Abstract
Children are less susceptible to coronavirus disease 2019 (COVID-19), and they have manifested lower morbidity and mortality after infection, for which a multitude of mechanisms may be considered. Whether the normal development of the gut-airway microbiome in children is affected by COVID-19 has not been evaluated. Here, we demonstrate that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection alters the upper respiratory tract and the gut microbiomes in nine children. The alteration of the microbiome is dominated by the genus Pseudomonas, and it sustains for up to 25–58 days in different individuals. Moreover, the patterns of alternation are different between the upper respiratory tract and the gut. Longitudinal investigation shows that the upper respiratory tract and the gut microbiomes are extremely variable among children during the course of COVID-19. The dysbiosis of microbiome persists in 7 of 8 children for at least 19–24 days after discharge from the hospital. Disturbed development of both the gut and the upper respiratory microbiomes and prolonged dysbiosis in these nine children imply possible long-term complications after clinical recovery from COVID-19, such as predisposition to the increased health risk in the post-COVID-19 era.
Keywords: SARS-CoV-2/COVID-19, Upper respiratory microbiome, Gut microbiota, Dysbiosis, Children
Introduction
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been diagnosed in more than 140 million people around the world (https://coronavirus.jhu.edu/data). Although children are often susceptible to respiratory virus infections, and experience recurrent respiratory virus infections (Santee et al., 2016; Dubourg et al., 2019; Li et al., 2019), compared with adults, they seem to be less susceptible to COVID-19 and have extremely low morbidity and mortality postinfection (Guan et al., 2020; Onder et al., 2020; Wu and McGoogan, 2020). In fact, children afflicted by COVID-19 usually have mild symptoms but a faster recovery and a better prognosis, for which the reasons are not clear. It is known that early-life development and maturation of human microbiomes shape health status in later life (Teo et al., 2015; Man et al., 2017; Derrien et al., 2019; Dubourg et al., 2019), and delayed development or dysbiosis of the microbiomes during childhood has been linked to predisposition to various health problems in adulthood (Cox et al., 2014; Blanton et al., 2016; Man et al., 2017; Derrien et al., 2019; Dubourg et al., 2019; Gehrig et al., 2019). The effect of COVID-19 on the gut microbiome had just began to be evaluated in adults (Gu et al., 2020; Zuo et al., 2020), but not in children. We recently evaluated the longitudinal effects of COVID-19 on both the upper respiratory tract and gut microbiomes in adults and revealed that the respiratory and the gut microbiomes presented a contemporaneous change from early dysbiosis toward late incomplete restoration during the course of disease (Xu et al., 2021). How COVID-19 impacts the respiratory tract and gut microbiomes in children is unknown. Here, we report the temporal dynamics of the upper respiratory tract and gut microbiome in children afflicted by COVID-19.
Results
Study cohort
Nine COVID-19 children aged between 7 and 139 months were enrolled, together with 14 age-matched healthy control children (Table S1). A total of 103 specimens, including 27 sets of paired specimens of throat swabs, nasal swabs, or feces, were collected from children with COVID-19 (Fig. S1). These children were followed for 25–58 days after symptom onset. All samples were subjected to high-throughput sequencing of the V4-region of bacterial 16S-rRNA gene.
Comparison of the microbiomes between COVID-19 children and healthy controls
We analyzed the 16S-rRNA gene sequences of all specimens from three body sites (throat, nose, and gut) and obtained 2187 amplicon sequence variants (ASVs) that represent 15 known phyla, including 200 known genera (Dataset S1). We first compared the diversity of the microbiomes in throat swabs, nasal swabs, and feces (Figs. S2A and S3A). The microbiomes in all three body sites were significantly separated between the COVID-19 children and the healthy controls (R: 0.431–0.761, P < 0.003; Fig. S2A) and characterized by distinct bacterial compositions (Figs. S2B and S3B). Interestingly, the microbiomes in the throat and nasal swabs had significantly lower richness in COVID-19 children than that of healthy controls. In addition, significantly higher evenness was observed in the gut microbiome of COVID-19 children than that of healthy controls (P < 0.05; Fig. S3A). At phylum level, Bacteroidetes (mean ± standard deviation: 34.9 ± 24.4%) and Firmicutes (37.5 ± 21.6%) were significantly enriched in the gut (feces) of COVID-19 children, whereas Proteobacteria (48.3 ± 30.2%) was enriched in the gut of healthy controls (Fig. S3B). In contrast to the gut, Bacteroidetes (throat: 29.5 ± 7.2%; nasal: 36.5 ± 11.4%) and Firmicutes (throat: 30.9 ± 20.8%; nasal: 39.3 ± 11.5%) were significantly enriched in the upper respiratory tract of healthy controls, whereas Proteobacteria appeared to be enriched in the upper respiratory tract (throat: 48.3 ± 28.4%; nasal: 49.3 ± 24.9%) of COVID-19 children (Fig. S3B). Although the enrichment of bacterial phyla was different between different groups, the variations appeared to be remarkably high among individuals within each group. To further evaluate the difference in bacterial composition between COVID-19 children and healthy controls, the relative abundance of each genus was analyzed. The vast majority of resident commensals showed a significantly higher abundance in both the upper respiratory tract and the gut of healthy controls than the COVID-19 children, and a significantly increased abundance of opportunistic pathogenic and environmental bacteria was observed in the latter (Fig. S2B). In particular, Pseudomonas, Herbaspirillum, and Burkholderia were significantly enriched in both the upper respiratory tract and the gut of the COVID-19 children, and Comamonadaceae_U was significantly enriched in the upper respiratory tract. These results strongly indicate that COVID-19 infection altered both the upper respiratory tract and the gut microbiomes in children.
Dynamics of the upper respiratory tract and the gut microbiomes in children with COVID-19
Because of the high variability in bacterial composition among individuals within each group, we further characterized the patterns of bacterial community composition using the Dirichlet multinomial mixtures (DMMs) method and identified eight community types (Fig. 1 A). The specimens from healthy children were clustered into 2 community types, one bearing the signature of stool samples (83.3%) and being named as H-GUT and another representing the collection of all 3 types of samples (6 throat swabs, 12 nasal swabs, and 6 feces) and being called H-MIX (Fig. S4). Because early development of the infant microbiome is influenced by maternal materials from multiple sites (gut, vagina, mouth, and skin), among which bacteria from maternal gut are the most important contributor to the microbiome of children (Ferretti et al., 2018); therefore, infants and children may share the same or very similar microbial community structures of the nasal cavity, throat, and gut of their mothers (Ding and Schloss, 2014; Stewart et al., 2018). Our data indicate that the community type H-MIX may reflect the characteristics of the microbiome development in childhood. The H-GUT was significantly separated from the H-MIX (Fig. 1B) and had significantly lower richness and evenness than the H-MIX (Fig. 1C). The separation of the H-GUT from the H-MIX possibly implies a divergence of the gut microbiome from the early symbiotic microbial community structure shared by various organs.
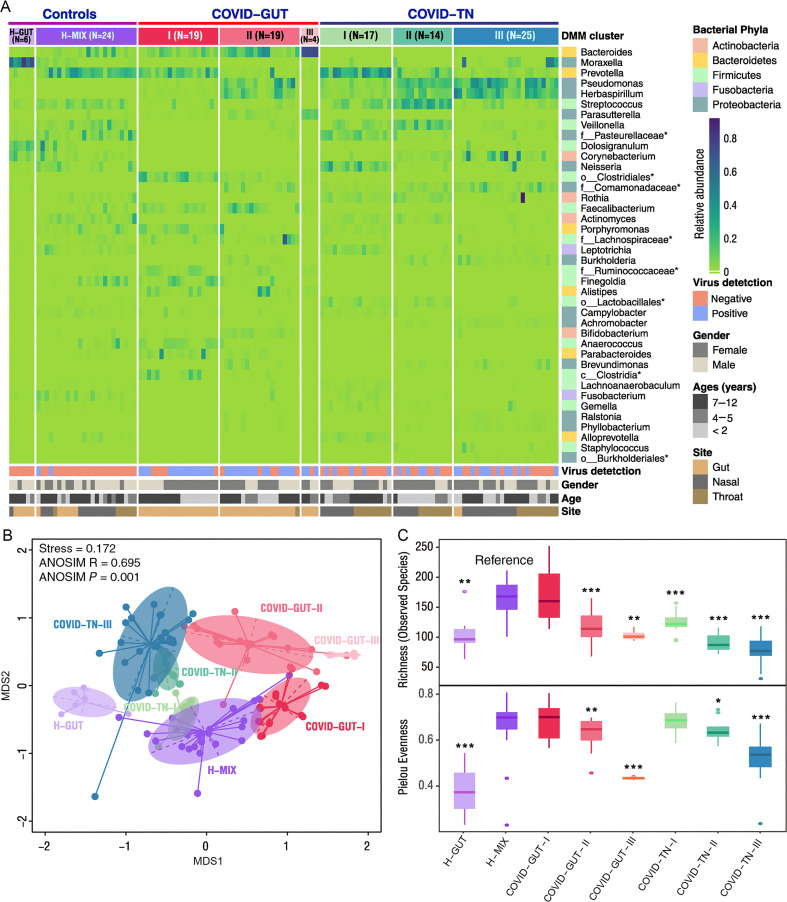
Fig. 1.
Dirichlet multinomial mixtures (DMMs) clustering analyses of 16S rRNA data separate COVID-19 children into groups with distinctive features (N = 128). A: Eight distinct clusters were identified based on lowest Laplace approximation for control and patient samples from gut or nasal and throat cavities. Heat map shows the relative abundance of top 40 bacterial genera contributing to 70% of cumulative difference for DMM clustering. The stars represent unclassified genera. Eight community clusters/types were identified. Type H-GUT indicates abnormal gut microbial community structure of healthy children. H-MIX contains a mixture of bacteria from fecal, nasal, and throat samples of healthy individuals, representing the normal microbial community structure. COVID-GUT types Ⅰ-Ⅲ are enriched in fecal samples of COVID-19 children, and COVID-TN types Ⅰ-Ⅲ are enriched in both nasal and throat samples of COVID-19 children. B: Nonmetric multidimensional scaling (NMDS) visualization of DMM clusters using Bray-Curtis distance of bacterial genera. Significant separation of microbial community structures was implicated by the ANOSIM statistic R closer to 1 with P value < 0.05. The stress value lower than 0.2 provides a good representation in reduced dimensions. C: Box plots show the alpha diversity (richness and evenness) of each DMM cluster. Wilcoxon test was used to compare the alpha diversity difference of each COVID-19-related community types with the reference community type H-MIX cluster. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001. TN, throat and nasal samples.
The vast majority (94.2%, 97/103) of the specimens of COVID-19 patients were divided into 6 bacterial community clusters except 1 in the H-GUT and 5 in the H-MIX (Fig. 1A). All COVID-19-related community types were significantly separated from that in the H-MIX and H-GUT groups (Fig. 1B) and had lower richness and evenness than that in the H-MIX (Fig. 1C), except one small cluster (COVID-TN-Ⅰ) that partly overlapped with the H-MIX and another cluster (COVID-GUT-Ⅰ) that had similar alpha diversity as the H-MIX. These results further support that COVID-19 infection altered both the upper respiratory tract and the gut microbiomes in children. Furthermore, the microbiomes of the upper respiratory tract and the gut were significantly separated from each other (Fig. 1B). Given that healthy children shared similar community type as the H-MIX in both organs, the separation of the respiratory and the gut microbiomes indicates that COVID-19 may affect the normal development of the microbiomes in both organs of children.
The vast majority (97.8%, 44/45) of stool specimens of COVID-19 children fell into 3 distinct community types, which were designated as COVID-GUT I–III (Figs. 1A and S4). Similarly, the vast majority (91.5%, 54/59) of nasal and throat swabs formed another 3 distinct types, which were designated as COVID-TN I-III (Figs. 1A and S4). Three upper respiratory tract-related types and three GUT-related types of COVID-19 children were significantly separated from each other (Fig. 1B). The community types from the same organ, regardless of the upper respiratory tract and the gut, were also separated from each other, and their alpha diversity orderly decreased from type I to type III (Fig. 1C). The decreased alpha diversity from type I to type III may represent different levels of microbiome homeostasis (dysbiosis) in both the upper respiratory tract and the gut.
Indicator genera of eight DMM clusters
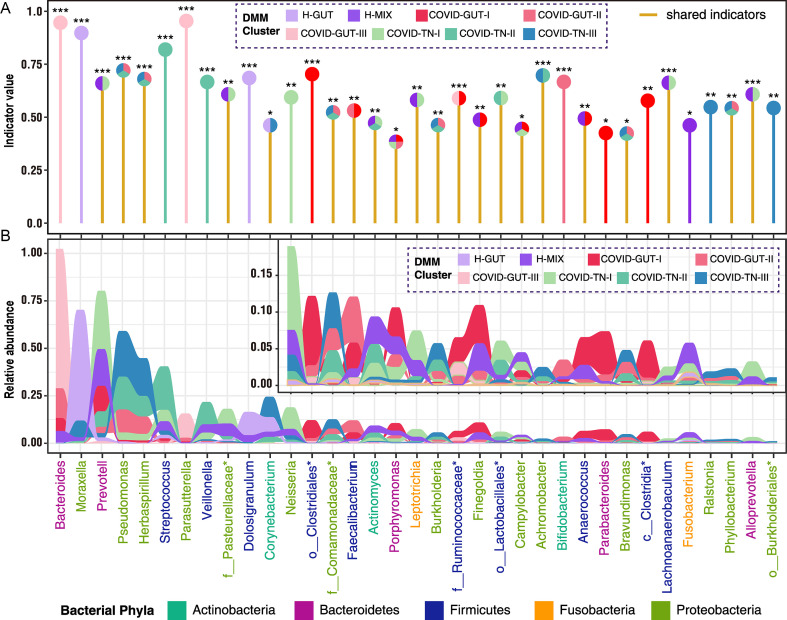
To characterize 8 microbial community types, we identified 35 indicator genera (Fig. 2 A). The H-MIX type was characterized by 11 genera with predominant commensal bacteria of Prevotella, Veillonella, Streptococcus, unclassified Pasteurellaceae, Actinomyces, Porphyromonas, Finegoldia, and Fusobacterium (Fig. 2A). The presence of some normal upper respiratory commensals (Prevotella, Veillonella) and the gut core functional bacteria (Finegoldia, Fusobacterium) suggests that H-MIX is a normal microbiome structure in healthy children. The H-GUT type was mainly featured by Moraxella with an abundance of 58.2 ± 18.4%. Because Moraxella is a common human respiratory tract pathogen, and it is often present in the individuals with chronic obstructive pulmonary disease (Murphy et al., 2005; Murphy and Parameswaran, 2009), we did not use the H-GUT as a healthy microbiome reference. Community type COVID-GUT-III had the lowest diversity and was dominated by Bacteroides and Parasutterella (Fig. 2B). Of note, Parasutterella is associated with irritable bowel syndrome and other intestinal chronic inflammation (Chen et al., 2018). Community type COVID-TN-III was dominated by highly abundant Pseudomonas and Herbaspirillum and had higher levels of genera of Corynebacterium, Comamonadaceae, Burkholderia, Achromobacter, Brevundimonas, Ralstonia, Phyllobacterium, and Burkholderiales than other community types (Fig. 2B). Among these genera, Pseudomonas is a notorious human pathogen that has been associated with various diseases (e.g., pneumonia). In our samples, there was an overrepresentation of the dominant species Pseudomonas veronii (100% sequence identity) (Cheuk et al., 2000). Achromobacter and Burkholderia are associated with cystic fibrosis (Jones et al., 2004; Spilker et al., 2013), whereas most other genera are environmental bacteria. The predominance of some opportunistic pathogenic bacteria with the colonization of various environmental bacteria in COVID-GUT-III and/or COVID-TN-III suggests a dysbiosis of the microbiome. Apart from COVID-TN-III, genus Pseudomonas also dominated community type COVID-TN-II together with Streptococcus, and COVID-GUT-II together with Bacteroides. Interestingly, we observed that some of the indicator bacteria in H-MIX were shared by the community type COVID-GUT-I (e.g., Prevotella, Porphyromonas, Finegoldia, Anaerococcus, etc.) and COVID-TN-I (e.g., Prevotella, Neisseria, Fusobacterium, unclassified Pasteurellaceae, Leptotrichia etc.; Fig. 2A). These suggest that COVID-GUT-I and COVID-TN-I were closer to the healthy microbiome type H-MIX than other COVID-19-related community types. Combining the results of the indicator bacteria and the alpha diversity characteristics, the microbial community types from I to III revealed a stepwise dysbiosis of microbiome regardless of the upper respiratory tract or the gut (Fig. 1, Fig. 2).
Fig. 2.
Identification and relative abundance distribution of indicator genera of eight DMM clusters. Indicators of 8 microbial community types (DMM clusters) were identified from top 40 genus contributing to 70% of cumulative difference for DMM clustering in Fig. 1A. Twenty-one indicator genera are shared by two or more community types. In panel (A), shared indicator genera by two or more DMM cluster are shown by yellow vertical lines. Eight DMM clusters (community types) are shown with circles of different colors above their corresponding indicator genera. Significance levels of indicators are as follows: ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001. In panel (B), the area of different colors shows the relative abundance of each indicator genus in corresponding DMM clusters.
The dynamic change of the microbiomes in three body sites of children during COVID-19
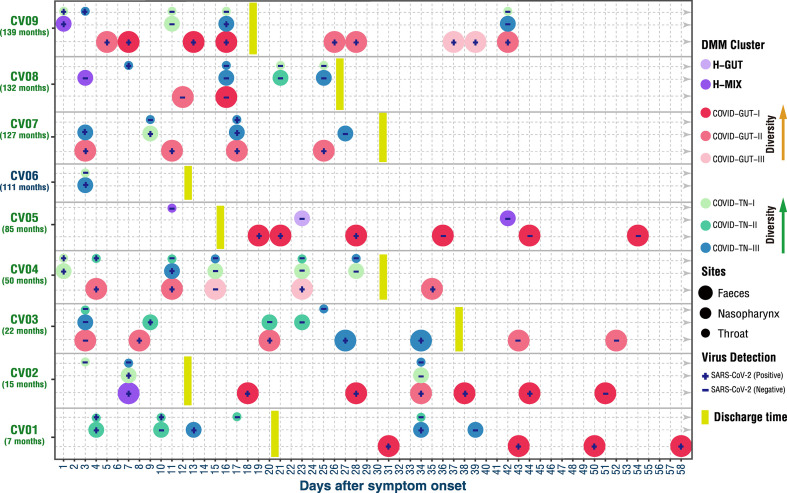
Recently, we observed synchronous restoration of the microbiomes of both the upper respiratory tract and the gut toward more diverse community structure in COVID-19 adults within a short time (6–17 days) after symptom onset (Xu et al., 2021). Distinct from adults, the microbiome community compositions were extremely variable in children during the course of COVID-19 disease, and the changes of the community types were not the same between the upper respiratory tract and the gut (Fig. 3 ). The upper respiratory (especially nasopharyngeal) microbiome of seven of eight children (except CV05) appeared to evolve from early healthy (H-MIX) or high-diversity community structure (COVID-TN-I) to late low-diversity dysbiosis structure (COVID-TN-III), indicating a steady deterioration in composition and function of the upper respiratory microbiome despite mild symptoms and clinical recovery (Fig. 3). In particular, the dysbiosis in the upper respiratory tract was observed to last at least 19–24 days after discharge (i.e., 42–58 days after symptom onset) in 3 children (CV01, CV02, and CV09).
Fig. 3.
Dynamic change of DMM clusters in three body sites of COVID-19 children. The community type levels were divergent between respiratory tract and gut (e.g., on Days 11, 15, and 23 in CV04, and Days 16 and 42 in CV09). The respiratory microbiome appeared to be progressively worsening in 7 children (CV01–CV04 and CV07–CV09) for 25–42 days after symptom onset. The worsening of the gut microbiome appeared in 4 children (CV03, CV04, CV07, and CV09) and sustained for 25–52 days. Age (months) of each COVID-19 child is shown in parenthesis below the patient code. Eight DMM clusters (community types) are shown with circles of different colors. Small, medium, and large circles represent the body sites of throat, nasopharynx, and feces, respectively. The discharge date of each COVID-19 child is highlighted by vertical yellow thick line. The status of SARS-CoV-2 RNA in each sample is shown in corresponding circle with “+” for positive and “−” for negative. The arrows indicate an increase of the alpha diversity of bacterial community type.
Compared with the upper respiratory tract microbiome, the gut microbiome alteration was more diverse among these COVID-19 children. Improvement or restoration in the gut microbiome was observed in three children (CV01, CV02, and CV05), but a deterioration occurred in another three children (CV03, CV04, and CV09) (Fig. 3). For example, the bacterial community type of CV09 improved from COVID-GUT-II to COVID-GUT-I on Day 7 after symptom onset but returned back to COVID-GUT-III on Day 37. For CV03, whose microbiome became worse from a gut community type COVID-GUT-II on Day 19 to a respiratory community type COVID-TN-III on Day 27, and reverted to COVID-GUT-II on Day 43. The appearance of the community type COVID-TN-III in the gut microbiome implies microbial translocations between the upper respiratory tract and the gut. The restoration or deterioration of the gut microbiome showed no association with clinical recovery (discharge from the hospital) or the presence or absence of SARS-CoV-2 RNA in the gut (Fig. 3). The detailed dynamic changes of bacterial compositions are shown in Fig. S5.
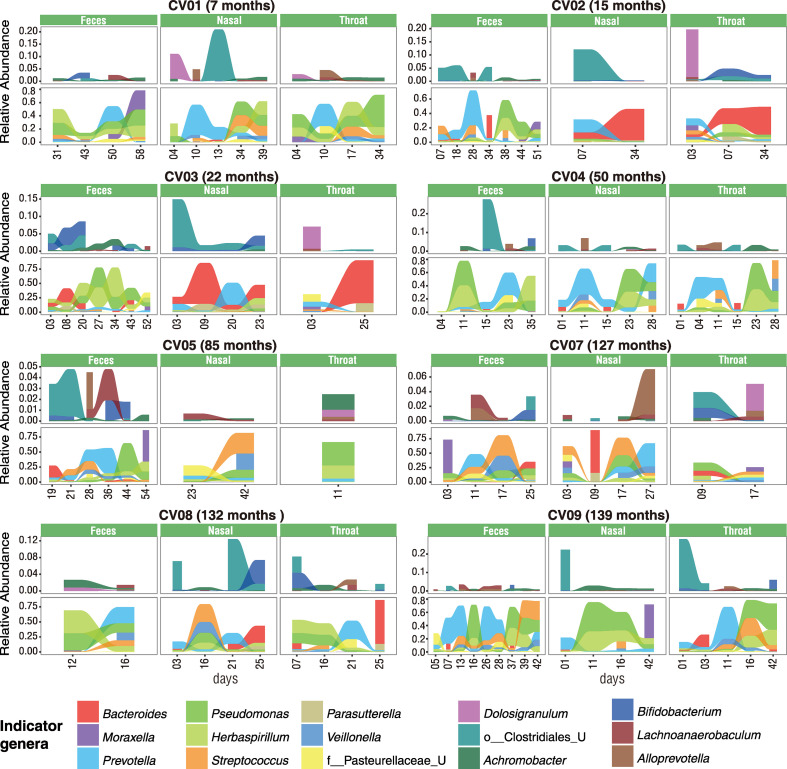
To further assess their dynamic change in relative abundance over time, the top indicator bacteria with >0.5 indicator values were selected from each community type as the representative bacteria. The abundance of representative bacteria appeared to be extremely dynamics in both the upper respiratory tract and the gut of these children during the course of COVID-19 disease, and some core enterotype identifiers were often detected in the upper respiratory tract (e.g., Bacteroides in CV02, CV03, and CV08; Prevotella in CV01, CV04, CV08, and CV09; Fig. 4 ). In particular, we found that pathogenic bacteria Pseudomonas persistently existed and dominated the microbiome in both the upper respiratory tract and the gut of the children during the whole course of COVID-19 (Figs. 4 and S5). The most prevalent Pseudomonas species was identified as P. veronii, which had a relative abundance of more than 20% in most COVID-19 children. Furthermore, pathogenic bacteria Streptococcus (mainly S.mitis) was also found in both organs of some COVID-19 children with relatively high abundance, which might contribute to the deterioration of the microbiome (Mitchell, 2011). These results supported a progressive deterioration of microbiome in both the respiratory tract and the gut of children during the course of COVID-19.
Fig. 4.
Dynamic changes of key taxa in three body sites of COVID-19 children. Indicator genera of DMM clusters with both > 0.5 indicator values and P values < 0.001 were selected as key taxa and shown in 8 COVID-19 children with at least 2 time points of sampling. Change of the relative abundance of each indicator genus is shown with the area of different colors. This figure is linked to Fig.S5.
Bacteria-bacteria co-occurrence network
We identified two main co-occurrence networks (Fig. S6). One is predominated by the members of phyla Proteobacteria. Most bacteria in this network are pathogenic or opportunistic pathogenic bacteria that mainly appeared in the low-diversity community types (e.g., COVID-GUT-III, COVID-GUT-II, and COVID-TN-III) and had positive correlations with each other (FDR-adjusted P < 0.001, Pearson correlation r > 0.5; Fig. S6 right panel). Achromobacter was the hub species that linked to nine other species in this network. Another network is mainly predominated by the members of phyla Firmicutes, Bacteroidetes, and unclassified Clostridiales, with Clostridiales being the hub to other species (Fig. S6 left panel). Most bacteria in this network are the indicator bacteria of COVID-GUT-I, H-MIX, and COVID-TN-I, and the species from the same organs often showed positive correlations. Negative correlations were mainly observed between bacteria from the upper respiratory tract and the gut, as well as between some core gut functional bacteria and opportunistic pathogenic bacteria in the upper respiratory tract.
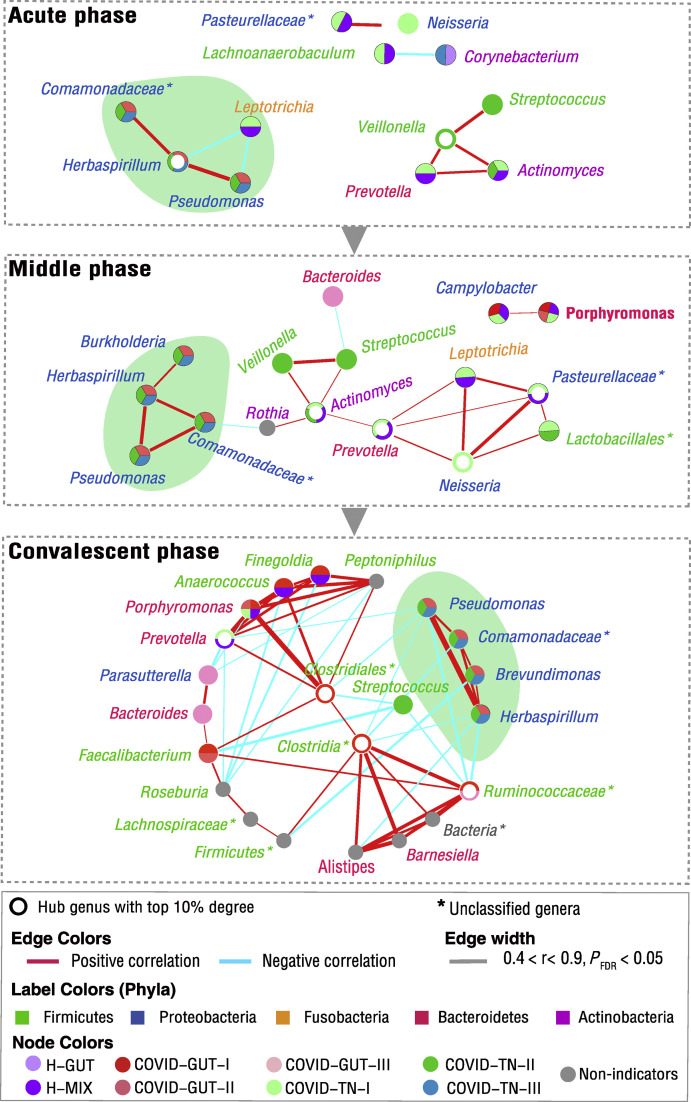
To reveal the progress of the microbiome in these patients, we further performed the co-occurrence network analysis using data from three disease stages: acute phase (the first three days since symptom onset), middle phase (from the fourth day to the day of discharge), and convalescent (after discharge) phase. We found that the bacteria-bacteria co-occurrence network enlarged and became more complex from early phase to convalescent phase, accompanied by the involvement of more commensals (Fig. 5 ). In each phase, two core co-occurrence networks were identified. The bacteria in the same core network were often linked by positive correlation, whereas the bacteria in different networks were linked by negative correlation, especially in the convalescent phase (Fig. 5). Of particular importance is that a core co-occurrence network was found to be mainly formed by Pseudomonas, Herbaspirillum, and Comamonadaceae_U in early phase, and it maintained to late convalescent phase (Fig. 5). Pseudomonas is a pathogenic bacteria; Herbaspirillum and Comamonadaceae_U are environmental bacteria. In the convalescent phase, all these bacteria had a negative correlation with some common commensals (including hub bacteria Prevotella and unclassified Clostridia, as well as non-hub bacteria Faecalibacterium and Roseburia). The persistent existence of these pathogenic and environmental bacteria implies a slow improvement of the microbiome in these COVID-19 children.
Fig. 5.
Co-occurrence network of the microbiome in the progression of COVID-19 children. We selected those microbial genera appearing in at least 30% of samples with at least 0.3% average abundance as core microbiomes to perform Pearson correlation analysis among microbial taxa. The microbial relative abundance was normalized by the centered log-ratio transformation, and correlated microbial pairs with both Pearson correlation r values higher than |0.5| and FDR-adjusted P values lower than 0.001 are shown. The acute, middle, and convalescent phases are defined as the first three days since symptom onset, the fourth day to the day of discharge, and the days after discharge, respectively. Each indicator genus is labeled by a circle that is filled with different colors showing the corresponding DMM clusters (community types), and its name is colored according to its phyla. The hub genus is highlighted by a half-open circle. The networks mainly formed by Pseudomonas, Herbaspirillum, and Comamonadaceae_U are highlighted by the cyan shadows. The line thickness is proportional to the strength of the correlation.
Functional profiling of microbial clusters associated with COVID-19 children
PIRCUST2 was used for the prediction of the microbial cluster functions. Based on KEGG ORTHOLOGY(KO) abundances predicted by PIRCUST2, we identified indicator KOs of each microbial cluster and then conducted KEGG pathway enrichment analysis per microbial cluster. A total of 66 KEGG pathways belonging to 19 KEGG functional categories were significantly enriched in 8 microbial community types (Fig. S7). The top three functional categories were amino acid metabolism, carbohydrate metabolism, and energy metabolism. Different from those of the H-MIX type cluster, we found that some enriched pathways in other seven community types might be associated with the virulence or infection/growth of opportunistic pathogenic bacteria. For example, the bacterial secretion system that contributes to the pathogenicity of pathogenic bacteria by secreting virulence factors from the cytosol of the bacteria into host cells or the host environment (Green and Mecsas, 2016) were significantly enriched in COVID-TN-II, COVID-TN-III, COVID-GUT-II, and COVID-GUT-III. Flagellum has been implicated in bacterial adhesion to and invasion into host cells (Haiko and Westerlund-Wikstrom, 2013). Consistently, we found that flagellar assembly pathways were significantly enriched in COVID-GUT-II, COVID-TN-II, and COVID-TN-III. Two-component system regulating the virulence of bacterial pathogens (Beier and Gross, 2006) were found to be significantly enriched in COVID-GUT-II, COVID-TN-II, COVID-TN-III, and H-GUT. The pathway “Biofilm formation-Pseudomonas aeruginosa” was significantly enriched in both COVID-TN-II and COVID-TN-III. Furfural can be used as carbon sources of Pseudomonas (Lee et al., 2016). Furfural degradation pathway was enriched in COVID-TN-III. This is consistent with the finding of Pseudomonas as being indicator bacteria of both COVID-TN-II and COVID-TN-III. It was noted that lipoarabinomannan (LAM) biosynthesis pathway was significantly enriched in both COVID-TN-III and H-GUT. LAM is the major glycolipid found in all mycobacteria, a virulence factor of Mycobacterium tuberculosis, and it is thought to play a role in the pathogenesis of tuberculosis (Fukuda et al., 2013). LAM might be essential for the growth of predominant bacteria in those two community types and/or the coordinated expression of virulence factors.
Discussion
SARS-CoV-2 infection causes damage to multiple organs in adults either through direct infection or indirect disruption of host homeostasis, including perturbation of the gut and the upper respiratory microbiota (Gu et al., 2020; Zuo et al., 2020). Here, we report a longitudinal investigation of the microbiome with sampling from multiple anatomical sites of children with COVID-19 and present evidence that children have a distinct dynamic process of the microbiota during the course of COVID-19 compared with adults (Xu et al., 2021). The study of children is a significant feature because early-life microbiome plays important roles in the development of host immunity, metabolism, and neural systems, affecting profoundly health status in later life (Derrien et al., 2019). The infant microbiome attains a relatively stable adult-like structure at the age 3 years after a highly dynamic initial developmental (Months 3–14) phase, a transitional (Months 15–30) phase, and finally a stable phase (Months 31–46) (Stewart et al., 2018). The development of infant microbiome is more easily influenced by various internal (genetic) and external factors (e.g., birth mode, feeding type, siblings, antibiotics, and infection) (Man et al., 2017; Derrien et al., 2019). Maternal gut bacterial strains provide the largest contribution to early-life microbiome composition (Ferretti et al., 2018). Disruption of early-life microbiome development caused by antibiotics and malnutrition was demonstrated to be associated with an increased risk of health problems later in childhood and even adulthood, such as developmental retardation, allergic diseases, obesity, diabetes, and immune dysfunction (Livanos et al., 2016; Grier et al., 2018; Guan et al., 2020; Onder et al., 2020; Wu and McGoogan, 2020). We found that SRAR-CoV-2 infection altered the upper respiratory tract and the gut microbiota of children and disturbed their normal development.
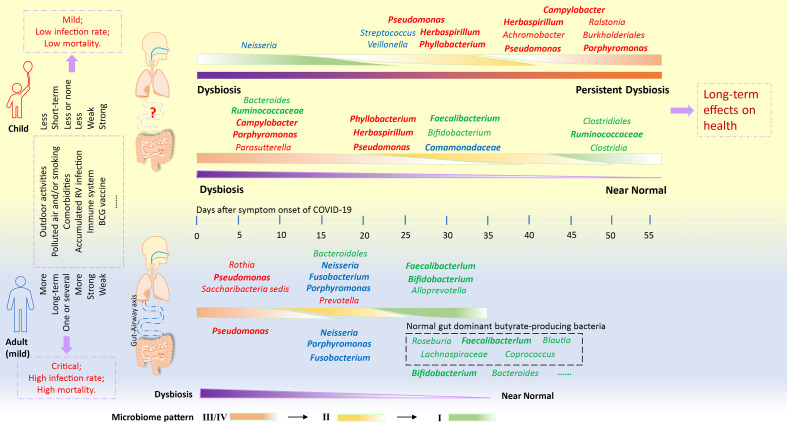
SARS-CoV-2 infection impaired the upper respiratory tract and the gut microbiome in both adults and children, but the microbiome in both organs faced diverging fates between children and adults (Fig. 6 ). In some adults with mild COVID-19 symptoms, the upper respiratory tract and the gut microbiomes showed a synchronous restoration from early dysbiosis toward later more diverse bacterial community structure within a short period (6–17 days), accompanied with their clinical recovery (Xu et al., 2021). In children, the microbiomes at both organs were extremely variable during COVID-19 and remained a long-term dyshomeostasis. Furthermore, the dynamic change of the microbiome was divergent between the upper respiratory tract and the gut. The microbiome, especially in the upper respiratory tract, appeared to maintain persistent dysbiosis for a long period (25–52 days) in spite of clinical recovery of these patients at 13–38 days after symptom onset (Fig. 3, Fig. 4, Fig. 6). In this cohort, six children who were aged older than three years were expected to have relatively stable adult-like microbiome. However, their impaired microbiomes by COVID-19 showed an extremely variable feature. These suggest that the microbiome is still particularly vulnerable and less resilient and is easily impaired by various respiratory viral infection during childhood even after attaining a stable phase (Man et al., 2017; Stewart et al., 2018; Derrien et al., 2019). In particular, the persistent dysbiosis of the microbiomes caused by COVID-19 might impart potential short-term and long-term health problems during childhood.
Fig. 6.
Distinct gut and respiratory microbiome mechanisms associated with the progress of COVID-19 in adults and children. The dynamic mechanism of the microbiome in adults was interpreted from our recent observation based on the longitudinal throat and anal swabs from 35 adults with COVID-19 (Xu et al., 2021). Similar community types from I to III/IV indicate a progressive dysbiosis of the microbiome. In mild adults with COVID-19, a synchronous shift of community type from early dysbiosis towards late incomplete restoration was found in both respiratory and gut microbiomes within a short time. In children, however, the changes of the community types were divergent between the respiratory tract and gut, possibly implying that the “airway-gut axis” is still not established during the childhood. Moreover, children's respiratory microbiome appeared to be progressively deteriorating for a long period despite a fast clinical recovery. RV, respiratory virus.
Previous studies have reported that alteration of the respiratory tract microbiome with lower bacterial diversity increased the susceptibility of children to acute respiratory tract infections (ARTIs) (Teo et al., 2015; Piters et al., 2016; Langevin et al., 2017; Dubourg et al., 2019; Man et al., 2019). The impaired microbiome in children with COVID-19 was characterized by Pseudomonas-dominated community types. In particular, a core bacteria-bacteria co-occurrence network driven by Pseudomonas, Herbaspirillum and Comamonadaceae_U was maintained from the early phase to the convalescent phase (25–58 days). The persistent existence of these opportunistic pathogens and environmental bacteria resulted in the enrichment of some specific metabolic pathways that might favor the colonization and growth of opportunistic pathogenic and environmental bacteria but inhibit the growth of some beneficial commensals. Probiotic (e.g., Bifidobacterium) and butyrate-producing bacteria (e.g., Faecalibacterium) that have anti-inflammatory ability were found to be extensively depleted from the gut and the upper respiratory tract microbiomes of some children, especially at the late stage of COVID-19 (Riviere et al., 2016; Lopez-Siles et al., 2017) (Fig. 6). Low abundance or lack of butyrate-producing bacteria might not tolerate low-level inflammation induced by SARS-CoV-2, rendering children more vulnerable to ARTIs and diarrheal diseases (Man et al., 2017; Dubourg et al., 2019; Li et al., 2019). In particular, the enrichment of genera Moraxella and Streptococcus in these COVID-19 children might predispose them to an increased risk of recurrent ARTIs (Teo et al., 2015).
One limitation of our study is that only nine COVID-19 children were recruited in this study. This is because of the fact that the epidemic was quickly controlled in Shanghai and surrounding areas, and very few children were infected and available for study. Although the relatively small patient number requires further validation in a larger cohort, our results clearly showed extremely dynamical and impaired microbiomes in both the upper respiratory tract and the gut in children with COVID-19. Although we followed the dynamic changes of the microbiomes in these children up to 58 days after symptom onset, the long-term effect of COVID-19 on the development of microbiome in childhood needs to be further investigated. In addition, the use of only 16S rRNA gene sequence data may limit our ability to more exactly identify bacteria and infer their function.
To the best of our knowledge, the complex dynamics of the upper respiratory and the gut microbiota in children with COVID-19 has yet been reported. SARS-CoV-2 infection altered the microbiomes of both organs in children and disturbed their development. The observation of a prolonged dyshomeostasis of the microbiomes in both organs of these children implies possible short-term and long-term complications after they have recovered from COVID-19 and predispose afflicted children to an increased health risk in later life. Although the long-term outcomes of COVID-19 on children need to be further studied with more extended follow-up and larger cohort, our data suggest that early implementation of various intervention strategies to modulate the microbiome development may provide clinical benefit to children in the post-COVID-19 era.
Materials and methods
Study population
Nine children were diagnosed as COVID-19 patients by Children's Hospital of Fudan University according to the National Guidelines for Diagnosis and Treatment of COVID-19. A total of 103 samples, including 31 nasal swabs, 28 throat swabs, and 44 feces, were collected from these patients (Fig. S1). Twenty-five samples from 14 age-matched healthy children were used as controls (Table S1). The upper respiratory samples were collected using flexible, sterile, dry swabs, which can reach the posterior nasopharynx and oropharynx easily (approximately 2 inches). About 2 mL spontaneous (unstimulated) fecal specimen (300 mg/tube) were collected into a sterile cryogenic vial (Corning, NY, USA), divided into aliquots and stored at −80°C until use. The sampling was performed by the professionals at the hospital.
The study was approved by Children's Hospital of Fudan University (2020–27). Informed consents were obtained from involved patients or their guardians.
Confirmation of COVID-19 children
The clinical and epidemiological characteristics and SARS-CoV-2 RNA shedding patterns of these patients have previously been reported (Cai et al., 2020; Liu et al., 2020). All nine COVID-19 children had mild symptoms. The virus RNA was extracted from all samples using a Mag-Bind RNA Extraction Kit (MACCURA, Sichuan, China) according to the manufacturer's instruction. The ORFlab and N genes of SARS-CoV-2 were detected using a Novel Coronavirus (2019-nCoV) RNA detection kit (PCR-Fluorescence Probing; DAAN gene, Guangzhou, China) according to the manufacturer's instruction.
16S rRNA gene sequencing
All samples, including nasal swabs, throat swabs, and stool samples, were subjected to the DNA extraction using a QIAamp DNA Microbiome Kit (QIAGEN, Düsseldorf, Germany). A novel triple-index amplicon sequencing strategy was used for 16S rRNA gene sequencing (D'Amore et al., 2016). In brief, a set of universal bacterial primers was used to amplify the V4 hypervariable region (515–806 nt) of the 16S rRNA gene. Two rounds of PCR amplification were performed (De Muinck et al., 2017). A reaction mixture containing 8 μL nuclease-free water, 0.5 μL KOD-Plus-Neo (TOYOBO, Osaka Boseki, Japan), 2.5 μL each of 1 μM PCR1 forward and reverse primers, and 5 μL DNA template was prepared for the first round of the PCR (PCR1) amplification. PCR1 products were purified using Monarch DNA Gel Extraction Kit (New England Biolabs, Ipswich, MA, USA) and quantified by a Qubit 4.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). Purified PCR1 products were pooled with equal amounts and then subjected to the secondary round of PCR (PCR2) amplification. The PCR2 mix contains 21 μL nuclease-free water, 1 μL KOD-Plus-Neo (TOYOBO, Osaka Boseki, Japan), 5 μL each of 1 μM PCR2 forward and reverse primers, and 5 μL pooled PCR1 products. The PCR2 products were purified using the same Gel Extraction Kit and qualified using the Qubit 4.0 Fluorometer. The specific products were further qualified using Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). The PCR2 products with equal moles of specific products were pooled and then mixed with AMPure XP beads (Beckman Coulter, Pasadena, CA, USA) in a ratio of 0.8:1. After purification, the amplicons were paired-end sequenced (2 × 250) using the Illumina-P250 sequencer. To control contamination from laboratory, negative controls with nuclease-free water (NC-1) were simultaneously performed with clinical samples during the two rounds of PCR amplification. Furthermore, nuclease-free water (NC-2) was also subjected to DNA extraction and subsequent two rounds of PCR amplification. No amplification was detected in these NCs, and therefore, no 16S data were obtained for NCs. These suggest that there was less possibility of contamination from laboratory and related reagents (including DNA extraction kit).
Bioinformatic analysis of 16S rRNA gene sequence data
Using USEARCH11 software (Edgar, 2013) and FASTX-Toolkit (Hannon, 2010), sequenced reads were merged, demultiplexed, and filtered. After trimming barcode, adapter and primer sequences using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), 14,702,896 sequences were retained with an average of 100,019 sequences per sample. Based on the QIIME 2 platform (https://qiime2.org) (Caporaso et al., 2010), the Deblur (Amir et al., 2017) was used to cluster sequence data into ASVs. ASVs are better than the traditional operational taxonomic unit picking usually according to 97% sequence similarity threshold, which may miss subtle and real biological sequence variation (Gilbert et al., 2018). In particular, we used the Deblur to perform quality filtering, dereplication, chimeras removal with default settings except for merged sequences truncated to 250 bp. An ASVs count table was generated (2187 ASVs). The taxonomic classification of ASVs representative sequences was assigned by using the RDP Naive Bayesian Classifier algorithm (Wang et al., 2007) based on the Ribosomal Database Project 16S rRNA training set (v16) database (Cole et al., 2014). Finally, the ASVs table was subsampled at an even depth of 3600 sequences per sample to eliminate the bias led by different sequencing depths among samples. The ASVs coverage of 87% was sufficient to capture microbial diversity.
Identification and characteristics of microbial community types
Based on the bacterial genus abundance, we used the DMMs (Holmes et al., 2012) algorithm implemented in mothur (v1.44.1) (Schloss et al., 2009) to identify microbial community types. The DMM algorithm can efficiently cluster samples based on microbial composition, and its sensitivity, reliability, and accuracy were widely confirmed in many microbiome studies (Ding and Schloss, 2014; Fujimura et al., 2016; Stewart et al., 2018). Based on the lowest Laplace approximation index, the appropriate microbial community type numbers (DMM clusters) were determined. Conjugated with the Analysis of Similarities (ANOSIM), the reliability of DMM clustering was further validated and then visualized by the nonmetric multidimensional scaling (NMDS) based on the Bray-Curtis distance under bacterial genus level. The ANOSIM statistic “R” compares the mean of ranked dissimilarities between groups to the mean of ranked dissimilarities within groups. An “R” value close to “1.0” suggests dissimilarity between groups, whereas an “R” value close to “0” suggests an even distribution of high and low ranks within and between groups (Clarke, 1993). ANOSIM P values lower than 0.05 suggest higher similarity within sites. Richness (observed ASVs) and Pielou's or Species evenness for each community type were calculated for estimating the difference of alpha diversity. The analyses described above were performed using “R” package “vegan” v2.5-6. Using “R” package “Pheatmap,” the dynamic change of microbial community types and compositions were visualized. Alpha diversity difference between groups was measured using the Wilcoxon Rank Sum Test in “R” package.
Identification of indicator taxa contributing to microbial community typing
To get the reliable indicator genus for community typing, we performed the Indicator Species Analysis using the indicspecies package (ver.1.7.8) (De Caceres et al., 2010) in “R” platform with top 40 genus contributing to DMM clustering that accounted for 71% cumulative difference. Dynamic changes of indicator genera corresponding to each community type were showed for all COVID-19 children using the pheatmap package in “R” language.
Co-occurrence network analysis of the microbiomes at the upper respiratory tract and the gut of the COVID-19 children
Based on microbial genus abundances normalized by the centered log-ratio transformation of gut, nasal, and throat samples collected from eight COVID-19 children, we calculated the Pearson correlation coefficient (Pearson's r) among bacterial genera of three body sites. Before performing correlation analysis, we selected the microbial genera appearing in at least 30% of samples with at least 0.3% average abundance as core microbiome composition to perform microbial occurrence analysis. The Pearson's “r” higher than 0.5 or lower than −0.5 with P value that was below 0.001 after the FDR adjustment was considered significant correlation. Co-occurrence network of significantly correlated bacterial genus pairs was visualized using Cytoscape v3.8.0 (Otasek et al., 2019).
Prediction of unique indicator functions from 16S marker sequences
To investigate the potential function enriched in the microbiome of the COVID-19 children, the microbial metabolic KEGG pathways were reconstructed using PIRUST2 (Douglas et al., 2020) to predict KOs (KEGG Orthologs) and produce KO relative abundances in each sample assigned into microbial clusters. Indicator KO analysis for each cluster was performed by the “indicspecies” package in R. Based on indicator KOs in each cluster, a hypergeometric test was used to determine significant enrichment of a specific KEGG pathway under holm adjusted P value lower than 0.05. All information of both KO and KEGG pathway were downloaded from KEGG API (https://www.kegg.jp/kegg/rest/keggapi.html).
Availability of data and materials
The raw data of 16S rRNA gene sequences are available at NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) at BioProject ID PRJNA642019. The data sets to generate the Figures are provided in Dataset S1. The scripts used for the analyses are provided in the Text S1.
CRediT authorship contribution statement
Rong Xu: Methodology, Data curation, Visualization, Writing - Original draft. Pengcheng Liu: Clinical samples and data collection, Clinical data analysis. Tao Zhang: Software, Validation, Investigation, Visualization, Writing - Original draft. Qunfu Wu: Software. Mei Zeng: Resources. Yingying Ma: Methodology. Xia Jin: Writing - Review & Editing. Jin Xu: Conceptualization, Resources, Funding acquisition. Zhigang Zhang: Software, Project administration, Data curation, Writing - Review & Editing, Funding acquisition. Chiyu Zhang: Conceptualization, Project administration, Data curation, Writing - Review & Editing, Funding acquisition.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We thank Mr. Kai Liu at Institut Pasteur of Shanghai, Chinese Academy of Sciences for his technical support. This work was supported by the grants from the National Key Research and Development Program of China (2018YFC2000500 and 2017ZX10103009-002), Major Science and Technology Project in Yunnan Province of China (202001BB050001), the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0503), the Key Research Program of the Chinese Academy of Sciences (FZDSW-219), the Chinese National Natural Science Foundation of China (31970571), and grants specific for coronavirus disease 2019 from the Children’s Hospital of Fudan University (EKXGZX006).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgg.2021.05.004.
Appendix A Supplementary data
The following are the Supplementary data to this article:
References
- Amir A., McDonald D., Navas-Molina J.A., Kopylova E., Morton J.T., Xu Z.Z., Kightley E.P., Thompson L.R., Hyde E.R., Gonzalez A., et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2017;2 doi: 10.1128/mSystems.00191-16. e00191-00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D., Gross R. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 2006;9:143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Blanton L.V., Charbonneau M.R., Salih T., Barratt M.J., Venkatesh S., Ilkaveya O., Subramanian S., Manary M.J., Trehan I., Jorgensen J.M., et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351:aad3311. doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Xu J., Lin D., Yang Z., Xu L., Qu Z., Zhang Y., Zhang H., Jia R., Liu P., et al. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin. Infect. Dis. 2020;71:1547–1551. doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.J., Wu H., Wu S.D., Lu N., Wang Y.T., Liu H.N., Dong L., Liu T.T., Shen X.Z. Parasutterella, in association with irritable bowel syndrome and intestinal chronic inflammation. J. Gastroenterol. Hepatol. 2018;33:1844–1852. doi: 10.1111/jgh.14281. [DOI] [PubMed] [Google Scholar]
- Cheuk W., Woo P.C.Y., Yuen K.Y., Yu P.H., Chan J.K.C. Intestinal inflammatory pseudotumour with regional lymph node involvement: identification of a new bacterium as the aetiological agent. J. Pathol. 2000;192:289–292. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH767>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Clarke K.R. Nonparametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993;18:117–143. [Google Scholar]
- Cole J.R., Wang Q., Fish J.A., Chai B.L., McGarrell D.M., Sun Y.N., Brown C.T., Porras-Alfaro A., Kuske C.R., Tiedje J.M. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox L.M., Yamanishi S., Sohn J., Alekseyenko A.V., Leung J.M., Cho I., Kim S.G., Li H., Gao Z., Mahana D., et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amore R., Ijaz U.Z., Schirmer M., Kenny J.G., Gregory R., Darby A.C., Shakya M., Podar M., Quince C., Hall N. A comprehensive benchmarking study of protocols and sequencing platforms for 16s rRNA community profiling. BMC Genom. 2016;17:55. doi: 10.1186/s12864-015-2194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Caceres M., Legendre P., Moretti M. Improving indicator species analysis by combining groups of sites. Oikos. 2010;119:1674–1684. [Google Scholar]
- De Muinck E.J., Trosvik P., Gilfillan G.D., Hov J.R., Sundaram A.Y.M. A novel ultra high-throughput 16S rRNA gene amplicon sequencing library preparation method for the Illumina HiSeq platform. Microbiome. 2017;5:68. doi: 10.1186/s40168-017-0279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., Alvarez A.S., de Vos W.M. The gut microbiota in the first decade of life. Trends Microbiol. 2019;27:997–1010. doi: 10.1016/j.tim.2019.08.001. [DOI] [PubMed] [Google Scholar]
- Ding T., Schloss P.D. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., Huttenhower C., Langille M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourg G., Edouard S., Raoult D. Relationship between nasopharyngeal microbiota and patient's susceptibility to viral infection. Expert Rev. Anti-Inf. 2019;17:437–447. doi: 10.1080/14787210.2019.1621168. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Ferretti P., Pasolli E., Tett A., Asnicar F., Gorfer V., Fedi S., Armanini F., Truong D.T., Manara S., Zolfo M., et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24:133–145. doi: 10.1016/j.chom.2018.06.005. e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K.E., Sitarik A.R., Haystad S., Lin D.L., Levan S., Fadrosh D., Panzer A.R., LaMere B., Rackaityte E., Lukacs N.W., et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016;22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T., Matsumura T., Ato M., Hamasaki M., Nishiuchi Y., Murakami Y., Maeda Y., Yoshimori T., Matsumoto S., Kobayashi K., et al. Critical roles for lipomannan and lipoarabinomannan in cell wall integrity of mycobacteria and pathogenesis of tuberculosis. Mbio. 2013;4 doi: 10.1128/mBio.00472-12. e00472-00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig J.L., Venkatesh S., Chang H.W., Hibberd M.C., Kung V.L., Cheng J.Y., Chen R.Y., Subramanian S., Cowardin C.A., Meier M.F., et al. Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science. 2019;365 doi: 10.1126/science.aau4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J.A., Blaser M.J., Caporaso J.G., Jansson J.K., Lynch S.V., Knight R. Current understanding of the human microbiome. Nat. Med. 2018;24:392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E.R., Mecsas J. Bacterial Secretion Systems: an Overview. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.VMBF-0012-2015. 10.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier A., McDavid A., Wang B.K., Qiu X., Java J., Bandyopadhyay S., Yang H.M., Holden-Wiltse J., Kessler H.A., Gill A.L., et al. Neonatal gut and respiratory microbiota: coordinated development through time and space. Microbiome. 2018;6:193. doi: 10.1186/s40168-018-0566-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S., Chen Y., Wu Z., Chen Y., Gao H., Lv L., Guo F., Zhang X., Luo R., Huang C., et al. Alterations of the gut microbiota in patients with Coronavirus Disease 2019 or H1N1 influenza. Clin. Infect. Dis. 2020;159:944–955. doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiko J., Westerlund-Wikstrom B. The role of the bacterial flagellum in adhesion and virulence. Biology (Basel) 2013;2:1242–1267. doi: 10.3390/biology2041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G.J. 2010. Fastx-toolkit.http://hannonlabcshledu/fastx_toolkit [Google Scholar]
- Holmes I., Harris K., Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One. 2012;7:e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M., Dodd M.E., Govan J.R.W., Barcus V., Doherty C.J., Morris J., Webb A.K. Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax. 2004;59:948–951. doi: 10.1136/thx.2003.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin S., Pichon M., Smith E., Morrison J., Bent Z., Green R., Barker K., Solberg O., Gillet Y., Javouhey E., et al. Early nasopharyngeal microbial signature associated with severe influenza in children: a retrospective pilot study. J. Gen. Virol. 2017;98:2425–2437. doi: 10.1099/jgv.0.000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.A., Wrona L.J., Cahoon A.B., Crigler J., Eiteman M.A., Altman E. Isolation and characterization of bacteria that use furans as the sole carbon source. Appl. Biochem. Biotechnol. 2016;178:76–90. doi: 10.1007/s12010-015-1859-9. [DOI] [PubMed] [Google Scholar]
- Li Y.P., Fu X.M., Ma J.M., Zhang J.H., Hu Y.H., Dong W., Wan Z.Z., Li Q.F., Kuang Y.Q., Lan K., et al. Altered respiratory virome and serum cytokine profile associated with recurrent respiratory tract infections in children. Nat. Commun. 2019;10:2288. doi: 10.1038/s41467-019-10294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Cai J., Jia R., Xia S., Wang X., Cao L., Zeng M., Xu J. Dynamic surveillance of SARS-CoV-2 shedding and neutralizing antibody in children with COVID-19. Emerg. Microb. Infect. 2020;9:1254–1258. doi: 10.1080/22221751.2020.1772677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livanos A.E., Greiner T.U., Vangay P., Pathmasiri W., Stewart D., McRitchie S., Li H.L., Chung J., Sohn J., Kim S., et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat. Microbiol. 2016;1:16140. doi: 10.1038/nmicrobiol.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M., Duncan S.H., Garcia-Gil L.J., Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017;11:841–852. doi: 10.1038/ismej.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man W.H., Piters W.A.A.D., Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man W.H., van Houten M.A., Merelle M.E., Vlieger A.M., Chu M.L.J.N., Jansen N.J.G., Sanders E.A.M., Bogaert D. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Resp. Med. 2019;7:417–426. doi: 10.1016/S2213-2600(18)30449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J. Italic walking the line between commensalism and pathogenesis. Mol. Oral Microbiol. 2011;26:89–98. doi: 10.1111/j.2041-1014.2010.00601.x. [DOI] [PubMed] [Google Scholar]
- Murphy T.F., Brauer A.L., Grant B.J., Sethi S. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am. J. Respir. Crit. Care Med. 2005;172:195–199. doi: 10.1164/rccm.200412-1747OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T.F., Parameswaran G.I. Moraxella catarrhalis a human respiratory tract pathogen. Clin. Infect. Dis. 2009;49:124–131. doi: 10.1086/599375. [DOI] [PubMed] [Google Scholar]
- Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA-J Am Med Assoc. 2020;323:1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- Otasek D., Morris J.H., Boucas J., Pico A.R., Demchak B. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019;20:185. doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piters W.A.A.D., Heinonen S., Hasrat R., Bunsow E., Smith B., Suarez-Arrabal M.C., Chaussabel D., Cohen D.M., Sanderl E.A.M., Ramilo O., et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am. J. Resp. Crit. Care. 2016;194:1104–1115. doi: 10.1164/rccm.201602-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere A., Selak M., Lantin D., Leroy F., De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front. Microbiol. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santee C.A., Nagalingam N.A., Faruqi A.A., DeMuri G.P., Gern J.E., Wald E.R., Lynch S.V. Nasopharyngeal microbiota composition of children is related to the frequency of upper respiratory infection and acute sinusitis. Microbiome. 2016;4:34. doi: 10.1186/s40168-016-0179-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilker T., Vandamme P., LiPuma J.J. Identification and distribution of achromobacter species in cystic fibrosis. J. Cyst. Fibros. 2013;12:298–301. doi: 10.1016/j.jcf.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Stewart C.J., Ajami N.J., O'Brien J.L., Hutchinson D.S., Smith D.P., Wong M.C., Ross M.C., Lloyd R.E., Doddapaneni H., Metcalf G.A., et al. Temporal development of the gut microbiome in early childhood from the teddy study. Nature. 2018;562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo S.M., Mok D., Pham K., Kusel M., Serralha M., Troy N., Holt B.J., Hales B.J., Walker M.L., Hollams E., et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.Y., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China smmary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA-J. Am. Med. Assoc. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xu R., Lu R.F., Zhang T., Wu Q.F., Cai W.H., Han X.D., Wan Z.Z., Jin X., Zhang Z.G., Zhang C.Y. Temporal association between human upper respiratory and gut bacterial microbiomes during the course of COVID-19 in adults. Commun. Biol. 2021;4:240. doi: 10.1038/s42003-021-01796-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T., Zhang F., Lui G.C.Y., Yeoh Y.K., Li A.Y.L., Zhan H., Wan Y., Chung A., Cheung C.P., Chen N., et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–955. doi: 10.1053/j.gastro.2020.05.048. e948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of 16S rRNA gene sequences are available at NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) at BioProject ID PRJNA642019. The data sets to generate the Figures are provided in Dataset S1. The scripts used for the analyses are provided in the Text S1.