Abstract
Mounting evidence has identified that impaired amyloid-β (Aβ) clearance might contribute to Alzheimer’s disease (AD) pathology. The lysosome-autophagy network plays an important role in protein homeostasis and cell health by removing abnormal protein aggregates via intracellular degradation. Therefore, stimulation of cellular degradative machinery for efficient removal of Aβ has emerged as a growing field in AD research. However, mechanisms controlling such pathways and drugs to promote such mechanisms are poorly understood. Aspirin is a widely used drug throughout the world and recent studies have identified a new function of this drug. At low doses, aspirin stimulates lysosomal biogenesis and autophagy to clear amyloid plaques in an animal model of AD. This review delineates such functions of aspirin and analyzes underlying mechanisms that involve peroxisome proliferator-activated receptor alpha (PPARα)-mediated transcription of transcription factor EB (TFEB), the master regulator of lysosomal biogenesis.
Keywords: Alzheimer’s disease, amyloid plaques, autophagy, lysosomal biogenesis, PPARα, TFEB
ALZHEIMER’S DISEASE
Alzheimer’s disease (AD), clinically characterized by progressive cognitive impairment, is the most common neurodegenerative disorder. At present, AD approximately affects 5.7 million Americans across all age groups. About 10% and total 5.5 million American people of age 65 and older have been reported to have AD, making it the fifth leading cause of death in older population. In the US, roughly two-thirds of the AD-affected individuals are women. It is estimated that by 2050, the number of individuals living with AD in the US alone will rise to 13.8 million. It was reported that between 2000 and 2015, mortality due to heart disease, stroke, and prostate cancer have reduced whereas mortality from AD has increased 123%.
At present, there is no effective treatment for preventing or halting the disease. In AD brain, hippocampus and the association cortices undergo atrophy which can be detected by MRI [1–4]. At the microscopic level, the major defining neuropathological features of AD are deposition of extracellular senile plaques, composed of toxic amyloid-β (Aβ) aggregates, and formation of intracellular neurofibrillary tangles originated from hyperphosphorylation of the microtubule-associated protein tau [5, 6]. The early onset familial forms of AD have genetic origins characterized by mutations in the gene encoding the amyloid precursor protein (APP), a neuronal trans-membrane protein, and the presenilins (PS1 and PS2), the catalytic subunit of the gamma secretase complex [7]. While the familial forms of AD are rare (1–5%), the major AD occurrences are sporadic in nature with etiology that still remains elusive.
Neuropathological Features of AD
Upon autopsy, significant hippocampal atrophy along with a distinctly dilated temporal horn region of the left ventricle constitutes substantial indication of AD which is further confirmed upon histological evidence of neuropathological lesions [8]. The two pathological alterations described by Dr. Alzheimer are still considered as the hallmarks of AD and are used as criteria for diagnosis. These major neuropathological characteristics of AD are the development of extracellular senile plaques containing Aβ and formation of intraneuronal neurofibrillary tangles [9]. Abnormal accumulation of Aβ (40 or 42) forms the senile plaques which can be broadly classified into two categories, diffuse and dense-core plaques, based on the morphological characteristic and staining using Thioflavin-S and Congo Red, dyes that specifically recognize the β-pleated sheet conformation [10].
Diffuse plaques are Thioflavin-S-negative, non-neuritic, and are frequently observed in the cognitively intact aged people. However, dense-core plaques, present in the brains of AD patients, are composed of fibrillar Aβ and stain positively for Thioflavin-S. These compact-core plaques are associated with dystrophic neurites or neuritic plaques, accumulation of reactive astrocytes and microglia surrounding the plaque [11, 12], neuronal degeneration, and synaptic loss [13, 14]. While deposition of amyloid plaques is believed to start before the signs of cognitive decline, neurofibrillary tangle development and neuronal and synaptic losses occur concomitantly with the cognitive impairment [10].
Proteolysis of amyloid-β protein precursor (AβPP) and generation of amyloid plaques
AβPP is a neuronal type I transmembrane protein, involved in physiological processes such as neuronal trafficking, neurotrophic pathways, cell adhesion, and signaling [15–17]. In the non-amyloidogenic pathway, on the membrane, proteolytic cleavage of AβPP within the Aβ domain by alpha secretases, ADAM10, ADAM9, and ADAM17 (ADAM: a disintegrin- and metalloproteinase) precludes Aβ formation and generates N-terminal sAβPPα (soluble AβPPα) fragment and membrane associated CTFα (C-terminal fragment α) (C83) [18, 19]. Alternatively, AβPP can be internalized into the early endosomes where it undergoes amyloidogenic processing. In the amyloidogenic pathway, sequential cleavage of AβPP by beta secretase BACE1 (β-site APP-cleaving enzyme 1) and gamma secretase complex generates Aβ. Cleavage of AβPP by BACE1 produces sAβPPβ, which is secreted and CTFβ fragment (C99), which remains membrane bound and contains the Aβ sequence. Further intramembrane cleavage of CTFβ by the gamma secretase complex containing presenilin (PS1 or 2), nicastrin, APH-1 (anterior pharynx defective-1), and PEN2 (presenilin enhancer 2) produces Aβ peptide comprised of 40 or 42 residues. Among these amyloid species, due to insolubility and a greater propensity to aggregate, Aβ42 is more toxic and abundantly found in the plaques [9, 10, 16]. Aggregation of Aβ peptides involves a nucleation and elongation phase which ultimately forms the Aβ protofibrils and fibrils [20–22].
Stimulation of lysosomal clearance as a therapeutic strategy in AD
Although AD involves a multifactorial etiology, the amyloid cascade hypothesis, proposed by Hardy and Higgins, is widely established as the underlying model of AD pathophysiology [23]. According to this hypothesis, deposition of Aβ is the initiating and driving event in AD pathogenesis and an imbalance between production and clearance of Aβ leads to other pathological events including neurofibrillary tangles, neuronal dysfunction, and dementia [23, 24]. Therefore, many strategies for development of novel therapeutics for AD are focused on targeting Aβ dyshomeostasis.
Emerging studies suggest that impaired clearance of Aβ is the underlying mechanism of the widespread sporadic AD which constitutes 95% of all AD cases [25]. Therefore, strategies targeted at effective clearance of Aβ from the brain have immense therapeutic implications for AD. Several studies have highlighted the potential of astrocytic lysosomal induction for efficient clearance of Aβ. Cultured adult astrocytes has been shown to migrate, bind, and degrade amyloid plaques in brain sections from an AD mouse model implicating inefficient astroglial clearance of Aβ in AD pathogenesis [26]. Further study indicated apolipoprotein E plays an important role in astrocyte-mediated Aβ clearance [27]. Another study demonstrated that attenuating astrocytic activation by deletion of genes encoding intermediate filament proteins, glial fibrillary acidic protein (GFAP) and vimentin, accelerates the amyloid plaque pathogenesis suggesting an important role of astrocyte activation in preventing AD pathogenesis [28]. Therefore, astrocytes play an important role in clearance of Aβ plaques in AD.
Attempts to enhance the lysosomal function and restore normal autophagy by modulating transcription factor EB (TFEB), the essential regulator of the lysosome system, have generated promising therapeutic results in rescuing the amyloid pathogenesis in AD. Enhancing lysosomal function with TFEB leads to increased lysosomal degradation of holo-AβPP in the neurons and thus reduces the amyloidogenic processing of AβPP and Aβ generation [29]. Recent studies have demonstrated that TFEB overexpression can alleviate AD pathology by regulation of the autophagy-lysosome pathway [30]. Targeted TFEB expression in astrocytes mediated by viral gene transfer promotes attenuation of the amyloid pathology by enhancing lysosomal biogenesis and facilitating Aβ uptake and lysosomal degradation by astrocytes [31].
ASPIRIN
Acetylsalicylic acid, commonly known as aspirin, is one of the most frequently used pharmaceutics in medical practice and is available over the counter [32]. As a member of the nonsteroidal anti-inflammatory drugs (NSAIDs) group, aspirin is known to exert its anti-inflammatory effects by inhibiting cycloxygenases and thereby suppressing the generation of proinflammatory molecules like prostaglandins [33]. Other than its extensive use as an analgesic and antipyretic, aspirin has also been demonstrated to have beneficial effects for atherosclerosis, cardiovascular diseases and several cancers [34–37]. Earlier studies have explored the neuroprotective effect of aspirin under different disease conditions. Aspirin was shown to have protective effects in an animal model of Parkinson’s disease, independent of its cycloxygenase inhibitory properties [38]. Recently we have seen that aspirin is capable of increasing the production of dopamine from dopaminergic neurons via upregulation of tyrosine hydroxylase [39]. In a previous study, we demonstrated that aspirin upregulates the expression of ciliary neurotrophic factor in astrocytes which could have beneficial role for remyelination in demyelinating disorders [40]. Accordingly,we have reported that low-dose aspirin reduces the disease process of experimental autoimmune encephalomyelitis, an animal model of the most common human demyelinating disease multiple sclerosis [41]. Memory enhancing effects of aspirin was observed in an AlCl3-induced mouse model of neurotoxicity [42].
Aspirin stimulates lysosomal biogenesis
One recent study highlights a new property of aspirin [43]. At low doses, aspirin increases lysosomal biogenesis in astrocytes and neurons as monitored by LysoTracker analysis and elevation of different lysosomal marker molecules (LAMP2, LIMP2, and NPC1) [43]. Transcription factor EB (TFEB), a member of the microphthalmia-transcription factor E (MiT/TFE) subfamily of basic helix-loop-helix factors, is considered as the master regulator of lysosomal biogenesis [44–46]. Aspirin also upregulates TFEB in brain cells in culture and in vivo [43]. Electron microscopic analyses show the presence of increased number of lysosomes as well as different stages of autophagic vesicles in aspirin-treated brain cells [43]. Low-dose aspirin augments the activity of lysosomal enzyme tri-peptidyl-peptidase 1 (TPP1), dysfunction of which causes late-infantile Batten disease or late-infantile neuronal ceroid lipofuscinosis (LINCL) [43], suggesting that aspirin may have therapeutic implications in LINCL. Aspirin treatment also upregulates the activity of CathepsinB, a cysteine protease, and CathepsinD, an aspartyl protease important for lysosomal proteolysis [43]. These results indicate that aspirin is capable of promoting lysosomal biogenesis and autophagy in brain cells.
PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR (PPAR)
PPARs are ligand-inducible transcription factors belonging to the class of nuclear hormone receptors that primarily act as lipid sensors [47, 48]. Three different isoforms of PPARs: PPARα, PPARβ/δ, and PPARγ, have distinct tissue expression patterns, physiological functions and ligand specificity [49].
The α isoform, PPARα, is principally involved in regulation of energy homeostasis of the cell through inducing fatty acid and cholesterol metabolism and decreasing serum triglyceride content [50, 51]. Prototype PPARα agonists include lipid-lowering fibrate drugs (gemfibrozil, fenofibrate), WY14643, and GW7647 [48].
The protein PPARα contains 466 amino acids and is comprised of following functional domains [52, 53]:
A zinc finger-containing highly conserved DNA binding domain that is responsible for binding to the conserved peroxisome proliferator response element (PPRE) site on the promoter of target gene
A hinge region
C-terminal ligand-binding domain (LBD) containing five helices (H-3, −5, −7, −11, −12) that form a large ligand binding pocket
E/F domain for dimerization with RXR and ligand-dependent transactivation of the receptor
N-terminal domain for ligand-independent receptor regulation
All PPARs control the expression of target genes via transcriptional regulation. In the cytosol, PPARα is kept in an inactive form by forming complex with heat-shock protein 90 (HSP-90) and hepatitis virus B-X-associated protein-2 (XAP-2) [48, 54, 55]. In the nucleus, PPARs form heterodimers with transcriptional partner retinoid-X-receptors (RXRs). The PPAR:RXR heterodimer binds to the promoter of the target gene at cis-acting regulatory sequence known as PPRE. The sequence of the canonical PPRE contains two direct repeats of AGGTCA, separated by a DR1 element [56, 57]. In the absence of stimulation by ligands, PPAR:RXR heterodimers are kept inactive via binding to corepressor molecules such as nuclear receptor co-repressor, silencing mediator for retinoid and thyroid hormone receptor (SMRT) which, via direct interaction with Sin3 complex, recruits a multicomponent repressor complex [58, 59]. Additionally, SMRT promotes the recruitment of histone deacetylases (HDACs) to the repressor complex. The corepressor complex containing HDACs suppresses the transcription of the target gene by causing histone deacetylation [60].
Upon ligand binding, a conformational change of the receptor leads to release of the co-repressors and facilitates recruitment of co-activators which enhance the transcription of target gene through histone acetylation/methylation and stabilization of basal transcription apparatus [57, 61]. Some of the coactivators associated with PPAR:RXR complex are CBP/P300, SRC, PGC-1α, SWI/SNF, PRIP/PIMT, etc.
ASPIRIN BINDS TO PPARα FOR ITS ACTIVATION
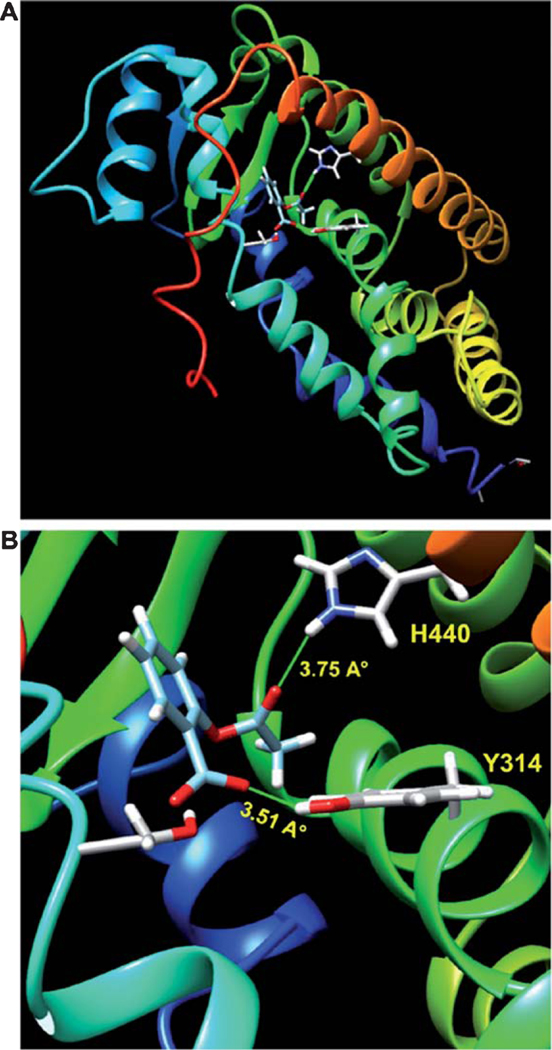
Although PPARα is a lipid-lowering transcription factor, we have demonstrated that PPARα is constitutively expressed in different brain regions and cells [62–66]. Surprisingly, PPARα stimulates lysosomal biogenesis in brain cells via direct transcriptional regulation of TFEB, the master regulator of lysosomal biogenesis [67]. Since aspirin stimulates lysosomal biogenesis [43], it makes sense to investigate whether aspirin induces the activation of PPARα. Increase in nuclear translocation, upregulation of DNA-binding activity and elevation in transcriptional venture of PPARα by aspirin suggest activation of PPARα by aspirin [43]. Moreover, aspirin induces PPRE-driven reporter activity in wild type and PPARβ (−/−), but not PPARα (−/−), astrocytes confirming activation of PPARα by aspirin [43]. How does aspirin activate PPARα? Recently, by thermal shift assay and TR-FRET analysis, Patel et al. [68] have shown direct binding of aspirin with PPARα. During ligand binding, a catalytic triad of H440, S280, and Y314 of PPARα LBD is involved in the complex formation of any ligand with PPARα [57,61]. In-silico analysis has identified that hydrogen atom of OH group located at the sidechain of Tyr314 residue forms a moderate H-bond with O3 of aspirin, whereas HE2 of H440 makes a weaker H-bond with O1 of aspirin (Fig. 1). Accordingly, we focused on the Y314 of PPARα LBD for interaction with aspirin [68]. This aspirin-to-PPARα LBD binding is further confirmed by site-directed mutagenesis as in-silico examination, thermal shift assay and TR-FRET analysis do not find any interaction of aspirin with mutated Y314D PPARα [68]. Aspirin also dose-dependently induces the luciferase reporter activity driven by wild type PPARα, but not by Y314D PPARα [68]. The anti-pyretic action of aspirin stems from its inhibition of cyclooxygenase 2 (COX2) on PGE1/PGE2 levels. However, celecoxib, ibuprofen, naproxen (potent inhibitors of COX1/2) display very weak interaction with PPARα [68], suggesting the specificity of this effect. Together, these results clearly indicate that aspirin binds to Y314 of PPARα ligand-binding domain for its activation.
Fig. 1.
In Silico docking analysis of aspirin in PPARα ligand-binding domain (LBD). A, B) A best-fit superposition of the LBD of PPARα (mouse) and aspirin was obtained with the help of SwissDock docking software and finally displayed in Chimera visualization interface. Ribbon representation of the complex has revealed that a catalytic triad of H440, S280, and Y314 is required for the complex formation of aspirin with PPARα. Interestingly, hydrogen atom of OH group located at the sidechain of Tyr314 residue forms a moderate H-bond with O3 of Aspirin, whereas HE2 of H440 makes a weaker H-bond with O1 of Aspirin.
ASPIRIN INCREASES TFEB AND LYSOSOMAL BIOGENESIS VIA PPARα
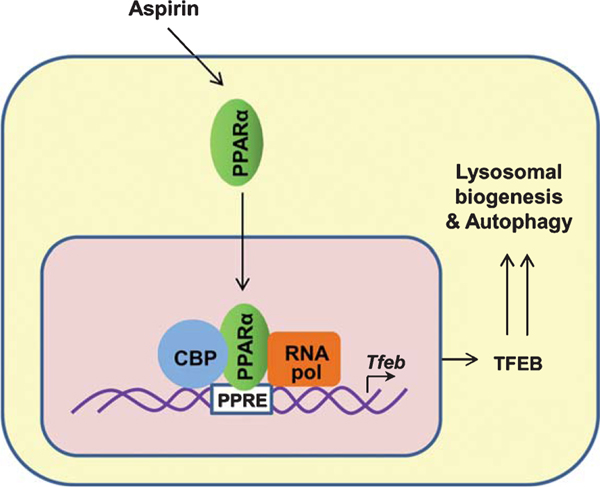
Since lysosomal biogenesis is controlled by PPARα, the promoter of Tfeb gene was analyzed and it was found to harbor a consensus PPRE site, 480 bp upstream of the transcription start site [67]. Accordingly, gemfibrozil, a prototype activator of PPARα, has been found to upregulate the transcription of Tfeb gene via PPARα [67]. Consistent to the activation of PPARα, aspirin induces TFEB-driven reporter activity in astrocytes via PPARα, but neither PPARβ nor PPARγ [43]. It is found that aspirin treatment stimulates the recruitment of PPARα, CBP, and RNA polymerase to the PPRE site on the TFEB promoter for its transcription (Fig. 2). Therefore, when the PPRE core sequence in the TFEB gene promoter is mutated, aspirin remains unable to induce mutated TFEB-driven reporter activity [43]. Consistent to the PPARα-dependent stimulation of TFEB by aspirin, this drug increases lysosomal biogenesis and autophagy in brain cells via PPAR (Fig. 2).
Fig. 2.
Transcriptional upregulation of Tfeb by aspirin via PPARα. Aspirin activates PPARα that binds to peroxisome proliferator response element (PPRE) present in Tfeb gene promoter and causes its transcription. Other members of the transcriptional complex are CREB-binding protein (CBP) and RNA polymerase II. TFEB is responsible for lysosomal biogenesis and autophagy.
ASPIRIN LOWERS AMYLOID PLAQUES VIA PPARα
Although AD involves a multifactorial pathophysiology, accumulation of Aβ is widely considered as the initiating and driving factor of AD pathology [23]. Therefore, lowering cerebral plaque load is an important area of research as it may have therapeutic implication for AD. Increase in lysosomal biogenesis and autophagy by aspirin in brain cells suggests possible beneficial effect of aspirin in this aspect. Accordingly, when FAD5X mice with established plaques are treated with aspirin, significant plaque clearance is seen from hippocampus and cortex [43]. Recent discovery has shed light on the mechanisms by which autophagy occurs [69]. Lysosomal adaptation under different physiological as well as pathological scenario is dependent on a regulatory gene network known as CLEAR (coordinated lysosomal expression and regulation), with the master regulator TFEB at its core [69, 70]. It is interesting to see that after oral administration, aspirin increases the level of TFEB in the hippocampus of 5XFAD mice.
Given the observation that aspirin can activate PPARα and thus drive TFEB mediated lysosomal enrichment, it is logical to explore if the activation of PPARα by aspirin is the underlying mechanism by which it exhibits the amyloid lowering effects. Findings that deletion of PPARα in FAD5X mice aggravates plaque pathology as compared to FAD5X mice and that aspirin lowers cerebral plaque load in FAD5X, but not FAD5X-PPARα (−/−), mice [43] clearly indicate an essential role of PPARα in plaque pathology and aspirin-mediated plaque lowering. Accordingly, aspirin also improves spatial learning and memory in 5XFAD mice in a PPARα-dependent manner [68].
WHAT DOES IT MEAN FOR AD PATIENTS?
Current state of knowledge goes both for beneficial and not favorable role of aspirin in AD. In the following discussion, we have tried to analyze such information with respect to PPARα-dependent plaque-lowering activity.
Evidence for a beneficial role of aspirin in AD
After a global, cross-sectional, and longitudinal (1991–2000) epidemiological analyses, one study [71] has reported that high-dose aspirin users exhibit lower prevalence of AD and better cognition than non-users.
In a study [72] to evaluate the association of anti-inflammatory drug use on the incidence of AD, it has been shown that aspirin can significantly lower the risk of AD.
Based on a systematic review and meta-analysis of observational studies published between 1966 and October 2002, Etminan et al. [73] have reported a protective effect of aspirin in AD patients.
According to a Nationwide Retrospective Cohort Study in Taiwan [74], a mean daily dosage of aspirin use within 40mg may decrease the risk of developing AD in patients with type 2 diabetes mellitus.
Evidence against a beneficial role of aspirin in AD
In the Reasons for Geographical and Racial Differences in Stroke (REGARDS) study, Kelly et al. [75] have seen no relationship between daily aspirin use and cognitive change over 2 to 6 years of follow-up after controlling for the impact of age.
Veronese et al. [76] has investigated whether low-dose aspirin (<300mg/d) can influence the onset of cognitive impairment or dementia in observational studies and improve cognitive test scores in randomized controlled trials in older participants without dementia. They have seen no evidence that low-dose aspirin delays in cognitive decline or dementia.
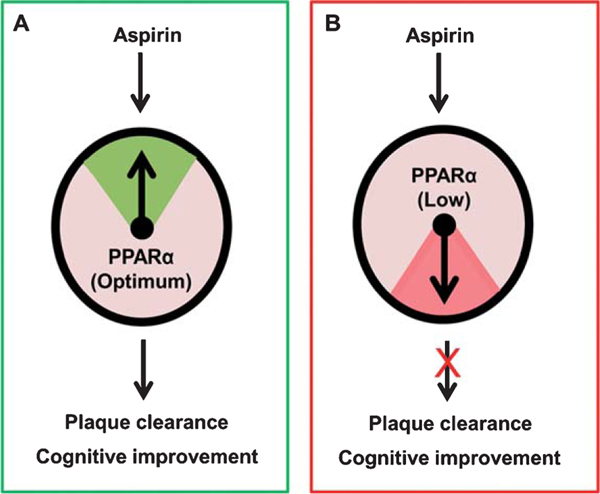
One of the possible explanations for this aspirin puzzle in AD could be that the level of PPARα might be less in the brains of older AD and dementia patients (Fig. 3). In addition to its central role in lipid metabolism, PPARα has also been established to regulate multiple physiological pathways [50, 65, 77] including lysosomal biogenesis [67, 78]. Since aspirin requires PPARα to stimulate lysosomal biogenesis and autophagy and clear amyloid plaques in AD mouse model, in the absence of a basal level of PPARα, aspirin may not exhibit an optimal effect on plaque lowering (Fig. 3). Mounting evidence also suggests that activation of PPARα improves memory and learning via upregulation of the CREB signaling pathway [63–65]. Accordingly, in another study [68], we have seen that aspirin is capable of stimulating hippocampal plasticity and improving memory and learning in an animal model of AD via PPARα. Taken together, aspirin requires PPARα for both plaque clearance and improvement in cognitive functions, two important stakeholders in AD therapy (Fig. 3).
Fig. 3.
PPARα controls aspirin’s ability to lower plaques and improve memory. A) When level of PPARα is optimum, aspirin treatment clears plaques and improves memory and learning. B) On the other hand, when PPARα level is low, aspirin treatment does not lead to plaque clearance and cognitive enhancement.
CONCLUSION
Currently, there is no valid option to reduce plaques and improve cognitive functions in AD patients. Therefore, it always stimulates enthusiasm and eagerness to learn the possibility that low dose of a household drug, aspirin, may clear plaques and boost memory in AD patients. If mouse results are translated, our aged medical companion may be repurposed to lower plaques and enhance memory in the most important age-related cognitive disorder via PPARα. However, since PPARα is the rate-limiting molecule (Fig. 3) in aspirin-mediated possible therapeutic efficacy in AD, future studies should be directed to understand the level of PPARα in AD patients and age-matched healthy individuals, which may help tailoring aspirin therapy inpatients with AD or mild cognitive impairment.
ACKNOWLEDGMENTS
This study was supported by a merit award from Veteran Affairs (I01BX002174), a grant (AG050431) from NIH and the Zenith Fellows Award (ZEN-17–438829) from Alzheimer’s Association.
Footnotes
Authors’ disclosures available online (https://www.j-alz.com/manuscript-disclosures/19–0586r1).
REFERENCES
- [1].Byun MS, Kim SE, Park J, Yi D, Choe YM, Sohn BK, Choi HJ, Baek H, Han JY, Woo JI, Lee DY, Alzheimer’s Disease Neuroimaging Initiative (2015) Heterogeneity of regional brain atrophy patterns associated with distinct progression rates in Alzheimer’s disease. PLoS One 10, e0142756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dickerson BC, Bakkour A, Salat DH, Feczko E, Pacheco J, Greve DN, Grodstein F, Wright CI, Blacker D, Rosas HD, Sperling RA, Atri A, Growdon JH, Hyman BT, Morris JC, Fischl B, Buckner RL (2009) The cortical signature of Alzheimer’s disease: regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb Cortex 19, 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Frisoni GB, Fox NC, Jack CR Jr., Scheltens P, Thompson PM (2010) The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol 6, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW (2003) Dynamics of gray matter loss in Alzheimer’s disease. J Neurosci 23, 994–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Querfurth HW, LaFerla FM (2010) Alzheimer’s disease. N Engl J Med 362, 329–344. [DOI] [PubMed] [Google Scholar]
- [6].Yoon SY, Kim DH (2016) Alzheimer’s disease genes and autophagy. Brain Res 1649, 201–209. [DOI] [PubMed] [Google Scholar]
- [7].Whyte LS, Lau AA, Hemsley KM, Hopwood JJ, Sargeant TJ (2017) Endo-lysosomal and autophagic dysfunction: a driving factor in Alzheimer’s disease? J Neurochem 140, 703–717. [DOI] [PubMed] [Google Scholar]
- [8].Perl DP (2010) Neuropathology of Alzheimer’s disease. Mt Sinai J Med 77, 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].LaFerla FM, Green KN, Oddo S (2007) Intracellular amyloid-beta in Alzheimer’s disease. Nat Rev Neurosci 8, 499–509. [DOI] [PubMed] [Google Scholar]
- [10].Serrano-Pozo A, Frosch MP, Masliah E, Hyman BT (2011) Neuropathological alterations in Alzheimer disease. Cold Spring Harb Perspect Med 1, a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Itagaki S, McGeer PL, Akiyama H, Zhu S, Selkoe D (1989) Relationship of microglia and astrocytes to amyloid deposits of Alzheimer disease. J Neuroimmunol 24, 173–182. [DOI] [PubMed] [Google Scholar]
- [12].Pike CJ, Cummings BJ, Cotman CW (1995) Early association of reactive astrocytes with senile plaques in Alzheimer’s disease. Exp Neurol 132, 172–179. [DOI] [PubMed] [Google Scholar]
- [13].Knowles RB, Wyart C, Buldyrev SV,Cruz L, Urbanc B, Hasselmo ME, Stanley HE, Hyman BT(1999) Plaque-induced neurite abnormalities: implications for disruption of neural networks in Alzheimer’s disease. Proc Natl Acad Sci U S A 96, 5274–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Masliah E, Mallory M, Hansen L, DeTeresa R, Alford M, Terry R (1994) Synaptic and neuritic alterations during the progression of Alzheimer’s disease. Neurosci Lett 174, 67–72. [DOI] [PubMed] [Google Scholar]
- [15].Hoareau C, Borrell V, Soriano E, Krebs MO, Prochiantz A, Allinquant B (2008) Amyloid precursor protein cytoplasmic domain antagonizes reelin neurite outgrowth inhibition of hippocampal neurons. Neurobiol Aging 29, 542–553. [DOI] [PubMed] [Google Scholar]
- [16].Nixon RA (2007) Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci 120, 4081–4091. [DOI] [PubMed] [Google Scholar]
- [17].Reinhard C, Hebert SS, De Strooper B (2005) The amyloid-beta precursor protein: integrating structure with biological function. EMBO J 24, 3996–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Asai M, Hattori C, Szabo B, Sasagawa N, Maruyama K, Tanuma S, Ishiura S (2003) Putative function of ADAM9, ADAM10, and ADAM17 as APP alpha-secretase. Biochem Biophys Res Commun 301, 231–235. [DOI] [PubMed] [Google Scholar]
- [19].Kojro E, Fahrenholz F (2005) The non-amyloidogenic pathway: structure and function of alpha-secretases. Subcell Biochem 38, 105–127. [DOI] [PubMed] [Google Scholar]
- [20].Lee CC, Nayak A, Sethuraman A, Belfort G, McRae GJ (2007) A three-stage kinetic model of amyloid fibrillation. Biophys J 92, 3448–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lomakin A, Chung DS, Benedek GB, Kirschner DA, Teplow DB (1996) On the nucleation and growth of amyloid beta-protein fibrils: detection of nuclei and quantitation of rate constants. Proc Natl Acad Sci U S A 93, 1125–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lomakin A, Teplow DB, Kirschner DA, Benedek GB (1997) Kinetic theory of fibrillogenesis of amyloid beta-protein. Proc Natl Acad Sci U S A 94, 7942–7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hardy JA, Higgins GA (1992) Alzheimer’s disease: the amyloid cascade hypothesis. Science 256, 184–185. [DOI] [PubMed] [Google Scholar]
- [24].Selkoe DJ, Hardy J (2016) The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med 8, 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mawuenyega KG, Sigurdson W, Ovod V, Munsell L, Kasten T, Morris JC, Yarasheski KE, Bateman RJ (2010) Decreased clearance of CNS beta-amyloid in Alzheimer’s disease. Science 330, 1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wyss-Coray T, Loike JD, Brionne TC, Lu E, Anankov R, Yan F, Silverstein SC, Husemann J (2003) Adult mouse astrocytes degrade amyloid-beta in vitro and in situ. Nat Med 9, 453–457. [DOI] [PubMed] [Google Scholar]
- [27].Koistinaho M, Lin S, Wu X, Esterman M, Koger D, Hanson J, Higgs R, Liu F, Malkani S, Bales KR, Paul SM (2004) Apolipoprotein E promotes astrocyte colocalization and degradation of deposited amyloid-beta peptides. Nat Med 10, 719–726. [DOI] [PubMed] [Google Scholar]
- [28].Kraft AW, Hu X, Yoon H, Yan P, Xiao Q, Wang Y, Gil SC, Brown J, Wilhelmsson U, Restivo JL, Cirrito JR, Holtzman DM, Kim J, Pekny M, Lee JM (2013) Attenuating astrocyte activation accelerates plaque pathogenesis in APP/PS1 mice. FASEB J 27, 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, Gonzales E, Tripoli DL, Czerniewski L, Ballabio A, Cirrito JR, Diwan A, Lee JM (2015) Neuronal-targeted TFEB accelerates lysosomal degradation of APP, reducing Abeta generation and amyloid plaque pathogenesis. J Neurosci 35, 12137–12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang YD, Zhao JJ (2015) TFEB participates in the abeta-induced pathogenesis of Alzheimer’s disease by regulating the autophagy-lysosome pathway. DNA Cell Biol 34, 661–668. [DOI] [PubMed] [Google Scholar]
- [31].Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, Gonzales E, Burchett JM, Schuler DR, Cirrito JR, Diwan A, Lee JM (2014) Enhancing astrocytic lysosome biogenesis facilitates Abeta clearance and attenuates amyloid plaque pathogenesis. J Neurosci 34, 9607–9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Green GA (2001) Understanding NSAIDs: from aspirin to COX-2. Clin Cornerstone 3, 50–60. [DOI] [PubMed] [Google Scholar]
- [33].Vane JR (1971) Inhibition of prostaglandin synthesis as a mechanism of action for aspirin-like drugs. Nat New Biol 231, 232–235. [DOI] [PubMed] [Google Scholar]
- [34].Berk M, Dean O, Drexhage H, McNeil JJ, Moylan S, O’Neil A, Davey CG, Sanna L, Maes M (2013) Aspirin: a review of its neurobiological properties and therapeutic potential for mental illness. BMC Med 11, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dai Y, Ge J (2012) Clinical use of aspirin in treatment and prevention of cardiovascular disease. Thrombosis 2012, 245037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Moyad MA (2001) An introduction to aspirin, NSAids, and COX-2 inhibitors for the primary prevention of cardiovascular events and cancer and their potential preventive role in bladder carcinogenesis: part II. Semin Urol Oncol 19, 306–316. [PubMed] [Google Scholar]
- [37].Rothwell PM, Wilson M, Price JF, Belch JF, Meade TW, Mehta Z (2012) Effect of daily aspirin on risk of cancer metastasis: a study of incident cancers during randomised controlled trials. Lancet 379, 1591–1601. [DOI] [PubMed] [Google Scholar]
- [38].Aubin N, Curet O, Deffois A, Carter C (1998) Aspirin and salicylate protect against MPTP-induced dopamine depletion in mice. J Neurochem 71, 1635–1642. [DOI] [PubMed] [Google Scholar]
- [39].Rangasamy SB, Dasarathi S, Pahan P, Jana M, Pahan K (2018) Low-dose aspirin upregulates tyrosine hydroxylase and increases dopamine production in dopaminergic neurons: implications for Parkinson’s disease. J Neuroimmune Pharmacol 14, 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Modi KK, Sendtner M, Pahan K (2013) Up-regulation of ciliary neurotrophic factor in astrocytes by aspirin: implications for remyelination in multiple sclerosis. J Biol Chem 288, 18533–18545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mondal S, Jana M, Dasarathi S, Roy A, Pahan K (2018) Aspirin ameliorates experimental autoimmune encephalomyelitis through interleukin-11-mediated protection of regulatory T cells. Sci Signal 11, eaar8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rizwan S, Idrees A, Ashraf M, Ahmed T (2016) Memory-enhancing effect of aspirin is mediated through opioid system modulation in an AlCl3-induced neurotoxicity mouse model. Exp Ther Med 11, 1961–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chandra S, Jana M, Pahan K (2018) Aspirin induces lysosomal biogenesis and attenuates amyloid plaque pathology in a mouse model of Alzheimer’s disease via PPARalpha. J Neurosci 38, 6682–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, Wollenberg AC, Di Bernardo D, Chan L, Irazoqui JE, Ballabio A (2013) TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 15, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A (2011) TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Settembre C, Fraldi A, Medina DL, Ballabio A (2013) Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Keller H, Mahfoudi A, Dreyer C, Hihi AK, Medin J, Ozato K,Wahli W (1993) Peroxisome proliferator-activated receptors and lipid metabolism. Ann N Y Acad Sci 684, 157–173. [DOI] [PubMed] [Google Scholar]
- [48].Reddy JK, Mannaerts GP (1994) Peroxisomal lipid metabolism. Annu Rev Nutr 14, 343–370. [DOI] [PubMed] [Google Scholar]
- [49].Zoete V, Grosdidier A, Michielin O (2007) Peroxisome proliferator-activated receptor structures: ligand specificity, molecular switch and interactions with regulators. Biochim Biophys Acta 1771, 915–925. [DOI] [PubMed] [Google Scholar]
- [50].Heneka MT, Landreth GE (2007) PPARs in the brain. Biochim Biophys Acta 1771, 1031–1045. [DOI] [PubMed] [Google Scholar]
- [51].Kummer MP, Heneka MT (2008) PPARs in Alzheimer’s disease. PPAR Res 2008, 403896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kersten S, Wahli W (2000) Peroxisome proliferator activated receptor agonists. EXS 89, 141–151. [DOI] [PubMed] [Google Scholar]
- [53].Motojima K (1993) Peroxisome proliferator-activated receptor (PPAR): structure, mechanisms of activation and diverse functions. Cell Struct Funct 18, 267–277. [DOI] [PubMed] [Google Scholar]
- [54].Roy A, Pahan K (2009) Gemfibrozil, stretching arms beyond lipid lowering. Immunopharmacol Immunotoxicol 31, 339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sumanasekera WK, Tien ES, Davis JW 2nd, Turpey R, Perdew GH, Vanden Heuvel JP (2003) Heat shock protein-90 (Hsp90) acts as a repressor of peroxisome proliferator-activated receptor-alpha (PPARalpha) and PPARbeta activity. Biochemistry 42, 10726–10735. [DOI] [PubMed] [Google Scholar]
- [56].Varanasi U, Chu R, Huang Q, Castellon R, Yeldandi AV, Reddy JK (1996) Identification of a peroxisome proliferator-responsive element upstream of the human peroxisomal fatty acyl coenzyme A oxidase gene. J Biol Chem 271, 2147–2155. [DOI] [PubMed] [Google Scholar]
- [57].Viswakarma N, Jia Y, Bai L, Vluggens A, Borensztajn J, Xu J, Reddy JK (2010) Coactivators in PPAR-regulated gene expression. PPAR Res 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chen JD, Evans RM (1995) A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377, 454–457. [DOI] [PubMed] [Google Scholar]
- [59].Desvergne B, Wahli W (1999) Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev 20, 649–688. [DOI] [PubMed] [Google Scholar]
- [60].Karagianni P, Wong J (2007) HDAC3: taking the SMRT-N-CoRrect road to repression. Oncogene 26, 5439–5449. [DOI] [PubMed] [Google Scholar]
- [61].Tetel MJ, Auger AP, Charlier TD (2009) Who’s in charge? Nuclear receptor coactivator and corepressor function in brain and behavior. Front Neuroendocrinol 30, 328–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Corbett GT, Gonzalez FJ, Pahan K (2015) Activation of peroxisome proliferator-activated receptor alpha stimulates ADAM10-mediated proteolysis of APP. Proc Natl Acad Sci U S A 112, 8445–8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Roy A, Jana M, Corbett GT, Ramaswamy S, Kordower JH, Gonzalez FJ, Pahan K (2013) Regulation of cyclic AMP response element binding and hippocampal plasticity-related genes by peroxisome proliferator-activated receptor alpha. Cell Rep 4, 724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Roy A, Kundu M, Jana M, Mishra RK, Yung Y, Luan CH, Gonzalez FJ, Pahan K (2016) Identification and characterization of PPARalpha ligands in the hippocampus. Nat Chem Biol 12, 1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Roy A, Pahan K (2015) PPARalpha signaling in the hippocampus: crosstalk between fat and memory. J Neuroimmune Pharmacol 10, 30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Roy A, Jana M, Kundu M, Corbett GT, Rangaswamy SB, Mishra RK, Luan CH, Gonzalez FJ, Pahan K (2015) HMG-CoA reductase inhibitors bind to PPARalpha to upregulate neurotrophin expression in the brain and improve memory in mice. Cell Metab 22, 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ghosh A, Jana M, Modi K, Gonzalez FJ, Sims KB, Berry-Kravis E, Pahan K (2015) Activation of peroxisome proliferator-activated receptor alpha induces lysosomal biogenesis in brain cells: implications for lysosomal storage disorders. J Biol Chem 290, 10309–10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Patel D, Roy A, Kundu M, Jana M, Luan CH, Gonzalez FJ, Pahan K (2018) Aspirin binds to PPARalpha to stimulate hippocampal plasticity and protect memory. Proc Natl Acad Sci U S A 115, E7408–E7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A (2009)A gene network regulating lysosomal biogenesis and function. Science 325, 473–477. [DOI] [PubMed] [Google Scholar]
- [70].Martini-Stoica H, Xu Y, Ballabio A, Zheng H (2016) The autophagy-lysosomal pathway in neurodegeneration: a TFEB perspective. Trends Neurosci 39, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Nilsson SE, Johansson B, Takkinen S, Berg S, Zarit S, McClearn G, Melander A (2003) Does aspirin protect against Alzheimer’s dementia? A study in a Swedish population-based sample aged>or=80 years. Eur J Clin Pharmacol 59, 313–319. [DOI] [PubMed] [Google Scholar]
- [72].Wang J, Tan L, Wang HF, Tan CC, Meng XF, Wang C, Tang SW, Yu JT (2015) Anti-inflammatory drugs and risk of Alzheimer’s disease: an updated systematic review and meta-analysis. J Alzheimers Dis 44, 385–396. [DOI] [PubMed] [Google Scholar]
- [73].Etminan M, Gill S, Samii A (2003) Effect of non-steroidal anti-inflammatory drugs on risk of Alzheimer’s disease: systematic review and meta-analysis of observational studies. BMJ 327, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Chang CW, Horng JT, Hsu CC, Chen JM (2016) Mean daily dosage of aspirin and the risk of incident Alzheimer’s dementia in patients with type 2 diabetes mellitus: a nationwide retrospective cohort study in Taiwan. J Diabetes Res 2016, 9027484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kelley BJ, McClure LA, Unverzagt FW, Kissela B, Klein-dorfer D, Howard G, Wadley VG (2015) Regular aspirin use does not reduce risk of cognitive decline. J Am Geriatr Soc 63, 390–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Veronese N, Stubbs B, Maggi S, Thompson T, Schofield P, Muller C, Tseng PT, Lin PY, Carvalho AF, Solmi M (2017) Low-dose aspirin use and cognitive function in older age: a systematic review and meta-analysis. J Am Geriatr Soc 65, 1763–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Heneka MT, Reyes-Irisarri E, Hull M, Kummer MP (2011) Impact and therapeutic potential of PPARs in Alzheimer’s disease. Curr Neuropharmacol 9, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ghosh A, Pahan K (2016) PPARalpha in lysosomal biogenesis: A perspective. Pharmacol Res 103, 144–148. [DOI] [PMC free article] [PubMed] [Google Scholar]