDHAD was concentrated using a 10-kDa centrifugal filter to 10.0 mg∙mL−1 for further analysis. Protein concentration was determined with Bradford assay (BSA as standard). The UV-visible spectra of DHADs (10.0 mg∙mL−1) were recorded using UV–visible spectrophotometer (Shimadzu UV-2700) from 300 to 800 nm at 20 °C. The storage buffer was used as the blank.
Characterization of enzyme activity
The reaction solutions (100 μL) of EcDHAD, SnDHAD and its mutants (C50S, C123S, S437A, D124L, D124E and Y477H) contained 50 mM Tris-Cl, pH 8.0, 1 mM DTT, 10 mM MgCl2 and 1 mM 2R-DHIV. The reactions were initiated by mixing with 0.25 μM of proteins, aerobically incubated at room temperature for 30 min, and then terminated by mixing with 25 μL of 2 M HCl. Enzymatically synthesized KIV in the solutions was then derivatized with 50 μL of saturated 2,4-dinitrophenylhydrazine (DNPH) in 2 M HCl at room temperature for 30 min. Next, 25 μL of 10 M NaOH were added to each reaction solution and mixed well. Centrifugation at 5,000 × g for 10 min was followed to remove precipitates. To detect KIV-DNPH adduct, 100 μL of the supernatant were transferred to a clear 96-well flat bottom microplate to record the absorbance at 540 nm. The standard curve of KIV-DNPH adduct showed an excellent linearity in the range of 0–1 mM. More details were included in Supporting Information. All enzyme activities were determined in triplicate.
Crystallization and structure determination of SnDHAD
Positive crystal hits of as-purified SnDHAD were observed from the Hampton Index screen (condition 83: 0.2 M Mg2Cl-hexahydrate, 0.1 M Bis-Tris pH 6.5, 25 % PEG 3,350) to yield crystals. Diffraction data sets were collected at LS-CAT, 21-ID-F beamline, Advanced Proton Source, Argonne National Laboratory. Details about data processing and model building were included in Supporting Information. Relevant data collection, phasing, and refinement statistics were included in Table S4. The atomic coordinates and structure factors have been deposited in the Protein Data Bank under the ID code 6NTE.
Supplementary Material
Acknowledgments
This work was supported by startup funds provided by the University of Florida (YD and SB) and NIH (R35 GM128742 to YD). We thank C.P. Wolk (Michigan State University) for the gift of Anabaena sp. PCC 7120 and C. Li and R.W. Huigens (UF) for materials and discussions. We thank the staff of LS-CAT, ANL for help with data collection.
Footnotes
Supporting Information includes additional experimental details, Tables S1 to S4 and Figures S1 to S20.
The authors declare no conflict of interest.
Reference:
- 1.Nelson MB, Martiny AC, and Martiny JB (2016) Global biogeography of microbial nitrogen-cycling traits in soil, Proc Natl Acad Sci U S A 113, 8033–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glassman SI, Weihe C, Li J, Albright MBN, Looby CI, Martiny AC, Treseder KK, Allison SD, and Martiny JBH (2018) Decomposition responses to climate depend on microbial community composition, Proc Natl Acad Sci U S A 115, 11994–11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemente JC, Ursell LK, Parfrey LW, and Knight R (2012) The impact of the gut microbiota on human health: an integrative view, Cell 148, 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg E, and Zilber-Rosenberg I (2016) Microbes drive evolution of animals and plants: the hologenome concept, MBio 7, e01395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paul VJ (2008) Global warming and cyanobacterial harmful algal blooms, Adv Exp Med Biol 619, 239–257. [DOI] [PubMed] [Google Scholar]
- 6.Hutchison CA 3rd, Chuang RY, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, Pelletier JF, Qi ZQ, Richter RA, Strychalski EA, Sun L, Suzuki Y, Tsvetanova B, Wise KS, Smith HO, Glass JI, Merryman C, Gibson DG, and Venter JC (2016) Design and synthesis of a minimal bacterial genome, Science 351, aad6253. [DOI] [PubMed] [Google Scholar]
- 7.Lewis K (2012) Antibiotics: Recover the lost art of drug discovery, Nature 485, 439–440. [DOI] [PubMed] [Google Scholar]
- 8.Murima P, McKinney JD, and Pethe K (2014) Targeting bacterial central metabolism for drug development, Chem Biol 21, 1423–1432. [DOI] [PubMed] [Google Scholar]
- 9.Nie C, He T, Zhang W, Zhang G, and Ma X (2018) Branched chain amino acids: Beyond nutrition metabolism, Int J Mol Sci 19, pii: E954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franco TMA, and Blanchard JS (2017) Bacterial branched-chain amino acid biosynthesis: Structures, mechanisms, and drugability, Biochemistry 56, 5849–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh BK, and Shaner DL (1995) Biosynthesis of branched-chain amino acid-from test tube to field, Plant Cell 7, 935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia MD, Nouwens A, Lonhienne TG, and Guddat LW (2017) Comprehensive understanding of acetohydroxyacid synthase inhibition by different herbicide families, Proc Natl Acad Sci U S A 114, E1091–E1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh V, Chandra D, Srivastava BS, and Srivastava R (2011) Downregulation of Rv0189c, encoding a dihydroxyacid dehydratase, affects growth of Mycobacterium tuberculosis in vitro and in mice, Microbiology 157, 38–46. [DOI] [PubMed] [Google Scholar]
- 14.Garcia MD, Chua SMH, Low YS, Lee YT, Agnew-Francis K, Wang JG, Nouwens A, Lonhienne T, Williams CM, Fraser JA, and Guddat LW (2018) Commercial AHAS-inhibiting herbicides are promising drug leads for the treatment of human fungal pathogenic infections, Proc Natl Acad Sci U S A 115, E9649–E9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.UniProt Consortium(2019) UniProt: a worldwide hub of protein knowledge, Nucleic Acids Res 47, D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flint DH, Emptages MH, Finnegann MG, Full W, and Johnson MK (1993) The role and properties of the iron-sulfur cluster in Escherichia coli dihydroxy-acid dehydratase, J Biol Chem 268, 14732–14742. [PubMed] [Google Scholar]
- 17.Flint DH, Smyk-Randalln E, Kinello JF, Draczynska-Lusiakn B, and Brownn OR (1993) The inactivation of dihydroxy-acid dehydratase in Escherichia coli treated with hyperbaric oxygen occurs because of the destruction of its Fe-S cluster, but the enzyme remains in the cell in a form that can be reactivate, J Biol Chem 268, 25547–25552. [PubMed] [Google Scholar]
- 18.Bashiri G, Grove TL, Hegde SS, Lagautriere T, Gerfen GJ, Almo SC, Squire CJ, Blanchard JS, and Baker EN (2019) The active site of the Mycobacterium tuberculosis branched-chain amino acid biosynthesis enzyme dihydroxyacid dehydratase contains a 2Fe-2S cluster, J Biol Chem 294, 13158–13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xing RY, and Whitman WB (1991) Characterization of enzymes of the branched-chain amino acid biosynthetic pathway in Methanococcus spp, J Bacteriol 173, 2086–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, and Lee SB (2006) Catalytic promiscuity in dihydroxy-acid dehydratase from the hermoacidophilic archaeon Sulfolobus solfataricus, J Biochem 139, 591–596. [DOI] [PubMed] [Google Scholar]
- 21.Oliver JD, Kaye SJ, Tuckwell D, Johns AE, Macdonald DA, Livermore J, Warn PA, Birch M, and Bromley MJ (2012) The Aspergillus fumigatus dihydroxyacid dehydratase Ilv3A/IlvC is required for full virulence, Plos One 7, e43559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satyanarayana T, and Radhakrishnan AN (1964) Biosynthesis of valine + isoleucine in plants .2. Dihydroxyacid dehydratase from Phaseolus Radiatus, Biochim Biophys Acta 92, 367–377. [DOI] [PubMed] [Google Scholar]
- 23.Flint DH, and Emptage MH (1988) Dihydroxy acid dehydratase from spinach contains a [2Fe-2S] cluster, J Biol Chem 263, 3558–3564. [PubMed] [Google Scholar]
- 24.Gao H, Azam T, Randeniya S, Couturier J, Rouhier N, and Johnson MK (2018) Function and maturation of the Fe-S center in dihydroxyacid dehydratase from Arabidopsis, J Biol Chem 293, 4422–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan Y, Liu Q, Zang X, Yuan S, Bat-Erdene U, Nguyen C, Gan J, Zhou J, Jacobsen SE, and Tang Y (2018) Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action, Nature 559, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knoot CJ, Ungerer J, Wangikar PP, and Pakrasi HB (2018) Cyanobacteria: Promising biocatalysts for sustainable chemical production, J Biol Chem 293, 5044–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huisman J, Codd GA, Paerl HW, Ibelings BW, Verspagen JMH, and Visser PM (2018) Cyanobacterial blooms, Nat Rev Microbiol 16, 471–483. [DOI] [PubMed] [Google Scholar]
- 28.Michalak AM, Anderson EJ, Beletsky D, Boland S, Bosch NS, Bridgeman TB, Chaffin JD, Cho K, Confesor R, Daloglu I, Depinto JV, Evans MA, Fahnenstiel GL, He L, Ho JC, Jenkins L, Johengen TH, Kuo KC, Laporte E, Liu X, McWilliams MR, Moore MR, Posselt DJ, Richards RP, Scavia D, Steiner AL, Verhamme E, Wright DM, and Zagorski MA (2013) Record-setting algal bloom in Lake Erie caused by agricultural and meteorological trends consistent with expected future conditions, Proc Natl Acad Sci U S A 110, 6448–6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Neil JM, Davis TW, Burford MA, and Gobler CJ (2012) The rise of harmful cyanobacteria blooms: The potential roles of eutrophication and climate change, Harmful Algae 14, 313–334. [Google Scholar]
- 30.Paerl HW, and Otten TG (2013) Harmful cyanobacterial blooms: causes, consequences, and controls, Microb Ecol 65, 995–1010. [DOI] [PubMed] [Google Scholar]
- 31.Ikeuchi M, and Tabata S (2001) Synechocystis sp. PCC 6803 - a useful tool in the study of the genetics of cyanobacteria, Photosynth Res 70, 73–83. [DOI] [PubMed] [Google Scholar]
- 32.Rahman MM, Andberg M, Thangaraj SK, Parkkinen T, Penttila M, Janis J, Koivula A, Rouvinen J, and Hakulinen N (2017) The crystal structure of a bacterial L-arabinonate dehydratase contains a [2Fe-2S] cluster, ACS Chem Biol 12, 1919–1927. [DOI] [PubMed] [Google Scholar]
- 33.Rahman MM, Andberg M, Koivula A, Rouvinen J, and Hakulinen N (2018) The crystal structure of D-xylonate dehydratase reveals functional features of enzymes from the Ilv/ED dehydratase family, Sci Rep 8, 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riccardi G, de Rossi E, and Milano A (1989) Amino acid biosynthesis and its regulation in cyanobacteria, Plant Sci 64, 135–151. [Google Scholar]
- 35.Kouhen OM, and Joset F (2002) Biosynthesis of the branched-chain amino acids in the cyanobacterium Synechocystis PCC6803: existence of compensatory pathways, Curr Microbiol 45, 94–98. [DOI] [PubMed] [Google Scholar]
- 36.Gupta V, Sendra M, Naik SG, Chahal HK, Huynh BH, Outten FW, Fontecave M, and de Choudens SO (2009) Native Escherichia coli SufA, coexpressed with SufBCDSE, purifies as a [2Fe-2S] protein and acts as an Fe-S transporter to Fe-S target enzymes, J Am Chem Soc 131, 6149–6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeitner TM (2014) Optimized ferrozine-based assay for dissolved iron, Anal Biochem 454, 36–37. [DOI] [PubMed] [Google Scholar]
- 38.Bridwell-Rabb J, Zhong A, Sun HG, Drennan CL, and Liu HW (2017) A B12-dependent radical SAM enzyme involved in oxetanocin A biosynthesis, Nature 544, 322–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mapolelo DT, Zhang B, Naik SG, Huynh BH, and Johnson MK (2012) Spectroscopic and functional characterization of iron-sulfur cluster-bound forms of Azotobacter vinelandii (Nif)IscA, Biochemistry 51, 8071–8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atkinson HJ, Morris JH, Ferrin TE, and Babbitt PC (2009) Using sequence similarity networks for visualization of relationships across diverse protein superfamilies, PLOS ONE 4, e4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian LL, Chen M, Ren CY, Wang YY, and Li L (2018) Anticyanobacterial effect of L-lysine on Microcystis aeruginosa, RSC Adv 8, 21606–21612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flint DH, and Nudelman A (1993) Studies on the active-site of dihydroxy-acid dehydratase, Bioorg Chem 21, 367–385. [Google Scholar]
- 43.Begemann MB, Zess EK, Walters EM, Schmitt EF, Markley AL, and Pfleger BF (2013) An organic acid based counter selection system for cyanobacteria, PLoS One 8, e76594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krissinel E, and Henrick K (2007) Inference of macromolecular assemblies from crystalline state, J Mol Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- 45.Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, and Baker D (2013) High-resolution comparative modeling with RosettaCM, Structure 21, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fleishman SJ, Leaver-Fay A, Corn JE, Strauch EM, Khare SD, Koga N, Ashworth J, Murphy P, Richter F, Lemmon G, Meiler J, and Baker D (2011) RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite, PLoS One 6, e20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feliciano PR, and Drennan CL (2019) Structural and biochemical investigations of the [4Fe-4S] cluster-containing fumarate hydratase from Leishmania major, Biochemistry 58, 5011–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


