Abstract
Targeted wastewater surveillance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been proposed by the United States Centers for Disease Control and Prevention's National Wastewater Surveillance System as a complementary approach to clinical surveillance to detect the presence of Coronavirus Disease 2019 (COVID-19) at high-density facilities and institutions such as university campuses, nursing homes, and correctional facilities. In this study we evaluated the efficacy of targeted wastewater surveillance of SARS-CoV-2 RNA together with individual-level testing for outbreak mitigation on a university campus during Fall 2020 semester. Wastewater samples (n = 117) were collected weekly from manholes or sewer cleanouts that receive wastewater inputs from dormitories, community-use buildings, and a COVID-19 isolation dormitory. Quantitative RT-PCR N1 and N2 assays were used to measure SARS-CoV-2 nucleocapsid genes in wastewater. Due to varying human waste input in different buildings, pepper mild mottle virus (PMMV) RNA was also measured in all samples and used to normalize SARS-CoV-2 N1 and N2 RNA wastewater concentrations. In this study, temporal trends of SARS-CoV-2 in wastewater samples mirrored trends in COVID-19 cases detected on campus. Normalizing SARS-CoV-2 RNA concentrations using human fecal indicator, PMMV enhanced the correlation between N1 and N2 gene abundances in wastewater with COVID-19 cases. N1 and N2 genes were significant predictors of COVID-19 cases in dormitories, and the N2 gene was significantly correlated with the number of detected COVID-19 cases in dormitories. By implementing several public health surveillance programs include targeted wastewater surveillance, individual-level testing, contact tracing, and quarantine/isolation facilities, university health administrators could act decisively during an outbreak on campus, resulting in rapid decline of newly detected COVID-19 cases. Wastewater surveillance of SARS-CoV-2 is a proactive outbreak monitoring tool for university campuses seeking to continue higher education practices in person during the COVID-19 pandemic.
Keywords: SARS-CoV-2, COVID-19, Targeted wastewater surveillance, Wastewater-based epidemiology, Outbreak mitigation, University campus
Graphical abstract
1. Introduction
The ongoing pandemic of Coronavirus Disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has impacted all aspects of the global community and individual daily function. Although public health professionals around the world are working to limit the spread the SARS-CoV-2, the disease containment has been outpaced by viral spread and limited resources for testing. The current clinical disease surveillance, which does not adequately test asymptomatic individuals, limits the ability of decision makers to determine when and where outbreaks are occurring within their communities. Wastewater surveillance of SARS-CoV-2 is an emerging public health tool to understand the spread of COVID-19 in communities and is currently being implemented worldwide (D'Aoust et al., 2021; Hasan et al., 2021; La Rosa et al., 2020; Medema et al., 2020; Peccia et al., 2020; Randazzo et al., 2020; Westhaus et al., 2021).
There is a long history of the use of environmental surveillance to monitor the circulation of enteric viruses such as noroviruses, rotaviruses, hepatitis A virus, as well as wild and vaccine strains of poliovirus in population (Hovi et al., 2012; Lodder et al., 2012; Majumdar et al., 2018; Metcalf et al., 1995; Wang et al., 2020a). SARS-CoV-2 is spread primarily by human-to-human transmission via aerosols and droplets, direct contact with an infected subject or indirect contact via hand-mediated transfer of the virus from contaminated fomites (Azimi et al., 2021; Edwards et al., 2021; Ferretti et al., 2020; Harrison et al., 2020; Wang et al., 2020c). However, SARS-CoV-2 also can infect gastrointestinal glandular epithelial cells and is excreted in feces of infected individuals including asymptomatic individuals (Xiao et al., 2020; Zhang et al., 2020). Therefore, the virus could enter the sewer system via human excretions. This suggests that wastewater may be used to monitor progression or abatement of viral spread at the community level complementary to individual testing. Additionally, wastewater surveillance is particularly useful to provide an early indication of re-emergence of SARS-CoV-2 in communities that contained an initial outbreak or with low disease prevalence. It has been demonstrated that SARS-CoV-2 genetic signal in wastewater precedes the detection of cases of COVID-19 in the population by 2–7 days (Medema et al., 2020).
To date, wastewater monitoring for SARS-CoV-2 has primarily focused on untreated wastewater influent received at centralized wastewater treatment plants (Carrillo-Reyes et al., 2020; D'Aoust et al., 2021; Gonzalez et al., 2020; Nemudryi et al., 2020; Sherchan et al., 2020; Wu et al., 2020). Although this downstream sampling approach provides broad municipality-level data, it cannot be directly used to guide more localized estimates and related interventions. Another approach is to focus on upstream wastewater sample collection, for example directly from sewer holes and wastewater lift stations, which allows for more community- or neighborhood-level granular resolution. This targeted community-level wastewater sampling can be deployed in settings with a high risk of COVID-19 transmission such as in nursing homes, university campuses, factory and industrial workplaces, and correctional facilities to inform early disease containment and mitigation measurements.
The ongoing pandemic provides an opportunity to field test the hypothesis that wastewater surveillance can be employed to monitor the temporal and spatial dynamics of infectious disease transmission within a given community. To date, there are few data on the effectiveness of targeted wastewater surveillance for COVID-19 to guide institutional operations and outbreak mitigation (Ahmed et al., 2020; Betancourt et al., 2021; Gibas et al., 2021; Hong et al., 2021; Wang et al., 2020b). Therefore, in this study, we examined the effectiveness of weekly wastewater surveillance of COVID-19 in conjunction with individual-level testing for outbreak mitigation on a university campus. The objectives of this study were to (i) monitor wastewater collected from individual buildings on a university campus for SARS-CoV-2 nucleocapsid 1 (N1) and nucleocapsid 2 (N2) genes using quantitative reverse transcription PCR (RT-qPCR); (ii) compare the SARS-CoV-2 RNA signals in wastewater to the reported COVID-19 cases in a university community; and (iii) determine if dormitory types, grouped by room style and bathroom access, affect the detection of SARS-CoV-2 genes in wastewater. The combination of frequent individual nasal swab tests of university constituents and weekly wastewater monitoring was a unique approach to outbreak surveillance on a university campus.
2. Materials and methods
2.1. Wastewater samples and sampling strategies
Tulane University, located in New Orleans, Louisiana, had an overall enrollment of 14,062 students in the 2020–2021 academic year with 8,610 undergraduate students and 5,452 graduate and professional students. The dormitories on the main campus were the primary focus of the COVID-19 wastewater surveillance. Wastewater samples from two community-use buildings, a student union and a library, were also collected to inform on the potential spread of the virus among university staff and students living off campus.
During the Fall 2020 semester (August 19, 2020–December 1, 2020), wastewater samples were collected from manholes serving single buildings, sewer cleanouts, and municipal manholes (Table S1). Sampling preference was given to buildings with single building manholes or 8″ diameter sewer cleanouts. Wastewater samples were also collected from a COVID-19 isolation dormitory to use as a positive sampling method control in this study. The isolation dormitory was used only as a living space for isolating campus residents after testing positive for COVID-19. Three dormitory types were sampled: dormitories with communal bathrooms (greater than 10 people sharing a bathroom), suite style dormitories (5–10 people sharing a bathroom), and apartment style dorms (fewer than 5 people sharing a bathroom). Two dormitories of each type were monitored weekly. Two community use buildings, a library and a student union, were also sampled weekly. Prior to the start of the Fall 2020 semester, wastewater from each building was sampled for background measures of SARS-CoV-2 RNA.
Approximately 500 ml of grab samples of wastewater were collected between 10:30 a.m. and 11:30 a.m. weekly, when wastewater levels in sampled manholes were observably higher. All samples were immediately processed or stored at 4 °C for up to 72 h until analysis. Turbidity and pH were measured immediately after sampling. A total of 117 wastewater samples were collected during the surveillance.
2.2. Concentration of wastewater samples for SARS-CoV-2
Viruses were concentrated from 200 ml of wastewater using a polyethylene glycol (PEG) precipitation (Borchardt et al., 2017). Briefly, the samples were mixed with 8% (w/vol) molecular biology grade PEG 8000 (Promega Corporation, Madison WI) and 0.2 M NaCl (w/v). The samples were mixed slowly on magnetic stirrer at 4 °C for 2 h and then held at 4 °C for 16 h. Following the overnight incubation, samples were centrifuged at 4700×g for 45 min at 4 °C. The supernatant was then removed, and the pellet was resuspended in the remaining liquid, approximately 2–4 ml. All sample concentrates were immediately proceeded to RNA extraction and RT-qPCR.
To determine the virus recovery of the PEG method, Pseudomonas phage Phi6 was inoculated in a subset of 13 wastewater samples. Phage Phi6 and its bacterial host Pseudomonas syringae were kindly provided by Dr. Krista Wigginton's lab at University of Michigan. Pseudomonas phage Phi6 has been used as a model enveloped virus in virus recovery and persistence studies in wastewater and microdroplets (Aquino de Carvalho et al., 2017; Fedorenko et al., 2020; Titcombe Lee et al., 2016; Ye et al., 2016). Phi6 was inoculated into 200 ml of wastewater samples at a concentration of 103 plaque forming unit per ml. Recovery efficiencies were determined by comparing the concentration of the spiked phage Phi6 in each sample before and after concentration using reverse transcription droplet digital PCR (RT-ddPCR).
2.3. Viral nucleic acid extraction
Viral ribonucleic acid (RNA) was extracted from wastewater concentrates using the Qiagen QIAmp Viral RNA Minikit (Qiagen, Germany) according to the manufacturer's protocol. In this study, a total of 200 μl of concentrate was used for RNA extraction resulting in a final elution volume of 80 μl.
2.4. Detection and quantification of SARS-CoV-2 and pepper mild mottle virus (PMMV) RNA using RT-qPCR
One-step RT-qPCR approach was used to quantify SARS-CoV-2 and PMMV gene markers in wastewater samples. All the primers and probes used in this study are listed in Table S2. For SARS-CoV-2, amplification reaction mixtures (final total volume of 20 μl) contained five μl template RNA, 15 μl of 2 × qScript one-step RT-qPCR ToughMix (QuantaBio), 300 nM and 500 nM of forward primer for N1 and N2, respectively, 500 nM and 800 nM of reverse primer for N1, and N2, respectively, and 200 nM of probe. The thermal cycling protocol was as follows: 10 min at 50 °C for cDNA synthesis, 3 min at 95 °C for initial denaturation, followed by 45 cycles of two steps consisting of 3 s at 95 °C and 30 s at 55 °C. Quantitative synthetic SARS-CoV-2 RNA included fragments from nucleocapsid and envelope regions (ATCC VR-3276SD) and was used to generate standard curves. qPCR amplification efficiencies for the quantification of the N1 and N2 gene assays were 92.6 ± 4.3% and 95.1 ± 3.4%, respectively. The correlation coefficients (R2) of the standard curves of N1 and N2 gene assays were 0.968 ± 0.002 and 0.982 ± 0.004, respectively. Limits of detection were determined based on the lowest copy number of the SARS-CoV-2 synthetic RNA templates with detectable CT values. The limit of detection for both gene targets was one gene copy per reaction.
Fecal indicator viral molecular target PMMV was analyzed to account for differences in building occupancy levels (human waste input) and changes in wastewater dilution (Haramoto et al., 2013). Amplification reaction mixtures for analysis of the PMMV gene (final total volume of 20 μl) contained five μl template RNA, 15 μl of 2 × qScript one-step RT-qPCR ToughMix (QuantaBio), 500 nM of forward primer for PMMV gene, 500 nM of reverse primer for PMMV gene, and 200 nM of probe. The thermal cycling protocol was as follows: 10 min at 50 °C for cDNA synthesis, 30 s at 95 °C for initial denaturation, followed by 45 cycles of two steps consisting of 5 s at 95 °C and 60 s at 60 °C. Quantitative synthetic DNA fragments (gBlock, Integrated DNA Technology) were used to generate standard curves. qPCR amplification efficiencies for the quantification of the PMMV gene was 100.39 ± 4.28% and the correlation coefficient (R2) of the standard curve was 0.997 ± 0.002. The limit of detection was one gene copy per reaction. Limits of detection were determined based on the lowest copy number of the PMMV synthetic DNA g-block templates with detectable CT values.
All RT-qPCR reactions were performed using a StepOne Plus™ real-time PCR sequence detector (Applied Biosystems, Foster City, CA). For each assay, a 10-fold diluted standard curve of at least five points, a non-template control, and samples were tested in triplicate. Sample processing blank and extraction blank using sterile water were also included for each assay as negative controls.
Samples tested negative for both SARS-CoV-2 and PMMV RNA were re-analyzed for RT-qPCR inhibition. Known copy numbers of SARS-CoV-2 synthetic RNA (1 × 103 gene copies per reaction) were added in new N1 RT-qPCR reactions (with molecular grade water) and the CT values obtained were used as reference points. The CT value for uninhibited samples was 33.7 ± 0.1. The same amount of SARS-CoV-2 synthetic RNA was also added in extracted RNA from wastewater samples and N1 RT-qPCR assay was repeated. Reactions were considered inhibited if the CT value of a wastewater RNA sample was greater than three CT value for the control sample (molecular grade water) (Staley et al., 2012). In this study, only one wastewater sample contained inhibitors of the RT-qPCR since the mean CT value for the spiked SARS-CoV-2 synthetic RNA was 39.7. The RNA sample was diluted 5-fold and 10-fold for re-analysis.
2.5. Detection and quantification of Phi6 phage RNA using RT-ddPCR
One-step RT-ddPCR approach with a QX200 ddPCR system (Bio-Rad, CA, USA) was used to quantify the Phi6 RNA to determine the recovery efficiencies for PEG concentration method. Each reaction contained a final concentration of 1 × Supermix (One-Step RT-ddPCR Advanced Kit for Probes, Bio-Rad, CA, USA), 20 U per ul of reverse transcriptase (RT) (Bio-Rad, CA, USA), 15 mM DTT, 900 nM of each primer, 250 nM of probe, 1 μl of molecular grade RNAse-free water, and 5.5 μl of template RNA for a final reaction volume of 22 μl. Droplet generation was performed by microfluidic mixing of 20 μl of each reaction mixture with 70 μl of droplet generation oil in a droplet generator (Bio-Rad, CA, USA) resulting in a final volume of 40 μl of reaction mixture-oil emulsions. The resulting droplets were then transferred to a 96-well PCR plate which was heat-sealed with foil and placed into a C1000 96-deep well thermocycler (Bio-Rad, CA, USA) for PCR amplification using the following parameters: 25 °C for 3 min, 50 °C for 60 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s and 60 °C for 1 min with ramp rate of 2 °C per second followed by a final cycle of 98 °C for 10 min. Following PCR, each 96-well plate was transferred to a QX200 Droplet Reader (Bio-Rad, CA, USA) for the concentration determination through the detection of positive droplets containing each gene target by spectrophotometric detection of the fluorescent probe signal.
2.6. Individual-level testing for students and university staff
During the Fall 2020 academic semester, Tulane University required COVID-19 nasal swab testing for all students, faculty, and staff. The frequency of testing varied among different groups of the community. Both on campus and off campus undergraduate students were required to get tested twice per week. Graduate and professional students, faculty, and university staff were required to get tested every two weeks. Frequency of testing was increased by the university when testing positivity rates were approximately 2% or higher than the positivity rate of the surrounding city of New Orleans.
2.7. Data analysis
All distributions of gene concentrations were non-normal. The ratio of SARS-CoV-2 to human fecal control (PMMV) concentrations were determined for each wastewater sample to account for differences in building occupancy and human waste input by dividing the concentration of N1 or N2 by the PMMV concentration for the same sample. If none of the gene targets, N1, N2, nor PMMV, were detected then sample was excluded from statistical analysis. To test the efficacy of the N1 and N2 genes in wastewater for indicating the number of COVID-19 cases, the gene concentrations with and without PMMV normalization were tested for correlation against the number of COVID-19 cases detected among residents in that dormitory for the same day as wastewater sampling occurred. For community-use buildings, the correlation was run against the total number of COVID-19 cases detected on campus for the same day as wastewater sampling. To assess correlation the Spearman R test was used. To assess the association of pH and turbidity with gene copy concentrations, pH and turbidity parameters were logarithmically transformed and a simple linear regression was run. For comparisons of gene copy concentrations between multiple buildings, the Kruskal-Wallis test was used. An alpha of 0.05 was used as the threshold of statistical significance for all tests. Statistical tests were performed using Prism (GraphPad).
3. Results
3.1. Wastewater characteristics and recovery of Phi6 in wastewater
The wastewater samples collected directly from manholes or sewer cleanouts of different buildings had an average pH of 8.68, and average turbidity of 125.66 NTU (Table S3). Wastewater samples collected in this study showed a wide range of turbidity, ranging from 2.52 NTU to 604 NTU. Little variation in pH was observed. Increasing pH was positively correlated with SARS-CoV-2 N2 gene concentrations in wastewater (p = 0.022, R2 = 0.0815) (Figure S1). pH was not correlated with N1 gene concentrations (p = 0.1353, R2 = 0.0387). Turbidity was not associated with N1 (p = 0.6097, R2 = 0.0046) or N2 (p = 0.4065, R2 = 0.0115) gene concentrations in wastewater.
The recovery of Pseudomonas phage Phi6 by PEG precipitation ranged from 2.1% to 33.7% (average of 15.2%) (Table S4). Viral recovery was not significantly affected by pH (P = 0.3995) or turbidity (P = 0.3801) of wastewater.
3.2. SARS-CoV-2 RNA in wastewater samples and COVID-19 cases on a university campus
Prior to the start of the semester, seven wastewater samples were collected. SARS-CoV-2 RNA was not detected in those samples (Table 1 ). During the Fall 2020 semester, 110 wastewater samples were collected from different buildings on campus. Three samples were negative for all viral gene targets (SARS-CoV-2 N1, N2 and PMMV genes). Of 107 viral positive wastewater samples, N1 and N2 genes were detected in 58 (54.2%) and 58 (54.2%) samples, respectively. Both N1 and N2 genes were detected in 53 (49.5%) samples. The average gene copies of N1 and N2 were 1.75 × 103 copies/100 mL and 6.76 × 103 copies/100 mL, respectively (Table 2 ). The average PMMV concentration detected in wastewater samples was 1.06 × 103 copies/100 mL (Table 2).
Table 1.
Monthly detection rates of SARS-CoV-2 RNA in campus wastewater samples and COVID-19 cases.
| Month | Number wastewater samples positive/Total number samples (%) |
COVID-19 Cases | |
|---|---|---|---|
| N1 | N2 | ||
| July | 0/7 (0%) | 0/7 (0%) | 0a |
| August | 2/5 (40%) | 2/5 (40%) | 269 |
| September | 20/35 (57%) | 18/35 (51%) | 478 |
| October | 15/27 (56%) | 15/27 (56%) | 183 |
| November | 18/35 (51%) | 20/35 (57%) | 526 |
| December | 3/8 (38%) | 3/8 (38%) | 20a |
aCOVID-19 individual-level testing began on July 28, 2020 and ceased on December 6, 2020.
Table 2.
Concentrations of SARS-CoV-2 (unnormalized N1, N2) and PMMV gene targets detected in wastewater from each building.
| Sample site | Site ID | N1 mean (range) Gene copies/100 mL | N2 mean (range) Gene copies/100 mL | PMMV mean (range) Gene copies/100 mL |
|---|---|---|---|---|
| Apartment Dorm 1 | AD1 (n = 9) | 2.67 × 102 (NDa-1.83 × 103) | 5.20 × 102 (ND-2.36 × 103) | 1.46 × 103 (ND-7.26 × 103) |
| Apartment Dorm 2 | AD2 (n = 13) | 2.32 × 101 (ND -3.00 × 102) | 8.16 × 101 (ND –1.06 × 103) | 2.07 × 103 (2.17 × 101–1.70 × 104) |
| Suite Dorm 1 | SD1 (n = 9) | 1.76 × 102 (ND – 9.42 × 102) | 1.17 × 102 (ND – 5.09 × 102) | 9.09 × 102 (1.12 × 102–2.40 × 103) |
| Suite Dorm 2 | SD2 (n = 8) | 8.25 × 101 (ND –5.07 × 102) | 1.30 × 102 (ND – 5.47 × 102) | 1.02 × 103 (2.22 × 102–1.65 × 103) |
| Communal Bathroom Dorm 1 | CD1 (n = 14) | 5.09 × 103 (ND – 3.68 × 104) | 7.90 × 103 (ND – 8.02 × 104) | 8.66 × 102 (ND – 5.74 × 104) |
| Communal Bathroom Dorm 2 | CD2 (n = 14) | 2.26 × 103 (ND – 3.02 × 104) | 1.47 × 103 (ND –1.39 × 104) | 4.66 × 102 (3.74 × 101–1.12 × 103) |
| Community Building 1 | CB1 (n = 13) | 2.25 × 101 (ND –1.16 × 102) | 9.70 × 101 (ND – 4.47 × 102) | 7.85 × 102 (ND –2.05 × 103) |
| Community Building 2 | CB2 (n = 13) | 6.24 × 102 (ND –6.04 × 102) | 2.75 × 103 (ND – 3.30 × 104) | 1.47 × 103 (2.83 × 103–7.10 × 103) |
| COVID-19 Isolation Dorm | ID (n = 14) | 5.27 × 103 (1.62 × 102–3.38 × 104) | 3.91 × 104 (ND – 3.56 × 105) | 7.50 × 102 (ND –4.29 × 104) |
ND denotes samples in which the gene targeted was below the limit of detection.
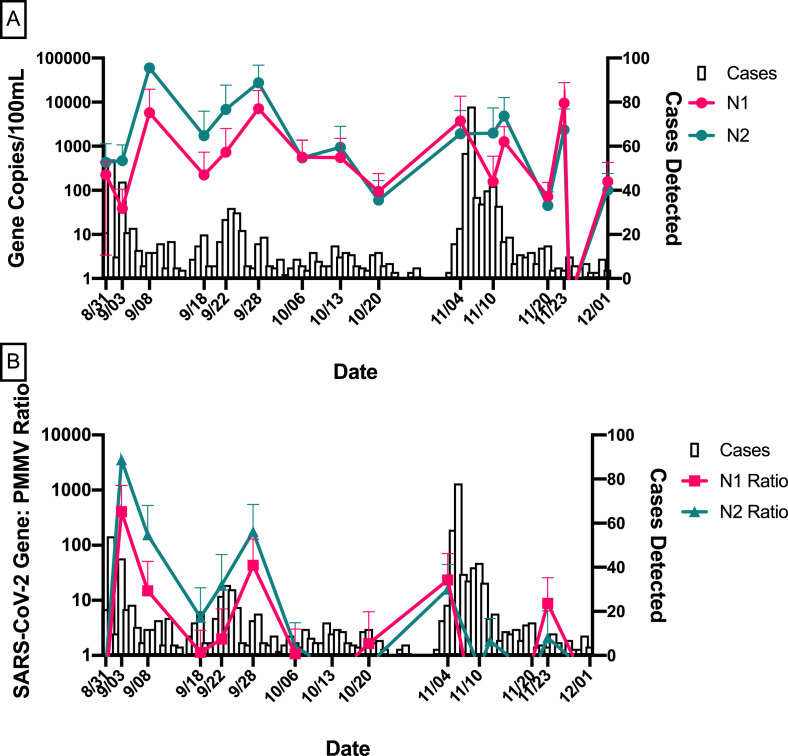
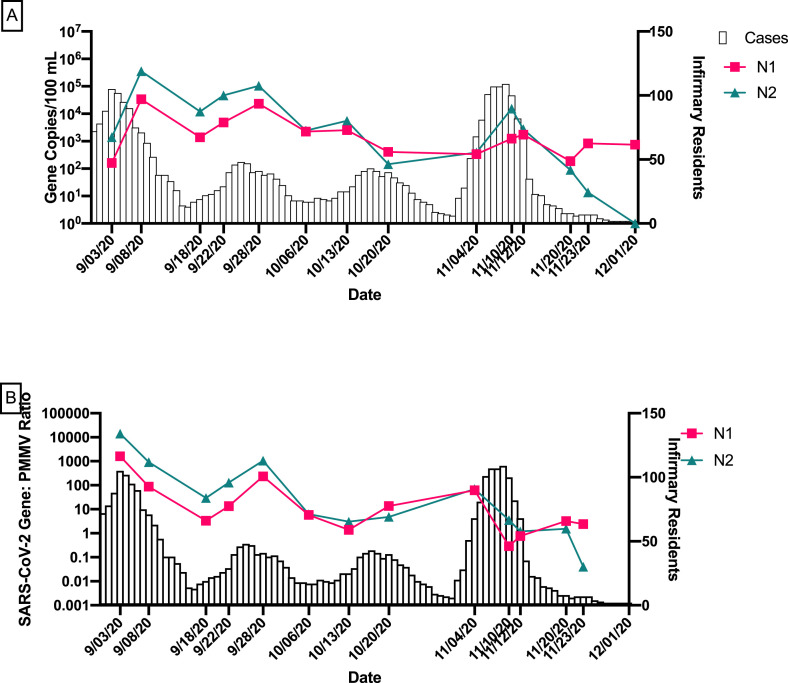
From August 1, 2020 to December 1, 2020, a total of 1476 cases of COVID-19 were detected among students, staff, and faculty at Tulane University (Table 1). In general, temporal trends of SARS-CoV-2 in wastewater, particularly, after human fecal marker normalization, mirrored trends in COVID-19 case data (Fig. 1 ).
Fig. 1.
Temporal trends in SARS-CoV-2 wastewater data and COVID-19 cases on campus from August to December 2020.
(A) Average N1 and N2 gene concentrations of all buildings for each sampling date with the number of COVID-19 cases detected on campus. (B) Average N1 and N2/PMMV ratios of all buildings for each sampling date with the number of COVID-19 cases detected on campus. Error bars indicate standard deviation.
3.3. The detection of SARS-CoV-2 RNA in wastewater and COVID-19 cases by dormitory and building use type
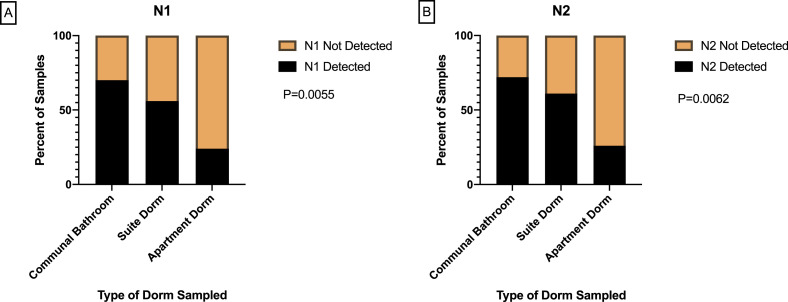
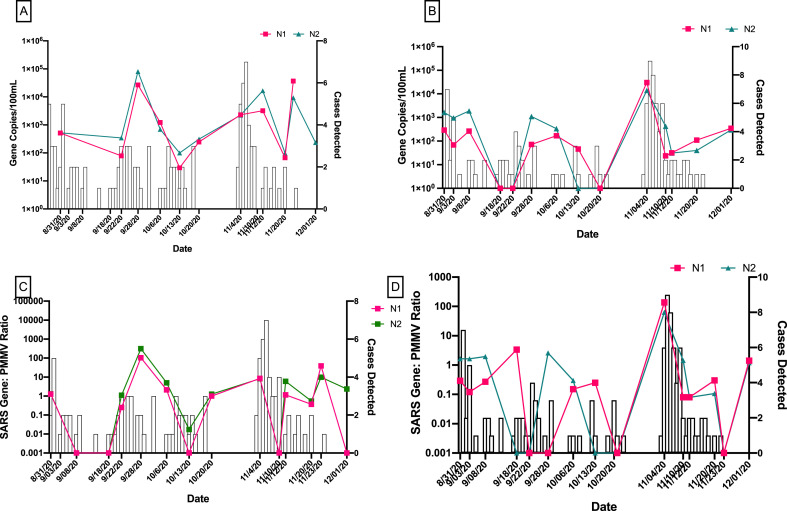
In this study, wastewater samples were collected from three dormitory types. The N1 gene was detected in 70% of wastewater samples taken from dormitories with communal bathrooms, 56% of samples from suite style dormitories, and 24% of apartment style dormitory samples (Fig. 2 A). The detection rates of N2 gene followed the similar trend as N1 with higher detection rate was observed in wastewater samples collected from dormitories with communal bathrooms (Fig. 2B). Type of dormitory was significantly associated with the frequency of the detection of N1 and N2 genes (P = 0.0055 and P = 0.0062, respectively). When comparing with other dormitory types, higher mean concentrations of SARS-CoV-2 RNA were detected in wastewater from dormitories with communal bathrooms (Table 2). For Communal Bathroom Dorm 2 in particular, both gene copies (P = 0.0006) and SARS-CoV-2/PMMV ratios (P = 0.0004) were significantly correlated with the number of COVID-19 cases detected among residents of that dormitory (Fig. 3 ).
Fig. 2.
Frequency of SARS-CoV-2 genes detection in wastewater by dormitory type.
(A) Frequency of N1 gene detection by dormitory types. (B) Frequency of N2 gene detection by dormitory types.
Fig. 3.
Temporal trends in SARS-CoV-2 wastewater data for dormitories with communal bathrooms and COVID-19 cases detected in those dormitories.
(A) N1 and N2 gene concentrations during Fall 2020 semester in CD1 wastewater (B) N1 and N2 gene concentrations during Fall 2020 semester in CD2 wastewater. (C) N1 and N2/PMMV ratios in CD1 wastewater. (D) N1 and N2/PMMV ratios in CD2 wastewater.
On average, N1 and N2 gene copy numbers in wastewater from both suite style and apartment style dormitories were lower than those in wastewater from dormitories with communal bathrooms (Table 2). The manholes used to collect wastewater from both suite style dormitories are municipal manholes, serving nearby neighborhoods. N1 and N2 genes were detected in both manholes when no COVID-19 cases were identified in the respective dormitories (Figure S2). N1 and N2 gene concentrations in wastewater from both suite style dormitories did not significantly correlate with the COVID-19 cases detected in those dormitories (Figure S2). A similar trend was observed for the apartment style dormitories (Figure S3).
The highest mean concentrations of SARS-CoV-2 RNA were detected in wastewater samples collected from COVID-19 isolation dormitory, a living space for students who have tested positive (Table 2). The N2 gene in wastewater was significantly correlated with the number of COVID-19 positive residents in the isolation dormitory when normalized with PMMV (P = 0.0480) (Fig. 4 ).
Fig. 4.
Temporal trends in SARS-CoV-2 wastewater data for COVID-19 isolation dormitory and the number of residents.
(A) N1 and N2 gene concentrations in COVID-19 isolation dormitory wastewater during Fall 2020 semester. (B) N1 and N2/PMMV ratios in COVID-19 isolation dormitory wastewater during Fall 2020 semester.
For community use buildings on campus, 10 out of the 26 (38.5%) analyzed wastewater samples tested positive for either one of the two nucleocapsid genes of SARS-CoV-2 (Table 2, Figure S4). The average concentrations of both genes ranged from 2.25 × 101 gene copies/100 ml to 2.75 × 103 gene copies/100 mL. These data suggest that infectious individuals, including those living off campus, utilized community spaces. Students who tested positive for COVID-19 were required to isolate.
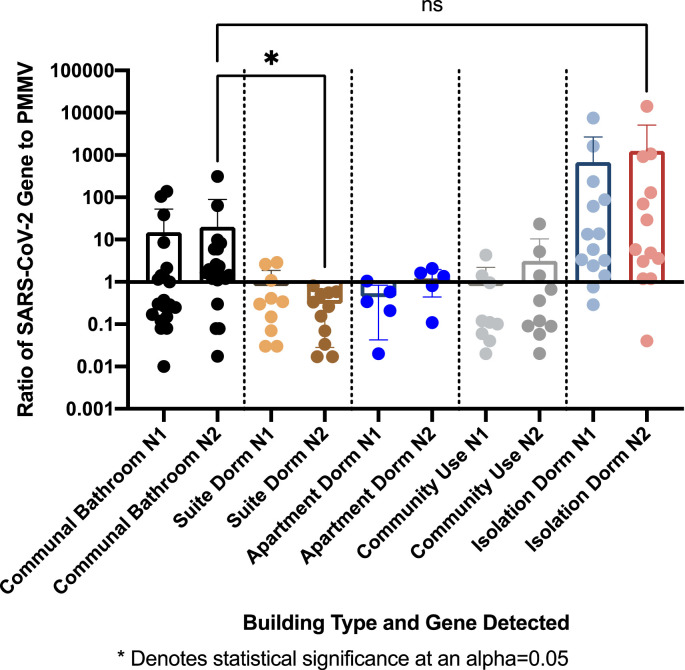
When normalizing SARS-CoV-2 concentrations using the PMMV control to account for changes in wastewater dilution and differences in human waste input in different buildings, the ratios of SARS-CoV-2 to PMMV varied significantly between all building types (Fig. 5 ). The N2/PMMV ratios were significantly higher in wastewater from dormitories with communal bathrooms than from suite style dormitories (P = 0.024). The N2/PMMV ratios were not significantly different in wastewater from dormitories with communal bathrooms and from the COVID-19 isolation dormitory (Fig. 5).
Fig. 5.
A comparison of SARS-CoV-2 gene/PMMV ratios by building use type.
3.4. The detection of SARS-CoV-2 and PMMV RNA in wastewater and COVID-19 cases by sewer access types
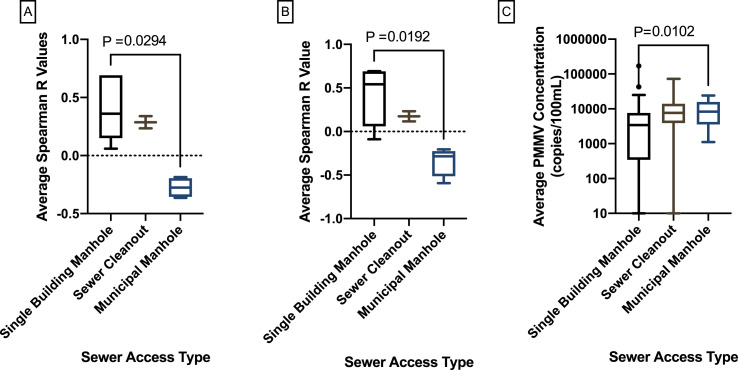
We also determined if the type of sewer access for sampling affects the detection of SARS-CoV-2 and PMMV RNA. The correlations between viral gene concentrations in wastewater from the type of sewer access (municipal manholes, manholes serving only a single building and sewer cleanout) and the respective number of COVID-19 cases were examined. Concentrations of PMMV RNA were significantly higher in wastewater samples collected from municipal manholes than those from single building manholes (P = 0.0102) (Fig. 6 ). Spearman R values for wastewater from each building and its associated sewer access type are shown in Table S5. Among tested dormitories, there was a significant stochastic difference in variation among Spearman R values for the three sewer access types (P = 0.0093) (Fig. 6). Spearman R values were significantly higher for N1 and N2 gene concentrations in wastewater samples from single building manholes than municipal manholes (P = 0.0294).
Fig. 6.
Comparison of Spearman R values for N1 and N2 genes by sewer access type.
(A) Comparison of Spearman R values for N1 and N2 gene concentrations vs. COVID-19 cases detected in dormitories on the same day for three sewer access types. (B) Comparison of Spearman R values for N1 and N2/PMMV ratios vs. COVID-19 cases detected in dormitories on the same day for three sewer access types. (C) Average PMMV RNA concentrations for each sewer access type for all dormitories.
3.5. Assessment of N1 and N2 gene assays in wastewater
The U.S. Centers for Disease Control and Prevention (CDC) currently recommends two different RT-PCR assays for the detection of SARS-CoV-2: N1 and N2 assays both targeting virus nucleocapsid gene. In this study, we compared the detection rates and gene concentrations using these assays to better understand if future wastewater surveillance studies require the analysis of two gene targets simultaneously. Across all wastewater samples, N1 and N2 gene concentrations did not significantly differ (P = 0.3572) (Figure S5). Concentrations of N1 (P < 0.0001) and N2 (P < 0.0001) genes in wastewater were significantly correlated with the number of COVID-19 cases detected in a dormitory (Table 3 ). The detection rates of N1 (OR = 3.958) and N2 (OR = 2.829) genes in wastewater significantly predicted the presence of COVID-19 cases in dormitories. This association was stronger when both genes were analyzed together (P = 0.0093). Together, the N1 and N2 genes had a negative predictive power of 61.3% and a positive predictive power of 71.4%.
Table 3.
Regression parameters and P-Values of regression analysis of N1 and N2 assays in wastewater.
| N1 (n = 81) |
N2 (n = 81) |
|||
|---|---|---|---|---|
| Statistical Parameter (95% CI) | P-Value | Statistical Parameter (95% CI) | P-Value | |
| Correlation R-value | 0.5067 (0.3170–0.6570) |
<0.0001 | 0.4790 (0.2837–0.6361) |
<0.0001 |
| Multiple Linear Regression R2 | −0.0002 (-0.0010–0.0006) |
0.6071 | 0.0002 (<0.0001–0.0003) |
0.0022 |
| Simple Logistic Regression ORa | 3.958 (1.556–10.55) |
0.0046 | 2.829 (1.132–7.299) |
0.0140 |
| Multiple Logistic Regression OR | 7.423 (1.014–150.2) |
0.0093 | 0.4943 (0.0244–3.597) |
0.0093 |
OR denotes Odds Ratio.
4. Discussion
The primary objective of this study was to evaluate the efficacy of targeted wastewater surveillance of SARS-CoV-2 RNA as an outbreak mitigation tool for COVID-19 on a university campus. Both SARS-CoV-2 N1 and N2 genes were detected in wastewater samples collected from dormitories and community-use buildings. Normalizing SARS-CoV-2 RNA concentrations using human fecal controls, PMMV increased the correlations between SARS-CoV-2 RNA concentrations in wastewater and the COVID-19 cases detected in the same building. This indicates that accounting for the quantity of human waste input is important when comparing virus concentrations between samples over time, particularly for grab sampling from sanitary sewers. PMMV was used because several studies have shown that PMMV is the most abundant RNA virus in human stool and a promising indicator of human fecal source (Colson et al., 2010; Rosario et al., 2009; Zhang et al., 2005). Unexpectedly, turbidity was not significantly associated with the detection of N1 and N2 genes in wastewater. Therefore, measurement of wastewater turbidity may not be as reliable of an indicator of fecal waste as fecal indicator viral molecular targets such as PMMV.
In this study, both SARS-CoV-2 N1 and N2 genes in wastewater were reliable markers for the presence of COVID-19 cases in dormitories. While highly correlated to each other, N2 gene was the more precise and reliable indicator of COVID-19 case numbers in a dormitory than N1 gene. Conversely, the N1 gene was the more reliable indicator of the presence or absence of any COVID-19 cases in a dormitory of the two genes. Combining N1 and N2 results from wastewater provided the most reliable prediction of the presence of COVID-19 cases in a dormitory, while N2 alone was the best tested predictor of the number of COVID-19 cases in a single dormitory. Depending on the action policy threshold for university outbreak surveillance teams, different gene target approaches could be taken. If the goal is to determine whether any cases of COVID-19 are present in a dormitory, both N1 and N2 assays could be used. If the desired metric is to estimate the number of COVID-19 cases present in a dormitory, the N2 assay alone could be employed. While N1 and N2 genes were not always significantly correlated with COVID-19 case numbers detected on the same day of wastewater sampling, it is clear that the trend of N1 and N2 gene concentrations in wastewater follow the trend of COVID-19 cases over time. The trend is best displayed in dormitories with greater resident density, such as CD1 and CD2, and the COVID-19 isolation dormitory. However, further research is needed to develop accurate and reliable wastewater models for the prediction of COVID-19 cases for a building-level by incorporating virus excretion rates in human stool and the persistence of SARS-CoV-2 in wastewater. Characterization of fecal load by demographic and temporally during infection will increase the power of future SARS-CoV-2 wastewater models. In this study, RT-qPCR assays targeting only the nucleocapsid gene of SARS-CoV-2 were used. Further wastewater monitoring to include the assessment of other gene targets, for example the envelope gene, to provide a complementary information regarding COVID-19 cases is warranted (Corman et al., 2020).
Manholes serving an individual building were the most efficient sewer access sites for targeted wastewater surveillance. Negative Spearman R values for municipal manholes and the number of COVID-19 cases detected in the same building may be due to background viral loads from surrounding neighborhood input. This is supported by the significantly higher PMMV concentrations found in municipal manholes as compared to single building manholes. The buildings SD1 and SD2, both served by municipal manholes, had positive detection of N1 and N2 genes when no cases of COVID-19 were detected in the respective dormitory. Sewer cleanouts presented another sampling challenge because of the pipe design and diameter. Despite the sampling constraints, PMMV concentrations in wastewater samples collected from the sewer cleanout did not significantly differ from either single building manholes or municipal manholes. In the development of targeted wastewater surveillance plan, we recommend identifying manholes or sewer cleanouts serving an individual building before resorting to sampling of municipal manholes.
We demonstrated that by combining wastewater surveillance, frequent testing of individuals, extensive contact tracing, and providing quarantine housing enabled us to monitor and mitigate COVID-19 outbreaks on a university campus. During this investigation, an outbreak was detected in the first week of November, shown both in case numbers of university-wide individual testing and wastewater data of dormitories with shared bathrooms (Fig. 3). The robust data offered by individual-level testing and wastewater surveillance allowed campus health officials to effectively mitigate the outbreak by enforcing more strict social distancing guidelines for the most affected dormitories. Case numbers in these dormitories, CD1 and CD2, dropped drastically after the implementation of these guidelines on November 5, 2020 (Fig. 3). The same outbreak was not detected in the wastewater for AD2 (Figure S3). We hypothesized that detected cases were contact traced, moved to a quarantine facility, and later tested positive. These detected cases would not have been staying in AD2 at the time of their case detection, but in a quarantine facility.
A secondary objective of this study was to determine the effects of different dormitory styles on the frequency of SARS-CoV-2 gene detection in wastewater and COVID-19 cases. While N1/PMMV ratios did not significantly differ across building types, N2/PMMV ratios were higher in communal bathroom dormitories than suite style dormitories. Additionally, N1 and N2 genes were detected more frequently in wastewater samples collected from communal bathroom dormitories than other dormitory types. Further investigation is required to determine if density of residents in a dormitory or behavioral quality of students in certain types of dormitories are associated with the increased COVID-19 transmission.
Ideally, for wastewater surveillance, composite samples should be collected using automated samplers, because grab samples may be less representative of community fecal contributions (Medema et al., 2020; Polo et al., 2020). However, the use of automated samplers is not always feasible particularly on an old university campus with different types of sewer access sites. Grab samples can be collected rapidly from manholes or sewer cleanouts. Further studies on the optimal sampling strategies and timing of sampling for targeted wastewater surveillance are warranted. In this study, we demonstrated that PEG precipitation could be a cost-effective concentration method for detecting SARS-CoV-2 in wastewater. The PEG method used in this study has been evaluated in a recent interlaboratory methods evaluation study and has shown a high degree of reproducibility across laboratories (Pecson et al., 2021). However, one of the limitations of the PEG method is that it is time consuming particularly with an overnight incubation. Our preliminary results showed that recovery efficiencies of PEG precipitation with and without an overnight incubation for Phi6 phage were not statistically significant (data not shown). This is also in agreement with other studies that reported a 2-h incubation for PEG method is sufficient for viruses (Deboosere et al., 2011; Polaczyk et al., 2008). Therefore, the wastewater sample processing time could be shortened to enable rapid public health response.
The true magnitude of asymptomatic cases of COVID-19 is currently unknown and limited diagnostic testing capacity results significantly in this uncertainty (Gillam et al., 2020; Li et al., 2020; Polo et al., 2020; Vang et al., 2021; Wilson et al., 2020). University campuses are considered hotbeds for COVID-19 transmission due to the prevalence of close-quarter social interactions within an age demographic associated with a low-risk of severe coronavirus infection (Fox, 2021; Venugopal et al., 2020; Wilson et al., 2020). While testing all members of a population on a regular basis is expensive, this method of outbreak surveillance has been essential for safe in-person university business to continue during the pandemic. As the vaccinated population increases and outbreaks become sparser, targeted wastewater surveillance could be a long-term, sustainable monitoring tool for universities to proactively detect disease prevalence in the community without constant, consistent individual testing. Wastewater surveillance for a combination of dormitories and mixed-use buildings could produce a comprehensive picture of the impact of COVID-19 on the university community.
In conclusion, we validated the use of targeted wastewater surveillance of SARS-CoV-2 RNA as a complementary method for detecting and managing COVID-19 transmission on a university campus. Wastewater surveillance can be conducted down to the resolution of a single dormitory, and community use buildings can be monitored as possible transmission hubs. Grab samples from various sewer access points can be utilized. The addition of human fecal indicator measurement enhances the correlation between SARS-CoV-2 N1 and N2 gene abundances in wastewater with COVID-19 cases. The combination of multiple surveillance tools with wastewater monitoring, such as individual testing, contact tracing, and quarantine facilities may enable university authorities to act quickly when facing with rapidly growing COVID-19 outbreaks.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by Tulane University School of Public Health and Tropical Medicine COVID-19 Rapid Response Grant. We thank Robert Dubroc, Chuck Fox, and Autin “Poochie” Willery, of the Tulane Campus Services, for assisting in weekly wastewater sampling. We also thank Ellie Kriner of the Tulane Campus Health for compiling data on COVID-19 cases.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envres.2021.111374.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K.A., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani H.T., Shi J., Shimko K.M., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J. Trav. Med. 2020;27 doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino de Carvalho N., Stachler E.N., Cimabue N., Bibby K. Evaluation of Phi6 persistence and suitability as an enveloped virus surrogate. Environ. Sci. Technol. 2017;51:8692–8700. doi: 10.1021/acs.est.7b01296. [DOI] [PubMed] [Google Scholar]
- Azimi P., Keshavarz Z., Laurent J.G.C., Stephens B.R., Allen J.G. Mechanistic transmission modeling of COVID-19 on the diamond princess cruise ship demonstrates the importance of aerosol transmission. Proc. Natl. Acad. Sci. 2021;118(8) doi: 10.1073/pnas.2015482118. p. e2015482118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt M.A., Spencer S.K., Hubbard L.E., Firnstahl A.D., Stokdyk J.P., Kolpin D.W. Avian influenza virus RNA in groundwater wells supplying poultry farms affected by the 2015 influenza outbreak. Environ. Sci. Technol. Lett. 2017;4:268–272. [Google Scholar]
- Carrillo-Reyes J., Barragán-Trinidad M., Buitrón G. Surveillance of SARS-CoV-2 in sewage and wastewater treatment plants in Mexico. J. Water Process Eng. 2020 doi: 10.1016/j.jwpe.2020.101815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Richet H., Desnues C., Balique F., Moal V., Grob J.-J., Berbis P., Lecoq H., Harlé J.-R., Berland Y., Raoult D. Pepper mild mottle virus, a plant virus associated with specific immune responses, fever, abdominal pains, and pruritus in humans. PloS One. 2010;5 doi: 10.1371/journal.pone.0010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. : Bull. Eur. sur les maladies transmissibles = Eur. Commun. Disease Bull. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Graber T.E., Mercier E., Montpetit D., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Servos M.R., Srikanthan N., Mackenzie M., Figeys D., Manuel D., Jüni P., Mackenzie A.E., Delatolla R. Catching a resurgence: increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deboosere N., Horm S.V., Pinon A., Gachet J., Coldefy C., Buchy P., Vialette M. Development and validation of a concentration method for the detection of influenza a viruses from large volumes of surface water. Appl. Environ. Microbiol. 2011;77:3802–3808. doi: 10.1128/AEM.02484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D.A., Ausiello D., Salzman J., Devlin T., Langer R., Beddingfield B.J., Fears A.C., Doyle-Meyers L.A., Redmann R.K., Killeen S.Z., Maness N.J., Roy C.J. Exhaled aerosol increases with COVID-19 infection, age, and obesity. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2021830118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorenko A., Grinberg M., Orevi T., Kashtan N. Survival of the enveloped bacteriophage Phi6 (a surrogate for SARS-CoV-2) in evaporated saliva microdroplets deposited on glass surfaces. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-79625-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti L., Wymant C., Kendall M., Zhao L., Nurtay A., Abeler-Dörner L., Parker M., Bonsall D., Fraser C. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368 doi: 10.1126/science.abb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox M.B., DC, Seamon M.D., Miranda M.L. MMWR; 2021. Response to a COVID-19 Outbreak on a University Campus-Indiana, August 2020; p. 118. Morbidity and Mortality Weekly Report. 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C., Lambirth K., Mittal N., Juel M.A.I., Barua V.B., Brazell L.R., Hinton K., Lontai J., Stark N., Young I., Quach C., Russ M., Kauer J., Nicolosi B., Akella S., Tang W., Chen D., Schlueter J., Munir M. 2021. Implementing Building-Level SARS-CoV-2 Wastewater Surveillance on a University Campus. medRxiv. 2020.12.31.20248843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam T.B., Cole J., Gharbi K., Angiolini E., Barker T., Bickerton P., Brabbs T., Chin J., Coen E., Cossey S., Davey R., Davidson R., Durrant A., Edwards D., Hall N., Henderson S., Hitchcock M., Irish N., Lipscombe J., Jones G., Parr G., Rushworth S., Shearer N., Smith R., Steel N. Norwich COVID-19 testing initiative pilot: evaluating the feasibility of asymptomatic testing on a university campus. J. Public Health. 2020;43(1):82–88. doi: 10.1093/pubmed/fdaa194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E, Kitajima M, Kishida N, Konno Y, Katayama H, Asami M, Akiba M. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Environ. Microbiol. 2013;79(23):7413–7418. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S.W., Ibrahim Y., Daou M., Kannout H., Jan N., Lopes A., Alsafar H., Yousef A.F. Detection and quantification of SARS-CoV-2 RNA in wastewater and treated effluents: surveillance of COVID-19 epidemic in the United Arab Emirates. Sci. Total Environ. 2021;764 doi: 10.1016/j.scitotenv.2020.142929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P.-Y., Rachmadi A.T., Mantilla-Calderon D., Alkahtani M., Bashawri Y.M., Al Qarni H., O'Reilly K.M., Zhou J. Estimating the minimum number of SARS-CoV-2 infected cases needed to detect viral RNA in wastewater: to what extent of the outbreak can surveillance of wastewater tell us? Environ. Res. 2021;195 doi: 10.1016/j.envres.2021.110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovi T., Shulman L.M., van der Avoort H., Deshpande J., Roivainen M., Em D.E.G. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol. Infect. 2012;140:1–13. doi: 10.1017/S095026881000316X. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W.J., Buisman A.M., Rutjes S.A., Heijne J.C., Teunis P.F., de Roda Husman A.M. Feasibility of quantitative environmental surveillance in poliovirus eradication strategies. Appl. Environ. Microbiol. 2012;78:3800–3805. doi: 10.1128/AEM.07972-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar M., Klapsa D., Wilton T., Akello J., Anscombe C., Allen D., Mee E.T., Minor P.D., Martin J. Isolation of vaccine-like poliovirus strains in sewage samples from the United Kingdom. J. Infect. Dis. 2018;217:1222–1230. doi: 10.1093/infdis/jix667. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Metcalf T.G., Melnick J.L., Estes M.K. Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology—a trip of over 50 years. Annu. Rev. Microbiol. 1995;49:461–487. doi: 10.1146/annurev.mi.49.100195.002333. [DOI] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell reports. Medicine. 2020;1 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., Darby E., Haas C.N., Amha Y.M., Bartolo M., Danielson R., Dearborn Y., Di Giovanni G., Ferguson C., Fevig S., Gaddis E., Gray D., Lukasik G., Mull B., Olivas L., Olivieri A., Qu Y., Sars-Cov-2 Interlaboratory C. Environmental Science: Water Research & Technology; 2021. Reproducibility and Sensitivity of 36 Methods to Quantify the SARS-CoV-2 Genetic Signal in Raw Wastewater: Findings from an Interlaboratory Methods Evaluation in the U.S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polaczyk A.L., Narayanan J., Cromeans T.L., Hahn D., Roberts J.M., Amburgey J.E., Hill V.R. Ultrafiltration-based techniques for rapid and simultaneous concentration of multiple microbe classes from 100-L tap water samples. J. Microbiol. Methods. 2008;73:92–99. doi: 10.1016/j.mimet.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., Romalde J.L. Making waves: wastewater-based epidemiology for COVID-19 - approaches and challenges for surveillance and prediction. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simon P., Allende A., Sanchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K., Symonds E.M., Sinigalliano C., Stewart J., Breitbart M. Pepper mild mottle virus as an indicator of fecal pollution. Appl. Environ. Microbiol. 2009;75:7261–7267. doi: 10.1128/AEM.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley C., Gordon K.V., Schoen M.E., Harwood V.J. Performance of two quantitative PCR methods for microbial source tracking of human sewage and implications for microbial risk assessment in recreational waters. Appl. Environ. Microbiol. 2012;78:7317–7326. doi: 10.1128/AEM.01430-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titcombe Lee M., Pruden A., Marr L.C. Partitioning of viruses in wastewater systems and potential for aerosolization. Environ. Sci. Technol. Lett. 2016;3:210–215. doi: 10.1021/acs.estlett.6b00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vang K.E., Krow-Lucal E.R., James A.E., Cima M.J., Kothari A., Zohoori N., Porter A., Campbell E.M. MMWR; 2021. Participation in Fraternity and Sorority Activities and the Spread of COVID-19 Among Residential University Communities — Arkansas, August 21–September 5, 2020; pp. 20–23. Morbidity and Mortality Weekly Report. 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal A., Ganesan H., Sudalaimuthu Raja S.S., Govindasamy V., Arunachalam M., Narayanasamy A., Sivaprakash P., Rahman P., Gopalakrishnan A.V., Siama Z., Vellingiri B. Novel wastewater surveillance strategy for early detection of coronavirus disease 2019 hotspots. Curr. Opin Environ. Sci. Health. 2020;17:8–13. doi: 10.1016/j.coesh.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Neyvaldt J., Enache L., Sikora P., Mattsson A., Johansson A., Lindh M., Bergstedt O., Norder H. Variations among viruses in influent water and effluent water at a wastewater plant over one year as assessed by quantitative PCR and metagenomics. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.02073-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Feng H., Zhang S., Ni Z., Ni L., Chen Y., Zhuo L., Zhong Z., Qu T. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the Coronavirus Disease 2019 outbreak in a Chinese hospital. Int. J. Infect. Dis. 2020;94:103–106. doi: 10.1016/j.ijid.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ma W., Zheng X., Wu G., Zhang R. Household transmission of SARS-CoV-2. J. Infect. 2020;81:179–182. doi: 10.1016/j.jinf.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany - suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E., Donovan C.V., Campbell M., Chai T., Pittman K., Seña A.C., Pettifor A., Weber D.J., Mallick A., Cope A., Porterfield D.S., Pettigrew E., Moore Z. Multiple COVID-19 clusters on a university campus — North Carolina, August 2020. MMWR. MMWR (Morb. Mortal. Wkly. Rep.) 2020;69:1416–1418. doi: 10.15585/mmwr.mm6939e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5 doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhang T., Breitbart M., Lee W.H., Run J.-Q., Wei C.L., Soh S.W.L., Hibberd M.L., Liu E.T., Rohwer F., Ruan Y. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2005;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., Wang Y.-Y., Xiao G.-F., Yan B., Shi Z.-L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microb. Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.