Abstract
The demographic shift toward an older population will increase the number of prostate cancer cases. A challenge in the treatment of prostate cancer is to avoid undertreatment of patients at high risk of progression following curative treatment. These men can benefit from early salvage treatment. An explorative cohort consisting of tissue from 16 patients who underwent radical prostatectomy, and were either alive or had died from prostate cancer within 10 years postsurgery, was analyzed by mass spectrometry analysis. Following proteomic and bioinformatic analyses, major vault protein (MVP) was identified as a putative prognostic biomarker. A publicly available tissue proteomics dataset and a retrospective cohort of 368 prostate cancer patients were used for validation. The prognostic value of the MVP was verified by scoring immunohistochemical staining of a tissue microarray. High level of MVP was associated with more than 4-fold higher risk for death from prostate cancer (hazard ratio = 4.41, 95% confidence interval: 1.45–13.38; P = 0.009) in a Cox proportional hazard models, adjusted for Cancer of the Prostate Risk Assessments Post-surgical (CAPRA-S) score and perineural invasion. Decision curve analyses suggested an improved standardized net benefit, ranging from 0.06 to 0.18, of adding MVP onto CAPRA-S score. This observation was confirmed by receiver operator characteristics curve analyses for the CAPRA-S score versus CAPRA-S and MVP score (area under the curve: 0.58 versus 0.73). From these analyses, one can infer that MVP levels in combination with CAPRA-S score might add onto established risk parameters to identify patients with lethal prostate cancer.
Mass spectrometry of an explorative cohort identified a promising biomarker, major vault protein. Using two independent validation cohorts, we verified an association between high MVP and adverse outcome. MVP was associated with prostate cancer-specific death independently of a composite risk model.
Introduction
The need for better risk stratification of prostate cancer patients at the time of diagnosis is pressing as the incidence and mortality of prostate cancer is predicted to increase due to a shift in the demographic distribution (1). There was an estimated 1.27 million new prostate cancer cases worldwide in 2018, and prostate cancer was the major cancer diagnosis among males in Western Europe and North America (2). Current nomograms often underestimate truly aggressive tumors as being indolent, leading to undertreatment of a subset of primary prostate cancer tumors. To improve the accuracy of nomograms, we aim to identify novel molecular biomarkers. Previously, biomarkers like PTEN, 4Kscore and Decipher have been reported to predict lethal prostate cancer, and Decipher can be used to change clinical practice for adjuvant and salvage treatment (3–6). In the era of precision medicine, the use of proteomic profiling might become an essential tool to stratify prostate cancer patients into optimal treatment groups (7,8).
In recent years, there has been an increase in the number of studies that use proteomic profiling of formalin fixed paraffin embedded (FFPE) tissue to identify new prognostic and predictive biomarker candidates (9,10). In the field of prostate cancer proteomics, recent publications have enlarge focused on establishing the shift in the proteomic landscape occurring during oncogenesis and progression, which is of great value in the process of establishing a complete overview of the changes in the proteome in the course of development and progression of prostate cancer (11,12). But very few of these studies have validated their findings in larger patient cohorts with well-annotated clinical information. These sorts of cohorts are needed to address some of the challenges that prostate cancer patients are facing with regard to distal spread of the disease and development of therapy resistance. As noted in the REMARK guidelines, verification using independent validation cohorts will positively impact the confidence in a potential biomarker (13).
The aim of this study was to generate a proteomic profile of a well-defined pilot cohort of prostate cancer patients which could be used to stratify patients into indolent or lethal disease after surgery, and further validation in a larger and well-annotated cohort with time to event data.
Materials and methods
Patients data
The patients included in this study had all been treated with radical prostatectomies at four different hospitals in Norway. The explorative cohort started with 23 patients, and 7 patients were excluded from further analysis due to low or no signal in the MS spectra (Supplementary Figure S1, available at Carcinogenesis Online). These patients were treated at Oslo University Hospital between 1995 and 2002. The patients were identified retrospectively based on the criteria that they either had died from prostate cancer or were alive 10 years post radical prostatectomy treatment. Very few intermediate- and high-risk patients were operated in this period, limiting the number cases available in the lethal patients’ group. The indolent group was selected to match the lethal group based on age, PSA (prostate-specific antigen), Gleason score [International Society of Urological Pathology (ISUP) grade] and pT-stage. Higher Gleason score is, however, observed in the group of lethal cases.
The validation cohort was retrospectively identified from the archives of the Departments of Pathology in the three northernmost health regions in Norway (14). Clinical records from 671 consecutive patients radically prostatectomized for prostate cancer adenocarcinoma between 01.01.1995 and 31.12.2005 were reviewed. After exclusion of 137 patients, 535 eligible patients with complete follow-up data and available tissues from St. Olav Hospital/Trondheim University Hospital (St. Olav) in the Central Norway region (n = 228), and Nordlandssykehuset Bodø (NLSH, n = 59) and the University Hospital of North Norway (UNN, n = 248) were included. In addition to standard clinicopathological variables, the composite CAPRA-S score (Cancer of the Prostate Risk Assessments Post-surgical) (15,16) was calculated for each patient as a validated postsurgical tool to thoroughly explore whether major vault protein (MVP) expression added prognostic value.
In the validation cohort, biochemical recurrence (BCR) was defined as a post radical prostatectomy PSA ≥0.4 ng/ml and rising according to Stephenson et al. (17). BCR-free survival was calculated as time from surgery to last follow-up date or date of BCR. Clinical failure (CF) was defined as symptomatic, locally advanced progression or metastasis to bone, visceral organs or lymph nodes verified by radiology. CF-free survival was calculated from date of surgery to last follow-up date without CF or to CF date. Prostate cancer-specific mortality (PCSM) was defined as death from progressive and disseminated castration-resistant prostate cancer despite therapy. More extensive information regarding patients, exclusion, definitions of variables and endpoints in the validation cohort has been published previously (14). Last clinical update was 1 December 2015. Median follow-up of survivors was 150 months.
Sample preparation for mass spectrometry
FFPE tissue samples were initially dehydrated using increasing concentration of ethanol (80, 96 then 100%) and paraffin was removed by adding Paraffin Removal Solution (BIOstic). Samples were washed with absolute xylene (Merck) and 100% ethanol before proteins were extracted by adding 40 µl of 0.2% ProteaseMax (Promega) in 50 mM of NH4HCO3 to a final volume of 187 µl, plus additional 2 µl of 0.5 M dithiothreitol. Samples were sonicated in a water bath (37°C) for 60 min, followed by sample heating at 98°C for 90 min on a heating block. For trypsin digestion, additional 2 µl of 1% ProteaseMax was added to the sample prior to adding 1 µg of trypsin (MS-grade, modified, Promega), and incubated overnight at 37°C. After tryptic digestion, 1% trifluoroacetic acid was added to the supernatant for ProteaseMax degradation for 5 min at room temperature, and then centrifuged at 14 000 rcf for 10 min for removal of degraded surfactant. Peptides were then further cleaned using STAGE-TIPS protocol using a C18 resin disk (3M Empore) (18).
Mass spectrometry analysis and processing
All experiments were performed on an Easy nLC1000 nano-LC system connected to a quadrupole—Orbitrap (QExactive) mass spectrometer (ThermoElectron, Bremen, Germany) equipped with a nanoelectrospray ion source (EasySpray/Thermo). For liquid chromatography separation an EasySpray column (C18, 2 µm beads, 100 Å, 75 µm inner diameter) (Thermo) capillary of 25 cm bead length was used. The flow rate used was 0.3 µl/min, and the solvent gradient was 5% B to 30% B in 240 min, followed by 90% B wash for 20 min. Solvent A was aqueous 0.1% formic acid, whereas solvent B was 100% acetonitrile in 0.1% formic acid. Column temperature was kept at 60°C. Peptide sequence information and peptide/protein identification were performed using mass spectrometer and search engine parameters as described previously (19).
Tissue microarray and immunohistochemistry
FFPE tissue blocks from all included patients were collected from the archives. For tissue microarray (TMA), one experienced uropathologist (ER) marked the dedifferentiated neoplastic cell compartment (tumor) from adjacent tumor stroma (stroma) in H&E slides. Then, two duplicate cores from the tumor and stroma were sampled and inserted to recipient TMA blocks using a tissue-arraying instrument (Beecher Instruments, Silver Springs, MD). The core diameter was 0.6 mm and a total of 12 TMA blocks were constructed and sectioned for immunohistochemistry (IHC) staining.
The TMA slides (4 µm sections) were stained by a fully automated Discovery Ultra (Ventana, Tucson). After onboard deparaffinization (three cycles, each 12 min) and antigen retrieval (48 min, CC1 buffer), the primary antibody was loaded onto the Discovery Ultra at a 1/25 dilution. The primary antibody was anti-MVP mouse monoclonal (clone: 1032) antibody from Abcam (cat no: Ab2376). Primary antibody incubation was 60 min at 37°C temperature, followed by 20 min incubation with an anti-mouse secondary antibody (OmniMap anti-Ms HRP; cat no: 760-4310, Ventana). The immune reaction was visualized using the DAB-Map detection kit (cat no: 760-124, Ventana). Finally, the chromogen was followed by hematoxylin (cat no: 790-2208, Ventana) as a counterstain and bluing reagent (cat no: 760-2037, Ventana) as post-counterstain. Slides were rinsed, dehydrated through alcohol and xylene and then cover-slipped. Prior the primary antibody adding, DAB CM-inhibitor (integrated with DAB-MAP kit) was incubated for 8 min to block the endogenous peroxidase activity. Breast cancer TMA slides and normal prostate and brain tissue sections were used as positive and negative controls, respectively (Supplementary Figure S4, available at Carcinogenesis Online). In addition, the primary antibody was validated using lysate of MVP transiently transfected HEK293 cells (OriGene, cat no: LY413762). The western blotting protocol and blot images are presented in Supplementary Figure S5, available at Carcinogenesis Online.
Scoring of IHC and cutoff
The MVP intensity of tumor cells was scored semiquantitatively in a four-tiered score as: 0 = negative, 1 = weak, 2 = moderate and 3 = strong. In fibroblasts, MVP intensity was scored as 0 = negative and 1 = positive if more than five fibroblasts were positive in a core. Two observers (ER and MR) independently reported the score for morphologically verified tumor cells and fibroblasts. They were blinded to each other’s score and to the clinicopathological follow-up data. In cases of disagreement of more than 2 values, cores were reassessed until a consensus was reached. Mean cutoffs for dichotomization, negative and weak staining intensities were denoted low MVP and moderate and strong staining intensities were denoted high MVP, were used for both tumor cell and fibroblast scores.
Analysis of publicly available dataset
Deposited proteomic mass spectrometry and clinical data from Sinha et al. were used to verify our initial findings from our processed mass spectrometry data and to evaluate MVP’s association with BCR-free survival (20).
Statistical analyses
Statistical analyses were done using the SPSS software version 25 (IBM SPSS, Chicago, IL), RStudio v.1.2.1335 and R version 3.5.3 (21). Interobserver reliability of pathologist scoring was tested by use of a two-way random effect model with absolute agreement. LIMMA (linear models for microarray data) was used to query for differential expressed proteins. The false discovery rate method (Benjamini–Hochberg) was used to adjust for multiple testing. LASSO (least absolute shrinkage and selection operator) regression was used for classification of the indolent and lethal groups. Associations between MVP and clinicopathological markers were analyzed using the Spearman’s correlation test. The Kaplan–Meier method was used to generate plots of CF-free survival, and statistical significance of differences in survival distributions were assessed by use of the log-rank tests. Variables from the univariable analyses with a P < 0.10 were assessed for significant and independent impact on endpoints in a stepwise backward multivariate Cox regression model with a probability at 0.05 for entry and 0.10 for removal. The concordance index (C-index) was used for prediction testing. A logistic regression model was use for the decision curve analysis. The chosen significance level for all analyses was P < 0.05.
Ethics
These studies were approved by the Regional Committees for Medical and Health Research Ethics (REC): REC North 2009/1393 and REC South East 2009/1028 and 2013/1713. The reporting of clinicopathological variables, survival data and biomarker expressions was conducted in accordance with the REMARK guidelines (13).
Data availability
The proteomics data in this study have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/) via the PRIDE partner repository (22) with the dataset identifier PXD17712.
Results
Identification of differentially expressed proteins by proteomic profiling
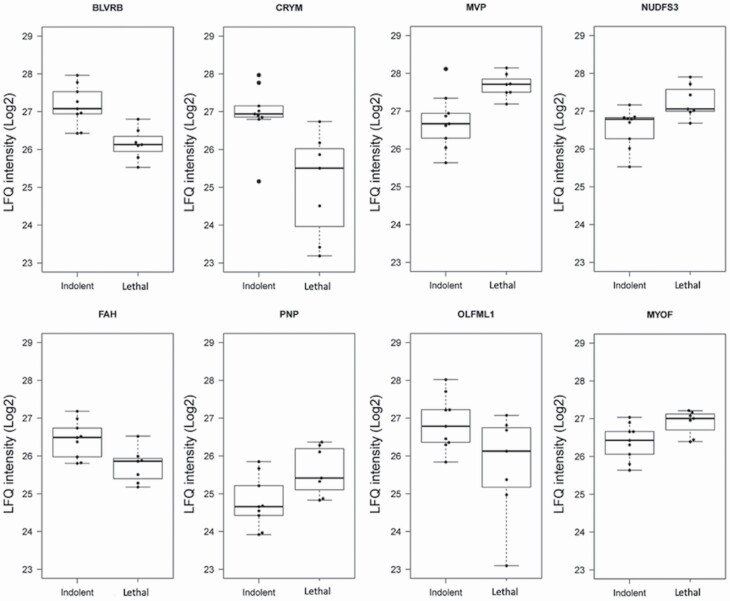
For the explorative pilot study, protein extracts from FFPE slices from 16 radical prostatectomy samples were prepared for high-resolution LC–MS/MS analysis. The clinical characteristics of the discovery cohort are presented in Supplementary Table S1, available at Carcinogenesis Online. The corresponding raw data files were processed for peptide and protein identification with MaxQuant resulting in the identification of 3657 protein groups (Supplementary Table S2, available at Carcinogenesis Online). To identify differentially expressed proteins between the indolent and lethal patient samples groups, we applied a conservative approach and used only protein groups that were detected in all the processed samples, resulting in a total of 766 proteins with label-free quantification (LFQ) intensity values. These 766 proteins were used in a LIMMA analysis to query for differential expressed proteins. The LIMMA analysis identified 56 proteins with an unadjusted P < 0.05, but none of these were significant after adjusting for multiple testing (false discovery rate < 0.05; Supplementary Table S2, available at Carcinogenesis Online). To strengthen our statistical analysis, we used LASSO regression to identify the proteins that best could predict the separation of the two groups (Supplementary Table S2, available at Carcinogenesis Online). The estimated lambda coefficient in the LASSO analysis is adjusted to minimize underfitting and overfitting of the model. The LIMMA and LASSO analyses identified eight overlapping proteins, and the top three proteins were Biliverdin reductase B (BLVRB), Ketimine reductase mu-crystallin (CRYM) and MVP (Figure 1 and Supplementary Table S3, available at Carcinogenesis Online).
Figure 1.
Differentially expressed proteins in indolent versus lethal PCa. Boxplot indicates median LFQ intensity value and the IQR, whereas the whiskers extend to min and max values. MVP, NDUFS3, PNP and MYOF were upregulated and BLVRB, CRYM, FAH and OLFML1 were downregulated in the lethal group compared with indolent group of patients.
We queried the ProteomicsDB database using the list of eight proteins to verify that these proteins had been detected in prostate tissue previously (Supplementary Figure S2, available at Carcinogenesis Online) (23). All eight proteins have been previously detected in prostate tissue. Seven (BLVRB, PNP, FAH, CRYM, MVP, MYOF and NDUFS3) and six (BLVRB, PNP, FAH, CRYM, MVP and MYOF) had been detected in urine and extracellular vesicles isolated from urine samples, respectively. MVP was selected for further analyses because it has been shown to be associated with disseminated prostate cancer and there are numerous publications reporting a possible prognostic and predictive value of MVP in other cancer types (24–27). BLVRB has been reported to be a diagnostic biomarker candidate for prostate cancer (28). CRYM was included in a list of proteins that could separate low and high stage prostate cancer based on Gleason score.
Prognostic significance of MVP using an independent mass spectrometry dataset
We investigated the association between MVP LFQ values and time to BCR by using a publicly available proteomic dataset (20). In this study, Sinha et al. analyzed prostate cancer tissue from 75 patients. High MVP LFQ values (above median) associated with shorter time to BCR (log-rank test; P = 0.024), as presented in the Kaplan–Meier plot in Supplementary Figure S3, available at Carcinogenesis Online.
Patients and distribution of MVP in validation cohort
The validation cohort with its 535 prostate cancer patients was used to further assess the prognostic value of MVP with higher statistical power. The whole cohort was stained with a validated MVP antibody. MVP was expressed in the cytoplasm with accentuated membranous staining and there was no nuclear expression (Supplementary Figure S4, available at Carcinogenesis Online), in accordance with previous studies (26,27). Within the stromal compartment, MVP stained positively in endothelial cells, lymphocytes and macrophages, but these stainings were not scored. Representative images of the MVP staining of four tissue cores with either low or high MVP staining scores, including negative and positive controls, are shown in Supplementary Figure S4, available at Carcinogenesis Online. A total of 368 patients, with a median follow-up time of 141 months, had morphologically verified tumor cells that were scored for MVP expression and the clinical characteristics of these patients are presented in Table 1. The patients (N = 167) with a missing tumor score did not have viable morphologically tumor cells present for scoring in either of the two tumor cores. Although this reduces the power, this is an expected finding in a multifocal cancer like prostate cancer and reduces bias compared with non-in situ analyses. The intraclass correlation coefficient between the pathologists who scored MVP was 0.92 (95% confidence interval: 0.90–0.93; P < 0.001). The cohort was dichotomized into low or high MVP expression using the mean IHC intensity score of tumor cells as the cutoff value. For the baseline clinical variables evaluated for association with MVP expression, it was only perineural invasion (PNI) that indicated a potential association (Table 1). Noteworthy, the frequency distribution of low and high MVP patients within the three different CAPRA-S risk groups (low, intermediate and high) were similar.
Table 1.
Baseline clinical characteristics of patients with MVP IHC scores
| MVP score | ||||
|---|---|---|---|---|
| Characteristic | Overall (n = 368) | Low (n = 250) | High (n = 118) | P value |
| Age, year, mean (SD) | 61.9 (5.22) | 61.7 (5.32) | 62.3 (4.99) | 0.29 |
| PSA (%) | 0.31 | |||
| ≤6 | 79 (21.5%) | 49 (19.6%) | 30 (25.4%) | |
| 6–10 | 139 (37.8%) | 96 (38.4%) | 43 (36.4%) | |
| 10–20 | 115 (31.2%) | 81 (32.4%) | 34 (28.8%) | |
| ≥20 | 35 (9.5%) | 24 (9.6%) | 11 (9.3%) | |
| ISUP grade | 0.44 | |||
| 1 | 110 (29.9%) | 74 (29.6%) | 36 (30.5%) | |
| 2 | 153 (41.6%) | 108 (43.2%) | 45 (38.1%) | |
| 3 | 64 (17.4%) | 48 (19.2%) | 16 (13.6%) | |
| 4 | 13 (3.5%) | 7 (2.8%) | 6 (5.1%) | |
| 5 | 28 (7.6%) | 13 (5.2%) | 15 (12.7%) | |
| Surgical margin | 0.27 | |||
| Negative | 172 (46.7%) | 112 (44.8%) | 60 (50.8%) | |
| Positive | 196 (53.3%) | 138 (55.2%) | 58 (49.2%) | |
| Seminal vesicle invasion | 0.35 | |||
| Negative | 332 (90.2%) | 228 (91.2%) | 104 (88.1%) | |
| Positive | 36 (9.8%) | 22 (8.8%) | 14 (11.9%) | |
| Extracapsular extension | 0.85 | |||
| Negative | 255 (69.3%) | 174 (69.6%) | 81 (68.7%) | |
| Positive | 113 (30.7%) | 76 (30.4%) | 37 (31.3%) | |
| Lymph node status | 0.45 | |||
| Negative | 186 (50.5%) | 128 (51.2%) | 58 (49.2%) | |
| Positive | 3 (0.8%) | 1 (0.4%) | 2 (1.7%) | |
| Not assessed | 179 (48.6%) | 121 (48.4%) | 58 (49.2%) | |
| Perineural invasion | 0.09 | |||
| Negative | 276 (75.0%) | 194 (77.6%) | 82 (69.5%) | |
| Positive | 92 (25.0%) | 56 (22.4%) | 36 (30.5%) | |
| CAPRA-S score | 0.96 | |||
| 0–2 | 108 (29.3%) | 71 (28.4%) | 37 (31.4%) | |
| 2–5 | 183 (49.7%) | 129 (51.6%) | 54 (45.8%) | |
| ≥6 | 77 (20.9%) | 50 (20.0%) | 27 (22.9%) | |
The prognostic value of MVP using IHC
We evaluated the association between the MVP dichotomous and BCR-free survival, clinical progression-free survival and PCSM using Kaplan–Meier analyses (Figure 2). For all endpoints, namely, high MVP levels associated with shorter time to the assessed events. The log-rank test for the different Kaplan–Meier curves gave P values of P = 0.004 for BCR-free survival, P = 0.04 for clinical progression-free survival and P = 0.005 for PCSM. Modeling with the cumulative incident function showed that there was no difference between the low and high MVP patients with regard to overall mortality, but the PCSM assumption still held true when including competing risk factors (Supplementary Figure S6, available at Carcinogenesis Online).
Figure 2.
High radical prostatectomy tumor MVP immunohistochemical staining is associated with poor prognosis. Low MVP denotes negative or weak MVP staining intensities in the tumor cores, whereas high MVP denotes moderate or strong MVP staining intensities. Kaplan–Meier curves, with the P values from the log-rank test, display event-free proportions using (A) BCR, (B) clinical progression and (C) PCSM as endpoints. Number of paints in each group of low and high expression of MVP at different time intervals is indicated underneath each Kaplan–Meier plot.
A high tumoral MVP was associated with increased risk of PCSM (hazard ratio for high MVP = 4.41: 95% confidence interval: 1.45–13.38; P = 0.009) in a multivariable model adjusted for CAPRA-S score and PNI (Table 2). The relative risk of the high MVP group was also increased using BCR and clinical progression as outcomes, but CAPRA-S score was a stronger prognostic variable for these two endpoints. Using the concordance index (C-index) and receiver operator characteristics with PCSM as endpoint, MVP and CAPRA-S score gave C-indexes of 0.84 and 0.71, and area under the curve values of 0.70 and 0.58, respectively (Supplementary Table S4 and Figure S6, available at Carcinogenesis Online). Combining MVP and CAPRA-S score increased the C-index from 0.71 to 0.78 for PCSM, and the area under the curve value increased from 0.58 to 0.73, which can be extrapolated to a sensitivity of 85.6% and specificity of 80.6% (Supplementary Table S4 and Figure S7, available at Carcinogenesis Online).
Table 2.
Uni- and multivariable hazard ratios for MVP score and BCR, clinical progression and PCSM
| Univariable analysis | Multivariable analysisa | ||||
|---|---|---|---|---|---|
| Covariate | No. of events/total | HR (95% CI) | P value | HR (95% CI) | P value |
| BCR | |||||
| MVP score | |||||
| Low | 84/250 | Reference | Reference | ||
| High | 59/118 | 1.62 (1.16–2.27) | 0.004 | 1.64 (1.17–2.29) | 0.004 |
| CAPRA-S score | |||||
| 0–2 | 21/108 | Reference | Reference | ||
| 3–5 | 65/183 | 1.95 (1.19–3.19) | 0.008 | 2.06 (1.25–3.37) | 0.004 |
| ≥6 | 57/77 | 6.19 (3.74–10.23) | <0.001 | 5.56 (3.34–9.28) | <0.001 |
| PNI | |||||
| Negative | 87/276 | Reference | Reference | ||
| Positive | 56/92 | 2.32 (1.65–3.24) | <0.001 | 1.70 (1.20–2.41) | 0.003 |
| Clinical progression | |||||
| MVP score | |||||
| Low | 22/250 | Reference | Reference | ||
| High | 20/118 | 1.88 (1.02–3.47) | 0.044 | 2.06 (1.11–3.82) | 0.021 |
| CAPRA-S score | |||||
| 0–2 | 3/108 | Reference | Reference | ||
| 3–5 | 20/183 | 4.01 (1.19–13.50) | 0.024 | 4.39 (1.30–14.83) | 0.017 |
| ≥6 | 19/77 | 8.21 (2.41–27.93) | <0.001 | 7.39 (2.14–25.48) | 0.001 |
| PNI | |||||
| Negative | 23/276 | Reference | Reference | ||
| Positive | 19/92 | 2.34 (1.26–4.34) | 0.007 | 1.85 (0.97–3.53) | 0.063 |
| PC-specific mortality | |||||
| MVP score | |||||
| Low | 5/250 | Reference | Reference | ||
| High | 9/118 | 4.22 (1.40–12.67) | 0.010 | 4.41 (1.45–13.38) | 0.009 |
| CAPRA-S score | |||||
| 0–2 | 2/108 | Reference | Reference | ||
| 3–5 | 5/183 | 1.27 (0.24–6.60) | 0.774 | 1.49 (0.28–7.75) | 0.637 |
| ≥6 | 7/77 | 4.05 (0.83–19.72) | 0.083 | 3.47 (0.69–17.44) | 0.130 |
| PNI | |||||
| Negative | 6/276 | Reference | Reference | ||
| Positive | 8/92 | 3.58 (1.23–10.43) | 0.019 | 2.70 (0.88–8.19) | 0.079 |
CI, confidence interval; HR, hazard ratio.
aAdjusted for CAPRA-S score and PNI (perineural invasion).
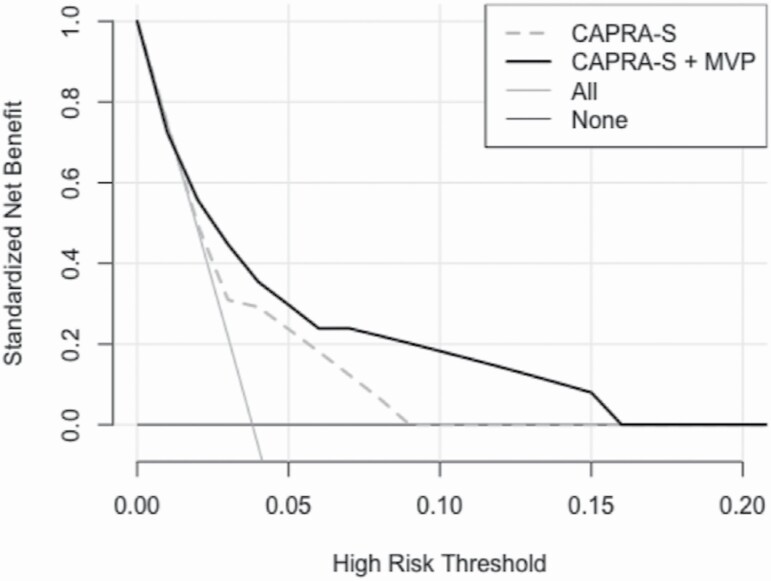
To further assess the effect of combining MVP with CAPRA-S, a decision curve analysis using a logistic regression model was performed to estimate a potential net benefit of adding MVP onto established diagnostic nomograms (Figure 3). In our validation cohort, we observed that combining MVP with the CAPRA-S score improved the standardized net benefit between 0.06 and 0.18 relative to the CAPRA-S score in the risk threshold range of 0.05–0.15, with PCSM as endpoint.
Figure 3.
Decision curves analysis indicating the standardized net benefit for PCSM using CAPRA-S (gray dashed line) and CAPRA-S + MVP (black line) as a function of the risk threshold. As comparison the default strategies of treating All patient (light gray line) or None (dark gray line) of the patients are indicated.
Discussion
In this study, we have shown that MVP was one of the top candidates in the list of proteins from mass spectrometry analysis that were differentially expressed in patients with indolent compared with fatal prostate cancer. This was validated in an independent mass spectrometry study by using publicly available data from 75 prostate cancer patients, using BCR as an endpoint (20). We further used IHC staining of TMAs from a validated independent cohort with commonly used endpoints like PCSM, and clinical variables used to adjust for confounding factors. The main finding from our analysis was that prostate cancer patients with high levels of MVP had a shorter time to PCSM, and by combining the CAPRA-S score and MVP level the net prognostic accuracy was improved over either one individually.
The use of mass spectrometry analysis of FFPE tissue poses many challenges when one’s aim is to identify potentially new biomarkers (29). There are the common challenges of sample preparation and technical issues with the mass spectrometry instruments (10). Another challenge relates to the analysis of the mass spectrometry results with regard to identifying protein groups and handling of missing values. To overcome some of these issues we only used proteins detected in all samples for further analysis in this study, which should reduce the chances of false positive at the cost of higher chance of false negative. Although our initial cohort is small, the use of different statistical methods and previously published mass spectrometry results to validate our findings strengthens our results (20).
When validating findings from an initial screen, it is essential that the cohort being used is well annotated. Our validation cohort, which was a large unselected retrospective cohort, included information for three different endpoints, BCR, clinical progression and PCSM, and the median follow-up time exceeded 10 years. A weakness of this cohort is that only 14 out of 535 patients had died from prostate cancer, which lowers the statistical power of our analysis when using PCSM as an endpoint. Although IHC scoring is a semiquantitative method, this was improved by using the average of two independent scorings of the TMA cores. The in situ analysis of MVP expression is a benefit when considering the multifocal nature of prostate cancer, and it enables one to analyze the expression of MVP in surroundings tissue like stroma.
MVP has previously been evaluated as a biomarker for therapy response and survival in melanoma, lung- and bladder carcinomas (30). The only study we have found that has investigated MVP in prostate cancer patients was the study by Van Brussel et al. (27). In this study, they observed an association between MVP expression and progression. When they compared a low stage group of patients with local disease to a group of patients with late stage advanced disease that had relapsed after antiandrogen treatment, they found a statistically significant difference in MVP levels between the groups. In this study, they only used immunohistochemical scoring to quantify MVP and it was a rather small cohort with only 6 and 12 patients in each of the groups. In two studies by Sánchez et al. where they investigated the expression of multidrug resistance proteins (MDRs) after chemotherapy exposure of prostate cancer cell lines, they observed an increased expression of MDRs, and one of them was MVP (31,32). It has been reported that single nucleotide polymorphisms in the MVP gene is associated with more aggressive prostate cancer (33). The authors were not able to assess if the single nucleotide polymorphism they detected in the intron of the MVP sequences had any functional consequences. In a recent breast cancer study, it was reported that a multidrug resistance phenotype was induced by adipocytes through upregulation of MVP, rendering the breast cancer cell lines resistant to doxorubicin (25). In patients with non-small-cell lung cancer, it has been reported that patients that were positive for MVP expression had a shorter overall survival compared with MVP-negative patients (34). There is still disagreement about the prognostic and predictive value of MVP since some studies have failed to show any association between MVP, therapy response and survival (30,35).
When the vault ribonucleoprotein complexes, which MVP is the major component of, was discovered some 30 years ago it was not know that it was the same protein that had been previously identified and named lung resistance-related protein (LRP) (36). The vaults complexes are barrel shaped structures that are made up of MVP, vault poly(ADP-ribose) polymerase (vPARP/PARP4), telomerase-associated protein 1 (TEP1) and small untranslated RNA (vault RNA) (37). It has been proposed that vaults are involved in drug resistance, and that the vault complex can open and close, and thereby shuttle molecules and proteins inside the hollow part of the barrel shaped structure (35). The dissociation of the two vault halves can be initiated by changes in intracellular pH fluxes, which is of relevance in hypoxic tumor microenvironments (38). The vault particles have also been reported to be involved in signal transmission and immune responses, but there is still some uncertainty around the cellular functional role of vaults (37).
Besides MVP potential function in drug resistance, there are studies that have linked MVP to DNA damage and DNA repair processes based on the increased expression of MVP after ionizing radiation (30). In a recent study of a small group of prostate cancer patients that were given radiation therapy, it was reported that MVP was upregulated in 6 out of 10 patients after radiation therapy (39). These findings are of interest in light of the radiation-induced DNA repair process and MVP potential role in this process. This raises the possibility of MVP as a predictive biomarker for response to radiotherapy.
In our study, the CARPA-S score was used to stratify the patients into risk groups and the prognostic value of the CAPRA-S score has been assessed against BCR and PCSM in other cohorts. The C-index for the CAPRA-S score was 0.77 in our cohort which is in accordance with previous studies that has reported C-index values of 0.77 and 0.73 (15,16). MVP had better accuracy compared with CAPRA-S score when PCSM was used as endpoint, and improved the prognostic value when combined with CAPRA-S score using BCR and clinical progression as endpoints.
This is the first study to evaluate MVP as a potential tissue biomarker for fatal prostate cancer. The observation was validated in an independent cohort, but the prognostic value of MVP should also be validated in other cohorts, preferably a prospective cohort with more lethal events. MVP has also the potential to become a noninvasive test since MVP is present in urine and extracellular vesicles from urine.
Based on the analyses in this study we propose that MVP is a highly relevant prognostics biomarker for subgrouping prostate cancer patients with increased risk of PCSM who may possibly benefit from early salvage treatment and closer monitoring. By combining MVP with an established stratification tool, the CAPRA-S score, we were able to improve the discriminatory accuracy of the validated postsurgical scoring assessment commonly used to predict PCSM.
Supplementary material
Supplementary data are available at Carcinogenesis online.
Supplementary Table S1. Baseline clinical characteristics of patients analyzed by mass spectrometry.
Supplementary Table S2. Supplementary excel file with list of proteins detected in all samples and the result list from the LIMMA and LASSO analyses.
Supplementary Table S3. Table of proteins that are common in both the LIMMA and LASSO analyses.
Supplementary Table S4. Concordance index (C-index) for MVP, CAPRA-S and CAPRA-S + MVP using BCR, clinical progression and prostate cancer-specific mortality as endpoints.
Supplementary Figure S1. Flow chart of sample processing.
Supplementary Figure S2. Data from the ProteomicsDB indicate that the eight identified proteins have been detected in prostate tissue previously. A list of other fluids where these proteins have been detected is also shown.
Supplementary Figure S3. Survival analysis of patients stratified as low or high MVP levels. MVP values from the Sinha cohort were used to generate a Kaplan–Meier plot using time to BCR as endpoint. The median MVP LFQ intensity value was used as cutoff for stratification of low and high MVP patients.
Supplementary Figure S4. Representative images of different major vault protein (MVP) staining intensities of prostate cancer tissue (TMA cores) and tissue controls. (A) No expression in prostate tumor tissue, positive expression on stromal fibroblasts, ISUP grade 2. (B) No expression in prostate tumor tissue, negative to weak expression on stromal fibroblasts. (C) Moderate expression in prostate tumor tissue, ISUP grade 1. (D) High expression in prostate tumor tissue, ISUP grade 2. (E) Negative tissue control of staining, normal brain tissue. (F) Positive tissue control of staining, breast cancer tissue (magnification ×15).
Supplementary Figure S5. Major vault protein (MVP) antibody validation. Western blotting was performed using overexpressed human HEK293T cell lysate for MVP and HEK293 as the negative vector. Equal amounts of protein lysates were loaded into a 12% Bis–Tris gel (Life Technologies, cat no: NP0341). A goat anti-mouse IRDye 800CW secondary antibody (LI-COR, cat no: 926-32210, dilution 1/10 000) was used for protein detection using Odyssey CLx Imaging System (Li-Cor Biosciences). (A) The observed molecular weight was corresponded with the predicted weight provided by the manufacturer (99.1 kDa). (B) As a loading control, the same membrane was reprobed with an anti-β-actin mAb (MilliporeSigma, cat no: A2066, dilution: 1/1000).
Supplementary Figure S6. Cumulative incidence curves of dichotomized MVP score for overall mortality (OM) and prostate cancer-specific survival (PCSS).
Supplementary Figure S7. Receiver operator characteristic curve analysis with the time variable set at 10 years of follow-up using BCR, clinical progression and prostate cancer-specific mortality as endpoints.
Acknowledgements
We would also like to thank Yngve Nordby and Nora Ness for help in data collection and Helena Bertilsson and Øystein Størkersen for help to identify patients and collection of tissue from St. Olav. We also would like to acknowledge the Prostate Biobank at Oslo University Hospital (REC 2013/1713) for providing the patients information and tissue in the OUS cohort. At last we would like to thank all the patients that have contributed with their tissue and clinical information.
Glossary
Abbreviations
- BCR
biochemical recurrence
- CAPRA-S
Cancer of the Prostate Risk Assessments Post-surgical score
- CF
clinical failure
- FFPE
formalin fixed paraffin embedded
- HR
hazard ratio
- MVP
major vault protein
- PCSM
prostate cancer-specific mortality
- PSA
prostate-specific antigen
Funding
We would like to thank MOVEMBER, the Norwegian Cancer Society and the Northern Norway Regional Health Authority for unrestricted grants enabling this work.
Conflict of Interest Statement: None declared.
References
- 1. Rawla, P. (2019) Epidemiology of prostate cancer. World J. Oncol., 10, 63–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Culp, M.B., et al. (2020) Recent global patterns in prostate cancer incidence and mortality rates. Eur. Urol., 77, 38–52. [DOI] [PubMed] [Google Scholar]
- 3. Zelic, R., et al. (2020) Predicting prostate cancer death with different pretreatment risk stratification tools: a head-to-head comparison in a nationwide cohort study. Eur. Urol., 77, 180–188. [DOI] [PubMed] [Google Scholar]
- 4. Hamid, A.A., et al. (2019) Loss of PTEN expression detected by fluorescence immunohistochemistry predicts lethal prostate cancer in men treated with prostatectomy. Eur. Urol. Oncol., 2, 475–482. [DOI] [PubMed] [Google Scholar]
- 5. Stattin, P., et al. (2015) Improving the specificity of screening for lethal prostate cancer using prostate-specific antigen and a panel of kallikrein markers: a nested case–control study. Eur. Urol., 68, 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kohaar, I., et al. (2019) A rich array of prostate cancer molecular biomarkers: opportunities and challenges. Int. J. Mol. Sci., 20, 1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murphy, K., et al. (2018) Integrating biomarkers across omic platforms: an approach to improve stratification of patients with indolent and aggressive prostate cancer. Mol. Oncol., 12, 1513–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tonry, C., et al. (2016) The role of proteomics in biomarker development for improved patient diagnosis and clinical decision making in prostate cancer. Diagnostics, 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaffney, E.F., et al. (2018) Factors that drive the increasing use of FFPE tissue in basic and translational cancer research. Biotech. Histochem., 93, 373–386. [DOI] [PubMed] [Google Scholar]
- 10. Giusti, L., et al. (2013) Proteomic studies of formalin-fixed paraffin-embedded tissues. Expert Rev. Proteomics, 10, 165–177. [DOI] [PubMed] [Google Scholar]
- 11. Iglesias-Gato, D., et al. (2016) The proteome of primary prostate cancer. Eur. Urol., 69, 942–952. [DOI] [PubMed] [Google Scholar]
- 12. Zhu, Y., et al. (2019) High-throughput proteomic analysis of FFPE tissue samples facilitates tumor stratification. Mol. Oncol., 13, 2305–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sauerbrei, W., et al. (2018) Reporting recommendations for tumor marker prognostic studies (REMARK): an abridged explanation and elaboration. J. Natl. Cancer Inst., 110, 803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Andersen, S., et al. (2014) Disease-specific outcomes of radical prostatectomies in Northern Norway; a case for the impact of perineural infiltration and postoperative PSA-doubling time. BMC Urol., 14, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cooperberg, M.R., et al. (2011) The CAPRA-S score: a straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer, 117, 5039–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Punnen, S., et al. (2014) Multi-institutional validation of the CAPRA-S score to predict disease recurrence and mortality after radical prostatectomy. Eur. Urol., 65, 1171–1177. [DOI] [PubMed] [Google Scholar]
- 17. Stephenson, A.J., et al. (2006) Defining biochemical recurrence of prostate cancer after radical prostatectomy: a proposal for a standardized definition. J. Clin. Oncol., 24, 3973–3978. [DOI] [PubMed] [Google Scholar]
- 18. Rappsilber, J., et al. (2003) Detection of arginine dimethylated peptides by parallel precursor ion scanning mass spectrometry in positive ion mode. Anal. Chem., 75, 3107–3114. [DOI] [PubMed] [Google Scholar]
- 19. Tutturen, A.E.V., et al. (2018) Characterization of the small intestinal lesion in celiac disease by label-free quantitative mass spectrometry. Am. J. Pathol., 188, 1563–1579. [DOI] [PubMed] [Google Scholar]
- 20. Sinha, A., et al. (2019) The proteogenomic landscape of curable prostate cancer. Cancer Cell, 35, 414–427.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. R Core Team (2019) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://R-project.org. [Google Scholar]
- 22. Vizcaíno, J.A., et al. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res., 44, D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Samaras, P., et al. (2020) ProteomicsDB: a multi-omics and multi-organism resource for life science research. Nucleic Acids Res., 48, D1153–D1163. doi: 10.1093/nar/gkz974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Diestra, J.E., et al. (2003) Expression of multidrug resistance proteins P-glycoprotein, multidrug resistance protein 1, breast cancer resistance protein and lung resistance related protein in locally advanced bladder cancer treated with neoadjuvant chemotherapy: biological and clinical implications. J. Urol., 170(4 Pt 1), 1383–1387. [DOI] [PubMed] [Google Scholar]
- 25. Lehuédé, C., et al. (2019) Adipocytes promote breast cancer resistance to chemotherapy, a process amplified by obesity: role of the major vault protein (MVP). Breast Cancer Res., 21, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Henríquez-Hernández, L.A., et al. (2012) MVP expression in the prediction of clinical outcome of locally advanced oral squamous cell carcinoma patients treated with radiotherapy. Radiat. Oncol., 7, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Van Brussel, J.P., et al. (2001) Expression of multidrug resistance related proteins and proliferative activity is increased in advanced clinical prostate cancer. J. Urol., 165, 130–135. [DOI] [PubMed] [Google Scholar]
- 28. Pallua, J.D., et al. (2013) MALDI-MS tissue imaging identification of biliverdin reductase B overexpression in prostate cancer. J. Proteomics, 91, 500–514. [DOI] [PubMed] [Google Scholar]
- 29. Hernández, B., et al. (2014) Why have so few proteomic biomarkers “survived” validation? (Sample size and independent validation considerations). Proteomics, 14, 1587–1592. [DOI] [PubMed] [Google Scholar]
- 30. Lara, P.C., et al. (2011) MVP and vaults: a role in the radiation response. Radiat. Oncol., 6, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sánchez, C., et al. (2011) Chemotherapy sensitivity recovery of prostate cancer cells by functional inhibition and knock down of multidrug resistance proteins. Prostate, 71, 1810–1817. [DOI] [PubMed] [Google Scholar]
- 32. Sánchez, C., et al. (2009) Expression of multidrug resistance proteins in prostate cancer is related with cell sensitivity to chemotherapeutic drugs. Prostate, 69, 1448–1459. [DOI] [PubMed] [Google Scholar]
- 33. Henríquez-Hernández, L.A., et al. (2016) Association between single-nucleotide polymorphisms in DNA double-strand break repair genes and prostate cancer aggressiveness in the Spanish population. Prostate Cancer Prostatic Dis., 19, 28–34. [DOI] [PubMed] [Google Scholar]
- 34. Chen, Z.J., et al. (2011) Lung resistance protein and multidrug resistance protein in non-small cell lung cancer and their clinical significance. J. Int. Med. Res., 39, 1693–1700. [DOI] [PubMed] [Google Scholar]
- 35. Mossink, M.H., et al. (2003) Vaults: a ribonucleoprotein particle involved in drug resistance? Oncogene, 22, 7458–7467. [DOI] [PubMed] [Google Scholar]
- 36. Kedersha, N.L., et al. (1986) Isolation and characterization of a novel ribonucleoprotein particle: large structures contain a single species of small RNA. J. Cell Biol., 103, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Berger, W., et al. (2009) Vaults and the major vault protein: novel roles in signal pathway regulation and immunity. Cell. Mol. Life Sci., 66, 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Querol-Audí, J., et al. (2009) The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP. EMBO J., 28, 3450–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Keam, S.P., et al. (2018) Exploring the oncoproteomic response of human prostate cancer to therapeutic radiation using data-independent acquisition (DIA) mass spectrometry. Prostate, 78, 563–575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomics data in this study have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/) via the PRIDE partner repository (22) with the dataset identifier PXD17712.