Abstract
Carbon turnover in aquatic environments is dependent on biochemical properties of organic matter (OM) and its degradability by the surrounding microbial community. Non-additive interactive effects represent a mechanism where the degradation of biochemically persistent OM is stimulated by the provision of bioavailable OM to the degrading microbial community. Whilst this is well established in terrestrial systems, whether it occurs in aquatic ecosystems remains subject to debate. We hypothesised that OM from zooplankton carcasses can stimulate the degradation of biochemically persistent leaf material, and that this effect is influenced by the daphnia:leaf OM ratio and the complexity of the degrading microbial community. Fresh Daphnia magna carcasses and 13C-labelled maize leaves (Zea mays) were incubated at different ratios (1:1, 1:3 and 1:5) alongside either a complex microbial community (<50 µm) or solely bacteria (<0.8 µm). 13C stable-isotope measurements of CO2 analyses were combined with phospholipid fatty acids (PLFA) analysis and DNA sequencing to link metabolic activities, biomass and taxonomic composition of the microbial community. Our experiments indicated a significantly higher respiration of leaf-derived C when daphnia-derived OM was most abundant (i.e. daphnia:leaf OM ratio of 1:1). This process was stronger in a complex microbial community, including eukaryotic microorganisms, than a solely bacterial community. We concluded that non-additive interactive effects were a function of increased C–N chemodiversity and microbial complexity, with the highest net respiration to be expected when chemodiversity is high and the degrading community complex. This study indicates that identifying the interactions and processes of OM degradation is one important key for a deeper understanding of aquatic and thus global carbon cycle.
Subject terms: Limnology, Biogeochemistry, Ecology, Microbiology
Introduction
Freshwaters are seasonally dynamic ecosystems that play an active role in biogeochemical cycles, such as organic matter (OM) sequestration or remineralisation. Even if lakes only make a small fraction of the global carbon (C) budget [1, 2], they are active ecosystems that transport, transform and store considerable amounts of C, and therefore have a significant effect on the global C cycle [3]. Thus, identifying interactions and processes impacting C fluxes is an important step towards a better understanding of global C cycling [4]. The C pool of aquatic environments is composed of autochthonous OM, such as algae, zooplankton and macrophytes, and terrestrially derived (i.e. allochthonous) OM, such as leaves and leaf leachate from the catchment area. Seasonally changing sources of OM (e.g., algae blooms, zooplankton successions, pollen deposition or litter fall) provide freshwaters with a continuously changing diversity of OM [5]. Aquatic OM turnover is mainly controlled by heterotrophic microbes, efficiently exploiting C and nutrients from different sources to meet their energy and growth demands [6–8].
The rates of microbial OM turnover depend on OM quantity and on the biochemical properties (e.g., molecular weight, chemical structure (monomeric, polymeric and colloidal) and stoichiometry) of the specific organic carbon compounds [9–11]. Autochthonous OM consists predominantly of low-molecular-weight compounds, including simple carbohydrates, amino acids and acetate [12]. Thus, autochthonous OM is generally easily accessible to microorganisms (i.e. bioavailable) ensuring a short turnover time in the environment [13]. In contrast, allochthonous OM (e.g., leaf litter) has a large fraction of insoluble, complex polymeric carbohydrates, such as lignin (15–40%), cellulose (10–50%) and hemicelluloses (30–40%) [14], which require specific, energetically expensive enzymes for microbial utilisation [15, 16]. Consequently, the insoluble fraction of terrestrially derived OM (i.e. mainly lignocellulose derivatives in leaf litter) accumulates in aquatic environments, forming a large pool of allochthonous OM [12]. Phytoplankton blooms, zooplankton successions and litter fall contribute to the seasonal dynamics of allochthonous and autochthonous OM pools in freshwater ecosystems and thus impact microbial auto- and heterotrophic activity [17]. This results in a complex series of cascading and partly unknown effects on OM bioavailability and chemodiversity influencing net C fluxes [18].
It has been hypothesised that an increase in chemodiversity, e.g., an addition of bioavailable OM to a system, provides the microbial community with additional energy and affects the net breakdown of biochemically more persistent OM, a concept termed the “priming effect” [19, 20]. Because priming effects can be positive (enhanced degradation of less bioavailable material upon the addition of easily accessible OM) or negative (decreased degradation of the less bioavailable fraction due to a preferential consumption of the more bioavailable OM), Bengtsson et al. [21] suggested to use the term “interactive effects” over “priming effect”, to avoid biasing the interpretation of results a priori. While the concept of interactive effects is widely accepted in soil sciences [22], its impact on net OM turnover in aquatic ecosystems is not obvious [21, 23–25]. Two meta-analyses concluded that interactive effects do not significantly increase net OM degradation rates in freshwater environments [21, 24]. The authors urge on the need to acknowledge microbial diversity and the range of different molecular compounds included in distinct OM sources [21], and to consider nutrient availability and stoichiometry as drivers of interactive effects [24]. Although numerous studies have evidenced interactive effects at the terrestrial–aquatic interface [26–29], there is little consensus about the role of interactive effects on net OM turnover, perhaps because of the lack of mechanistic understanding to predict where and when they can be expected [23].
Zooplankton carcasses are an abundant but poorly studied OM source in freshwaters [30]. While the exuviae of daphnids consists of relatively persistent chitin (3–7% of total daphnia biomass), most of the biomass comprises proteins (30% upward), lipids (up to 20%) and carbohydrates (10–30%) and hence represents a mainly easy-to-degrade and bioavailable OM source [31, 32]. Zooplankton experiences non-predatory mortality, resulting from pathogen infections, starvation or environmental stress [30, 33], with a significant fraction of carcasses persisting for several days in the water column before sedimentation [34]. Zooplankton carcasses represent hotspots of OM turnover, providing the microbial community with a growth structure (i.e., the chitin exuviae) facilitating attachment and colonisation, and a diversity of nutrient-rich OM compounds [32, 35, 36]. Seasonal fluctuations in zooplankton-derived OM trigger shifts in microbial community composition [37], but whether it can stimulate the degradation of less bioavailable, leaf-derived OM remains unknown.
Eukaryotic microorganisms (e.g., fungi, oomycetes and protozoa) play an important role in the degradation of terrestrially derived OM and stimulate the turnover of OM [10, 38, 39]. Fungi are well-characterised degraders of various terrestrially derived OM, via the production of extracellular ligninolytic oxidative enzymes, which can physically penetrate and break down colloidal and particulate OM [40–42]. Although the microbial degradation of lignocellulose has been intensively studied in fungi [15, 39, 43], there is emerging evidence that bacteria are involved in delignification [44, 45]. Protozoa do not directly consume particulate OM or complex macromolecules, like lignocellulose derivatives, yet, by grazing on bacteria and excreting secondary metabolites they have significant effects on the bacterial community structure, the composition of the C and nutrient pools and, consecutively, on net degradation rates of OM [38, 46]. Therefore, the complexity of microbial communities and interactions should be considered when studying aquatic OM turnover and C fluxes.
Using a stable-isotope and sequencing approach, we incubated a complex microbial lake community and a simplified microbial (mostly small bacteria) community with Daphnia magna carcasses (acting as a predominantly bioavailable OM pool) and Zea mays-derived lignocellulose (as a biochemically persistent OM pool) in different concentrations. We selected a gradient of daphnia- to leaf-derived OM ratios (1:1, 1:3 and 1:5), to simulate interactive effects occurring at different seasonal states [30, 33, 34]. These range from high zooplankton-derived OM abundances during the clear-water phase (i.e., 1:1 ratio) to lower concentrations (i.e., ratios of 1:3 and 1:5) resulting from the subsequent decline of zooplankton populations. The maize leaves were 13C-labelled to trace its microbial mineralisation via 13CO2 measurements and microbial assimilation by membrane phospholipid fatty acid (PLFA) analysis. PLFAs, found in cell membranes of all living organisms, rapidly degrade upon cell death [47], and therefore serve as an indicator for living biomass. Isotope analyses of PLFAs follow the assimilation of C from different substrates (distinct in their isotopic signature) into the biomass of different microbial groups (e.g., eukaryotic vs. prokaryotic). Based on previous findings [10, 38, 48], we expect that a complex microbial community (including eukaryotic microorganisms and bacteria) will exploit leaf-derived C more efficiently compared to a simplified, bacteria-dominated community. We hypothesised that a complex and chemodiverse system combining both, a predominantly bioavailable source of OM (i.e., zooplankton carcasses) and a rather persistent OM source (i.e., maize leaf-derived lignocellulose), would lead to a higher utilisation of the persistent OM source. Furthermore, we examined how seasonal dynamics of zooplankton OM availability, reflected by the ratios of daphnia- to leaf-derived OM (D:L), influence the direction and magnitude of interactive effects. Zooplankton-derived OM is often overlooked as it is short-lived and comprises only a small proportion of the total OM pool. However, mass mortality events, much like phytoplankton blooms, can result in drastic shifts in OM composition and stimulate microbial activity which, in the short term, promotes the turnover of persistent OM.
Methods
Experimental setup
Incubated microbial communities originated from the north-eastern (NE), less acidic basin of Lake Große Fuchskuhle (Northeastern Germany, 53°06′N 12°59′E). This bog lake receives large amounts of terrestrially derived OM and its microbial community is therefore adapted to persistent terrestrial OM [49]. The water for inoculum preparation was sampled from the shore, where the microbial community is in close contact with the surrounding accumulated leaf litter and, thus, expected to be well adapted to leaf-litter degradation. Two inoculum solutions were prepared: one with a complex microbial community (including eukaryotic microorganisms like protozoa and fungi) and another with a solely bacterial community (see protocol in the Supplementary). Incubations were performed on a roller apparatus in the dark at temperatures fluctuating between 16 and 18 °C. One litre acid-washed and muffled glass bottles were filled (air-bubble free) with artificial acidic lake water [50] adjusted to the pH of the NE basin of Lake Große Fuchskuhle (i.e., 5.7) and inoculated with 1 ml of bacterial or total microbial community concentrate accordingly. We estimated the initial cell density in the total microbial community treatments to be in the range of 2.5 × 106 cells ml−1 and roughly 106 cells ml−1 in the bacterial treatment (see Supplementary Information for calculations). Even though these estimates are lower than natural bacterial concentrations in Lake Große Fuchskuhle (~5 × 107 cells ml−1), they are still in the range of natural bacterial densities.
Cultured Daphnia magna was freeze-dried, to allow a precise determination of the added dry weight, and used as a source for zooplankton-derived OM. We selected maize leaves (Zea mays, ~10 atom% 13C, IsoLife bv) as a model substrate for lignocellulose derivatives, which are the predominating C compounds in leaf litter and the 13C enrichment makes it possible to determine its specific utilisation. The maize leaves were shredded and leached for 48 h in the dark in distilled water at room temperature to remove all soluble compounds [51], and ensure that the remaining leaf material consisted mainly of insoluble lignocellulosic compounds. All treatments were amended with the 6 mg l−1 of OM, but using three different daphnia- to leaf-derived OM ratios: 1:1, 1:3 and 1:5 (see Table 1). Three additional treatments with only daphnia, only leaves and a blank served as controls. For each treatment, four initial replicates (ninitials = 4), four final replicates (nfinals = 4) and one additional bottle for daily monitoring were prepared, sealed with PTFE-coated silicone septa and placed horizontally on a roller apparatus at the above-mentioned incubating conditions. The four initial replicates were sacrificed on the same day to measure initial values. The daily monitoring bottles served to measure dissolved gas concentrations (i.e., O2, 12CO2 and 13CO2) to ensure that the incubations remained oxic and to follow the daily microbial utilisation of daphnia- and leaf-derived OM.
Table 1.
Overview on the different treatments.
| Treatment | b-DL1:1 | DL1:1 | DL1:3 | DL1:5 | Daphnia | Leaves | Blank |
|---|---|---|---|---|---|---|---|
| Inoculum | Bacterial community < 0.8 µm | Complex microbial community < 50 µm | |||||
| Daphnia:leaves ratio | 1:1 | 1:1 | 1:3 | 1:5 | Controls | ||
| Daphnia (mg l−1) | 3 | 3 | 1.5 | 1 | 3 | – | – |
| 13C leaves (mg l−1) | 3 | 3 | 4.5 | 5 | – | 3 | – |
Replicates: 4 initials, 4 finals and 1 additional bottle for daily gas measurements (O2, CO2). Blank: water with complex microbial community, but without addition of daphnia or leaves; Daphnia: daphnia only; D:L: daphnia and leaves in a given ratio with a complex microbial community; Leaves: leaves only; b-DL1:1: daphnia and leaves (ratio 1:1) in bacterial community only.
Dissolved O2 content was measured daily in one bottle of each treatment (monitoring bottle) by inserting a microfibre optic oxygen sensor (Microx TX3, PreSense, Regensburg, Germany) through the septa. Subsequently, dissolved O2, 12CO2 and 13CO2 were measured via a membrane inlet mass spectrometer (MIMS, Bay Instruments, Maryland, US). MIMS data were expressed as ratios to argon (Ar), as Ar is an inert gas that is not affected by any biochemical process. The daily O2, 12CO2 and 13CO2 measurements served as criteria to evaluate at which time point interactive effects would be most evident and to stop the incubations at that moment. The experiment was finalised after 11 incubation days. On the last incubation day, samples for isotopic analyses of dissolved CO2 were taken before sacrificing the bottles for final sampling. For analyses of particulate organic carbon (POC), PLFA and DNA, the bottles were shaken thoroughly and the whole content was filtered on muffled GF/75 filters (Advantec, 47-mm diameter, nominal pore size of 0.3 µm). The filters were then freeze-dried and subsampled for 13C, DNA and PLFA analyses. The flow-through was collected to measure dissolved organic carbon (DOC) and dissolved nitrogen with a TOC-L analyser (Shimadzu, Kyoto, Japan).
Stable carbon isotope analyses and mixing model
Stable carbon isotopes were measured as gaseous (i.e., dissolved inorganic C) and particulate phase at the UC Davis Stable Isotope Facility. Water for dissolved CO2 analyses was sampled on the initial and final day, filled in 12-ml Labco Exetainer vials and fixed with ZnCl2. The samples were analysed for δ13C and ppm CO2 using a GasBench II system interfaced to a Delta V Plus isotope ratio mass spectrometer (Thermo Scientific, Bremen, Germany). Concentrations of CO2 in the water samples were calculated as described for headspace equilibration techniques by Oil and Halbedel [52].
For solid 13C analyses, freeze-dried GF/75 filters were encapsulated in tin capsules (EA Consumables, Inc.) and measured using an Elementar Vario EL Cube elemental analyser (Elementar Analysensysteme GmbH, Hanau, Germany) interfaced to a PDZ Europa 20–20 isotope ratio mass spectrometer (Sercon Ltd., Cheshire, UK). The daphnia carcasses and leaves used for the incubations were also sent for 13C measurement. The measuring accuracy is 0.2 per mill.
The formula for calculations of the direction and magnitude of non-additive interactive effects was adapted from Bengtsson et al. [21] as follows:
| 1, |
where IE refers to interactive effects (in %), CO2(treatment) is the CO2 production in the treatment of interest (i.e., where interactive effects are expected) and CO2(controls) is the sum of produced CO2 in the daphnia and leaf controls. For treatments DL1:3 and DL1:5 for which we did not have controls with the same amount of daphnia and leaves, CO2(controls) was calculated from CO2 production per mg daphnia and leaves.
To estimate the amount of daphnia- and leaf-derived POC and to trace the C source of the measured CO2, we used a two-source mixing model approach proposed by Cheng [53]. In treatments where daphnia and leaves were mixed, we calculated the CO2 fraction of the desired source as
| 2 |
When source A is daphnia, source B is leaves and vice versa. For daphnia- and leaf-derived POC, we used δ13C instead of δ13CO2 in Eq. 2. We did not apply the mixing model to the daphnia and leaf controls, but used total POC and CO2 concentrations as they contain only one C source. Based on Eq. 2, we calculated the normalised concentration of CO2 of the C source of interest as
| 3 |
A summary of the calculation steps for all samples is provided in Supplementary Table 1.
Phospholipid-derived fatty acids analyses (PLFA)
Microbial cell enumeration can be difficult, particularly when considering partially particulate and colloidal OM sources such as daphnia carcasses. Previous studies demonstrated a clear suitability of PLFAs for this purpose [10, 54, 55]. Thus, we used PLFAs as an indicator for microbial biomass. Furthermore, PLFA can differentiate between major groups of organisms (e.g., prokaryotes vs. eukaryotes) at low taxonomic resolution [56]. Linking PLFAs with 13C analyses (i.e., compound-specific isotope analyses) reveals the C sources utilised by the different organisms to build up biomass. PLFAs were extracted from GF/75 filters for initial and final samples [57] and measured as fatty acid methyl esters (FAMEs) on a gas chromatographer (Agilent 6890, Germany) equipped with a silica capillary column (CP Sil 88 for FAME, Agilent) coupled to a mass-selective detector (Agilent 5973-N, Germany). FAMEs were identified by comparing retention times and mass spectra with a standard mixture of FAME (Bacterial Acid Methyl Esters Mixture, Matreya LLC, State College, US) and quantified using FAME-specific calibration curves. For the compound-specific isotope analyses, the four replicates were pooled due to the low concentrations of PLFAs in the samples. Individual PLFAs were analysed using a gas chromatographic system (Agilent 7890 Series GC) coupled via a combustion interface to a Finnigan MAT 253 isotope ratio mass spectrometer. Isotope ratios of individual biomarkers were expressed as 13C values in per mill relative to the V-PDB standard. The omega nomenclature was used to describe the detected FAMEs.
DNA extraction and sequencing
Total DNA was extracted according to a modified protocol described by Nercessian et al. [58]. Briefly, filters and zirconia–silica beads were suspended in extraction buffer (CTAB), to which sodium dodecyl sulfate, lauroyl sarcosine and phenol–chloroform–isoamylalcohol were added before homogenising and centrifuging the samples. The aqueous phase was first transferred and washed with chloroform–isoamylalcohol and subsequently with polyethylene glycol. Nucleic acids were precipitated at 4 °C, washed with ethanol, air-dried and finally dissolved in ultra-pure water. The detailed protocol is available in the Supplementary Information. PCR, library preparation and sequencing was done by MrDNA (USA). The V3–V4 region of the 16S rRNA gene was amplified using bacterial primers 341F-785R [59], followed by library preparation (2 × 300 bp) and sequencing on a MiSeq (Illumina) platform. Fungal primers ITS4ngs (forward) and LF402 (reverse) were used for fungal community analysis [60].
Statistical tests and bioinformatic approaches
Statistical significances and t values of O2, 13CO2 and PLFA concentrations were calculated using a two-way ANOVA test (Bonferonni’s multiple comparisons). Sequences were processed with the dada2 (ver. 3.6) R package [61] and taxonomy was assigned using the R package DECIPHER (ver. 2.16.1) [62]. Sequences were run against the SILVAv132 and LSUv11 database for identification of bacterial and fungal taxa. We tested for treatment-specific effects on both fungal and bacterial community composition using PERMANOVA following Hellinger transformation of the OTU matrix, using the Primer6 software package (Primer-E Ltd, Plymouth). Treatment-specific OTU enrichment was assessed using the multipatt function of the indicspecies (ver. 1.7.1) R package [63]. Specifically, relative OTU abundances were used to identify treatment-specific indicator species, allowing for multigroup comparisons. False discovery was minimised by applying a Bonferroni adjustment.
We used the WGCNA (ver. 1.68) R package for weighted correlation network analysis [64] to construct a topological overlap matrix (TOM) and test for the effects of external variables on the presence of different taxonomic groups. Briefly, rare OTUs (<0.01%) were excluded and samples were hierarchically clustered using the hclust function to identify outliers. We related external variables (such as daphnia or leaf-derived CO2 concentrations, O2, presence/absence of leaves or daphnia carcasses) to the samples and used this relationship as input for network construction and module detection. Network construction was performed using a soft-thresholding power of 6 and modules were detected using the TOM type unsigned and a minimum module size of 30. Modules were related to external variables by calculating module eigengenes and correlating those with the external variables. The results were visualised in a heatmap, where values of 1 correspond to a positive module-variable correlation and −1 to a negative correlation. The OTUs comprised within a module of interest were extracted and, thus, allowed to identify the microbial community related to specific variables.
Results
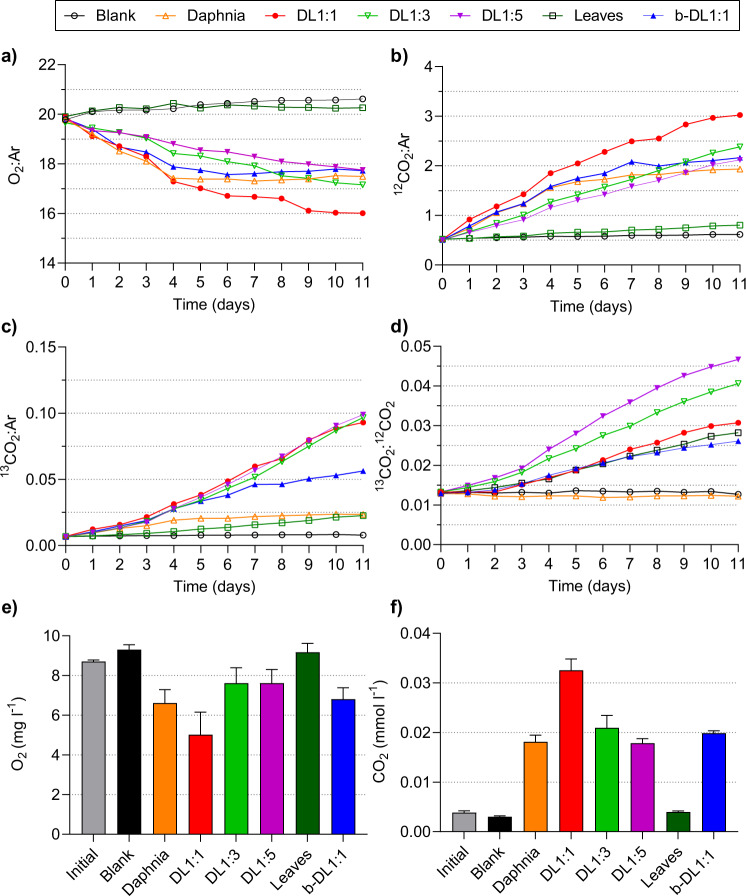
Throughout 11 days of incubation, respiration rates of the microbial community, expressed either as O2 consumption or CO2 production, increased with increasing D:L ratios. The highest respiration rates were measured in the complex community treatment DL1:1, followed by treatments DL1:3 and DL1:5 (Fig. 1, Supplementary Fig. 1). O2 concentrations in the leaf controls were not significantly different from the blanks or the initials (Supplementary Table 2). In the daphnia control and bacterial treatment (b-DL1:1), respiration rates were high until day 4 but stagnated until the end of the incubation (Fig. 1a, b, Supplementary Fig. 1). The final O2 and CO2 concentrations measured on the last day in the replicated treatments (Fig. 1 e, f) supported the measurements in the daily monitoring bottles, i.e., the highest respiration in the complex community treatment DL1:1, comparable O2 and CO2 concentrations between the bacterial treatment (b-DL1:1) and the daphnia controls as well as between the blanks and the leaf controls. The initial C/N ratio of daphnia carcasses was 3.5 ± 0.15 (n = 5) and of the leaves 25.2 ± 2.2 (n = 5). DOC was (on purpose) very low and was completely consumed during the incubation (i.e., 0–0.5 mg l−1 in the finals, Supplementary Table 3). Hence, C was either incorporated into biomass (PLFAs) or respired (CO2).
Fig. 1. Microbial respiration across treatments during the incubation.
Respiration measured daily in the monitoring bottles (not replicated) as ion currents (m/z ratios) and expressed as ratios of a O2 to Ar, b 12CO2 to Ar, c 13CO2 to Ar and d 13CO2 to 12CO2. Concentrations of O2 (e) and CO2 (f) measured in replicates (n = 4) at the beginning and at the end (day 11) of the experiment for each treatment. Blank: water with complex microbial community, but without addition of daphnia or leaves; Daphnia: daphnia only; D:L: daphnia and leaves in a given ratio with a complex microbial community; Leaves: leaves only; b-DL1:1: daphnia and leaves (ratio 1:1) in the bacterial community only.
The turnover of POC increased with higher abundance of Daphnia carcasses, i.e., the final POC concentrations were the lowest in the daphnia-rich treatments (b-DL1:1, DL1:1) (for more information see the POC section of the Supplementary Information, Supplementary Fig. 2, Supplementary Tables 4 and 5). When comparing additive (i.e., sum of CO2 produced in separated daphnia and leaf controls) to non-additive CO2 production (i.e., the amount of CO2 produced in treatments where daphnia and leaves were mixed) in the DL1:1 treatment, we observed non-additive (interactive) effects of 98% (Fig. 2a). Because we only had controls for the DL1:1 treatment (i.e., 3 mg of leaves and 3 mg of daphnia in separated bottles), we calculated interactive effects normalised to the POC amounts as pseudocontrols for DL1:3 and DL1:5. The calculated interactive effect for DL1:3 was 131% and for DL1:5 178%. In the bacterial treatment (b-DL1:1), the non-additive effect accounted for 10% of CO2 production. However, since we only had controls with a complex microbial community, the magnitude of this effect might vary when comparing to controls with a reduced community complexity (bacterial community). CO2 derived from daphnia degradation was always significantly higher than leaf-derived CO2 of the same treatment (Fig. 2b, Supplementary Table 6). Absolute and normalised respiration rates from both daphnia- and leaf-derived CO2 were always significantly higher in the complex community treatment (DL1:1) compared to bacteria (b-DL1:1) (Fig. 2b, c, Supplementary Table 6). The highest absolute values of daphnia-derived CO2 were measured in the DL1:1 treatment (Fig. 2b). However, after normalising CO2 concentrations to the amount of daphnia added to the treatments, utilisation efficiency of daphnia-derived C increased with decreasing amounts of daphnia material in the treatments (i.e., the highest daphnia-derived CO2 concentrations in the DL1:5 treatment) (Fig. 2c). The highest leaf-derived CO2 concentrations per amount of leaves added were measured in the treatment with a high D:L ratio (i.e., DL1:1).
Fig. 2. Interactive effects and source-specfic CO2 production.
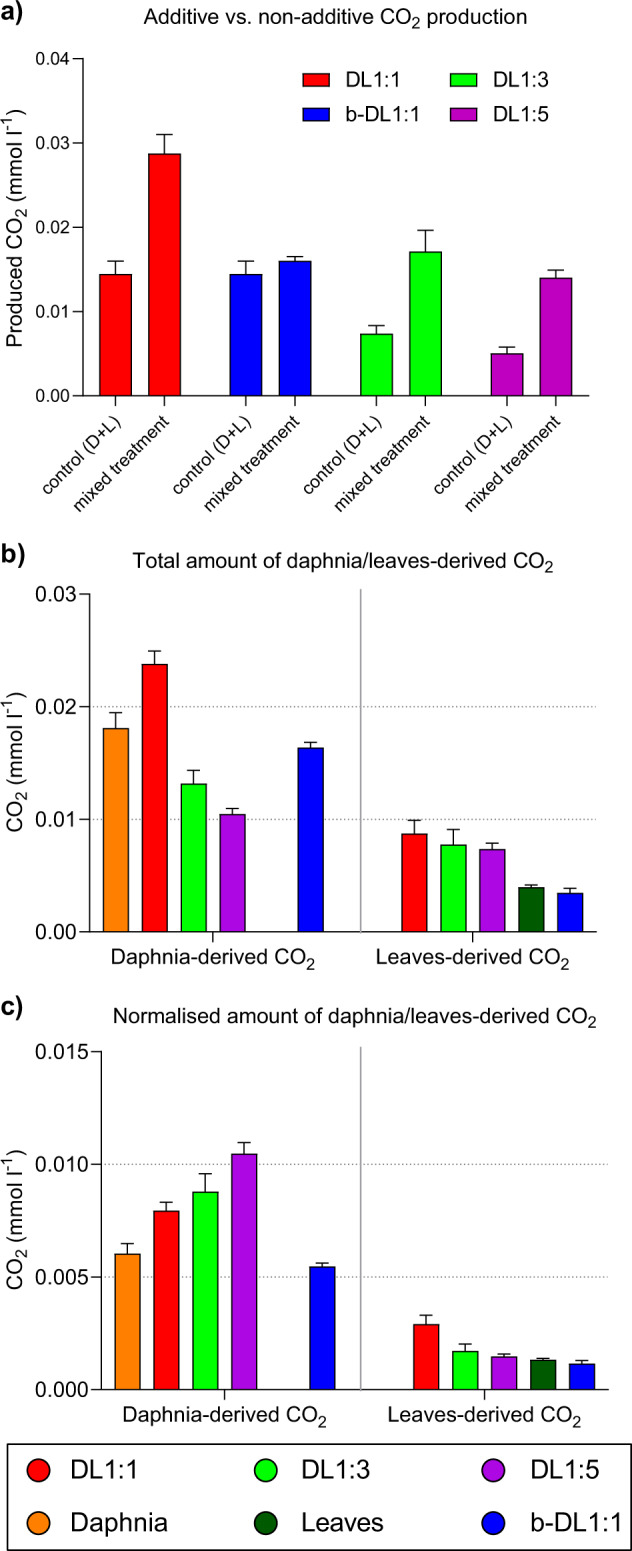
a Non-additive interactive effects measured as CO2 production in daphnia and leaves combined treatments compared to the sum of CO2 produced when daphnia and leaves were isolated (for DL1:3 and DL1:5, controls were calculated based on the CO2 production per mg daphnia or leaves in the controls for treatment DL1:1). a CO2 concentrations derived from the different carbon sources calculated with the two-source mixing model for the total amount of respired CO2 and b normalised by the amount of daphnia and/or leaf material in each treatment (n = 4). Daphnia: daphnia only; D:L: daphnia and leaves in a given ratio with a complex microbial community; Leaves: leaves only; b-DL1:1: daphnia and leaves (ratio 1:1) with a bacterial community only.
Some PLFAs have been commonly used as markers for fungal (e.g., 18:2ω6,9) or bacterial biomass (e.g., i15:0), while other unspecific PLFAs are generally used as universal biomarkers, covering prokaryotes and eukaryotes. By applying a cluster analysis to all PLFAs, we obtained two groups, which we assigned to eukaryotic and bacterial markers accordingly (Supplementary Fig. 3). The PLFAs 18:1ω9t and 18:1ω7c could not be separated chromatographically, thus they will be referred to as 18:1ω9t/7c further on. One cluster was composed of the PLFAs 18:1ω9t/7c and 18:2ω6,9, which are known to be present in daphnia [65] and fungal biomass [54]. Thus, these two PLFAs were used as indicators of eukaryotic biomass. The second cluster consisted mostly of bacteria-specific PLFAs (i15:0, a15:0, cy-17:0) and some unspecific PLFAs (16:1ω9, 18:1ω9c). Since the unspecific PLFAs clustered with bacterial-specific PLFAs and also showed a significant increase in the bacterial treatment, they were used as markers of bacterial biomass.
The initial samples revealed high abundances of eukaryotic PLFAs corresponding to the amendment of daphnia biomass to the treatments (Fig. 3a). Thus, the highest eukaryotic PLFA (i.e., mainly daphnia-derived PLFAs) concentrations were measured in the DL1:1 treatments (bacterial and complex microbial community) and the daphnia control (which corresponds to treatments where 3 mg of daphnia were added), but decreased with a lower D:L ratio. The final eukaryotic PLFA concentrations were not significantly different across treatments (daphnia, DL1:1, DL1:3, DL1:5 and b-DL1:1). When we consider the large contribution of daphnia carcasses to the eukaryotic PLFAs, the significantly lower final concentrations compared to the initial concentrations (Supplementary Table 7), indicate an efficient degradation of the initial daphnia biomass. Initial bacterial PLFA concentrations decreased with decreasing amounts of daphnia carcasses in the treatments (Fig. 3b), suggesting that the daphnia carcasses had an associated bacterial community. The final bacterial PLFA concentrations increased significantly in all DL treatments during the incubation period (Fig. 3b, Supplementary Table 7). A particularly steep increase of bacterial PLFA was observed in the bacteria-only treatment, which is mainly attributed to the enrichment of the bacteria-specific PLFAs i15:0 and cy-17:0 and the unspecific PLFAs 16:1ω9 and 18:1ω9 (Fig. 3c).
Fig. 3. Changes in PLFA compositions during the incubation.
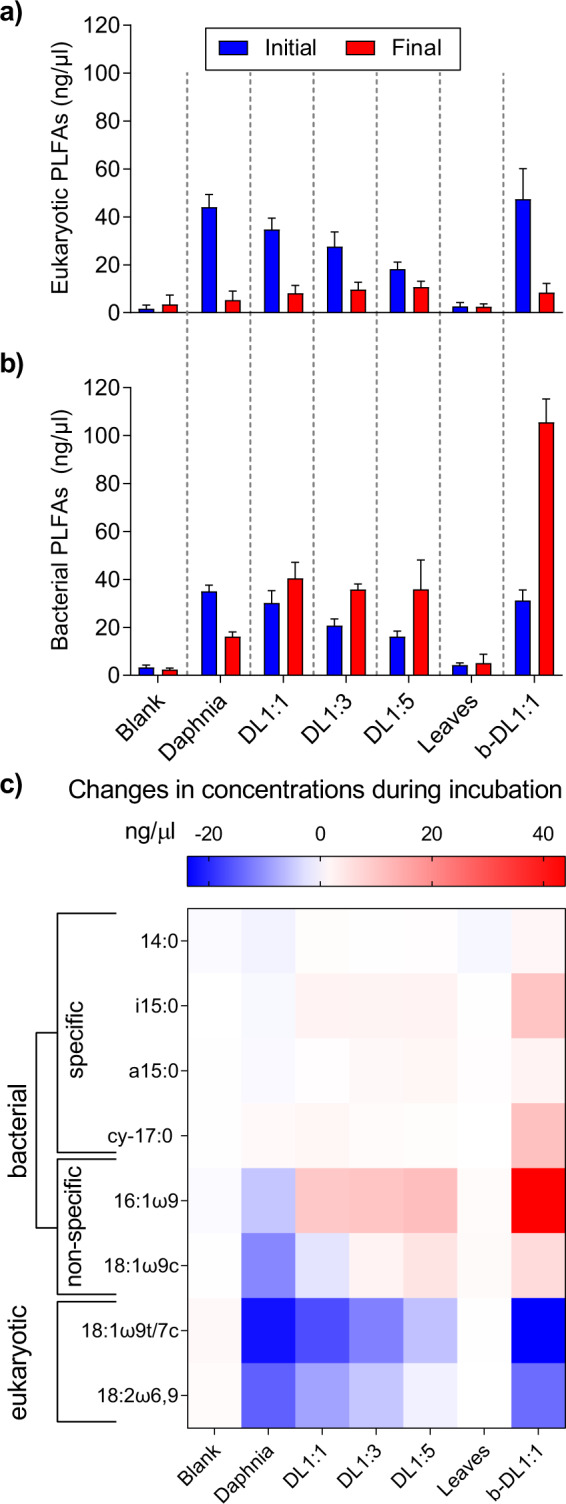
Initial and final concentrations of a eukaryotic and b bacterial PLFAs; c concentration changes of specific PLFA across treatments during the incubation. Blank: water with complex microbial community, but without addition of daphnia or leaves; Daphnia: daphnia only; D:L: daphnia and leaves in a given ratio with a complex microbial community; Leaves: leaves only; b-DL1:1: daphnia and leaves (ratio 1:1) with a bacterial community only.
We generally observed a higher enrichment of the 13C label in the final samples compared to initials (Fig. 4). The amount of daphnia-derived C in two PLFAs (i.e., eukaryotic PLFA 18:2ω6,9 and bacterial PLFA 16:1ω9) decreased from initial values of 73–96% to 27–64% in the final samples (Supplementary Table 8). For these two PLFAs, the leaf-derived C increased from 3–27 % in the initials, to 35–72% in the finals.
Fig. 4. PLFA-specific isotope analyses.
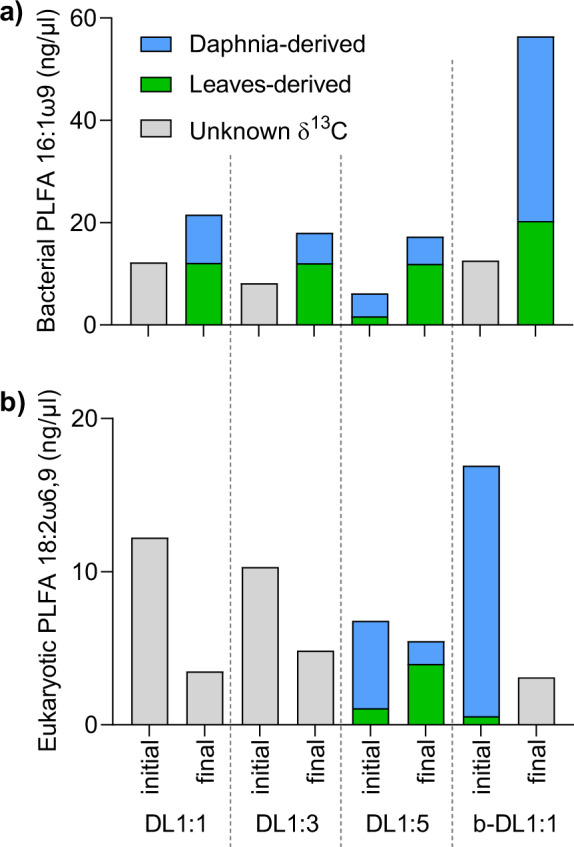
Amount of daphnia- and leaf-derived carbon of a bacterial PLFA 16:1ω9 and b eukaryotic PLFA 18:2ω6,9 based on the PLFA-specific 13C isotopic analysis. A two-source isotope-mixing model was applied to estimate for single PLFAs the proportion of carbon derived from either daphnia or leaves. Isotopic values could not be measured for every sample due to the low concentrations of PLFAs. Thus, grey bars indicate samples for which we have no 13C values. D:L: daphnia and leaves in a given ratio with a complex microbial community; b-DL1:1: daphnia and leaves (ratio 1:1) with a bacterial community only.
Microbial community composition
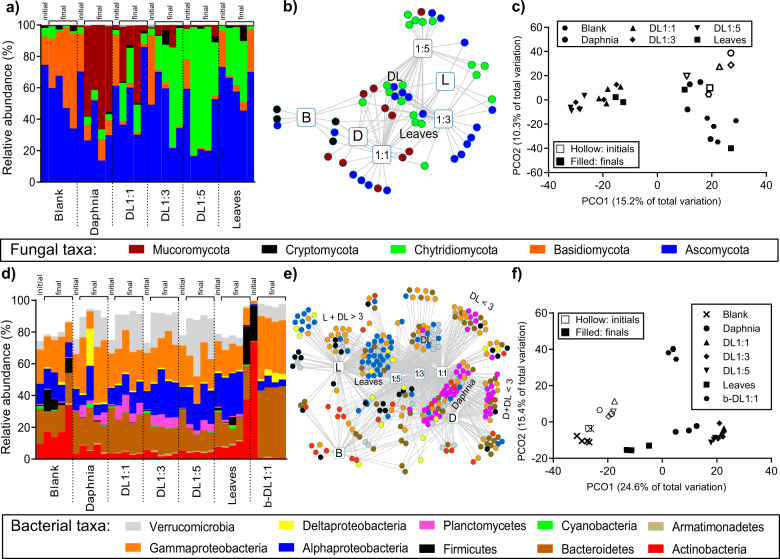
The fungal community composition in the roller bottles was initially dominated by Ascomycota and Basidiomycota. Addition of daphnia carcasses resulted in an increase in the relative abundance of Mucoromycota, particularly in the daphnia and DL1:1 treatments (Fig. 5a). Addition of leaf material resulted in an increase in the relative abundance of Chytridiomycota, an effect which was amplified in the DL1:3 and DL1:5 treatments (Fig. 5a). We found a significant difference amongst treatments (Fperm = 2.0123, p = 0.0001). More specifically, we found significant differences between treatments with high D:L ratios (1:1) and low DL ratios (1:5), or controls (Supplementary Table 9). Indicator species analyses identified only a few fungal indicators of individual treatments. We found Mucoromycota as indicators of high daphnia-to-leaf ratio treatments and Chytridiomycota as indicators of low daphnia-to-leaf ratio treatments (Fig. 5b). Treatments with daphnia and leaf addition formed a separate cluster (Fig. 5c).
Fig. 5. Microbial community analyses.
Upper graphs: fungal taxonomic analyses: a relative abundances of taxa, b network analysis of indicator species and c PCO plots of fungal (ITS) sequences. Lower graphs: bacterial taxonomic analyses, d relative abundances of taxa. One sample from the daphnia controls and one from the leaf controls were identified as outliers (bar charts marked with a star) and excluded from the downstream analysis (i.e., network analysis, PCO plot); e network analysis of indicator species; f PCO plots of bacterial (16s) sequences. Blank: water with a complex microbial community, but without addition of daphnia or leaves; Daphnia: daphnia only; D:L: daphnia and leaves in a given ratio with a complex microbial community; Leaves: leaves only; b-DL1:1: daphnia and leaves (ratio 1:1) with a bacterial community only.
The complex microbial community initially contained similar abundances of Actinobacteria, Bacteroidetes, Alphaproteobacteria and Gammaproteobacteria, whereas the bacteria-only treatment was dominated by small Actinobacteria and Firmicutes (Fig. 5d). With the addition of daphnia carcasses, we observed increases in the relative abundance of Bacteroidetes, Gammaproteobacteria, Planctomycetes and Verrucomicrobia in the complex community treatments (Fig. 5d). While Verrucomicrobia and a few Alphaproteobacteria emerged as indicators for the mixed treatments, Gammaproteobacteria were more likely to be indicators for low daphnia-to-leaf ratios and Planctomycetes for high daphnia-to-leaf ratios (Fig. 5e). Alphaproteobacteria increased in treatments containing leaf material (Fig. 5d), which was also reflected as strong indicator species for leaves (Fig. 5e). In the bacteria-only treatment (b-DL1:1), we saw a dominance of both Bacteroidetes and Gammaproteobacteria. More generally, the bacterial community composition was defined by the presence of the daphnia carcasses. Complex community treatments containing daphnia clustered together, while the bacteria-only treatment (b-DL1:1) formed a separate cluster (Fig. 5f). The community containing only leaf material did not differ substantially from the community where no POC source was provided (blank), or from initial samples (Fig. 5f).
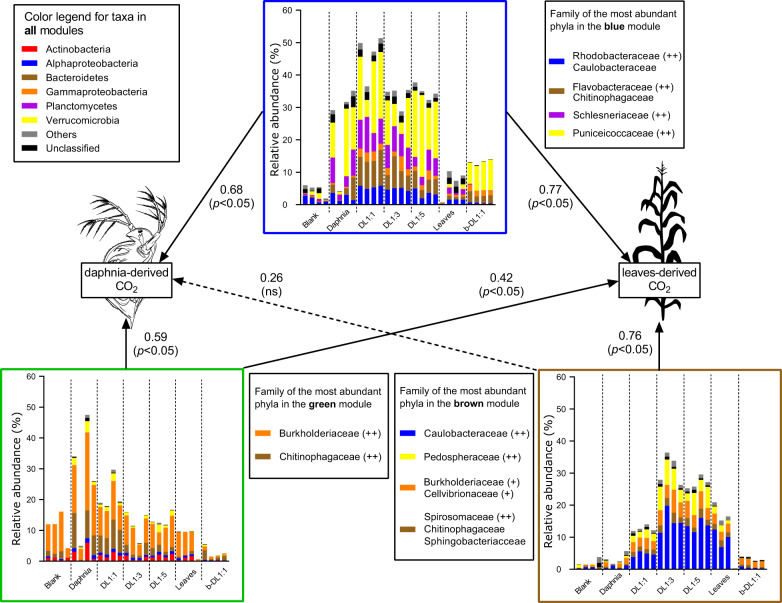
Based on a WGCNA analysis of the bacterial 16S rRNA gene sequences, we explored modules that were positively correlated with the variables “daphnia-derived CO2” or “leaves-derived CO2” (Supplementary Fig. 4). Some modules comprised only few sequences or sequences that were dominant in single samples only (e.g., salmon module). We identified three modules that reflected best the bacterial communities linked to daphnia and/or leaf degradation (i.e., blue, brown and green modules, Fig. 6). Gammaproteobacteria (Burkholderiaceae) and Bacteroidetes (Chitinophagaceae) were common taxa in the green module, which was correlated to daphnia-derived CO2 (ρ = 0.59, p = 0.001) (Fig. 6). The brown module was correlated with leaf-derived respiration (ρ = 0.76, p = 3e−6) and was dominated by Alphaproteobacteria (exclusively Caulobacteraceae). Verrucomicrobia (Pedospheraceae), Gammaproteobacteria and some Bacteroidetes (mainly Spirosomaceae) were also present in this module, although at lower abundance (Fig. 6). The blue module was correlated with mixed treatments, i.e., leaf- (ρ = 0.77, p = 2e−6) and daphnia- (ρ = 0.68, p = 7e−5) derived CO2. The bacterial taxa within this module were predominantly composed of Verrucomicrobia (Puniceicoccaceae), Bacteroidetes (Flavobacteraceae), Alphaproteobacteria (Rhodobacteraceae) and Planctomycetes (Fig. 6).
Fig. 6. Relative abundance of bacterial taxa.
Relative abundance of bacterial tax comprised within three selected WGCNA-based modules that were positively correlated with the external variables “daphnia-derived CO2” and/or “leaves-derived CO2”. External variables (i.e., CO2–) were used as input for network construction and module detection. The OTUs comprised within a module of interest were extracted and, thus, allowed to identify the microbial community related to specific variables. Arrows show the correlations between a module and the variables; continuous lines correspond to statistically significant correlations, dashed lines non-significant ones. Plus (+) symbols represent abundance of a certain family in a phylum (++: dominant family, +: abundant but not dominant, no sign: present but not abundant). Blank: water with a complex microbial community, but without addition of daphnia or leaves; Daphnia: daphnia only; D:L: daphnia and leaves in a given ratio with a complex microbial community; Leaves: leaves only; b-DL1:1: daphnia and leaves (ratio 1:1) with the bacterial community only.
Discussion
Chemodiversity and microbial community complexity are a key component in OM degradation. We could show that the occurrence of zooplankton carcasses has the potential to stimulate the decomposition of leaf litter in aquatic environments. Furthermore, the magnitude of this enhanced decomposition appears to be also dependent on the degrading microbial community composition (i.e., bacteria only vs. bacteria and eukaryotic microorganisms).
Interactions between OM sources and microbial activity
High respiration rates in treatments amended with daphnia (Fig. 1, Supplementary Fig. 1) and a very efficient utilisation of daphnia-derived OM (Fig. 2) supported our previous findings that daphnia carcasses are a bioavailable source of OM [37], which can easily be accessed and utilised by the microbial community. Trends in respiration were consistent with the microbial communities initially utilising OM derived from daphnia carcasses and progressively shifting their energy demands towards the less bioavailable leaf material.
Although slightly delayed, we observed a similar response to the daphnia controls in the simplified microbial community (b-DL1:1) in the presence of leaves (Fig. 1, Supplementary Fig. 1). This suggests that this simplified community was not as efficient in exploiting daphnia-derived OM as the complex community, or that whilst leaf-derived OM was available, it was not sufficient to sustain respiration beyond day 6–7. Higher 13C values of the very abundant bacterial PLFA 16:1ω9 (unspecific, Fig. 4), which contained 36% leaf-derived C (Supplementary Table 8), support a partial utilisation of the leaf-derived OM. If a small fraction of the soluble and bioavailable leaf compounds remained in the leaf material following leaching, the observed partial utilisation of leaf material could be attributed to the utilisation of leaf-derived (non-leached) compounds such as simple sugars. However, this would be inconsistent with the rapid leaching of the soluble fraction from leaf litter [51] and respiration rates in the leaf controls, which were only slightly higher than in the blanks. It is thus safe to assume a partial utilisation of leaf-derived lignin by the simplified community (b-DL1:1, Fig. 1).
Interactions between eukaryotic microorganisms and bacteria in degrading OM
The removal of eukaryotes and large particles with their associated microbiota had manifold effects on the bacterial community structure and on OM cycling processes. These differences were expressed in the turnover rates of daphnia and leaf-derived C (Figs. 1, 2), levels of bacterial biomass (Fig. 3b) and in the bacterial community composition (Fig. 5d, f). Despite a much higher bacterial biomass in the bacteria-only treatment b-DL1:1 (Fig. 3b), the complex microbial community was more active (Figs. 1, 2). The initial community in the simplified community was less diverse than in the other treatments and was dominated by small bacteria (Actinobacteria, Firmicutes) (Fig. 5d). However, while the additional OM increased the diversity of the complex microbial community, the diversity of the simplified community remained low, instead dominated by copiotrophic Gammaproteobacteria and Bacteroidetes (Fig. 5d). Thus, the addition of the OM pools impacted the final microbial community as much, if not more, as the initial community composition. These differences indicate that eukaryotic microorganisms are important for shaping the bacterial community composition and sustaining a more diverse microbial community structure. We suggest two mechanisms that are responsible for this response: the presence of bacterial grazers is expected to control the growth of bacterial communities by predator-prey interactions, hindering the ability of single bacterial taxa to outcompete other groups and dominate the community [66]. This could partly explain the high bacterial PLFA concentrations (Fig. 3b) and the predominance of Gammaproteobacteria and Bacteroidetes (Fig. 5d) in the bacteria-only treatment, as these two phyla were associated with a high concentration of daphnia-derived OM (Fig. 5e). Consequently, the presence of daphnia-derived OM and the absence of bacterial predators could have led to an abundant copiotrophic bacterial community, lacking functional diversity and, therefore, with restricted capabilities to access the less bioavailable C of the leaf material [67, 68].
While bacteria might outcompete fungi in the fast consumption of easily accessible compounds (e.g., simple carbohydrates and amino acids) due to higher metabolic activities [9, 69], their ability to break down complex lignocellulosic compounds is more restricted [48]. Thus, bacteria are likely to become limited in energy and nutrients once most bioavailable resources have been used up. Saprotrophic fungi are known to play a decisive role in the decomposition of terrestrially derived OM [10, 40], as they can release extracellular enzymes or reactive oxygen species that break down or modify complex molecular structures like lignocellulosic compounds [70, 71]. This chemical transformation and degradation of large colloidal molecules releases smaller molecular products and by-products that, in turn, can become accessible to bacteria. Hence, even though fungi might be key organisms in breaking down the leaf material, bacteria might be as important when it comes to metabolising these degradation products [48]. Furthermore, increasing the relative abundances of Caulobacteraceae (Alphaproteobacteria) with increasing leaf content (Figs. 5d, 6) is consistent with the adaptation of Caulobacteraceae to low-nutrient conditions and their chemotactic capacity to trace and degrade lignocellulosic compounds even in very diluted environments [45, 72]. Both pathways would lead to an incorporation of leaf-derived C into bacterial biomass and explain the higher 13C enrichment in the bacterial PLFA 16:1ω9 in the complex microbial community treatments (56–69% leaf-derived C) compared to the bacterial treatment (35% leaf-derived C) (Fig. 4), highlighting the role of bacteria in processing (either directly or indirectly) leaf-derived C.
The fungal phylum Mucoromycota, which was enriched in daphnia controls, lacks laccases, required for the enzymatic breakdown of lignin derivatives, but exhibits high leucine aminopeptidase activity, and is therefore well suited for the breakdown of daphnia-derived OM [73]. Mucoromycota have also faster growth rates than Basidiomycota and Ascomycota [73], with their copiotrophic growth mirroring that of the Gammaproteobacteria and Bacteroidetes. Chytridiomycota, which are known to possess cellulose and hemicellulose-degrading enzymes [74, 75], were most abundant in the leaf-rich treatments (i.e., DL1:3, DL1:5 and leaf control) despite low initial abundances (Fig. 5a). Even though Ascomycota and Basidiomycota are prominent degraders of lignin derivatives [73, 74, 76], Basidiomycota were almost absent in the leaf-rich treatments and, while Ascomycota were still abundant, they generally decreased in relative abundances compared to the initials.
Experimental and methodological restrictions limit our capabilities to clearly disentangle single pathways of energy and nutrient flows from microbial cross-feeding or other feedback processes. Higher bacterial abundances, lower diversity and microbial activity in the bacteria-only treatment could be equally explained by the absence of bacterial grazers, leading to a low bacterial and functional diversity in regard to OM utilisation [67, 68], as by the absence of fungi or lignocellulose-degrading bacteria, which could initiate the degradation of complex compounds and trigger cascading processes of OM turnover. Therefore, we cannot evaluate the role of specific processes or interactions, since each of them seems equally important to the biogeochemical function. Equally, the relative impact of these processes throughout the year, as bacterial, fungal and grazing abundances fluctuate, would likely vary and have impacts on turnover of both OM pools. Yet, this study provides evidence for close and complex links between the different levels of microbial interactions, chemodiversity and OM cycling processes.
Interactive effects induced by daphnia carcasses and leaf OM
The combined presence of both daphnia carcasses and leaves resulted in increased microbial respiration and a more efficient turnover of both OM sources, than we observed for each individual OM source (Fig. 2). While this effect almost doubled (increase of 98%) in the complex community treatment DL1:1, it only increased by 10% in the bacterial community b-DL1:1 compared to the controls. However, we only had controls with a complex microbial community, and thus interactive effects of the bacterial community are likely to be higher than 10% when comparing to bacterial controls. This implies that twice as much CO2 was produced by the complex community with both OM sources combined, than was when both OM sources were degraded in isolation (Fig. 2a). Even though in absolute terms non-additive CO2 production was the highest in the DL1:1 treatment, in relative terms, non-additive interactive effects were the highest in the low daphnia-to-leaf ratios (130% in DL1:3, 178% in DL1:5), indicating a more efficient utilisation when bioavailable daphnia-derived OM is limiting. This was also reflected in higher daphnia-derived CO2 concentrations (normalised) with decreasing abundances of daphnia carcasses in the treatments (Fig. 2c). These findings are consistent with the presence of a positive non-additive effect in aquatic ecosystems, similar to that observed in terrestrial ecosystems [19, 20]. Previous studies addressing this topic either supported [26, 27, 29] or refuted [77, 78] the presence of non-additive interactive effects in an experimental setup. A substantial part of the discrepancy in the results stems from differences in experimental and methodological approaches [21]. The approaches differed mainly in the microbial inoculum (single bacterial isolates, bacterial communities, fungal isolates, mixed or natural microbial communities), in the C substrates used (pure or synthetic C compounds vs. different natural OM substrates) and in the response variables used (CO2, DOC, POC and bacterial and/or fungal growth). By combining a wide array of methodological approaches to different experimental setups (chemodiversity vs. community complexity), we were able to identify some key factors and processes affecting non-additive interactive effects. We have shown that the responses differed between complex microbial and bacterial communities only, and that using a different community would lead to other or variable results, even in the same experimental setup (Fig. 1). Furthermore, only few studies have addressed the effects of different fractions of bioavailable OM for the degradation rates of more persistent OM pools [9, 77]. Whilst the highest respiration rates of leaf-derived material were measured in the treatment with the highest abundances of daphnia carcasses (i.e., DL1:1), the presence of leaves also stimulated the degradation of daphnia-derived OM (Fig. 2c, d), indicating that chemodiversity and the ratio of bioavailable to more persistent OM sources is an important factor that needs to be considered in studies targeting processes involved in OM turnover. We strongly encourage future studies, aiming to further explore the underlying mechanisms of non-additive interactive effects, to combine biogeochemical approaches (e.g., double-source labelling: 13C and 15N) to innovative analytical techniques (e.g., nanoSIMS, CARD-FISH and DNA-SIP) and bioinformatics (e.g., metagenomics, transcriptomics and proteomics). Such interdisciplinary approaches have the potential to clearly link OM utilisation to specific taxa and community responses via gene expression and functional enzymes amongst others.
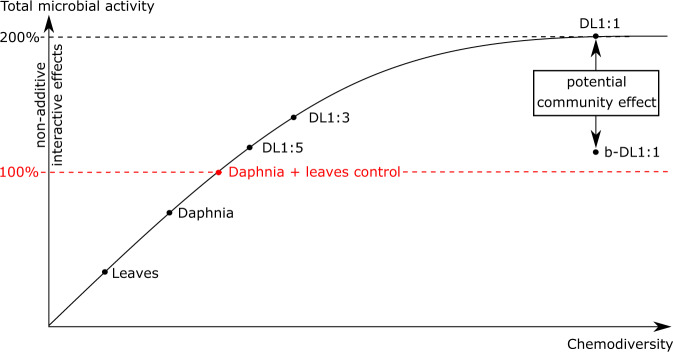
The more bioavailable and chemically diverse compounds (e.g., proteins, lipids and chitin from zooplankton carcasses) are available to provide the degrading community with sufficient energy and diverse nutrients, the more it will allow a complex microbial community to exploit C from additional persistent OM sources to sustain growth. While leaves were relatively poor in nutrients (C/N = 25), they provided abundant C to complement the nutrient-rich, daphnia-derived OM (C/N = 3.5) and maintain a stoichiometric balance for microbial growth. Based on these results, we conclude that non-additive interactive effects are a function of OM chemodiversity and microbial community complexity (Fig. 7). This is consistent with a previous study by Tanentzap et al. [79], addressing the relationship between chemodiversity and microbial diversity on the turnover rates of OM in lakes. Our experimental approach revealed that if OM sources do not provide enough biochemical diversity (e.g., leaves and daphnia controls), even a complex microbial community will face C or nutrient limitations. On the other hand, a system with high chemodiversity but a low microbial community complexity (e.g., bacterial treatment, b-DL1:1), is also restricted in growth as a result of limited access to C and nutrients due to lacking functional diversity. While chemodiversity is expected to have a stronger effect on the microbial complexity than vice versa, the degrading microbial community also has an impact on the complexity of the surrounding OM pool [79, 80]. Consequently, only a system combining both, a complex microbial community and a chemically diverse OM pool, is likely to sustain microbial growth and maximise net OM turnover rates.
Fig. 7. Total microbial activity and non-additive interactive effects as a function of organic matter chemodiversity.
Non-additive interactive effects start above the red line, which visualises the sum of microbial activity (i.e., CO2 production) when carbon sources are isolated. In the zone of non-additive interactive effects, the increase of microbial activity is not linear and reaches a steady state when chemodiversity is high. Furthermore, we suggest a community effect that influences the slope and the maxima of the curve: while DL1:1 and b-DL1:1 had the same conditions in regard to organic matter chemodiversity, DL1:1 had the highest microbial activity resulting from a high community complexity, compared to b-DL1:1- simplified bacterial community composition. Daphnia: daphnia only; D:L: daphnia and leaves in a given ratio with a complex microbial community; Leaves: leaves only; b-DL1:1: daphnia and leaves (ratio 1:1) with the bacterial community only.
Freshwater environments worldwide are facing increasing C and nutrient loads, changes in phytoplankton compositions [81] and declining zooplankton abundances [82, 83]. The tight interaction between chemical and microbial diversity, highlighted in this study and elsewhere, should be considered in the context of overall C turnover in aquatic ecosystems, since long-term changes and seasonal pulses of allochthonous and autochthonous OM input can affect both aquatic chemodiversity and microbial diversity, impacting net remineralisation rates in response [79]. Zooplankton carcasses are an important and up to now largely neglected piece of the puzzle with regard to OM degradation in marine and freshwater ecosystems, and should be considered in more detail in future studies of C cycling.
Supplementary information
Acknowledgements
We thank Luca Zoccarato for help with bioinformatical methods and all members of the Aquatic Microbial Ecology group (IGB, Stechlin) for insightful discussions during this study. From the Department of Chemical Analytics and Biogeochemistry (IGB, Berlin), we thank Angela Krüger for her help with the analyses of the PLFAs, Tobias Goldhammer and the technicians for their support in chemical analyses. Furthermore, we would like to thank Cornelia Krager and Doreen Noack from the Department of Organic Geochemistry (GFZ, Potsdam) for the stable-isotope analyses of the PLFAs. We thank Thomas Mehner and the participants of the workshop ‘Scientific Writing’ in March 2019 at the IGB for helpful discussions on an earlier version of the paper. We also thank the two anonymous reviewers for their meticulous revisions, criticisms and recommendations on the previous version of this paper. This research was conducted within the DFG projects “Microprime” and “Zooflux” (GR 1540/23-1 and 29-1, respectively) funded by the German Research Foundation (DFG). Olesya Kolmakova’s research at IGB was supported by the German Academic Exchange Service (DAAD), Research Grants for Doctoral Candidates and Young Academics and Scientists (more than 6 months), 2017/18 (57299291). The work (Michail Gladyshev) was also supported by Federal Tasks for Institute of Biophysics SB RAS No. 51.1.1 and Federal Tasks for Siberian Federal University No. FSRG-2020-0019.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Darshan Neubauer, Email: neubauer@igb-berlin.de.
Hans-Peter Grossart, Email: hgrossart@igb-berlin.de.
Supplementary information
The online version of this article (10.1038/s41396-020-00883-w) contains supplementary material, which is available to authorized users.
References
- 1.Chen M, Zeng G, Zhang J, Xu P, Chen A, Lu L. Global landscape of total organic carbon, nitrogen and phosphorus in lake water. Sci Rep. 2015;5:15043. doi: 10.1038/srep15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toming K, Kotta J, Uuemaa E, Sobek S, Kutser T, Tranvik LJ. Predicting lake dissolved organic carbon at a global scale. Sci Rep. 2020;10:8471. doi: 10.1038/s41598-020-65010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tranvik LJ, Downing JA, Cotner JB, Loiselle SA, Striegl RG, Ballatore TJ, et al. Lakes and reservoirs as regulators of carbon cycling and climate. Limnol Oceanogr. 2009;54:2298–314.. [Google Scholar]
- 4.Tranvik LJ, Cole JJ, Prairie YT. The study of carbon in inland waters-from isolated ecosystems to players in the global carbon cycle. Limnol Oceanogr Lett. 2018;3:41–48. [Google Scholar]
- 5.Jaffé R, McKnight D, Maie N, Cory R, McDowell WH, Campbell JL. Spatial and temporal variations in DOM composition in ecosystems: the importance of long-term monitoring of optical properties. J Geophys Res Biogeosci. 2008;113:G04032. [Google Scholar]
- 6.Crump BC, Kling GW, Bahr M, Hobbie JE. Bacterioplankton community shifts in an Arctic Lake correlate with seasonal changes in organic matter source. Appl Environ Microbiol. 2003;69:2253–68.. doi: 10.1128/AEM.69.4.2253-2268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fasching C, Behounek B, Singer GA, Battin TJ. Microbial degradation of terrigenous dissolved organic matter and potential consequences for carbon cycling in brown-water streams. Sci Rep. 2014;4:4981. doi: 10.1038/srep04981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yakimovich KM, Emilson EJS, Carson MA, Tanentzap AJ, Basiliko N, Mykytczuk NCS. Plant litter type dictates microbial communities responsible for greenhouse gas production in amended lake sediments. Front Microbiol. 2018;9:2662. doi: 10.3389/fmicb.2018.02662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attermeyer K, Hornick T, Kayler ZE, Bahr A, Zwirnmann E, Grossart HP, et al. Enhanced bacterial decomposition with increasing addition of autochthonous to allochthonous carbon without any effect on bacterial community composition. Biogeosciences. 2014;11:1479–89.. [Google Scholar]
- 10.Fabian J, Zlatanovic S, Mutz M, Premke K. Fungal-bacterial dynamics and their contribution to terrigenous carbon turnover in relation to organic matter quality. ISME J. 2017;11:415–25.. doi: 10.1038/ismej.2016.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quigley LNM, Edwards A, Steen AD, Buchan A. Characterization of the interactive effects of labile and recalcitrant organic matter on microbial growth and metabolism. Front Microbiol. 2019;10:493. doi: 10.3389/fmicb.2019.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tranvik LJ. Degradation of dissolved organic matter in humic waters by bacteria. In: Hessen DOTLJ, editor. Aquatic Humic Substances. Berlin, Heidelberg: Springer; 1998.
- 13.Søndergaard M, Borch NH, Riemann B. Dynamics of biodegradable DOC produced by freshwater plankton communities. Aquat Micro Ecol. 2000;23:73–83. [Google Scholar]
- 14.Berg B, McClaugherty C. Initial litter chemical composition. Plant Litter. 2014;3:53–83. [Google Scholar]
- 15.Bugg TD, Ahmad M, Hardiman EM, Rahmanpour R. Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep. 2011;28:1883–96. doi: 10.1039/c1np00042j. [DOI] [PubMed] [Google Scholar]
- 16.Klotzbücher T, Kaiser K, Guggenberger G, Gatzek C, Kalbitz K. A new conceptual model for the fate of lignin in decomposing plant litter. Ecology. 2011;92:1052–62.. doi: 10.1890/10-1307.1. [DOI] [PubMed] [Google Scholar]
- 17.Grey J, Jones RI, Sleep D. Seasonal changes in the importance of the source of organic matter to the diet of zooplankton in Loch Ness, as indicated by stable isotope analysis. Limnol Oceanogr. 2001;46:505–13.. [Google Scholar]
- 18.Cole JJ, Carpenter SR, Kitchell JF, Pace ML. Pathways of organic carbon utilization in small lakes: results from a whole-lake 13C addition and coupled model. Limnol Oceanogr. 2002;47:1664–75. [Google Scholar]
- 19.Guenet B, Danger M, Abbadie L, Lacroix G. Priming effect: bridging the gap between terrestrial and aquatic ecology. Ecology. 2010;91:2850–61.. doi: 10.1890/09-1968.1. [DOI] [PubMed] [Google Scholar]
- 20.Bianchi TS. The role of terrestrially derived organic carbon in the coastal ocean: a changing paradigm and the priming effect. Proc Natl Acad Sci USA. 2011;108:19473–81.. doi: 10.1073/pnas.1017982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bengtsson MM, Attermeyer K, Catalán N. Interactive effects on organic matter processing from soils to the ocean: are priming effects relevant in aquatic ecosystems? Hydrobiologia. 2018;822:1–17. [Google Scholar]
- 22.Kuzyakov Y, Friedel JK, Stahr K. Review of mechanisms and quantification of priming effects. Soil Biol Biochem. 2000;32:1485–98.. [Google Scholar]
- 23.Bianchi TS, Ward ND. Editorial: the role of priming in terrestrial and aquatic ecosystems. Front Earth Sci. 2019;7:321. [Google Scholar]
- 24.Halvorson HM, Francoeur SN, Findlay RH, Kuehn KA. Algal-mediated priming effects on the ecological stoichiometry of leaf litter decomposition: a meta-analysis. Front Earth Sci. 2019;7:76. [Google Scholar]
- 25.Kayler ZE, Premke K, Gessler A, Gessner MO, Griebler C, Hilt S, et al. Integrating aquatic and terrestrial perspectives to improve insights into organic matter cycling at the landscape scale. Front Earth Sci. 2019;7:127. [Google Scholar]
- 26.Danger M, Cornut J, Chauvet E, Chavez P, Elger A, Lecerf A. Benthic algae stimulate leaf litter decomposition in detritus-based headwater streams: a case of aquatic priming effect? Ecology. 2013;94:1604–13.. doi: 10.1890/12-0606.1. [DOI] [PubMed] [Google Scholar]
- 27.Guenet B, Danger M, Harrault L, Allard B, Jauset-Alcala M, Bardoux G, et al. Fast mineralization of land-born C in inland waters: first experimental evidences of aquatic priming effect. Hydrobiologia. 2013;721:35–44. [Google Scholar]
- 28.Ward ND, Sawakuchi HO, Richey JE, Keil RG, Bianchi TS. Enhanced aquatic respiration associated with mixing of clearwater tributary and turbid Amazon river waters. Front Earth Sci. 2019;7:101. [Google Scholar]
- 29.Bianchi TS, Thornton DCO, Yvon-Lewis SA, King GM, Eglinton TI, Shields MR, et al. Positive priming of terrestrially derived dissolved organic matter in a freshwater microcosm system. Geophys Res Lett. 2015;42:5460–67.. [Google Scholar]
- 30.Tang KW, Gladyshev MI, Dubovskaya OP, Kirillin G, Grossart H-P. Zooplankton carcasses and non-predatory mortality in freshwater and inland sea environments. J Plankton Res. 2014;36:597–612. [Google Scholar]
- 31.Cauchie HM, Jaspar-Versali MF, Hoffmann L, Thomé JP. Analysis of the seasonal variation in biochemical composition of Daphnia magna Straus (Crustacea: Branchiopoda: Anomopoda) from an aerated wastewater stabilisation pond. Ann Limnol. 1999;35:223–31.. [Google Scholar]
- 32.Smirnov NN. Physiology of the Cladocera, 2nd ed., London, Academic Press: Elsevier; 2017.
- 33.Dubovskaya OP, Tang KW, Gladyshev MI, Kirillin G, Buseva Z, Kasprzak P, et al. Estimating in situ zooplankton non-predation mortality in an oligo-mesotrophic lake from sediment trap data: caveats and reality check. PLoS ONE. 2015;10:e0131431. doi: 10.1371/journal.pone.0131431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirillin G, Grossart H-P, Tang KW. Modeling sinking rate of zooplankton carcasses: effects of stratification and mixing. Limnol Oceanogr. 2012;57:881–94.. [Google Scholar]
- 35.Tang KW, Hutalle KML, Grossart HP. Microbial abundance, composition and enzymatic activity during decomposition of copepod carcasses. Aquat Micro Ecol. 2006;45:219–27.. [Google Scholar]
- 36.Tang KW, Bickel SL, Dziallas C, Grossart HP. Microbial activities accompanying decomposition of cladoceran and copepod carcasses under different environmental conditions. Aquat Micro Ecol. 2009;57:89–100. [Google Scholar]
- 37.Kolmakova OV, Gladyshev MI, Fonvielle JA, Ganzert L, Hornick T, Grossart HP. Effects of zooplankton carcasses degradation on freshwater bacterial community composition and implications for carbon cycling. Environ Microbiol. 2019;21:34–49. doi: 10.1111/1462-2920.14418. [DOI] [PubMed] [Google Scholar]
- 38.Corno G, Salka I, Pohlmann K, Hall AR, Grossart HP. Interspecific interactions drive chitin and cellulose degradation by aquatic microorganisms. Aquat Micro Ecol. 2015;76:27–37. [Google Scholar]
- 39.Masigol H, Khodaparast SA, Woodhouse JN, Rojas‐Jimenez K, Fonvielle J, Rezakhani F, et al. The contrasting roles of aquatic fungi and oomycetes in the degradation and transformation of polymeric organic matter. Limnol Oceanogr. 2019;64:2662–78.. [Google Scholar]
- 40.Gessner MO, Chauvet E. Importance of stream microfungi in controlling breakdown rates of leaf litter. Ecology. 1994;75:1807–17.. [Google Scholar]
- 41.Grossart HP, Van den Wyngaert S, Kagami M, Wurzbacher C, Cunliffe M, Rojas-Jimenez K. Fungi in aquatic ecosystems. Nat Rev Microbiol. 2019;17:339–54.. doi: 10.1038/s41579-019-0175-8. [DOI] [PubMed] [Google Scholar]
- 42.Osono T. Functional diversity of ligninolytic fungi associated with leaf litter decomposition. Ecol Res. 2019;35:30–43. [Google Scholar]
- 43.Taube R, Ganzert L, Grossart HP, Gleixner G, Premke K. Organic matter quality structures benthic fatty acid patterns and the abundance of fungi and bacteria in temperate lakes. Sci Total Environ. 2018;610-611:469–81.. doi: 10.1016/j.scitotenv.2017.07.256. [DOI] [PubMed] [Google Scholar]
- 44.Cragg SM, Beckham GT, Bruce NC, Bugg TD, Distel DL, Dupree P, et al. Lignocellulose degradation mechanisms across the Tree of Life. Curr Opin Chem Biol. 2015;29:108–19.. doi: 10.1016/j.cbpa.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilhelm RC, Singh R, Eltis LD, Mohn WW. Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J. 2019;13:413–29.. doi: 10.1038/s41396-018-0279-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuikman PJ, Jansen AG, van Veen JA, Zehnder AJB. Protozoan predation and the turnover of soil organic carbon and nitrogen in the presence of plants. Biol Fertil Soils. 1990;10:22–28. [Google Scholar]
- 47.White DC, Davis WM, Nickels JS, King JD, Bobbie RJ. Determination of the sedimentary microbial biomass by extractible lipid phosphate. Oecologia. 1979;40:51–62. doi: 10.1007/BF00388810. [DOI] [PubMed] [Google Scholar]
- 48.Romaní AM, Fischer H, Mille-Lindblom C, Tranvik LJ. Interactions of bacteria and fungi on decomposing litter: differential extracellular enzyme activities. Ecology. 2006;87:2559–69.. doi: 10.1890/0012-9658(2006)87[2559:iobafo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 49.Hutalle-Schmelzer KM, Zwirnmann E, Kruger A, Grossart HP. Changes in pelagic bacteria communities due to leaf litter addition. Micro Ecol. 2010;60:462–75.. doi: 10.1007/s00248-010-9639-0. [DOI] [PubMed] [Google Scholar]
- 50.Smith EJ, Davison W, Hamilton-Taylor J. Methods for preparing synthetic freshwaters. Water Res. 2002;36:1286–96.. doi: 10.1016/s0043-1354(01)00341-4. [DOI] [PubMed] [Google Scholar]
- 51.Attermeyer K, Premke K, Hornick T, Hilt S, Grossart HP. Ecosystem-level studies of terrestrial carbon reveal contrasting bacterial metabolism in different aquatic habitats. Ecology. 2013;94:2754–66.. doi: 10.1890/13-0420.1. [DOI] [PubMed] [Google Scholar]
- 52.Halbedel S (2015) Protocol for CO2 sampling in waters by the use of the headspaceequilibration technique, based on the simple gas equation; second update. Protoc Exch. https://assets.researchsquare.com/files/nprot-4275/v1/manuscript.pdf
- 53.Cheng W. Measurement of rhizosphere respiration and organic matter decomposition using natural 13C. Plant Soil. 1996;183:263–68.. [Google Scholar]
- 54.Taube R, Fabian J, Van den Wyngaert S, Agha R, Baschien C, Gerphagnon M, et al. Potentials and limitations of quantification of fungi in freshwater environments based on PLFA profiles. Fungal Ecol. 2019;41:256–68.. [Google Scholar]
- 55.Zhang Z, Qu Y, Li S, Feng K, Wang S, Cai W, et al. Soil bacterial quantification approaches coupling with relative abundances reflecting the changes of taxa. Sci Rep. 2017;7:4837. doi: 10.1038/s41598-017-05260-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mangelsdorf K, Karger C, Zink K-G. Phospholipids as life markers in geological habitats. Hydrocarbons, oils and lipids: diversity, origin, chemistry and fate. 2019. pp. 1–29.
- 57.Frostegård Å, Tunlid A, Bååth E. Microbial biomass measured as total lipid phosphate in soils of different organic content. J Microbiol Methods. 1991;14:151–63.. [Google Scholar]
- 58.Nercessian O, Noyes E, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Bacterial populations active in metabolism of C1 compounds in the sediment of Lake Washington, a freshwater lake. Appl Environ Microbiol. 2005;71:6885–99.. doi: 10.1128/AEM.71.11.6885-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tedersoo L, Anslan S, Bahram M, Põlme S, Riit T, Liiv I, et al. Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys. 2015;10:1–43. [Google Scholar]
- 61.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murali A, Bhargava A, Wright ES. IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome. 2018;6:140. doi: 10.1186/s40168-018-0521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Caceres M, Legendre P. Associations between species and groups of sites: indices and statistical inference. Ecology. 2009;90:3566–74.. doi: 10.1890/08-1823.1. [DOI] [PubMed] [Google Scholar]
- 64.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taipale SJ, Kainz MJ, Brett MT. Diet-switching experiments show rapid accumulation and preferential retention of highly unsaturated fatty acids in Daphnia. Oikos. 2011;120:1674–82.. [Google Scholar]
- 66.Corno G, Jurgens K. Structural and functional patterns of bacterial communities in response to protist predation along an experimental productivity gradient. Environ Microbiol. 2008;10:2857–71.. doi: 10.1111/j.1462-2920.2008.01713.x. [DOI] [PubMed] [Google Scholar]
- 67.Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012;6:1007–17.. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song HK, Song W, Kim M, Tripathi BM, Kim H, Jablonski P, et al. Bacterial strategies along nutrient and time gradients, revealed by metagenomic analysis of laboratory microcosms. FEMS Microbiol Ecol. 2017;93:fix114. doi: 10.1093/femsec/fix114. [DOI] [PubMed] [Google Scholar]
- 69.Bardgett RD, Kandeler E, Tscherko D, Hobbs PJ, Bezemer TM, Jones TH, et al. Below-ground microbial community development in a high temperature world. Oikos. 1999;85:193–203. [Google Scholar]
- 70.Hammel KE, Kapich AN, Jensen KA, Ryan ZC. Reactive oxygen species as agents of wood decay by fungi. Enzym Micro Technol. 2002;30:445–53.. [Google Scholar]
- 71.Rojas-Jimenez K, Fonvielle JA, Ma H, Grossart H-P. Transformation of humic substances by the freshwater Ascomycete Cladosporium sp. Limnol Oceanogr. 2017;62:1955–62.. [Google Scholar]
- 72.Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen JA, et al. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci USA. 2001;98:4136–41.. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng W, Lehmann A, Ryo M, Valyi KK, Rillig MC. Growth rate trades off with enzymatic investment in soil filamentous fungi. Sci Rep. 2020;10:11013. doi: 10.1038/s41598-020-68099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bärlocher F, Boddy L. Aquatic fungal ecology—How does it differ from terrestrial? Fungal Ecol. 2016;19:5–13. [Google Scholar]
- 75.Lange L, Barrett K, Pilgaard B, Gleason F, Tsang A. Enzymes of early-diverging, zoosporic fungi. Appl Microbiol Biotechnol. 2019;103:6885–902.. doi: 10.1007/s00253-019-09983-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Janusz G, Pawlik A, Swiderska-Burek U, Polak J, Sulej J, Jarosz-Wilkolazka A, et al. Laccase properties, physiological functions, and evolution. Int J Mol Sci. 2020;21:966. doi: 10.3390/ijms21030966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Catalán N, Kellerman AM, Peter H, Carmona F, Tranvik LJ. Absence of a priming effect on dissolved organic carbon degradation in lake water. Limnol Oceanogr. 2015;60:159–68.. [Google Scholar]
- 78.Bengtsson MM, Wagner K, Burns NR, Herberg ER, Wanek W, Kaplan LA, et al. No evidence of aquatic priming effects in hyporheic zone microcosms. Sci Rep. 2014;4:5187. doi: 10.1038/srep05187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanentzap AJ, Fitch A, Orland C, Emilson EJS, Yakimovich KM, Osterholz H, et al. Chemical and microbial diversity covary in fresh water to influence ecosystem functioning. Proc Natl Acad Sci USA. 2019;116:24689–95.. doi: 10.1073/pnas.1904896116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Orland C, Emilson EJS, Basiliko N, Mykytczuk NCS, Gunn JM, Tanentzap AJ. Microbiome functioning depends on individual and interactive effects of the environment and community structure. ISME J. 2019;13:1–11. doi: 10.1038/s41396-018-0230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winder M, Sommer U. Phytoplankton response to a changing climate. Hydrobiologia. 2012;698:5–16. [Google Scholar]
- 82.Pothoven SA, Fahnenstiel GL. Spatial and temporal trends in zooplankton assemblages along a nearshore to offshore transect in southeastern Lake Michigan from 2007 to 2012. J Gt Lakes Res. 2015;41:95–103. [Google Scholar]
- 83.Selmeczy GB, Abonyi A, Krienitz L, Kasprzak P, Casper P, Telcs A, et al. Old sins have long shadows: climate change weakens efficiency of trophic coupling of phyto- and zooplankton in a deep oligo-mesotrophic lowland lake (Stechlin, Germany)—a causality analysis. Hydrobiologia. 2018;831:101–17.. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.