ABSTRACT
Proper base-pairing of a miRNA with its target mRNA is a key step in miRNA-mediated mRNA repression. RNA remodelling by RNA-binding proteins (RBPs) can improve access of miRNAs to their target mRNAs. The largest isoform p45 of the RBP AUF1 has previously been shown to remodel viral or AU-rich RNA elements. Here, we show that AUF1 is capable of directly promoting the binding of the miRNA let-7b to its target site within the 3ʹUTR of the POLR2D mRNA. Our data suggest this occurs in two ways. First, the helix-destabilizing RNA chaperone activity of AUF1 disrupts a stem-loop structure of the target mRNA and thus exposes the miRNA target site. Second, the RNA annealing activity of AUF1 drives hybridization of the miRNA and its target site within the mRNA. Interestingly, the RNA remodelling activities of AUF1 were found to be isoform-specific. AUF1 isoforms containing a YGG motif are competent RNA chaperones, whereas isoforms lacking the YGG motif are not. Overall, our study demonstrates that AUF1 has the ability to modulate a miRNA-target site interaction, thus revealing a new regulatory function for AUF1 proteins during post-transcriptional control of gene expression. Moreover, tests with other RBPs suggest the YGG motif acts as a key element of RNA chaperone activity.
KEYWORDS: AUF1, RNA chaperone, RNA remodeling, YGG motif, miRNA-mediated gene silencing
Introduction
MicroRNAs (miRNAs) are small, non-coding RNAs that assemble with Argonaute (AGO) and other proteins into miRNA-induced silencing complexes (miRISCs), which regulate post-transcriptional silencing of target mRNAs. Binding of miRISCs to partially complementary target sites, which are mostly located in the mRNA’s 3ʹUTR, may lead to translation repression and/or mRNA destabilization [1,2]. Furthermore, there are indications that as an alternative to the canonical model of RISC loading miRNAs may be bound to mRNAs even in the absence of AGO proteins and that these preformed miRNA-mRNA complexes can be detected and repressed by AGO in cells [3].
Usually, functional miRNA–mRNA interactions require complementarity of the mRNA target site and the miRNA’s seed region (nt 2–8 from the 5ʹend of the miRNA) [4]. Furthermore, target site accessibility can modulate the activity of miRNAs. For example, miRNA target sites can be located in stem-loop structures, thus hindering access of the miRNA to the mRNA [5,6].
RNA-binding proteins (RBPs) can modulate miRNA–target interaction through remodelling of miRNA target sites. For example, binding of the RBP PUM1 induces a local change in RNA structure within the 3ʹUTR of the mRNA encoding the tumour suppressor p27 thereby favouring association with miRNAs [7]. Competitive or cooperate regulation of mRNAs by RBPs and miRNAs appears to be widespread as miRNA-binding sites are preferentially located in or close to AU-rich elements (AREs) in the 3ʹUTR of mRNAs [8,9]. Furthermore, miRNA targeting of AREs in cooperation with ARE-binding proteins, like Tristetraprolin, appears to be essential for ARE-mediated mRNA degradation [10]. The ARE-binding protein HuR displays both positive and negative effects on miRNA-mediated repression [11–14]. In addition, HuR and AU-rich element binding factor 1 (AUF1) bind to many AU-rich target mRNAs on common sites in a competitive fashion [15,16]. Altogether, these findings indicate a complex interplay between miRNA and RBPs when interacting with mRNA’s 3ʹUTRs.
The RBP AUF1, also known as hnRNPD, is involved in many steps of RNA metabolism including the regulation of mRNA stability (reviewed in [17]), mRNA translation [18] and RNA splicing [19]. A relatively new role for AUF1 was proposed during RISC-mediated regulation of gene expression. AUF1 was found to bind miRNA let-7b with high affinity and to promote loading of let-7b onto AGO2, thus triggering AGO2–miRNA-mediated mRNA decay [20,21]. AUF1 is a family of four isoforms that are generated by alternative splicing of a common pre-mRNA resulting in the exclusion (AUF1 p37) or inclusion of isoform-specific sequences encoded by exons 2 (AUF1 p40 and p45) and 7 (AUF1 p42 and p45) (Fig. 1A). The sequence encoded by exon 7 is rich in tyrosine and glycine residues, and thus is termed a YGG motif [22]. The largest isoform AUF1 p45 was previously shown to mediate the flaviviral RNA switch, a critical step during flavivirus RNA replication, by two RNA remodelling activities, a helix-destabilizing RNA chaperone and an RNA annealing activity [23–25]. This observation prompted us to investigate whether AUF1’s RNA remodelling activities induced structural switches in mRNAs, thus modulating miRNA-mediated regulation of gene silencing.
Figure 1.
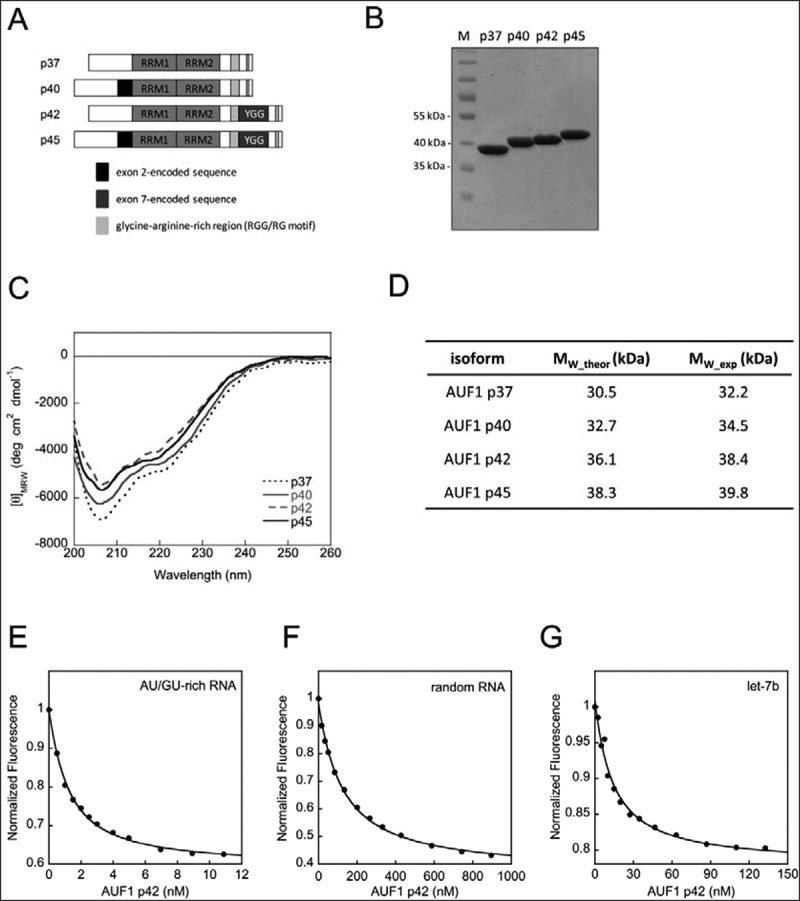
Characterization of AUF1 isoforms. (A) Domain organization of AUF1 isoforms. The RNA recognition motifs (RRM) are indicated. (B) AUF1 isoforms were produced in, and purified from, E. coli. A 4 μg sample of protein was analysed on a Coomassie-stained SDS-gel in parallel with a molecular mass marker (M). (C) Far-UV circular dichroism (CD) spectra of AUF1 isoforms were recorded. The acquired data were normalized to mean residue weight (MRW) ellipticities. (D) Summary of the analytical ultracentrifugation experiments demonstrating that all AUF1 isoforms are monomeric proteins (see Supplementary Fig. S1). The data for AUF1 p45 were taken from [24]. (E) Representative binding isotherm with AU/GU-rich RNA and increasing concentrations of AUF1 p42. The data were normalized to the initial intensity of the unbound RNA. The binding curves were fitted to a single-site binding model (Equation (1) in materials and methods). (F) Same as in (E) but with random RNA. (G) Same as in (E) but with let-7b-RNA
Applying the miRNA let-7b and its target site within the 3ʹUTR of the POLR2D mRNA, we report here that AUF1 in fact promotes RNA remodelling of the miRNA target site as well as subsequent mRNA–miRNA interaction. Remarkably, the RNA remodelling activities of AUF1 turn out to be isoform-specific, with the isoforms containing the YGG motif being considerably more active as RNA chaperones. We demonstrate that aromatic residues within the YGG motif are critical for RNA remodelling. With this first report showing that AUF1 directly promotes association of a miRNA to its target site, we propose a new role for the RNA remodelling activities of AUF1 in miRNA-mediated gene silencing. The analysis of other RBPs revealed that YGG motif-containing proteins preferentially act as RNA chaperones.
Materials and methods
Plasmids encoding SUMO-fusion proteins
The pETSUMO expression system was used for the expression of recombinant proteins in E. coli. The plasmid encoding human AUF1 p45 is described elsewhere [24]. The cDNAs encoding AUF1 p37, p40 and p42 were amplified from pcDNA5/TO-FLAG-AUF1 p37, -p40 and -p42 [24] and cloned into the pETSUMO expression vector. Plasmids encoding the tryptophan substitution-variants of AUF1 p45 were generated by site-directed mutagenesis using appropriately designed primer pairs. The plasmid encoding AUF1 p45 YYGGA was generated by inserting a pre-annealed oligonucleotide fragment, which contains all tyrosine-to-alanine mutations into PCR-amplified pETSUMO-AUF1 p45. The cDNAs encoding the hnRNPA1 isoform p34 and HuR were generated by reverse-transcription-PCR of human total mRNA isolated from Huh7 cells and cloned into the pETSUMO expression vector. The plasmid encoding hnRNPA1 isoform p39 was generated by inserting a fragment encoding the alternatively spliced exon into PCR-amplified pETSUMO-hnRNPA1 p34. All primer sequences are supplied in Supplementary Table S1.
Expression and purification of recombinant proteins
Expression and purification of all proteins were performed as described [24]. Briefly, proteins were purified from the soluble fraction of Escherichia coli BL21-CodonPlus® (DE3)-RP cells using nickel-agarose affinity chromatography and, after cleavage with SUMO-protease, by heparin-sepharose affinity chromatography and gel-filtration (HiLoadTM 16/60 Superdex 75TM, GE Healthcare). UV absorption spectra were measured using a JASCO V-550 spectrometer. The protein concentration was determined by measuring the absorbance at 280 nm using extinction coefficients supplied in Supplementary Table S2. The proteins were stored at −80°C in 20 mM Tris/HCl, pH 7.6, 150 mM KCl, 1 mM Tris (2-carboxyethyl)phosphine (TCEP) except for HuR and hnRNPA1 p39, which were stored at −80°C in 20 mM Tris/HCl, pH 7.6, 250 mM KCl, 1 mM Tris (2-carboxyethyl)phosphine (TCEP).
Measurement of RNA-binding constants
Proteins were added to 1–25 nM 5ʹ-FAM-EX-labelled RNA (EX – extended linker arm between oligonucleotide and dye; purchased from IBA, Göttingen, Germany) in assay buffer (50 mM Hepes/NaOH, pH 8.0, 100 mM KCl, 5 mM MgCl2). Fluorescence changes were monitored using a Fluoromax-4 spectrofluorometer (Jobin Yvon) at 22°C. The signal amplitudes of the 5ʹ-FAM-EX-labelled RNAs (Supplementary Table S3) were measured (excitation at 491 nm, emission at 515 nm) and corrected for the volume change. Fluorescence intensities relative to the starting fluorescence were plotted against the protein concentration. Fitting the binding isotherms to a single-site binding model according to Equation (1) [26] with KaleidaGraph (Synergy Software) yielded the KD values of the interaction of the protein and the labelled RNA.
| (1) |
ΔF – relative change in fluorescence intensity, γ – signal amplitude, b – total concentration of the RNA, c – total concentration of the protein, KD – dissociation constant.
Fluorescence-based POLR2D-let-7b interaction assay
To study protein-mediated RNA remodelling of the POLR2D SL, which contains the let7-b target site, an RNA chaperone assay was performed. The purified, recombinant proteins were added to 5 nM of 5ʹ-Cy5-labelled and 3ʹ-BHQ-labelled SLPOLR2D-RNA (purchased from IBA, Göttingen, Germany; Fig. 4A and Supplementary Table S3) at different concentrations in assay buffer (50 mM Hepes/NaOH, pH 8.0, 100 mM KCl, 5 mM MgCl2). Then, 10 nM of non-labelled let-7b miRNA was added and readings were taken for another 200 s. Alternatively, protein and non-labelled let-7b miRNA were added simultaneously. Changes in the fluorescence signals were monitored in a Fluoromax-4 Spectrofluorometer (Jobin Yvon) at 22°C with the following parameters: excitation at 643 nm, emission at 667 nm. Fluorescence intensities relative to the starting fluorescence were plotted against the time and fitted by KaleidaGraph (Synergy) to first-order reaction when protein was omitted (Equation (2)) and second-order reaction when protein was included (Equation (3)) yielding the corresponding rate constants kobs.
| (2) |
| (3) |
Figure 4.
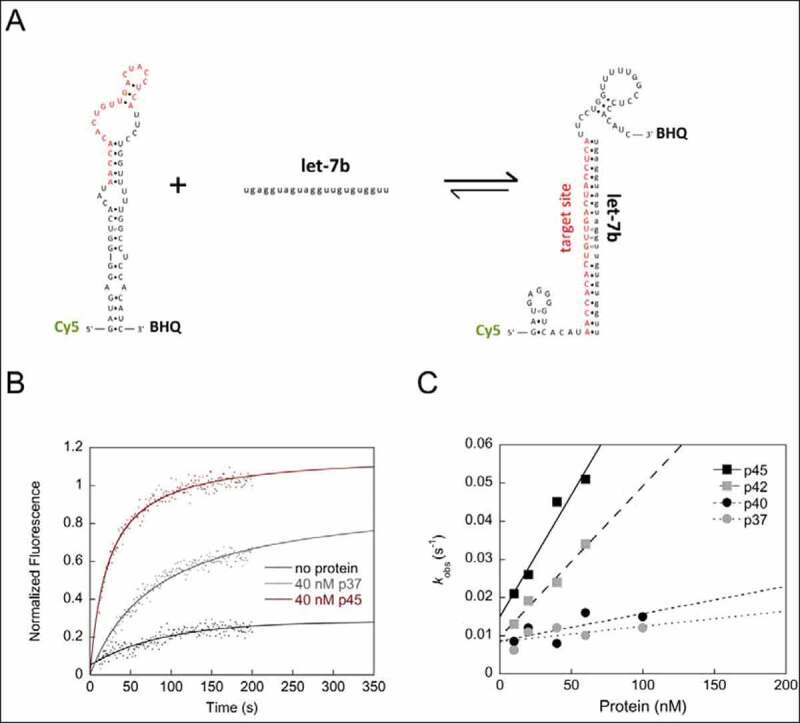
AUF1 remodels the let-7b target site-containing SL structure within the POLR2D 3ʹUTR and thus promotes let-7b binding. (A) Scheme of the fluorescence-based chaperone assay to detect protein-mediated conformational rearrangement of the SLPOLR2D-RNA containing the let-7b target site. Rearrangement of the RNA is measured by de-quenching of Cy5. (B) Examples of kinetic traces with Cy5- and BHQ (black hole quencher)-labelled SLPOLR2D-RNA incubated with or without 40 nM of AUF1 p37 or p45 are shown. Following the addition of let-7b the fluorescence signals were measured, plotted as a function of time, and fitted according to a first-order reaction (no protein; Equation 2) or second-order reaction (in the presence of protein; Equation 3). (C) The observed rate constants kobs (s−1) that were measured for the RNA chaperone reaction in the presence of AUF1 isoforms were plotted as a function of the protein concentration
ΔF – total change of relative fluorescence amplitude, Foffset – fluorescence intensity at the start point of the reaction, Fmax – maximum signal amplitude, kobs – observed rate constant, t – time. Errors of the rate constant values obtained from the fitting routine were in the range of 15%.
FRET-based RNA annealing of let-7b target site and let-7b
To study the influence of proteins on the interaction of miRNA let-7b and its target site within the mRNA of POLR2D, an RNA annealing assay was performed. The purified, recombinant proteins were added at different concentrations to 10 nM of 5ʹ-Cy3-labelled let-7b miRNA (purchased from IBA, Göttingen, Germany; Fig. 3B and Supplementary Table S3) in assay buffer (50 mM Hepes/NaOH, pH 8.0, 100 mM KCl, 5 mM MgCl2). Then, 10 nM of 5ʹ-Cy5-labelled let-7b target site of POLR2D mRNA was added and readings were taken for another 200 s. Changes in the fluorescence signals were monitored in a Fluoromax-4 Spectrofluorometer (Jobin Yvon) at 22°C. The Cy3 fluorophore was excited at 535 nm wavelength and readings were taken at the Cy5 emission wavelength 680 nm. Fluorescence intensities relative to the starting fluorescence were plotted against the time and fitted by KaleidaGraph (Synergy software) to a second-order reaction (Equation (3), see above) yielding the corresponding rate constants kobs. Errors of the rate constant values obtained from the fitting routine were in the range of 5%.
Figure 3.
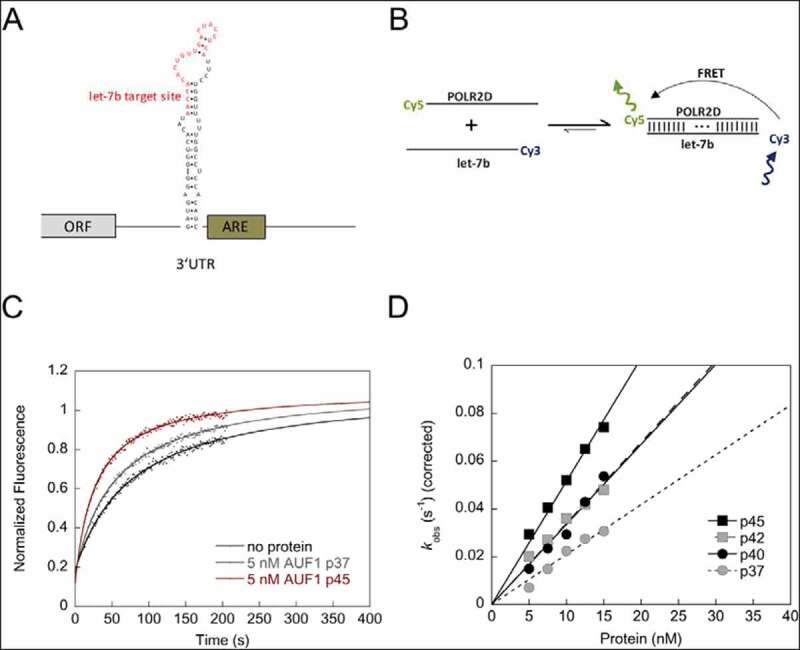
AUF1 promotes hybridization of let-7b and its target site in the POLR2D mRNA (A) Schematic view of the POLR2D mRNA. The nucleotide sequence and secondary structure of the stem-loop containing the let-7b target site is indicated. The let-7b target site is shown in red. ARE = AU-rich element. (B) Scheme of the FRET-based RNA annealing assay to analyse the hybridization of the miRNA let-7b and its target site of the POLR2D mRNA. Annealing of the let-7b target site of POLR2D mRNA and let-7b, which are fluorescently labelled with Cy5 and Cy3, respectively, leads to a detectable FRET signal. (C) Examples of kinetic traces of the RNA-RNA interaction in the absence or presence of 5 nM AUF1 p37 or AUF1 p45 are shown. The fluorescence signals were analysed according to a second-order reaction (Equation 3). (D) The rate constants measured for the RNA annealing reaction were normalized by subtracting the non-enzymatic rate from the total rate and plotted as a function of the protein concentration
Analytical ultracentrifugation
Sedimentation equilibrium measurements were performed as previously described [25]. Analysis was carried out at a protein concentration of 14 µM (AUF1 p37), 11 µM (AUF1 p40) or 5.6 µM (AUF1 p42) at 10000 rpm at 20°C in 20 mM Tris/HCl, pH 7.6, 150 mM KCl, 1 mM TCEP at 20°C.
Circular dichroism
Measurements of dichroic properties of proteins were performed using a JASCO J-810 spectropolarimeter. Far-UV circular dichroism spectra were recorded at a protein concentration of 2 µM in 20 mM Tris/HCl, pH 7.6, 150 mM KCl, 1 mM TCEP at 22°C using cuvettes with optical pathlengths of 1 mm. Acquired protein spectra were corrected for buffer contribution and smoothed using the Spectra Manager I software (JASCO). The data were converted to mean residue ellipticity ΘMRW.
Native mass spectrometry
AUF1 p37, p40, p42 or p45 samples were transferred into 200 mM aqueous ammonium acetate solution using 10 K MWCO cut-off centrifugal filters (Vivaspin 500, Sartorius, Göttingen, Germany). This step was repeated four times to ensure removal of interfering components of the purification buffer. Protein concentration in the filtrate was determined by UV absorbance at 280 nm. A total of 3 µl sample was loaded into an in-house prepared gold-coated borosilicate capillary [27] and native MS was performed on a Waters Synapt G1 instrument modified for transmission of high masses. Instrument parameters were 1.5 kV capillary voltage, 80 V cone voltage, 5.3 mbar backing pressure, 20 V trap collision voltage, 1.6 × 10−2 mbar trap pressure, 10 V transfer collision voltage, 500–15000 m/z scan range. Data were calibrated externally using 1 mg/ml caesium iodide solution.
For analysis of let-7b binding to AUF1, RNA was prepared at 10 µM concentration in 200 mM ammonium acetate and added to the protein samples in ammonium acetate solution at a 1:1 molar ratio. Analyses were performed as described above.
Results
The AUF1 isoforms are monomeric proteins
We recently analysed the largest AUF1 isoform p45 in terms of its RNA remodelling activities and deciphered the RGG/RG motif to be a key regulator for RNA chaperoning and RNA annealing [26]. To characterize isoform-specific functions we performed a comparative analysis of all four AUF1 isoforms. The proteins were heterologously synthesized in E. coli and purified to homogeneity (Fig. 1B). The far-UV CD spectra of all purified AUF1 isoforms were very similar and demonstrated a high content of disordered regions emphasized by a low CD signal (Fig. 1C). The largest isoform p45 was previously shown to be monomeric in solution [24]. In close agreement with this, we demonstrated the monomeric state of all other AUF1 isoforms by analytical ultracentrifugation in this study (Fig. 1D, Supplementary Fig. S1). While these data are in contrast to previous publications that reported AUF1 isoforms to be dimers [28,29], we explained our findings by using native, i.e. with no additional sequences, and nucleic acid-free protein. The absence of co-purifying nucleic acids was demonstrated by a 280/260 nm ratio, which was very close to the theoretical value (Supplementary Fig. S2).
The AUF1 isoforms bind RNA with similar affinity
In order to compare RNA-binding activities of all AUF1 isoforms, we analysed the binding to a 16 nt-long AU/GU-rich, single-stranded RNA, and to a randomly composed single-stranded RNA of the same length. With these fluorescently-labelled RNA substrates, we could previously confirm the specificity of AUF1 p45 for AU/GU-rich sequences [26]. For all AUF1 isoforms, there was a clear substrate preference for AU/GU-rich RNA as expected (Table 1; 60–120-fold more efficient compared to random RNA; representative binding isotherms with AUF1 p42 shown in Fig. 1E, F). The smaller isoforms p37 and p40 showed a two- to three-fold stronger binding activity for both RNA substrates compared to the larger isoforms AUF1 p42 and p45. These results indicate that the presence of the exon 7-encoded sequence within the C-terminus of p42 and p45 slightly reduces the RNA-binding efficiency.
Table 1.
RNA-binding affinity of AUF1 isoforms and AUF1 p45 variants
| |
AU/GU-rich RNA |
Random RNA |
|---|---|---|
| Protein | KD (nM)a | KD (nM)a |
| AUF1 p37 | 0.8 ± 0.2 | 52 ± 2 |
| AUF1 p40 | 0.7 ± 0.1 | 48 ± 4 |
| AUF1 p42 | 1.1 ± 0.2 | 131 ± 27 |
| AUF1 p45b | 1.6 ± 0.5 | 157 ± 24 |
| AUF1 p45 W290A | 1.8 ± 0.2 | 122 ± 24 |
| AUF1 p45 W298A | 2.5 ± 1.1 | 97 ± 11 |
| AUF1 p45 W290, 298A | 1.5 ± 0.3 | 152 ± 15 |
| AUF1 p45 YYGGA | 1.7 ± 0.7 | 50 ± 15 |
| hnRNPA1 p34 | 1.9 ± 0.5 | 464 ± 105 |
| hnRNPA1 p39 | 3.5 ± 1.2 | 708 ± 245 |
| HuR | 17 ± 3.4 | 789 ± 150 |
aDissociation constants and standard deviations derived from at least three measurements.
bData were taken from Meyer et al. 2019.
AUF1 isoforms bind miRNA let-7b as monomers
AUF1 was previously shown to promote the loading of the miRNA let-7b onto Argonaute 2 [20]. We wondered if binding of let-7b by AUF1 also directly affects miRNA–mRNA interaction. To characterize the interaction of the AUF1 isoforms to let-7b we first analysed their binding affinity and behaviour. All isoforms showed strong binding to a fluorescently-labelled let-7b RNA with KD-values ranging from approximately 4 to 10 nM (Table 2). These results confirm binding of AUF1 to let-7b in the low nanomolar range; however, the differences between the isoforms observed here were not as pronounced as previously reported [20]. In order to test the binding behaviour, let-7b was then titrated to the AUF1 isoforms and analysed by native mass spectrometry. In the absence of let-7b, all AUF1 isoforms were found to be monomers and thus confirmed the data obtained by analytical ultracentrifugation (Fig. 2A). After binding to the 22 nt-long miRNA let-7b, the detected masses corresponded to one monomer bound to one RNA molecule indicating that all AUF1 isoforms maintain their monomeric state upon binding to let-7b (Fig. 2B). These data show that there is no large difference between the AUF1 isoforms when considering binding affinity and behaviour to the miRNA let-7b.
Table 2.
RNA-binding affinity of AUF1 isoforms to let-7b
| Protein | KD (nM)a |
|---|---|
| AUF1 p37 | 6.7 ± 1.0 |
| AUF1 p40 | 4.2 ± 0.6 |
| AUF1 p42 | 9.9 ± 2.0 |
| AUF1 p45 | 9.3 ± 3.5 |
aDissociation constants and standard deviations derived from at least
three measurements.
Figure 2.
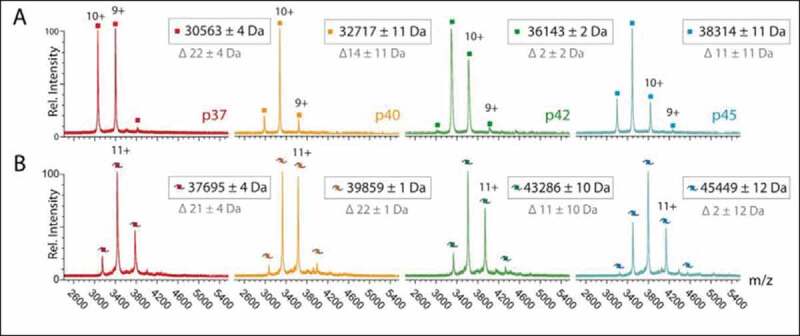
Binding behaviour of AUF1 monomers to let-7b. (A) Native mass spectra of variants p37 (4.2 µM), p40 (5.0 µM), p42 (4.0 µM) and p45 (5.0 µM) in the absence of RNA show peak distributions corresponding in mass to the monomeric protein variants. (B) Addition of equimolar amounts of let-7b RNA (Molecular mass: 7133.3 Da) to p37 (3.0 µM), p40 (3.3 µM), p42 (2.9 µM) and p45 (3.3 µM) shifts peaks to masses corresponding to RNA-bound monomers. Masses assigned to peak distributions (black) and mass differences of theoretical masses (grey) are indicated
AUF1 isoforms promote let-7b-target site RNA annealing with different efficiencies
To test whether AUF1 participates in miRNA–mRNA interaction we first tested whether AUF1 can promote the hybridization of the let-7b miRNA with its target site within the 3ʹUTR of the POLR2D mRNA. The POLR2D mRNA was chosen here as a model system because it was previously shown to be jointly regulated by AUF1 and let-7b [20]. For this purpose, we applied a FRET-based RNA annealing assay using the let-7b miRNA and its target site, which were labelled with a Cy3 and a Cy5 fluorophore, respectively (Fig. 3B). RNA hybridization of both RNAs resulted in an increase of fluorescence and this can be measured time-dependently to determine the rate constants of the reaction. The largest isoform p45 accelerated the interaction of let-7b and its target site of the POLR2D mRNA, thus confirming its activity as an RNA annealer (Fig. 3C, D). Interestingly, when testing the other isoforms, we observed isoform-specific RNA annealing activities. AUF1 isoforms p40 and p42 showed similar but lower activities than that of isoform p45, whereas the smallest isoform AUF1 p37 was the least active (Fig. 3C, D). These data demonstrate that all AUF1 isoforms are capable of promoting let-7b interaction with its target site but do so with different efficiencies.
The AUF1 exon7-encoded protein sequence promotes destabilization of a let-7b target site-containing stem-loop element
The let-7b target site within the POLR2D mRNA is located just upstream of the ARE within the 3ʹUTR and is part of a stem-loop (SL) structure (Fig. 3A). Hence, we speculated that AUF1 with its RNA chaperone activity promotes the opening of this SL, allowing let-7b to gain access to the target site. To address this, we established a fluorescence-based assay to follow the rearrangement of the POLR2D SL upon binding of let-7b. In this assay, the POLR2D SL RNA, containing the let-7b target site, is labelled with a Cy5 fluorophore and a black-hole quencher. Hybridization of the target site with let-7b accordingly requires a rearrangement of the stem structure, which dislocates the Cy5 fluorophore from the BHQ (Fig. 4A). The RNA–RNA interaction can be measured in a time-dependent manner in the presence of increasing protein concentrations to determine the rate constant for each reaction. In this approach, we observed an evident acceleration of the RNA–RNA interaction in the presence of protein. This was most effective with AUF1 isoform p45, while p42 was slightly less active. Interestingly, the smaller isoforms p37 and p40 showed almost no ability to promote the rearrangement of the POLR2D SL and thus the binding of let-7b to its target site (Fig. 4B, C). These data suggest that the exon7-encoded YGG motif, which is only present in the larger isoforms p42 and p45, drives the RNA chaperone function of AUF1. Moreover, the data demonstrate that AUF1 is not only able to restructure viral but also a cellular SL RNA suggesting a new role for AUF1 in miRNA-mediated gene repression.
Aromatic amino acids of the YGG motif are critical for RNA chaperone activity of AUF1
The next set of experiments addressed the importance of the YGG motif in the ability of AUF1 p45 to function as a RNA chaperone. Stacking interactions between aromatic amino acids and bases are common in RNA–protein interactions [30]. Accordingly, to analyse possible contributions of aromatic amino acids we generated protein variants by substituting all tyrosine residues as well as single or double tryptophane residues to alanine residues within the YGG motif of AUF1 p45 (Fig. 5A). The protein variants AUF1 p45 W290A, W298A, W290, 298A and YYGGA were purified from E. coli using the same protocol as for the wild-type (WT) protein. The purified variants had the same characteristics as the WT protein with respect to them being free of impurities and contaminating nucleic acids (Fig. 5B, Supplementary Fig. 2B). Importantly, the far-UV CD spectra of the YGG variants closely resembled the characteristics of the WT AUF1 p45 confirming the structural integrity of the variants (Fig. 5C).
Figure 5.
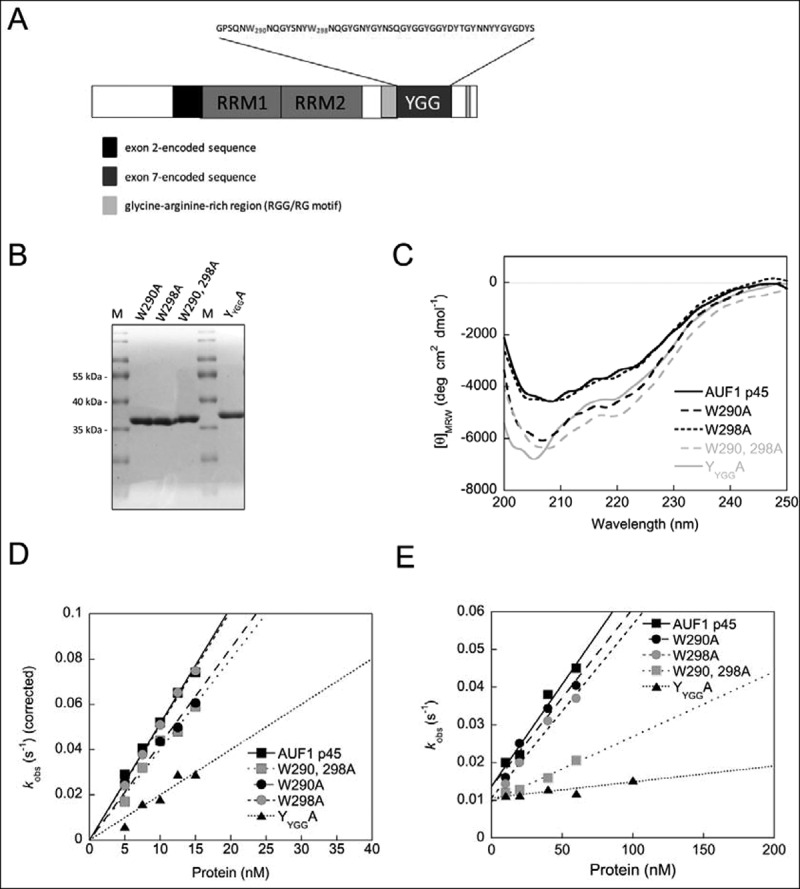
The YGG motif of AUF1 isoforms p42 and p45 essentially drives RNA chaperone activity. (A) Domain organization of AUF1 isoform p45. The RNA recognition motifs (RRM) are indicated. The amino acid sequence of the tyrosine-glycine rich region (YGG motif) is shown above. The tryptophan residues at position 290 and 298, as well as all tyrosine residues are emphasized. (B) YGG motif substitution variants of AUF1 p45 were produced in, and purified from, E. coli. A 4 μg sample of protein was analysed on a Coomassie-stained SDS-gel in parallel with a molecular mass marker (M). (C) Far-UV circular dichroism (CD) spectra of AUF1 p45 and its YGG motif substitution variants were recorded. The acquired data were normalized to mean residue weight (MRW) ellipticities. (D) RNA annealing assay to analyse the hybridization of the miRNA let-7b and its target site of the POLR2D mRNA (for more details see text and Fig. 3). The rate constants that were measured for the RNA annealing reaction were normalized by subtracting the non-enzymatic rate from the total rate and plotted as a function of the protein concentration. (E) RNA chaperone assay to detect protein-mediated conformational rearrangement of the SLPOLR2D-RNA containing the let-7b target site (for more details see text and Fig. 4). The observed rate constants kobs (s−1) that were measured for the RNA chaperone reaction in the presence of AUF1 p45 and its variants were plotted as a function of the protein concentration
RNA annealing by the YGG variants was not, or only mildly, affected, with variant YYGGA showing the strongest decrease in activity (Fig. 5D). In contrast, RNA chaperone activity was severely affected. Whereas the single-substitution variants AUF1 p45 W290A and W298A showed a moderate decrease in activity, the double-substitution variant W290, 298A was significantly impaired and the YYGGA variant displayed almost no ability to promote the rearrangement of the POLR2D SL (Fig. 5E). Note that the loss of RNA chaperone activity was not due to a loss of RNA-binding activity as all YGG variants showed RNA-binding affinities that were similar to those of the WT AUF1 p45 (Table 1). These data demonstrate aromatic amino acids of the YGG motif of AUF1 p45 are critical for the protein’s RNA remodelling activities, in particular for its RNA chaperone activity.
The YGG motif is critical for RNA chaperone activity of different RBPs
The fact that the YGG motif, in particular its aromatic residues, was found to be essential for the RNA chaperone activity of AUF1 p45, strongly suggested that this motif was an essential element of RBPs showing this activity. To test this hypothesis, we extended our analysis to the RBPs hnRNPA1 and HuR, which differ specifically in the presence or size of a YGG motif. The RBP hnRNPA1 is closely related to AUF1 and exists in two isoforms, which we named p34 and p39 according to their molecular mass (Fig. 6A). Both isoforms share two RRM domains, a RGG motif and a YGG motif. However, the YGG motifs of both isoforms differ in size, with isoform p39 having a larger YGG motif compared to hnRNPA1 p34 (Fig. 6A). The RBP HuR contains three RRMs but has neither a RGG nor a YGG motif (Fig. 6A). HuR, hnRNPA1 p34 and p39 were purified using the same protocol as used for AUF1 p45 (Supplementary SFig. 3A). The absence of co-purifying nucleic acids was confirmed (Supplementary SFig. 2B) and the far-UV CD spectra demonstrated the structural integrity of the proteins (Supplementary SFig. 3B). Considering previous reports, HuR [31] as well as both isoforms of hnRNPA1 [32] showed, as expected, strong binding preferences for AU/GU-rich RNA (Table 1).
Figure 6.
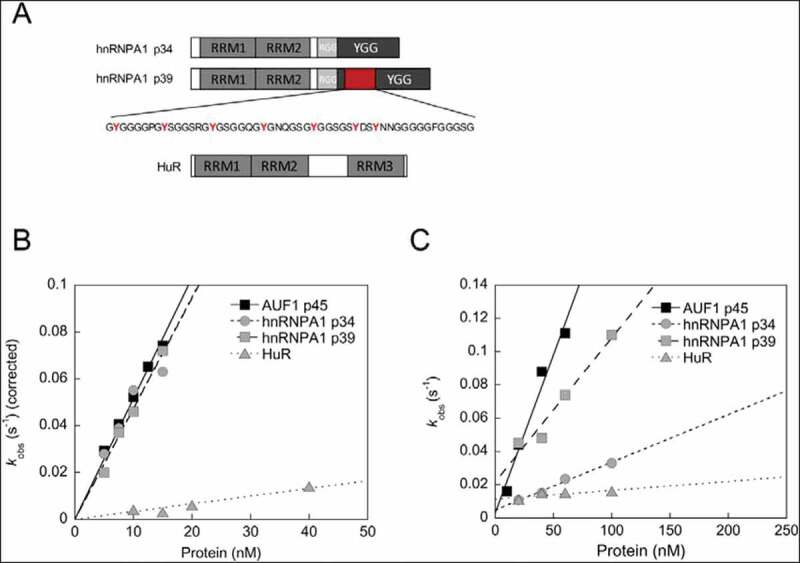
The presence of a YGG motif in an RBP is associated with a high RNA chaperone activity. (A) Domain organization of hnRNPA1 isoforms p34 and p39 and HuR. The RNA recognition motifs (RRM), the RGG as well as the YGG motifs are indicated. The isoform-specific sequence in hnRNPA1 p39 encoded by exon 8 is shown in red. (B) RNA annealing assay to analyse the hybridization of the miRNA let-7b and its target site of the POLR2D mRNA (for more details see text and Fig. 3). The rate constants that were measured for the RNA annealing reaction in the presence of AUF1 p45, hnRNPA1 isoforms p34 and p39 and HuR were normalized by subtracting the non-enzymatic rate from the total rate and plotted as a function of the protein concentration. Data for AUF1 p45 are from Fig. 3. (C) RNA chaperone assay to detect protein-mediated conformational rearrangement of the SLPOLR2D-RNA containing the let-7b target site (for more details see text and Fig. 4). The observed rate constants kobs (s−1) that were measured for the RNA chaperone reaction in the presence of AUF1 p45 hnRNPA1 isoforms p34 and p39 and HuR were plotted as a function of the protein concentration. Please note that the assay setup was slightly modified, i.e. the protein and the unlabelled let-7b miRNA were added simultaneously. This resulted in larger kobs values for AUF1 p45 in comparison to Fig. 4
In the RNA annealing assay, the hnRNPA1 isoforms p34 and p39 promoted RNA–RNA interaction of let-7b and its target site and were as active as AUF1 p45 (Fig. 6B). The results confirm the RNA annealing activity for the smaller hnRNPA1 isoform p34 [33] and reveal that the larger isoform p39 shares the same activity. Conversely, HuR promoted RNA annealing to only a small extent (Fig. 6B).
The RNA chaperone activity of the hnRNPA1 isoforms was promoted with different efficiencies. Whereas the larger isoform p39, which contains a longer YGG motif, promoted RNA chaperone function almost as effectively as AUF1 p45, the smaller isoform p34 showed considerably less ability to destabilize the POLR2D SL (Fig. 6C). Interestingly, HuR turned out to be inactive in supporting the rearrangement of the POLR2D SL and thus binding of let-7b to its target site (Fig. 6C). Taken together, these results indicate that the YGG motif is a key modulator of RNA chaperone activity of RBPs.
Discussion
AUF1 has been shown to remodel viral or AU-rich RNA elements [23,24,34] and to effectively bind specific miRNAs like let-7b [20]. Given these facts, the goal of this study was to investigate the capability of AUF1 to modulate miRNA binding to its mRNA target site. We could demonstrate that AUF1 is able to remodel a let-7b-binding site-containing stem-loop of the POLR2D target mRNA, and, in this way, promotes the binding of let-7b to its target site. Thus, our findings have revealed a new mechanism by which AUF1-induced RNA remodelling of mRNA target sites facilitates miRNA binding. Interestingly, the helix-destabilizing RNA chaperone activity was isoform-specific, with the larger isoforms p42 and p45, which contain the sequence encoded by exon 7, being active, while the smaller isoforms were essentially inactive (Fig. 4).
Previously, we identified the RGG/RG motif to be a key regulator of RNA remodelling by the largest AUF1 isoform p45. Our data supported a model in which the arginines of the RGG/RG motif promote RNA remodelling. Arginine-mediated stacking interactions were proposed to promote AUF1’s helix-destabilizing RNA chaperone activity, while the electropositive character of the arginine residues is the major determinant for the RNA annealing activity [26]. Here, we show that AUF1 isoforms p37 and p40 exhibit almost no RNA chaperone activity, although both isoforms have an RGG/RG motif and display RNA-binding activities comparable to those of the larger isoforms p42 and p45. This indicates that for AUF1 the RGG/RG motif alone is not sufficient to mediate RNA chaperone activity and that additional protein determinants encoded by exon 7 are required. We were able to demonstrate that the YGG motif of AUF1 isoforms p42 and p45 can be considered as an additional determinant of RNA chaperone activity.
YGG motifs (also called [G/S]Y[G/S]) are frequently found in combination with classical RNA-binding domains and RGG motifs [22,35]. YGG motifs are characterized by intrinsic disorder (low complexity) and are believed to be determinants of RBPs to allow dynamic entry in and out of RNA granules [36–40]. Here, we could show that the length or presence of a YGG motif determines RNA chaperone activity (Figs. 4C and 6C), indicating that the YGG motif is a key modulator of an RBP’s RNA chaperone activity.
The mechanisms by which the YGG motif contributes to RNA chaperone activity might involve RNA-stacking interactions, which are mediated by aromatic amino acid residues of the YGG motif in a similar manner to the role of arginine residues of the RGG/RG motif. In view of the fact that the exon 7-encoded YGG motif was previously indicated to enhance RNA-dependent protein oligomerization [29] as well as homodimeric interactions in nuclear bodies [41], the higher activity of AUF1 isoforms p42 and p45 could be further explained by RNA-induced protein oligomerization. YGG motif-dependent oligomerization of AUF1 would result in an increase in its local concentration and thus may also enhance RNA chaperone activity of the protein.
Another important aspect to consider is the flexibility of the protein structure, which appears to be crucial for the activity of RNA chaperones and RNA annealers. Within this context, the intrinsically disordered C-terminal domain of the RNA chaperone Hfq has been shown to be required for the release of the dsRNA product [42]. In analogy to the Hfq C-terminal domain, it is conceivable that the YGG motif of AUF1 p42 and p45 could support the release of the RNA substrate after the RNA chaperone activity is no longer required. In the case of p37 and p40, they remain attached to the RNA and either inhibit RNA remodelling or are simply not available for further rounds of RNA remodelling. Interestingly, p37 and p40 show 2- to 3-fold stronger RNA-binding compared to p42 and p45, suggesting that a strong RNA-binding activity might not be beneficial to a RNA chaperone function. A similar situation exists for AUF1-homologous proteins from mosquitos, named squid p30 and p32, that differ in the length of their YGG motifs and RNA-binding affinities. Although p32 displays a lower RNA binding affinity, its RNA chaperone activity is higher compared to that of squid p30 [43].
Differential inclusion of alternative exons, as found in the RBPs hnRNPA1 and AUF1, controls the formation of tyrosine-dependent higher-order protein assemblies that function to regulate splicing events [19]. In addition to the formation of higher-order hnRNP complexes, regulation of these exon-skipping events requires formation of long-range RNA duplexes. AUF1 p45’s RNA remodelling activities were previously implicated to participate in long-range RNA–RNA interactions that are required for flavivirus RNA replication [23,24]. Thus, it is tempting to speculate that not only YGG-mediated protein oligomerization but also RNA remodelling by AUF1 is required for the regulation of alternative splicing events.
The fact that miRNAs are expressed in excess relative to AGO proteins implies the existence of miRNA-mRNA duplexes that can be bound and repressed by AGOs, thus supporting a catalytic model of miRNA-mediated repression [3]. It is not yet known how miRNAs can hybridize to mRNAs without miRISC. Binding of miRNAs by RBPs could be one mechanism to stabilize miRNAs in the absence of AGO proteins, and co-regulation of mRNA degradation by RBPs and miRNAs is becoming increasingly important [44]. The formation of miRNA-mRNA complexes is made more difficult because miRNA-binding sites are often trapped within RNA secondary structures. DICER has recently been implicated to remodel RNA structures and to support RISC-targeting of sequence elements located within stable secondary structures of mRNA transcripts [45]. Here, we show for the first time that the miRNA-binding protein AUF1 promotes association of a miRNA to its target mRNA by (1) remodelling the secondary structure of the mRNA thus releasing the miRNA-binding site and (2) hybridizing the miRNA to its complementary target site. Improving the accessibility of miRNA-binding sites through remodelling of target RNA secondary structure by RBPs accordingly may be an important aspect of post-transcriptional control of gene silencing.
The production of alternatively spliced forms of AUF1 has recently been identified as mammalian specific [19], and previously, it has already been shown that the expression of AUF1 is tissue- and isoform-specific [46]. This supports the idea that different amounts of AUF1 isoforms, which have different levels of RNA remodelling activity, are a means of regulating mRNA stability. Accordingly, one would expect that high abundances of the RNA chaperone-active AUF1 isoforms p42 and p45 would cause destabilization of certain mRNAs, e.g. by recruitment of let-7b to the respective target sites and subsequent repression by AGO proteins. Such isoform-specific functions on the local RNA structure may therefore contribute to different regulatory consequences and serve as an explanation for the distinct functional roles of AUF1 isoforms on different RNA targets (reviewed in 47). However, the situation is further complicated by the fact that post-transcriptional control of gene silencing by AUF1 is highly context-dependent and is additionally influenced by competitive or cooperative binding of other trans-acting factors [47].
Even though we did not find a strong RNA annealing or RNA chaperone activity for the miRNA-binding protein HuR, this protein obviously participates in miRISC activity in other ways. For example, Kundu et al. demonstrated that HuR attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA although HuR and miRISC binding sites are positioned at a distance [13]. Another study observed cooperativity of a miRNA mimic and HuR-binding at adjacent sites and suggested binding of HuR to the MYC mRNA would enhance access of let-7 to the let-7-binding site [48], thus providing a possible mechanism as to how HuR recruits let-7 to repress MYC expression [12].
In summary, this study provides evidence for a further function of AUF1 in post-transcriptional control of gene expression. Future studies should examine whether exposing miRNA-binding sites in target mRNAs is also facilitated by other RBPs and whether this represents a general mechanism in miRNA-mediated gene silencing.
Supplementary Material
Acknowledgments
We thank Gary Sawers for critically reading the manuscript. This research was funded by the Deutsche Forschungsgemeinschaft (DFG) under grant number BE1885/12-1 (awarded to S.F. and S.-E.B.). C.S. and J.B. acknowledge funding from the Federal Ministry for Education and Research (BMBF, ZIK programme, 03Z22HN22), the European Regional Development Funds (EFRE, ZS/2016/04/78115), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation: project number 391498659, RTG 2467 “Intrinsically Disordered Proteins – Molecular Principles, Cellular Functions, and Diseases”) and the MLU Halle-Wittenberg. J.B. acknowledges funding from the Studienstiftung des deutschen Volkes.
Funding Statement
This work was supported by the Bundesministerium für Bildung und Forschung [03Z22HN22]; Deutsche Forschungsgemeinschaft [BE1885/12-1]; Deutsche Forschungsgemeinschaft [391498659, RTG 2467]; European Regional Development Fund [ZS/2016/04/78115]; Studienstiftung des Deutschen Volkes.
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Jonas S, Izaurralde E.. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–433. [DOI] [PubMed] [Google Scholar]
- [2].Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Janas MM, Wang B, Harris AS, et al. Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins. RNA. 2012;18:2041–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. [DOI] [PubMed] [Google Scholar]
- [6].Long D, Lee R, Williams P, et al. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–294. [DOI] [PubMed] [Google Scholar]
- [7].Kedde M, van Kouwenhove M, Zwart W, et al. A pumilio-induced RNA structure switch in p27-3ʹ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–1020. [DOI] [PubMed] [Google Scholar]
- [8].Mukherjee N, Corcoran DL, Nusbaum JD, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yoon JH, De S, Srikantan S, et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun. 2014;5:5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jing Q, Huang S, Guth S, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. [DOI] [PubMed] [Google Scholar]
- [11].Bhattacharyya SN, Habermacher R, Martine U, et al. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. [DOI] [PubMed] [Google Scholar]
- [12].Kim HH, Kuwano Y, Srikantan S, et al. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kundu P, Fabian MR, Sonenberg N, et al. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 2012;40:5088–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yoon JH, Abdelmohsen K, Kim J, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lal A, Mazan-Mamczarz K, Kawai T, et al. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. Embo J. 2004;23:3092–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fotinos A, Fritz DT, Lisica S, et al. Competing repressive factors control bone morphogenetic protein 2 (BMP2) in mesenchymal cells. J Cell Biochem. 2015;117:439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].White EJ, Brewer G, Wilson GM. Post-transcriptional control of gene expression by AUF1: mechanisms, physiological targets, and regulation. Biochim Biophys Acta. 2013;1829:680–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol. 2007;14:511–518. [DOI] [PubMed] [Google Scholar]
- [19].Gueroussov S, Weatheritt RJ, O’Hanlon D, et al. Regulatory expansion in mammals of multivalent hnrnp assemblies that globally control alternative splicing. Cell. 2017;170:324–339 e323. [DOI] [PubMed] [Google Scholar]
- [20].Yoon JH, Jo MH, White EJ, et al. AUF1 promotes let-7b loading on argonaute 2. Genes Dev. 2015;29:1599–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Min KW, Jo MH, Shin S, et al. AUF1 facilitates microRNA-mediated gene silencing. Nucleic Acids Res. 2017;45:6064–6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Castello A, Fischer B, Eichelbaum K, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. [DOI] [PubMed] [Google Scholar]
- [23].Friedrich S, Engelmann S, Schmidt T, et al. The host factor AUF1 p45 supports flavivirus propagation by triggering the RNA switch required for viral genome cyclization. J Virol. 2018;92:e01647–01617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Friedrich S, Schmidt T, Geissler R, et al. AUF1 p45 promotes West Nile virus replication by an RNA chaperone activity that supports cyclization of the viral genome. J Virol. 2014;88:11586–11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Friedrich S, Schmidt T, Schierhorn A, et al. Arginine methylation enhances the RNA chaperone activity of the West Nile virus host factor AUF1 p45. RNA. 2016;22:1574–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Meyer A, Golbik RP, Sanger L, et al. The RGG/RG motif of AUF1 isoform p45 is a key modulator of the protein’s RNA chaperone and RNA annealing activities. RNA Biol. 2019;16:960–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. [DOI] [PubMed] [Google Scholar]
- [28].DeMaria CT, Sun Y, Long L, et al. Structural determinants in AUF1 required for high affinity binding to A + U-rich elements. J Biol Chem. 1997;272:27635–27643. [DOI] [PubMed] [Google Scholar]
- [29].Zucconi BE, Ballin JD, Brewer BY, et al. Alternatively expressed domains of AU-rich element RNA-binding protein 1 (AUF1) regulate RNA-binding affinity, RNA-induced protein oligomerization, and the local conformation of bound RNA ligands. J Biol Chem. 2010;285:39127–39139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. Febs J. 2005;272:2118–2131. [DOI] [PubMed] [Google Scholar]
- [31].Ma W-J, Cheng S, Campbell C, et al. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271(14):8144–8151. [DOI] [PubMed] [Google Scholar]
- [32].Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. Embo J. 1994;13:1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kumar A, Wilson SH. Studies of the strand-annealing activity of mammalian hnRNP complex protein A1. Biochemistry. 1990;29:10717–10722. [DOI] [PubMed] [Google Scholar]
- [34].Zucconi BE, Wilson GM. Assembly of functional ribonucleoprotein complexes by AU-rich element RNA-binding protein 1 (AUF1) requires base-dependent and -independent RNA contacts. J Biol Chem. 2013;288:28034–28048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Castello A, Fischer B, Frese CK, et al. Comprehensive identification of RNA-binding domains in human cells. Mol Cell. 2016;63:696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Han TW, Kato M, Xie S, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. [DOI] [PubMed] [Google Scholar]
- [37].Kato M, Han TW, Xie S, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. [DOI] [PubMed] [Google Scholar]
- [39].Calabretta S, Richard S. Emerging roles of disordered sequences in RNA-binding proteins. Trends Biochem Sci. 2015;40(11):662–672. [DOI] [PubMed] [Google Scholar]
- [40].Wu H, Fuxreiter M. The structure and dynamics of higher-order assemblies: amyloids, signalosomes, and granules. Cell. 2016;165:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mannen T, Yamashita S, Tomita K, et al. The Sam68 nuclear body is composed of two RNase-sensitive substructures joined by the adaptor HNRNPL. J Cell Biol. 2016;214:45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Santiago-Frangos A, Kavita K, Schu DJ, et al. C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc Natl Acad Sci U S A. 2016;113:E6089–E6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Meyer A, Freier M, Schmidt T, et al. An RNA thermometer activity of the west nile virus genomic 3ʹ-terminal stem-loop element modulates viral replication efficiency during host switching. Viruses. 2020;12:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zealy RW, Wrenn SP, Davila S, et al. microRNA-binding proteins: specificity and function. Wiley Interdiscip Rev RNA. 2017;8:e1414. [DOI] [PubMed] [Google Scholar]
- [45].Pokornowska M, Milewski MC, Ciechanowska K, et al. The RNA-RNA base pairing potential of human Dicer and Ago2 proteins. Cell Mol Life Sci. 2019;2797:3231–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lu JY, Schneider RJ. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J Biol Chem. 2004;279:12974–12979. [DOI] [PubMed] [Google Scholar]
- [47].White EJ, Matsangos AE, Wilson GM. AUF1 regulation of coding and noncoding RNA. Wiley Interdiscip Rev RNA. 2017;8:e1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gunzburg MJ, Sivakumaran A, Pendini NR, et al. Cooperative interplay of let-7 mimic and HuR with MYC RNA. Cell Cycle. 2015;14:2729–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


