Abstract
The use of intact cells in medical research offers a number of advantages over employing cell-free systems. In diagnostics, cells isolated from liquid biopsies can be directly used, speeding up the time of analysis and diminishing the risk of protein degradation by sample manipulation. In drug discovery, studies in live cells take into account aspects neglected in cell-free systems, such as uptake, metabolization, and subcellular concentration by compartmentalization of potential drug candidates. Therefore, probes for studies in cellulo are of paramount importance. Acid ceramidase (AC) is a lysosomal enzyme that hydrolyses ceramides into sphingoid bases and fatty acids. The essential role of this enzyme in the outburst and progress of several diseases, some of them still incurable, is well sustained. Despite the great clinical relevance of AC as a biomarker and therapeutic target, the specific monitoring of AC activity in live cells has remained elusive due to the concomitant existence of neutral and alkaline ceramidases. In this work, we report that 1-deoxydihydroceramides are exclusively hydrolysed by AC. Using N-octanoyl-18-azidodeoxysphinganine as a probe and a BODIPY-substituted bicyclononyne, we show the click-reliant predominant staining of lysosomes, with extra-lysosomal labeling also occurring in some cells. Importantly, using pharmacological and genetic tools together with high resolution mass spectrometry, we demonstrate that both lysosomal and extra-lysosomal staining are AC-dependent. These findings are translated into the specific flow cytometry monitoring of AC activity in intact cells, which fills an important gap in the field of diseases linked to altered AC activity.
The use of intact cells in medical research offers a number of advantages over employing cell-free systems.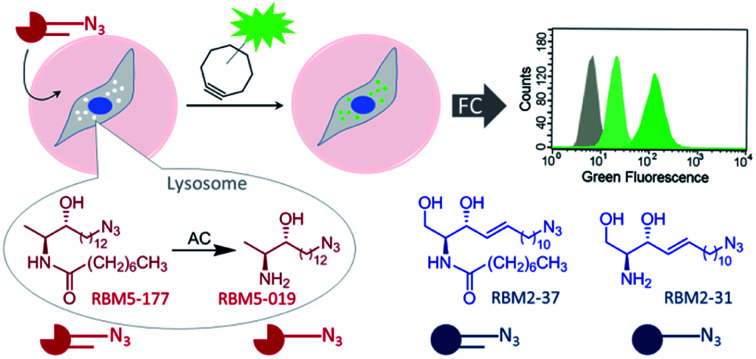
Introduction
Ceramidases are lipid hydrolases involved in the deamidation of ceramides and phytoceramides to the free base and fatty acids. Ceramidases are encoded by five different genes and are differentiated by the pH required for optimal activity (acid, neutral and alkaline).1,2 Several reports sustain the involvement of ceramidases in the development of malignancies and in the failure of their treatment. Some examples include the role of acid ceramidase (AC) in melanoma,3–6 prostate cancer7,8 and acute myeloid leukemia,9,10 the implication of neutral ceramidase (NC) in colorectal cancer11 and the contribution of alkaline ceramidase 3 (ACER3) to hepatocellular carcinoma12 and acute myeloid leukemia.13 On the other hand, mutations in the genes encoding for AC14 and ACER3 15 have been discovered to cause rare inherited diseases. One such condition is Farber disease, linked to mutations in the AC-encoding gene.14 The clinical interest of these amidases highlights the importance of developing specific probes for application in diagnostic and library screening for drug discovery. In both cases, the use of intact cells offers a number of advantages over employing cell-free systems. In the case of diagnostics, cells isolated from liquid biopsies can be directly used, speeding up the time of analysis and diminishing the risk of protein degradation. In the field of drug discovery, phenotypic screening in living cells has emerged as a promising strategy as it takes into account aspects neglected in cell-free systems, such as cell uptake, metabolization, and subcellular compartmentalization and concentration of potential drug candidates.
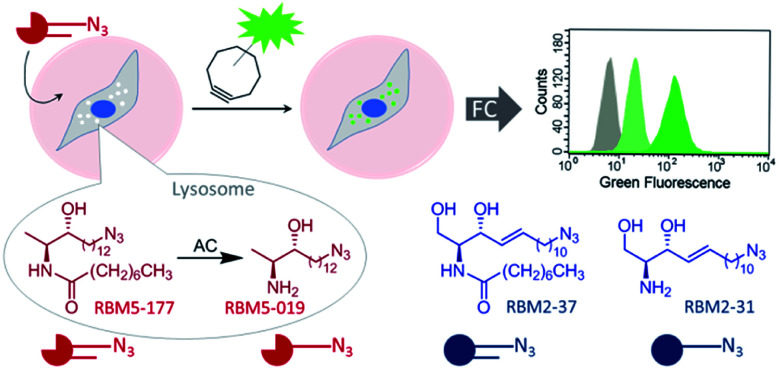
Although several assays have been reported to measure ceramidase activity in live cells, none of them is specific for a particular enzyme. In this article we report on the specific monitoring of AC activity in cellulo by flow cytometry counting of cells labeled by a click reaction with a fluorescent partner of the AC product (Fig. 1). The key point was the discovery that deoxydihydroceramides are specifically hydrolyzed by AC, remaining inert to neutral and alkaline ceramidases.
Fig. 1. Acid ceramidase activity monitoring by flow cytometry. AC-catalyzed hydrolysis of the specific AC substrate (RBM5-177) occurs in the lysosome, wherein the reaction product (RBM5-019) accumulates and leads to lysosome staining after the click reaction with a BODIPY-substituted bicyclononyne. Comparative studies with the corresponding canonical counterparts (RBM2-37 and RBM2-31) are illustrative. FC, flow cytometry.
Deoxydihydroceramides are produced in cells bearing mutated forms of serine palmitoyltransferase (SPT). SPT is the first and rate-determining enzyme in de novo ceramide biosynthesis and it catalyzes the conjugation of l-serine with palmitoyl-CoA to produce 3-ketosphinganine, which is subsequently reduced to sphinganine and N-acylated to dihydroceramides by ceramide synthases (CerS) (ESI Fig. S1†).16 Besides producing sphingolipids with the classical 2-amino-1,3-diol structure, several mutated forms of SPT show an alternative activity towards other amino acids, such as l-alanine, resulting in the formation of 1-deoxysphinganine or spisulosine (DOXSa).17 These mutations and the consequent accumulation of DOXSa and its downstream metabolites are the underlying cause of the rare Hereditary Sensory and Autonomic Neuropathy type 1 (HSAN1).18 By lacking the C1-hydroxyl group of typical sphingolipids, 1-deoxydihydroceramides (DOXDHC), formed by CerS-catalyzed N-acylation of DOXSa, cannot be desaturated by the canonical dihydroceramide desaturase 1. Furthermore, the absence of the C1-hydroxyl group precludes the formation of complex sphingolipids and of sphingosine-1-phosphate, the essential catabolic intermediate in the canonical degradation of sphingolipids. Therefore, although metabolic reactions of DOXSa and DOXDHC have been reported19,20 their metabolic stability is higher than that of the typical C1-hydroxylated counterparts. Such metabolic stability should confer robustness to assays based on the use of DOXSa and DOXDHC derived probes.
Results and discussion
As mentioned in the Introduction, ceramides and phytoceramides are hydrolyzed by ceramidases in the first step of the sphingolipid catabolic pathway. Whether the amide hydrolysis by ceramidases occurs also in DOXDHC had not been examined. To investigate this issue, we envisaged that the azido labeled derivative N-octanoyl-18-azidodeoxysphinganine (RBM5-177) could be a suitable probe, as it would allow the unambiguous identification of metabolites by ultraperformance liquid chromatography coupled to mass spectrometry (UPLC-HRMS) and their intracellular distribution by fluorescence confocal microscopy upon labeling with a fluorescent click reagent.
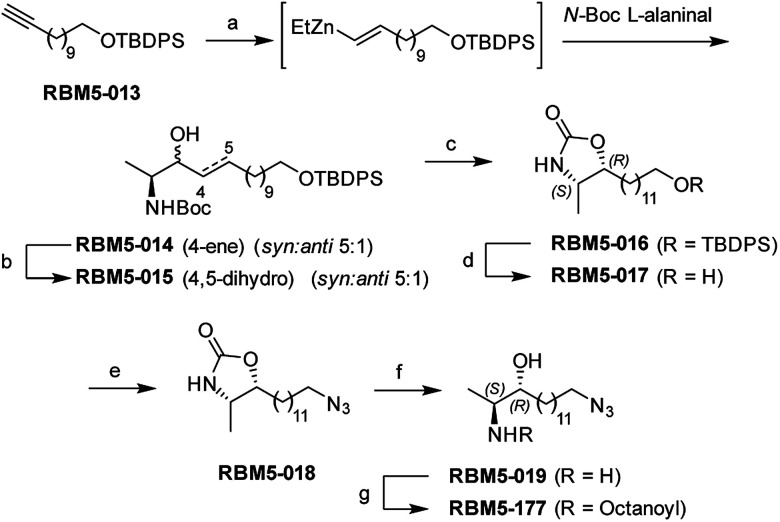
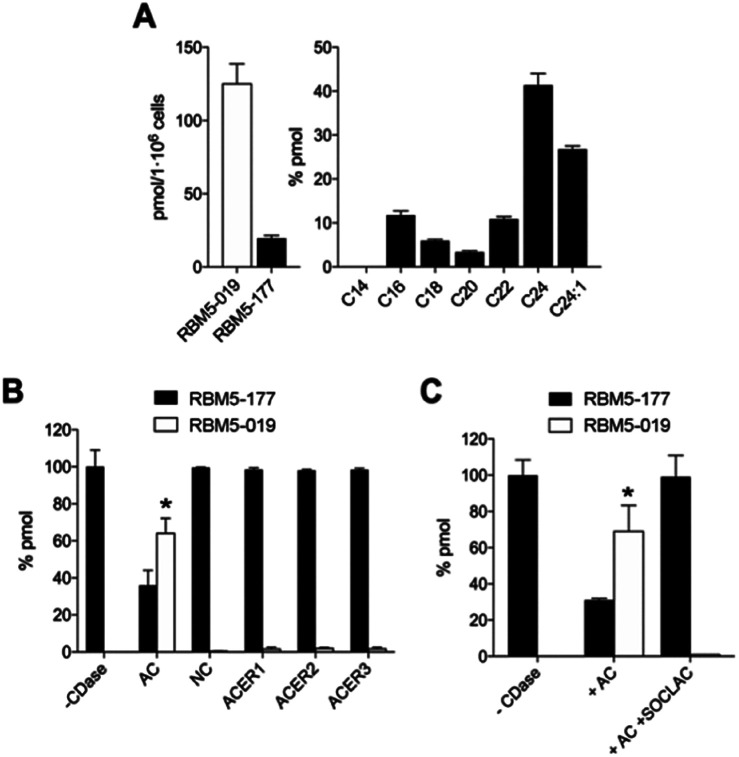
Probes RBM5-019 and RBM5-177 were obtained following the route depicted in Scheme 1. Hydrozirconation/transmetallation21 of RBM5-013, followed by condensation with N-Boc alaninal,22 afforded RBM5-014 as a syn-enriched inseparable mixture of diastereoisomers (5 : 1 syn : anti, all-E),21 which was hydrogenated to the mixture of isomers RBM5-015. The major diastereomer was separated and configurationally assigned through the oxazolidinone RBM5-016, resulting from the intramolecular N-Boc promoted cyclization of RBM5-015 with inversion of the configuration at the C3.21 Silyl deprotection, installation of the azide group and alkaline hydrolysis led to RBM5-019, whose acylation afforded RBM5-177. Putative hydrolysis of RBM5-177 was first examined in A549 lung adenocarcinoma cells. As shown in Fig. 2, RBM5-177 (5 μM, 30 min) underwent an almost complete metabolization to both the free base (RBM5-019) and differently transacylated products, as concluded by UPLC-HRMS lipid analysis (Fig. 2A). Together with previously published data,21 these results suggested that DOXDHC are hydrolyzed by at least one ceramidase. To confirm this hypothesis, human recombinant neutral ceramidase (NC) or lysates (AC) and microsomes (alkaline ceramidases 1, 2 and 3) from cells overexpressing these 4 enzymes were incubated with RBM5-177 and the RBM5-019 formation was quantified by UPLC-HRMS. As shown in Fig. 2B, RBM5-177 was transformed into the free amine only by AC. Moreover, pre-incubation with SOCLAC, a specific irreversible AC inhibitor,23 totally prevented the formation of RBM5-019 from RBM5-177 in AC-enriched cell lysates (Fig. 2C). The finding that RBM5-177 is only hydrolysed by AC suggests that the AC content may be relevant to the different affection of nervous tissues (peripheral vs. central) in HSAN1.
Scheme 1. Synthesis of probes RBM5-019 and RBM5-177. Reagents and conditions: (a) (i) Cp2Zr(H)Cl, CH2Cl2, 0 °C, 30 min (ii) Et2Zn, CH2Cl2, −40 °C, 10 min (iii) N-Boc l-alaninal, −40 °C to rt, 3 h, 19%, dr = 5 : 1 (syn : anti), only E isomer; (b) H2, 5 wt% Rh on Al2O3, EtOAc, rt, 3 h, 86%; (c) (i) MsCl, NEt3, CH2Cl2, rt, 2 h (ii) NEt3, ClCH2CH2Cl, reflux, overnight, 61% (anti diastereoisomer); (d) TBAF, THF, 0 °C, 2 h, 86%; (e) (i) NBS, PPh3, DMF, 0 °C to rt, 1 h, (ii) NaN3, DMF, 80 °C, 3 h, 71% (over two steps); (f) NaOH (aq.) : EtOH (1 : 1), reflux, 8 h, 66%; (g) octanoic acid, EDC, HOBt, Et3N, CH2Cl2, 89%.
Fig. 2. Metabolization of RBM5-177. (A) A549 cells were incubated with RBM5-177 (5 μM, 30 min) and the lipid extracts were analyzed by UPLC-HRMS. Data (mean ± SD) were obtained from two experiments in triplicate. (B) Lysates from A375/+Dox cells (120 μg), recombinant NC (5 ng) or microsomes from ACER1 (100 μg from HeLa TRex ACER1 cells induced with tetracycline), ACER2 (100 μg from HeLa TRex ACER2 cells induced with tetracycline) or ACER3 (140 μg of Asah2-null mouse embryonic fibroblasts)24 overexpressing cells were incubated with RBM5-177 (5 μM, 30 min) in the appropriate buffer, and the lipid extracts were analyzed by UPLC-HRMS. Data (mean ± SD) were obtained from two different experiments in duplicate. Data corresponding to RBM5-019 were analysed by one way ANOVA followed by Dunnett's multiple comparison post-test if ANOVA < 0.05 (*, P < 0.001 from control without enzyme, −CDase). (C) Lysates from A375/+Dox cells were incubated with RBM5-177 (5 μM, 30 min) prior to treatment with SOCLAC (5 μM, 1 h) and the lipid extracts were analyzed by UPLC-HRMS. Data (mean ± SD) were obtained from two experiments in duplicate. Data corresponding to RBM5-019 were analysed by one way ANOVA followed by Dunnett's multiple comparison post-test if ANOVA P < 0.05 (*, P < 0.001 from −CDase). The right panel in (A) corresponds to the acyl chain of reacylated RBM5-019 (C14, tetradecanoyl, etc.).
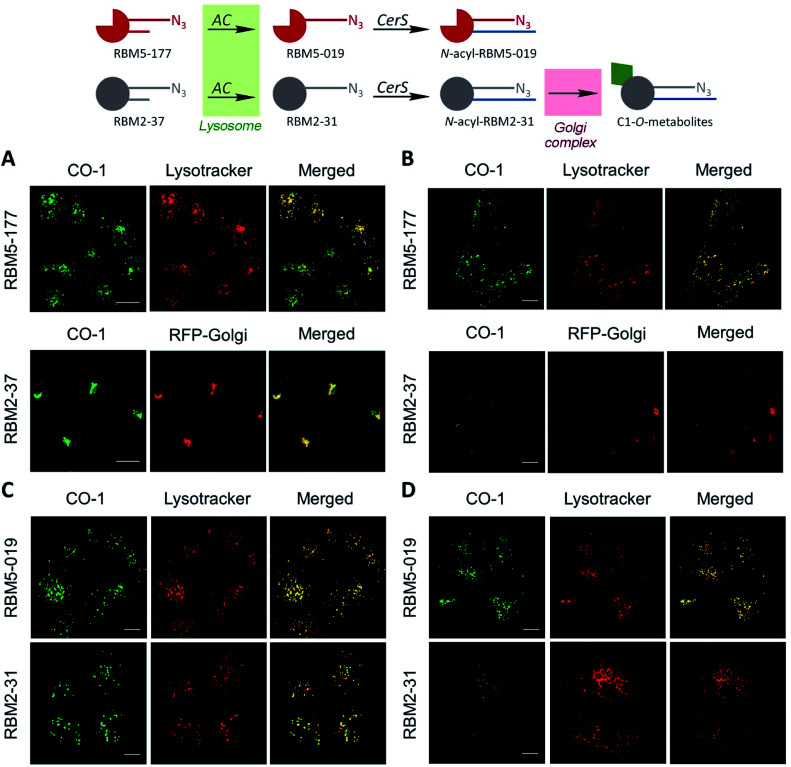
These results suggested that, since RBM5-177 is specifically hydrolysed by AC, this probe could enable the specific monitoring of AC activity in intact cells by flow cytometry prior to the click reaction with the BODIPY-labelled bicyclo[6.1.0]nonyne reagent CO-1.25 To validate this putative approach, the AC-activity dependence of the subcellular staining distribution by the RBM5-177/CO-1 system was first examined by fluorescence confocal microscopy. Thus, A549 cells, previously found to contain AC activity,23 were pulsed with RBM5-177 (5 μM, 30 min)§ and then the click reaction with CO-1 (1 μM) was carried out (10 min). As shown in Fig. 3A, fluorescence confocal microscopy revealed that the labelling from RBM5-177/CO-1 co-localized with Lysotracker, but not with the Golgi apparatus (ESI Fig. S3†). The Pearson correlation coefficient and the Manders's (M) overlap coefficient,26 both used to quantify the degree of colocalization between fluorophores, afforded very good values (Pearson = 0.802; M1 = 0.866; M2 = 0.744), indicating that the probe was directed to the lysosome, consistent with the lysosomal localization of AC. To verify that the RBM5-177/CO-1 labelling system was reliable, the experiment was performed in parallel with the canonical ceramide sibling RBM2-37.21 In accordance with the use of fluorescent ceramides as Golgi trackers,27 Golgi staining was observed with the RBM2-37/CO-1 system (Fig. 3A) (Pearson = 0.615; M1 = 0.588; M2 = 0.804). Because of poor RFP-Golgi staining, the co-localization coefficients were not as good as expected. High M2 indicated that almost all RFP-Golgi co-localized with RBM2-37, which stained more effectively than the RFP-Golgi marker. Importantly, when a 5 h incubation step in fresh culture medium was introduced after the RBM5-177 pulse prior to CO-1 addition, staining remained stable with high co-localization with the Lysotracker (Fig. 3B) (Pearson = 0.746; M1 = 0.822; M2 = 0.711). In contrast, the co-localization of the RBM2-37/CO-1 staining with the Golgi tracker decreased considerably and was found in other compartments (Fig. 3B) (Pearson = 0.346; M1 = 0.210; M2 = 0.206), in agreement with the probe metabolization as a ceramide analog. These results supported the metabolic stability of RBM5-177. A similar outcome was found with the free base, RBM5-019, which largely remained in the lysosome after the 5 h chase (compare Fig. 3C with Fig. 3D) (Pearson = 0.762; M1 = 0.735; M2 = 0.688) while labelling intensity from the canonical base, RBM2-31, which can be further metabolized via the ceramide catabolic pathway, decreased considerably (compare Fig. 3C with Fig. 3D). Besides confirming the metabolic stability of RBM5-019, these results also suggested that the AC-generated RBM5-019, which would be protonated at the acidic lysosomal pH, is largely retained inside the lysosome, as reported for other lipophilic amino alcohols such as propranolol,28 while sphingosine egress from the lysosome is not so restricted.29
Fig. 3. Co-localization studies of the indicated probes in A549 cells. (A and C) A549 cells incubated or not with RFP-Golgi were treated with the indicated probe (5 μM, 20 min), washed and incubated with CO-1 (1 μM, 10 min). (B and D) After the probe treatment, cells were washed and incubated with medium for 5 h, washed and incubated with CO-1 (1 μM, 10 min). Lysotracker (75 nM) was added 30 minutes before fluorescence confocal microscopy (scale bar: 10 μm). Co-localization coefficients were measured on the different stacks of images (at least n = 4) with each stack containing 3–10 cells. Pearson's (P) and Manders' (M1 and M2) coefficients: (A) RBM5-177-Lysotracker (P = 0.802; M1 = 0.866; M2 = 0.744) and RBM2-37-RFP-Golgi (P = 0.615; M1 = 0.588; M2 = 0.804). (B) RBM5-177-Lysotracker (P = 0.746; M1 = 0.822; M2 = 0.711) and RBM2-37-RFP-Golgi (P = 0.346; M1 = 0.210; M2 = 0.206). (C) RBM5-019-Lysotracker (P = 0.713; M1 = 0.670; M2 = 0.774) and RBM2-31-Lysotracker (P = 0.731; M1 = 0.768; M2 = 0.570). (D) RBM5-019-Lysotracker (P = 0.762; M1 = 0.735; M2 = 0.688).
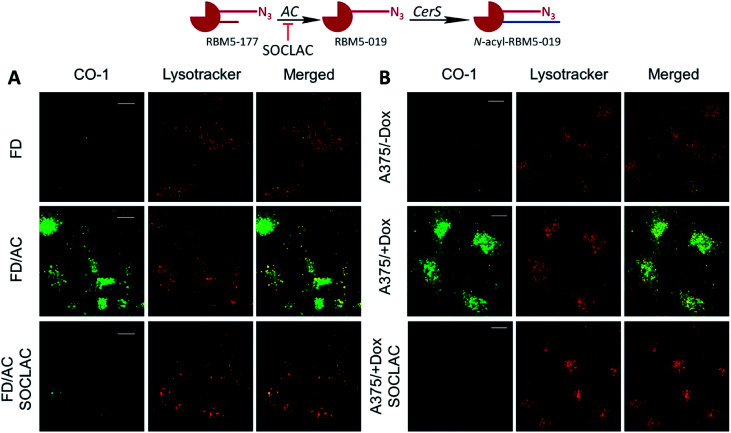
The above results suggested that the in situ generated RBM5-019 might be responsible for the observed lysosome staining. In order to prove this possibility, the effect of AC-overexpression and AC activity deficiency/blockade on lysosome labelling by RBM5-177 was analyzed using Farber disease fibroblasts lacking AC (FD) and the same cells transduced to overexpress AC (FD/AC), and AC-overexpressing melanoma A375 cells (A375/+Dox) and their WT pairs (A375/−Dox). As depicted in Fig. 4A, lysosome staining was observed in FD/AC cells after RBM5-177/CO-1 treatment (Pearson = 0.725; M1 = 0.631; M2 = 0.760), while no lysosomal labelling occurred in either FD cells or in SOCLAC pretreated FD/AC cells. In this line, UPLC/HRMS showed the presence of RBM5-019 and reacylated products in FD/AC cells, but not in FD and SOCLAC pretreated FD/AC cells (ESI Fig. S2A†), thus correlating the lysosome staining with AC activity. Furthermore, lysosome labelling intensity was higher in A375/+Dox than in A375/−Dox cells with very high co-localization with lysosomes (Pearson = 0.754; M1 = 0.723; M2 = 0.713) and the organelle staining in the latter was completely abolished by preincubation with SOCLAC (Fig. 4B). In agreement, UPLC-HRMS analysis of lipid extracts from cells at the same experimental conditions showed that the levels of RBM5-019 and reacylated products were significantly higher in A375/+Dox than in A375/−Dox cells, while they dropped dramatically in the former upon SOCLAC pretreatment (ESI Fig. S2B†). In contrast, lysosome labelling with RBM5-019 was not sensitive to AC overexpression or blockade (ESI Fig. S4A†) with very high co-localization with lysosomes in all cases (Pearson = 0.856; M1 = 0.831; M2 = 0.765 on average). Accordingly, the levels of RBM5-019 and transacylated products were not affected either by differences in AC-activity (ESI Fig. S4B†). These results indicated that RBM5-019, but not RBM5-177 was responsible for lysosome labelling, and that labelling by RBM5-177, was AC-activity dependent.
Fig. 4. AC activity-dependence of labeling with RBM5-177. (A) FD and FD/AC cells were incubated with RBM5-177 (5 μM, 4 h) prior to treatment with SOCLAC 5 μM or vehicle for 1 h in FD/AC. (B) A375 cells stimulated (+Dox) or not (−Dox) for AC overexpression were incubated for 20 min with RBM5-177 (5 μM, 20 min) prior to treatment with SOCLAC 5 μM or vehicle for 1 h. Cells were then treated with CO-1 (1 μM, 10 min), washed and analyzed by fluorescence confocal microscopy (scale bar: 10 μm). Lysotracker (75 nM) was used to stain lysosomes. Co-localization coefficients were measured on the different stacks of images (at least n = 4) with each stack containing 3–10 cells. Pearson's (P) and Manders' (M1 and M2) coefficients: (A) RBM5-177-Lysotracker (P = 0.725; M1 = 0.631; M2 = 0.760). (B) RBM5-177-Lysotracker (P = 0.754; M1 = 0.723; M2 = 0.713).
Importantly, UPLC-HRMS analysis showed that while no RBM5-019 was detected and the levels of transacylated products decreased dramatically upon SOCLAC treatment, the expected increase in the amounts of RBM5-177 was not observed in the cell lipids. However, significantly higher levels of RBM5-177 were found in the medium of SOCLAC pretreated cells than in controls (ESI Fig. S2D.† Only the data for A549 cells are shown, but similar results were found with the other cell lines). This proved that the untransformed RBM5-177 was pumped out from cells, which resulted to be the reason of the negligible background observed in cells without AC activity.
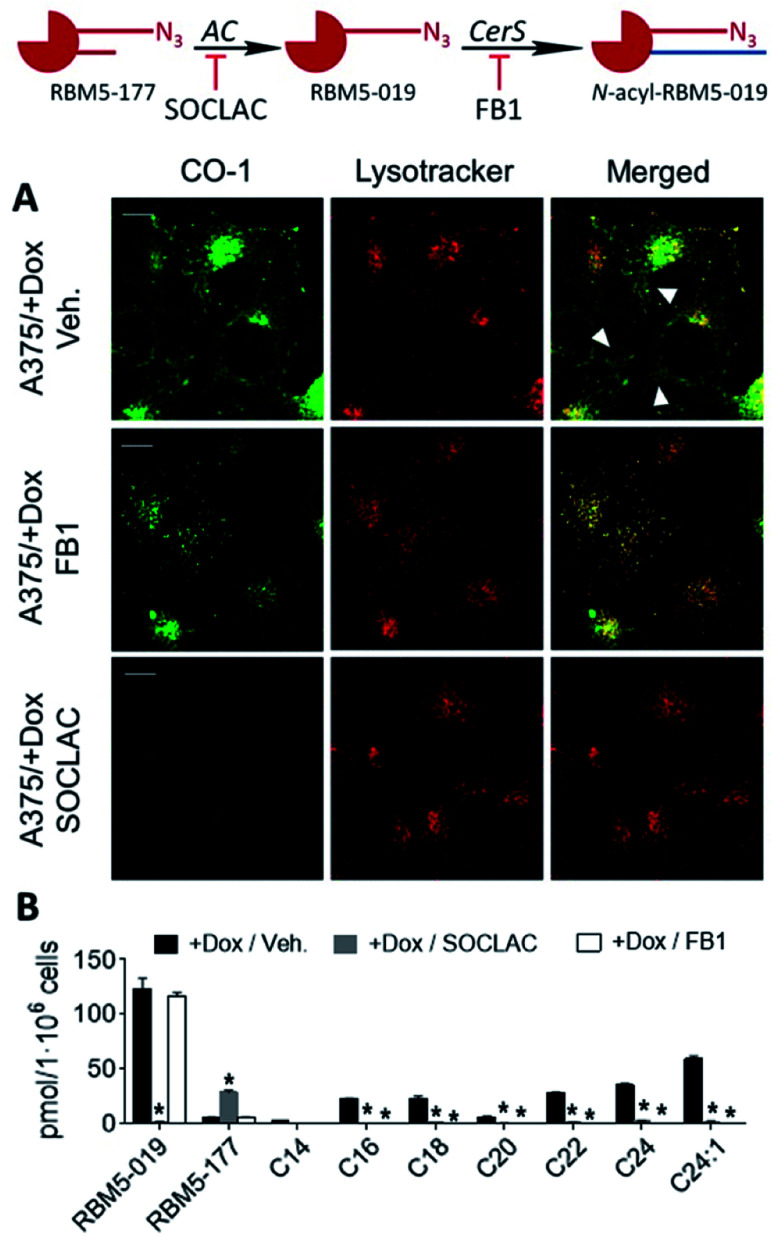
Interestingly, in both AC-overexpressing cells (FD/AC and A375/+Dox), incubation with RBM5-177 produced labelling also outside the lysosomes (Fig. 4), which might be indicative of the extralysosomal CerS-mediated re-acylation of the RBM5-019 fraction released from the acidic compartment. In order to confirm this hypothesis, the effect of the CerS inhibitor FB1 (ref. 30) on the intracellular stain distribution was tested. As shown in Fig. 5A, the extralysosomal staining found in RBM5-177/CO-1 treated A375/+Dox cells was prevented by preincubation with FB1 (100 μM, 24 h), while the lysosome labelling remained intact (Pearson = 0.841; M1 = 0.717; M2 = 0.716). UPLC-HRMS lipid analysis confirmed that transacylation was blocked with FB1, since no RBM5-019 reacylated products were detected and RBM5-019 levels remained unaltered (Fig. 5B). That the extralysosomal staining was due to the in situ generated base re-acylation was further confirmed by treatment with SOCLAC, which avoided the staining of both lysosomes and extralysosomal structures (Fig. 5A) and precluded the formation of both RBM5-019 and transacylated products, as concluded from their absence in the lipid extracts from SOCLAC pretreated/RBM5-177 treated cells (UPLC-HRMS) (Fig. 5B). Therefore, all these results proved that lysosome staining was due to RBM5-019 accumulation while the re-acylated products were responsible for the labelling outside the lysosomes, which was shown to be CerS-dependent. Importantly, these results also confirmed that not only hydrolysis, but also the formation of RBM5-177 transacylation products was dependent on the cell's AC activity.
Fig. 5. CerS activity-dependence of labeling with RBM5-177. A375 cells stimulated (+Dox) for AC overexpression were incubated for 4 h with RBM5-177 (5 μM, 4 h), prior to treatment with vehicle, SOCLAC (5 μM, 1 h) or FB1 (100 μM, 24 h). The click reaction was carried out by incubation with CO-1 (1 μM, 10 min). Cells were analysed by fluorescence confocal microscopy (scale bar: 10 μm) (A) or the lipid extracts were analyzed by UPLC-HRMS (B). Lysotracker (75 nM) was used to stain lysosomes. White arrowheads point out putative mitochondrial staining. Co-localization coefficients were measured on the different stacks of images (at least n = 4) with each stack containing 3–10 cells. Pearson's (P) and Manders' (M1 and M2) coefficients: RBM5-177-Lysotracker with vehicle (P = 0.816; M1 = 0.791; M2 = 0.778) and RBM5-177-Lysotracker with FB1 (P = 0.841; M1 = 0.717; M2 = 0.761). Data in (B) (mean ± SD) were obtained from two different experiments in triplicate. Data were analysed by one-way ANOVA followed by Dunnett's multiple comparison post-test if ANOVA P < 0.05 (*, P < 0.001 from vehicle). The right panel in (B) corresponds to the acyl chain of reacylated RBM5-019 (C14, tetradecanoyl, etc.).
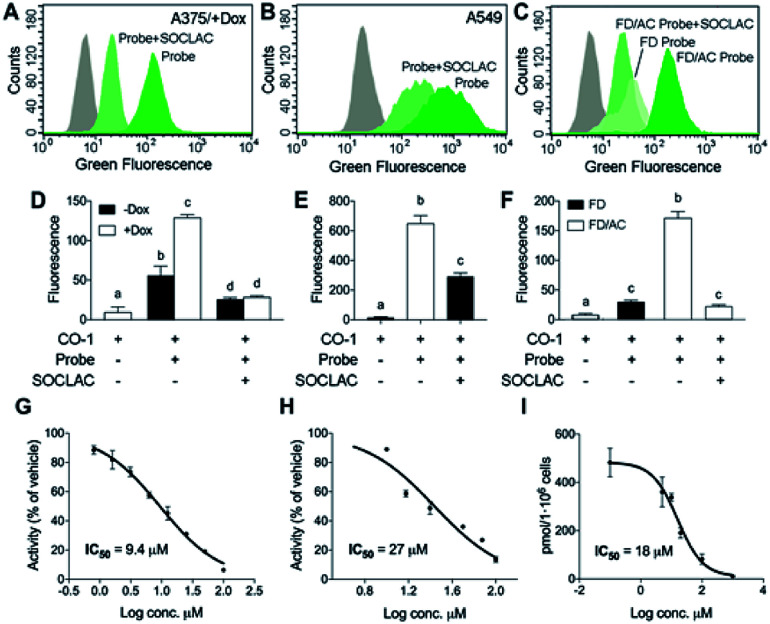
After establishing that both lysosome and extralysosome staining with RBM5-177 were dependent on AC activity and that the staining was robust and long lasting, the flow cytometry-based monitoring of AC activity was investigated. As shown in Fig. 6, the number of cells labelled with RBM5-177/CO-1 highly correlated with both the confocal microscopy and the UPLC/HRMS data mentioned above. Thus, in A375 cells (Fig. 6A and D) the number of fluorescent counts was significantly higher in AC-overexpressing cells than in non-induced cells. Furthermore, the signal significantly decreased in the former with SOCLAC pre-incubation. Moreover, the ability of SOCLAC to significantly decrease the number of labelled cells was also confirmed in the A549 cell line (Fig. 6B and E), consistent with the absence of RBM5-019 and re-acylated products in the lipid extracts (ESI Fig. S2C†). In the case of the two Farber cell models, the AC deficient FD cells were negligibly labelled with RBM5-177/CO-1, while a significant 7-fold increase in the number of fluorescent counts over FD was measured in the AC-transduced counterparts. In the latter, the number of labelled cells was reduced to FD levels upon SOCLAC preincubation (Fig. 6C and F).
Fig. 6. AC activity determination by flow cytometry with RBM5-177/CO-1. A375 cells stimulated (+Dox) or not (−Dox) with doxycycline for AC overexpression (A and D), A549 cells (B and E) or FD and FD/AC cells (C and F) were incubated with RBM5-177 (probe) (5 μM, 4 h), prior to treatment with SOCLAC (5 μM, 1 h) or vehicle, and then with CO-1 (1 μM, 10 min). Cells were analyzed by flow cytometry measuring green fluorescence. The grey curves correspond to a CO-1 control without RBM5-177. Data (mean ± SD) were obtained from two or three different experiments in triplicate. Data were analysed by two-way (D) or one-way (E, F) ANOVA followed by Bonferroni's multiple comparison post-test if ANOVA P < 0.05. Different letters denote a statistical significance (P < 0.001). Representative histograms of panel (D–F) are given in panels (A–C). (G–I) A375/+Dox were incubated for 4 h with different concentrations of DM120 in the presence of fluorogenic substrate RBM14C12 (40 μM)32 (G), or RBM5-177 (H, I). AC activity was determined as reported (G),32 by flow cytometry after labelling with CO-1 (1 μM, 10 min) (H) or by UPLC-HRMS quantification of the amounts of RBM5-019 (I). Data (mean ± SD) were obtained from two to three experiments in triplicate. Curve fitting with the sigmoidal dose–response (variable slope) equation afforded the IC50 values detailed in figure. (A–F) Fluorescence at excitation/emission 488/525 nm.
Finally, the applicability of the flow cytometry method to test for AC-inhibitory activity was examined in order to assess its reliability for genotypic library screening. Although its usefulness for this purpose was already inferred from the results found with SOCLAC, we expanded the study to a reversible inhibitor, using the previously reported compound DM120 (ref. 31) in AC-overexpressing A375 cells. First, DM120 AC inhibitory activity was examined using a fluorogenic assay,32 which afforded an IC50 value of 9.4 μM (Fig. 6G) without toxicity in the whole range of concentrations examined. Using RBM5-177 as the substrate and flow cytometry analysis after cell labelling with CO-1, the concentration–response curve yielded an IC50 of 27 μM (Fig. 6H). A similar value (IC50 = 18 μM) was obtained by measuring the decrease in RBM5-019 amounts as the DM120 concentration increased by UPLC-HRMS (Fig. 6I). The slightly lower IC50 value obtained with the fluorogenic assay can be explained considering that while RBM5-177 is AC-specific, the fluorogenic substrate is also accepted by NC and ACER3.24 Therefore, putative inhibition of these ceramidases by DM120 (its specificity for the different ceramidases has not been investigated) might contribute to the inhibition when monitored with the fluorogenic substrate. It is worth noting that some fluorescent counts are detected under conditions where AC is not active (treatment with SOCLAC or FD cells). These low readouts are likely due to the fluorescence coming from the small fraction of RBM5-177 not expelled by cells or to the incomplete AC inhibition by SOCLAC.
Finally, we measured the AC activity of cells with different amounts of AC (A375/+Dox, A375/−Dox, FD and FD/AC) by both the fluorogenic enzyme assay32 and the flow cytometry assay reported here. The results correlated well for cells with the same background (A375/+Dox vs. A375/−Dox and FD vs. FD/AC), but correlation was not so good between cell types (ESI Fig. S6†). This is due to the fact that no specific substrate for AC activity determination in live cells is available (RBM5-177 is the first one reported) and therefore, other ceramidases (neutral and alkaline 3) may contribute to the final readout in the enzyme activity assay performed here.24
Overall, these results demonstrate that RBM5-177 is a good probe for the flow cytometry monitoring of AC activity in intact cells using fluorescently labelled bicyclo[6.1.0]nonyne reagents such as CO-1 as click reaction partners.
Conclusions
In summary, we have reported a simple, robust, fast and sensitive assay for the specific determination of AC activity in intact cells by flow cytometry. The main advantage of this method over reported fluorogenic assays32–34 is the probe specificity for AC, allowing the reliable determination of AC activity in intact cells. Furthermore, the metabolic stability of the AC reaction product allows a very persistent readout, which makes the assay very robust. Moreover, the availability of CO-1 analogs with varying emission colors35 widens the range of possible combinations with other organelle stains. Finally, the possibility of using the system reported in massive phenotypic screening (high throughput flow cytometry) should allow progress in drug discovery to fight conditions related to altered AC activity, which include both diseases of a high socioeconomic impact and rare genetic illnesses.
Experimental section
Full experimental procedures are detailed in the ESI;† a summary of key experimental methods is given below.
Confocal analysis
Cells were seeded at 1 × 105 cells per mL on glass-bottom plates and allowed to adhere for 24 h. For Golgi staining, cells were incubated with 40 μL per well of CellLight Golgi-RFP BacMam 2.0 for 24 h. The medium was replaced with fresh medium containing RBM5-177, RBM5-019, RBM2-37 or RBM2-31 (5 μM final concentration) and cells were incubated for 20 min at 37 °C. After this time, the medium was replaced with fresh medium containing CO-1 (1 μM final concentration), and cells were incubated for 10 min. Then cells were washed 3 times with DMEM for 5 min at 37 °C, 75 nM LysoTracker Red DND-99 or 75 nM Mitotracker in phenol red-free DMEM was added to stain lysosomes or mitochondria, and cells were kept in the incubator for microscope analysis. Staining was viewed with a Zeiss LSM 880 confocal microscope. Image analysis was performed using Fiji software.36 To measure the Manders's and Pearson's coefficients a threshold including organelles and discarding the background was set. Co-localization coefficients were measured using the JaCoP plugin37 on the different stacks of images (at least n = 4) with each stack containing 3–10 cells. The Pearson's coefficient is a well-established measure of correlation and has a range of +1 (perfect correlation) to −1 (perfect but negative correlation) with 0 denoting the absence of a relationship. On the other hand, Manders's coefficients M1 and M2, ranging from 0 to 1, report the fraction of the total intensity for one channel that co-localizes.
AC activity by flow cytometry
Cells were seeded at 1 × 105 cells per mL into 6 well plates (1 mL per well) and were allowed to adhere for 24 h. The medium was replaced with fresh medium containing SOCLAC (5 μM final concentration) and incubated for 1 h. After this time, the medium was replaced with fresh medium containing RBM5-177, RBM5-019 or RBM2-37 (5 μM final concentration) ± SOCLAC and incubated for 20 min. In the experiments with DM120, the compound, SOCLAC and RBM5-177 were incubated together for 4 h. After incubation with the probes, the medium was replaced with fresh medium containing CO-1 (1 μM final concentration), and cells were further incubated for 10 min. Then cells were washed 3 times with DMEM for 5 min at 37 °C, and finally washed with 400 μL PBS and harvested with 400 μL trypsin–EDTA and 600 μL of medium. Pellets were resuspended with 200 μL of PBS and stained cells were analyzed by using a Guava EasyCyte™ flow cytometer (Merck Millipore, Billerica, MA) measuring green fluorescence (488/525 nm). Data analysis was performed using the Multicycle AV program (Phoenix Flow Systems, San Diego, CA).
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This article is dedicated to the memory of Professor Lina M. Obeid. We are very grateful to Dr Ignacio Alfonso (IQAC-CSIC) and Dr Gemma Triola (IQAC-CSIC) for their thorough critical reading of a previous version of the manuscript. We thank Alexandre Garcia (IQAC-CSIC), Ignacio Pérez-Pomeda (IQAC-CSIC), Eva Dalmau (IQAC-CSIC), and Dr Manel Bosch (CCiT-UB) for their excellent technical assistance.
Electronic supplementary information (ESI) available: Supplementary experimental details; supplementary figures; NMR spectra. See DOI: 10.1039/d0sc03166f
Footnotes
Concentration–response curves for RBM5-177 carried out at 4 h indicate that the readout is linear from 1.25 to 5 μM (ESI Fig. S8†), which is the concentration used in the assays (maximum response within the linear range).
Notes and references
- Fabriàs G. Bedia C. Casas J. Abad J. L. Delgado A. Anti-Cancer Agents Med. Chem. 2011;11:830–843. doi: 10.2174/187152011797655104. [DOI] [PubMed] [Google Scholar]
- Coant N. Sakamoto W. Mao C. Hannun Y. A. Adv. Biol. Regul. 2017;63:122–131. doi: 10.1016/j.jbior.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc J. Garandeau D. Pandiani C. Gaudel C. Bille K. Nottet N. Garcia V. Colosetti P. Pagnotta S. Bahadoran P. Tondeur G. Mograbi B. Dalle S. Caramel J. Levade T. Ballotti R. Andrieu-Abadie N. Bertolotto C. Oncogene. 2018;38:1282–1295. doi: 10.1038/s41388-018-0500-0. [DOI] [PubMed] [Google Scholar]
- Lai M. Realini N. La Ferla M. Passalacqua I. Matteoli G. Ganesan A. Pistello M. Mazzanti C. M. Piomelli D. Sci. Rep. 2017;7:7411. doi: 10.1038/s41598-017-07606-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Realini N. Palese F. Pizzirani D. Pontis S. Basit A. Bach A. Ganesan A. Piomelli D. J. Biol. Chem. 2016;291:2422–2434. doi: 10.1074/jbc.M115.666909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedia C. Casas J. Andrieu-Abadie N. Fabriàs G. Levade T. J. Biol. Chem. 2011;286:28200–28209. doi: 10.1074/jbc.M110.216382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. C. Bai A. Beckham T. H. Marrison S. T. Yount C. L. Young K. Lu P. Bartlett A. M. Wu B. X. Keane B. J. Armeson K. E. Marshall D. T. Keane T. E. Smith M. T. Jones E. E. Drake Jr R. R. Bielawska A. Norris J. S. Liu X. J. Clin. Invest. 2013;123:4344–4358. doi: 10.1172/JCI64791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho L. Meca-Cortes O. Abad J. L. Garcia S. Rubio N. Diaz A. Celia-Terrassa T. Cingolani F. Bermudo R. Fernandez P. L. Blanco J. Delgado A. Casas J. Fabrias G. Thomson T. M. J. Lipid Res. 2013;54:1207–1220. doi: 10.1194/jlr.M032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S.-F. Liu X. Fox T. E. Barth B. M. Sharma A. Turner S. D. Awwad A. Dewey A. Doi K. Spitzer B. Shah M. V. Morad S. A. F. Desai D. Amin S. Zhu J. Liao J. Yun J. Kester M. Claxton D. F. Wang H.-G. Cabot M. C. Schuchman E. H. Levine R. L. Feith D. J. Loughran T. P. Oncotarget. 2016;7:83208–83222. doi: 10.18632/oncotarget.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. M. Tan S.-F. Sharma A. Annageldiyev C. Fox T. E. Abad J. L. Fabrias G. Desai D. Amin S. Wang H.-G. Cabot M. C. Claxton D. F. Kester M. Feith D. J. Loughran T. P. Mol. Cancer Res. 2020;18:352–363. doi: 10.1158/1541-7786.MCR-19-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barros M. Coant N. Kawamori T. Wada M. Snider A. J. Truman J.-P. Wu B. X. Furuya H. Clarke C. J. Bialkowska A. B. Ghaleb A. Yang V. W. Obeid L. M. Hannun Y. A. FASEB J. 2016;30:4159–4171. doi: 10.1096/fj.201600611R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y. Xu M. Gao J. Li M. Pathol., Res. Pract. 2018;214:1381–1387. doi: 10.1016/j.prp.2018.07.029. [DOI] [PubMed] [Google Scholar]
- Chen C. Yin Y. Li C. Chen J. Xie J. Lu Z. Li M. Wang Y. Zhang C. C. Biochem. Biophys. Res. Commun. 2016;478:33–38. doi: 10.1016/j.bbrc.2016.07.099. [DOI] [PubMed] [Google Scholar]
- Yu F. P. S. Amintas S. Levade T. Medin J. A. Orphanet J. Rare Dis. 2018;13:121. doi: 10.1186/s13023-018-0845-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvardson S. Yi J. K. Jalas C. Xu R. Webb B. D. Snider J. Fedick A. Kleinman E. Treff N. R. Mao C. Elpeleg O. J. Med. Genet. 2016;53:389–396. doi: 10.1136/jmedgenet-2015-103457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangoiti P. Camacho L. Arana L. Ouro A. Granado M. H. Brizuela L. Casas J. Fabriás G. Abad J. L. Delgado A. Gómez-Muñoz A. Prog. Lipid Res. 2010;49:316–334. doi: 10.1016/j.plipres.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Duan J. Merrill A. H. J. Biol. Chem. 2015;290:15380–15389. doi: 10.1074/jbc.R115.658823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone M. A. Santos T. Alecu I. Silva L. C. Hornemann T. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids. 2019;1864:512–521. doi: 10.1016/j.bbalip.2018.12.013. [DOI] [PubMed] [Google Scholar]
- Karsai G. Lone M. Kutalik Z. Brenna J. T. Li H. Pan D. von Eckardstein A. Hornemann T. J. Biol. Chem. 2020;295:1889–1897. doi: 10.1074/jbc.AC119.011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alecu I. Othman A. Penno A. Saied E. M. Arenz C. von Eckardstein A. Hornemann T. J. Lipid Res. 2017;58:60–71. doi: 10.1194/jlr.M072421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abad J. L. Nieves I. Rayo P. Casas J. Fabrias G. Delgado A. J. Org. Chem. 2013;78:5858–5866. doi: 10.1021/jo400440z. [DOI] [PubMed] [Google Scholar]
- Ocejo M. Vicario J. L. Badía D. Carrillo L. Reyes E. Synlett. 2005;2005:2110–2112. [Google Scholar]
- Ordóñez Y. F. Abad J. L. Aseeri M. Casas J. Garcia V. Casasampere M. Schuchman E. H. Levade T. Delgado A. Triola G. Fabrias G. J. Am. Chem. Soc. 2019;141:7736–7742. doi: 10.1021/jacs.8b11687. [DOI] [PubMed] [Google Scholar]
- Casasampere M. Camacho L. Cingolani F. Casas J. Egido-Gabás M. Abad J. L. Bedia C. Xu R. Wang K. Canals D. Hannun Y. A. Mao C. Fabriàs G. J. Lipid Res. 2015;56:2019–2028. doi: 10.1194/jlr.D061564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamudi S. H. Satapathy R. Kim J. Su D. Ren H. Das R. Hu L. Alvarado-Martínez E. Lee J. Y. Hoppmann C. Peña-Cabrera E. Ha H.-H. Park H.-S. Wang L. Chang Y.-T. Nat. Commun. 2016;7:11964. doi: 10.1038/ncomms11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler J. Parmryd I. Cytometry, Part A. 2010;77:733–742. doi: 10.1002/cyto.a.20896. [DOI] [PubMed] [Google Scholar]
- Chazotte B. Cold Spring Harb. Protoc. 2012;2012:913–915. doi: 10.1101/pdb.prot070599. [DOI] [PubMed] [Google Scholar]
- Kazmi F. Hensley T. Pope C. Funk R. S. Loewen G. J. Buckley D. B. Parkinson A. Drug Metab. Dispos. 2013;41:897–905. doi: 10.1124/dmd.112.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom T. Li Z. Bittman R. Somerharju P. Ikonen E. Traffic. 2012;13:1234–1243. doi: 10.1111/j.1600-0854.2012.01379.x. [DOI] [PubMed] [Google Scholar]
- Riley R. T. Merrill A. H. J. Lipid Res. 2019;60:1183–1189. doi: 10.1194/jlr.S093815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedia C. Canals D. Matabosch X. Harrak Y. Casas J. Llebaria A. Delgado A. Fabriás G. Chem. Phys. Lipids. 2008;156:33–40. doi: 10.1016/j.chemphyslip.2008.07.012. [DOI] [PubMed] [Google Scholar]
- Bedia C. Camacho L. Abad J. L. Fabrias G. Levade T. J. Lipid Res. 2010;51:3542–3547. doi: 10.1194/jlr.D010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhabak K. P. Proksch D. Redmer S. Arenz C. Bioorg. Med. Chem. 2012;20:6154–6161. doi: 10.1016/j.bmc.2012.08.035. [DOI] [PubMed] [Google Scholar]
- Bhabak K. P. Hauser A. Redmer S. Banhart S. Heuer D. Arenz C. ChemBioChem. 2013;14:1049–1052. doi: 10.1002/cbic.201300207. [DOI] [PubMed] [Google Scholar]
- Alamudi S. H. Su D. Lee K. J. Lee J. Y. Belmonte-Vázquez J. L. Park H.-S. Peña-Cabrera E. Chang Y.-T. Chem. Sci. 2018;9:2376–2383. doi: 10.1039/C7SC04716A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J. Arganda-Carreras I. Frise E. Kaynig V. Longair M. Pietzsch T. Preibisch S. Rueden C. Saalfeld S. Schmid B. Tinevez J. Y. White D. J. Hartenstein V. Eliceiri K. Tomancak P. Cardona A. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S. Cordelières F. P. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.