A synthetic protocell with subcompartments was designed to mimic cellular signal processing.
Abstract
As the basic unit of life, cells are compartmentalized microreactors with molecularly crowded microenvironments. The quest to understand the cell origin inspires the design of synthetic analogs to mimic their functionality and structural complexity. In this work, we integrate membraneless coacervate microdroplets, a prototype of artificial organelles, into a proteinosome to build hierarchical protocells that may serve as a more realistic model of cellular organization. The protocell subcompartments can sense extracellular signals, take actions in response to these stimuli, and adapt their physicochemical behaviors. The tiered protocells are also capable of enriching biomolecular reactants within the confined organelles, thereby accelerating enzymatic reactions. The ability of signal processing inside protocells allows us to design the Boolean logic gates (NOR and NAND) using biochemical inputs. Our results highlight possible exploration of protocell-community signaling and render a flexible synthetic platform to study complex metabolic reaction networks and embodied chemical computation.
INTRODUCTION
Living cells are highly dynamic and structurally complex with the capability to integrate a series of spatially and temporally ordered chemical reactions and biological processes (1–3). The construction of their synthetic analogs often starts with rudimentary cell-like entities having compartmentalized structures that can be used for protocell modeling to unravel mechanisms underlying prebiotic organization and the origin of life (4–6). Existing membrane-bound protocells are mostly prepared through the self-assembly of amphiphilic molecules [e.g., surfactants (7), lipids (8–10), and block copolymers (11, 12)], protein-polymer conjugates (13, 14), functionalized inorganic colloids (15, 16), and layer-by-layer polyelectrolyte structures (17). Alternatively, membraneless protocells can be synthesized by spontaneous coacervation leading to liquid-liquid phase separation (LLPS) (18–21). Notwithstanding versatile protocells currently available, considerable challenges exist in bottom-up synthesis of artificial cells with tiered structures and advanced functions (22–27).
An important feature of modern cells lies in their capability of growth, division, migration, and metabolism; key to these functionalities is information processing that encompasses cellular responses to both internal and external signals such as chemical/physical stimuli and biological cues (28, 29). One example is intracellular processes that use primary information carrier molecules such as nucleic acids and proteins to take actions in response to extracellular cues (30). Cells can not only sense the presence of a range of signals at the interface of plasma membranes as involved in extracellular communications but also adapt themselves in terms of morphology and behaviors (31). These signals include physical stimuli (e.g., light and temperature) and the concentrations of chemical compounds (e.g., H+ and OH−) and biomacromolecules (e.g., nucleic acids and proteins) (32–34).
Recent years witness a growing interest in mimicking the way that cells coordinate inter- and extracellular regulatory signals to generate responses and control cellular fates (35–40). In this work, we design hierarchical protocells with modern cell–like compartmentalization and, more importantly, the associated behavior and functionality. These protocells allow regulated chemical exchanges with the surroundings and information transduction across the membrane. Specifically, we construct tiered compartmentalized protocell systems in which membraneless coacervate microdroplets are encapsulated as artificial organelles by a proteinosome. The procedure is similar to the cellular condensation of proteins and RNA under physiological conditions into membraneless droplets via LLPS (41). Although the demixing of polyelectrolytes and oligopeptides has been reported before (42), we here demonstrate the adaptive behavior of confined coacervate microdroplets (artificial organelles) in a protocell, as well as the signal processing and information transduction between the protocells and the extracellular environment across the semipermeable membrane constructed by protein-polymer nanoconjugates. We anticipate that these protocells will inspire further work on the design of protocell-based complex metabolic reaction networks and embodied chemical computation.
RESULTS
Construction of coacervate microdroplets
To construct artificial organelles, we synthesized a bola-shaped amphiphile with two negatively charged glutamic acids appended to a rigid azobenzene [4,4′-glutamic acid azobenzene (AzoGlu2)] (fig. S1). The latter served as a photosensitive ingredient of the artificial organelles and was paired with a pH-responsive cationic polyelectrolyte, diethylaminoethyl-dextran (DEAE-dextran) (Fig. 1A). The azobenzene main stem undergoes reversible and efficient photoisomerization between planar trans-AzoGlu2 and nonplanar metastable cis-AzoGlu2 states by irradiation with ultraviolet (UV) and blue light (table S1 and figs. S2 and S3). The carboxylic groups on AzoGlu2 and tertiary amine groups on DEAE-dextran showed protonation/deprotonation transitions in response to changes in the environmental pH. Direct mixing of trans-AzoGlu2 and DEAE-dextran led to a turbid suspension of coacervate microdroplets over a broad range of solution conditions (Fig. 1B and fig. S4A). At a total concentration of macromolecules equal to 20 mM and 1:1 monomer molar ratio, the system showed spherical coacervates (~3.6 μm in diameter) with a good optical contrast in the aqueous phase (Fig. 1, C and D, and fig. S4, B and C).
Fig. 1. AzoGlu2/DEAE-dextran coacervate microdroplets.
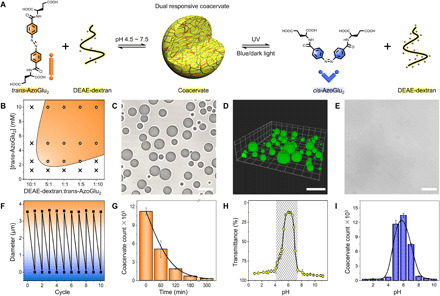
(A) Schematic of active AzoGlu2/DEAE-dextran complexation to produce microdroplet condensation via LLPS. The process is responsive to wavelength-dependent light irradiation, or subtle pH changes triggered disassembly-assembly of coacervates. (B) Phase diagram showing the presence of microdroplets (orange region) at different trans-AzoGlu2/DEAE-dextran molar ratio and trans-AzoGlu2 concentrations. (C) Optical microscopy image and (D) 3D confocal fluorescence microscopy image (loaded with HPTS) showing the formation of coacervate microdroplets in a mixture of trans-AzoGlu2 (10 mM) and DEAE-dextran (10 mM monomer). (E) Optical microscopy image showing disassembly of microdroplets after UV light irradiation for 7 min. (F) Count number of coacervates in the mixture of trans-AzoGlu2 (10 mM) and DEAE-dextran (10 mM monomer) as detected by flow cytometry with different durations of UV light irradiation. Error bars represent the SDs of three independent measurements. (G) Reversible diameter changes of trans-AzoGlu2/DEAE-dextran microdroplets with UV/blue light irradiation for 10 cycles. (H) Transmittance of trans-AzoGlu2/DEAE-dextran mixtures and (I) their coacervate counts suggesting the existence of coacervate microdroplets at a narrow pH window. Scale bars, 10 μm.
We explored the sequestration of incoming molecules to trans-AzoGlu2/DEAE-dextran coacervates with several model compounds (fig. S5). Small compounds like 8-hydroxypyrene-1,3,6-trisulfonic acid trisodium (HPTS; negatively charged), rhodamine 6G (positively charged), and Nile Red (neutral) could be readily loaded into the microdroplets. However, the sequestration of macromolecules was more complicated. While highly charged polyelectrolytes such as rhodamine isothiocyanate–tagged polyelectrolyte (RITC-PDDA) and single-stranded oligonucleotide (TAMRA-ssDNA) could be sequestered, nonionic dextran (FITC-dextran) was excluded from the coacervates. The dependence on electrostatic interactions was reasonable because polyelectrolytes had the tendency to complex with ionic species even when the zeta potential of the coacervate microdroplets was close to zero. Notably, the coacervate microdroplets were observed to recruit both hydrophilic (HPTS) and hydrophobic (Nile Red) fluorophores, indicating the existence of hydrophobic microdomains within hydrophilic droplets.
The coacervate microdroplets can serve as a protocell model that sensitively responds to external stimuli such as light and pH. Immediately after exposing to UV light, trans-AzoGlu2/DEAE-dextran microdroplets underwent size shrinkage and eventually disassembled (Fig. 1, E and F, and figs. S6 and S7). The demixing process was also indicated by the changes in macroscopic appearance from turbid to clear solution (fig. S8). The microdroplet dissociation can be reversed by thermal relaxation of cis-azobenzene in dark (fig. S9) or exposure to blue light for approximately 8 min (movie S1 and fig. S10). These assembly/disassembly transitions were fully reversible for over 10 cycles (Fig. 1G and fig. S11). We explored the molecular origin of macroscopic LLPS with density functional theory (DFT; fig. S12), which suggests an increase of the dipole moment of azobenzene from 5.27 D (trans state) to 11.20 D (cis state) after photoisomerization. This would reduce hydrophobicity of azobenzene and, eventually, lower the tendency of cis-AzoGlu2 to complex with DEAE-dextran to form coacervate microdroplets. The theoretical predictions were confirmed by a control experiment with glutamylglutamic acid/DEAE-dextran complexation that did not produce microdroplets, highlighting the indispensable role of hydrophobic interaction in coacervation. Furthermore, AzoGlu2 and DEAE-dextran are known to have pH-sensitive functional groups [e.g., –COOH and –N(C2H5)2], with the pKa (where Ka is the acid dissociation constant) reported to be 3 to 4 (43) and 9.2 (44, 45), respectively. The carboxylic acids on AzoGlu2 are protonated when the solution pH is below 4, whereas the number of positive charges on DEAE-dextran would be reduced under basic (pH > 9.2) conditions. As a consequence, the electrostatically driven trans-AzoGlu2/DEAE-dextran complexation was highly pH sensitive, and stable coacervate microdroplets exist within a narrow pH window ranging from 4.5 to 7.5 (Fig. 1, H and I). The chemical signal-responsive coacervation enables us to build hierarchical protocells with information processing function.
Engineering artificial organelles within a confined space
We then transferred the trans-AzoGlu2/DEAE-dextran complexation into a membrane-confined cell-sized cavity. To this end, we fabricated semipermeable proteinosomes using a crosslinked monolayer of bovine serum albumin/poly(N-isopropylacrylamide) (BSA-NH2/PNIPAAm) nanoconjugates (Fig. 2A). To prepare coacervate microdroplets inside proteinosomes (coacervate-in-proteinosome protocells), we initially encapsulated DEAE-dextran in the proteinosome cavity (Supplementary Materials) and introduced trans-AzoGlu2 at a monomer molar ratio of 1:1 to the bulk solution. Because of their small size, trans-AzoGlu2 molecules are able to penetrate through the porous protein membrane, while large DEAE-dextran molecules are retained inside proteinosomes, resulting in the electrostatic complexation of trans-AzoGlu2/DEAE-dextran through LLPS (Fig. 2A). The presence of small dots with intensified green fluorescence suggested the formation of coacervate droplets with condensed fluorescently labeled DEAE-dextran, and the surrounding red fluorescence lumen indicated the intactness of protein membrane (Fig. 2, B and C).
Fig. 2. Engineering artificial organelles within a proteinosome.
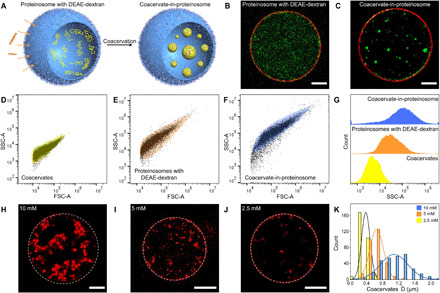
(A) Schematic of engineered artificial organelles inside a proteinosome. Fluorescence microscopic images showing (B) uncondensed FITC-labeled DEAE-dextran (green) evenly dispersed inside an RITC-labeled proteinosome (red) and (C) condensed microdroplets due to complexation of trans-AzoGlu2 and DEAE-dextran. (D to F) Flow cytometry analysis of FSC-A versus SSC-A for (D) coacervates, (E) DEAE-dextran–encapsulated proteinosomes, and (F) proteinosomes containing condensed trans-AzoGlu2/DEAE-dextran microdroplets. (G) Comparison of SSC-A distributions showing the increase of internal complexity as the polymer was condensed inside proteinosomes. (H to K) Fluorescence microscopy images showing distinct size distribution of microdroplet grown inside proteinosomes. The microdroplet diameter is strongly dependent on the DEAE-dextran concentration. Scale bars, 10 μm.
Flow cytometry is a powerful tool to detect and identify different cell populations. The analysis is especially sensitive to cellular granularity or complexity changes during cell differentiation or between different phenotypes. Here, we applied flow cytometry to characterize the granularity and complexity of our model protocells (i.e., coacervates, DEAE-dextran encapsulated proteinosomes, and coacervate-in-proteinosome protocells) with varied hierarchical levels. Figure 2 (D to G) presents the dot plot of forward-scattered light area (FSC-A) versus side-scattered light area (SSC-A) of the above systems. In these figures, each dot represents the results for an individual protocell analyzed by a flow cytometer. The characteristic distribution of different protocell populations was determined by different physical properties such as cell size and granularity. We noticed that proteinosomes exhibited stronger FSC-A signals relative to trans-AzoGlu2/DEAE-dextran coacervate microdroplets owing to the larger size of proteinosomes (~40 μm) in comparison with coacervates (~3.6 μm). The broad distribution of SSC-A signal indicates the varied internal granularity and complexity of different protocells. As shown in Fig. 2G, DEAE-dextran–loaded proteinosomes displayed higher SSC-A values than coacervate droplets, implying the increased complexity of internal structures. When DEAE-dextran was further condensed into droplets inside proteinosomes, the hierarchical system exhibited an even higher SSC-A signal intensity. Overall, the forward and side-scattered light profiles confirmed the encapsulation and condensation of DEAE-dextran inside proteinosomes.
We noticed that the condensed microdroplets inside proteinosomes (~1 μm; Fig. 2H) are much smaller than those formed in the free space (~3.6 μm; fig. S4, B and C). In principle, the size of microdroplets was determined by the amount of polyelectrolyte available from the surrounding. In a bulk solution, the microdroplets grow over time due to continuous polyelectrolyte recruitment and droplet coalescence (fig. S13). However, this was not the case inside proteinosomes because the protein layer served as a semipermeable membrane that prevented DEAE-dextran from diffusing out and harvested additional trans-AzoGlu2 from environment (fig. S14). As a result, the microdroplets can only recruit polyelectrolyte (DEAE-dextran) trapped inside each proteinosome and the size will be much smaller than that of those formed in open environments. To validate this hypothesis, we determined the size of proteinosome-bound microdroplets by changing the concentration of DEAE-dextran initially trapped inside proteinosomes. We observed microdroplet growth dependent on DEAE-dextran concentration (Fig. 2, H to J). As expected, a higher polyelectrolyte concentration resulted in larger microdroplets. The diameter of proteinosome-bound microdroplets increased from 0.4 μm to 0.7 and 1.1 μm when the concentration of DEAE-dextran rose from 2.5 mM to 5 and 10 mM, respectively.
Signal processing and biological computing of complex protocell
The protocells made of trans-AzoGlu2/DEAE-dextran microdroplets in proteinosomes offer a versatile platform to condense biomacromolecules (e.g., protein, enzyme, and DNA) under confinement and recruit small molecules [e.g., adenosine 5′-triphosphate (ATP)] from outside the protein membranes (Fig. 3A). Especially, the coacervation upon external stimuli (e.g., light and pH) in bulk solution can be transferred to an intelligent, miniaturized coacervate-in-proteinosome protocell system within a confined space. We took advantage of this system to exert control over biomolecular condensation and thereby to regulate their biological activities, which was regarded as a biomimetic process of microdroplet-mediated biological functions.
Fig. 3. Active control over biological condensation in membraneless artificial organelles.
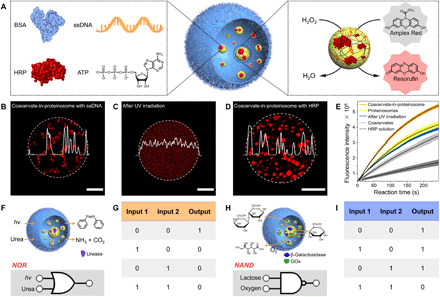
(A) Left: Schematic of biomolecules harvested by trans-AzoGlu2/DEAE-dextran coacervate microdroplets within a confined space. Right: Simultaneous enrichment of enzyme (e.g., HRP) and the corresponding substrate accelerates the catalytic reaction. Fluorescence microscopy images showing that (B) TAMRA-ssDNA was captured by trans-AzoGlu2/DEAE-dextran coacervates inside a proteinosome, (C) TAMRA-ssDNA was liberated from the coacervate microdroplets upon UV light irradiation, and (D) RITC-HRP was sequestrated from trans-AzoGlu2/DEAE-dextran coacervates in a proteinosome. (E) Comparison of HRP-mediated peroxidation reaction under different conditions of Amplex Red (substrate) oxidization by H2O2. The reaction rate was measured according to the fluorescence intensity at 590 ± 10 nm emitting from resorufin (oxidation product). The catalytic reaction was studied under various conditions: (i) HRP bulk solution (gray), (ii) HRP-loaded coacervates (light gray), (iii) HRP-loaded coacervate-in-proteinosome protocells (after light irradiation, blue), (iv) HRP-loaded coacervate-free proteinosomes (yellow), and (v) HRP-loaded coacervate-in-proteinosome protocells (orange). (F to I) Truth table and schematic representation of NOR and NAND logic gates constructed from coacervate-in-proteinosome complex systems. Scale bars, 10 μm.
We demonstrated that the coacervate-in-proteinosome protocells were able to spatially control the condensation of genetic materials such as a single-stranded DNA. To this end, fluorescence-labeled ssDNA (TAMRA-ssDNA) chains were introduced into proteinosome together with DEAE-dextran. The homogeneous red fluorescence indicated that ssDNA was not condensed at the initial stage (movie S2 and fig. S15). Immediately after the addition of trans-AzoGlu2, a series of bright fluorescent clusters emerged inside the proteinosome, suggesting rapid molecular diffusion and permeation across the protein membrane. The coacervation facilitates the efficient partition of ssDNA into the artificial organelles (Fig. 3B and movies S3 and S4). ssDNA condensation can be actively mediated with external light irradiation. Upon exposure to UV light, the strength of fluorescence from microdroplets gradually decreased and disappeared after 24 s, indicating the disassembly of coacervates and the release of ssDNA (Fig. 3C, movie S5, and figs. S16 and S17). When switching to blue light, coacervate microdroplets emerged along with the condensation of ssDNA chains (movie S6 and fig. S18). The actively controlled membraneless condensation of nucleic acid represents a model system mimicking the spatiotemporal organization of eukaryotic chromosomes.
The harvesting of biomolecules by coacervate microdroplets in a confined proteinosome may be regarded as a model process that mimics organelle-coordinated biochemical reactions. To elucidate this functionality, we exploited coacervate-in-proteinosome protocells to simultaneously encapsulate enzymes and corresponding substrates such that the catalytic reactions can be accelerated. To this end, we enclosed horseradish peroxidase (HRP; Fig. 3D) inside coacervate-in-proteinosome protocells and recruited the enzyme substrate, 10-acetyl-3,7-dihydroxyphenoxazine (Amplex Red), from the external surroundings. The substrate diffused across the semipermeable proteinosome membranes and was subsequently harvested by trans-AzoGlu2/DEAE-dextran microdroplets within the protocells. The addition of H2O2 to the surrounding solution initiated HRP-catalyzed oxidation reaction of nonfluorescent Amplex Red and converted it into a fluorescent product (resorufin). In comparison with that for free enzymes in a bulk solution, the oxidation reaction was accelerated due to the accumulation of HRP inside the coacervates within proteinosomes (Fig. 3E). The confinement effect was further amplified when HRP was condensed by coacervate-in-proteinosomes. By contrast, when the coacervate-in-proteinosome protocells were exposed to UV light, the microdroplets were disaggregated (fig. S19), leading to a reduction of the catalytic reaction rate (Fig. 3E). Similarly, co-encapsulation of glucose oxidase (GOx) and HRP by coacervate microdroplets within proteinosomes (fig. S20) was also found to increase the cascade reaction rate (fig. S21). The acceleration of catalytic reactions inside coacervates was likely due to the macromolecular crowding from coacervation, as well as the spatial proximity effect from co-encapsulation of enzymes and substrates (46, 47). Therefore, trans-AzoGlu2/DEAE-dextran microdroplets confined within proteinosomes serve as artificial organelles that can harvest biomolecules and coordinate enzymatic reactions.
We further explored the possibility of constructing Boolean logic gates using the artificial organelles presented above. To develop a NOR gate (Fig. 3, F and G), we prepared urease-loaded trans-AzoGlu2/DEAE-dextran coacervates within proteinosomes and interfaced this biosystem with UV light (Input 1) and urea (Input 2). The presence of UV light is known to cause trans-to-cis transition of azobenzene group and disassembles the coacervate microdroplets (0 for Output). Similarly, the addition of urea initiated the hydrolysis of urea into carbon dioxide and ammonia, which triggered a rapid increase in the pH of the solution (fig. S22). This subsequently caused the dissociation (0 for Output) of coacervate microdroplets inside proteinosomes (fig. S23). This system can thus be described as a NOR logic gate, where the activity (viz., coacervate formation, 1 for Output) was observed only in the absence of both signals (UV light and urea) (fig. S24). In another system, we developed a NAND gate (Fig. 3, H and I) by integrating a cascade enzyme reaction within coacervate microdroplets. To this end, we incorporated β-galactosidase (β-gal) and GOx into the coacervates by co-condensation (fig. S25) and used lactose and oxygen as two inputs. β-Gal is able to catalyze the conversion of lactose into galactose and glucose; the latter can be further oxidized into gluconic acid by glucose oxidase and oxygen. This cascade reaction reduced the solution pH (below ~4; fig. S26), which consequently disassembled the coacervates (Fig. 1, H and I). Hence, the coacervate-in-proteinosome system can be described as a NAND logic gate (Fig. 3, H and I), where the activity (viz., coacervate formation within proteinosomes, 1 for Output) was observed in the absence of either lactose (Input 1) or oxygen (Input 2) (fig. S27). We expect that the protocells can be integrated in any artificially designed complex reacting processes mimicking networks for multistep information processing.
DISCUSSION
In summary, we reported a tiered protocell system that consists of membraneless coacervate microdroplets (viz., artificial organelles) enclosed with a protein membrane. These subcompartments of the protocells were able to sense a variety of extracellular signals (e.g., photons, protons, and small molecular weight compounds) and adapted their behaviors through a series of biochemical reactions. The hybrid protocells have the ability to harvest biomacromolecules (e.g., DNA and proteins) by condensing them into liquid droplets and recruit small molecules from surroundings by penetrating through the semipermeable protein membrane. The active control allows effective acceleration of enzyme-catalyzed reactions in cell-like settings. Last, we demonstrated that the Boolean logic gates (NOR and NAND) can be implemented within the protocell system by integrating active coacervate formation/disassembly behavior with enzyme-involved biocatalytic (cascade) reactions. Our results open opportunities for the construction of hierarchical protocell compartments and flexible synthetic platforms that can be used to investigate complex metabolic reaction networks and embodied chemical computation.
METHODS
Preparation of DEAE-dextran coacervates
Trans-AzoGlu2 solution was stored for at least 3 days in the dark before use to ensure complete isomerization to the trans state, and protected from light during use by wrapping the vial with aluminum foil. Coacervate suspension was prepared by directly mixing trans-AzoGlu2 (20 mM, pH 6.0) and DEAE-dextran (20 mM, pH 6.0) to reach a total concentration of 20 mM. The freshly prepared coacervate microdroplets were incubated for ca. 15 min before use.
Phase diagram
Coacervate solutions were prepared by mixing trans-AzoGlu2 (20 mM, pH 6.0) and DEAE-dextran (20 mM, pH 6.0) solutions with different ratios. If necessary, the solutions were further diluted with deionized water before addition of the second component in Eppendorf tubes to reach desired concentrations. The samples were incubated for ca. 15 min, and their coacervation behaviors were investigated on an optical microscope. The results were obtained from three independent experiments.
Sequestration properties of the coacervate microdroplets
Trans-AzoGlu2/DEAE-dextran droplet suspensions (50 μl) were prepared in Eppendorf tubes, and then HPTS (1 μl, 5 μM in water), rhodamine 6G (1 μl, 5 μM in water), Nile Red (1 μl, 2 μM in methanol), TAMRA-ssDNA (0.3 μl, 20 μM in 50 mM tris-HCl containing 100 mM NaCl, pH 7.4), RITC-PDDA (0.3 μl, 1.5 mM in water), and FITC-dextran (2.5 μl, 0.5 mg/ml in water) were added.
Light-induced trans/cis-AzoGlu2 isomerization and reversible disassembly/reassembly of microdroplets
Trans/cis-AzoGlu2 isomerization was achieved with UV (365 nm, 30-W light-emitting diode light) and blue light (450 nm, 20-W high-intensity discharge lamp) irradiation. For the isomerization experiments characterized by 1H nuclear magnetic resonance (NMR) spectra, 0.6 ml of trans-AzoGlu2 solution [6 mM in dimethyl sulfoxide (DMSO)–d6] was used. Trans-to-cis isomerization occurred with 20-min UV light irradiation, while the cis-to-trans isomerization was induced after 60-min blue light exposure. The isomerization process of AzoGlu2 (0.6 mM in water) was also investigated on a UV-vis spectrophotometer with 30-s UV and 10-min blue light irradiation. For light-induced disassembly/reassembly experiments studied by flow cytometry, 2 ml of trans-AzoGlu2/DEAE-dextran coacervate suspension (1:1, 20 mM total concentration, pH 6.0) or AzoGlu2/DEAE-dextran solution [1:1, 20 mM total concentration, trans/cis states of AzoGlu2 molar ratio = 31:69 (see table S1), pH 6.0] was irradiated with 300-min UV and 60-min blue light, respectively.
Light-induced disassembly/formation processes of microdroplets were realized by an optical microscope with UV (325 < λ < 375 nm, 120-W short-arc Hg light source) and blue light (460 < λ < 500 nm, 120-W short-arc Hg light source) irradiation, respectively.
For confocal laser scanning microscopy (CLSM) experiments, disassembly/reassembly processes of microdroplets were realized by a laser on CLSM with UV (405 nm, 30-mW diode laser) and blue light (488 nm, 25-mW argon laser) irradiation, respectively. The microdroplets were disassembled with 7-min UV light irradiation, which were again condensed into the trans-AzoGlu2/DEAE-dextran coacervates after 8-min blue light irradiation.
pH-induced disassembly and reassembly of the microdroplets
The disassembly of the trans-AzoGlu2/DEAE-dextran coacervates was observed when the pH was adjusted to acidic (pH < 4.5) or basic (pH > 7.5) conditions with HCl (0.1 M) or NaOH (0.1 M).
Transmittance studies
To a freshly made trans-AzoGlu2/DEAE-dextran droplet suspension (1:1, 20 mM total concentration, pH 6.0) in a UV-vis cuvette (3-ml volume, 1-cm path length), a solution of HCl (0.1 M) or NaOH (0.1 M) was added to adjust the pH values. The transmittance of suspensions was measured at 650 nm as a function of pH on a UV-vis spectrophotometer (PerkinElmer, Lambda 950).
Preparation of proteinosomes and coacervate-in-proteinosome protocells
BSA-NH2/PNIPAAm proteinosomes were prepared according to a previously reported method (13). Proteinosomes encapsulated with DEAE-dextran/TAMRA-ssDNA or DEAE-dextran/RITC-HRP were prepared following the above procedures. For DEAE-dextran/TAMRA-ssDNA–containing proteinosomes, DEAE-dextran (10 μl, 120 mM monomer concentration) and TAMRA-ssDNA (2.5 μl, 100 μM) were added to BSA-NH2/PNIPAAm aqueous solution to get a 60-μl aqueous solution, which was further added to the oil phase to form Pickering emulsion. For DEAE-dextran/RITC-HRP–containing proteinosomes, DEAE-dextran (10 μl, 120 mM monomer concentration) and RITC-HRP (2.5 μl, 10 mg/ml) were added to BSA-NH2/PNIPAAm aqueous solution to obtain a 60-μl aqueous solution. To make coacervate-in-proteinosome protocells, trans-AzoGlu2 solution (20 μl, 20 mM, pH 6.0) was added to 20 μl of BSA-NH2/PNIPAAm proteinosome suspension at a trans-AzoGlu2/DEAE-dextran molar ratio of 1:1.
RITC-labeled, DEAE-dextran–containing proteinosomes were prepared as above by using a mixture of BSA-NH2/PNIPAAm and RITC-BSA-NH2/PNIPAAm nanoconjugates (90 and 10 weight %, respectively).
Light-induced release and capture of TAMRA-ssDNA inside the proteinosomes
The coacervate-in-proteinosome protocells (trans-AzoGlu2/DEAE-dextran monomer molar ratio = 1:1, 20 mM total concentration) were loaded with TAMRA-ssDNA (4.2 μM). The mixture was exposed to UV light (405 nm, 30-mW diode laser) for 24 s to disassemble the microdroplets and release TAMRA-ssDNA inside the proteinosomes. To condense TAMRA-ssDNA within coacervates, the mixture was irradiated with blue light (488 nm, 25-mW argon laser) for 40 s.
Enzyme-mediated peroxidation in coacervate-in-proteinosome protocells
The HRP was encapsulated inside coacervate-in-proteinosome protocell following the protocol as below. Typically, 12 μl of trans-AzoGlu2 solution (20 mM) was added to 80 μl of proteinosome suspension [50 mM phosphate-buffered saline (PBS) buffer, pH 6.0] with encapsulated HRP (6.25 μg/ml) and DEAE-dextran (3 mM). The mixture was further diluted with 8 μl of PBS buffer (50 mM, pH 6.0). To test the catalytic reaction, 100 μl of HRP-encapsulated coacervate-in-proteinosome protocell suspension and 1 μl of Amplex Red solution (250 μM in DMSO) were added to 96-well plates (Thermo Fisher Scientific; lighttight, flat bottom, nonsterile). The mixture was incubated at room temperature to ensure that the sequestration of the substrate into the coacervate phase had reached equilibrium. Subsequently, a solution of H2O2 (42.5 μl, 14.7 μM) was added to initiate the HRP-mediated peroxidation reaction. The fluorescence intensity (excitation = 530 ± 10 nm, emission = 590 ± 10 nm) was monitored on a microplate reader (CLARIOstar Plus) over 250 s.
In a control experiment, 100 μl of coacervate-in-proteinosome protocell dispersion was irradiated with UV light (365 nm, 30-W light-emitting diode light) for 24 s to induce trans-to-cis transition of AzoGlu2 inside proteinosomes. The catalytic reaction was investigated following the above protocol.
In other control experiments, 100 μl of HRP-encapsulated proteinosome (coacervate-free) suspension containing DEAE-dextran (2.4 mM) and HRP (5 μg/ml), 100 μl of HRP-contained trans-AzoGlu2/DEAE-dextran coacervate suspension (4.8 mM total concentration, 5 μg/ml HRP), and 100 μl of freshly prepared HRP solution (5 μg/ml) were prepared separately to study the catalytic reaction following the protocol as described above.
HRP/GOx-mediated cascade reaction in coacervate-in-proteinosome protocells
Coacervate-in-proteinosome protocells doped with HRP (6.25 μg/ml) and GOx (3.2 μg/ml) were prepared in a similar manner. The cascade enzymatic reaction was initiated by addition of glucose solution (0.4 mM), and the catalytic reaction was investigated following the above protocol.
Urease-mediated pH changes
The urease solution (50 μl, 0.4 mg/ml) was adjusted to pH 6.0, and a solution of urea (1.3 μl, 0.5 M) was added to initiate the urease-mediated catalytic reaction.
β-Gal/GOx-mediated pH changes
Fifty microliters of a mixture solution of β-gal (0.05 mg/ml) and GOx (0.2 mg/ml) was adjusted to pH 6.0, and a solution of lactose (5 μl, 0.5 M) was added to initiate the cascade enzymatic reaction.
Constructing Boolean logic gates in coacervate-in-proteinosome protocells
To construct a NOR logic gate, coacervate-in-proteinosome protocells (trans-AzoGlu2/DEAE-dextran, 1:1, 20 mM total concentration) doped with urease (0.4 mg/ml) were prepared following the above protocol. To this mixture, a solution of urea (1.3 μl, 0.5 M) was added to initiate the catalytic reaction. The dissociation of microdroplets was imaged by CLSM.
To construct a NAND gate, coacervate-in-proteinosome protocells doped with β-gal (0.05 mg/ml) and GOx (0.2 mg/ml) were prepared in a similar manner. The cascade enzymatic reaction was initiated by addition of lactose solution (5 μl, 0.5 M), and the stability of coacervate microdroplets inside proteinosomes was recorded on a CLSM.
DFT calculations
NWChem was used for the quantum chemical calculation (48). Similar to literature (49, 50), DFT calculations used the B3LYP exchange-correlation and 6-31G basis set. The solvent effect was considered by using “conductor-like screening model” (COSMO) (51). The dielectric constant used for water was 78.4. The radius of atoms was taken from Stefanovich and Truong’s work (52). In the plot of electrostatic potential, red stood for negative potential, while blue represented positive potential.
Optical and fluorescence microscopy
Optical and fluorescence microscopy experiments were recorded on a Leica DMI8 inverted microscope using a ×100 oil immersion lens [HCX PL APO, 1.4 numerical aperture (NA)]. Fluorescence imaging was performed using a Leica DFC9000 GT setup. In situ UV and blue light irradiation of the microdroplets were recorded on an optical microscope by using a 120-W short-arc Hg light source (OSRAM Licht AG, Germany) equipped with bandpass filters to select the desired wavelength (UV light: 325 < λ < 375 nm; blue light: 460 < λ < 500 nm). Coacervate microdroplets were imaged at ca. 15 min after formation by loading an aliquot of the suspension onto a custom-made polyethylene glycol (PEG)–functionalized capillary slide. The droplets were left to settle for ca. 5 min on the glass coverslip before imaging.
Confocal laser scanning microscopy
CLSM measurements were carried out on a Zeiss LSM 880 using a ×63 oil immersion lens with a diode laser (405 nm for Hoechst 33258 excitation), an argon laser (488 nm for FITC and HPTS excitation, 514 nm for rhodamine 6G excitation), and a HeNe543 laser (543 nm for Nile Red and RITC excitation).
Flow cytometry
All measurements were performed by using a Novo Cyte 2060R flow cytometer. The samples were analyzed immediately after preparation to minimize the effect of coalescence.
For light-induced disassembly experiments, 2 ml of trans-AzoGlu2/DEAE-dextran coacervate suspension (1:1, 20 mM total concentration, pH 6.0) in a UV-vis cuvette (3-ml volume, 1-cm path length) was irradiated with UV light (365 nm, 30-W light-emitting diode light). The samples were analyzed by flow cytometry after 0-, 60-, 120-, 180-, and 300-min UV light exposure, respectively.
For light-triggered reassembly experiments, 2 ml of AzoGlu2/DEAE-dextran solution (1:1, 20 mM total concentration, pH 6.0) was irradiated with blue light (450 nm, 20-W high-intensity discharge lamp). The samples were analyzed by flow cytometry after 0-, 30-, and 60-min blue light irradiation, respectively.
For pH-induced disassembly/reassembly experiments, 2 ml of trans-AzoGlu2/DEAE-dextran (1:1) coacervate suspension was analyzed after adjusting the pH values between 1.0 and 10.0.
All the flow cytometry measurements were recorded at a flow speed of 10 μl/min for 2 min, and the particle number was counted in 30 s.
2D pseudo-color dot plots of the FSC and SSC light for 100-μl aqueous dispersions of trans-AzoGlu2/DEAE-dextran coacervates, DEAE-dextran–containing proteinosomes, and coacervate-in-proteinosome protocells were determined for a total of 20,000 events at a flow speed of 6 μl/min.
Acknowledgments
Funding: We thank the National Natural Science Foundation of China (22072159) and the Fundamental Research Funds for the Central Universities (buctrc202015) for financial support. Author contributions: W.M., Y.L., and Y.Q. conceived the experiments. W.M. performed the experiments. M.Z. and J.W. undertook the DFT calculations. W.M., Z.J., Y.L., and Y.Q. undertook the data analysis and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/22/eabf9000/DC1
REFERENCES AND NOTES
- 1.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Nakashima K. K., Vibhute M. A., Spruijt E., Biomolecular chemistry in liquid phase separated compartments. Front. Mol. Biosci. 6, 21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buddingh B. C., van Hest J. C. M., Artificial cells: Synthetic compartments with life-like functionality and adaptivity. Acc. Chem. Res. 50, 769–777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin Y., Niu L., Zhu X., Zhao M., Zhang Z., Mann S., Liang D., Non-equilibrium behaviour in coacervate-based protocells under electric-field-induced excitation. Nat. Commun. 7, 10658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin N., Dynamic synthetic cells based on liquid–liquid phase separation. Chembiochem 20, 2553–2568 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Brea R. J., Bhattacharya A., Bhattacharya R., Song J. J., Sinha S. K., Devaraj N. K., Highly stable artificial cells from galactopyranose-derived single-chain amphiphiles. J. Am. Chem. Soc. 140, 17356–17360 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Deng N. N., Vibhute M. A., Zheng L., Zhao H., Yelleswarapu M., Huck W. T. S., Macromolecularly crowded protocells from reversibly shrinking monodisperse liposomes. J. Am. Chem. Soc. 140, 7399–7402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L., Zou Y., Bhattacharya A., Zhang D., Lang S. Q., Houk K. N., Devaraj N. K., Enzyme-free synthesis of natural phospholipids in water. Nat. Chem. 12, 1029–1034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seoane A., Brea R. J., Fuertes A., Podolsky K. A., Devaraj N. K., Biomimetic generation and remodeling of phospholipid membranes by dynamic imine chemistry. J. Am. Chem. Soc. 140, 8388–8391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters R. J., Marguet M., Marais S., Fraaije M. W., van Hest J. C., Lecommandoux S., Cascade reactions in multicompartmentalized polymersomes. Angew. Chem. Int. Ed. 53, 146–150 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Amstad E., Kim S.-H., Weitz D. A., Photo- and thermoresponsive polymersomes for triggered release. Angew. Chem. Int. Ed. 51, 12499–12503 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Huang X., Li M., Green D. C., Williams D. S., Patil A. J., Mann S., Interfacial assembly of protein-polymer nano-conjugates into stimulus-responsive biomimetic protocells. Nat. Commun. 4, 2239 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Huang X., Patil A. J., Li M., Mann S., Design and construction of higher-order structure and function in proteinosome-based protocells. J. Am. Chem. Soc. 136, 9225–9234 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Arco L., Li M., Mann S., Phagocytosis-inspired behaviour in synthetic protocell communities of compartmentalized colloidal objects. Nat. Mater. 16, 857–863 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Li M., Harbron R. L., Weaver J. V. M., Binks B. P., Mann S., Electrostatically gated membrane permeability in inorganic protocells. Nat. Chem. 5, 529–536 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Liu X., Appelhans D., Voit B., Hollow capsules with multiresponsive valves for controlled enzymatic reactions. J. Am. Chem. Soc. 140, 16106–16114 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Vieregg J. R., Lueckheide M., Marciel A. B., Leon L., Bologna A. J., Rivera J. R., Tirrell M. V., Oligonucleotide-peptide complexes: Phase control by hybridization. J. Am. Chem. Soc. 140, 1632–1638 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Schuster B. S., Reed E. H., Parthasarathy R., Jahnke C. N., Caldwell R. M., Bermudez J. G., Ramage H., Good M. C., Hammer D. A., Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun. 9, 2985 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin N., Tian L., Spencer D., Coutable-Pennarun A., Anderson J. L. R., Mann S., Photoswitchable phase separation and oligonucleotide trafficking in DNA coacervate microdroplets. Angew. Chem. Int. Ed. 58, 14594–14598 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Jia L., Ji Z., Ji Y. M., Zhou C., Xing G. W., Qiao Y., Design and fluorescence localization of lipid-rich domains in multiphase coacervate droplets based on AIE-active molecules. ChemSystemsChem 2, e2000044 (2020). [Google Scholar]
- 22.Deng N. N., Huck W. T. S., Microfluidic formation of monodisperse coacervate organelles in liposomes. Angew. Chem. Int. Ed. 56, 9736–9740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Booth R., Qiao Y., Li M., Mann S., Spatial positioning and chemical coupling in coacervate-in-proteinosome protocells. Angew. Chem. Int. Ed. 58, 9120–9124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Last M. G. F., Deshpande S., Dekker C., pH-controlled coacervate-membrane interactions within liposomes. ACS Nano 14, 4487–4498 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jing H., Bai Q., Lin Y., Chang H., Yin D., Liang D., Fission and internal fusion of protocell with membraneless “organelles” formed by liquid-liquid phase separation. Langmuir 36, 8017–8026 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Merindol R., Loescher S., Samanta A., Walther A., Pathway-controlled formation of mesostructured all-DNA colloids and superstructures. Nat. Nanotechnol. 13, 730–738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samanta A., Sabatino V., Ward T. R., Walther A., Functional and morphological adaptation in DNA protocells via signal processing prompted by artificial metalloenzymes. Nat. Nanotechnol. 15, 914–921 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai E. C., Notch signaling: Control of cell communication and cell fate. Development 131, 965–973 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Waters C. M., Bassler B. L., Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Pawson T., Nash P., Assembly of cell regulatory systems through protein interaction domains. Science 300, 445–452 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Li Y. R., King O. D., Shorter J., Gitler A. D., Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin Y., Berry J., Pannucci N., Haataja M. P., Toettcher J. E., Brangwynne C. P., Spatiotemporal control of intracellular phase transitions using light-activated optodroplets. Cell 168, 159–171.e114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilks J. C., Slonczewski J. L., pH of the cytoplasm and periplasm of Escherichia coli: Rapid measurement by green fluorescent protein fluorimetry. J. Bacteriol. 189, 5601–5607 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toparlak Ö. D., Zasso J., Bridi S., Serra M. D., Macchi P., Conti L., Baudet M. L., Mansy S. S., Artificial cells drive neural differentiation. Sci. Adv. 6, eabb4920 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang T. D., Cecchi D., Fracasso G., Accardi D., Coutable-Pennarun A., Mansy S. S., Perriman A. W., Anderson J. L. R., Mann S., Gene-mediated chemical communication in synthetic protocell communities. ACS Synth. Biol. 7, 339–346 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Qiao Y., Li M., Booth R., Mann S., Predatory behaviour in synthetic protocell communities. Nat. Chem. 9, 110–119 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Qiao Y., Li M., Qiu D., Mann S., Response-retaliation behavior in synthetic protocell communities. Angew. Chem. Int. Ed. 58, 17758–17763 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Martin N., Sharma K. P., Harniman R. L., Richardson R. M., Hutchings R. J., Alibhai D., Li M., Mann S., Light-induced dynamic shaping and self-division of multipodal polyelectrolyte-surfactant microarchitectures via azobenzene photomechanics. Sci. Rep. 7, 41327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love C., Steinkühler J., Gonzales D. T., Yandrapalli N., Robinson T., Dimova R., Tang T. D., Reversible pH-responsive coacervate formation in lipid vesicles activates dormant enzymatic reactions. Angew. Chem. Int. Ed. 59, 5950–5957 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deshpande S., Brandenburg F., Lau A., Last M. G. F., Spoelstra W. K., Reese L., Wunnava S., Dogterom M., Dekker C., Spatiotemporal control of coacervate formation within liposomes. Nat. Commun. 10, 1800 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bracha D., Walls M. T., Wei M.-T., Zhu L., Kurian M., Avalos J. L., Toettcher J. E., Brangwynne C. P., Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell 175, 1467–1480.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koga S., Williams D. S., Perriman A. W., Mann S., Peptide-nucleotide microdroplets as a step towards a membrane-free protocell model. Nat. Chem. 3, 720–724 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Yin D., Hu X., Liu D., Du W., Wang H., Guo M., Tang D., Enhanced detection of amino acids in hydrophilic interaction chromatography electrospray tandem mass spectrometry with carboxylic acids as mobile phase additives. Eur. J. Mass Spectrom. 23, 98–104 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Kalina M., Kargerova A., Pekar M., DEAE-dextran hydrochloride behaviour in aqueous solution-The effect of ionic strength and concentration. Carbohydr. Polym. 220, 163–169 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Huguet M. L., Neufeld R. J., Dellacherie E., Calcium-alginate beads coated with polycationic polymers: Comparison of chitosan and DEAE-dextran. Process Biochem. 31, 347–353 (1996). [Google Scholar]
- 46.Deng J., Walther A., Programmable ATP-fueled DNA coacervates by transient liquid-liquid phase separation. Chem 6, 3329–3343 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ellis R. J., Macromolecular crowding: Obvious but underappreciated. Trends Biochem. Sci. 26, 597–604 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Valiev M., Bylaska E. J., Govind N., Kowalski K., Straatsma T. P., Van Dam H. J. J., Wang D., Nieplocha J., Apra E., Windus T. L., de Jong W. A., NWChem: A comprehensive and scalable open-source solution for large scale molecular simulations. Comput. Phys. Commun. 181, 1477–1489 (2010). [Google Scholar]
- 49.Rego L. G. C., Bortolini G., Modulating the photoisomerization mechanism of semiconductor-bound azobenzene-functionalized compounds. J. Phys. Chem. C 123, 5692–5698 (2019). [Google Scholar]
- 50.Benmensour M. A., Ayadi A., Akdas-Kilig H., Boucekkine A., Fillaut J.-L., El-Ghayoury A., Azobased iminopyridine ligands and their rhenium metal complexes: Syntheses, spectroscopic, trans-cis photoisomerization and theoretical studies. J. Photochem. Photobiol. A Chem. 368, 78–84 (2019). [Google Scholar]
- 51.Klamt A., Schüürmann G., COSMO: A new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J. Chem. Soc. Perkin Trans. 2, 799–805 (1993). [Google Scholar]
- 52.Stefanovich E. V., Truong T. N., Optimized atomic radii for quantum dielectric continuum solvation models. Chem. Phys. Lett. 244, 65–74 (1995). [Google Scholar]
- 53.He L.-h., Wang G.-m., Tang Q., Fu X.-k., Gong C.-b., Synthesis and characterization of novel electrochromic and photoresponsive materials based on azobenzene-4,4′-dicarboxylic acid dialkyl ester. J. Mater. Chem. C 2, 8162–8169 (2014). [Google Scholar]
- 54.Zhou Y., Zhao M., Wu Y., Li C., Wu J., Zheng M., Peng L., Peng S., A class of novel Schiff’s bases: Synthesis, therapeutic action for chronic pain, anti-inflammation and 3D QSAR analysis. Bioorg. Med. Chem. 18, 2165–2172 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/22/eabf9000/DC1


