Abstract
Estrogen receptor alpha (ERα) belongs to the nuclear hormone receptor family of ligand-inducible transcription factors and regulates gene networks in biological processes such as cell growth and proliferation. Disruption of these networks by chemical compounds with estrogenic activity can result in adverse outcomes such as unscheduled cell proliferation, ultimately culminating in tumor formation. To distinguish disruptive activation from normal physiological responses, it is essential to quantify relationships between different key events leading to a particular adverse outcome. For this purpose, we established fluorescent protein MCF7 reporter cell lines for ERα-induced proliferation by bacterial artificial chromosome-based tagging of 3 ERα target genes: GREB1, PGR, and TFF1. These target genes are inducible by the non-genotoxic carcinogen and ERα agonist 17β-estradiol in an ERα-dependent manner and are essential for ERα-dependent cell-cycle progression and proliferation. The 3 GFP reporter cell lines were characterized in detail and showed different activation dynamics upon exposure to 17β-estradiol. In addition, they demonstrated specific activation in response to other established reference estrogenic compounds of different potencies, with similar sensitivities as validated OECD test methods. This study shows that these fluorescent reporter cell lines can be used to monitor the spatial and temporal dynamics of ERα pathway activation at the single-cell level for more mechanistic insight, thereby allowing a detailed assessment of the potential carcinogenic activity of estrogenic compounds in humans.
Keywords: Estrogen receptor alpha, fluorescent reporters, live-cell imaging, single cell
Carcinogens can be divided in 2 different groups based on their mode of action: genotoxic and non-genotoxic carcinogens. Genotoxic carcinogens directly interact with DNA, thereby inducing mutations that can eventually lead to tumor formation. On the other hand, non-genotoxic carcinogens affect tumor formation via, for example, inflammatory responses or endocrine mediators without direct interaction with the DNA (Hernández et al., 2009; Silva Lima and Van der Laan, 2000). The regulatory standard assay for the detection of any carcinogenic compound is a 2-year bioassay in rodents (von Wittenau and Estes, 1983). However, the translational relevance of the findings in this rodent bioassay to humans remains unclear (Haseman, 2000). For instance, nearly half of all chronically administered pharmaceuticals act as carcinogens in rodents, but do not pose a threat for human therapeutic usage (Friedrich and Olejniczak, 2011; Van Oosterhout et al., 1997). Several nonanimal alternatives to the rodent bioassay have been globally accepted or are currently under review by the Organization for Economic Co-Operation and Development (OECD). However, these alternatives primarily focus on the detection of genotoxic carcinogens, while leaving the non-genotoxic carcinogens undetected (Lilienblum et al., 2008; Luijten et al., 2016). Because Hérnandez et al. classified 16.9% of the known human carcinogens, as defined by the International Agency for Research on Cancer (IARC), as non-genotoxic carcinogens (Hernández et al., 2009), there is a need for improved methods to assess the carcinogenic risk for humans.
The list of proven (IARC Group 1), probable (IARC Group 2A), and possible (IARC Group 2B) human non-genotoxic carcinogens contains several endocrine modifiers which bind to nuclear hormone receptors (Hernández et al., 2009). These endocrine-disrupting chemicals (EDCs) impair the human endocrine and reproductive systems by affecting hormone signaling, biosynthesis, and metabolism, which might contribute to a potential non-genotoxic carcinogenic effect (Diamanti-Kandarakis et al., 2009). Frequently, the estrogen receptor alpha (ERα) is the primary target of these EDCs (Schug et al., 2011). Estrogenic EDCs have been identified in different classes of compounds, such as pharmaceuticals (ie, diethylstilbestrol), organochlorine pesticides (ie, methoxychlor and dichlorodiphenyltrichloroethane), industrial phenolics (ie, bisphenol A and alkylphenols), and phytoestrogens (ie liquiritigenin and genistein) (Maqbool et al., 2016; Rodgers et al., 2018; Shanle and Xu, 2011). Although these estrogenic EDCs are not all classified as proven human carcinogens, evidence on hormonal modulation, that is, ER activation, is considered a potential risk factor in carcinogenic risk assessment (Hernández et al., 2009; van der Laan et al., 2017).
Overactivation or deregulation of ERα is a known non-genotoxic carcinogen mode of action (Birnbaum and Fenton, 2003). Upon binding of a ligand, ERα undergoes conformational changes, displacing inhibitory heat shock proteins. The receptor will form dimers, which translocate to the nucleus and bind to the estrogen response element (ERE) in the DNA (Aranda and Pascual, 2001; Chen et al., 2018; McDonnell and Norris, 2002). This enables regulation of gene networks involved in various biological processes such as cell growth, motility, evasion of apoptosis, and proliferation (Couse and Korach, 1999; Jia et al., 2015). Disruption of these networks, for instance by administration of the natural non-genotoxic carcinogen and ERα agonist 17β-estradiol (E2), can result in adverse outcomes such as unanticipated cell proliferation ultimately culminating in tumor formation (Liehr, 2000; Yue et al., 2013). Because activation of the ERα pathway is a normal physiological program and does not necessarily lead to adverse outcomes, it is essential to quantify relationships between different key events induced by nonphysiological ERα pathway activation in the context of enhancement of cell proliferation.
Although there are several in vitro assays for measuring the effect of estrogenic compounds on ERα pathway activation and proliferation, such as the MCF7-based E-Screen (Soto et al., 1995), the ERα CALUX assay (Sonneveld et al., 2005; van der Burg et al., 2010), or the GeneBLAzer assay (Huang et al., 2011), these assays are not suitable for a dynamic analysis of key events in ERα pathway activation. Moreover, these assays are generally based on nonphysiological constructs that harbor multiple copies of the minimal promoter region for ERα binding linked to, for example, luciferase reporter. In this study, we have used the human ERα-positive MCF7 breast cancer cell line to identify E2-inducible ERα target genes that play a role in proliferation. For 3 of these genes, growth-regulating estrogen receptor binding 1 (GREB1), progesterone receptor (PGR), and trefoil factor 1 (TFF1), we established fluorescent protein reporters using bacterial artificial chromosome (BAC) cloning technology. These in vitro fluorescent reporters allow monitoring of dynamic pro-proliferative ERα pathway activation on a single-cell level using live-cell imaging.
MATERIALS AND METHODS
Cell culture and compounds
MCF7 cells (ATTC) were routinely maintained in RPMI-1640 medium modified with l-glutamine, HEPES, and phenol red (no. 22400089, Gibco, ThermoFisher Scientific) and supplemented with 10% fetal bovine serum (no. 10270106, Gibco, ThermoFisher Scientific), 25 U/ml penicillin, and 25 µg/ml streptomycin (no. 15070063, Gibco, ThermoFisher Scientific), further referred to as “complete medium,” at 37°C under 5% CO2 atmosphere. Unless stated otherwise, cells were seeded in complete medium and after 16–24 h the medium was replaced by phenol red-free RPMI1640 medium modified with l-glutamine (no. 11835105, Gibco, ThermoFisher Scientific) and supplemented with 5% charcoal/dextran-treated fetal bovine serum (cdFBS) (no. SH30068.03, HyClone, GE Healthcare), further referred to as “experimental medium.” After a 16–24-h starvation period in experimental medium, cells were treated with 17β-estradiol (E2) (no. E1024, Sigma-Aldrich) and/or 4-hydroxytamoxifen (4-OHT) (no. H6278, Sigma-Aldrich). 17α-ethinylestradiol, diethylstilbestrol, genistein, 4-cumylphenol, ethyl paraben, p,p′-methoxychlor, linuron, and spironolactone were kindly provided by BioDetection Systems B.V. All compounds were dissolved in DMSO (VWR International) and further diluted in experimental medium with a maximum concentration of 0.2% (v/v) DMSO.
RT-qPCR
To examine the mRNA expression of the candidate target genes, cells were seeded in a 12-well plate (no. 3512, Corning Costar). The medium was replaced after 16–24 h by phenol red-free RPMI supplemented with 1% cdFBS. Cells were exposed after 24 h starvation to E2 and/or 4-OHT in serum-free phenol red-free RPMI. After different time points, RNA was isolated using the NucleoSpin RNA Kit (Macherery-Nagel, Bioké) according to the manufacturer’s protocol. The yield and quality of the RNA were determined with a Nanodrop ND-1000 spectrophotometer (ThermoFisher Scientific). cDNA was synthesized with 800 ng RNA using the RevertAid H minus first-strand cDNA synthesis kit (ThermoFisher Scientific) according to the manufacturer’s protocol, using an Arktik Thermal Cycler (ThermoFisher Scientific). The final reverse transcriptional sample was diluted 5 times in RNAse-free water. RT-qPCR was performed with SYBR Green PCR Master Mix (Thermo Fisher Scientific) in a Quantstudio 6 Flex Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific). The following KiCqStart primers (Sigma-Aldrich) were used: ESR1 (forward 5′-GGAGTGTACACATTTCTGTC-3′, reverse: 5′-CAAAGTGTCTGTGATCTTGTC-3′), CCND1 (forward: 5′-GCCTCTAAGATGAAGGAGAC-3′, reverse: 5′-CCATTTGCAGCAGCTC-3′), GREB1 (forward: 5′-CTTGGTTTCTCTGGGAATTG-3′, reverse: 5′-TTCCAACAGATTAAAGGTCC-3′), PGR (forward: 5′-ATTCACTTTTTCACCAGGTC-3′, reverse: 5′-AACCTGGCAATGATTTAGAC-3′), PTGES (forward: 5′-CAAAAACATCACTCCCTCTC-3′, reverse: 5′-AAAAGTCTGCATTCTTAGCC-3′), and TFF1 (forward: 5′-CAGAATTGTGGTTTTCCTGG-3′, reverse: 5′AATTCACACTCCTCTTCTGG3′). The following primers (Sigma-Aldrich) were used for the internal standard: actin (forward: 5′-TCAAGATCATTGCTCCTCCTGAG-3′, reverse: 5′-ACATCTGCTGGAAGGTGGACA-3′), GAPDH (forward: 5′-CTGGTAAAGTGGATATTGTTGCCAT-3′, reverse: 5′-TGGAATCATATTGGAACATGTAAACC-3′) and TBP (forward: 5′-CATGACTCCCATGACCC-3′, reverse: 5′-TGGTTCGTGGCTCTCTTA-3′). Each sample was measured in triplicate and the fold change in mRNA expression relative to the average of the internal standard and the reference sample was calculated using the 2−ΔΔCT method. Three biological independent replicates were performed per experiment, unless stated otherwise. Statistical analysis (ordinary 1-way ANOVA, Dunnett’s multiple comparisons test) and graphs (mean ± SD) were made with GraphPad Prism 8 software.
Transient siRNA transfection
siGENOME SMARTpool (containing 4 single siRNAs) siRNAs for ESR1 (M-003401-02), GREB1 (M-008187-01), PGR (M-003433-01), and TFF1 (M-003715-02) were purchased from Dharmacon. Reverse transfection with 50 nM siRNA was performed in MCF7 cells in a 12-well or 96-well format at ∼70% confluency using transfection reagent INTERFERin (Polyplus) according to the manufacturer’s protocol in complete medium. Mock and siControl (cocktail of 720 SMARTpool siRNAs from the siGENOME human protein kinase library) were used as negative controls. To determine the mRNA expression by RT-qPCR and protein expression by Western blot after siRNA-mediated knockdown, the transfection medium was replaced by phenol red-free RPMI supplemented with 1% cdFBS after 16–24 h. Cells were exposed in serum-free phenol red-free RPMI after 16–24 h starvation. To determine the proliferation rate or to perform live-cell imaging, the transfection medium was replaced by experimental medium after 16–24 h and cells were exposed in experimental medium after a 16–24 h starvation period.
Cell proliferation assay
Cells were seeded in a 96-well plate (no. 3599, Corning) in complete medium. After 16–24 h, the medium was replaced by experimental medium. The next day, cells were exposed to E2 and/or 4-OHT in experimental medium. After 48, 72, and/or 96 h exposure, the cells were fixed and the sulforhodamine B (SRB) colometric assay was performed as described previously by our group (Zhang et al., 2011). Three biological independent replicates were performed per experiment, unless stated otherwise. Nonlinear regression with a fixed slope (Hill slope = 1.0) was used to determine EC50 values ± SEM with GraphPad Prism 8 software.
Generation of stable FUCCI-H2B reporter cell line
MCF7 cells were seeded in a 6-well plate (3516, Corning Costar) in complete medium and were allowed to attach overnight. The next day, the cells were transduced with a lentiviral construct containing the pLL3.7m-Clover-Geminin(1-110)-IRES-mKO2-Cdt1(30-120) (FUCCI) plasmid (no. 83841, Addgene) (Bajar et al., 2016). After 24 h the transduction medium was replaced by complete medium. After expansion, cells were FACS sorted for GFP and/or RFP expression to eliminate wild-type (WT) cells. Subsequently, MCF FUCCI cells were transduced with a lentiviral construct containing a pLentiPGK DEST H2B-iRFP670 plasmid (no. 90237, Addgene). After expansion, cells were FACS sorted for iRFP expression to eliminate iRFP-negative FUCCI cells.
Generation of stable BAC-GFP reporter cell lines
Human GREB1 (RP11-117O19), PGR (RP11-418N14), and TFF1 (RP11-550K14) BAC clones were selected and tagged at the C-terminal with GFP as described previously (Poser et al., 2008). MCF7 WT cells were seeded in a 6-well plate (no. 3599, Corning Costar) in complete medium and were allowed to attach overnight. The next day, cells were transfected with 4 µg BAC-GFP construct using Lipofectamine 2000 transfection reagent (Thermofisher Scientific) according to the manufacturer’s protocol. After 24 h the transfection medium was replaced by selection medium (complete medium + 500 µg/ml G418; Sigma-Aldrich). After selection, 6–24 (monoclonal) BAC transfected MCF7 colonies per target were separately grown out and evaluated by live-cell imaging. Subsequently, several GFP-positive clones per target were FACS sorted once (TFF1-GFP) or twice (GREB1-GFP and PGR-GFP) on GFP expression to eliminate MCF7 WT cells. After evaluation of the different clones, eventually one clone per target was fully characterized.
Western blot
To examine the protein expression in the reporter cell lines, cells were seeded in a 12-well plate (no. 3512, Corning Costar). The medium was replaced after 16–24 h by phenol red-free RPMI supplemented with 1% cdFBS. Cells were exposed after 24 h starvation to E2 in serum-free phenol red-free RPMI. Cells were lysed in sample buffer (6 times diluted bromophenol blue solution with β-mercaptoethanol) and separated by SDS-polyacrylamide gel electrophoresis and electrophoretically transferred to an Immobilon-P membrane (Millipore). Membranes were blocked for 1 h at room temperature in 5% (w/v) nonfat milk powder or 5% (w/v) BSA in Tris-buffered saline/Tween 20 and subsequently incubated overnight at 4°C with the following primary antibodies: Rabbit anti-GFP (no. 11814460001, Roche), Rabbit anti-GREB1 (no. ab72999, Abcam), Rabbit anti-PGR (no. 8757S, Cell Signaling), or Rabbit anti-TFF1 (no. ab92377, Abcam). The next day, the blots were incubated 1 h at room temperature with a goat anti-Rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (no. 111-035-003, Jackson ImmunoResearch), and protein signals were detected with ECL prime (GE Healthcare). Tubulin was used as a loading control: mouse anti-tubulin antibody (no. T-9026, Sigma-Aldrich) followed by goat anti-Mouse Alexa 647-labeled secondary antibody (no. 115-605-146, Jackson ImmunoResearch). Blots were visualized with an Amersham Imager 600 (GE Healthcare). Per experiment, 2 biological independent replicates were performed.
Live cell imaging
Cells were seeded in a 96-well (no. 655866, Greiner Bio-One) or 384-well (no. 781866, Greiner Bio-One) black screenstar imaging plate in complete medium and after 16–24 h the medium was replaced by experimental medium. After a 16–24-h starvation period, and 2 h prior to exposure to E2 and/or 4-OHT, cells were loaded with 100 ng/ml Hoechst 33342 (ThermoFisher Scientific) to visualize the nuclei. To avoid Hoechst phototoxicity, Hoechst-containing medium was removed before exposure. Hoechst, GFP, RFP, and iRFP levels were monitored using a Nikon TiE2000 confocal laser microscope (lasers: 408, 488, 561, 647 nm), equipped with an automated stage, perfect focus system, and climate chamber (at 37°C under 5% CO2 atmosphere). Imaging was done with a Nikon Plan Apo 20× magnification objective lens (1×, 2× or 4× optical zoom) using NIS elements software (Nikon).
Image analysis
Quantitative image analysis was done with CellProfiler version 2.1.1 with an in-house developed module for segmentation, as described by Wink et al. (2017). A binary mask based on the segmented nuclei served as primary objects. These primary nuclear objects served as seeds for the secondary-objects module to determine the cytoplasmic signal with a N-distance method (8 pixels). Features of interest were extracted from HDF5 files obtained from CellProfiler using an in-house developed R script (Wink et al., 2017). For the GREB1-GFP and TFF1-GFP reporter, we used the integrated GFP intensity in the cytoplasm and for the PGR-GFP reporter, we used the integrated GFP intensity in the nucleus on a single-cell level. The GFP-positive fraction of the cells was determined as GFP intensity higher than the mean GFP intensity of the control wells (DMSO) plus 2 times the standard deviation (SD) of control wells. Normalization to DMSO was done by subtracting the mean DMSO value. For visualization purposes, we have scaled some imaging data for each reporter between 0 and 1. Three biological independent replicates were performed per experiment, unless stated otherwise. Nonlinear regression with a fixed slope (Hill slope = 1.0) was used to determine EC50 values ± SEM with GraphPad Prism 8 software.
RESULTS
GREB1, PGR, and TFF1 Are E2-Inducible ERα Transcriptional Targets
To establish fluorescent reporter cell lines for ERα pathway stimulated proliferation, we selected the ERα-positive MCF7 breast cancer cell line. When treated with E2 for 48–96 h, MCF7 cells show a concentration-dependent increase in proliferation (Supplementary Figure 1A). To verify that this increased proliferation is mediated via the ERα pathway, we co-treated the cells with the breast tissue-specific ERα antagonist 4-hydroxytamoxifen (4-OHT). Pharmacological inhibition of ERα signaling by 100 nM 4-OHT resulted in a clear shift of the E2 concentration-response curve, thereby demonstrating the validity of our experimental model system (Supplementary Figure 1A). Subsequently, we set out to identify suitable inducible ERα target genes that could serve as candidate ERα activity reporters. We selected 5 potential candidates for our reporter cell lines-based transcriptomics information from our own work and other studies: CCND1, GREB1, PGR, PTGES, and TFF1 (Frasor et al., 2003; Lin et al., 2004; Moerkens et al., 2014; Vantangoli et al., 2016; Zhang et al., 2011). Except for CCND1, these genes have previously been described as primary ERα transcriptional target genes rather than indirect or secondary targets, because they contain EREs in their regulatory regions (Berry et al., 1989; Deschênes et al., 2007; Frasor et al., 2008; Lin et al., 2004; Sun et al., 2007). The CCND1 promoter does not contain a classical ERE, however, estrogen-responsive transcription is mediated via a c-Jun/c-Fos/ERα-complex (Cicatiello et al., 2004). These targets are described in literature to be upregulated after E2 exposure and they are (in)directly linked to cell proliferation (Amiry et al., 2009; Butt et al., 2005; Cenciarini and Proietti, 2019; Frasor et al., 2003; Lin et al., 2004; Rae et al., 2005; Vantangoli et al., 2016). To verify whether these targets are indeed inducible by E2 in our MCF7 cell line, cells were exposed to 1 nM E2 up to 24 h, showing different temporal dynamics of mRNA expression (Figure 1A). The mRNA expression of PGR, PTGES, and TFF1 reached maximum levels after 4–8 h, whereas GREB1 mRNA levels kept increasing up to the 24 h time point. We observed no significant changes in CCND1 mRNA expression under these treatment conditions. After 24 h of E2 exposure, only GREB1, PGR, PTGES, and TFF1 showed concentration-dependent induction (Figure 1B).
Figure 1.
GREB1, PGR, and TFF1 are E2-inducible ERα transcriptional targets. A, Expression of candidate target genes after 1 nM E2 exposure up to 24 h. Relative mRNA expression compared with t = 0 h. B, Expression of candidate target genes after 24 h exposure to different concentrations of E2. Relative mRNA expression compared with DMSO (1-way ANOVA: * = p < .05; ** = p < .01; *** = p < .001). C, Expression of candidate target genes after 24 h exposure to different concentrations of E2 with or without 100 nM 4-OHT. Relative mRNA expression compared with DMSO (1 representative biological replicate). D, Expression of candidate target genes after siRNA knockdown of ESR1 and 1 nM E2 exposure up to 24 h. Relative mRNA expression compared with siControl at t = 0 h.
To determine ERα-dependent induction, MCF7 cells were exposed to a concentration range of E2 with or without 100 nM 4-OHT for 24 h. 4-OHT exposure decreased basal mRNA expression levels of CCND1, GREB1, PGR, and TFF1 (Supplementary Figure 1B). In addition, these targets showed pharmacological inhibition after co-exposure to E2 and 100 nM 4-OHT, demonstrating ERα-dependent induction (Figure 1C, Supplementary Figure 1C). In contrast, PTGES was upregulated by 4-OHT under the same experimental conditions, which is in line with the previous observation that 4-OHT can also have agonistic effects on PTGES expression (Frasor et al., 2008). Besides pharmacological inhibition, we performed siRNA knockdown of ESR1 in MCF7 cells. A ∼60% knockdown efficiency of ESR1 was achieved (Supplementary Figure 1D), resulting in downregulation of the basal expression level of all five target genes (Supplementary Figure 1E). Also, E2 exposure did not induce target gene expression under ESR1 knockdown conditions (Figure 1D, Supplementary Figure 1F). These data identify GREB1, PGR, and TFF1 as promising candidate ERα-reporter genes.
GREB1, PGR, and TFF1 Play a Role in ERα-Mediated Cell-Cycle Progression and Proliferation
We were seeking ERα target genes that can mechanistically and quantitatively link ERα activation to cell proliferation. To verify if GREB1, PGR, and TFF1 are indeed involved in ERα-induced proliferation, we performed a series of siRNA knockdown experiments. Knockdown of ESR1 significantly reduced MCF7 proliferation and knockdown of ERα target genes GREB1, PGR, and TFF1 had similar, but less severe, effects (Supplementary Figs. 2A and 2B). Thus, besides ERα itself, also all 3 ERα target genes support proliferation of MCF7 cells.
To further examine the role of these target genes in cell-cycle progression after E2 exposure, we employed a Fluorescent Ubiquitination-based Cell Cycle Indicator (FUCCI) sensor in a live-cell imaging setup. This FUCCI sensor consists of fluorescently labeled Geminin-GFP (green) and Cdt1-RFP (red) to mark different stages of the cell cycle (Bajar et al., 2016) (Supplementary Figure 2C). Because the FUCCI reporter cells have a colorless phase directly after mitosis, we incorporated histone 2B (H2B)-iRFP (infrared fluorescent protein) as a nuclear marker for all cells during live cell imaging. Stable lentiviral integration of both the FUCCI and H2B-iRFP construct did not affect the proliferative behavior, nor did it affect ERα-mediated proliferation up to 72 h compared with the parental MCF7 WT cells (Supplementary Figs. 2D and 2E). In the absence of E2, MCF7 FUCCI-H2B cells maintained in experimental medium with charcoal-stripped serum showed reduced proliferation. This is reflected by an accumulation of cells in the G1 and early S phase of the cell cycle, and concomitant decreasing numbers of G2-M-S and early G1 cells (Supplementary Figure 2F). These changes in cell-cycle distribution are reverted upon E2 exposure as a stimulus to proliferate. We observed a concentration-dependent increase in the S-G2-M fraction with a concomitant decrease in the G1-S transition phase fraction (Figure 2A). Co-exposure to 100 nM 4-OHT resulted in pharmacological inhibition, demonstrating an ERα-mediated response (Figure 2A). In line with ERα dependency, knockdown of ESR1, GREB1, PGR, or TFF1 suppressed E2-induced proliferation (Figure 2B) and the percentage of S-G2-M cells, while increasing the percentage of cells in the G1-S transition phase of the cell cycle (Figure 2C). However, we also observed some notable differences, such as a relatively strong increase in the G1-S fraction of GREB1 depleted cells (Supplementary Figure 2G), which suggests an ERα-independent role for GREB1 in proliferation. Altogether, our results indicate that the ERα target genes GREB1, PGR, and TFF1 are important for ERα-mediated cell-cycle progression and proliferation.
Figure 2.
GREB1, PGR, and TFF1 play a role in ERα-mediated cell-cycle progression and proliferation. A, Different fractions of FUCCI-H2B reporter upon 48 h E2 exposure with or without 100 nM 4-OHT. B, Proliferation of MCF7 FUCCI-H2B cells after siRNA knockdown, 2 days after transfection followed by 48 h E2 exposure. Relative proliferation based on the amount of H2B+ cells compared with t = 0 h per knockdown condition. C, Different fractions of FUCCI-H2B reporter upon 48 h DMSO or 100 nM E2 exposure, 2 days after siRNA transfection. D, FUCCI-H2B reporter upon 48 h 100 nM E2 exposure, 2 days after siRNA transfection. Cells right after mitosis are visualized in blue, cells in the G1 phase in red, cells in the G1 to S transition phase are yellow, and cells in the S-G2-M phase are green.
Characterization of GREB1-GFP, PGR-GFP, and TFF1-GFP Reporters in MCF7 Cells
Fluorescent reporters for GREB1, PGR, and TFF1 were generated in MCF7 cells with BAC technology as described by Wink et al. (2017). After BAC transfection and selection, we obtained several candidate MCF7 colonies per target gene. For each target gene, clones were picked based on fluorescence intensity and anticipated localization of the GFP signal. Subsequently, GFP-positive cell populations were collected by fluorescence-activated cell sorting. The newly developed reporters were characterized based on the induction and localization of the GFP fusion protein, correct fusion and size of the fusion protein, and the proliferation rate compared with WT cells (Figure 3A).
Figure 3.
Characterization of GREB1-GFP, PGR-GFP, and TFF1-GFP reporters in MCF7 cells. A, Schematic overview of BAC-GFP reporter generation and selection. Adapted from Wink et al. (2017). B, Localization of GFP fusion protein of reporters. Blue = Hoechst; green = GFP (representative image for 3 independent biological replicates). C, Western blot of reporters 2 days after siRNA transfection, followed by 24 h 1 nM E2 exposure (representative image for 2 independent biological replicates). D, Reporters 2 days after siRNA transfection, followed by 24 h 1 nM E2 exposure (representative image for 3 independent biological replicates).
To determine the localization and induction of the GFP fusion protein, the reporter cell lines were exposed to 1 nM E2 and imaged after 24 h. In line with previously published data for GREB1 (Pellegrini et al., 2012) and TFF1 (Levak et al., 2018), GREB1-GFP and TFF1-GFP are mainly expressed in the cytoplasm (Figure 3B). In contrast, PGR-GFP shows predominantly nuclear localization that is expected for the PGR transcription factor (Lim et al., 1999) (Figure 3B). To verify correct GFP tagging, sizes of the GFP fusion proteins were evaluated by Western blot analysis. We observed the expected size shifts for all reporters (Supplementary Figure 3A). In addition, we performed siRNA knockdown experiments to validate our reporter cell lines. Knockdown of the target genes led to reduced expression of both the endogenous target protein and the GFP fusion protein for all reporter cell lines (Figure 3C). Knockdown of GFP only led to reduced GFP fusion protein expression in the reporter cell lines and did not alter the expression of the endogenous protein. Live-cell imaging confirmed that GFP fluorescence reports target gene expression (Figure 3D). Integration of the BAC-GFP construct did not affect proliferation for the GREB1-GFP and TFF1-GFP reporter cell line, but the PGR-GFP reporter cells showed a slightly slower proliferation rate (Supplementary Figure 3B). However, E2-induced proliferation was similar in all GFP reporter cell lines and the parental MCF7 cell line (Supplementary Figure 3C). In addition to proliferation, we assessed the E2-induced target gene expression on the protein level in the GFP reporter cell lines. All reporter cell lines demonstrated similar trends of target gene induction compared with the parental cell line, which were closely mimicked by the expression of the GFP-fusion proteins (Supplementary Figure 3D). Thus, we confirmed correct GFP tagging and observed no interference of the BAC-GFP incorporation with the ERα signaling pathway in our newly established reporter cell lines.
GREB1-GFP, PGR-GFP, and TFF1-GFP Reporters Demonstrate Different Activation Dynamics With Similar Sensitivity Towards Estrogenic Compounds
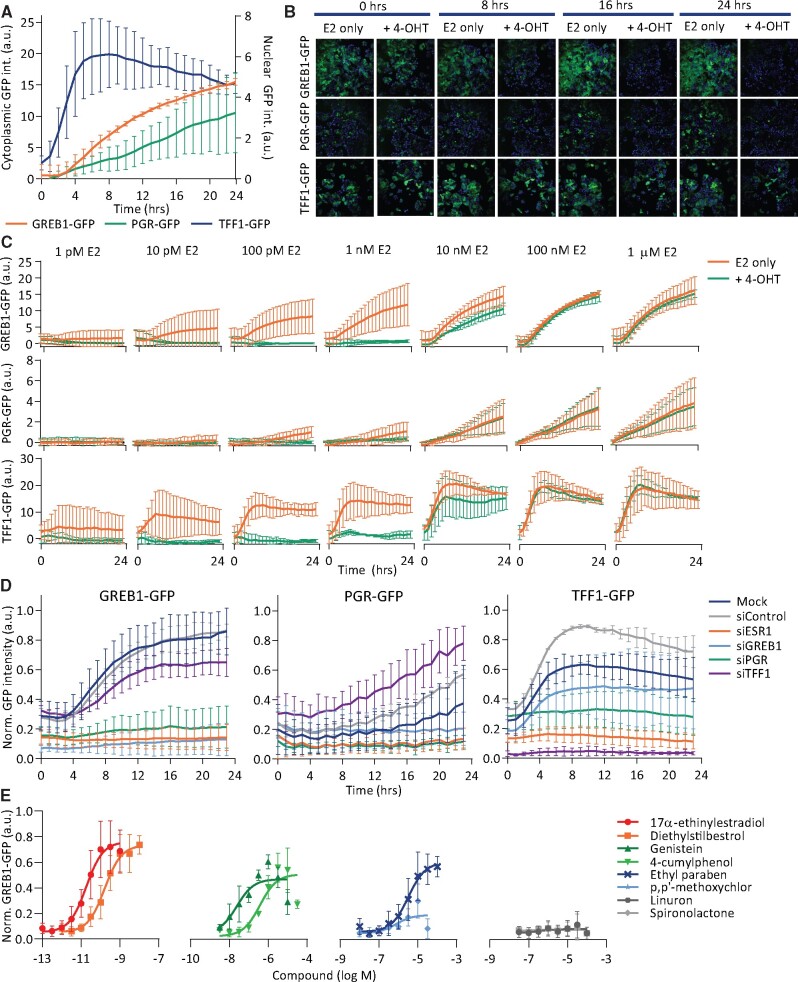
To study the dynamics of pathway activation, the GFP reporters were exposed to E2 and imaged every hour up to 24 h, demonstrating different activation dynamics. We evaluated both the nuclear and the cytoplasmic GFP signals for all 3 reporters (Figure 4A, Supplementary Figure 4A). Cytoplasmic GREB1-GFP expression was induced after 3 h E2 and kept increasing over time, whereas cytoplasmic TFF1-GFP showed fast activation with peak levels after approximately 7 h. The nuclear signal for these reporters followed the same trend as the GFP signal (Supplementary Figure 4A). Nuclear PGR-GFP was only expressed at low levels directly after exposure and steadily increased over time, whereas higher E2 concentrations led to a faster response (Figure 4C). Interestingly, the ratio of nuclear versus cytoplasmic PGR-GFP signal increased in a concentration-dependent manner (Supplementary Figure 4B). We used the cytoplasmic GFP signal for GREB1-GFP and TFF1 and the nuclear signal for the PGR-GFP reporter for further analysis.
Figure 4.
GREB1-GFP, PGR-GFP, and TFF1-GFP reporters demonstrate different activation dynamics upon exposure to estrogenic compounds. A, Mean integrated GFP intensity in the cytoplasm for GREB1 and TFF1 (left Y-axis) and mean integrated nuclear PGR-GFP intensity (right Y-axis) of reporters exposed to 100 nM E2 and imaged every hour up to 24 h. Normalized to DMSO. B and C, Mean integrated GFP intensity of reporters exposed to different concentrations E2 with or without 100 nM 4-OHT and imaged every hour up to 24 h. Normalized to DMSO. D, Min-max normalized integrated GFP intensity of reporters, 48 h after siRNA transfection followed by DMSO or 1 nM E2 exposure and imaged every hour up to 24 h. E, Min-max normalized integrated GFP intensity of GREB1-GFP reporter after 48 h exposure to chemicals with different estrogenic activity (n = 3 − 6).
To confirm that E2-induced reporter activation depends on ERα, we suppressed ERα signaling in all 3 reporter cell lines. Upon co-exposure to 100 nM 4-OHT, all reporters showed pharmacological inhibition of the response to up to 1 nM E2 over time (Figs. 4B and 4C). Furthermore, knockdown of ESR1 or the reporter gene itself led to reduced basal GFP expression in all reporter lines, and E2 exposure only increased GFP expression under control conditions (Figure 4D, Supplementary Figure 4C). Interestingly, PGR knockdown significantly reduced GREB1 expression, which may be explained by crosstalk between PGR and ERα in transcription regulation (Camden et al., 2017; Mohammed et al., 2015).
We evaluated the response to E2 with or without the presence of 100 nM 4-OHT in all 3 reporters up to 72 h. Independent of the duration of exposure, mean GFP intensity reached a plateau around 1 nM E2 (Supplementary Figure 4D). However, an overall increase in maximum mean GFP intensity was observed over time. Sensitivity to E2 and 4-OHT remained similar over time, as reflected by the EC50 values (Table 1). In addition to whole population-based analysis, we analyzed single-cell data by measuring GFP intensities per cell. We determined the overall commitment of individual cells to the response, by setting a background GFP threshold under control conditions. Cells were considered committed, that is, GFP positive, when the GFP intensity exceeded this threshold. When evaluating the GFP-positive fraction, a similar concentration-dependent response as the mean GFP signal was observed (Supplementary Figure 4E). Independent of the duration of exposure, the fraction of GFP-positive cells reached a plateau around 1 nM E2. However, a larger fraction of cells was committed to the response over time, indicating a delay in the response of some cells. Sensitivity to E2 based on the GFP-positive fraction, reflected by EC50 values in the low picomolar range (Table 2), were similar to the EC50 values based on the mean GFP intensity.
Table 1.
EC50 ± SEM Values (M) for E2 With or Without 100 nM 4-OHT Based on Mean GFP Intensity
| 24 h |
48 h |
72 h |
||||
|---|---|---|---|---|---|---|
| E2 | E2 + 4-OHT | E2 | E2 + 4-OHT | E2 | E2 + 4-OHT | |
| GREB1-GFP | 3.4 ± 1.0 e-11 | 8.1 ± 1.1 e-9 | 3.9 ± 1.0 e-11 | 6.0 ± 1.3 e-9 | 6.8 ± 1.6 e-11 | 6.9 ± 1.3 e-9 |
| PGR-GFP | 4.4 ± 3.8 e-11 | 3.1 ± 2.1 e-9 | 4.6 ± 2.2 e-11 | 3.6 ± 1.6 e-9 | 3.4 ± 1.9 e-11 | 2.7 ± 1.1 e-9 |
| TFF1-GFP | 2.2 ± 1.4 e-11 | 3.0 ± 1.5 e-9 | 2.5 ± 1.0 e-11 | 2.4 ± 0.7 e-9 | 2.7 ± 0.8 e-11 | 2.1 ± 0.7 e-9 |
Table 2.
EC50 ± SEM Values (M) for E2 With or Without 100 nM 4-OHT Based on GFP Positive Fraction
| 24 h |
48 h |
72 h |
||||
|---|---|---|---|---|---|---|
| E2 | E2 + 4-OHT | E2 | E2 + 4-OHT | E2 | E2 + 4-OHT | |
| GREB1-GFP | 2.3 ± 0.6 e-11 | 6.2 ± 1.0 e-9 | 1.7 ± 0.3 e-11 | 4.1 ± 0.7 e-9 | 2.2 ± 0.4 e-11 | 4.0 ± 0.7 e-9 |
| PGR-GFP | 3.8 ± 3.1 e-11 | 3.2 ± 2.0 e-9 | 3.5 ± 1.6 e-11 | 3.1 ± 1.1 e-9 | 2.0 ± 1.0 e-11 | 2.4 ± 0.9 e-9 |
| TFF1-GFP | 3.5 ± 2.3 e-11 | 3.2 ± 2.0 e-9 | 3.5 ± 1.6 e-11 | 2.5 ± 0.9 e-9 | 3.3 ± 0.8 e-11 | 2.5 ± 0.8 e-9 |
In addition to E2, we also evaluated the potency and magnitude of the response of our reporters toward a number of known estrogenic or non-estrogenic compounds. These compounds represent different chemical classes, varying from the highly potent pharmaceutical estrogenics 17α-ethinylestradiol and diethylstilbestrol and weaker industrial phenolic 4-cumylphenol or natural phytoestrogen genistein to OECD TG-455 approved negative controls spironolactone and linuron (OECD, 2016). Our reporters showed comparable responses, depending on the potencies of the estrogenic compounds, and no activation was observed upon exposure to the negative controls (Figure 4E, Supplementary Figure 4F). Compared with the validated OECD reference test methods, all 3 reporters showed similar sensitivity (Table 3). In conclusion, these reporters respond accurately to estrogenic compounds and can monitor dynamic pathway activation for a population of cells. In addition, the sensitive fluorescent readout allows the evaluation of reporter activity at a single-cell level.
Table 3.
EC50 ± SEM Values (M) for Estrogenic Compounds Obtained With Our GFP Reporters Versus OECD Data
| GREB1-GFPa | PGR-GFPa | TFF1-GFPa | STTAb | VM7Luc ERb | |
|---|---|---|---|---|---|
| 17β-Estradiol | 3.9 ± 1.0 e-11 | 4.6 ± 2.2 e-11 | 2.5 ± 1.0 e-11 | <1.0 e-11 | 5.63 e-12 |
| 17α-Ethinylestradiol | 1.8 ± 0.6 e-11 | 2.0 ± 0.8 e-11 | 9.0 ± 3.3 e-12 | <1.0 e-11 | 7,31 e-12 |
| Diethylstilbestrol | 1.6 ± 0.4 e-10 | 2.2 ± 0.8 e-10 | 1.5 ± 0.5 e-10 | 2.04 e-11 | 3.34 e-11 |
| Genistein | 2.4 ± 2.0 e-8 | 1.0 ± 0.6 e-7 | 8.1 ± 5.1 e-8 | 2.45 e-8 | 2.71 e-7 |
| 4-Cumylphenol | 3.7 ± 1.7 e-7 | 4.8 ± 2.6 e-7 | 3.6 ± 1.0 e-7 | 1.60 e-6 | 3.20 e-7 |
| Ethyl paraben | 3.2 ± 1.2 e-6 | 4.9 ± 3.2 e-6 | 7.5 ± 4.0 e-6 | c | 2.48 e-5 |
| p,p′-Methoxychlor | 7.5 ± 10 e-7 | 4.4 ± 3.8 e-6 | 8.9 ± 7.2 e-7 | c | 1.92 e-6 |
| Linuron | Negative | Negative | Negative | Negative | Negative |
| Spironolactone | Negative | Negative | Negative | Negative | Negative |
EC50 values were determined based on imaging data obtained 48 h after exposure.
Data obtained from OECD TG-455 for the stably transfected transactivation assay (STTA) and VM7Luc ER TA assay (OECD, 2016).
No EC50 determined.
DISCUSSION
There is a need for improved nonanimal approaches to assess the carcinogenic risk of pharmaceuticals and chemicals. To specifically evaluate the non-genotoxic carcinogenic effects of estrogenic compounds, we established MCF7 reporter cell lines for pro-proliferative ERα pathway activation. Reporters were generated by the introduction of GFP tags in different transcriptional target genes of ERα using BAC transgenesis. Three of these reporters, GREB1-GFP, PGR-GFP, and TFF1-GFP, showed ERα-dependent activation of fluorescent protein expression by the natural ERα agonist and known non-genotoxic carcinogen E2, and were able to detect other estrogenic compounds.
This novel panel of ERα reporter cell lines offers an important addition to the currently available assay systems for the assessment of estrogenic activity. Our fluorescent reporter cell lines can detect known agonistic estrogenic compounds with similar sensitivity compared with validated OECD reference test methods. In addition, these reporters can be used to evaluate estrogen antagonists such as 4-OHT. Because basal expression of these reporters is low, co-exposure to E2 is required for the detection of antagonistic estrogenic activity for chemicals. Because the development of the MCF7-based E-screen proliferation test 25 years ago (Soto et al., 1995), several biochemical and cell-based assays for estrogenicity have been developed. The U.S. Environment Protection Agency ToxCast program harbors 18 different in vitro assays that study various aspects of ERα biology, such as receptor binding, gene expression, and cell growth (Browne et al., 2015; Judson et al., 2015). However, the currently available assays cannot directly link activation of the ERα pathway to physiological outcomes such as proliferation. Moreover, these assays are not designed to monitor the dynamics of pathway activation over time or provide information at a single-cell level. A major drawback of established assays measuring the induction of ERα-mediated transcription is the lack of endogenous regulation. For instance, the GeneBLAzer ERalpha-UAS-bla GripTite assay depends on the induction of a transgenic β-lactamase reporter gene by a human ERα ligand-binding domain fused to the DNA-binding domain of the yeast transcription factor GAL4 in human kidney HEK293T cells (Huang et al., 2011). Another widely used assay, the ERα CALUX assay, is based on a transgenic luciferase reporter gene established in the U-2 OS human osteosarcoma cell line (Sonneveld et al., 2005; van der Burg et al., 2010). Thus, both assays make use of an artificial ERα pathway reporter in cell lines that are normally not estrogen responsive.
In contrast, our BAC transgenic reporters are based on natural full-length ERα target genes, expressed in functional ERα-positive MCF7 breast cancer cells. A critical advantage of BAC transgene-based reporter assays is that BACs contain a genomic copy of the target gene of interest, including its natural promotor region and other regulatory elements (Poser et al., 2008). This allows a highly physiologically relevant regulation of reporter activity that can directly quantitatively be related to cell proliferation in the same MCF7 test system. Moreover, our fluorescent reporters allow for the spatial and temporal recovery of ERα activation at the single-cell level, provide more detailed insight on the timing of activation, the potency of the activity with respect to the cell population dynamics of the activation, and the sustainability of the activity. When incorporated in a quantitative adverse outcome pathway (qAOP) modeling framework, these reporters can provide quantitative relationships between key event activation leading up to a defined adverse outcome. In our particular case, interaction of an estrogenic compound with ERα serves as the molecular initiating event, followed by consecutive key events including the target gene transcription and translation followed by cell-cycle progression leading to increased cell proliferation. Altogether, our different reporters will allow quantitative mapping of these key events for ultimate application in a qAOP framework.
The role of GREB1, PGR, and TFF1 in non-genotoxic carcinogenicity mediated by ERα signaling remains to be determined. We observed a clear decrease in MCF7 cell proliferation after siRNA-mediated knockdown of these individual target genes. By employing the FUCCI cell-cycle reporter in combination with siRNA knockdown and E2 exposure, we showed an increase of the number of cells residing in the G1 and G1-S fraction after knockdown of PGR and TFF1. Interestingly, knockdown of GREB1 resulted in an increase of cells residing in the G1-S phase, suggesting a role for GREB1 in the G1-S transition. GREB1 was identified as a gene upregulated by E2 and repressed by the ERα antagonist 4-OHT in MCF7 cells (Ghosh et al., 2000), has been described as a potential co-factor for ERα (Mohammed et al., 2013), and is required for hormone-induced proliferation in breast cancer cells (Rae et al., 2005). Besides its function in the mammary gland, GREB1 may also play a role in other hormone-responsive organs such as ovaries, prostate, uterus, and testes (Cheng et al., 2018; Hodgkinson and Vanderhyden, 2014; Laviolette et al., 2014; Pellegrini et al., 2012; Rae et al., 2006). Of the 3 target genes, PGR is by far the most widely studied. PGR is a ligand-inducible transcription factor belonging to the same superfamily of nuclear hormone receptors as ERα (Elizalde and Proietti, 2012; Lu et al., 2006) and mouse models have demonstrated an important role of PGR in promoting spontaneous and chemically induced mammary tumors (Cenciarini and Proietti, 2019). PGR is an ERα-associated protein that can modulate the transcriptional behavior of ERα (Mohammed et al., 2015; Singhal et al., 2016), but the exact mechanism of this crosstalk between ERα and PGR in relation to proliferation is not completely clear. TFF1 is an autocrine/paracrine factor which is frequently overexpressed in several cancers, including breast cancer (Perry et al., 2008), and forced expression of TFF1 results in increased cell proliferation in mammary carcinoma cells (Amiry et al., 2009). However, the role of TFF1 in cancer is still under debate, as both oncogenic and tumor-suppressive roles have been described in various organ systems (Jahan et al., 2020). The reporter cell lines we generated can be used to gain more insight in the roles of GREB1, PGR, and TFF1 in the estrogenic response. Other studies have demonstrated differences in ERα activation dynamics and target responses between individual cells using single-molecule RNA FISH (Stossi et al., 2020), or single-cell RNA sequencing (Zhu et al., 2018), highlighting the added value of single-cell analysis. In combination with advanced live-cell imaging, our MCF7 GFP-fusion reporters can visualize protein induction or translocation over time at a single-cell level. The tracking of single cells over time can provide quantitative relationships between pathway activation and cell-cycle progression and proliferation. This quantitative information is particularly suitable for dynamic computational modeling of pathway activation. Furthermore, in combination with RNA interference-based approaches, our reporters can be used as mechanistic biomarkers for a better understanding of ERα pathway regulation.
More insight in ERα pathway regulation is important in view of the initiative of the International Council for Harmonization of Technical Requirements for Pharmaceuticals for Human Use (ICH) to reduce the number of rodent studies for the prediction of human carcinogenicity. The ICH S1 Expert Working Group is evaluating the possibility to predict carcinogenic potential in an early stage of drug development, that is, at the end of clinical Phase II. At this stage, most of the nonclinical data are present, except for the 2-year bioassays in rodents that are currently required for the assessment of carcinogenicity (ICH, 2016). By integrating the knowledge on the mode of action, that is, direct and indirect pharmacology data (van der Laan et al., 2016), with the outcomes of 6-month repeat-dose in vivo toxicity studies, compounds can be classified using a weight-of-evidence approach to determine the likelihood of human carcinogenicity. Only in case, the outcome is uncertain, 2-year bioassays in rodents may have added value (van der Laan et al., 2017). When incorporated in qAOPs, in vitro toxicity assays can provide essential information for weight-of-evidence-based risk assessment. This is especially relevant for non-genotoxic mechanisms of carcinogenicity such as pro-proliferative ERα signaling, that may show differences between species. The detailed mechanistic insight and quantitative data that can be acquired with our ERα reporter cell lines make them well suited for this approach.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
Dutch Medicines Evaluation Board/College ter Beoordeling van Geneesmiddelen (CBG-MEB) and the European Commission Horizon2020 EU-ToxRisk project (681002).
DECLARATION OF CONFLICTING INTERESTS
The views expressed in this article are personal views of the authors and may not be understood or quoted as being made on behalf of, or reflecting the position of the Medicines Evaluations Board or one of its committees.
Supplementary Material
REFERENCES
- Amiry N., Kong X., Muniraj N., Kannan N., Grandison P. M., Lin J., Yang Y., Vouyovitch C. M., Borges S., Perry J. K., et al. (2009). Trefoil factor-1 (TFF1) enhances oncogenicity of mammary carcinoma cells. Endocrinology 150, 4473–4483. [DOI] [PubMed] [Google Scholar]
- Aranda A., Pascual A. (2001). Nuclear hormone receptors and gene expression. Physiol. Rev. 81, 1269–1304. [DOI] [PubMed] [Google Scholar]
- Bajar B. T., Lam A. J., Badiee R. K., Oh Y.-H., Chu J., Zhou X. X., Kim N., Kim B. B., Chung M., Yablonovitch A. L., et al. (2016). Fluorescent indicators for simultaneous reporting of all four cell cycle phases. Nat. Methods 13, 993–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M., Nunez A. M., Chambon P. (1989). Estrogen-responsive element of the human PS2 gene is an imperfectly palindromic sequence. Proc. Natl. Acad. Sci. U.S.A. 86, 1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum L. S., Fenton S. E. (2003). Cancer and developmental exposure to endocrine disruptors. Environ. Health Perspect. 111, 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne P., Judson R. S., Casey W. M., Kleinstreuer N. C., Thomas R. S. (2015). Screening chemicals for estrogen receptor bioactivity using a computational model. Environ. Sci. Technol. 49, 8804–8814. [DOI] [PubMed] [Google Scholar]
- Butt A. J., McNeil C. M., Musgrove E. A., Sutherland R. L. (2005). Downstream targets of growth factor and oestrogen signalling and endocrine resistance: The potential roles of c-Myc, cyclin D1 and cyclin E. Endocrine Relat. Cancer 12, S47–59. [DOI] [PubMed] [Google Scholar]
- Camden A. J., Szwarc M. M., Chadchan S. B., DeMayo F. J., O'Malley B. W., Lydon J. P., Kommagani R. (2017). Growth regulation by estrogen in breast cancer 1 (GREB1) is a novel progesterone-responsive gene required for human endometrial stromal decidualization. Mol. Human Reprod. 23, 646–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenciarini M. E., Proietti C. J. (2019). Molecular mechanisms underlying progesterone receptor action in breast cancer: Insights into cell proliferation and stem cell regulation. Steroids 152, 108503. [DOI] [PubMed] [Google Scholar]
- Chen Q., Tan H., Yu H., Shi W. (2018). Activation of steroid hormone receptors: Shed light on the in silico evaluation of endocrine disrupting chemicals. Sci. Total Environ. 631–632, 27–39. [DOI] [PubMed] [Google Scholar]
- Cheng M., Michalski S., Kommagani R. (2018). Role for growth regulation by estrogen in breast cancer 1 (GREB1) in hormone-dependent cancers. Int. J. Mol. Sci. 19, 2543–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicatiello L., Addeo R., Sasso A., Altucci L., Petrizzi V. B., Borgo R., Cancemi M., Caporali S., Caristi S., Scafoglio C., et al. (2004). Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via C-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 T. Mol. Cell. Biol. 24, 7260–7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couse J. F., Korach K. S. (1999). Estrogen receptor null mice: What have we learned and where will they lead us? Endocrine Rev. 20, 358–417. [DOI] [PubMed] [Google Scholar]
- Deschênes J., Bourdeau V., White J. H., Mader S. (2007). Regulation of GREB1 transcription by estrogen receptor α through a multipartite enhancer spread over 20 Kb of upstream flanking sequences. J. Biol. Chem. 282, 17335–17339. [DOI] [PubMed] [Google Scholar]
- Diamanti-Kandarakis E., Bourguignon J-p., Giudice L. C., Hauser R., Prins G. S., Soto A. M., Zoeller R. T., Gore A. C. (2009). Endocrine-disrupting chemicals: An Endocrine Society scientific statement. Endocrine Rev. 30, 293–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizalde P. V., Proietti C. J. (2012). The molecular basis of progesterone receptor action in breast carcinogenesis. Hormone Mol. Biol. Clin. Invest. 9, 105–117. [DOI] [PubMed] [Google Scholar]
- Frasor J., Danes J. M., Komm B., Chang K. C. N., Richard Lyttle C., Katzenellenbogen B. S. (2003). Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: Insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144, 4562–4574. [DOI] [PubMed] [Google Scholar]
- Frasor J., Weaver A. E., Pradhan M., Mehta K. (2008). Synergistic up-regulation of prostaglandin E synthase expression in breast cancer cells by 17beta-Estradiol and proinflammatory cytokines. Endocrinology 149, 6272–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich A., Olejniczak K. (2011). Evaluation of carcinogenicity studies of medicinal products for human use authorised via the European centralised procedure (1995–2009). Regul. Toxicol. Pharmacol. 60, 225–248. [DOI] [PubMed] [Google Scholar]
- Ghosh M. G., Thompson D. A., Weigel R. J. (2000). PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 60, 6367–6375. [PubMed] [Google Scholar]
- Haseman J. K. (2000). Using the NTP database to assess the value of rodent carcinogenicity studies for determining human cancer risk. Drug Metab. Rev. 32, 169–186. [DOI] [PubMed] [Google Scholar]
- Hernández L. G., van Steeg H., Luijten M., van Benthem J. (2009). Mechanisms of non-genotoxic carcinogens and importance of a weight of evidence approach. Mutat. Res. Rev. Mutat. Res. 682, 94–109. [DOI] [PubMed] [Google Scholar]
- Hodgkinson K. M., Vanderhyden B. C. (2014). Consideration of GREB1 as a potential therapeutic target for hormone-responsive or endocrine-resistant cancers. Expert Opin. Ther. Targets 18, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Huang R., Xia M., Cho M.-H., Sakamuru S., Shinn P., Houck K. A., Dix D. J., Judson R. S., Witt K. L., Kavlock R. J., et al. (2011). Chemical genomics profiling of environmental chemical modulation of human nuclear receptors. Environ. Health Perspect. 119, 1142–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICH. (2016). Regulatory notice on changes to core guideline on rodent carcinogenicity testing of pharmaceuticals. 2016. http://www.ema.europa.eu/docs/en_GB/document_library/Regulatory_and_procedural_guideline/2012/12/WC500136405.pdf. Accessed January 3, 2021.
- Jahan R., Shah A., Kisling S. G., Macha M. A., Thayer S., Batra S. K., Kaur S. (2020). Odyssey of trefoil factors in cancer: Diagnostic and therapeutic implications. Biochim. Biophys. Acta Rev. Cancer 1873, 188362. [DOI] [PubMed] [Google Scholar]
- Jia M., Dahlman-Wright K., Gustafsson J.-Å. (2015). Estrogen receptor alpha and beta in health and disease. Best Pract. Res. Clin. Endocrinol. Metab. 29, 557–568. [DOI] [PubMed] [Google Scholar]
- Judson R. S., Magpantay F. M., Chickarmane V., Haskell C., Tania N., Taylor J., Xia M., Huang R., Rotroff D. M., Filer D. L., et al. (2015). Integrated model of chemical perturbations of a biological pathway using 18 in vitro high-throughput screening assays for the estrogen receptor. Toxicol. Sci. 148, 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laviolette L. A., Hodgkinson K. M., Minhas N., Perez-Iratxeta C., Vanderhyden B. C. (2014). 17β-estradiol upregulates GREB1 and accelerates ovarian tumor progression in vivo. Int. J. Cancer 135, 1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levak M. T., Mihalj M., Koprivčić I., Lovrić I., Novak S., Bijelić N., Baus-Lončar M., Belovari T., Kralik K., Pauzar B. (2018). Differential expression of TFF genes and proteins in breast tumors. Acta Clin. Croat. 57, 264–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liehr J. G. (2000). Is estradiol a genotoxic mutagenic carcinogen? Endocrine Rev. 21, 40–54. [DOI] [PubMed] [Google Scholar]
- Lilienblum W., Dekant W., Foth H., Gebel T., Hengstler J. G., Kahl R., Kramer P.-J., Schweinfurth H., Wollin K.-M. (2008). Alternative methods to safety studies in experimental animals: Role in the risk assessment of chemicals under the new European Chemicals Legislation (REACH). Arch. Toxicol. 82, 211–236. [DOI] [PubMed] [Google Scholar]
- Lim C. S., Baumann C. T., Htun H., Xian W., Irie M., Smith C. L., Hager G. L. (1999). Differential localization and activity of the A- and B-forms of the human progesterone receptor using green fluorescent protein chimeras. Mol. Endocrinol. 13, 366–375. [DOI] [PubMed] [Google Scholar]
- Lin C.-Y., Ström A., Vega V. B., Kong S. L., Yeo A. L., Thomsen J. S., Chan W. C., Doray B., Bangarusamy D. K., Ramasamy A., et al. (2004). Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 5, R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N. Z., Wardell S. E., Burnstein K. L., Defranco D., Fuller P. J., Giguere V., Hochberg R. B., McKay L., Renoir J.-M., Weigel N. L., et al. (2006). International Union of Pharmacology. LXV. The pharmacology and classification of the nuclear receptor superfamily: Glucocorticoid, mineralocorticoid, progesterone, and androgen receptors. Pharmacol. Rev. 58, 782–797. [DOI] [PubMed] [Google Scholar]
- Luijten M., Olthof E. D., Hakkert B. C., Rorije E., van der Laan J.-W., Woutersen R. A., van Benthem J. (2016). An integrative test strategy for cancer hazard identification. Crit. Rev. Toxicol. 46, 615–639. [DOI] [PubMed] [Google Scholar]
- Maqbool F., Mostafalou S., Bahadar H., Abdollahi M. (2016). Review of endocrine disorders associated with environmental toxicants and possible involved mechanisms. Life Sci. 145, 265–273. [DOI] [PubMed] [Google Scholar]
- McDonnell D. P., Norris J. D. (2002). Connections and regulation of the human estrogen receptor. Science 296, 1642–1644. [DOI] [PubMed] [Google Scholar]
- Moerkens M., Zhang Y., Wester L., van de Water B., Meerman J. H. N. (2014). Epidermal growth factor receptor signalling in human breast cancer cells operates parallel to estrogen receptor α signalling and results in tamoxifen insensitive proliferation. BMC Cancer 14, 283–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H., D’Santos C., Serandour A. A., Ali H. R., Brown G. D., Atkins A., Rueda O. M., Holmes K. A., Theodorou V., Robinson J. L. L., et al. (2013). Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 3, 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H., Russell I. A., Stark R., Rueda O. M., Hickey T. E., Tarulli G. A., Serandour A. A., Serandour A. A. A., Birrell S. N., Bruna A., et al. (2015). Progesterone receptor modulates ERα action in breast cancer. Nature 523, 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD. (2016). OECD-455 performance-based test guideline for stably transfected transactivation in vitro assays to detect estrogen receptor agonists and antagonists, no. July. https://read.oecd-ilibrary.org/environment/test-no-455-performance-based-test-guideline-for-stably-transfected-transactivation-in-vitro-assays-to-detect-estrogen-receptor-agonists-and-antagonists_9789264265295-en#page1. Accessed January 3, 2021.
- Pellegrini C., Gori I., Achtari C., Hornung D., Chardonnens E., Wunder D., Fiche M., Canny G. O. (2012). The expression of estrogen receptors as well as GREB1, c-MYC, and cyclin D1, estrogen-regulated genes implicated in proliferation, is increased in peritoneal endometriosis. Fertil. Steril. 98, 1200–1208. [DOI] [PubMed] [Google Scholar]
- Perry J. K., Kannan N., Grandison P. M., Mitchell M. D., Lobie P. E. (2008). Are trefoil factors oncogenic? Trends Endocrinol. Metab. 19, 74–81. [DOI] [PubMed] [Google Scholar]
- Poser I., Sarov M., Hutchins J. R. A., Hériché J.-K., Toyoda Y., Pozniakovsky A., Weigl D., Nitzsche A., Hegemann B., Bird A. W., et al. (2008). BAC transgeneomics: A high-throughput method for exploration of protein function in mammals. Nat. Methods 5, 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae J. M., Johnson M. D., Cordero K. E., Scheys J. O., Larios J. M., Gottardis M. M., Pienta K. J., Lippman M. E. (2006). GREB1 is a novel androgen-regulated gene required for prostate cancer growth. Prostate 66, 886–894. [DOI] [PubMed] [Google Scholar]
- Rae J. M., Johnson M. D., Scheys J. O., Cordero K. E., Larios J. M., Lippman M. E. (2005). GREB1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res. Treat. 92, 141–149. [DOI] [PubMed] [Google Scholar]
- Rodgers K. M., Udesky J. O., Rudel R. A., Brody J. G. (2018). Environmental chemicals and breast cancer: An updated review of epidemiological literature informed by biological mechanisms. Environ. Res. 160, 152–182. [DOI] [PubMed] [Google Scholar]
- Schug T. T., Janesick A., Blumberg B., Heindel J. J. (2011). Endocrine disrupting chemicals and disease susceptibility. J. Steroid Biochem. Mol. Biol. 127, 204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanle E. K., Xu W. (2011). Endocrine disrupting chemicals targeting estrogen receptor signaling: Identification and mechanisms of action. Chem. Res. Toxicol. 24, 6–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva Lima B., Van der Laan J. W. (2000). Mechanisms of nongenotoxic carcinogenesis and assessment of the human hazard. Regul. Toxicol. Pharmacol. 32, 135–143. [DOI] [PubMed] [Google Scholar]
- Singhal H., Greene M. E., Tarulli G., Zarnke A. L., Bourgo R. J., Laine M., Chang Y.-F., Ma S., Dembo A. G., Raj G. V., et al. (2016). Genomic agonism and phenotypic antagonism between estrogen and progesterone receptors in breast cancer. Sci. Adv. 2, e1501924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneveld E., Jansen H. J., Riteco J. A. C., Brouwer A., van der Burg B. (2005). Development of androgen- and estrogen-responsive bioassays members of a panel of human cell line-based highly selective steroid-responsive bioassays. Toxicol. Sci. 83, 136–148. [DOI] [PubMed] [Google Scholar]
- Soto A. M., Sonnenschein C., Chung K. L., Fernandez M. F., Olea N., Olea Serrano F. (1995). The E-SCREEN assay as a tool to identify estrogens: An update on estrogenic environmental pollutants. Environ. Health Perspect. 103, 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossi F., Dandekar R. D., Mancini M. G., Gu G., Fuqua S. A. W., Nardone A., De Angelis C., Fu X., Schiff R., Bedford M. T., et al. (2020). Estrogen-induced transcription at individual alleles is independent of receptor level and active conformation but can be modulated by coactivators activity. Nucleic Acids Res. 48, 1800–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Nawaz Z., Slingerland J. M. (2007). Long-range activation of GREB1 by estrogen receptor via three distal consensus estrogen-responsive elements in breast cancer cells. Mol. Endocrinol. 21, 2651–2662. [DOI] [PubMed] [Google Scholar]
- van der Burg B., Winter R., Weimer M., Berckmans P., Suzuki G., Gijsbers L., Jonas A., van der Linden S., Witters H., Aarts J., et al. (2010). Optimization and prevalidation of the in vitro ERα CALUX method to test estrogenic and antiestrogenic activity of compounds. Reprod. Toxicol. 30, 73–80. [DOI] [PubMed] [Google Scholar]
- van der Laan J. W., Duijndam B., van den Hoorn T., Woutersen R., van de Water B. (2017). Changing the field of carcinogenicity testing of human pharmaceuticals by emphasizing mode of action. Current Opin. Toxicol. 3, 55–61. [Google Scholar]
- van der Laan J. W., Kasper P., Silva Lima B., Jones D. R., Pasanen M. (2016). Critical analysis of carcinogenicity study outcomes. Relationship with pharmacological properties. Crit. Rev. Toxicol. 46, 587–614. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout J. P., Van Der Laan J. W., De Waal E. J., Olejniczak K., Hilgenfeld M., Schmidt V., Bass R. (1997). The utility of two rodent species in carcinogenic risk assessment of pharmaceuticals in Europe. Regul. Toxicol. Pharmacol. 25, 6–17. [DOI] [PubMed] [Google Scholar]
- Vantangoli M. M., Madnick S. J., Wilson S., Boekelheide K. (2016). Estradiol exposure differentially alters monolayer versus microtissue MCF-7 human breast carcinoma cultures. PLoS One 11, e0157997-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wittenau M. S., Estes P. C. (1983). The redundancy of mouse carcinogenicity bioassays. Toxicol. Sci. 3, 631–639. [PubMed] [Google Scholar]
- Wink S., Hiemstra S., Herpers B., van de Water B. (2017). High-content imaging-based BAC-GFP toxicity pathway reporters to assess chemical adversity liabilities. Arch. Toxicol. 91, 1367–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W., Yager J. D., Wang J.-P., Jupe E. R., Santen R. J. (2013). Estrogen receptor-dependent and independent mechanisms of breast cancer carcinogenesis. Steroids 78, 161–170. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Moerkens M., Ramaiahgari S., de Bont H., Price L., Meerman J., van de Water B. (2011). Elevated insulin-like growth factor 1 receptor signaling induces antiestrogen resistance through the MAPK/ERK and PI3K/Akt signaling routes. Breast Cancer Res. 13, R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Zhao Z., Cui G., Chang S., Hu L., See Y. X., Lim M. G. L., Guo D., Chen X., Poudel B., et al. (2018). Single-cell transcriptome analysis reveals estrogen signaling coordinately augments one-carbon, polyamine, and purine synthesis in breast cancer. Cell Rep. 25, 2285–2298. e4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.