Abstract
What factors influence the evolution of a heavily selected functional trait in a diverse clade? This study adopts rattlesnakes as a model group to investigate the evolutionary history of venom coagulotoxicity in the wider context of phylogenetics, natural history, and biology. Venom-induced clotting of human plasma and fibrinogen was determined and mapped onto the rattlesnake phylogenetic tree to reconstruct the evolution of coagulotoxicity across the group. Our results indicate that venom phenotype is often independent of phylogenetic relationships in rattlesnakes, suggesting the importance of diet and/or other environmental variables in driving venom evolution. Moreover, the striking inter- and intraspecific variability in venom activity on human blood highlights the considerable variability faced by physicians treating envenomation. This study is the most comprehensive effort to date to describe and characterize the evolutionary and biological aspects of coagulotoxins in rattlesnake venom. Further research at finer taxonomic levels is recommended to elucidate patterns of variation within species and lineages.
Keywords: Rattlesnakes, Venom, Evolution, Blood, Coagulotoxicity, Snakebite
1. Introduction
Rattlesnakes (Viperidae: Crotalinae: Crotalus and Sistrurus) have long fascinated mankind (Klauber, 1972; Reiserer, 2016), and are one of the most famous and well-studied groups of snakes (Beaman and Hayes, 2008). Distributed from southern Canada south into northern Argentina (Campbell et al., 2005), rattlesnake species are adapted to a variety of different ecosystems ranging from below sea level deserts up to at least 4400 m above sea level where they inhabit bunchgrass grasslands above tree line (Klauber, 1972, Campbell et al., 2005).
The monophyly of New World pit-vipers as descendants of a single colonization event by an ancestral Asian pit-viper species that invaded the New World via the Bering Land Bridge (Kraus et al., 1996; Parkinson, 1999; Parkinson et al., 2002; Alencar et al., 2018) during the late Oligocene or Early Miocene (Wüster et al., 2008; Alencar et al., 2016) is well supported. Interrelationships within Crotalus however are less understood (see review by Wüster, 2016 and references therein). Two competing hypotheses exist for the origin and evolution of rattlesnakes and differ in geographical and ecological setting. One hypothesis proposes a Mexican origin in the pine-oak woodlands of the Sierra Madre Occidental (Gloyd, 1940, Campbell et al., 2005, Place and Abramson, 2004, Blair and Sánchez-Ramírez, 2016) whereas the other proposes an origin in the temperate mixed woodland and grassland communities of the continental United States (Setser et al., 2011; Meik and Schuett, 2016; Reiserer and Schuett, 2016). Regardless of the location of rattlesnake geographic origin, the clade rapidly diversified (Blair and Sánchez-Ramírez, 2016) likely due to a combination of orogenesis and climatic events (Pook et al., 2000; Douglas et al., 2006; Bryson et al., 2011a; Bryson et al., 2011b; Myers et al., 2017).
The variations in phylogeography and natural history present in rattlesnakes reflect the convoluted patterns of evolutionary pressures experienced by these pit-vipers in their diversification. This divergence is arguably best represented by the high variability in venom composition found across the clade (Klauber, 1972; Calvete et al., 2010; Mackessy, 2010; Saviola et al., 2015; Saviola et al., 2017). Mackessy (2008) broadly grouped rattlesnake venoms into two categories based on their snake venom metalloprotease (SVMP) content: high SVMP content and low toxicity (Type I venoms) vs. low SVMP content and high toxicity (Type II venoms). More specifically, the Type I phenotype is markedly haemorrhagic, with copious blood loss often accompanied by localized cytotoxicity due to SVMPs lysing the membrane of blood vessels and/or inhibiting platelet aggregation (Gutiérrez et al., 2005; Gutíerrez et al., 2016).
Conversely, the Type II category refers to venoms with a high percentage of presynaptic neurotoxins of the phospholipase A2 (PLA2) family (and significantly lower content of SVMPs), which generally cause paralysis and systemic myotoxicity rather than haemorrhage (Hendon and Fraenkel-Conrat, 1971; Klauber, 1972; Dobson et al., 2018a; Neri-Castro et al., 2019). Remarkably, the two phenotypes are not mutually exclusive at the species level, as separate populations of species such as the Mohave rattlesnake (Crotalus scutulatus ssp.) and the South American rattlesnake (Crotalus durissus ssp.) are known to fall into either category and/or even to combine the two toxic arsenals (Glenn and Straight, 1978; Glenn and Straight, 1989; Saravia et al., 2002; Calvete et al., 2010; Strickland et al., 2018). From an evolutionary perspective, recent research supported the presence of neurotoxic PLA2s in the last common ancestor of rattlesnakes (Yang et al., 2015; Dowell et al., 2016). The genetic pattern behind such a diverse array of venom compositions within and between rattlesnake species has been the subject of extensive research (Giorgianni et al., 2020; Gibbs and Rossiter, 2008; Dowell et al., 2016; Margres et al., 2017; Dowell et al., 2018).
Other recurrent components of rattlesnake venoms include kallikrein-type serine proteases (SVSP) L-amino acid oxidases (LAAOs), phosphodiesterases (PDEs), disintegrins, and C-type lectin-like proteins (Mackessy, 2010; Saviola et al., 2015; Mackessy, 2008; Durban et al., 2017), all of which include representatives known to induce coagulopathy (Fry, 2015). Overall, coagulotoxins represent a major component of the toxic arsenal of most rattlesnake species, therefore warranting great research attention from an ecological and a clinical perspective alike (Markland, 1983; Mackessy, 2008). Of the non-SVMP toxins, the SVSP contribute most strongly to the coagulopathy resulting from envenomation. Like SVMPs, SVSPs directly interfere with the blood clotting cascade by depleting fibrinogen via two mechanisms: the direct anticoagulant mechanism destructively cleaves fibrinogen, while the pseudo-procoagulant (aka: thrombin-like) mechanism cleaves fibrinogen to form transient, weak, unstable fibrin clots that rapidly break down (Debono et al., 2019a; Debono et al., 2019b; Debono et al., 2019c; Youngman et al., 2019; Bourke et al., 2020). Conspicuously, the presence and prevalence of SVSPs in rattlesnake venoms does not strictly follow the Type I vs Type II dichotomy, as serine protease abundance is variable in species displaying either phenotype (Mackessy, 2008). Thus the utility of this simplistic division of venoms is limited in predicting functional variation.
From an animal biology perspective, the link between venom variations and shifts in dietary habits between species and populations in snakes is well established (Daltry et al., 1996; Barlow et al., 2009; Cipriani et al., 2017; Lyons et al., 2020), with venom and resistance to toxins in prey animals continuously driving each other’s evolution in what has often been described as an arms race (Casewell et al., 2013; Fry et al., 2008; Jackson et al., 2016). Examples of such a marked influence of diet on venom abound in rattlesnakes (Mackessy, 1988; Sanz et al., 2006; Mackessy, 2010; Sunagar et al., 2014; Saviola et al., 2017; Holding et al., 2018). However, prey specificity is unlikely to be the only driver of venom diversification in this clade, since significant differences in venom composition can be observed in populations and species characterized by similar prey preferences (Mackessy, 2008; Zancolli et al., 2019). Furthermore, venom variation in several species of rattlesnakes is known to be ontogenetic, with juveniles generally possessing a Type II venom phenotype that gradually transitions to Type I in adults (Mackessy et al., 2003; Calvete et al., 2010). While this process often reflects ontogenetic dietary shifts (Mackessy, 1988; Gibbs et al., 2011), age-related modifications in venom composition have been documented in species that likely feed on the same prey type throughout their life (Calvete et al., 2010; Seneci et al., 2021). Consequently, a concatenation of multiple factors, from environmental variables ranging from local climate and elevation (Zancolli et al., 2019) to interspecific and even intergeneric hybridization (Dowell et al., 2016, Dowell et al., 2018, but see Zancolli et al., 2016), is likely at the root of inter- and intraspecific variation in venom composition in rattlesnakes.
In this study, we aim to elucidate the evolutionary history and diversity of coagulotoxic activity in rattlesnake venom by assessing venom-induced clotting times through a comprehensive phylogenetic approach on a scale never attempted to date. We extensively cover the intersection of biological and environmental factors driving the specialization of venom activity to integrate our findings in a broader evolutionary and biological context, whereby rattlesnakes represent an ideal model clade. Based on current knowledge (Mackessy, 2008; Gibbs et al., 2013), we predict that phylogenetic relationships alone will not explain patterns of variability at the inter- and intraspecific level. We also investigate whether the Type I vs Type II dichotomy described by Mackessy (2008) potentially overshadows patterns of variability in coagulotoxic activity within and beyond the boundaries of each type.
2. Materials and methods
2.1. Venom selection and preparation
All venom work was undertaken under the auspices of UQ IBSC approval #IBC134BSBS2015. 55 lyophilized venom samples were selected from the long-term, cryogenic collection available at the Venom Evolution Lab. Furthermore, the NNTRC (an NIH/ORIP funded center) provided 23 venoms, and MToxins provided 14 venoms. Priority was given to venom samples associated with a specific locality and/or information on the animal’s age, size, and sex in order to account for multiple factors that might influence venom activity. ddH2O (ultra pure water) was added to the powdered venom sample before vortexing for 5 s and centrifuging at 14,000 rcf for 10 min. After centrifugation, the supernatant was transferred to a 1.5 mL Eppendorf tube and vortexed in preparation for testing in triplicate on a Nanodrop 2000 spectrophotometer (ThermoFisher Scientific) to measure concentration of venom proteins in the diluted sample at a 280 nm absorbance wavelength. The resulting concentration values were used to calculate the amount of ddH2O and venom solution to add in order to bring the concentration to 2 mg/mL. Subsequently, glycerol was added to obtain 50% ddH2O:50% glycerol final stock with a venom concentration of 1 mg/mL.
2.2. Plasma and fibrinogen coagulation assays
Healthy human plasma (citrate 3.2%, Lot# A540020142331 and A5400201137021) was provided by the Australian Red Cross (44 Musk Street, Kelvin Grove, Queensland 4059). Both plasma lots were pooled and the combined stock underwent the same preparation and experimental procedures. More specifically, plasma stocks were aliquoted into 1.5 mL Eppendorf tubes under sterile conditions before flash-freezing in liquid nitrogen and storing at −80 °C until use. All human plasma work was performed under University of Queensland Biosafety Approval #IBC134BSBS2015 and Human Ethics Approval #2016000256. Human fibrinogen was purchased from Sigma Aldrich (St. Louis, Missouri, United States) and aliquoted into 1.5 mL Eppendorf tubes after reconstitution into Fluoroskan running buffer (150 mM NaCl + 50 mM TrisHCl in 1 L ddH2O, pH 7.4) to a concentration of 4 mg/mL, flash-freezing in liquid nitrogen for 10 s and storing at −80 °C until use. Plasma and fibrinogen clotting times were measured on a Stago STA-R Max coagulation analyzer using Stago Analyzer software v. 0.00.04 (Stago, Asníeres sur Seine, France) upon thawing of aliquots at 37 °C for 5 min. More specifically, this robot determines clotting time as the interval required for a magnetic sphere inside a 250 μL cuvette to cease oscillating due to blockage caused by a clot in the incubated solution of venom and plasma/fibrinogen combined with several co-factors. A positive control for plasma was run by performing a Kaolin-activated Partial Thromboplastin Time assay (aPTT). For the fibrinogen positive control, 50 μL 50% ddH2O + glycerol, 25 μL STA Owren-Koller buffer (Stago catalog #00360) + CaCl2 (Stago catalog #00367, 25 mM, 2:1 dilution), 50 μL phospholipid from STA C.K. Prest Standard Kit (Stago Catalog # 00597, diluted in 5 mL Owren-Koller buffer), and 75 μL human fibrinogen were incubated for 120 s before adding 50 μL thrombin (STA Liquid-FIB) for a total volume of 250 μL. All venoms were run on STA-R Max in a 250 μL solution consisting of 25 μL Owren-Koller buffer, 50 μL phospholipid, 50 μL venom solution (i.e. venom diluted to 100 μg/mL in Owren-Koller buffer), and 50 μL CaCl2.
2.3. Phylogenetic comparative analysis
Ancestral state reconstruction for coagulotoxicity was conducted using maximum likelihood as implemented in the ContMap function of the package phytools (Revell, 2012) on RStudio v. 3.5.1. Two phylogenetic trees (one each for plasma and fibrinogen clotting times) were constructed based on the phylogeny of rattlesnakes published by Alencar et al. (2016), and trees obtained from timetree.org, which were manually recreated in Mesquite (version 3.2) and subsequently imported in RStudio in Newick format via the APE package.
3. Results
Mean plasma and fibrinogen clotting times were mapped over the organismal phylogenetic tree to highlight the diversification of coagulotoxicity across the rattlesnake clade (Fig. 1 for effects on whole plasma, Fig. 2 for specific pseudo-procoagulant effect upon fibrinogen). While clotting times above spontaneous control levels are indicative of anticoagulant venom activity on plasma (Fig. 1), this assay could not determine whether clotting times below that of the spontaneous control represented true procoagulant action (activation of the zymogens of clotting factors such as Factor X or prothrombin to result in the generation of endogenous thrombin, leading to the formation of strong, stable fibrin clot) or pseudo-procoagulant (the direct action upon fibrinogen to produce abberant fibrin clots that are weak, and short-lived, thus contributing to net anticoagulation by depleting the levels of intact fibrinogen). Thus, we subsequently tested for the ability to directly act upon fibrinogen and form clots in a pseudo-procoagulant manner (Fig. 2). Our ancestral state reconstruction for coagulotoxicity on fibrinogen (Fig. 2) shows that the vast majority of taxa that clotted plasma faster than the spontaneous control time (Fig. 1) also directly clotted fibrinogen (Fig. 2) thus indicating a pseudo-procoagulant action centered on cleaving fibrinogen to produce weak, unstable clots. This was particularly pronounced in the Neotropical rattlesnake lineage (C. mictlantecuhtli, C. tzabcan, C. durissus, C. vegrandis), which clotted fibrinogen in a pseudo-procoagulant manner in <100 s. A notable exception was C. culminatus, with a pool of neonate venoms clotting plasma in <15 s as opposed to >90 s for fibrinogen. This is due to the neonate life-stage species possessing a true procoagulant phenotype, by potently activating Factor X (Seneci et al., 2021). The clade containing C. stephensi, C. pyrrhus, C. adamanteus, and C. tigris was strongly pseudo-procoagulant fibrinogen clotting, while this trait was weak or absent in the sister clade containing the C. viridis complex. Such a dynamic variation was evident in other closely related species with C. enyo being pseudo-procoagulant while the sister species C. cerastes lacked this trait, with the same extreme variation for C. catalinensis (pseudo-procoagulant) compared to C. atrox (not pseudo-procoagulant). Among the small-sized montane rattlesnakes, only the basal-most C. ravus displayed a strong pseudo-procoagulant action on fibrinogen, whereas this trait was not present in C. aquilus, C. armstrongi, C. polystictus, C. pricei, or C. lepidus. Such extreme variation was also noted between localities of the wide-ranging species C. horridus, which was potently pseudo-procoagulant in the western part of its range (Texas), but not for the eastern population (Florida, Kentucky, and South Carolina) which prevented spontaneous clotting. The genus Sistrurus, which constitutes the earliest split in the rattlesnake phylogenetic tree, mostly lacked a strong coagulating activity on fibrinogen, with neither S. miliarius nor S. tergeminus achieving clotting times <341 s.
Fig. 1.
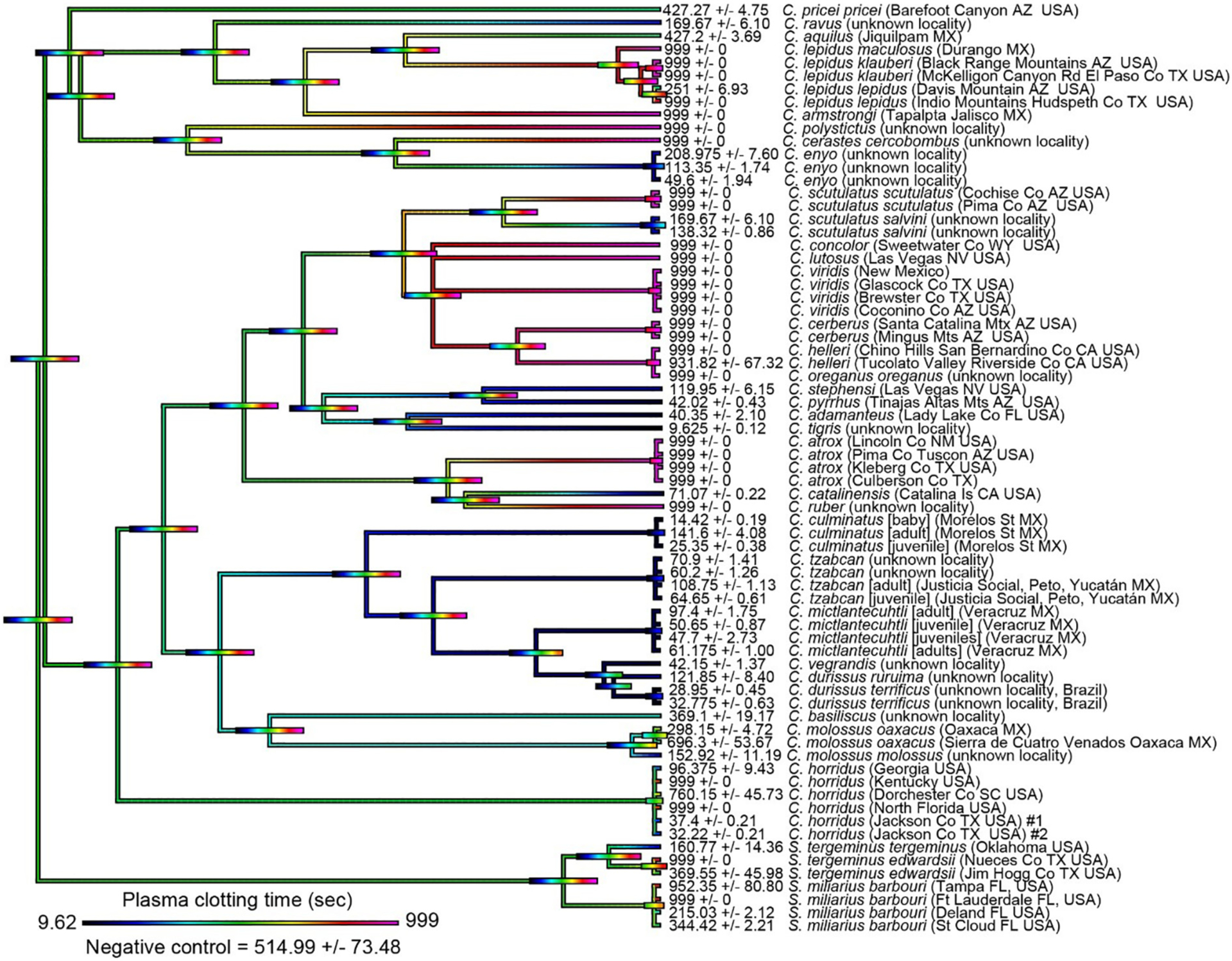
Ancestral state reconstruction of coagulotoxicity in rattlesnakes (Crotalus + Sistrurus) as tested on human plasma. Warmer colors indicate longer clotting times (anticoagulant effect) whereas colder colors represent shorter clotting times (procoagulant effect) relative to the negative control (514 s). Bars indicate 95% confidence intervals for the estimate at each node. Note: due to the high dynamicity of venom evolution, the node bar ranges quickly become broad as one moves down the tree. Maximum clotting time = 999 s. Values are mean (n = 4 ± SD). Organismal phylogenetic arrangements from Alencar et al. (2016) and timetree.org.
Fig. 2.
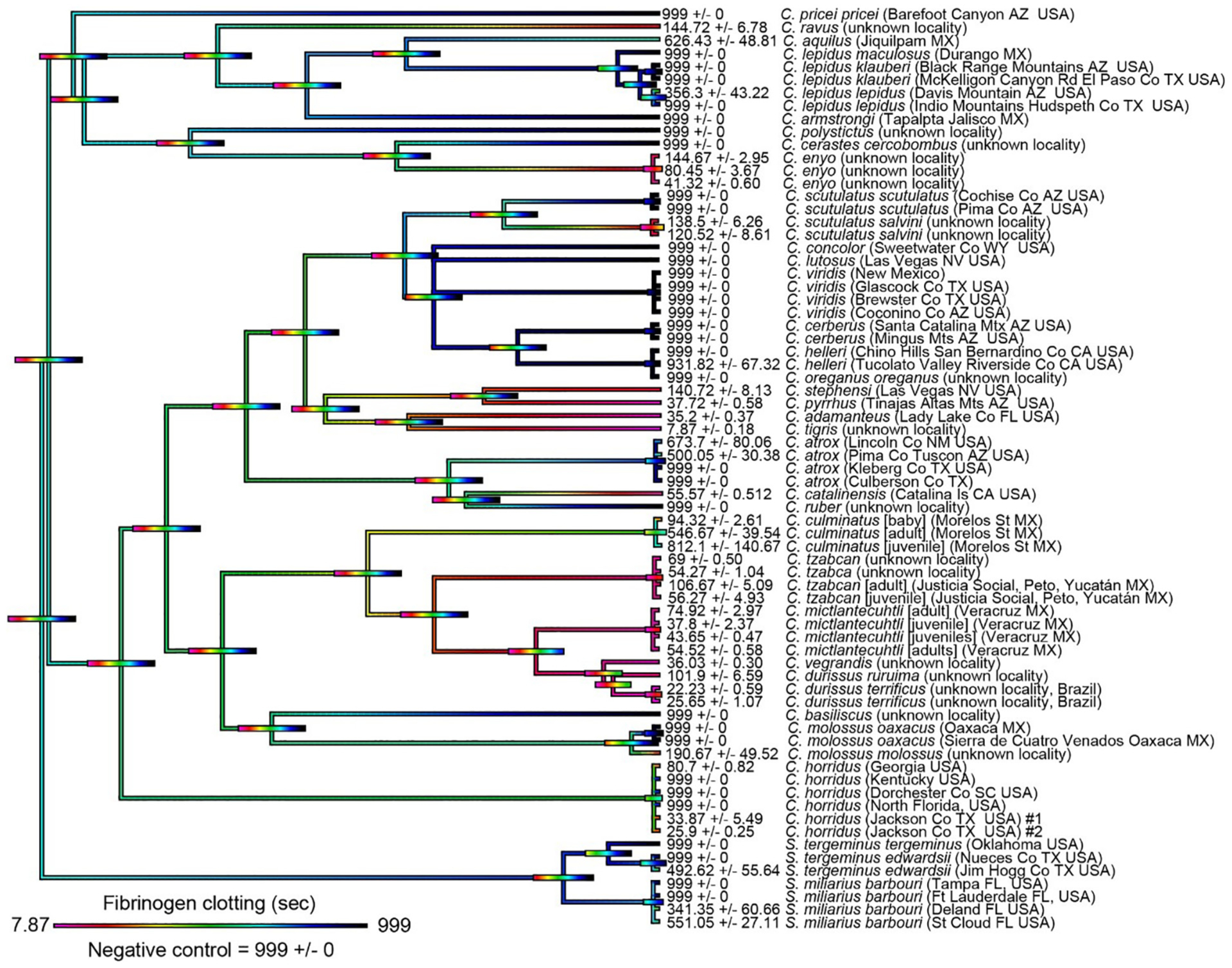
Ancestral state reconstruction of coagulotoxicity in rattlesnakes (Crotalus + Sistrurus) as tested on human fibrinogen. Warmer colors indicate shorter clotting times, whereby fibrinogen was clotted in a pseudo-procoagulant manner whereas colder colors represent longer clotting times (no effect) relative to the negative control (999 s). Bars indicate 95% confidence intervals for the estimate at each node. Note: due to the high dynamicity of venom evolution, the node bar ranges quickly become broad as one moves down the tree. Maximum clotting time = 999 s. Values are mean (n = 4 ± SD). Organismal phylogenetic arrangements from Alencar et al. (2016) and timetree.org.
Comparing the ancestral state reconstructions between plasma and fibrinogen showed dramatic disparity between phylogenetic relationships across taxa and coagulant activity of venom (Fig. 1). In fact, it was not uncommon to observe sister species (e.g. C. ruber and C. atrox compared to C. catalinensis, or C. cerastes and C. enyo) characterized by anticoagulant and pseudo-procoagulant (aka ‘thrombin-like’) phenotypes, respectively. This is true even at the intraspecific level, with specimens from different localities sometimes revealing evident differences in clotting times as apparent in C. horridus (extremes of 32 s to 999 s), S. m. barbouri (215 s to 999 s), and S. t. edwardsii (160 s to 999 s). The montane species C. armstrongi and C. polystictus were distinctly anticoagulant, as was the case for C. lepidus save for a single C. l. lepidus individual from the Davis Mountain in Arizona (251 s on plasma) and was confirmed as weakly pseudo-procoagulant (356 s on fibrinogen). More homogeneous patterns were found in other clades, such as a shared anticoagulant phenotype across the Western rattlesnake complex (C. viridis, C. oreganus, C. helleri, C. lutosus, C. concolor, and C. Cerberus, all of which reached the 999 s time limit on plasma) and a marked prevalence of pseudo-procoagulant venoms in the Neotropical rattlesnakes with the exception of C. culminatus (Figs. 1 & 2). The sister lineage to the Neotropical rattlesnakes, comprising C. molossus and C. basiliscus, was weakly pseudo-procoagulant, with one C. m. oaxacus sample showing anticoagulant activity. The clotting action of C. tigris venom was the fastest pseudo-procoagulant venom ever observed (9.62 s), greatly surpassing that of closely related C. pyrrhus (42.02 s), C. stephensi (119.95 s), and C. adamanteus (40.35 s). Overall, our data suggest that the ancestral condition for rattlesnakes regarding coagulotoxicity was a lack of significant pseudo-procoagulant anticoagulant or classic anticoagulant activity of venom upon plasma or fibrinogen clotting, as exemplified by the colder colors in the early-branching nodes of the tree. This condition is retained only in a small minority of the analyzed taxa, such as C. aquilus and C. pricei (Figs. 1 & 2). For the species which displayed strong classic anticoagulant activity on plasma (Fig. 1) it was beyond the scope of this study to ascertain if this was due to inhibition of clotting enzymes or destruction of fibrinogen, and this will be the subject of future work.
4. Discussion
The present study aimed to investigate the evolutionary history of coagulotoxicity in the venoms of a highly diverse pit-viper clade, the rattlesnakes (Crotalus and Sistrurus). Rattlesnakes are responsible for a considerable number of human envenomation cases both in the United States (Ruha et al., 2017) and throughout Latin America (Chippaux, 2017). Although efficient antivenom products are widely available, extensive variation in venom activity in these snakes is known to affect the efficacy of antivenom (Sunagar et al., 2014; Dobson et al., 2018b; Neri-Castro et al., 2020). Even considering coagulotoxicity alone, our results underscore extremely dynamic venom variation throughout the rattlesnake clade, with potentially notable clinical implications for the treatment of envenomed patients. Thorough testing of antivenom products against species not included in the immunizing mixture is therefore crucial to assess cross-reactivity against local medically relevant rattlesnakes. The dynamic variation in anticoagulant mechanisms may dramatically influence the efficacy of available antivenoms. Such antivenom testing was beyond the scope of this work, but will be the subject of a followup study.
The results presented herein indicate that the last common ancestor of rattlesnakes had fibrinogen-acting toxins or coagulation-enzyme inhibiting toxins expressed in only low levels in the venom (Figs. 1 and 2). Thus the ability to deplete fibrinogen levels through pseudo-procoagulant direct actions on fibrinogen or the ability to inhibit clotting enzymes were both traits that were convergently amplified on multiple independent occasions within this clade of American pitvipers. This suggests that the functional relevance and relative abundance of coagulotoxins in early rattlesnakes was possibly minor in comparison to other proteins such as neurotoxins, which are likely ancestral to the group (Yang et al., 2015; Dowell et al., 2016; Dowell et al., 2018). Extreme variation in coagulotoxic activity is a trait seen in the Asian pit-vipers of the genera Deinagkistrodon (Debono et al., 2019a), Gloydius (Debono et al., 2019b), Protobothrops (Debono et al., 2019c), and Trimeresurus (Debono et al., 2019d; Bourke et al., 2020). Among these genera, Gloydius is likely the closest relative to the American pitvipers. Combined with the lack of marked coagulotoxicity in the genus Agkistrodon (Lomonte et al., 2014; Nielsen et al., 2019), which is commonly retrieved as the sister group to the rattlesnakes (Wüster et al., 2008; Castoe et al., 2009; Alencar et al., 2016), this pattern emphasizes the need for further research on the evolution of coagulotoxins and their biological function in New World pitvipers and the selection pressures leading to such dynamic convergent amplifications of ancestral traits.
Our ancestral state reconstruction further shows that considerable differences in plasma and fibrinogen clotting can be observed between closely related lineages, sister species, and even at the intraspecific level (Figs. 1 and 2). This corroborates the findings reported by Mackessy (2008) and Gibbs et al. (2013) whereby venom composition across the rattlesnake tree was only weakly linked to phylogeny. The case of C. catalinensis, whose markedly pseudo-procoagulant venom phenotype contrasts with the anticoagulant action seen in its sister species C. atrox and C. ruber, is noteworthy. C. catalinensis is endemic to the island of Santa Catalina, where lack of natural predators has led to complete loss of the rattle in this species. The unique environmental conditions this species has adapted to could have driven prey-specific venom adaptations as well, a possibility that invokes further work. We highlight the cases of C. helleri, C. horridus and C. scutulatus ssp., which are known for considerable intraspecific variability regarding the neurotoxic PLA2 component of their venom (Bonilla et al., 1973; Glenn and Straight, 1978; Glenn et al., 1983; Weinstein et al., 1985; Massey et al., 2012; Sunagar et al., 2014; Rokyta et al., 2015; Dobson et al., 2018a; Strickland et al., 2018) and showed great variation in coagulotoxicity too in the present study.
Our observations possibly align with the findings of Yang et al. (2017) and Calvete et al. (2010), who reported a generally higher content of kallikrein-like serine proteases (many of which have pseudo-procoagulant activity) in neurotoxic individuals compared to non-neurotoxic ones in Gloydius intermedius and Crotalus simus, respectively. Rattlesnake species with evident disparities in venom composition and activity could therefore serve as ideal subjects to determine whether the same pattern is prevalent in this highly diverse clade. Another indication in this direction is the extremely short clotting time observed in C. tigris on plasma and especially fibrinogen (Figs. 1–2). Despite its unusually simple venom composition, this species is widely considered the most toxic Nearctic rattlesnake (Calvete et al., 2013). This is usually attributed to the high content of a neurotoxic PLA2 isoform in the venom, but our results indicate that coagulotoxicity (likely SVSP-driven) is a prominent feature of C. tigris as well. Interestingly, none of the species most closely related to C. tigris included in our sample (C. pyrrhus, C. stephensi, and C. adamanteus) came close to the exceptionally fast fibrinogen-clotting activity seen in this species, possibly suggesting different evolutionary trajectories. The recent sequencing of the C. tigris genome by Margres et al. (2021) corroborates our findings in showing that multiple SVSP genes are present and expressed in the venom glands of this species, which further underscores opportunities for novel research on such a peculiar venom.
A generally conserved pattern of strongly pseudo-procoagulant venom activity was also apparent in the Neotropical rattlesnakes (C. durissus ssp., C. mictlantecuhtli, C. tzabcan, and C. vegrandis, with the exception of C. culminatus), which represent a successful radiation in Central and South America and are known for the widespread presence of neurotoxic PLA2 isoforms in their venoms (Saravia et al., 2002; Calvete et al., 2010; Neri-Castro et al., 2019). Current knowledge indicates that SVSPs are the preponderant coagulotoxic venom component in this lineage, further supporting the possibility of a parallel evolutionary history with neurotoxic PLA2s. Whittington et al. (2018) framed this as a direct role of SVSPs in the evolution of Mojave toxin (MTX, a potently neurotoxic PLA2 from the venom of C. scutulatus) via cleaving of specific residues that opened new sites for the acquisition of novel toxic activities in the snake venom phospholipase family. While our observations on a limited sample do not allow for conclusive statements on the matter, the results presented herein warrant further investigation on the shared evolutionary history of serine proteases and phospholipases in rattlesnake venom in accordance with previous studies.
Other clades displayed a homogeneous venom profile that is likely ancestral to their lineage. A prime example is the Western rattlesnake complex, which includes seven species (C. viridis, C. oreganus, C. helleri, C. cerberus, C. concolor, C. lutosus, and C. abyssus) and is one of the most researched and debated rattlesnake clades in terms of systematics and biology (Pook et al., 2000; Ashton and De Queiroz, 2001; Douglas et al., 2002; Davis, 2016). While significant differences in venom composition are well documented within the complex (Mackessy, 2010; Saviola et al., 2015), our findings reveal a consistent anticoagulant action for the entire group (Fig. 1). This observation underlines the need for assays of venom activity to complement proteomic-centered findings, as -omics approaches only reveal the protein composition of venoms but does not indicate function as each toxin class may be multi-functional.
It is important to note that our findings only concern the coagulotoxic venom components, which do not amount to the total of the venom proteome in these snakes. Furthermore, our sample size for the Western rattlesnake complex only consisted of one or two representatives per species, the only exception being C. viridis itself with four samples and C. helleri with seven. The latter species displayed strikingly variable activities in our analysis, ranging from marked anticoagulant to strongly pseudo-procoagulant across individuals. Such dynamic variations have been documented previously in C. helleri (Sunagar et al., 2014), although the Factor X activating feature attributed to at least some populations (Denson et al., 1972; Suntravat et al., 2010) were not observed in the present work. Thus, we recommend further research on coagulotoxicity in these snakes to include multiple samples from various localities across the range of each species. Such studies should also test for ontogenetic variations in coagulotoxicity, as extreme variations have already been noted for at least one species, with C. culminatus being potently procoagulant through Factor X activation as neonates, but net anticoagulant in activity as adults (Seneci et al., 2021).
The evolutionary pattern behind such a diverse array of venom compositions and activities within and between species has been the object of extensive research. For instance, Giorgianni et al. (2020) selected rattlesnakes as model organisms to demonstrate how SVMPs evolved by neofunctionalization of pre-existent proteins via gene duplication and/or loss. A similar endeavor was undertaken by Gibbs and Rossiter (2008) and Dowell et al. (2018) to describe the evolution of PLA2 genes in Sistrurus and Crotalus, respectively. Schield et al. (2019a) reported that venom genes are grouped into several independent clusters along them genome of C. viridis, while Margres et al. (2021) showed how the remarkably simple venom phenotype observed in C. tigris actually arose from a complex genotype following several events of gene duplication and loss. Taken together, these observations are consistent with the well-supported hypothesis that describes gene duplication and subsequent neofunctionalization in the venom gland as the main genetic mechanism underlying venom evolution in snakes (Fry, 2005; Fry et al., 2009). Further research into snake genome sequencing is required to test whether this results in differences in mutation rates and evolvability of venom genes between different snake clades. Furthermore, the complex evolutionary dynamics of rattlesnakes- exemplified by the widespread occurrence of incomplete lineage sorting, introgression, and incipient speciation across the clade (Anderson and Greenbaum, 2012; Schield et al., 2015; Davis, 2016; Schield et al., 2019b; Watson et al., 2019; Carbajal-Márquez et al., 2020) - are likely to be reflected in such a highly evolvable and positively selected phenotypic trait as venom (Clark, 2016; Schuett et al., 2016; Rokyta et al., 2011; Schield et al., 2017).
Several studies have highlighted how venom studies cannot neglect ecology and natural history as venom is a functional adaptation that can only be comprehensively understood in a broader biological and ecological context (Lyons et al., 2020; Murray et al., 2020). Rattlesnakes are a prime example of this, as they display remarkably high genetic and species diversity and have successfully colonized a variety of ecosystems throughout their vast range. Specialization of venom activity towards a specific prey item is documented for numerous species such as C. pricei (Grabowsky and Mackessy, 2019), C. helleri (Mackessy, 1988; Holding et al., 2016; Holding et al., 2018; Gibbs et al., 2020), C. lepidus, C. willardi (Holycross et al., 2012; Martínez-Romero et al., 2013; Saviola et al., 2017) and Sistrurus as a whole (Sanz et al., 2006; Gibbs and Mackessy, 2009; Gibbs et al., 2011; Gibbs et al., 2013). Our findings did reveal widespread intraspecific and/or intra-lineage variation in venom activity in many clades (Figs. 1 & 2), although it must be emphasized that we performed our analysis entirely on human plasma and fibrinogen when several rattlesnakes specialize on non-mammalian prey whose physiology may be radically different from ours. We are indeed undertaking such followup studies examining taxon-specific effects upon coagulation. Aside from prey specificity, other diet-related factors can influence rattlesnake venom activity. Several studies have proposed that high levels of tissue-degrading toxins such as metalloproteases play an important role in digestion by initiating the breakdown of tissues even before the prey item is ingested (Klauber, 1972; Thomas and Pough, 1979; Mackessy, 1988; Mackessy, 2008). However, the digestive function of venom was disputed by LaBonte et al. (2011) in light of lack of differences in digestive performance between high- and low-SVMP venoms (i.e. Type II and Type I, respectively) in C. helleri.
While predation-related selection pressures have arguably received the most consideration in terms of research on snake venom evolution, the impacts of other factors on the dynamics of venom should not be underestimated (Sasa, 1999). This likely holds true for rattlesnakes as well. In fact, Strickland et al. (2018) and Zancolli et al. (2019) report a significant influence of climatic variables such as temperature and precipitation on venom composition in C. scutulatus scutulatus, even greater in importance than dietary preferences according to the latter study. Furthermore, Saviola et al. (2017) found that although prey specificity certainly was an important factor in explaining variation among subspecies of C. lepidus and C. willardi, temperature also played a considerable role. Our results are based on a sample too limited in size and diversity of locales to draw conclusions on whether any specific environmental factors have influenced venom activity in the analyzed species. Nonetheless, it is reasonable to believe that synthesis, storage, and action of certain proteins and/or toxin classes might be either facilitated or hampered by local temperature conditions. Individual venom variation at the intraspecific level not directly traceable to any specific driver is also widely documented in snakes, including rattlesnakes (Gregory-Dwyer et al., 1986; Chippaux et al., 1991; Smiley-Walters et al., 2019). Prey escape potential is another driver of venom variation. For example, lowland populations of C. helleri that feed in open scrub habitats where prey tracking is not problematic have a radically different venom phenotype than populations living in highlands characterized by abundant cracked rocks, where prey escape potential is high (Sunagar et al., 2014).
In conclusion, our study is the most comprehensive effort to date in investigating the evolutionary history and biology of rattlesnake venom coagulotoxins. More specifically, our observations support the findings of Mackessy (2008) and Gibbs et al. (2013) in that variation in coagulotoxic activity across the clade is often not related to phylogeny. Thus, it is likely that diet-related selection pressures and environmental factors including prey escape potential are at least as influential as phylogenetic relationships in shaping the evolution of rattlesnake venom activity. Furthermore, dynamic variation in clotting speed and/or clotting strength across taxa that are usually lumped into Type I or Type II categories is reported, indicating that this broad classification scheme overshadows differences not related to the two activities which form its basis, namely presynaptic neurotoxicity by PLA2 isoforms and extracellular matrix destruction by metalloproteases. This study showed that such actions upon the blood may be found in either Type I or Type II venoms and this classification scheme is limited in its utility when studying venom effects on coagulation. We are aware of the limitations affecting the current study, particularly low sample size for several of the species hereby examined and potential bias resulting from the use of human plasma and fibrinogen, which is useful for predicting potential clinical effects, rather than blood from animals more akin to common rattlesnake prey items (e.g. rodents, lizards) which would be essential for reconstructing evolutionary shaping pressures. Such natural history variables will be addressed in future studies. Nonetheless, we hope our results will provide valuable input for further research on specific clades and species whereby the influence of geographical and environmental drivers would be explored in greater detail. To this end, we point out the need to expand our scarce knowledge of the natural history and biology of many rattlesnake species to gain a comprehensive understanding of venom as a functional trait that evolves in response to biological and ecological selection pressures. Such dynamic variation in venom phenotype has immediate, tangible, real-world implications, as they directly influence clinically relevant pathophysiological effects and the relative efficacy of antivenom treatments as divergent effects upon the blood are caused by divergent toxin types, that may in turn be differentially neutralisesd by antivenom.
Acknowledgments
This study was funded by LUF Internationaal Studie Fonds (LISF), grant number L19104-1-45 and a LUSTRA+ scholarship to LS. This work was also funded by an Australian Research Council Discovery Project DP190100304 to BGF. We thank Dr. Jeroen Kool and Julien Slagboom from the Vrije Universiteit Amsterdam for helping us in preparing the venoms.
Footnotes
Declaration of competing interest
The authors declare no conflict of interest.
References
- Alencar LRV, Quental TB, Grazziotin FG, Alfaro ML, Martins M, Venzon M, Zaher H, 2016. Diversification in vipers: phylogenetic relationships, time of divergence and shifts in speciation rates. Mol. Phylogenet. Evol 105, 50–62. [DOI] [PubMed] [Google Scholar]
- Alencar LR, Martins M, Greene HW, 2018. Evolutionary history of vipers. ELS 1–10. [Google Scholar]
- Anderson CG, Greenbaum E, 2012. Phylogeography of northern populations of the black-tailed rattlesnake (Crotalus molossus Baird and Girard, 1853), with the revalidation of C. ornatus Hallowell, 1854. Herpetol. Monogr 26 (1), 19–57. [Google Scholar]
- Ashton KG, De Queiroz A, 2001. Molecular systematics of the western rattlesnake, Crotalus viridis (Viperidae), with comments on the utility of the D-loop in phylogenetic studies of snakes. Mol. Phylogenet. Evol 21 (2), 176–189. [DOI] [PubMed] [Google Scholar]
- Barlow A, Pook CE, Harrison RA, Wüster W, 2009. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. Biol. Sci. B: Biological Sciences 276 (1666), 2443–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaman KR, Hayes WK, 2008. Rattlesnakes: research trends and annotated checklist. In: Hayes WK, Beaman KR, Cardwell MD, Bush SP (Eds.), The Biology of Rattlesnakes. Loma Linda University Press, Loma Linda, CA, pp. 5–16. [Google Scholar]
- Blair C, Sánchez-Ramírez S, 2016. Diversity-dependent cladogenesis throughout western Mexico: evolutionary biogeography of rattlesnakes (Viperidae: Crotalinae: Crotalus and Sistrurus). Mol. Phylogenet. Evol 97, 145–154. [DOI] [PubMed] [Google Scholar]
- Bonilla CA, Faith MR, Minton SA, 1973. L-amino acid oxidase, phosphodiesterase, total protein and other properties of juvenile timber rattlesnake (C. h. horridus) venom at different stages of growth. Toxicon 11 (3), 301–302. [DOI] [PubMed] [Google Scholar]
- Bourke LA, Youngman NJ, Zdenek CN, op den Brouw B, Violette A, Fourmy R, Fry BG, 2020. Trimeresurus albolabris snakebite treatment implications arising from ontogenetic venom comparisons of anticoagulant function, and antivenom efficacy. Toxicol. Lett 327 (2–8). [DOI] [PubMed] [Google Scholar]
- Bryson RW, Murphy RW, Graham MR, Lathrop A, Lazcano D, 2011a. Ephemeral Pleistocene woodlands connect the dots for highland rattlesnakes of the Crotalus intermedius group. J. Biogeogr 38 (12), 2299–2310. 10.1111/j.1365-2699.2011.02565.x. [DOI] [Google Scholar]
- Bryson RW, Murphy RW, Lathrop A, Lazcano-Villareal D, 2011b. Evolutionary drivers of phylogeographical diversity in the highlands of Mexico: A case study of the Crotalus triseriatus species group of montane rattlesnakes. J. Biogeogr 38 (4), 697–710. [Google Scholar]
- Calvete JJ, Sanz L, Cid P, De La Torre P, Flores-Díaz M, Dos Santos MC, Gutíerrez JM, 2010. Snake venomics of the Central American Rattlesnake Crotalus simus and the South American Crotalus durissus complex points to neurotoxicity as an adaptive paedomorphic trend along Crotalus dispersal in South America. J. Proteome Res 9 (1), 528–544. [DOI] [PubMed] [Google Scholar]
- Calvete JJ, Pérez A, Lomonte B, Sánchez EE, Sanz L, 2013. Snake venomics of Crotalus tigris: the minimalist toxin arsenal of the deadliest neartic rattlesnake venom. J. Proteome Res 11 (2), 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JA, Lamar WW, Brodie ED, 2005. The Venomous Reptiles of the Western Hemisphere. Comstock Publishing Associates, Ithaca, NY. [Google Scholar]
- Carbajal-Márquez RA, Cedeño-Vázquez JR, Martínez-Arce A, Neri-Castro E, Machkour-M’Rabet SC, 2020. Accessing cryptic diversity in Neotropical rattlesnakes (Serpentes: Viperidae: Crotalus) with the description of two new species. Zootaxa 4729 (4), 451–481. [DOI] [PubMed] [Google Scholar]
- Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG, 2013. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol. Evol 28 (4), 219–229. [DOI] [PubMed] [Google Scholar]
- Castoe TA, Daza JM, Smith EN, Sasa MM, Kuch U, Campbell JA, Parkinson CL, 2009. Comparative phylogeography of pit-vipers suggests a consensus of ancient Middle American highland biogeography. J. Biogeogr 36 (1), 88–103. [Google Scholar]
- Chippaux JP, 2017. Incidence and mortality due to snakebite in the Americas. PLoS Negl. Trop. Dis 11 (6), e0005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippaux JP, Williams V, White J, 1991. Snake venom variability: methods of study. Toxicon 29 (11), 1279–1303. [DOI] [PubMed] [Google Scholar]
- Cipriani V, Debono J, Goldenberg J, Jackson TN, Arbuckle K, Dobson J, Fry BG, 2017. Correlation between ontogenetic dietary shifts and venom variation in Australian brown snakes (Pseudonaja). Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol 197, 53–60. [DOI] [PubMed] [Google Scholar]
- Clark RW, 2016. The hunting and feeding behavior of wild rattlesnakes. In: Schuett GW, Feldner MJ, Smith CF, Reiserer RS (Eds.), Rattlesnakes of Arizona. ECO Publishing, Rodeo, NM, pp. 93–118. [Google Scholar]
- Daltry JC, Wüster W, Thorpe RS, 1996. Diet and snake venom evolution. Nature 379 (6565), 537–540. [DOI] [PubMed] [Google Scholar]
- Davis MA, 2016. The western rattlesnake complex: 200 years of intrigue and change. In: Schuett GW, Feldner MJ, Smith CF, Reiserer RS (Eds.), Rattlesnakes of Arizona. ECO Publishing, Rodeo, NM, pp. 39–43. [Google Scholar]
- Debono J, Bos MHA, Coimbra F, Ge L, Frank N, Kwok HF, Fry BG, 2019a. Basal but divergent: clinical implications of differential coagulotoxicity in a clade of Asian vipers. Toxicol. in Vitro 58, 195–206. [DOI] [PubMed] [Google Scholar]
- Debono J, Bos MHA, Do MS, Fry BG, 2019b. Clinical implications of coagulotoxic variations in Mamushi (Viperidae: Gloydius) snake venoms. Comp. Biochem. Physiol. C Toxicol. Pharmacol 225, 108567. [DOI] [PubMed] [Google Scholar]
- Debono J, Bos MHA, Nouwens A, Ge L, Frank N, Kwok HF, Fry BG, 2019c. Habu coagulotoxicity: clinical implications of the functional diversification of Protobothrops snake venoms upon blood clotting factors. Toxicol. in Vitro 55, 62–74. [DOI] [PubMed] [Google Scholar]
- Debono J, Bos MH, Frank N, Fry B, 2019d. Clinical implications of differential antivenom efficacy in neutralising coagulotoxicity produced by venoms from species within the arboreal viperid snake genus Trimeresurus. Toxicol. Lett 316, 35–48. [DOI] [PubMed] [Google Scholar]
- Denson KWE, Russell FE, Almagro D, Bishop RC, 1972. Characterization of the coagulant activity of some snake venoms. Toxicon 10 (6), 557–562. [DOI] [PubMed] [Google Scholar]
- Dobson J, Yang DC, op den Brouw B, Cochran C, Huynh T, Kurrupu S, Fry BG, 2018a. Rattling the border wall: pathophysiological implications of functional and proteomic venom variation between Mexican and US subspecies of the desert rattlesnake Crotalus scutulatus. Comp. Biochem. Physiol. C Toxicol. Pharmacol 205, 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson J, Yang DC, Op den Brouw B, Cochran C, Huynh T, Kurrupu S, Fry BG, 2018b. Rattling the border wall: pathophysiological implications of functional and proteomic venom variation between Mexican and US subspecies of the desert rattlesnake Crotalus scutulatus. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol 205, 62–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas ME, Douglas MR, Schuett GW, Porras LW, Holycross AT, 2002. Phylogeography of the western rattlesnake (Crotalus viridis) complex, with emphasis on the Colorado Plateau. In: Schuett GW, Hoggren M, Douglas ME, Greene HW (Eds.), Biology of the Vipers. Eagle Mountain Publishing, pp. 11–50. [Google Scholar]
- Douglas MEMR, Douglas MEMR, Schuett GW, Porras LW, 2006. Evolution of rattlesnakes (Viperidae; Crotalus) in the warm deserts of western North America shaped by Neogene vicariance and Quaternary climate change. Mol. Ecol 15 (11), 3353–3374. [DOI] [PubMed] [Google Scholar]
- Dowell NL, Giorgianni MW, Kassner VA, Selegue JE, Sanchez EE, Carroll SB, 2016. The deep origin and recent loss of venom toxin genes in rattlesnakes. Curr. Biol 26 (18), 2434–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell NL, Giorgianni MW, Griffin S, Kassner VA, Selegue JE, Sanchez EE, Carroll SB, 2018. Extremely divergent haplotypes in two toxin gene complexes encode alternative venom types within rattlesnake species. Curr. Biol 28 (7), 1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durban J, Sanz L, Trevisan-Silva D, Neri-Castro E, Alagon A, Calvete JJ, 2017. Integrated venomics and venom gland transcriptome analysis of juvenile and adult mexican rattlesnakes Crotalus simus, C. tzabcan, and C. culminatus revealed miRNA-modulated ontogenetic shifts. J. Proteome. Res 16 (9), 3370–3390. [DOI] [PubMed] [Google Scholar]
- Fry BG, 2005. From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Re 15 (3), 403–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry BG, 2015. Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery. Oxford University Press, New York, NY: 10016. [Google Scholar]
- Fry BG, Scheib H, van der Weerd L, Young B, McNaughtan J, Ryan Ramjan SF, Norman JA, 2008. Evolution of an arsenal: structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol. Cell. Proteomics 7 (2), 215–246. [DOI] [PubMed] [Google Scholar]
- Fry BG, Roelants K, Champagne DE, Scheib H, Tyndall JDA, King GF, de la Vega RCR, 2009. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu. Rev. Genomics Hum. Genet 10 (1), 483–511. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Mackessy SP, 2009. Functional basis of a molecular adaptation: prey-specific toxic effects of venom from Sistrurus rattlesnakes. Toxicon 53 (6), 672–679. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Rossiter W, 2008. Rapid evolution by positive selection and gene gain and loss: PLA 2 venom genes in closely related Sistrurus rattlesnakes with divergent diets. J. Mol. Evol 66 (2), 151–166. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Sanz L, Chiucchi JE, Farrell TM, Calvete JJ, 2011. Proteomic analysis of ontogenetic and diet-related changes in venom composition of juvenile and adult Dusky Pigmy rattlesnakes (Sistrurus miliarius barbouri). J. Proteom 74 (10), 2169–2179. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Sanz L, Sovic MG, Calvete JJ, 2013. Phylogeny-based comparative analysis of venom proteome variation in a clade of rattlesnakes (Sistrurus sp. ). PLoS One 8 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs HL, Sanz L, Pérez A, Ochoa A, Hassinger AT, Holding ML, Calvete JJ, 2020. The molecular basis of venom resistance in a rattlesnake-squirrel predator-prey system. Mol. Ecol 29 (15), 2871–2888. [DOI] [PubMed] [Google Scholar]
- Giorgianni MW, Dowell NL, Griffin S, Kassner VA, Selegue JE, 2020. The origin and diversification of a novel protein family in venomous snakes. P. Natl. Acad. Sci 117 (20), 10911–10920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn JL, Straight R, 1978. Mojave rattlesnake Crotalus scutulatus scutulatus venom: variation in toxicity with geographical origin. Toxicon 16 (1), 81–84. [DOI] [PubMed] [Google Scholar]
- Glenn JL, Straight RC, 1989. Intergradation of two different venom populations of the Mojave rattlesnake (Crotalus scutulatus scutulatus) in Arizona . Toxicon 27 (4), 411–418. [DOI] [PubMed] [Google Scholar]
- Glenn JL, Straight RC, Wolfe MC, Hardy DL, 1983. Geographical variation in Crotalus scutulatus scutulatus (Mojave rattlesnake) venom properties. Toxicon 21 (1), 119–130. [DOI] [PubMed] [Google Scholar]
- Gloyd HK, 1940. The rattlesnakes. Genera Sistrurus and Crotalus: a study in zoogeography and evolution. Spec. Publ. Chicago Acad. Sci 4, 1–270. [Google Scholar]
- Grabowsky ER, Mackessy SP, 2019. Predator-prey interactions and venom composition in a high elevation lizard specialist, Crotalus pricei (Twin-spotted Rattlesnake). Toxicon 170, 29–40. [DOI] [PubMed] [Google Scholar]
- Gregory-Dwyer VM, Egen NB, Bosisio AB, Righetti PG, Russell FE, 1986. An isoelectric focusing study of seasonal variation in rattlesnake venom proteins. Toxicon 24 (10), 995–1000. [DOI] [PubMed] [Google Scholar]
- Gutíerrez JM, Rucavado A, Escalante T, Díaz C, 2005. Hemorrhage induced by snake venom metalloproteinases: biochemical and biophysical mechanisms involved in microvessel damage. Toxicon 45 (8), 997–1011. [DOI] [PubMed] [Google Scholar]
- Gutíerrez JM, Escalante T, Rucavado A, Herrera C, Fox JW, 2016. A comprehensive view of the structural and functional alterations of extracellular matrix by snake venom metalloproteinases (SVMPs): novel perspectives on the pathophysiology of envenoming. Toxins 8 (10), 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendon RA, Fraenkel-Conrat H, 1971. Biological roles of the two components of crotoxin. P. Natl. Acad. Sci 68 (7), 1560–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding ML, Biardi JE, Gibbs HL, 2016. Coevolution of venom function and venom resistance in a rattlesnake predator and its squirrel prey. Proc. Biol. Sci. B: Biological Sciences 283 (1829). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding ML, Margres MJ, Rokyta DR, Gibbs HL, 2018. Local prey community composition and genetic distance predict venom divergence among populations of the northern Pacific rattlesnake (Crotalus oreganus). J. Evol. Biol 31 (10), 1513–1528. [DOI] [PubMed] [Google Scholar]
- Holycross AT, Painter CW, Prival DB, Swann DE, Michael J, Edwards T, Schroff MJ, 2012. Diet of Crotalus lepidus klauberi (banded rock rattlesnake). J. Herpetol 36 (4), 589–597. [Google Scholar]
- Jackson TNW, Koludarov I, Ali SA, Dobson J, Zdenek CN, Dashevsky D, Fry BG, 2016. Rapid radiations and the race to redundancy: an investigation of the evolution of Australian elapid snake venoms. Toxins 8 (11), 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauber LM, 1972. Rattlesnakes: Their Habits, Life Histories, and Influence on Mankind, 2nd ed. University of California Press, Berkeley, CA. [Google Scholar]
- Kraus F, Mink DG, Brown WM, 1996. Crotaline intergeneric relationships based on mitochondrial DNA sequence data. Copeia 1996 (4), 763–773. [Google Scholar]
- LaBonte JP, Welch JC, Suarez RK, 2011. Digestive performance in neonatal Southern Pacific rattlesnakes (Crotalus oreganus helleri). Can. J. Zool 89 (8), 705–713. [Google Scholar]
- Lomonte B, Tsai WC, Ureña-Diaz JM, Sanz L, Mora-Obando D, Sánchez EE, Calvete JJ, 2014. Venomics of New World pit vipers: genus-wide comparisons of venom proteomes across Agkistrodon. J. Proteome 96, 103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons K, Dugon MM, Healy K, 2020. Diet breadth mediates the prey specificity of venom potency in snakes. Toxins 12 (2), 15–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackessy SP, 1988. Venom ontogeny in the Pacific rattlesnakes Crotalus viridis helleri and C. v. oreganus. Copeia 1988 (1), 92–101. [Google Scholar]
- Mackessy SP, 2008. Venom composition in rattlesnakes: trends and biological significance. In: Hayes WK, Beaman KR, Cardwell MD, Bush SP (Eds.), The Biology of Rattlesnakes. Loma Linda University Press, Loma Linda, CA, pp. 495–510. [Google Scholar]
- Mackessy SP, 2010. Evolutionary trends in venom composition in the Western Rattlesnakes (Crotalus viridis sensu lato): toxicity vs. tenderizers. Toxicon 55 (8), 1463–1474. [DOI] [PubMed] [Google Scholar]
- Mackessy SP, Williams K, Ashton KG, 2003. Ontogenetic variation in venom composition and diet of Crotalus oreganus concolor: a case of venom paedomorphosis? Copeia 2003 (4), 769–782. [Google Scholar]
- Margres MJ, Bigelow AT, Lemmon EM, Lemmon AR, Rokyta DR, 2017. Selection to increase expression, not sequence diversity, precedes gene family origin and expansion in rattlesnake venom. Genetics 206 (3), 1569–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres MJ, Rautsaw RM, Strickland JL, Mason AJ, Schramer TD, Hofmann EP, Parkinson CL, 2021. The Tiger Rattlesnake genome reveals a complex genotype underlying a simple venom phenotype. Proc. Nat. Acad. Sci 118 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markland FS, 1983. Rattlesnake venom enzymes that interact with components of the hemostatic system. Toxin Rev. 2 (2), 119–160. [Google Scholar]
- Martínez-Romero G, Rucavado A, Lazcano D, Gutíerrez JM, Borja M, Lomonte B, Zugasti-Cruz A, 2013. Comparison of venom composition and biological activities of the subspecies Crotalus lepidus lepidus, Crotalus lepidus klauberi and Crotalus lepidus morulus from Mexico. Toxicon 71, 84–95. [DOI] [PubMed] [Google Scholar]
- Massey DJ, Calvete JJ, Sánchez EE, Sanz L, Richards K, Curtis R, Boesen K, 2012. Venom variability and envenoming severity outcomes of the Crotalus scutulatus scutulatus (Mojave rattlesnake) from Southern Arizona. J. Proteome 75 (9), 2576–2587. [DOI] [PubMed] [Google Scholar]
- Meik J, Schuett GW, 2016. Structure, ontogeny, and evolutionary development of the rattlesnake rattle. In: Schuett GW, Feldner MJ, Smith CF, Reiserer RS (Eds.), Rattlesnakes of Arizona. ECO Publishing, Rodeo, NM, pp. 277–298. [Google Scholar]
- Murray KA, Martin G, T. I, 2020. Focus on snake ecology to fight snakebite. Lancet 395 (10220), e14. [DOI] [PubMed] [Google Scholar]
- Myers EA, Hickerson MJ, Burbrink FT, 2017. Asynchronous diversification of snakes in the North American warm deserts. J. Biogeogr 44 (2), 461–474. [Google Scholar]
- Neri-Castro E, Hernández-Dávila A, Olvera-Rodríguez A, Cardoso-Torres H, Bénard-Valle M, Bastiaans E, Alagón A, 2019. Detection and quantification of a β-neurotoxin (crotoxin homologs) in the venom of the rattlesnakes Crotalus simus, C. culminatus and C. tzabcan from Mexico. Toxicon X (2), 100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri-Castro EE, Bénard-Valle M, Gil G, Borja M, López de León J, Alagón A, 2020. Serpientes venenosas en México: revisión al estudio de los venenos, los antivenenos y la epidemiología. Revista Latinoamericana Herpetol. 3, 5–22. [Google Scholar]
- Nielsen VG, Frank N, Afshar S, 2019. De novo assessment and review of pan-American pit viper anticoagulant and procoagulant venom activities via kinetomic analyses. Toxins 11 (2), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson CL, 1999. Molecular systematics and biogeographical history of pit-vipers as determined by mitochondrial ribosomal DNA sequences. Copeia 1999 (3), 576–586. 10.2307/1447591. [DOI] [Google Scholar]
- Parkinson CL, Campbell JA, Chippindale PT, 2002. Multigene phylogenetic analysis of pit-vipers, with comments on their biogeography. In: Schuett GW, Hoggren M, Douglas ME, Greene HW (Eds.), Biology of the Vipers. Eagle Mountain Publishing, pp. 93–110. [Google Scholar]
- Place AJ, Abramson CI, 2004. A quantitative analysis of the ancestral area of rattlesnakes. J. Herpetol 38 (1), 152–156. [Google Scholar]
- Pook CE, Wüster W, Thorpe RS, 2000. Historical biogeography of the Western Rattlesnake (Serpentes: Viperidae: Crotalus viridis), inferred from mitochondrial DNA sequence information. Mol. Phylogenet. Evol 15 (2), 269–282. [DOI] [PubMed] [Google Scholar]
- Reiserer RS, 2016. Art and rattlesnakes: introduction to rattlesnakes in art. In: Schuett GW, Feldner MJ, Smith CF, Reiserer RS (Eds.), Rattlesnakes of Arizona. ECO Publishing, Rodeo, NM, pp. 21–38. [Google Scholar]
- Reiserer RS, Schuett GW, 2016. The origin and evolution of the rattlesnake rattle: misdirection, clarification, theory, and progress. In: Schuett GW, Feldner MJ, Smith CF, Reiserer RS (Eds.), Rattlesnakes of Arizona. ECO Publishing, Rodeo, NM, pp. 245–274. [Google Scholar]
- Revell LJ, 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol 3 (2), 217–223. [Google Scholar]
- Rokyta DR, Wray KP, Lemmon AR, Lemmon EM, Caudle SB, 2011. A high-throughput venom-gland transcriptome for the Eastern Diamondback Rattlesnake (Crotalus adamanteus) and evidence for pervasive positive selection across toxin classes. Toxicon 57 (5), 657–671. [DOI] [PubMed] [Google Scholar]
- Rokyta DR, Wray KP, McGivern JJ, Margres MJ, 2015. The transcriptomic and proteomic basis for the evolution of a novel venom phenotype within the Timber Rattlesnake (Crotalus horridus). Toxicon 98, 34–48. [DOI] [PubMed] [Google Scholar]
- Ruha AM, Kleinschmidt KC, Greene S, Spyres MB, Brent J, Wax P, Campleman S, 2017. The epidemiology, clinical course, and management of snakebites in the North American Snakebite Registry. J. Med. Toxicol 13, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, Lisle Gibbs H, Mackessy SP, Calvete JJ, 2006. Venom proteomes of closely related Sistrurus rattlesnakes with divergent diets. J. Proteome Res 5 (9), 2098–2112. [DOI] [PubMed] [Google Scholar]
- Saravia P, Rojas E, Arce V, Guevara C, López JC, Chaves E, Gutíerrez JM, 2002. Geographic and ontogenic variability in the venom of the neotropical rattlesnake Crotalus durissus: pathophysiological and therapeutic implications. Rev. Biol. Trop 50 (1), 337–346. [PubMed] [Google Scholar]
- Sasa M, 1999. Diet and snake venom evolution: can local selection alone explain intraspecific venom variation? Toxicon 37 (2), 249–252. [DOI] [PubMed] [Google Scholar]
- Saviola AJ, Pla D, Sanz L, Castoe TA, Calvete JJ, Mackessy SP, 2015. Comparative venomics of the Prairie Rattlesnake (Crotalus viridis viridis) from Colorado: identification of a novel pattern of ontogenetic changes in venom composition and assessment of the immunoreactivity of the commercial antivenom CroFab®. J. Proteomics 121, 28–43. [DOI] [PubMed] [Google Scholar]
- Saviola AJ, Gandara AJ, Bryson RW, Mackessy SP, 2017. Venom phenotypes of the Rock Rattlesnake (Crotalus lepidus) and the Ridge-nosed Rattlesnake (Crotalus willardi) from México and the United States. Toxicon 138, 119–129. [DOI] [PubMed] [Google Scholar]
- Schield DR, Card DC, Adams RH, Jezkova T, Reyes-Velasco J, Proctor FN, Castoe TA, 2015. Incipient speciation with biased gene flow between two lineages of the Western Diamondback Rattlesnake (Crotalus atrox). Mol. Phylogenet. Evol 83, 213–223. [DOI] [PubMed] [Google Scholar]
- Schield DR, Adams RH, Card DC, Perry BW, Pasquesi GM, Jezkova T, Castoe TA, 2017. Insight into the roles of selection in speciation from genomic patterns of divergence and introgression in secondary contact in venomous rattlesnakes. Ecol. Evol 7 (11), 3951–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schield DR, Card DC, Hales NR, Perry BW, Pasquesi GM, Blackmon H, Castoe TA, 2019a. The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 29 (4), 590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schield DR, Perry BW, Adams RH, Card DC, Jezkova T, Pasquesi GIM, Castoe TA, 2019b. Allopatric divergence and secondary contact with gene flow: a recurring theme in rattlesnake speciation. Biol. J. Linn. Soc 128 (1), 149–169. [Google Scholar]
- Schuett GW, Clark RW, Repp RA, Amarello M, Smith CF, Greene HW, 2016. Social behavior of rattlesnakes: a shifting paradigm. In: Schuett GW, Feldner MJ, Smith CF, Reiserer RS (Eds.), Rattlesnakes of Arizona. ECO Publishing, Rodeo, NM, pp. 161–242. [Google Scholar]
- Seneci L, Zdenek CN, Chowdhury A, Rodrigues CFB, Neri-Castro E, Bénard-Valle M, Alagón A, Fry BG, 2021. A clot twist: extreme variations in coagulotoxicity mechanisms in Mexican neotropical rattlesnake venoms. Front. Immunol 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setser K, Mociño-Deloya E, Meik MJ, 2011. Rattle button loss in juvenile Ridge-nosed Rattlesnakes (Crotalus willardi): a novel mechanism for the developmental delay of the rattle. J. Herpetol 45, 333–335. [Google Scholar]
- Smiley-Walters SA, Farrell TM, Lisle Gibbs H, 2019. High levels of functional divergence in toxicity towards prey among the venoms of individual pigmy rattlesnakes. Biol. Lett 15 (2), 10–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland JL, Mason AJ, Rokyta DR, Parkinson CL, 2018. Phenotypic variation in Mojave rattlesnake (Crotalus scutulatus) venom is driven by four toxin families. Toxins 10 (4), 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagar K, Undheim EAB, Scheib H, Gren ECK, Cochran C, Person CE, Fry BG, 2014. Intraspecific venom variation in the medically significant Southern Pacific Rattlesnake (Crotalus oreganus helleri): biodiscovery, clinical and evolutionary implications. J. Proteome 99, 68–83. [DOI] [PubMed] [Google Scholar]
- Suntravat M, Nuchprayoon I, Pérez JC, 2010. Comparative study of anticoagulant and procoagulant properties of 28 snake venoms from families Elapidae, Viperidae, and purified Russell’s viper venom-factor X activator (RVV-X). Toxicon 56 (4), 544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RG, Pough FH, 1979. The effect of rattlesnake venom on digestion of prey. Toxicon 17 (3), 221–228. [DOI] [PubMed] [Google Scholar]
- Whittington AC, Mason AJ, Rokyta DR, 2018. A single mutation unlocks cascading exaptations in the origin of a potent pitviper neurotoxin. Mol. Biol. Evol 35 (4), 887–898. [DOI] [PubMed] [Google Scholar]
- Watson JA, Spencer CL, Schield DR, Butler BO, Smith LL, Flores-Villela O, Meik JM, 2019. Geographic variation in morphology in the Mohave Rattlesnake (Crotalus scutulatus Kennicott 1861) (Serpentes: Viperidae): implications for species boundaries. Zootaxa 4683 (1), 129–143. [DOI] [PubMed] [Google Scholar]
- Weinstein SA, Minton SA, Wilde CE, 1985. The distribution among ophidian venoms of a toxin isolated from the venom of the Mojave rattlesnake (Crotalus scutulatus scutulatus). Toxicon 23 (5), 825–844. [DOI] [PubMed] [Google Scholar]
- Wüster W, 2016. Recent developments in rattlesnake phylogenetics, phylogeography and species delimitation models. In: Schuett GW, Feldner MJ, Smith CF, Reiserer RS (Eds.), Rattlesnakes of Arizona. ECO Publishing, Rodeo, NM. [Google Scholar]
- Wüster W, Peppin L, Pook CE, Walker DE, 2008. A nesting of vipers: phylogeny and historical biogeography of the Viperidae (Squamata: Serpentes). Mol. Phylogenet. Evol 49 (2), 445–459. [DOI] [PubMed] [Google Scholar]
- Yang ZM, Guo Q, Ma ZR, Chen Y, Wang ZZ, Wang XM, Tsai IH, 2015. Structures and functions of crotoxin-like heterodimers and acidic phospholipases A2 from Gloydius intermedius venom: insights into the origin of neurotoxic-type rattlesnakes. J. Proteome 112, 210–223. [DOI] [PubMed] [Google Scholar]
- Yang ZM, Yu H, Liu ZZ, Pei JZ, Yang YE, Yan SX, Tsai IH, 2017. Serine protease isoforms in Gloydius intermedius venom: full sequences, molecular phylogeny and evolutionary implications. J. Proteome 164, 19–32. [DOI] [PubMed] [Google Scholar]
- Youngman NJ, Debono J, Dobson JS, Zdenek CN, Harris RJ, den Brouw B. op, Fry BG, 2019. Venomous landmines: clinical implications of extreme coagulotoxic diversification and differential neutralization by antivenom of venoms within the viperid snake genus Bitis. Toxins 11 (7), 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancolli G, Baker TG, Barlow A, Bradley RK, Calvete JJ, Carter KC, Wüster W, 2016. Is hybridization a source of adaptive venom variation in rattlesnakes? A test, using a Crotalus scutulatus × viridis hybrid zone in Southwestern New Mexico. Toxins 8 (6), 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancolli G, Calvete JJ, Cardwell MD, Greene HW, Hayes WK, Hegarty MJ, Wüster W, 2019. When one phenotype is not enough: divergent evolutionary trajectories govern venom variation in a widespread rattlesnake species. Proc. Biol. Sci. B: Biological Sciences 286 (1898), 20182735. [DOI] [PMC free article] [PubMed] [Google Scholar]


