Abstract
Dimethylsulfoniopropionate (DMSP) is an abundant and ubiquitous organosulfur molecule in marine environments with important roles in global sulfur and nutrient cycling. Diverse DMSP lyases in some algae, bacteria, and fungi cleave DMSP to yield gaseous dimethyl sulfide (DMS), an infochemical with important roles in atmospheric chemistry. Here, we identified a novel ATP-dependent DMSP lyase, DddX. DddX belongs to the acyl-CoA synthetase superfamily and is distinct from the eight other known DMSP lyases. DddX catalyses the conversion of DMSP to DMS via a two-step reaction: the ligation of DMSP with CoA to form the intermediate DMSP-CoA, which is then cleaved to DMS and acryloyl-CoA. The novel catalytic mechanism was elucidated by structural and biochemical analyses. DddX is found in several Alphaproteobacteria, Gammaproteobacteria, and Firmicutes, suggesting that this new DMSP lyase may play an overlooked role in DMSP/DMS cycles.
Research organism: Other
eLife digest
The global sulfur cycle is a collection of geological and biological processes that circulate sulfur-containing compounds through the oceans, rocks and atmosphere. Sulfur itself is essential for life and important for plant growth, hence its widespread use in fertilizers.
Marine organisms such as bacteria, algae and phytoplankton produce one particular sulfur compound, called dimethylsulfoniopropionate, or DMSP, in massive amounts. DMSP made in the oceans gets readily converted into a gas called dimethyl sulfide (DMS), which is the largest natural source of sulfur entering the atmosphere. In the air, DMS is converted to sulfate and other by-products that can act as cloud condensation nuclei, which, as the name suggests, are involved in cloud formation. In this way, DMS can influence weather and climate, so it is often referred to as ‘climate-active’ gas.
At least eight enzymes are known to cleave DMSP into DMS gas with a few by-products. These enzymes are found in algae, bacteria and fungi, and are referred to as lyases, for the way they breakdown their target compounds (DMSP, in this case). Recently, researchers have identified some bacteria that produce DMS from DMSP without using known DMSP lyases. This suggests there are other, unidentified enzymes that act on DMSP in nature, and likely contribute to global sulfur cycling.
Li, Wang et al. set out to uncover new enzymes responsible for converting the DMSP that marine bacteria produce into gaseous DMS. One new enzyme called DddX was identified and found to belong to a superfamily of enzymes quite separate to other known DMSP lyases. Li, Wang et al. also showed how DddX drives the conversion of DMSP to DMS in a two-step reaction, and that the enzyme is found across several classes of bacteria. Further experiments to characterise the protein structure of DddX also revealed the molecular mechanism for its catalytic action.
This study offers important insights into how marine bacteria generate the climatically important gas DMS from DMSP, leading to a better understanding of the global sulfur cycle. It gives microbial ecologists a more comprehensive perspective of these environmental processes, and provides biochemists with data on a family of enzymes not previously known to act on sulfur-containing compounds.
Introduction
The organosulfur molecule dimethylsulfoniopropionate (DMSP) is produced in massive amounts by many marine phytoplankton, macroalgae, angiosperms, bacteria, and animals (Curson et al., 2018; Stefels, 2000; Otte et al., 2004; Curson et al., 2017; Raina et al., 2013). DMSP can function as an antioxidant, osmoprotectant, predator deterrent, cryoprotectant, protectant against hydrostatic pressure, chemoattractant and may enhance the production of quorum-sensing molecules (Sunda et al., 2002; Cosquer et al., 1999; Wolfe et al., 1997; Karsten et al., 1996; Zheng et al., 2020; Seymour et al., 2010; Johnson et al., 2016). DMSP also has important roles in global sulfur and nutrient cycling (Kiene et al., 2000; Charlson et al., 1987). Environmental DMSP can be taken up and catabolised as a carbon and/or sulfur source by diverse microbes, particularly bacteria (Curson et al., 2011b). DMSP catabolism can release volatile dimethyl sulfide (DMS) and/or methanethiol (MeSH) (Reisch et al., 2011a). DMS is a potent foraging cue for diverse organisms (Nevitt, 2011) and the primary biological source of sulfur transferred from oceans to the atmosphere (Andreae, 1990), which may participate in the formation of cloud condensation nuclei, and influence the global climate (Vallina and Simó, 2007).
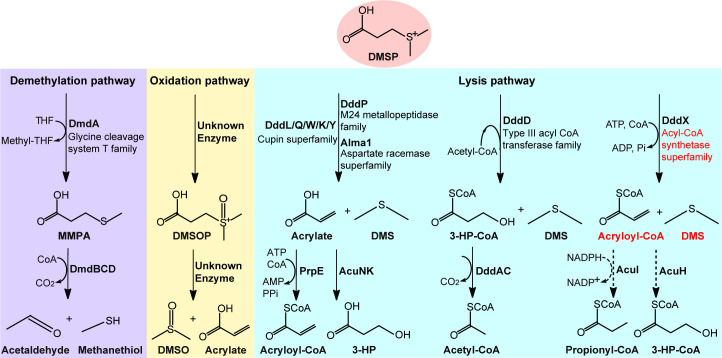
Bacteria can metabolize DMSP via three known pathways, the demethylation pathway (Howard et al., 2006), the recently reported oxidation pathway (Thume et al., 2018), and the lysis pathway (Curson et al., 2011b; Figure 1). The nomenclature of these pathways is based on the reaction type of the enzyme catalyzing the first step of DMSP catabolism. In the demethylation pathway, DMSP demethylase DmdA first demethylates DMSP to produce methylmercaptopropionate (MMPA) (Howard et al., 2006; Reisch et al., 2008), which can be further catabolized to MeSH and acetaldehyde (Figure 1; Reisch et al., 2011b; Bullock et al., 2017; Shao et al., 2019). In the oxidation pathway, DMSP is oxidized to dimethylsulfoxonium propionate (DMSOP), which is further metabolized to dimethylsulfoxide (DMSO) and acrylate; however, enzymes involved in this pathway are unknown (Thume et al., 2018; Figure 1).
Figure 1. Metabolic pathways for DMSP degradation.
Different pathways are shown in different colors. The demethylation of DMSP by DmdA produces MMPA (in purple). The oxidation of DMSP produces DMSOP (in yellow). In the lysis pathway (in blue), DMSP lyase DddP, DddL, DddQ, DddW, DddK, DddY, or Alma1 converts DMSP to acrylate and DMS, DddD converts DMSP to 3-HP-CoA and DMS, using acetyl-CoA as a CoA donor, and the newly identified DddX in this study converts DMSP to acryloyl-CoA and DMS, with ATP and CoA as co-substrates. Dotted lines represent unconfirmed steps of the DddX DMSP lysis pathway that we propose in this study. The protein families of enzymes involved in the first step of each pathway are indicated. The protein family of DddX and the products of its catalysis are highlighted in red color. THF, tetrahydrofolate; MMPA, methylmercaptopropionate; 3-HP, 3-hydroxypropionate; DMSOP, dimethylsulfoxonium propionate; DMSO, dimethylsulfoxide.
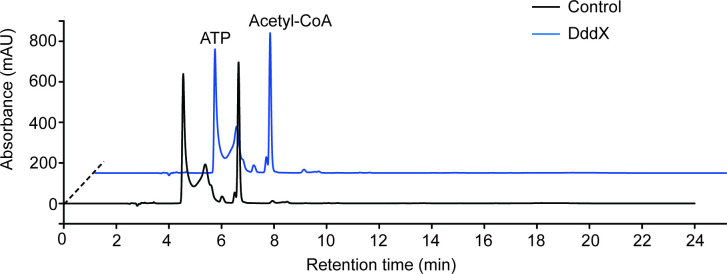
Figure 1—figure supplement 1. Enzymatic activity analysis of the recombinant DddX using acetyl-CoA as a CoA donor.
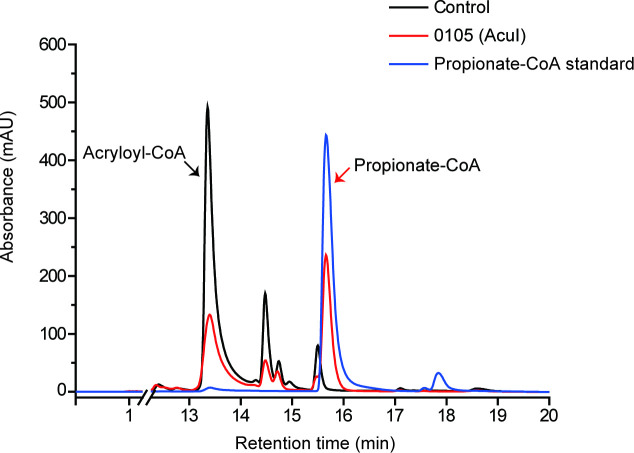
Figure 1—figure supplement 2. HPLC assay of the enzymatic activity of 0105 protein on acryloyl-CoA at 260 nm.
In the lysis pathway, diverse lyases cleave DMSP to produce DMS and acrylate or 3-hydroxypropionate-CoA (3-HP-CoA), which are further metabolized by ancillary enzymes (Curson et al., 2011b; Johnston et al., 2016; Figure 1). There is large biodiversity in DMSP lysis, with eight different known DMSP lyases that encompass four distinct protein families (DddD a CoA-transferase; DddP a metallopeptidase; cupin containing DddL, DddQ, DddW, DddK, and DddY; and Alma1 an aspartate racemase) functioning in diverse marine bacteria, algae, and fungi (Figure 1; Curson et al., 2011b; Johnston et al., 2016). With the exception of DddD, which catalyzes an acetyl-CoA-dependent CoA transfer reaction, all other DMSP lyases directly cleave DMSP (Bullock et al., 2017; Todd et al., 2007; Alcolombri et al., 2014; Lei et al., 2018; Li et al., 2014). Recently, several bacterial isolates were reported to produce DMS from DMSP but lack known DMSP lyases in their genomes (Liu et al., 2018; Zhang et al., 2019), suggesting the presence of novel enzyme(s) for DMSP degradation in nature.
A common feature of previously characterized DMSP metabolic pathways is that the metabolites (i.e. MMPA, acrylate) need to be ligated with CoA for further catabolism (Figure 1; Curson et al., 2011b; Reisch et al., 2011b). Currently, there is no known pathway whereby DMSP is ligated with free CoA, and it is tempting to speculate that there may be such a novel DMSP metabolic pathway. In this study, we screened DMSP-catabolizing bacteria from Antarctic samples, and obtained a strain Psychrobacter sp. D2 that grew on DMSP and produced DMS. Genetic and biochemical work showed that Psychrobacter sp. D2 possesses a novel DMSP lyase termed DddX for DMSP catabolism (Figure 1). DddX is an ATP-dependent DMSP lyase which catalyzes a two-step reaction: the ligation of DMSP and CoA, and the cleavage of DMSP-CoA to produce DMS and acryloyl-CoA. We further solved the crystal structure of DddX and elucidated the molecular mechanism for its catalysis based on structural and biochemical analyses. DddX is found in both Gram-negative and Gram-positive bacteria. Our results provide novel insights into the microbial metabolism of DMSP by this novel enzyme.
Results
A potentially novel DMSP lyase in a conventional DMSP catabolic gene cluster
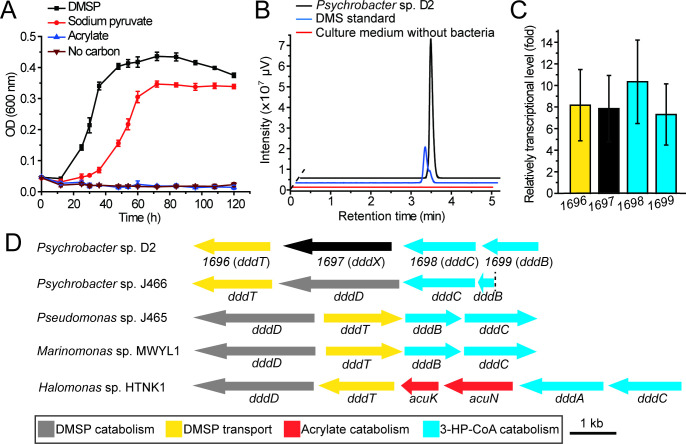
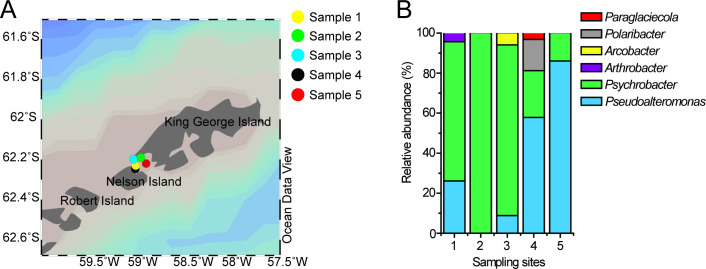
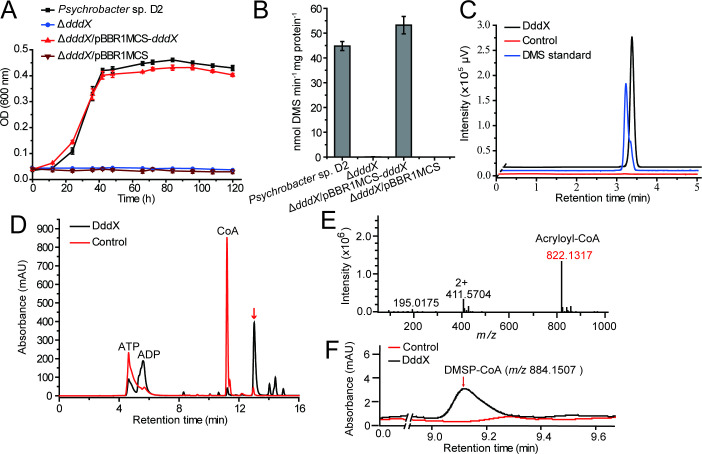
Using DMSP (5 mM) as the sole carbon source, DMSP-catabolizing bacteria were isolated from five Antarctic samples including alga, sediments, and seawaters (Figure 2—figure supplement 1, Supplementary file 1a). In total, 175 bacterial strains were obtained (Figure 2—figure supplement 1B). Among these bacterial strains, Psychrobacter sp. D2, a marine gammaproteobacterium, grew well in the medium containing DMSP as the sole carbon source, but not acrylate (Figure 2A). Moreover, gas chromatography (GC) analysis showed that Psychrobacter sp. D2 could catabolize DMSP and produce DMS (44.8 ± 1.8 nmol DMS min–1 mg protein–1) (Figure 2B).
Figure 2. The utilization of DMSP by Psychrobacter sp. D2 and the putative DMSP-catabolizing gene cluster in its genome.
(A) The growth curve of Psychrobacter sp. D2 on DMSP, sodium pyruvate or acrylate as sole carbon source (5 mM) at 15°C. The error bar represents standard deviation of triplicate experiments. (B), GC detection of DMS production from DMSP by strain D2. The culture medium without bacteria was used as the control. The DMS standard was used as a positive control. Psychrobacter sp. D2 could catabolize DMSP and produce DMS (44.8 ± 1.8 nmol DMS min–1 mg protein–1). (C), RT-qPCR assay of the transcriptions of the genes 1696, 1697, 1,698, and 1,699 in Psychrobacter sp. D2 in response to DMSP in the marine broth 2,216 medium. The bacterium cultured without DMSP in the same medium was used as the control. The recA gene was used as an internal reference. The error bar represents standard deviation of triplicate experiments. The locus tags of 1696, 1697, 1,698, and 1,699 are H0262_08195, H0262_08200, H0262_08205, and H0262_08210, respectively. (D), Genetic organization of the putative DMSP-catabolizing gene cluster. Reported DMSP catabolic/transport gene clusters from Psychrobacter sp. J466, Pseudomonas sp. J465, Marinomonas sp. MWYL1, and Halomonas sp. HTNK1 are shown (Todd et al., 2007; Todd et al., 2010; Curson et al., 2010; Curson et al., 2011b). The dashed vertical line indicates a breakpoint in dddB in the cosmid library of Pseudomonas sp. J466 (Curson et al., 2010).
Figure 2—figure supplement 1. Locations of the sampling sites and the relative abundance of DMSP-catabolizing bacteria isolated from the samples.
Figure 2—figure supplement 2. Transcriptomic analysis of the putative genes involved in DMSP metabolism in strain D2.
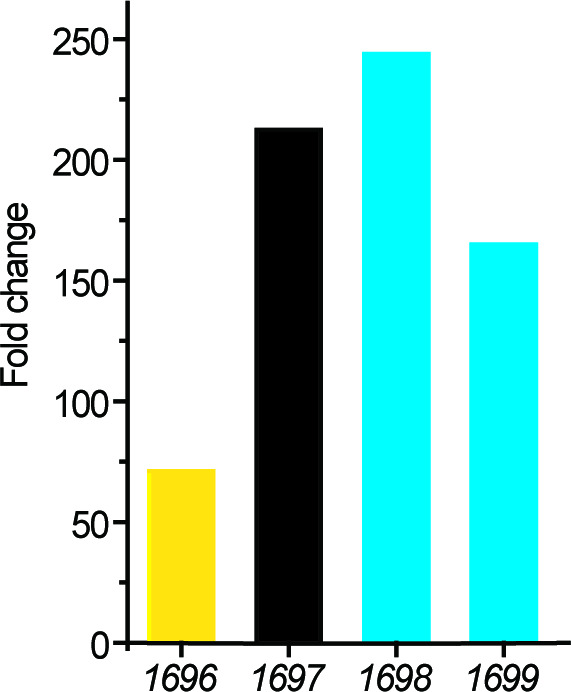
Figure 2—figure supplement 3. Confirmation of the deletion of the dddX gene from Psychrobacter sp.
To identify the genes involved in DMSP degradation in Psychrobacter sp. D2, we sequenced its genome and searched homologs of known DMSP lyases. However, no homologs of known DMSP lyases with amino acid sequence identity higher than 30% were found in its genome (Supplementary file 1b), implying that this strain may possess a novel enzyme or a novel pathway for DMSP catabolism. We then sequenced the transcriptomes of this strain when grown with and without DMSP as the sole carbon source. Transcriptional data analyses showed that the transcripts of four genes (1696, 1697, 1,698, and 1699) that compose a gene cluster were all highly upregulated (Figure 2—figure supplement 2) when DMSP was supplied as the sole carbon source, which was further confirmed by RT-qPCR analysis (Figure 2C). These results suggest that this gene cluster may participate in DMSP catabolism within Psychrobacter sp. D2.
In the gene cluster, 1696 is annotated as a betaine-carnitine-choline transporter (BCCT), sharing 32% amino acid identity with DddT, the predicted DMSP transporter in Marinomonas sp. MWYL1 (Sun et al., 2012; Todd et al., 2007); 1,697 is annotated as an acetate-CoA ligase, and shares 26% sequence identity with the acetyl-CoA synthetase (ACS) in Giardia lamblia (Sánchez et al., 2000); 1,698 is annotated as an aldehyde dehydrogenase, sharing 72% sequence identity with DddC in Marinomonas sp. MWYL1 (Todd et al., 2007); and 1,699 is annotated as an alcohol dehydrogenase, sharing 65% sequence identity with DddB in Marinomonas sp. MWYL1 (Todd et al., 2007). DddT, DddC, and DddB have been reported to be involved in DMSP import and catabolism (Sun et al., 2012; Todd et al., 2007; Todd et al., 2010). The pattern of the identified gene cluster 1696–1699 in Psychrobacter sp. D2 is similar to the patterns of those DMSP-catabolizing clusters reported in Pseudomonas, Marinomonas, and Halomonas, in which dddT, dddB and dddC are clustered with the DMSP lyase gene dddD¸ but which is missing in 1696–1699 and is replaced by 1,697 (Todd et al., 2007; Todd et al., 2010; Curson et al., 2010; Figure 2D). These data further support that the 1696–1699 gene cluster is involved in Psychrobacter sp. D2 DMSP catabolism and 1697 encodes a DMSP lyase equivalent to DddD. However, the sequence identity between 1,697 and DddD is less than 15%, suggesting that 1,697 is unlikely a DddD homolog. With these data we predicted that 1,697 encodes a novel DMSP lyase in Psychrobacter sp. D2, which we term as DddX hereafter.
The essential role of DddX in DMSP degradation in Psychrobacter Sp. D2
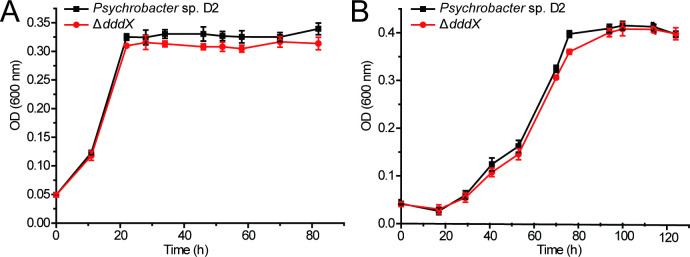
To identify the possible function of dddX in DMSP catabolism, we first deleted the majority of the dddX gene within the Psychrobacter sp. D2 genome to generate a ΔdddX mutant strain (Figure 2—figure supplement 3). The ΔdddX mutant was unable to grow on DMSP as the sole carbon source, but its ability to utilize DMSP was fully restored to wild type levels by cloned of dddX (in pBBR1MCS-dddX) (Figure 3A), indicating that dddX is essential for strain D2 to utilize DMSP. Furthermore, the ΔdddX mutant lost DMSP lyase activity, that is it no longer produced DMS when cultured in marine broth 2,216 medium with DMSP. DMSP lyase activity was fully restored to wild type levels in the complemented strain (ΔdddX/pBBR1MCS-dddX) (Figure 3B), indicating that dddX encodes a functional DMSP lyase enzyme degrading DMSP to DMS.
Figure 3. The function of Psychrobacter sp. D2 dddX in DMSP metabolism.
(A) Growth curves of the wild-type strain D2, the ΔdddX mutant, the complemented mutant (ΔdddX/pBBR1MCS-dddX), and the ΔdddX mutant complemented with an empty vector (ΔdddX/pBBR1MCS). All strains were grown with DMSP (5 mM) as the sole carbon source. The error bar represents standard deviation of triplicate experiments. (B), Detection of DMS production from DMSP degradation by the wild-type strain D2, the ΔdddX mutant, the complemented mutant ΔdddX/pBBR1MCS-dddX, and the mutant complimented with an empty vector ΔdddX/pBBR1MCS. The error bar represents standard deviation of triplicate experiments. (C), GC detection of DMS production from DMSP lysis catalyzed by the recombinant DddX. The reaction system without DddX was used as the control. DddX maintained a specific activity of ~8.0 μmol min–1 mg protein–1 at 20°C, pH 8.0.( D), HPLC analysis of the enzymatic activity of the recombinant DddX on DMSP at 260 nm. The peak of the unknown product is indicated with a red arrow. The reaction system without DddX was used as the control. (E), LC-MS analysis of the unknown product. (F), HPLC analysis of the intermediate of DddX catalysis at 260 nm. The HPLC system was coupled to a mass spectrometer for m/z determination. The reaction system without DddX was used as the control.
Figure 3—figure supplement 1. SDS-PAGE analysis of the recombinant DddX.

Figure 3—figure supplement 2. Two alternative mechanisms for DMSP degradation catalyzed by DddX.
Figure 3—figure supplement 3. Characterization of recombinant DddX.
Figure 3—figure supplement 4. HPLC assay of the enzymatic activity of DddX toward DMSP, sodium acetate, and sodium propionate at 260 nm.
Figure 3—figure supplement 5. The effects of potential inhibitors on the enzymatic activity of DddX.
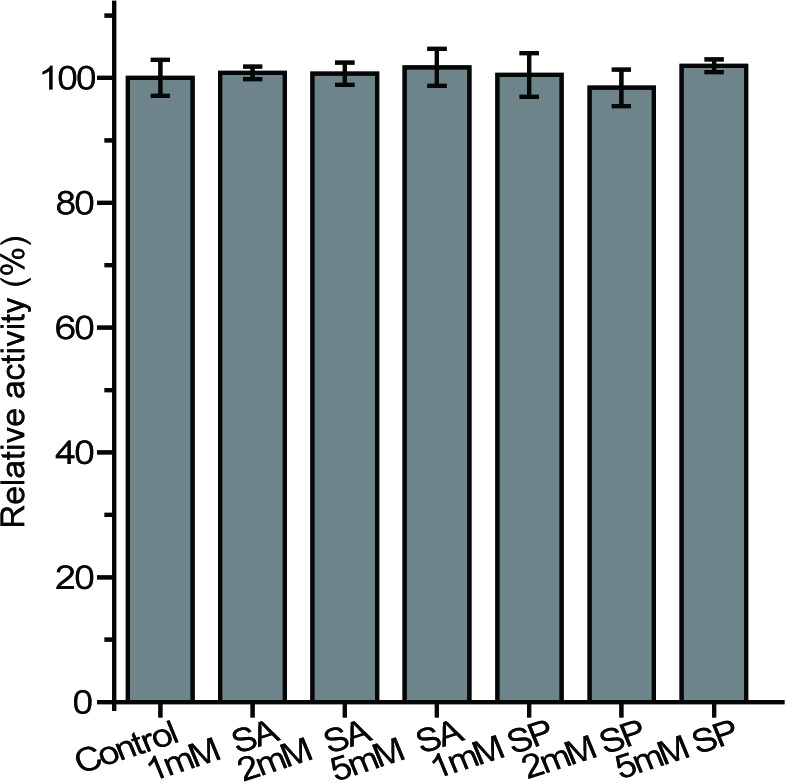
Figure 3—figure supplement 6. The growth curves of Psychrobacter sp.
DddX is an ATP-dependent DMSP lyase and its kinetic analysis
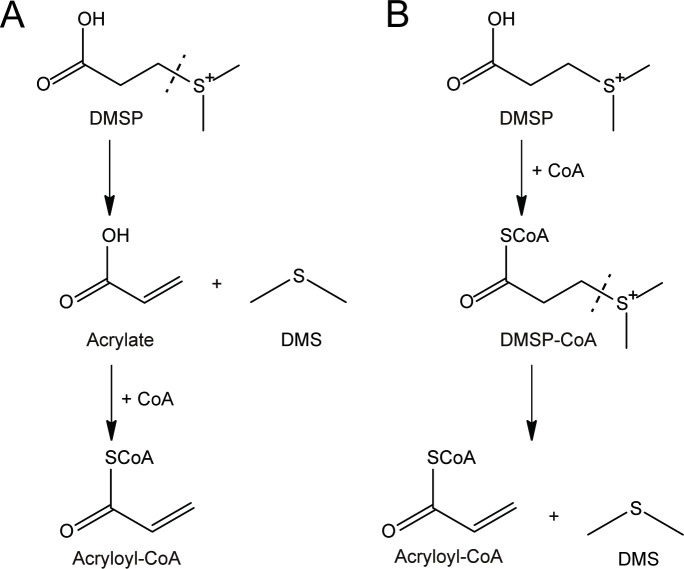
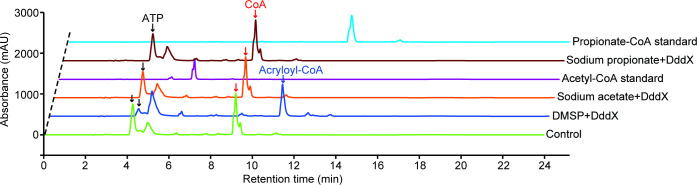
To verify the enzymatic activity of DddX on DMSP, we cloned the dddX gene, overexpressed it in Escherichia coli BL21 (DE3), and purified the recombinant DddX (Figure 3—figure supplement 1). Sequence analysis suggests that DddX is an acetate-CoA ligase, which belongs to the acyl-CoA synthetase (ACD) superfamily and requires CoA and ATP as co-substrates for catalysis (Musfeldt and Schonheit, 2002; Mai and Adams, 1996). Thus, we added CoA and ATP into the reaction system when measuring the enzymatic activity of the recombinant DddX on DMSP. GC analysis showed that the recombinant DddX directly acted on DMSP and produce DMS (Figure 3C). HPLC analysis uncovered ADP and an unknown product as DMS co-products (Figure 3D). The chromatographic retention time of the unknown product was consistent with it being acryloyl-CoA (Wang et al., 2017; Cao et al., 2017). Indeed, liquid chromatography-mass spectrometry (LC-MS) analysis found the molecular weight (MW) of the unknown product to be 822.1317, exactly matching acryloyl-CoA (Figure 3E). These data demonstrate that DddX is a functional ATP-dependent DMSP lyase that can catalyze DMSP degradation to DMS and acryloyl-CoA.
The biochemical results above suggest that DddX catalyzes a two-step degradation of DMSP, a CoA ligation reaction and a cleavage reaction. To perform this two-step reaction, there are two alternative pathways: (i), DMSP is first cleaved to form DMS and acrylate, and subsequently CoA is ligated with acrylate (Figure 3—figure supplement 2A). In this case, the intermediate acrylate is produced. (ii), CoA is primarily ligated with DMSP to form DMSP-CoA. Then, DMSP-CoA is cleaved, producing DMS and acryloyl-CoA (Figure 3—figure supplement 2B). In this scenario, the intermediate DMSP-CoA is produced. To determine the catalytic process of DddX, we monitored the occurrence of acrylate and/or DMSP-CoA in the reaction system via LC-MS. While acrylate was not detectable in the reaction system, a small peak of DMSP-CoA emerged after a 2 min reaction (Figure 3F), indicating that DMSP-CoA is primarily formed in the catalytic reaction of DddX, which is then cleaved to generate DMS and acryloyl-CoA.
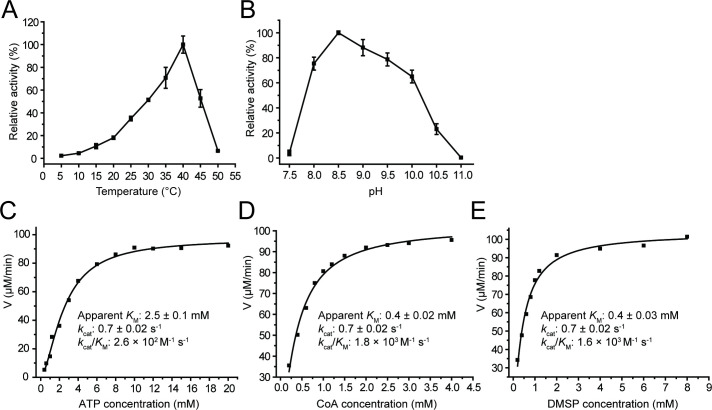
Knowing the DddX enzyme activity, we examined its in vitro properties. The DddX enzyme had an optimal temperature and pH of 40°C and 8.5, respectively (Figure 3—figure supplement 3A and B). The apparent KM of DddX for ATP and CoA was 2.5 mM (Figure 3—figure supplement 3C) and 0.4 mM (Figure 3—figure supplement 3D), respectively. DddX had an apparent KM value of 0.4 mM for DMSP (Figure 3—figure supplement 3E), which is lower than that of most other reported DMSP lyases and the DMSP demethylase DmdA (Supplementary file 1c). The kcat of DddX for DMSP was 0.7 s–1, with an apparent kcat/KM of 1.6 × 103 M–1 s–1. The catalytic efficiency of DddX toward DMSP is higher than known DMSP lyases DddK, DddP, DddD, but lower than DddY and Alma1 (Supplementary file 1c).
Despite DddX belongs to the ACD superfamily, the amino acid identity between DddX and known ACD enzymes is relatively low, with the highest being 26 % between DddX and the Giardia lamblia ACS (Sánchez et al., 2000). The kcat/KM value of DddX towards DMSP is lower than several reported ACS enzymes towards acetate (Chan et al., 2011; You et al., 2017). Because ACS enzymes were reported to have promiscuous activity toward different short chain fatty acids, such as acetate and propionate (Patel and Walt, 1987), we tested the substrate specificity of DddX. The recombinant DddX exhibited no activity towards acetate or propionate (Figure 3—figure supplement 4), and the presence of acetate or propionate had little effects on the enzymatic activity of DddX toward DMSP (Figure 3—figure supplement 5), indicating that DddX cannot utilize acetate or propionate as a substrate. Furthermore, we tested the ability of the strain D2 to grow with acetate or propionate as the sole carbon source. The wild-type strain D2 could use acetate or propionate as sole carbon source but deletion of dddX has little effect on the growth of strain D2 on these substrates (Figure 3—figure supplement 6), suggesting that dddX is unlikely to be involved in acetate and propionate catabolism. Together, these results indicate that DddX does not function as an acetate-CoA ligase.
The crystal structure and the catalytic mechanism of DddX
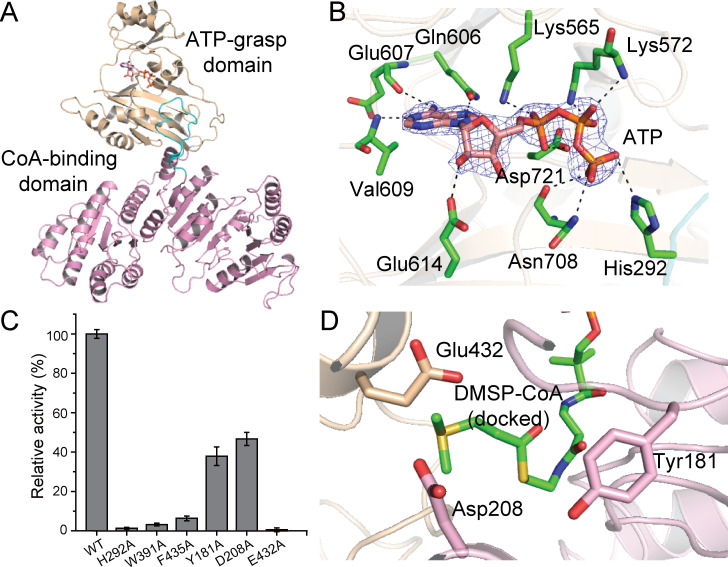
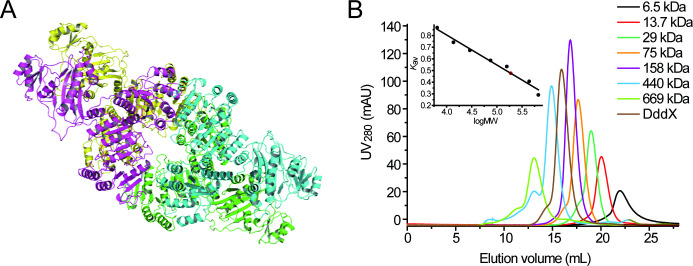
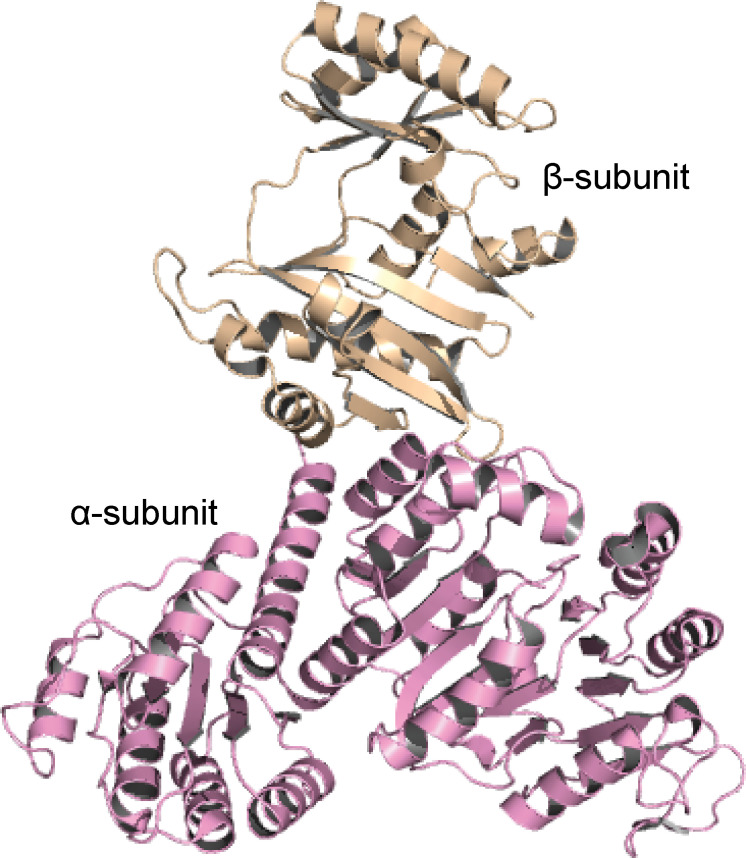
To elucidate the structural basis of DddX catalysis, we solved the crystal structure of DddX in complex with ATP by the single-wavelength anomalous dispersion method using a selenomethionine derivative (Se-derivative) (Supplementary file 1d). Although there are four DddX monomers arranged as a tetramer in an asymmetric unit (Figure 4—figure supplement 1A), gel filtration analysis indicated that DddX maintains a dimer in solution (Figure 4—figure supplement 1B). Each DddX monomer contains a CoA-binding domain and an ATP-grasp domain (Figure 4A), with one loop (Gly280-Tyr300) of the CoA-binding domain inserting into the ATP-grasp domain. ATP is bound in DddX mainly via hydrophilic interactions, including hydrogen bonds and salt bridges (Figure 4B). The overall structure of DddX is similar to that of NDP-forming acetyl-CoA synthetase ACD1 (Weiße et al., 2016; Figure 4—figure supplement 2), with a root mean square deviation (RMSD) between these two structures of 4.6 Å over 581 Cα atoms. ACD1 consists of separate α- and β-subunits (Weiße et al., 2016), which corresponds to the CoA-binding domain and the ATP-grasp domain of DddX, respectively.
Figure 4. Structural and mutational analyses of DddX.
(A) The overall structure of the DddX monomer. The DddX molecule contains a CoA-binding domain (colored in pink) and an ATP-grasp domain (colored in wheat). The loop region from the CoA-binding domain inserting into the ATP-grasp domain is colored in cyan. The ATP molecule is shown as sticks. (B) Residues of DddX involved in binding ATP. The 2Fo - Fc densities for ATP are contoured in blue at 2.0σ. Residues of DddX involved in binding ATP are colored in green. (C) Enzymatic activities of DddX and its mutants. The activity of WT DddX was taken as 100%. (D) Structural analysis of the possible catalytic residues for the cleavage of DMSP-CoA. The docked DMSP-CoA molecule and the probable catalytic residues of DddX are shown as sticks.
Figure 4—figure supplement 1. Structural and gel filtration analysis of DddX state of aggregation.
Figure 4—figure supplement 2. The overall structure of ACD1.
Figure 4—figure supplement 3. Sequence alignment of DddX homologs, acetyl-CoA synthetases (ACS), and ATP-citrate lyases (ACLY).
Figure 4—figure supplement 4. CD spectra of WT DddX and its mutants.
Figure 4—figure supplement 5. Structural analysis of DddX docked with DMSP and CoA, and DMSP-CoA.
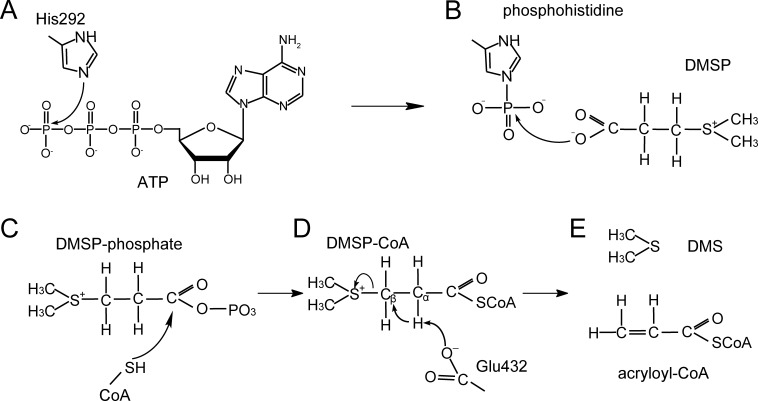
Both DddX and ACS belong to the ACD superfamily, which also contains the well-studied ATP citrate lyases (ACLY) (Weiße et al., 2016; Verschueren et al., 2019; Hu et al., 2017). The biochemistry of DddX catalysis is similar to that of ACLY, which converts citrate to acetyl-CoA and oxaloacetate with ATP and CoA as co-substrates (Verschueren et al., 2019; Hu et al., 2017). The catalytic processes of enzymes in the ACD superfamily involve a conformational change of a ‘swinging loop’ or ‘phosphohistidine segment’, in which a conserved histidine is phosphorylated (Weiße et al., 2016; Verschueren et al., 2019; Hu et al., 2017). Sequence alignment indicated that His292 of DddX is likely the conserved histidine residue to be phosphorylated, and Gly280-Tyr300 is likely the ‘swinging loop’ (Figure 4—figure supplement 3). In the crystal structure of DddX, His292 from loop Gly280-Tyr300 directly forms a hydrogen bond with the γ-phosphate of ATP (Figure 4B), suggesting a potential for phosphorylation, which is further supported by mutational analysis. Mutation of His292 to alanine abolished the activity of DddX (Figure 4C), indicating the key role of His292 during catalysis. Circular-dichroism (CD) spectroscopy analysis showed that the secondary structure of His292Ala exhibits little deviation from that of wild-type (WT) DddX (Figure 4—figure supplement 4), indicating that the enzymatic activity loss was caused by amino acid replacement rather than by structural change. Altogether, these data suggest that His292 is phosphorylated in the catalysis of DddX on DMSP.
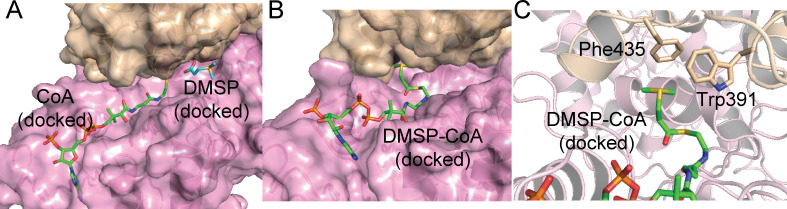
Having solved the crystal structure of the DddX-ATP complex, we next sought to determine the crystal structures of DddX in complex with CoA and DMSP. However, the diffractions of these crystals were poor and all attempts to solve the structures failed. Thus, we docked DMSP and CoA into the structure of DddX. In the docked structure, the CoA molecule is bound in the CoA-binding domain, while the DMSP molecule is bound in the interface between two DddX monomers (Figure 4—figure supplement 5A). Because our biochemical results demonstrated that DMSP-CoA is an intermediate of DddX catalysis (Figure 3F), we further docked DMSP-CoA into DddX. DMSP-CoA also locates between two DddX monomers (Figure 4—figure supplement 5B), and two aromatic residues (Trp391 and Phe435) form cation-π interactions with the sulfonium group of DMSP-CoA (Figure 4—figure supplement 5C). Mutations of these two residues significantly decreased the enzymatic activities of DddX (Figure 4C), suggesting that these residues play important roles in DddX catalysis. To cleave DMSP-CoA into DMS and acryloyl-CoA, a catalytic base is necessary to deprotonate DMSP-CoA. Structure analysis showed that Tyr181, Asp208, and Glu432 are close to the DMSP moiety (Figure 4D) and may function as the general base. Mutational analysis showed that the mutation of Glu432 to alanine abolished the enzymatic activity of DddX, while mutants Tyr181Ala and Asp208Ala still maintained ~40% activities (Figure 4C), indicating that Glu432 is the most probable catalytic residue for the final cleavage of DMSP-CoA. CD spectra of these mutants were indistinguishable from that of WT DddX (Figure 4—figure supplement 4), suggesting that the decrease in the enzymatic activities of the mutants were caused by residue replacement rather than structural alteration of the enzyme.
Based on structural and mutational analyses of DddX, and the reported molecular mechanisms of the ACD superfamily (Weiße et al., 2016; Verschueren et al., 2019; Hu et al., 2017), we proposed the molecular mechanism of DddX catalysis on DMSP (Figure 5). Firstly, His292 is phosphorylated by ATP, forming phosphohistidine (Figure 5A), which will be brought to the CoA-binding domain through the conformational change of the swinging loop Gly280-Tyr300. Next, the phosphoryl group is most likely transferred to DMSP to generate DMSP-phosphate (Figure 5B), which is subsequently attacked by CoA to form DMSP-CoA intermediate (Figure 5C). The last step is the cleavage of DMSP-CoA probably initiated by the base-catalyzed deprotonation of Glu432 (Figure 5D). Finally, acryloyl-CoA and DMS are generated (Figure 5E) and released from the catalytic pocket of DddX.
Figure 5. A proposed mechanism for DMSP cleavage to generate DMS and acryloyl-CoA catalyzed by DddX.
(A) The residue His292 attacks the γ-phosphate of ATP. (B), The phosphoryl group is transferred from phosphohistidine to the DMSP molecule. (C), DMSP-phosphate is attacked by CoA. (D), The residue Glu432 acts as a general base to attack DMSP-CoA. (E), DMS and acryloyl-CoA are generated.
Distribution of DddX in bacteria
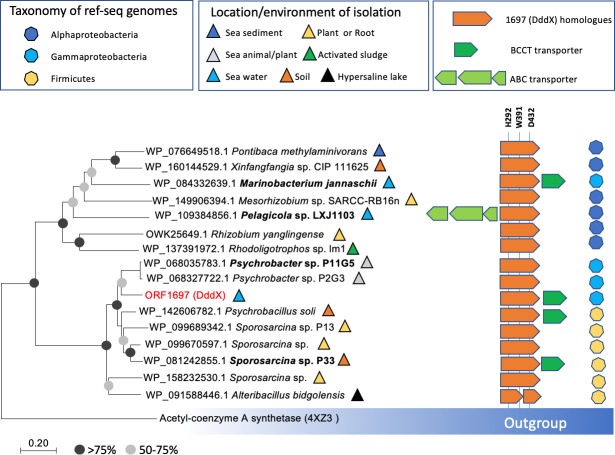
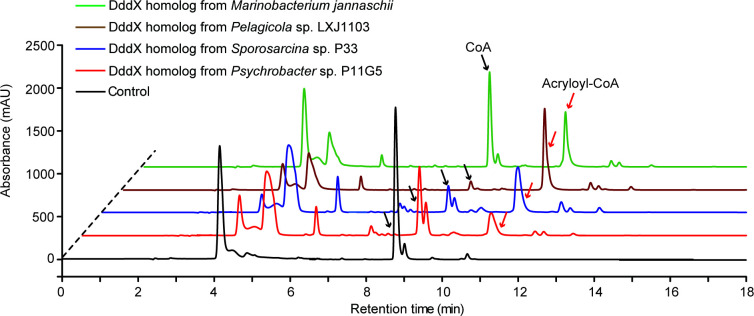
We next set out to determine the diversity and distribution of DddX in bacteria with sequenced genomes. We searched the NCBI Reference Sequence Database using the DddX sequence of Psychrobacter sp. D2 as the query. The data presented in Figure 6 showed that DddX homologs are present in several diverse groups of bacteria, including Alphaproteobacteria, Gammaproteobacteria, and Firmicutes. Multiple sequence alignment showed the presence of the key residues involved in phosphorylation (H292), co-ordination of the substrate (e.g. W391) and catalysis (D432), suggesting that these DddX homologs are likely functional in bacterial DMSP catabolism. To further validate that these DddX homologs are indeed functional DMSP degrading enzymes, we chemically synthesized representative dddX sequences from Alphaproteobacteria (Pelagicola sp. LXJ1103), Gammaproteobacteria (Psychrobacter sp. P11G5; Marinobacterium jannaschii), and Firmicutes (Sporosarcina sp. P33). These candidate DddX enzymes were purified and all were shown to degrade DMSP and produce acryloyl-CoA confirming their predicted activity (Figure 6—figure supplement 1). We predict that bacteria containing DddX will have DMSP lyase activity, but this will depend on the expression of this enzyme in the host and substrate availability.
Figure 6. Distribution of DddX in bacterial genomes.
The phylogenetic tree was constructed using neighbor-joining method in MEGA7. The acetyl-coenzyme A synthetase (ACS) (Weiße et al., 2016) was used as the outgroup. Sequence alignment was inspected for the presence of the key histidine residue (His292) involved in histidine phosphorylation that is known to be important for enzyme activity. A conserved Tyr391 is also found which is involved in cation-pi interaction with DMSP. The BCCT-type or ABC-type transporters for betaine-carnitine-choline-DMSP were found in the neighborhood of DddX in several genomes. Those DddX homologs that are functionally characterized (Figure 6—figure supplement 1) are highlighted in bold.
Figure 6—figure supplement 1. HPLC assay of the enzymatic activity of DddX homologs on DMSP at 260 nm.
Discussion
The cleavage of DMSP to produce DMS is a globally important biogeochemical reaction. Although all known DMSP lyases liberate DMS, they belong to different families, and likely evolved independently (Bullock et al., 2017). DddD belongs to the type III acyl CoA transferase family (Todd et al., 2007), DddP to the M24 metallopeptidase family enzyme (Todd et al., 2009), DddL/Q/W/K/Y to the cupin superfamily enzymes (Lei et al., 2018; Li et al., 2017) and Alma1 to the aspartate racemase superfamily (Alcolombri et al., 2015). To the best of our knowledge, DddX represents the first DMSP lyase of the ACD superfamily.
Of the reported DMSP lyases, only DddD catalyzes a two-step reaction which comprises a CoA transfer reaction and a cleavage reaction (Alcolombri et al., 2014). It is deduced that DMSP-CoA will be generated in the catalytic process of DddD (Alcolombri et al., 2014; Curson et al., 2011b; Todd et al., 2007). Despite this similarity, DddX is fundamentally different to DddD. Firstly, the co-substrates of DddX and DddD are different. ATP and CoA are essential co-substrates for the enzymatic activity of DddX, while for DddD catalysis, acetyl-CoA is used as a CoA donor, and ATP is not required (Johnston et al., 2016; Alcolombri et al., 2014). When CoA was replaced by acetyl-CoA in the reaction system, DddX failed to catalyze the cleavage of DMSP (Figure 1—figure supplement 1). Secondly, the products of DddD and DddX are different. DddD converts DMSP to DMS and 3-HP-CoA, whereas DddX produces DMS and acryloyl-CoA from DMSP. Except for DddD and DddX, all the other DMSP lyases cleave DMSP to DMS and acrylate.
It has been reported that accumulation of acryloyl-CoA is toxic to bacteria (Reisch et al., 2013; Wang et al., 2017; Cao et al., 2017; Todd et al., 2012). Thus, Psychrobacter sp. D2 requires an efficient system to metabolize the acryloyl-CoA produced from DMSP lysis by DddX. With the transcription of genes 1,698 and 1699,, directly downstream of dddX and likely co-transcribed with dddX, being significantly enhanced by growth on DMSP, their enzyme products (DddC and DddB) likely participate in the metabolism and detoxification of acryloyl-CoA or downstream metabolites. However, the recombinant 1698 and However, 1699 exhibited no enzymatic activity on acryloyl-CoA.
The Psychrobacter sp. D2 genome also contains acuI and acuH homologs (2674, 0105, 1810, 1,692, and 1695) (Supplementary file 1e), which may directly act on acryloyl-CoA to produce propionate-CoA or 3-HP-CoA (Reisch et al., 2013; Wang et al., 2017; Cao et al., 2017; Todd et al., 2012). If Psychrobacter sp. D2 employs its AcuH homolog to convert acryloyl-CoA to 3-HP-CoA (Cao et al., 2017), then, given the high-sequence identity of 1,698 to DddC and 1699 to DddB, it is possible that these enzymes further catabolize 3-HP-CoA to acetyl-CoA (Alcolombri et al., 2014; Curson et al., 2011b). Furthermore, we showed that the recombinant 0105, an AcuI homolog, could act on acryloyl-CoA to produce propionate-CoA with NADPH as a cofactor (Figure 1—figure supplement 2). Thus, Psychrobacter sp. D2 may also employ an AcuI (i.ei.e. 0105) to convert acryloyl-CoA to propionate-CoA (Figure 1), which would be metabolized through the methylmalonyl-CoA pathway (Reisch et al., 2013).
Several DMSP catabolizing bacteria, e.g. Halomonas HTNK1 with DddD, are reported to utilize acrylate as the carbon source for growth via e.g. acuN, acuK, acuI, acuH, and prpE gene products (Curson et al., 2011a; Reisch et al., 2013; Todd et al., 2010). Despite the presence of several acuN, acuK, acuI, acuH and prpE homologs in its genome (Supplementary file 1e), Psychrobacter sp. D2 could not use acrylate as a sole carbon source (Figure 2A). Thus, Psychrobacter sp. D2 either (i), lacks a functional acrylate transporter; (ii), these homologs that are predicted to be involved in acrylate metabolism are not functional in vivo; or (iii), these genes are not induced by acrylate. Clearly further biochemical and genetic experiments are required to establish the how acryloyl-CoA is catabolized in this bacterium.
Many marine bacteria, especially roseobacters, are reported to metabolize DMSP via more than one pathway (Curson et al., 2011b; Bullock et al., 2017). For example, Ruegeria pomeroyi DSS-3, one of the type strains of the marine Roseobacter clade, possesses both the demethylation and the lysis pathway for DMSP metabolism (Reisch et al., 2013). Moreover, it contains multiple ddd genes (dddQ, dddP and dddW) (Reisch et al., 2013; Todd et al., 2011). DmdA homologs were not identified in the genome of Psychrobacter sp. D2, indicating that the demethylation pathway is absent in strain D2. The fact that the mutant ΔdddX could not produce DMS from DMSP and was unable to grow on DMSP as the sole carbon source suggests that Psychrobacter sp. D2 only possesses one DMSP lysis pathway for DMSP degradation. Why some bacteria have evolved multiple DMSP utilization pathways and some bacteria only possess one pathway awaits further investigation.
Here, we demonstrate that DddX is a functional DMSP lyase present in several isolates of Gammaproteobacteria, Alphaproteobacteria and, notably, Gram-positive Firmicutes, for example in Sporosarcina sp. P33. The distribution of DddX in these bacterial lineages points to the role of horizontal gene transfer (HGT) in the dissemination of dddX in environmental bacteria and this certainly warrants further investigation. Interestingly, DddX is found in several bacterial isolates which were isolated from soil or plant roots, suggesting that DMSP may also be produced in these ecosystems. Finally, it has been reported that many other Gram-positive actinobacteria can make DMS from DMSP (Liu et al., 2018). Interestingly, these Actinobacteria lack dddX and any other known DMSP lyase genes. Thus, there is still more biodviversity in microbial DMSP lyases to be uncovered.
Conclusion
DMSP is widespread in nature and cleavage of DMSP produces DMS, an important mediator in the global sulfur cycle. In this study, we report the identification of a novel ATP-dependent DMSP lyase DddX from marine bacteria. DddX belongs to the ACD superfamily, and catalyzes the conversion of DMSP to DMS and acryloyl-CoA, with CoA and ATP as co-substrates. DddX homologs are found in both Gram-positive and Gram-negative bacterial lineages. This study offers new insights into how diverse bacteria cleave DMSP to generate the climatically important gas DMS.
Materials and methods
Key resources table.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Psychrobacter sp.) | D2 | This study;Zhang Laboratory | Wild-type isolate; Available from Zhang lab | |
| Strain, strain background (Psychrobacter sp.) | ΔdddX | This study;Zhang Laboratory | the dddX gene deletion mutant of Psychrobacter sp. D2; Available from Zhang lab | |
| Strain, strain background (Psychrobacter sp.) | ΔdddX/pBBR1MCS-dddX | This study;Zhang Laboratory | ΔdddX containing pBBR1MCS-dddX plasmid; Available from Zhang lab | |
| Strain, strain background (Psychrobacter sp.) | ΔdddX/pBBR1MCS | This study;Zhang Laboratory | ΔdddX containing pBBR1MCS plasmid; Available from Zhang lab | |
| Strain, strain background (Escherichia coli) | WM3064 | Dehio and Meyer, 1997 | Conjugation donor strain | |
| Strain, strain background (Escherichia coli) | DH5α | Vazyme Biotech company (China) | Transformed cells for gene cloning | |
| Strain, strain background (Escherichia coli) | BL21(DE3) | Vazyme Biotech company (China) | Transformed cells for gene expression | |
| Recombinant DNA reagent | pK18mobsacB-Ery | Wang et al., 2015b | Gene knockout vector | |
| Recombinant DNA reagent | pK18Ery-dddX | This study;Zhang Laboratory | pK18mobsacB-Ery containing the homologous arms of the dddX gene of Psychrobacter. sp. D2; Available from Zhang lab | |
| Recombinant DNA reagent | pBBR1MCS | Kovach et al., 1995 | Broad-host-range cloning vector | |
| Recombinant DNA reagent | pBBR1MCS-dddX | This study;Zhang Laboratory | pBBR1MCS containing the dddX gene and its promoter of Psychrobacter. sp. D2; Available from Zhang lab | |
| Recombinant DNA reagent | pET-22b-dddX | This study;Zhang Laboratory | Used for dddX expression; Available from Zhang lab | |
| Commercial assay or kit | Pierce BCA Protein Assay Kit | Thermo, USA | Protein assay | |
| Commercial assay or kit | Bacterial genomic DNA isolation kit | BioTeke Corporation, China | DNA extraction | |
| Commercial assay or kit | RNeasy Mini Kit | QIAGEN, America | RNA extraction | |
| Commercial assay or kit | PrimeScript RT reagent Kit | Takara, Japan | Reverse transcription | |
| Commercial assay or kit | Genome sequencing of Psychrobacter sp. D2 | Biozeron Biotechnology Co., Ltd, China | NCBI: JACDXZ000000000 | |
| Commercial assay or kit | Transcriptome sequencing of Psychrobacter sp. D2 | BGI Tech Solutions Co., Ltd, China | NCBI: PRJNA646786 | |
| Software, algorithm | HKL3000 program | Minor et al., 2006 | Diffraction data analysis | |
| Software, algorithm | CCP4 program Phaser | Winn et al., 2011 | Diffraction data analysis | |
| Software, algorithm | Coot | Emsley et al., 2010 | Diffraction data analysis | |
| Software, algorithm | Phenix | Adams et al., 2010 | Diffraction data analysis | |
| Software, algorithm | PyMOL | Schrödinger, LLC | http://www.pymol.org/ | |
| Software, algorithm | MEGA 7 | Kumar et al., 2016 | Phylogenetic analysis |
Bacterial strains, plasmids, and growth conditions
Strains and plasmids used in this study are shown in Supplementary file 1f. Isolates were cultured in the marine broth 2,216 medium or the basal medium (Supplementary file 1g) with 5 mM DMSP as the sole carbon source at 15°C–25°C. Psychrobacter sp. D2 was cultured in the marine broth 2,216 medium or the basal medium (Supplementary file 1g) supplied with different carbon sources (sodium pyruvate, acrylate or DMSP at a final concentration of 5 mM) at 15–25°C. The E. coli strains DH5α and BL21(DE3) were grown in the Lysogeny Broth (LB) medium at 37°C. Diaminopimelic acid (0.3 mM) was added to culture the E. coli WM3064 strain.
Isolation of bacterial strains from Antarctic samples
A total of five samples were collected from the Great Wall Station of Antarctica during the Chinese Antarctic Great Wall Station Expedition in January, 2017. Information of samples is shown in Figure 2—figure supplement 1 and Supplementary file 1a. Algae and sediments were collected using a grab sampler and stored in airtight sterile plastic bags at 4°C. Seawater samples were filtered through polycarbonate membranes with 0.22 μm pores (Millipore Co., United States). The filtered membranes were stored in sterile tubes (Corning Inc, United States) at 4°C. All samples were transferred into a 50 ml flask containing 20 ml 3% (w/v) seasalt solution (SS) and shaken at 100 rpm at 15°C for 2 hr. The suspension obtained was subsequently diluted to 10–6 with sterile SS. An aliquot (200 μl) of each dilution was spread on the basal medium (Supplementary file 1g) plates with 5 mM DMSP as the sole carbon source. The plates were then incubated at 15°C in the dark for 2–3 weeks. Colonies with different appearances were picked up and were further purified by streaking on the marine 2,216 agar plates for at least three passages. The abilities of the colonies for DMSP catabolism were verified in a liquid basal medium with DMSP (5 mM) as the sole carbon source. The isolates were stored at –80°C in the marine broth 2,216 medium containing 20 % (v/v) glycerol.
Sequence analysis of bacterial 16s rRNA genes
Genomic DNA of the isolates was extracted using a bacterial genomic DNA isolation kit (BioTeke Corporation, China) according to the manufacturer’s instructions. The 16 S rRNA genes of these strains were amplified using the primers 27 F/1492 R (Supplementary file 1h) and sequenced to determine their taxonomy. Pairwise similarity values for the 16 S rRNA gene of the cultivated strains were calculated through the EzBiocloud server (http://www.ezbiocloud.net/) (Yoon et al., 2017).
Bacterial growth assay with DMSP as the sole carbon source
Cells were grown in the marine broth 2,216 medium, harvested after incubation at 15 °C for 24 hr, and then washed three times with sterile SS. The washed cells were diluted to the same density of OD600 ≈ 2.0, and then 1 % (v/v) cells were inoculated into the basal medium with DMSP, sodium acetate, or sodium propionate (5 mM) as the sole carbon source. The bacteria were cultured in the dark at 15°C. The growth of the bacteria was measured by detecting the OD600 of the cultures at different time points using a spectrophotometer V-550 (Jasco Corporation, Japan).
Quantification of DMS by GC
To measure the production of DMS, cells were first cultured overnight in the marine broth 2,216 medium, and then washed three times with sterile SS. The washed cells were diluted to the same density of OD600 ≈ 0.3, then diluted 1:10 into vials (Anpel, China) containing the basal medium supplied with 5 mM DMSP as the sole carbon source. The vials were crimp sealed with rubber bungs and incubated for 2 hr at 25°C. The cultures were then assayed for DMS production on a gas chromatograph (GC-2030, Shimadzu, Japan) equipped with a flame photometric detector (Liu et al., 2018). An eight-point calibration curve of DMS standards was used (Curson et al., 2017). Abiotic controls of the basal medium amended with 5 mM DMSP were set up and incubated under the same conditions to monitor the background lysis of DMSP to DMS. Following growth of all bacteria strains in the marine broth 2,216 medium, cells were collected by centrifugation, resuspended in the lysis buffer (50 mM Tris-HCl, 100 mM NaCl, 0.5% glycerol, pH 8.0), and lysed by sonicated. The protein content in the cells was measured by Pierce BCA Protein Assay Kit (Thermo, USA). DMS production is expressed as nmol min–1 mg protein–1.
Transcriptome sequencing of Psychrobacter sp. D2
Cells of strain D2 were cultured in the marine broth 2,216 medium at 180 rpm at 15°C for 24 hr. The cells were collected and washed three times with sterile SS, and then cultured in sterile SS at 180 rpm at 15°C for 24 hr. Subsequently, the cells were washed twice with sterile SS, and incubated at 4°C for 24 hr. After incubation, the cells were harvested and resuspended in sterile SS, which were used as the resting cells. The resting cells were inoculated into the basal medium with DMSP (5 mM) as the sole carbon source, and incubated at 180 rpm at 15°C. When the OD600 of the cultures reached 0.3, the cells were harvested. The resting cells and those cultured in the basal medium with sodium pyruvate (5 mM) as the sole carbon source were set up as controls. Total RNA was extracted using a RNeasy Mini Kit (QIAGEN, America) according to the manufacturer’s protocol. After validating the quality, RNA samples were sent to BGI Tech Solutions Co., Ltd (China) for transcriptome sequencing and subsequent bioinformatic analysis.
Real-time qPCR analysis
Cells of Psychrobacter sp. D2 were cultured in the marine broth 2,216 medium at 180 rpm at 15°C to an OD600 of 0.8. Then, cells were induced by 5 mM DMSP, and the control group without DMSP was also set up. After 20 min’s induction, total RNA was extracted using a RNeasy Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. Genomic DNA was removed using gDNA Eraser (TaKaRa, Japan) and cDNA was synthesized using a PrimeScript RT reagent Kit. The qPCR was performed on the Light Cycler II 480 System (Roche, Switzerland) using a SYBR Premix Ex Taq (TaKaRa, Japan). Relative expression levels of target genes were calculated using the LightCycler480 software with the ‘Advanced Relative Quantification’ method. The recA gene was used as an internal reference gene. The primers used in this study are shown in Supplementary file 1h.
Genetic manipulations of Psychrobacter sp. D2
Deletion of the dddX gene was performed via pK18mobsacB-Ery-based homologue recombination (Wang et al., 2015a). The upstream and downstream homologous sequences of the dddX gene were amplified with primer sets dddX-UP-F/dddX-UP-R and dddX-Down-F/dddX-Down-R, respectively. Next, the PCR fragments were inserted to the vector pK18mobsacB-Ery with HindIII/BamHI as the restriction sites to generate pK18Ery-dddX, which was transferred into E. coli WM3064. The plasmid pK18Ery-dddX was then mobilized into Psychrobacter sp. D2 by intergeneric conjugation with E. coli WM3064. To select for colonies in which the pK18Ery-dddX had integrated into the Psychrobacter sp. D2 genome by a single crossover event, cells were plated on the marine 2,216 agar plates containing erythromycin (25 μg/ml). Subsequently, the resultant mutant was cultured in the marine broth 2,216 medium and plated on the marine 2,216 agar plates containing 10% (w/v) sucrose to select for colonies in which the second recombination event occurred. Single colonies appeared on the plates were streaked on the marine 2,216 agar plates containing erythromycin (25 μg/ml), and colonies sensitive to erythromycin were further validated to be the dddX gene deletion mutants by PCR with primer pairs of dddX-1000-F/dddX-1000-R and dddX-300Up-F/dddX-700Down-R.
For complementation of the ΔdddX mutant, the dddX gene with its native promoter was amplified using the primers set dddX-pBBR1-PF/dddX-pBBR1-PR. The PCR fragment was digested with KpnI and XhoI, and then inserted into the vector pBBR1MCS to generate pBBR1MCS-dddX. This plasmid was then transformed into E. coli WM3064, and mobilized into the ΔdddX mutant by intergeneric conjugation. After mating, the cells were plated on the marine 2,216 agar plates containing kanamycin (80 μg/ml) to select for the complemented mutant. The empty vector pBBR1MCS was mobilized into the ΔdddX mutant using the same protocol. Colony PCR was used to confirm the presence of the transferred plasmid. The strains, plasmids and primers used in this study are shown in Supplementary file 1f and h.
Gene cloning, point mutation, and protein expression and purification
The 2247 bp full-length dddX gene was amplified from the genome of Psychrobacter sp. D2 by PCR using FastPfu DNA polymerase (TransGen Biotech, China). The amplified gene was then inserted to the NdeI/XhoI restriction sites of the pET-22b vector (Novagen, Germany) with a C-terminal His tag. All the point mutations in DddX were introduced using the PCR-based method and verified by DNA sequencing. The DddX protein and its mutants were expressed in E. coli BL21 (DE3). The cells were cultured in the LB medium with 0.1 mg/ml ampicillin at 37 °C to an OD600 of 0.8–1.0 and then induced at 18°C for 16 hr with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). After induction, cells were collected by centrifugation, resuspended in the lysis buffer (50 mM Tris-HCl, 100 mM NaCl, 0.5% glycerol, pH 8.0), and lysed by pressure crusher. The proteins were first purified by affinity chromatography on a Ni2+-NTA column (GE healthcare, America), and then fractionated by anion exchange chromatography on a Source 15Q column (GE healthcare, America) and gel filtration on a Superdex G200 column (GE healthcare, America). The Se-derivative of DddX was overexpressed in E. coli BL21 (DE3) under 0.5 mM IPTG induction in the M9 minimal medium supplemented with selenomethionine, lysine, valine, threonine, leucine, isoleucine, and phenylalanine. The recombinant Se-derivative was purified using the aforementioned protocol for the wild-type DddX.
Enzyme assay and product identification
For the routine enzymatic activity assay of the DddX protein, the purified DddX protein (at a final concentration of 0.1 mM) was incubated with 1 mM DMSP, 1 mM CoA, 1 mM ATP, 2 mM MgCl2 and 100 mM Tris-HCl (pH 8.0). The reaction was performed at 37°C for 0.5 hr, and terminated by adding 10% (v/v) hydrochloric acid. The control groups had the same reaction system except that the DddX protein was not added. DMS was detected by GC as described above. Products of acryloyl-CoA and DMSP-CoA were analyzed using LC-MS. Components of the reaction system were separated on a reversed-phase SunFire C18 column (Waters, Ireland) connected to a high performance liquid chromatography (HPLC) system (Dionex, United States). The ultraviolet absorbance of samples was detected by HPLC under 260 nm. The samples were eluted with a linear gradient of 1–20% (v/v) acetonitrile in 50 mM ammonium acetate (pH 5.5) over 24 min. The HPLC system was coupled to an impact HD mass spectrometer (Bruker, Germany) for m/z determination. To determine the optimal temperature for DddX enzymatic activity, reaction mixtures containing 5 mM DMSP, 5 mM CoA, 5 mM ATP, 6 mM MgCl2, 100 mM Tris-HCl (pH 8.5), and 10 μM DddX were incubated at 5–50°C (with a 5°C interval) for 15 min. The optimum pH for DddX enzymatic activity was examined at 40°C (the optimal temperature for DddX enzymatic activity) using Britton-Robinson Buffer (Britton, 1952) with pH from 7.5 to 11.0, with a 0.5 interval. The kinetic parameters of DddX were measured by determining the production of DMS with nonlinear analysis based on the initial rates, and all the measurements were performed at the optimal pH and temperature.
The enzymatic activity of DddX toward sodium acetate or sodium propionate was measured by determining the production of acetyl-CoA or propionyl-CoA using HPLC as described above with DMSP replaced by sodium acetate or sodium propionate. To determine the effects of sodium acetate or sodium propionate on the enzymatic activity of DddX toward DMSP, sodium acetate or sodium propionate at a final concentration of 1 mM, 2 mM or 5 mM were individually added to the reaction mixture. All the measurements were performed at the optimum pH and temperature for DddX.
The enzymatic activity of 0105 (AcuI) toward acryloyl-CoA was measured by determining the production of propionate-CoA using HPLC as described above. The reaction mixture contained 2 mM DMSP, 2 mM CoA, 2 mM ATP, 10 mM MgCl2, 1 mM NADPH, 100 mM Tris-HCl (pH 8.5), 0.1 mM DddX, and 0.9 mM 0105. The reaction was performed at 40°C, pH 8.5 for 2 hr, and terminated by adding 10 % (v/v) hydrochloric acid.
Crystallization and data collection
The purified DddX protein was concentrated to ~8 mg/ml in 10 mM Tris–HCl (pH 8.0) and 100 mM NaCl. The DddX protein was mixed with ATP (1 mM), and the mixtures were incubated at 0°C for 1 hr. Initial crystallization trials for DddX/ATP complex were performed at 18°C using the sitting-drop vapor diffusion method. Diffraction-quality crystals of DddX/ATP complex were obtained in hanging drops containing 0.1 M lithium sulfate monohydrate, 0.1 M sodium citrate tribasic dihydrate (pH 5.5) and 20 % (w/v) polyethylene glycol (PEG) 1000 at 18°C after 2-week incubation. Crystals of the DddX Se-derivative were obtained in hanging drops containing 0.1 M HEPES (pH 7.5), 10% PEG 6000% and 5% (v/v) (+/-)–2-Methyl-2,4-pentanediol at 18°C after 2-week incubation. X-ray diffraction data were collected on the BL18U1 and BL19U1 beamlines at the Shanghai Synchrotron Radiation Facility. The initial diffraction data sets were processed using the HKL3000 program with its default settings (Minor et al., 2006).
Structure determination and refinement
The crystals of DddX/ATP complex belong to the C2 space group, and Se-derivative of DddX belong to the P212121 space group. The structure of DddX Se-derivative was determined by single-wavelength anomalous dispersion phasing. The crystal structure of DddX/ATP complex was determined by molecular replacement using the CCP4 program Phaser (Winn et al., 2011) with the structure of DddX Se-derivative as the search model. The refinements of these structures were performed using Coot (Emsley et al., 2010) and Phenix (Adams et al., 2010). All structure figures were processed using the program PyMOL (http://www.pymol.org/).
Circular dichroism (CD) spectroscopy
CD spectra for WT DddX and its mutants were carried out in a 0.1 cm-path length cell on a JASCO J-1500 Spectrometer (Japan). All proteins were adjusted to a final concentration of 0.2 mg/ml in 10 mM Tris-HCl (pH 8.0) and 100 mM NaCl. Spectra were recorded from 250 to 200 nm at a scan speed of 200 nm/min.
Molecular docking simulations
The structure of the DddX/ATP complex containing a pair of subunits, α and β was loaded and energy minimised in Flare (v3.0, Cresset) involving 11,248 moving heavy atoms (Chain A: 5312, Chain B: 5312, Chain G: 10 and Chain S Water: 614). The molecule minimized with 2000 iterations using a gradient of 0.657 kcal/A. The minimised structure had an RMSD 0.82 Å relative to the starting structure and a decrease in starting energy from 134999.58 kcal/mol to a final energy of 6888.60 kcal/mol. The DMSP, CoA and DMSP-CoA molecules were drawn in MarvinSketch (v19.10.0, 2019, ChemAxon for Mac) and exported as a Mol SDF format. The molecules were imported into Flare and docked into the proposed CoA/DMSP binding site using the software’s default docking parameters for intensive pose searching and scoring.
Identification of DddX homologs in bacteria and phylogenetic analysis
DddX (1697) of Psychrobacter sp. D2 was used as the query sequence to search for homologs in genome-sequenced bacteria in the NCBI Reference Sequence Database (RefSeq, https://www.ncbi.nlm.nih.gov/refseq/) using BLastP with a stringent setting with an e-value cut-off < –75, sequence coverage >70% and percentage identity >30%. These high stringency settings are necessary to exclude other acetyl-CoA synthetase family proteins (ACS) which are unlikely to be involved in DMSP catabolism. Multiple sequence alignment was carried out using MEGA 7 (Kumar et al., 2016) and the presence of histidine 292, tryptophan 391 and glutamate 432 was manually inspected. To confirm the activity of DddX homologs from retrieved sequences from these genome-sequenced bacteria, four sequences (Sporosarcina sp. P33; Psychrobacter sp. P11G5; Marinobacterium jannaschii; Pelagicola sp. LXJ1103) were chemically synthesized and their enzyme activity for DMSP degradation was confirmed experimentally (Figure 6—figure supplement 1). The phylogenetic tree was constructed using the neighbour-joining method with 500 bootstraps using MEGA 7 (Kumar et al., 2016). The characterized ACS ACD1 (Weiße et al., 2016) was used as the outgroup.
Acknowledgements
We thank the staffs from BL18U1 & BL19U1 beamlines of National Facility for Protein Sciences Shanghai (NFPS) and Shanghai Synchrotron Radiation Facility, for assistance during data collection. We thank Caiyun Sun and Jingyao Qu from State Key laboratory of Microbial Technology of Shandong University for their help in HPLC and LC-MS. This work was supported by the National Key Research and Development Program of China (2018YFC1406700, 2016YFA0601303), the National Science Foundation of China (grants 91851205, 31630012, U1706207, 42076229, 31870052, 31800107), Major Scientific and Technological Innovation Project (MSTIP) of Shandong Province (2019JZZY010817), the Program of Shandong for Taishan Scholars (tspd20181203) and Natural Environment Research Council Standard grants (NE/N002385, NE/P012671 and NE/S001352).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Yu-Zhong Zhang, Email: zhangyz@sdu.edu.cn.
Georg Pohnert, University of Jena, Germany.
Meredith C Schuman, University of Zurich, Switzerland.
Funding Information
This paper was supported by the following grants:
National Key Research and Development Program of China 2018YFC1406700 to Yu-Zhong Zhang.
National Science Foundation of China 91851205 to Yu-Zhong Zhang.
National Science Foundation of China 31630012 to Yu-Zhong Zhang.
National Key Research and Development Program of China 2016YFA0601303 to Yu-Zhong Zhang.
National Science Foundation of China U1706207 to Yu-Zhong Zhang.
National Science Foundation of China 42076229 to Chun-Yang Li.
National Science Foundation of China 31870052 to Xiu-Lan Chen.
National Science Foundation of China 31800107 to Peng Wang.
Major Scientific and Technological Innovation Project of Shandong Province 2019JZZY010817 to Yu-Zhong Zhang.
Program of Shandong for Taishan Scholars tspd20181203 to Yu-Zhong Zhang.
Natural Environment Research Council NE/N002385 to Jonathan D Todd.
Natural Environment Research Council NE/P012671 to Jonathan D Todd.
Natural Environment Research Council NE/S001352 to Jonathan D Todd.
Additional information
Competing interests
No competing interests declared.
Author contributions
investigation, validation, writing-original-draft.
investigation, writing-original-draft.
supervision, validation, writing-review-and-editing.
investigation.
investigation.
investigation.
investigation, writing-original-draft.
investigation, writing-original-draft.
investigation.
investigation.
writing-review-and-editing.
writing-review-and-editing.
writing-review-and-editing.
writing-review-and-editing.
writing-review-and-editing.
writing-review-and-editing.
investigation, writing-original-draft, writing-review-and-editing.
conceptualization, funding-acquisition, supervision.
Additional files
(a) Information of the Antarctic samples used in this study. (b) Homology alignment of proteins in Psychrobacter sp. D2 with known DMSP lyases. (c) Kinetic parameters of DMSP lyases and DMSP demethylase DmdA. (d) Crystallographic data collection and refinement parameters of DddX. (e) Homology alignment of proteins in Psychrobacter sp. D2 with known enzymes involved in acrylate catabolism. (f) Strains and plasmids used in this study. (g) Composition of the basal medium (lacking the carbon source). (h) Primers used in this study.
Data availability
The draft genome sequences of Psychrobacter sp. D2 have been deposited in the National Center for Biotechnology Information (NCBI) Genome database under accession number JACDXZ000000000. All the RNA-seq read data have been deposited in NCBI's sequence read archive (SRA) under project accession number PRJNA646786. The structure of DddX/ATP complex has been deposited in the PDB under the accession code 7CM9.
The following dataset was generated:
Wang X. 2020. The draft genome sequence of Psychrobacter sp.D2. NCBI GenBank. https://www.ncbi.nlm.nih.gov/nuccore/JACDXZ000000000JACDXZ000000000
Wang X. 2020. RNA-Seq of Psychrobacter sp. D2. NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/?term=prjna646786PRJNA646786
Li CY, Zhang YZ. 2020. DMSP lyase DddX. RCSB Protein Data Bank. https://www.rcsb.org/structure/7CM97CM9
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallographica. Section D, Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcolombri U, Laurino P, Lara-Astiaso P, Vardi A, Tawfik DS. DddD is a CoA-transferase/lyase producing dimethyl sulfide in the marine environment. Biochemistry. 2014;53:5473–5475. doi: 10.1021/bi500853s. [DOI] [PubMed] [Google Scholar]
- Alcolombri U, Ben-Dor S, Feldmesser E, Levin Y, Tawfik DS, Vardi A. Identification of the algal dimethyl sulfide-releasing enzyme: A missing link in the marine sulfur cycle. Science. 2015;348:1466–1469. doi: 10.1126/science.aab1586. [DOI] [PubMed] [Google Scholar]
- Andreae MO. Ocean-atmosphere interactions in the global biogeochemical sulfur cycle. Marine Chemistry. 1990;30:1–29. doi: 10.1016/0304-4203(90)90059-l. [DOI] [Google Scholar]
- Britton HT. Hydrogen Lons. 4th edn. London: Chapman and Hall; 1952. [Google Scholar]
- Bullock HA, Luo H, Whitman WB. Evolution of dimethylsulfoniopropionate metabolism in marine phytoplankton and bacteria. Frontiers in Microbiology. 2017;8:637. doi: 10.3389/fmicb.2017.00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao HY, Wang P, Xu F, Li PY, Xie BB, Qin QL, Zhang YZ, Li CY, Chen XL. Molecular insight into the acryloyl-CoA hydration by AcuH for acrylate detoxification in dimethylsulfoniopropionate-catabolizing bacteria. Frontiers in Microbiology. 2017;8:2034. doi: 10.3389/fmicb.2017.02034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Garrity J, Crosby HA, Escalante-Semerena JC. In Salmonella enterica, the sirtuin-dependent protein acylation/deacylation system (SDPADS) maintains energy homeostasis during growth on low concentrations of acetate. Molecular Microbiology. 2011;80:168–183. doi: 10.1111/j.1365-2958.2011.07566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson RJ, Lovelock JE, Andreae MO, Warren SG. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature. 1987;326:655–661. doi: 10.1038/326655a0. [DOI] [Google Scholar]
- Cosquer A, Pichereau V, Pocard JA, Minet J, Cormier M, Bernard T. Nanomolar levels of dimethylsulfoniopropionate, dimethylsulfonioacetate, and glycine betaine are sufficient to confer osmoprotection to Escherichia coli. Applied and Environmental Microbiology. 1999;65:3304–3311. doi: 10.1128/AEM.65.8.3304-3311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curson ARJ, Sullivan MJ, Todd JD, Johnston AWB. Identification of genes for dimethyl sulfide production in bacteria in the gut of Atlantic Herring (Clupea harengus. The ISME Journal. 2010;4:144–146. doi: 10.1038/ismej.2009.93. [DOI] [PubMed] [Google Scholar]
- Curson ARJ, Sullivan MJ, Todd JD, Johnston AWB. DddY, a periplasmic dimethylsulfoniopropionate lyase found in taxonomically diverse species of Proteobacteria. The ISME Journal. 2011a;5:1191–1200. doi: 10.1038/ismej.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curson ARJ, Todd JD, Sullivan MJ, Johnston AWB. Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nature Reviews Microbiology. 2011b;9:849–859. doi: 10.1038/nrmicro2653. [DOI] [PubMed] [Google Scholar]
- Curson ARJ, Liu J, Martinez AB, Green RT, Chan Y, Carrión O, Williams BT, Zhang SH, Yang GP, Bulman Page PC, Zhang XH, Todd JD. Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nature Microbiology. 2017;2:17009. doi: 10.1038/nmicrobiol.2017.9. [DOI] [PubMed] [Google Scholar]
- Curson ARJ, Williams BT, Pinchbeck BJ, Sims LP, Martínez AB, Rivera PPL, Kumaresan D, Mercadé E, Spurgin LG, Carrión O, Moxon S, Cattolico RA, Kuzhiumparambil U, Guagliardo P, Clode PL, Raina JB, Todd JD. DSYB catalyses the key step of dimethylsulfoniopropionate biosynthesis in many phytoplankton. Nature Microbiology. 2018;3:430–439. doi: 10.1038/s41564-018-0119-5. [DOI] [PubMed] [Google Scholar]
- Dehio C, Meyer M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. Journal of. Bacteriology. 1997;179:538–540. doi: 10.1128/jb.179.2.538-540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D-Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Bairoch A, Appel RD. In: The Proteomics Protocols Handbook. Walker JM, editor. Springer Protocols Handbooks. Humana Press; 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [DOI] [Google Scholar]
- Howard EC, Henriksen JR, Buchan A, Reisch CR, Bürgmann H, Welsh R, Ye W, González JM, Mace K, Joye SB, Kiene RP, Whitman WB, Moran MA. Bacterial taxa that limit sulfur flux from the ocean. Science. 2006;314:649–652. doi: 10.1126/science.1130657. [DOI] [PubMed] [Google Scholar]
- Hu J, Komakula A, Fraser ME. Binding of hydroxycitrate to human atp-citrate lyase. Acta Crystallographica Section D Structural Biology. 2017;73:660–671. doi: 10.1107/S2059798317009871. [DOI] [PubMed] [Google Scholar]
- Johnson WM, Kido Soule MC, Kujawinski EB. Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. The ISME Journal. 2016;10:2304–2316. doi: 10.1038/ismej.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AWB, Green RT, Todd JD. Enzymatic breakage of dimethylsulfoniopropionate-a signature molecule for life at sea. Current Opinion in Chemical Biology. 2016;31:58–65. doi: 10.1016/j.cbpa.2016.01.011. [DOI] [PubMed] [Google Scholar]
- Karsten U, Kück K, Vogt C, Kirst GO. In: Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds. Kiene RP, Visscher PT, Keller MD, Kirst GO, editors. Boston, MA: Springer; 1996. Dimethylsulfoniopropionate production in phototrophic organisms and its physiological functions as a cryoprotectant; pp. 143–153. [DOI] [Google Scholar]
- Kiene RP, Linn LJ, Bruton JA. New and important roles for DMSP in marine microbial communities. Journal of Sea Research. 2000;43:209–224. doi: 10.1016/S1385-1101(00)00023-X. [DOI] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. Four new derivatives of the broad-host-range cloning vector PBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Cherukuri KP, Alcolombri U, Meltzer D, Tawfik DS. The dimethylsulfoniopropionate (DMSP) lyase and lyase-like cupin family consists of bona fide DMSP lyases as well as other enzymes with unknown function. Biochemistry. 2018;57:3364–3377. doi: 10.1021/acs.biochem.8b00097. [DOI] [PubMed] [Google Scholar]
- Li CY, Wei TD, Zhang SH, Chen XL, Gao X, Wang P, Xie BB, Su HN, Qin QL, Zhang XY, Yu J, Zhang HH, Zhou BC, Yang GP, Zhang YZ. Molecular insight into bacterial cleavage of oceanic dimethylsulfoniopropionate into demethyl sulfide. PNAS. 2014;111:1026–1031. doi: 10.1073/pnas.1312354111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-Y, Zhang D, Chen X-L, Wang P, Shi W-L, Li P-Y, Zhang X-Y, Qin Q-L, Todd JD, Zhang Y-Z. Mechanistic insights into dimethylsulfoniopropionate lyase DDDY, a new member of the Cupin superfamily. Journal of Molecular Biology. 2017;429:3850–3862. doi: 10.1016/j.jmb.2017.10.022. [DOI] [PubMed] [Google Scholar]
- Liu J, Liu J, Zhang SH, Liang J, Lin H, Song D, Yang GP, Todd JD, Zhang XH. Novel insights into bacterial dimethylsulfoniopropionate catabolism in the East China Sea. Frontiers in Microbiology. 2018;9:3206. doi: 10.3389/fmicb.2018.03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai X, Adams MWW. Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. Journal of Bacteriology. 1996;178:5897–5903. doi: 10.1128/jb.178.20.5897-5903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution - from diffraction images to an initial model in minutes. Acta Crystallographica Section D-Structural Biology. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Musfeldt M, Schonheit P. Novel type of ADP-forming acetyl coenzyme A synthetase in hyperthermophilic archaea: heterologous expression and characterization of isoenzymes from the sulfate reducer Archaeoglobus fulgidus and the methanogen Methanococcus jannaschii. Journal of Bacteriology. 2002;184:636–644. doi: 10.1128/jb.184.3.636-644.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevitt GA. The neuroecology of dimethyl sulfide: a global-climate regulator turned marine infochemical. Integrative and Comparative Biology. 2011;51:819–825. doi: 10.1093/icb/icr093. [DOI] [PubMed] [Google Scholar]
- Otte ML, Wilson G, Morris JT, Moran BM. Dimethylsulphoniopropionate (DMSP) and related compounds in higher plants. Journal of Experimental Botany. 2004;55:1919–1925. doi: 10.1093/jxb/erh178. [DOI] [PubMed] [Google Scholar]
- Patel SS, Walt DR. Substrate specificity of acetyl coenzyme A synthetase. Journal of Biological Chemistry. 1987;262:7132–7134. doi: 10.0000/PMID2884217. [DOI] [PubMed] [Google Scholar]
- Raina JB, Tapiolas DM, Forêt S, Lutz A, Abrego D, Ceh J, Seneca FO, Clode PL, Bourne DG, Willis BL, Motti CA. DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature. 2013;502:677–680. doi: 10.1038/nature12677. [DOI] [PubMed] [Google Scholar]
- Reisch CR, Moran MA, Whitman WB. Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. Journal of Bacteriology. 2008;190:8018–8024. doi: 10.1128/JB.00770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisch CR, Moran MA, Whitman WB. Bacterial Catabolism of Dimethylsulfoniopropionate (DMSP. Frontiers in Microbiology. 2011a;2:172. doi: 10.3389/fmicb.2011.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisch CR, Stoudemayer MJ, Varaljay VA, Amster IJ, Moran MA, Whitman WB. Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature. 2011b;473:208–211. doi: 10.1038/nature10078. [DOI] [PubMed] [Google Scholar]
- Reisch CR, Crabb WM, Gifford SM, Teng Q, Stoudemayer MJ, Moran MA, Whitman WB. Metabolism of dimethylsulphoniopropionate by Ruegeria pomeroyi DSS-3. Molecular Microbiology. 2013;89:774–791. doi: 10.1111/mmi.12314. [DOI] [PubMed] [Google Scholar]
- Sánchez LB, Galperin MY, Müller M. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (Nucleoside diphosphate-forming. Journal of Biological Chemistry. 2000;275:5794–5803. doi: 10.1074/jbc.275.8.5794. [DOI] [PubMed] [Google Scholar]
- Schlitzer R. Interactive analysis and visualization of geoscience data with Ocean Data View. Computers & Geosciences. 2002;28:1211–1218. doi: 10.1016/S0098-3004(02)00040-7. [DOI] [Google Scholar]
- Seymour JR, Simó R, Ahmed T, Stocker R. Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science. 2010;329:342–345. doi: 10.1126/science.1188418. [DOI] [PubMed] [Google Scholar]
- Shao X, Cao H-Y, Zhao F, Peng M, Wang P, Li C-Y, Shi W-L, Wei T-D, Yuan Z, Zhang X-H, Chen X-L, Todd JD, Zhang Y-Z. Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria. Molecular Microbiology. 2019;111:1057–1073. doi: 10.1111/mmi.14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefels JP. Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. Journal of Sea Research. 2000;43:183–197. doi: 10.1016/S1385-1101(00)00030-7. [DOI] [Google Scholar]
- Sun L, Curson ARJ, Todd JD, Johnston AWB. Diversity of DMSP transport in marine bacteria, revealed by genetic analyses. Biogeochemistry. 2012;110:121–130. doi: 10.1007/s10533-011-9666-z. [DOI] [Google Scholar]
- Sunda W, Kieber DJ, Kiene RP, Huntsman S. An antioxidant function for DMSP and DMS in marine algae. Nature. 2002;418:317–320. doi: 10.1038/nature00851. [DOI] [PubMed] [Google Scholar]
- Thume K, Gebser B, Chen L, Meyer N, Kieber DJ, Pohnert G. The metabolite dimethylsulfoxonium propionate extends the marine organosulfur cycle. Nature. 2018;563:412–415. doi: 10.1038/s41586-018-0675-0. [DOI] [PubMed] [Google Scholar]
- Todd JD, Rogers R, Li YG, Wexler M, Bond PL, Sun L, Curson ARJ, Malin G, Steinke M, Johnston AWB. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science. 2007;315:666–669. doi: 10.1126/science.1135370. [DOI] [PubMed] [Google Scholar]
- Todd JD, Curson ARJ, Dupont CL, Nicholson P, Johnston AWB. The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi. Environmental Microbiology. 2009;11:1376–1385. doi: 10.1111/j.1462-2920.2009.01864.x. [DOI] [PubMed] [Google Scholar]
- Todd JD, Curson ARJ, Nikolaidou-Katsaraidou N, Brearley CA, Watmough NJ, Chan Y, Page PCB, Sun L, Johnston AWB. Molecular dissection of bacterial acrylate catabolism-unexpected links with dimethylsulfoniopropionate catabolism and dimethyl sulfide production. Environmental Microbiology. 2010;12:327–343. doi: 10.1111/j.1462-2920.2009.02071.x. [DOI] [PubMed] [Google Scholar]
- Todd JD, Curson ARJ, Kirkwood M, Sullivan MJ, Green RT, Johnston AWB. DddQ, a novel, cupin-containing, dimethylsulfoniopropionate lyase in marine roseobacters and in uncultured marine bacteria. Environmental Microbiology. 2011;13:427–438. doi: 10.1111/j.1462-2920.2010.02348.x. [DOI] [PubMed] [Google Scholar]
- Todd JD, Curson ARJ, Sullivan MJ, Kirkwood M, Johnston AWB. The Ruegeria pomeroyi acuI gene has a role in DMSP catabolism and resembles yhdH of E. coli and other bacteria in conferring resistance to acrylate. PLOS ONE. 2012;7:e35947. doi: 10.1371/journal.pone.0035947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallina SM, Simó R. Strong relationship between DMS and the solar radiation dose over the global surface ocean. Science. 2007;315:506–508. doi: 10.1126/science.1133680. [DOI] [PubMed] [Google Scholar]
- Verschueren KHG, Blanchet C, Felix J, Dansercoer A, Vos DD, Bloch Y, Beeumen JV, Svergun D, Gutsche I, Savvides SN, Verstraete K. Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle. Nature. 2019;568:571–575. doi: 10.1038/s41586-019-1095-5. [DOI] [PubMed] [Google Scholar]
- Wang P, Yu Z, Li B, Cai X, Zeng Z, Chen X, Wang X. Development of an efficient conjugation-based genetic manipulation system for pseudoalteromonas. Microbial Cell Factories. 2015a;14:11. doi: 10.1186/s12934-015-0194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Chen X, Li C, Gao X, Zhu D, Xie B, Qin Q, Zhang X, Su H, Zhou B, Xun L, Zhang Y. Structural and molecular basis for the novel catalytic mechanism and evolution of DddP , an abundant peptidase‐like bacterial Dimethylsulfoniopropionate lyase: a new enzyme from an old fold. Molecular Microbiology. 2015b;98:289–301. doi: 10.1111/mmi.13119. [DOI] [PubMed] [Google Scholar]
- Wang P, Cao H-Y, Chen X-L, Li C-Y, Li P-Y, Zhang X-Y, Qin Q-L, Todd JD, Zhang Y-Z. Mechanistic insight into acrylate metabolism and detoxification in marine dimethylsulfoniopropionate-catabolizing bacteria. Molecular Microbiology. 2017;105:674–688. doi: 10.1111/mmi.13727. [DOI] [PubMed] [Google Scholar]
- Weiße RHJ, Faust A, Schmidt M, Schönheit P, Scheidig AJ. Structure of ndp-forming acetyl-coa synthetase acd1 reveals a large rearrangement for phosphoryl transfer. PNAS. 2016;113:E519–E528. doi: 10.1073/pnas.1518614113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallographica Section D-Structural Biology. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe GV, Steinke M, Kirst GO. Grazing-activated chemical defence in a unicellular marine alga. Nature. 1997;387:894–897. doi: 10.1038/43168. [DOI] [Google Scholar]
- Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing Ezbiocloud: A taxonomically united database of 16s rrna gene sequences and whole-genome assemblies. International Journal of Systematic and Evolutionary Microbiology. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You D, Wang MM, Ye BC. Acetyl-coa synthetases of saccharopolyspora erythraea are regulated by the nitrogen response regulator GLNR at both transcriptional and post-translational levels. Molecular Microbiology. 2017;103:845–859. doi: 10.1111/mmi.13595. [DOI] [PubMed] [Google Scholar]
- Zhang X-H, Liu J, Liu J, Yang G, Xue C-X, Curson ARJ, Todd JD. Biogenic production of DMSP and its degradation to dms-their roles in the global sulfur cycle. Science China. Life Sciences. 2019;62:1296–1319. doi: 10.1007/s11427-018-9524-y. [DOI] [PubMed] [Google Scholar]
- Zheng YF, Wang JY, Zhou S, Zhang YH, Liu J, Xue CX, Williams BT, Zhao XX, Zhao L, Zhu XY, Sun C, Zhang HH, Xiao T, Yang GP, Todd TD, Zhang XH. Bacteria are important dimethylsulfoniopropionate producers in marine aphotic and high-pressure environments. Nature Communications. 2020;11:4658. doi: 10.1038/s41467-020-18434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]