Abstract
Commonly found flavonols in plants are synthesized from dihydroflavonols by flavonol synthase (FLS). The genome of Arabidopsis thaliana contains six FLS genes, among which FLS1 encodes a functional enzyme. Previous work has demonstrated that the R2R3-MYB subgroup 7 transcription factors MYB11, MYB12, and MYB111 redundantly regulate flavonol biosynthesis. However, flavonol accumulation in pollen grains was unaffected in the myb11myb12myb111 triple mutant. Here we show that MYB21 and its homologs MYB24 and MYB57, which belong to subgroup 19, promote flavonol biosynthesis through regulation of FLS1 gene expression. We used a combination of genetic and metabolite analysis to identify the role of MYB21 in regulating flavonol biosynthesis through direct binding to the GARE cis-element in the FLS1 promoter. Treatment with kaempferol or overexpression of FLS1 rescued stamen defects in the myb21 mutant. We also observed that excess reactive oxygen species (ROS) accumulated in the myb21 stamen, and that treatment with the ROS inhibitor diphenyleneiodonium chloride partly rescued the reduced fertility of the myb21 mutant. Furthermore, drought increased ROS abundance and impaired fertility in myb21, myb21myb24myb57, and chs, but not in the wild type or myb11myb12myb111, suggesting that pollen-specific flavonol accumulation contributes to drought-induced male fertility by ROS scavenging in Arabidopsis.
Keywords: Arabidopsis, flavonol, FLS1 promoter, MYB21, transcription factors, ROS scavenging
MYB21 modulates the accumulation of flavonols through directly binding to the GARE cis-element of the FLS1 promoter, which is essential for the reduced-fertility phenotype of the myb21 mutant.
Introduction
Flavonoids are synthesized via the phenylpropanoid pathway and include flavonols, anthocyanins, and proanthocyanidins (Falcone Ferreyra et al., 2012). Among them, the colorless flavonols are well known for their physiological functions and serve as protectants against UV radiation, regulators of fertility and auxin transport, and signals for pollinators and other organisms (Mo et al., 1992; Brown et al., 2001; Kuhn et al., 2011; Lewis et al., 2011; Emiliani et al., 2013). In plants, kaempferol, quercetin, and myricetin (and their derivatives) are often the major flavonol components, the composition and distribution of which are influenced by developmental and environmental factors (Winkel-Shirley, 2002).
The biosynthesis of flavonols follows the flavonoid pathway, where at the flavonol branch the oxidation of dihydroflavonols to flavonols is catalyzed by flavonol synthase (FLS) (Wisman et al., 1998; Owens et al., 2008). In Arabidopsis thaliana, there are six FLS genes, referred to as FLS1 to FLS6. Among their products, FLS1 is functional and shows strong enzymatic activity (Owens et al., 2008; Stracke et al., 2009). FLS3 is responsible for the formation of flavonols in the fls1 mutant (Preuss et al., 2009). The FLS1 gene is highly expressed in flowers, which also show flavonol accumulation (Peer et al., 2001; Nguyen et al., 2016). The transcription factors MYB11, MYB12, and MYB111, which belong to subgroup 7 of the R2R3-MYB family, have been reported to regulate flavonol biosynthesis (Stracke et al., 2007, 2010). Among them, MYB12 and MYB111 play a major role in the regulation of flavonol biosynthesis. While MYB12 acts mainly in the root, MYB111 acts primarily in the cotyledons. In fact, although flavonol accumulation was drastically reduced in the myb11myb12myb111 triple mutant, the amounts of flavonol products in pollen grains were equal to that in the wild type (Stracke et al., 2007, 2010). This phenotype suggests that there might be additional transcription factors that regulate FLS1 expression in the stamen.
Many transcription factors have been reported to influence stamen development; among these, MYB21 and its homologs MYB24 and MYB57, which belong to the R2R3-MYB subgroup 19, were either enriched or highly expressed in the stamen (Cheng et al., 2009; Dubos et al., 2010). Mutations of these three MYBs lead to indehiscent anthers, unviable pollen grains, and shorter stamen filaments (Cheng et al., 2009; Song et al., 2011; Qi et al., 2015). It has been demonstrated that gibberellin (GA) induces the expression of MYB21, MYB24, and MYB57 by increasing jasmonate production, resulting in the promotion of stamen development (Cheng et al., 2009). Later research revealed that the jasmonate-ZIM domain (JAZ) and DELLA proteins could interact with MYB21 and MYB24 to attenuate their regulatory activity in jasmonate/GA-regulated anther development and filament elongation, respectively (Song et al., 2011; Huang et al., 2020). Most recently, it has been reported that MYB99 modulates phenylpropanoid biosynthesis in a regulatory triad with MYB21 and MYB24 (Battat et al., 2019). Furthermore, Shan et al. (2020) reported that FhMYB21L1 and FhMYB21L2 activated FhFLS1 expression by directly binding to its promoter regions in Freesia hybrida. Given that AtMYB21 and AtMYB24 are the homologs of FhMYB21L1 and FhMYB21L2 in Arabidopsis, both have been predicted to regulate AtFLS1 expression (Shan et al., 2020). However, the molecular mechanism and biological function of MYB21/24/57 in the regulation of flavonol biosynthesis require further investigation in vitro and in vivo.
Here, we report that MYB21 and its homologs promote the expression of FLS1 and increase the accumulation of flavonols in Arabidopsis. Furthermore, we show that the representative transcription factor MYB21 regulates FLS1 expression by directly binding to GARE elements in its promoter, and that Arabidopsis plants overexpressing MYB21 accumulate flavonols in their flowers. More interestingly, when treated with kaempferol (one of the primary flavonols present in reproductive tissues) or when overexpressing FLS1, the siliques of the myb21 mutant contained more seeds. Consistent with the accumulation of excess reactive oxygen species (ROS) in the myb21 mutant, treatment with the ROS inhibitor diphenyleneiodonium chloride (DPI) could partly rescue the sterility phenotype of myb21. Although the mechanism requires further investigation, we further hypothesized that a MYB21-mediated flavonoid metabolism in pollen grains was essential for stamen development and male fertility, and this is probably because flavonols can prevent ROS from reaching damaging levels.
Materials and methods
Plant materials, constructs, and primers
Plants of Arabidopsis thaliana ecotype Columbia-0 (Col-0) were grown in a growth room at 22 °C with a 16 h light/8 h dark cycle unless otherwise indicated. The Arabidopsis Genome Initiative identifiers of the three MYBs under study are as follows: MYB21 (At3g27810), MYB24 (At5g40350), and MYB57 (At3g01530). The myb21 (SALK_042711), myb24 (SALK_017221), myb57 (SALK_065776), and myb21myb24myb57 mutants were kind gifts from D. X. Xie. The seeds of chs/tt4-11 (SALK_020583) and myb11myb12my111 mutant were a gift from J. R. Huang.
Transgenic plants overexpressing MYB21-FLAG (three copies of FLAG in tandem) in Col-0 and the myb21 mutant were generated by cloning MYB21 cDNA into the JW819 vector under the control of the 35S promoter. The overexpression of MYB21-FLAG was able to complement the phenotype of the myb21 mutant, including its shortened stamen and reduced fertility (Supplementary Fig. S1). To produce the promoter–GUS reporter construct, the upstream DNA fragments of FLS1 (837 bp) were amplified by PCR and ligated into the pBI121 vector. β-Glucuronidase (GUS) histochemical analysis was performed as described previously (Jefferson et al., 1987). At least five independent transformants were selected. To produce ProAlcA:MYB21, the expression of MYB21 was driven by the ethanol-inducible AlcA promoter, and the resultant fragment was inserted into the pMLBart vector harboring a Pro35S:AlcA cassette (Roslan et al., 2001). To produce FLS1-overexpressing (FLS1OE) myb21, the FLS1 cDNA fragment was inserted into pCAMBIA1300 under the control of the 35S promoter. The overexpression of FLS1 in the myb21 mutant was able to rescue the infertility phenotype of myb21 in 16 of 21 individual plants. These vectors were introduced into Arabidopsis plants by the Agrobacterium tumefaciens-mediated floral dip method (Clough et al., 2010). The sequences of primers used in this investigation are listed in Supplementary Table S1.
Plant treatments
For ethanol treatment, the ProAlcA:MYB21 plants were grown in soil until flowering (5 weeks old). The plants were then sprayed with 1% ethanol or water (mock control). One hour later, plant materials were harvested and total RNA was extracted.
For the kaempferol and DPI treatments, 10 mM kaempferol (Sigma-Aldrich) and DPI (Sigma-Aldrich) in DMSO solution were used. Before floral stage 13 (flower in full bloom), the plants were sprayed with 2 μM kaempferol and 20 μM DPI five times at intervals of 2 days. The same concentration of DMSO was used as a mock treatment. The phenotypes of inflorescences and siliques were observed and analyzed.
Expression analyses
For total RNA extraction, plant materials were homogenized in liquid nitrogen and mixed with Trizol reagent (Invitrogen) following the manufacturer’s instructions. First-strand cDNA was synthesized from 1 μg of total RNA using a reverse transcription system (TaKaRa). The fragments of interest were amplified by real-time PCR (qRT–PCR) using sequence-specific primers (Supplementary Table S1). qRT–PCR was conducted on an ABI7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) using GoTaq qPCR master mix (Promega) in accordance with the manufacturer’s instructions, and the gene expression level was normalized to that of β-TUBULIN2 (At5g62690). The primers used in this study are listed in Supplementary Table S1.
Dual-luciferase assay
Transient expression experiments and dual-luciferase assays were carried out as described previously (Liu et al., 2008). The promoter region of FLS1 was cloned into the pGreenII 0800-LUC vector. The 35S::REN gene (Renilla luciferase) in the vector was used as an internal control. Equal concentrations and volumes of transformed Agrobacterium strains were mixed and co-infiltrated into Nicotiana benthamiana leaves by using a needleless syringe. After culturing for 3 days, the infiltrated leaves were harvested and the fluorescent values of firefly luciferase (LUC) and Renilla luciferase (REN) were analyzed using a Dual-Luciferase Reporter Assay System (Promega). The ratio of LUC to REN represented the activity of the FLS1 promoter with or without the effect of the transcription factors MYB21, MYB24, and MYB57, as well as MYB12 and MYB99.
Yeast one-hybrid assay
The PCR products of MYB21 were amplified and purified, and full-length and truncated derivatives of the FLS1 promoter were inserted into a pHIS2.1 vector. MYB21-pGADT7-Rec (pGADT7-Rec plasmid digested by SmaI) was then co-transformed with bait vectors into yeast strain Y187 and plated on SD/-Leu/-Trp medium. Colonies were then transferred to SD/-Leu/-Trp/-His deficient medium with 25 mM 3-amino-1,2,4-triazole for 3 days.
Chromatin immunoprecipitation
Approximately 1 g of seedlings of 10-day-old Pro35S:MYB21-FLAG and wild-type plants were harvested for chromatin immunoprecipitation (ChIP)–qRT assays. The ChIP experiments were performed using an EpiQuikTM Plant ChIP Kit (EpiGentek, P-2014–48). According to the manufacturer’s instructions, the chromatin from the plant cells is extracted and sheared; the length of sheared DNA fragments is 200–500 bp. One-third of the crude chromatin extracts was saved for use as an input control, and the other two-thirds were treated with anti-FLAG antibody or (as a negative control) normal mouse IgG, separately. DNA was released from the antibody-captured protein–DNA complex and purified. Eluted DNA was further analyzed by qRT–PCR, with the Actin8 promoter as a reference, as described (Yu et al., 2010). The relative enrichment of each fragment was calculated first by normalizing the value for anti-FLAG against the value for the normal IgG control and then by comparing the result for Pro35S:MYB21-FLAG with that for the wild-type plants (Yu et al. 2015).
Electrophoretic mobility shift assay
The full-length coding sequence of MYB21 was cloned into the GST fusion vector (pGEX-4T-1) and then transformed into BL21 (DE3) Escherichia coli. The recombinant protein GST-MYB21 was purified using a Glutathione Sepharose 4 Fast Flow kit (GE Healthcare) according to the manufacturer’s protocol. The DNA fragments containing GARE motifs in the FLS1 promoter were amplified using biotin-labeled primers (Supplementary Table S1) and purified using a PCR purification kit (Qiagen). The unlabeled wild-type (TTGTTA) and mutant (ATCTCC) probes were then tested for binding to GARE in competitive electrophoretic mobility shift assays (EMSAs). EMSA was performed using the Light Shift Chemiluminescent EMSA kit (Thermo Scientific) according to the manufacturer’s instructions. The migration of biotin-labeled probes was detected using an enhanced chemiluminescence substrate (Thermo Scientific) and the ChemDoc XRS system (Bio-Rad).
In situ DPBA staining
For in situ visualization of flavonols, a method from Sheahan and Rechnitz (1992) was adapted. Inflorescences of plants were bleached with ethanol overnight at room temperature, and pollen grains were harvested from 10 flowers at late floral stage 12 to early stage 13 (just beginning to open) and stained with a freshly prepared aqueous solution of 0.25% (w/v) diphenylboric acid 2-aminoethylester (DPBA) and 0.00375% (v/v) Triton X-100 for at least 20 min. A Zeiss LSM LSM710 confocal laser scanning microscope was used to excite the pollen grains with 30% maximum laser power at 458 nm, and the fluorescence was collected at 475–504 nm for kaempferol and 577–619 nm for quercetin (Lewis et al., 2011). Fluorescence was visualized on a Leica DM5500 B epifluorescence microscope (Leica, Wetzlar, Germany) with an excitation wavelength of 340–380 nm and a 425 nm long-pass splitter (Starcke et al., 2010).
Flavonol extracts
Flavonol extracts were produced from ~100 mg of plant material in 2 ml reaction tubes by the addition of 0.5 ml of 80% methanol. Samples were frozen with liquid nitrogen and then homogenized. Homogenized samples were incubated for 15 min at 70 °C and centrifuged for 10 min at 15 000 g. The supernatants were transferred to new tubes and the extraction was repeated once. After the second extraction, the supernatants were combined and vacuum-dried at 35 °C.
HPTLC with subsequent DPBA staining
A 1 μl volume of flavonol extract was spotted on to 10 cm ×10 cm silica-60 high-performance thin-layer chromatography (HPTLC) glass plates (Merck, Darmstadt, Germany). A system of ethyl acetate/formic acid/acetic acid/water (100:26:12:12, v/v/v/v) was chosen as the mobile phase for the chromatography (Starcke et al., 2010) in a closed glass tank. Separated flavonols were visualized by spraying the plates with a solution of 0.25% DPBA (w/v) in ethanol, followed by fast drying. The stained chromatograms were observed under UV light (Lewis et al., 2011). The quantitative ultra-performance liquid chromatography-quadrupole-time of flight mass spectrometry (UPLC/Q-TOF MS) data of different flavonol derivatives are shown in Supplementary Table S2.
Metabolic extraction determined by LC-MS
The profiling of the flavonols was implemented by an UPLC system (Agilent 1290) coupled to a Q-TOF mass spectrometer (Agilent 6545). The methods used were as described in previous studies (Yonekura-Sakakibara et al., 2008; Stracke et al., 2010) with modifications. Briefly, the dried pellets of flavonol extracts (prepared as described above) were dissolved in 300 μl of 80% methanol containing 3 μg of isovitexin as the internal standard. UPLC was performed on an Agilent Extend 300-C18 column (4.6 mm×150 mm×3.5 μm) with a flow rate of 0.8 ml min–1 at 30 °C. Compounds were separated by gradient elution with solvent A (0.2% acetic acid in water) and solvent B (0.2% acetic acid in acetonitrile) with the following elution profile: 0 min 98% A, 2% B; 1 min 98% A, 2% B; 2 min 85% A, 15% B; 15 min 78% A, 22% B; 17.5 min 50% A, 50% B; 19 min 100% B; 21.5 min 100% B; 22 min 98% A, 2% B.
The flavonol glycosides were determined by using a Q-TOF mass analyzer with a Dual AJS electrospray ionization (ESI) ion source. Full-scan mass spectra from 100 to 1000 m/z were used for the detection of [M + H]+ and the peak of fragment ions at one scan per second in a positive mode. Nitrogen gas was used as the sheath gas. The ESI-MS was performed at a gas temperature of 300 °C with a flow of 6 l min–1, nebulizer pressure 206 kPa, and spray voltage 3.5 kV.
The qualitative analyses were confirmed by comparison with reported data (Tohge et al., 2005, 2007; Routaboul et al., 2006; Yonekura-Sakakibara et al., 2007, 2008), mainly including the retention time of the peak and the m/z values of flavonoid glycosides [M + H]+ with 10 V cone voltage and patterns of MS2 fragmentation under 20 V cone voltage (Supplementary Table S3).
CM-H2DCFDA staining of pollen grains
Anthers were detached from 10 flowers during late flower stage 12 to early stage 13 (just beginning to open) and placed in a 2 ml conical tube. The pollen was then released from the anthers by vortexing (Muhlemann et al., 2018; Luria et al., 2019) and resuspended in pollen viability solution (PVS; 290 mM sucrose, 1.27 mM Ca(NO3)2, 0.16 mM boric acid, 1 mM KNO3) containing 5 μM 5-(and 6)-chroromethyl-2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA) and 0.005% (v/v) Triton X-100. The PVS containing CM-H2DCFDA was then replaced with fresh PVS, and the stained pollen grains and stamen were placed on a microscope slide. Fluorescence was visualized on an inverted fluorescence microscope (ZEISS Axio Vert.A1) after excitation with a 488 nm laser. The quantification of viable pollen grains and stamens was performed using Fiji (Schindelin et al., 2012).
Statistical analysis
Differences were analyzed using Student’s t-test when comparing two variables and ANOVA with Fisher’s least significant difference (LSD) test when comparing three or more conditions. Values of P<0.05 were considered statistically significant. All analyses were performed using ORIGIN 8 software.
Results
MYB21 and its homologs positively regulate flavonol biosynthesis
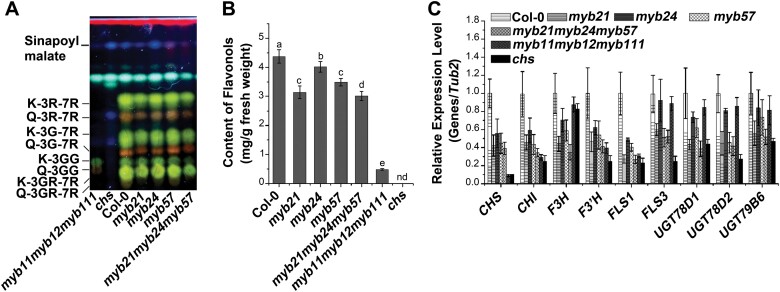
To explore whether the Arabidopsis stamen-enriched gene MYB21 and its homologs (MYB24 and MYB57) participate in flavonol biosynthesis, we quantified the flavonol content of inflorescences in the myb single mutants myb21, myb24, and myb57 and in the triple mutant myb21myb24myb57. Two flavonoid-deficient mutants, myb11myb12myb111 (lacking flavonols, except in the pollen grains) and chs (lacking all flavonoids), were used as positive controls. Analysis by HPTLC with subsequent DPBA staining showed that the inflorescences of the three myb single mutants accumulated less flavonol than the wild type, and the reduction in myb21myb24myb57 was more drastic (Fig. 1A). These results were consistent with the results of LC-MS analysis (Fig. 1B).
Fig. 1.
MYB mutations lead to reduced flavonol accumulation and flavonol synthase gene expression. (A) Flavonol glycoside accumulation patterns of the inflorescences of wild-type (Col-0) plants and myb21, myb24, myb57, myb21myb24myb57, myb11myb12myb111, and chs mutants. The flavonol accumulation of both myb11myb12myb111 and chs mutants is included as a reference. Methanolic extracts were separated by HPTLC followed by DPBA staining and UV illumination. (B) Quantification of flavonols in the inflorescences of wild-type plants and myb21, myb24, myb57, myb21myb24myb57, myb11myb12myb111, and chs mutants. The flavonol accumulation of both myb11myb12myb111 and chs mutants is included as a reference. nd, not detected. Error bars indicate the SD of three biological replicates. Different letters above the bars indicate significant differences in flavonol content between the wild type and other genotypes (P<0.05; ANOVA with Fisher’s LSD test). (C) Gene expression of flavonol biosynthesis genes in inflorescences of the wild type and the myb21, myb24, myb57, and myb21myb24myb57 mutants. The related gene expression of both myb11myb12myb111 and chs mutants is included as a reference. Transcripts were analyzed by qRT–PCR and β-TUBULIN2 was used as the internal standard. Error bars indicate the SD of three biological replicates.
We then investigated whether the transcription of flavonol biosynthesis genes was down-regulated in these myb mutants. Among the enzymes of flavonol biosynthesis, FLS is responsible for flavonol production, and A. thaliana At5g08640 was previously characterized to encode a functional FLS, designated FLS1, whose transcripts were found to be enriched in developing inflorescences, floral buds, flowers, and siliques (Owens et al., 2008). The expression of flavonol biosynthesis genes in Arabidopsis inflorescences was examined by qRT–PCR, which showed that the transcript levels of CHS (AT5G13930), CHI (AT3G55120), F3H (AT3G51240), F3′H (AT5G07990), FLS1, FLS3 (AT5G63590), UGT78D1 (AT1G30530), UGT78D2 (AT5G17050), and UGT79B6 (AT5G54010) were lower in the myb21, myb24, and myb57 mutants, and down-regulated to a greater extent in the myb21myb24myb57 triple mutant (Fig. 1C).
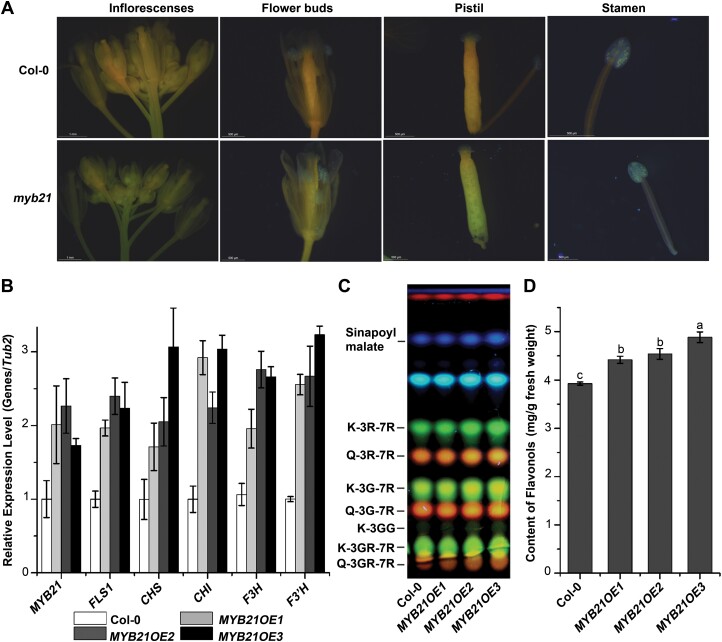
Redundant function and similar behavior have been reported among MYB21, MYB24, and MYB57 (Song et al., 2011; Huang et al., 2020). Given that the mutation of MYB21 alone causes defects in stamen development, leading to reduced fertility (Cheng et al., 2009; Song et al., 2011), we then focused on the role of MYB21 in FLS1 expression, as a representative flavonol biosynthesis gene. After staining with DPBA, the developing flowers of wild-type plants exhibited yellow fluorescence, indicating the accumulation of flavonols. By contrast, in parts of myb21 mutant flowers, including petals, pistils, filaments, and pollen, the flavonol accumulation was reduced (Fig. 2A). We found that the expression levels of FLS1 were higher in MYB21OE transgenic plants (Fig. 2B). In addition, the transcript levels of CHS, CHI, F3H, and F3′H were up-regulated in the inflorescences of MYB21OE plants, indicating that MYB21 might control the whole flavonol biosynthetic pathway (Fig. 2B). Consistently, the MYB21OE plants accumulated more flavonols than wild-type plants (Fig. 2C, D).
Fig. 2.
MYB21 and its homologs mediate flavonol biosynthesis through the regulation of flavonol synthase gene expression. (A) In situ flavonol staining of wild-type (Col-0) and myb21 inflorescences. Flavonols in ethanol-bleached inflorescences were stained with DPBA to saturation and imaged by epifluorescence microscopy. Bars=1 mm (for inflorescences) or 500 μm (for buds, pistils, and stamens). (B) Gene expression of FLS1, MYB21, CHS, CHI, F3H, and F3′H in inflorescences of wild-type (Col-0) and Pro35S:MYB21-FLAG (MYB21OE1–3) plants. Transcripts were analyzed by qRT–PCR. Error bars indicate the SD of three biological replicates. (C) Flavonol glycoside accumulation patterns of the inflorescences of wild-type and Pro35S:MYB21-FLAG (MYB21OE1–3) plants. Methanolic extracts were separated by HPTLC followed by DPBA staining and UV illumination. (D) Quantification of flavonols in the inflorescences of wild-type and Pro35S:MYB21-FLAG (MYB21OE1–3) plants. Error bars indicate the SD of three biological replicates. Different letters above the bars indicate significant differences in flavonol content between the wild type and other genotypes (P<0.05; ANOVA with Fisher’s LSD test).
MYB21 directly targets the FLS1 promoter
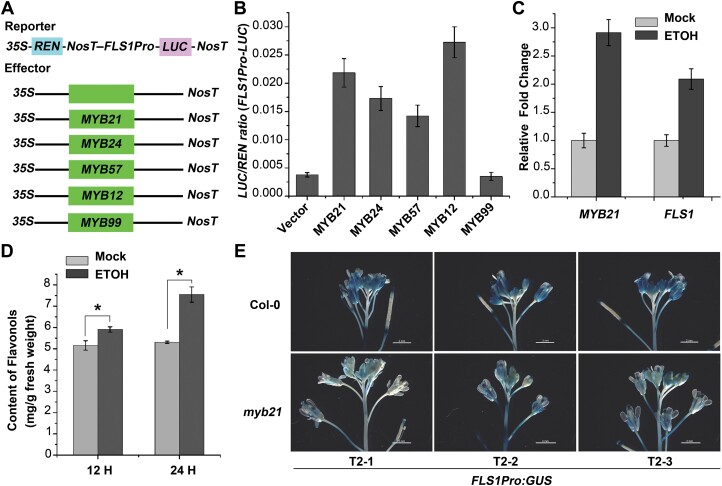
To determine how MYB21 and its homologs regulate the expression of FLS1, we performed a dual-luciferase assay by generating an FLS1Pro-LUC reporter. Here, we used MYB21, MYB24, and MYB57, as well as two other MYB transcription factors involved in flavonol biosynthesis (MYB12 and MYB99), under the control of the CaMV 35S promoter as effectors (Fig. 3A). The results showed that MYB21, MYB24, and MYB57 were functional in activating the FLS1 promoter, and among these the activity of MYB21 was relatively higher (Fig. 3B). We also found that MYB12, but not MYB99, could promote the transcription of FLS1 (Fig. 3B), which is consistent with a previous report (Mehrtens et al. 2005). We then used an alcohol-responsive promoter to generate MYB21 transient-induction plants (ProAlcA:MYB21). A clear (nearly 2-fold) increase in the abundance of FLS1 transcripts was observed 1 h after the application of 1% ethanol to flowers of the 5-week-old ProAlcA:MYB21 plants (Fig. 3C). We found that the concentration of flavonols also increased after prolonged ethanol treatment (Fig. 3D). To further examine the regulatory role of MYB21 in FLS1 expression, a ProFLS1:GUS plant was crossed with the myb21 mutant. In the wild-type background, ProFLS1:GUS exhibited strong GUS activity in inflorescences, a pattern consistent with a previous report (Owens et al., 2008). In the myb21 background, however, the GUS staining in inflorescences was much fainter (Fig. 3E), which suggested that MYB21 regulates FLS1 expression by acting on its promoter.
Fig. 3.
MYB21 directly regulates FLS1 gene expression and final products by increasing promoter activity. (A) Schematic of the structures of effector (35S:MYB21, 35S:MYB24, 35S:MYB57, 35S:MYB12, and 35S:MYB99) and reporter (FLS1Pro:LUC) constructions for the dual-luciferase transient expression assay. The 35S:REN gene (Renilla luciferase) in the pGreenII 0800-LUC vector was used as an internal control. (B) Relative LUC/REN ratio from the transient expression assays. The ratio of firefly luciferase (LUC) to REN represents the activity of the FLS1 promoter in the absence or presence of MYB21 and its homologs. MYB12 was included as a positive control. Error bars indicate the SD of three biological replicates. (C) Expression of MYB21 and FLS1 in ProAlcA:MYB21 inflorescences. The plants were sprayed with 1% ethanol, and the total RNA was isolated for analysis 1 h later. Error bars indicate the SD of three biological replicates. (D) Quantification of flavonols in the inflorescences of ProAlcA:MYB21. The plants were sprayed with 1% ethanol, and the flavonols were isolated after 12 h and 24 h of treatment. Error bars indicate the SD of three biological replicates. *P<0.05 (Student’s t-test). (E) GUS staining of inflorescences of ProFLS1:GUS in the wild-type (Col-0) and the myb21 background. ProFLS1:GUS T2-1/2/3 represents T2 generations of three individual transgenic lines, whereas ProFLS1:GUS/myb21 plants were produced via crossing a single ProFLS1:GUS line with myb21. The florescence samples were incubated in GUS reaction mixture at 37 °C for 4 h. Bars=2 mm.
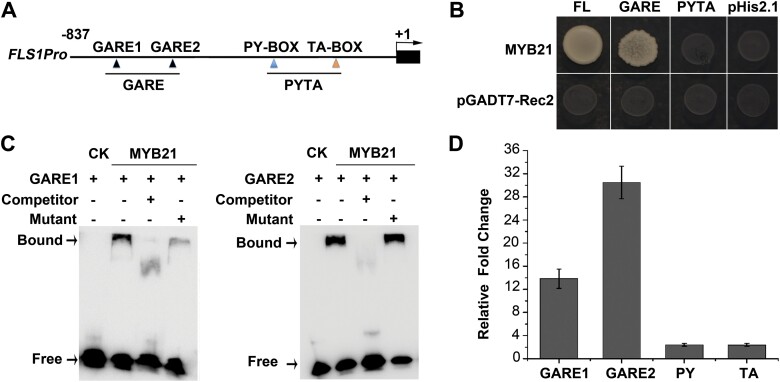
Since MYB21 is repressed by DELLA proteins and acts downstream of the GA pathway (Cheng et al., 2009), we investigated whether it functions as GAMYB and directly binds to the GAREs in the FLS1 promoter. Sequence analysis by PLACE (http://www.dna.affrc.go.jp/htdocs/PLACE/) revealed that the FLS1 promoter (–837 bp) contains two GARE cis-elements (Fig. 4A), which could be recognized by GAMYB (Gubler et al., 1995, 1999). In a yeast one-hybrid assay, the full-length and truncated derivatives of the FLS1 promoter were used as bait. We found that MYB21 bound to the full-length and GARE cis-elements of the FLS1 promoter (Fig. 4B). We then performed EMSA using a recombinant GST-MYB21 protein produced in bacterial cells. We found that GST-MYB21 was able to bind to both GARE1 and GARE2 of the FLS1 promoter. The specificity of this binding was further confirmed in a competition assay with excess amounts of unlabeled wild-type or mutant probes (Fig. 4C). To further demonstrate whether MYB21 could bind to the FLS1 promoter in vivo, we performed a ChIP assay using a transgenic line that expressed a fusion of MYB21-FLAG under the control of the 35S promoter. Chromatin extracted from 10-day-old seedlings was immunoprecipitated with an antibody against FLAG. The presence of FLS1 promoter sequences was analyzed by qPCR (Fig. 4D). Among the four regions we analyzed, two (GARE1 and GARE2) containing the GARE boxes were amplified in 35S:MYB21-FLAG samples after pulldown with anti-FLAG antibodies. This suggests that MYB21 is able to bind to the promoter region of FLS1. Together, these results demonstrate that MYB21 promotes flavonol biosynthesis mainly through positive regulation of FLS1 at the transcriptional level via the GARE domains in its promoter.
Fig. 4.
MYB21 recognizes and binds to the GARE elements of the FLS1 promoter. (A) Schematic diagrams of the FLS1 promoter constructs in yeast one-hybrid assays. Black triangles indicate GARE cis-elements (TTGTTA), and blue and orange triangles indicate the PY-BOX (TTTTTTCC) and TA-BOX (TATCCA), respectively. (B) Yeast one-hybrid assays of the interactions between MYB21 and FLS1 promoter fragments. The empty vectors of pGADT7-Rec were used as a negative control. (C) EMSA indicating that MYB21 binds to the GARE motifs. Recombinant GST-MYB21 was purified from E. coli. The promoter fragment containing the two GARE motifs of the FLS1 promoter was labeled with biotin. A competition assay for the protein–DNA binding was performed using 100× unlabeled wild-type (Competitor) and mutant probes. (D) ChIP enrichment of FLS1 promoter regions bound by MYB21-FLAG. Ten-day-old Pro35S:MYB21-FLAG and wild-type (Col-0) seedlings were used; DNA fragments were analyzed by quantitative PCR, with the Actin8 promoter as a reference. Enrichments in Pro35S:MYB21-FLAG were compared with wild-type seedlings. Error bars indicate the SD of three PCR repeats of four separate samples.
MYB21-regulated flavonol accumulation is essential for stamen development
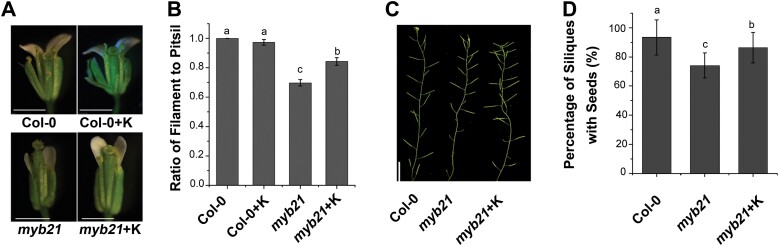
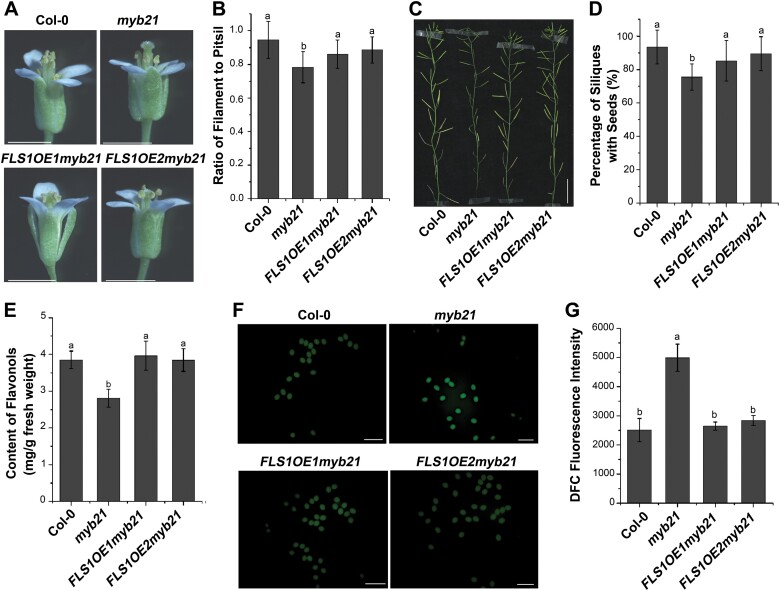
The myb21 mutants have defects in filament elongation and pollen maturation, leading to stamen infertility (Song et al., 2011; Qi et al., 2015). Lack of flavonol biosynthesis has been reported to influence the germination of petunia pollen grains, which could be rescued by the addition of kaempferol, the main product of floral flavonols (Mo et al., 1992). The regulation of the FLS1 gene by MYB21 further encouraged us to investigate the effect of flavonol biosynthesis on stamen development. We therefore treated myb21 flowers with 2 μM kaempferol before floral stage 13. The stamen development of myb21 was distinguishable before and after kaempferol treatment (Fig. 5A). In myb21, the ratio of filament to pistil length was ~0.70 on average, but increased to ~0.84 after kaempferol application (Fig. 5B). We also evaluated the effect of kaempferol on seed production in the myb21 mutant. After treatment with kaempferol, a higher proportion of the siliques in the myb21 mutant contained seeds (Fig. 5C, D). To further confirm the contribution of flavonols to stamen development in the myb21 mutant, we over-expressed FLS1 driven by the 35S promoter in the myb21 mutant. As expected, both transgenic plants (FLS1OE1myb21 and FLS1OE2myb21) showed rescue of the defects in stamen development of myb21 (Fig. 6A, B). Correspondingly, the seed production of the myb21 mutant was increased to wild-type levels when FLS1 was overexpressed in the transgenic plants (Fig. 6C, D).
Fig. 5.
Treatment with kaempferol partially rescues stamen filament growth and fertility in the myb21 mutant. (A) Phenotype of wild-type (Col-0) and myb21 flowers with or without kaempferol (K) treatment. Bar=1 mm. (B) Ratio of filament length to pistil length in wild-type and myb21 plants before and after kaempferol treatment. Error bars indicate the SD of three biological replicates. Different letters above the bars indicate significant differences between groups (P<0.05; ANOVA with Fisher’s LSD test). (C) Main shoot bearing siliques of wild-type and myb21 plants with and without kaempferol treatment. (D) Percentage of siliques with seeds in wild-type and myb21 plants with and without kaempferol treatment. Error bars indicate the SD of three biological replicates. Different letters above the bars indicate significant differences between groups (P<0.05; ANOVA with Fisher’s LSD test).
Fig. 6.
Overexpression of Pro35S:FLS1 partially complements the phenotype of the myb21 mutant. (A) Phenotype of wild-type (Col-0), myb21, FLS1OE1myb21, and FLS1OE2myb21 flowers. Bar=1 mm. (B) Ratio of filament length to pistil length in wild-type, myb21, FLS1OE1myb21, and FLS1OE2myb21 plants. Error bars indicate the SD of three biological replicates. Different letters above the bars indicate significant differences between groups (P<0.05; ANOVA with Fisher’s LSD test). (C) Main shoot bearing siliques of wild-type, myb21, FLS1OE1myb21, and FLS1OE2myb21 plants. Bar=2 cm. (D) Percentage of siliques with seeds in wild-type, myb21, FLS1OE1myb21, and FLS1OE2myb21 plants. Error bars indicate the SD of three biological replicates. Different letters above the bars indicate significant differences between groups (P<0.05; ANOVA with Fisher’s LSD test).(E) Quantification of flavonols in the inflorescences of wild-type, myb21, FLS1OE1myb21, and FLS1OE2myb21 plants. Error bars indicate the SD of three biological replicates. Different letters above the bars indicate significant differences between groups (P<0.05; ANOVA with Fisher’s LSD test). (F) Images of pollen grains of wild-type, myb21, FLS1OE1myb21, and FLS1OE2myb21 plants, stained with CM-H2DCFDA. Bar=50 μm. (G) Quantification of DCF fluorescence in pollen grains of wild-type plants and flavonoid biosynthesis mutants. Error bars indicate the SD of three biological replicates. Different letters above the bars indicate significant differences between groups (P<0.05; ANOVA with Fisher’s LSD test).
Excess ROS accumulation is involved in the stamen defects of the myb21 mutant
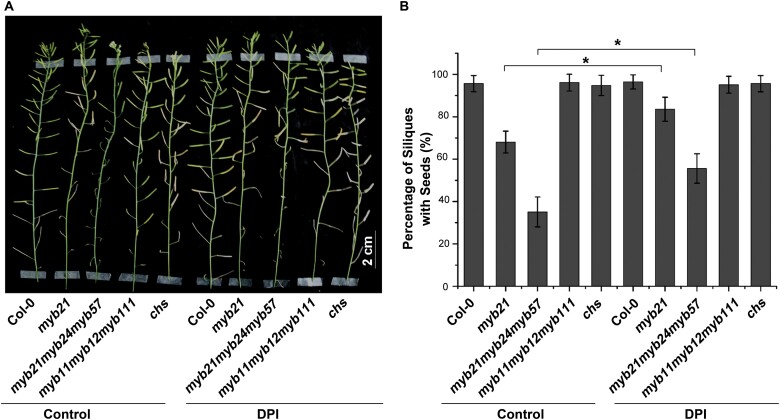
Although further research is needed to reveal how flavonol synthesis mediates stamen development, it has been hypothesized that flavonol accumulation contributes to plant fertility, probably due to involvement in the maintenance of ROS homeostasis (Potocký et al., 2007; Duan et al., 2014; Muhlemann et al., 2018). As shown in Fig 6E, the concentration of flavonols in both FLS1OE1myb21 and FLS1OE2myb21 was higher than that in myb21, and was comparable to that in the wild type. To visualize and quantify total ROS, we treated the inflorescences with CM-H2DCFDA. The dichlorodihydrofluorescein (DCF) fluorescence intensity (a measure of ROS content) of pollen grains was higher in the myb21 mutant than in the wild type (Fig. 6F, G). Importantly, DCF fluorescence intensity was reduced to wild-type levels in the myb21 mutant when FLS1 was overexpressed (i.e. in the transgenic plants FLS1OE1myb21 and FLS1OE2myb21). To further verify the effect of ROS accumulation on stamen development in myb mutants, we treated myb21 and myb21myb24myb57 flowers with 20 μM DPI (a ROS inhibitor) before floral stage 13 (Huang et al., 2019). After treatment with DPI, the siliques of the myb21 and myb21myb24myb57 mutants contained more seeds (Fig. 7). Although the cause of the excess ROS in myb21 and myb21myb24myb57, but not in myb11myb12myb111 or chs, needs further study, these data suggest that flavonols could prevent ROS from reaching damaging concentrations and restore impaired fertility.
Fig. 7.
Treatment with DPI partially rescues stamen filament growth and fertility in the myb21 and myb21myb24myb57 mutants. (A) Main shoot bearing siliques of wild-type (Col-0), myb21, myb21myb24myb57, myb11myb12myb111, and chs plants with and without DPI treatment. (B) Percentage of siliques with seeds in wild-type, myb21, myb21myb24myb57, myb11myb12myb111, and chs plants with and without DPI treatment. Error bars indicate the SD of three biological replicates. *P<0.05 (Student’s t-test).
Pollen-specific flavonol accumulation contributes to male fertility by ROS scavenging in Arabidopsis
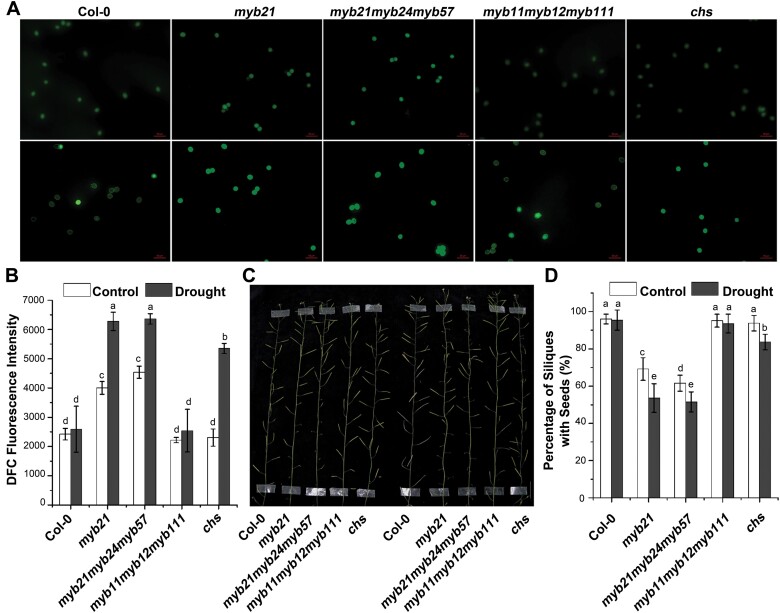
Plant reproductive organs are known to be highly sensitive to drought, which might result from the accumulation of excess ROS (Su et al., 2013). To further support a plausible hypothesis that flavonols mediate stamen development by regulating ROS homeostasis in Arabidopsis, we treated plants including wild type, myb21, myb21myb24myb57, myb11myb12myb111, and chs with drought, as described by Su et al., 2013. The results showed that drought increased the DCF fluorescence of pollen grains 1.5-fold in myb21, 1.4-fold in myb21myb24myb57, and 2-fold in chs. However, the pollen grains in both the wild type and the myb11myb12myb111 mutant maintained relatively stable ROS with and without drought (Fig. 8A, B). To investigate whether the higher ROS levels in myb21, myb21myb24myb57, and chs than those in myb11myb12myb111 and chs resulted in impaired fertility, we estimated the percentage of siliques with seeds in wild type, myb21, myb21myb24myb57, myb11myb12myb111, and chs plants. As expected, myb21, myb21myb24myb57, and chs showed a reduction of nearly 10% of siliques with seeds in response to drought, whereas drought did not affect fertility in the wild type or in myb11myb12myb111 (Fig. 8C, D). These results suggest that the enhanced sensitivity of flavonol biosynthesis mutants to drought -induced infertility was due to elevated ROS.
Fig. 8.
Pollen-specific flavonol accumulation contributes to male fertility by ROS scavenging in Arabidopsis. (A) Images of CM-H2DCFDA-stained pollen grains of wild-type (Col-0), myb21, myb21myb24myb57, myb11myb12myb111, and chs with and without drought treatment. Bars=50 μm. (B) Quantification of DCF fluorescence in pollen grains of the wild type and the flavonoid biosynthesis mutants. Different letters above the bars indicate significant differences between groups (P<0.05; ANOVA with Fisher’s LSD test). (C) Main shoot bearing siliques of wild-type, myb21, myb21myb24myb57, myb11myb12myb111, and chs plants with and without drought treatment. (D) Percentage of siliques with seeds in wild-type, myb21, myb21myb24myb57, myb11myb12myb111, and chs plants with and without drought treatment. Error bars indicate the SD of three biological replicates. Different letters above the bars indicate significant differences between groups (P<0.05; ANOVA with Fisher’s LSD test).
Together, our data demonstrate that the transcription factor MYB21 and its homologs (MYB24 and MYB57) regulate the expression of FLS1 at the transcriptional level. Furthermore, we propose that MYB21-mediated flavonol accumulation is involved in stamen development and seed production by ROS scavenging in Arabidopsis, which provides a new insight into the biological functions of flavonols.
Discussion
Recently, members of subgroup 7 of the R2R3-MYB transcription factors, namely MYB11, MYB12, and MYB111, were identified as flavonol-specific regulators of flavonoid biosynthesis in Arabidopsis (Mehrtens et al., 2005; Stracke et al., 2007, 2010). However, the expression pattern of FLS1 does not follow those of the subgroup 7 MYBs (Stracke et al., 2010). The flower-enriched pattern of FLS1 expression implies the involvement of other, tissue-specific, transcription factors. MYB21 and its homologs, which belong to subgroup 19 of the R2R3-MYB family, are enriched in the stamen and are involved in the regulation of stamen development (Cheng et al., 2009). However, the downstream pathways of these MYBs need further investigation in Arabidopsis. In this report, we identified that MYB21 and its homologs regulate the gene expression of FLS1, the key enzyme in the catalysis of dihydroflavonols into flavonols (Wisman 1998; Owens et al., 2008). In myb21 mutants, flavonol accumulation in inflorescences was reduced, and the reduction was even more pronounced in myb21myb24myb57 mutants (Fig. 1A, B). Considering the characteristic stamen phenotype of myb21, the enrichment of MYB21 transcripts in sepals, petals, stamen, and stigma, but not in roots or leaves (Supplementary Fig. S2), further convinced us to focus on MYB21-mediated flavonol biosynthesis in inflorescence. The flavonol content of myb21 suggested a relatively minor regulatory role of MYB21 on flavonol biosynthesis (Fig. 1). However, we found that MYB24 transcripts were enriched in the pistils and stamen of myb21, reaching expression levels that paralleled those of the wild type (Supplementary Fig. S3). As we have pointed out, MYB24 is homologous to MYB21, and mediated flavonol accumulation in Arabidopsis flowers. In addition, MYB21 transcripts were still detectable in the myb21 mutant, although at a much lower level than that in the wild type. Given that myb21 is likely a leaky allele, an additional possibility is that the residual function of MYB21 could partially promote FLS1 expression as well as flavonol accumulation. Furthermore, the myb21 mutants still contained some flavonols in pollen, implying that the SG19 Myb regulators might not be the exclusive regulators involved in pollen flavonol biosynthesis.
Previously, MYB21 and its homologs were characterized as GAMYB transcription factors and were thought to be involved in GA responses. The development of floral organs is impaired in GA-deficient plants, which indicates that GA influences the biosynthesis of flavonols (Koorneef et al., 1980; Cheng et al., 2009). Previous microarray data support our hypothesis, since they showed that paclobutrazol, an inhibitor of GA biosynthesis, decreased the transcript levels of FLS1 (Owens et al., 2008). Providing further support, it was also reported that the DELLA proteins, which are repressors of the GA pathway, interact with MYB21 and MYB24 to regulate filament elongation in Arabidopsis (Huang et al., 2020). Here, we present evidence that, following the mode of classic GAMYB, MYB21 can directly bind to the GAREs within the FLS1 promoter and regulate the expression of this gene (Fig. 4). UGT73B2, another flavonol biosynthetic gene, has been proposed to be under the control of MYB21 (Kim et al., 2006; Battat et al., 2019). This gene contains GARE elements in its promoter region, which supports our finding. Although it has been reported that MYB99 could act in a regulatory triad with MYB21 and MYB24 to mediate phenylpropanoid biosynthesis (Battat et al., 2019), the failure of MYB99 to activate the FLS1 promoter (Fig. 3A, B) suggests that MYB21/MYB24/MYB57 and MYB99 have different specific direct target genes.
The GARE element also exists in the promoters of other genes encoding enzymes involved in the phenylpropanoid pathway, such as CAD6 (AT4G37970), F5H (AT4G36220), and CCoAoMT (AT4G34050), which is responsible for lignin monolignol biosynthesis (Weng et al. 2010). The defect in lignified thickening leads to male sterility (Thévenin et al., 2011). We found that the expression of these three lignin biosynthetic genes was repressed in myb mutants, whereas it was induced in MYB21OE plants (Supplementary Fig. S4). HPTLC staining showed a consistent pattern of sinapoyl malate accumulation (blue spots; Figs 1A, 2C) and further suggested an effect of MYB21 on lignin deposition. In land plants, the phenylpropanoid pathway provides the precursor 4-coumaroyl CoA for the biosynthesis of lignins and flavonoids. The fine mediation of the branched phenylpropanoid biosynthetic metabolic flux is essential for plant growth and development, where when lignin biosynthesis is overactivated flavonoid biosynthesis is repressed (Besseau et al., 2007; Owens et al., 2008; Peng et al., 2008, Stracke et al., 2009; Gou et al., 2011). In this case, the transgenic plants (MYB21OE1–3) overexpressing MYB21 exhibited different induction effects on FLS1 and F5H (Fig. 2BSupplementary Fig. S4B). Hence, it was reasonable that the highest expression of FLS1 was detected in MYB21OE3, while the highest expression of lignin biosynthesis genes was detected in MYB21OE1. Furthermore, the excessive expression of MYB21 leads to a reduction in fertility (Song et al., 2011). Consistently, we also found that in FLS1OEmyb21 transgenic plants with rescued fertility, the moderate expression level of FLS1 was approximately twice that of myb21 and almost equal to that of the wild-type (Supplementary Fig. S5). Additionally, we analyzed the expression of lignin biosynthetic genes in chs and myb11myb12myb111. We observed that the filament length of chs and myb11my12myb111, two flavonol biosynthesis mutants with normal fertility, resembled that of the wild type (Supplementary Fig. S6). The qRT–PCR results indicated that lignin production in chs and myb11my12myb111 mutants was not affected (Supplementary Fig. S4A). All these results suggested that suitable contents and proportions of flavonols and of these other metabolites are important for stamen development.
The difference in lignin biosynthesis between myb21myb24myb57 and myb11myb12myb111 reminds us of the coordination between these MYBs in regulating flavonol biosynthesis. First, the qRT–PCR results showed that the expression level of MYB11, MYB12, and MYB111 was not altered significantly in the myb21 (or myb21myb24myb57) mutant, and neither was the level of MYB21 transcripts in myb11myb12myb111 (Supplementary Fig. S7A). It has been pointed out that flavonol accumulation is impaired in the pollen grains of myb21 and myb21myb24myb57, but not in myb11myb12myb111 (Supplementary Fig. S7B). Finally, we also noticed that the down-regulation of early biosynthesis enzyme genes, such as CHS and CHI, was more pronounced in myb11myb12myb111 than in myb21myb24myb57, whereas the decrease of late biosynthesis enzyme gene expression was equivalent between myb11myb12myb111 and myb21myb24myb57 (Fig. 1C). Since all flavonols are ultimately derived from chalcone and subsequent flavanones, the limit of a precursor reasonably leads to a defect in flavonol production that is much more severe in myb11myb12myb111 than that in myb21myb24myb57 (Fig. 1A, B). The flavonol biosynthetic pathway, from CHS to FLS1, which leads to the formation of the flavonol aglycone, is common throughout the whole plant. In addition, MYB11-, MYB12-, and MYB111-independent synthesis of the pollen-specific metabolites Q-3GG and K-3GG further indicate that UGTs, which are responsible for the synthesis of flavonol glycosides, are under the control of an unknown regulator (Stracke et al., 2010; Yonekura-Sakakibara et al. 2014). Interestingly, the transcripts of UGTs decreased more significantly in myb21myb24myb57 than in myb11myb12myb111 (Fig. 1C), implying a major regulatory effect of MYB21/MYB24/MYB57 on the expression of UGTs. All these observations hint that the regulatory action of MYB21/MYB24/MYB57 on flavonoid biosynthesis was at least partly independent of MYB11/MYB12/MYB111.
These flavonols are ubiquitous specialized metabolites of plants with varying degrees of hydroxylation, which possess antioxidant activity and thereby act as ROS scavengers (Winkel-Shirley et al., 2001; Pourcel et al., 2007; Hernández et al., 2009). Consistently, mutants with decreased levels of flavonols contain elevated levels of ROS in root hairs and guard cells (Buer et al., 2009; Watkins et al., 2017). Additionally, ROS have been implicated in reproduction, with effects on stamen development (Potocký et al., 2007; Kaya et al., 2014; Xie et al., 2014). Furthermore, several studies have made a link between reduced flavonol levels and impaired fertility in petunia, maize, tobacco, and tomato, but not in Arabidopsis (Mo et al., 1992; Ylstra et al., 1992; Pollak et al., 1993; Burbulis et al., 1996; Ylstra et al., 1996). In maize and petunia, the exogenous application of flavonols could rescue the infertility of a chs mutant (Mo et al., 1992; Taylor et al., 1992; Pollak et al., 1995). Consistently, the suppression of FLS expression reduced seed set in tobacco (Mahajan et al., 2011). More recently, the tomato are mutant, which is defective in flavonol biosynthesis, was reported to exhibit impaired pollen grains and reduced seed yield (Muhlemann et al., 2018). An OsCHS1 T-DNA insertion mutant (oschs1) with no seed formation was characterized, and results indicated that flavonols were essential for male fertility in rice (Wang et al., 2020). Here, we showed that the pollen grains of the myb21 mutant accumulated less flavonols and had a higher abundance of ROS than pollen grains of the wild type. These findings suggested that flavonols act as ROS scavengers in reproductive tissues (Fig. 6). Treatment with DPI, which blocks ROS synthesis, rescued fertility defects in the myb21 mutant (Fig. 7), consistent with the mediation of stamen development by flavonols through their ROS-scavenging properties. To uncover why myb11myb12myb111 and chs, which have pronounced defects in flavonol biosynthesis (Fig. 1), exhibited normal stamen development, we treated the flavonol-biosynthesis mutants myb21, myb21myb24myb57, myb11myb12myb111, and chs with drought. The results showed that myb21, myb21myb24myb57, and chs exhibited defects in pollen-specific flavonol biosynthesis along with higher drought-induced ROS accumulation, which resulted in greater sensitivity of male fertility to drought (Fig. 8). Although the cause of the overaccumulation of ROS in myb21 grown in normal conditions, but not in chs or myb11myb12myb111, is unclear, our work has enabled the development of a plausible and testable hypothesis that flavonols mediate stamen development by regulating ROS homeostasis in Arabidopsis. There are other hypotheses that flavonols regulate auxin concentration and transport, which is essential for reproductive development (Kuhn et al., 2011; Lewis et al., 2011; Zhao et al., 2013; Cardarelli et al., 2018; Tan et al., 2019). In conclusion, our study reveals that the regulation of flavonol biosynthesis is orchestrated by MYB21 or its homologs, and contributes to stamen development and male sterility in Arabidopsis.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Overexpression of Pro35S:MYB21-FLAG complements the phenotype of the myb21 mutant.
Fig. S2. Transcription of MYB21 is enriched in Arabidopsis flowers.
Fig. S3. Transcripts of MYB21 and MYB24 are detectable in the stamen and pistils of myb21.
Fig. S4. MYB21 is involved in the regulation of lignin biosynthesis genes.
Fig. S5. Gene expression of FLS1 in inflorescences of wild type (Col-0), myb21, FLS1OE1myb21, and FLS1OE2myb21.
Fig. S6. myb11myb12myb111 and chs plants show normal stamen development.
Fig. S7. MYB21 and MYB11/MYB12/MYB111 probably mediate the biosynthesis of phenylpropanoid metabolites in their own distinct way.
Table S1. Oligonucleotide primer sequences.
Table S2. Quantitative UPLC/Q-TOF MS data of different flavonol derivatives.
Table S3. Flavonol profiles in methanol–water extracts.
Acknowledgements
We thank Xiaoya Chen for manuscript preparation, Daoxin Xie and Ralf Stracke for sharing materials, and Yining Liu at the Core Facility Centre of the Institute of Plant Physiology and Ecology for mass spectrometry assistance. This work was funded by the National Natural Science Foundation of China (31500242, 31670291, 31800249, 32000234) and the State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products (2010DS700124-ZZ1901, 2010DS700124-ZZ2017).
Author contributions
XZ and YH designed the research and performed most of the experiments; LL and HL performed part of the experiments; XZ and YH were responsible for data analysis; GH wrote the manuscript. The authors agree with the content of the manuscript.
Conflict of interest
The authors have no conflict of interest to declare.
Data availability
The data supporting the findings of this study are available from the corresponding author, Gaojie Hong, upon request.
References
- Battat M, Eitan A, Rogachev I, Hanhineva K, Fernie A, Tohge T, Beekwilder J, Aharoni A. 2019. A MYB triad controls primary and phenylpropanoid metabolites for pollen coat patterning. Plant Physiology 180, 87–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M. 2007. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. The Plant Cell 19, 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK. 2001. Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiology 126, 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Djordjevic MA. 2009. Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana. Journal of Experimental Botany 60, 751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulis IE, Iacobucci M, Shirley BW. 1996. A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. The Plant Cell 8, 1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardarelli M, Costantino P. 2018. An auxin switch for male fertility. Nature Plants 4, 408–409. [DOI] [PubMed] [Google Scholar]
- Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J. 2009. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genetics 5, e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 2010. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Johnson EA, Aggarwal M, Gates L, Wu HM, Cheung AY. 2014. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nature Communications 5, 3129. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis. Trends in Plant Science 15, 573–581. [DOI] [PubMed] [Google Scholar]
- Emiliani J, Grotewold E, Ferreyra ML, Casati P. 2013. Flavonols protect Arabidopsis plants against UV-B deleterious effects. Molecular Plant 6, 1376–1379. [DOI] [PubMed] [Google Scholar]
- Falcone FML, Rius SP, Paula C. 2012. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front in Plant Science 3, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. 2011. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. The Plant Cell 23, 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV. 1995. Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI α-amylase gene promoter. The Plant Cell 7, 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV. 1999. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant Journal 17, 1–9. [DOI] [PubMed] [Google Scholar]
- Hernández I, Alegre L, Van Breusegem F, Munné-Bosch S. 2009. How relevant are flavonoids as antioxidants in plants? Trends in Plant Science 14, 125–132. [DOI] [PubMed] [Google Scholar]
- Huang Y, Cao H, Yang L, et al. 2019. Tissue-specific respiratory burst oxidase homolog-dependent H2O2 signaling to the plasma membrane H+-ATPase confers potassium uptake and salinity tolerance in Cucurbitaceae. Journal of Experimental Botany 70, 5879–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Gong Y, Liu B, Wu D, Zhang M, Xie D, Song S. 2020. The DELLA proteins interact with MYB21 and MYB24 to regulate filament elongation in Arabidopsis. BMC Plant Biology 20, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H, Nakajima R, Iwano M, et al. 2014. Ca2+-activated reactive oxygen species production by Arabidopsis RbohH and RbohJ is essential for proper pollen tube tip growth. The Plant Cell 26, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim BG, Ko JH, Lee Y, Hur HG, Lim Y, Ahn JH. 2006. Molecular cloning, expression, and characterization of a flavonoid glycosyltransferase from Arabidopsis thaliana. Plant Science 170, 897–903 [Google Scholar]
- Koornneef M, van der Veen JH. 1980. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theoretical and Applied Genetics 58, 257–263. [DOI] [PubMed] [Google Scholar]
- Kuhn BM, Geisler M, Bigler L, Ringli C. 2011. Flavonols accumulate asymmetrically and affect auxin transport in Arabidopsis. Plant Physiology 156, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Ramirez MV, Miller ND, Vallabhaneni P, Ray WK, Helm RF, Winkel BS, Muday GK. 2011. Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiology 156, 144–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, and Lin C. 2008. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539. [DOI] [PubMed] [Google Scholar]
- Luria G, Rutley N, Lazar I, Harper JF, Miller G. 2019. Direct analysis of pollen fitness by flow cytometry: implications for pollen response to stress. Plant Journal 98, 942–952. [DOI] [PubMed] [Google Scholar]
- Mahajan M, Ahuja PS, Yadav SK. 2011. Post-transcriptional silencing of flavonol synthase mRNA in tobacco leads to fruits with arrested seed set. PLoS ONE 6, e28315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. 2005. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiology 138, 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y, Nagel C, Taylor LP. 1992. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proceedings of the National Academy of Sciences, USA 89, 7213–7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlemann JK, Younts TLB, Muday GK. 2018. Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress. Proceedings of the National Academy of Sciences, USA 115, E11188–E11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NH, Kim JH, Kwon J, Jeong CY, Lee W, Lee D, Hong SW, Lee H. 2016. Characterization of Arabidopsis thaliana FLAVONOL SYNTHASE 1 (FLS1)-overexpression plants in response to abiotic stress. Plant Physiology and Biochemistry 103, 133–142. [DOI] [PubMed] [Google Scholar]
- Owens DK, Alerding AB, Crosby KC, Bandara AB, Westwood JH, Winkel BS. 2008. Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiology 147, 1046–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Tague BW, Muday GK, Taiz L, Murphy AS. 2001. Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiology 126, 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Hudson D, Schofield A, Tsao R, Yang R, Gu H, Bi YM, Rothstein SJ. 2008. Adaptation of Arabidopsis to nitrogen limitation involves induction of anthocyanin synthesis which is controlled by the NLA gene. Journal of Experimental Botany 59, 2933–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak PE, Hansen K, Astwood JD, Taylor LP. 1995. Conditional male fertility in maize. Sexual Plant Reproduction 8:231–241. [Google Scholar]
- Pollak PE, Vogt T, Mo Y, Taylor LP. 1993. Chalcone synthase and flavonol accumulation in stigmas and anthers of Petunia hybrida. Plant Physiology 102, 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocký M, Jones MA, Bezvoda R, Smirnoff N, Zárský V. 2007. Reactive oxygen species produced by NADPH oxidase are involved in pollen tube growth. New Phytologist 174, 742–751. [DOI] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Cheynier V, Lepiniec L, Debeaujon I. 2007. Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends in Plant Science 12, 29–36. [DOI] [PubMed] [Google Scholar]
- Preuss A, Stracke R, Weisshaar B, Hillebrecht A, Matern U, Martens S. 2009. Arabidopsis thaliana expresses a second functional flavonol synthase. FEBS Letters 583, 1981–1986. [DOI] [PubMed] [Google Scholar]
- Qi T, Huang H, Song S, Xie D. 2015. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. The Plant Cell 27, 1620–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roslan HA, Salter MG, Wood CD, et al. 2001. Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. The Plant Journal 28, 225–235. [DOI] [PubMed] [Google Scholar]
- Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L. 2006. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224, 96–107. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Li Y, Yang S, et al. 2020. The spatio-temporal biosynthesis of floral flavonols is controlled by differential phylogenetic MYB regulators in Freesia hybrida. New Phytologist 228, 1864–1879. [DOI] [PubMed] [Google Scholar]
- Sheahan JJ, Rechnitz GA. 1992. Flavonoid-specific staining of Arabidopsis thaliana. Biotechniques 13, 880–883. [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. 2011. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. The Plant Cell 23, 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, De Vos RC, Bartelniewoehner L, Ishihara H, Sagasser M, Martens S, Weisshaar B. 2009. Metabolomic and genetic analyses of flavonol synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta 229, 427–445. [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. 2007. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. The Plant Journal 50, 660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B. 2010. Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytologist 188, 985–1000. [DOI] [PubMed] [Google Scholar]
- Su Z, Ma X, Guo H, Sukiran NL, Guo B, Assmann SM, Ma H. 2013. Flower development under drought stress: morphological and transcriptomic analyses reveal acute responses and long-term acclimation in Arabidopsis. The Plant Cell 25, 3785–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Man C, Xie Y, Yan J, Chu J, Huang J. 2019. A crucial role of GA-regulated flavonol biosynthesis in root growth of Arabidopsis. Molecular Plant 12, 521–537. [DOI] [PubMed] [Google Scholar]
- Taylor LP, Jorgensen R. 1992. Conditional male fertility in chalcone synthase-deficient petunia. Journal of Heredity 83:11–17. [Google Scholar]
- Thévenin J, Pollet B, Letarnec B, Saulnier L, Gissot L, Maia-Grondard A, Lapierre C, Jouanin L. 2011. The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Molecular Plant 4, 70–82. [DOI] [PubMed] [Google Scholar]
- Tohge T, Matsui K, Ohme-Takagi M, Yamazaki M, Saito K. 2005. Enhanced radical scavenging activity of genetically modified Arabidopsis seeds. Biotechnology Letters 27, 297–303. [DOI] [PubMed] [Google Scholar]
- Tohge T, Yonekura-Sakakibara K, Niida R, Watanabe-Takahashi A, and Saito K. 2007. Phytochemical genomics in Arabidopsis thaliana: a case study for functional identification of flavonoid biosynthesis genes. Pure and Applied Chemistry 79, 811–823. [Google Scholar]
- Wang L, Lam PY, Lui ACW, Zhu FY, Chen MX, Liu H, Zhang J, Lo C. 2020. Flavonoids are indispensable for complete male fertility in rice. Journal of Experimental Botany 71, 4715–4728. [DOI] [PubMed] [Google Scholar]
- Watkins JM, Chapman JM, Muday GK. 2017. Abscisic acid-induced reactive oxygen species are modulated by flavonols to control stomata aperture. Plant Physiology 175, 1807–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Mo H, Chapple C. 2010. Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. The Plant Journal 64, 898–911. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. 2001. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology 126, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. 2002. Biosynthesis of flavonoids and effects of stress. Current Opinion in Plant Biology 5, 218–223. [DOI] [PubMed] [Google Scholar]
- Wisman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B. 1998. Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proceedings of the National Academy of Sciences, USA 95, 12432–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie HT, Wan ZY, Li S, Zhang Y. 2014. Spatiotemporal production of reactive oxygen species by NADPH oxidase is critical for tapetal programmed cell death and pollen development in Arabidopsis. The Plant Cell 26, 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylstra B, Muskens M, Van Tunen AJ. 1996. Flavonols are not essential for fertilization in Arabidopsis thaliana. Plant Molecular Biology 32, 1155–1158. [DOI] [PubMed] [Google Scholar]
- Ylstra B, Touraev A, Moreno RM, Stöger E, van Tunen AJ, Vicente O, Mol JN, Heberle-Bors E. 1992. Flavonols stimulate development, germination, and tube growth of tobacco pollen. Plant Physiology 100, 902–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Nakabayashi R, Sugawara S, Tohge T, Ito T, Koyanagi M, Kitajima M, Takayama H, Saito K. 2014. A flavonoid 3-O-glucoside:2″-O-glucosyltransferase responsible for terminal modification of pollen-specific flavonols in Arabidopsis thaliana. Plant Journal 79, 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. 2008. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene–metabolite correlations in Arabidopsis. The Plant cell 20, 2160–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Niida R, Saito K. 2007. Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. Journal of Biological Chemistry 282, 14932–14941. [DOI] [PubMed] [Google Scholar]
- Yu N, Cai WJ, Wang S, Shan CM, Wang LJ, Chen XY. 2010. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. The Plant Cell 22, 2322–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZX, Wang LJ, Zhao B, Shan CM, Zhang YH, Chen DF, Chen XY. 2015. Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and patchouli (Pogostemon cablin) by the miR156-targeted SPL transcription factors. Molecular Plant 8, 98–110. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhang Y, Liu X, et al. 2013. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Developmental Cell 27, 113–122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author, Gaojie Hong, upon request.