Abstract
Background
Autism spectrum disorders (ASDs) are a set of complex neurobiological disorders. Growing evidence has shown that the microbiota that resides in the gut can modulate brain development via the gut–brain axis. However, direct clinical evidence of the role of the microbiota–gut–brain axis in ASD is relatively limited.
Methods
A case-control study of 71 boys with ASD and 18 neurotypical controls was conducted at China-Japan Friendship Hospital. Demographic information and fecal samples were collected, and the gut microbiome was evaluated and compared by 16S ribosomal RNA gene sequencing and metagenomic sequencing.
Results
A higher abundance of operational taxonomic units (OTUs) based on fecal bacterial profiling was observed in the ASD group. Significantly different microbiome profiles were observed between the two groups. At the genus level, we observed a decrease in the relative abundance of Escherichia, Shigella, Veillonella, Akkermansia, Provindencia, Dialister, Bifidobacterium, Streptococcus, Ruminococcaceae UCG_002, Megasphaera, Eubacterium_coprostanol, Citrobacter, Ruminiclostridium_5, and Ruminiclostridium_6 in the ASD cohort, while Eisenbergiella, Klebsiella, Faecalibacterium, and Blautia were significantly increased. Ten bacterial strains were selected for clinical discrimination between those with ASD and the neurotypical controls. The highest AUC value of the model was 0.947.
Conclusion
Significant differences were observed in the composition of the gut microbiome between boys with ASD and neurotypical controls. These findings contribute to the knowledge of the alteration of the gut microbiome in ASD patients, which opens the possibility for early identification of this disease.
Keywords: Autism spectrum disorders, Gut microbiome, China
Introduction
Autism spectrum disorders (ASDs) are a set of complex neurobiological disorders that impair social interactions and communication and lead to restricted, repetitive, and stereotyped patterns of behavior, interests, and activities [1]. The prevalence of ASD has been steadily increasing in recent years, which may to some extent be due to greater awareness of the disease by health and education professionals and changes in diagnostic criteria. Treatments and educational interventions usually last for the entire lives of those who are diagnosed with ASD [2]. This represents a serious public health problem due to its increasing prevalence and huge financial costs.
ASD are etiologically heterogeneous. However, the exact causes of ASD are unclear and are believed to involve a combination of genetic and environmental risk factors. Many studies highlight the possibility of environmental risk factors and associated medical comorbidities that contribute to the disease. Some studies have even observed an association between ASD severity and gastrointestinal (GI) symptoms, such as constipation, diarrhea, and alternating constipation/diarrhea [3,4].
In recent years, growing evidence has shown that the microbiota that resides in the gut can modulate brain development via the gut–brain axis [5]. As far as this concept is concerned, increasing interest has developed focusing on the potential effects of the microbiota–gut–brain axis in neurodevelopmental disorders, especially in the etiology and pathogenesis of ASD.
In an attempt to clarify the role of gut microbiota in the appearance and development of ASD, some clinical studies have reported that compared with neurotypical controls, subjects with autism suffer from altered profiles and abundances of gut microbiota [[6], [7], [8]]. The human gut harbors up to 100 trillion microorganisms, including at least 1000 different species of known bacteria [9]. Colonization of the gut microbiota starts at the moment of birth, when the newborn is exposed to a complex microbiota during delivery [10]. Many ASD-affected children undergo extensive oral antibiotic treatment before 3 years of age [11], which is thought to cause the proliferation of anaerobic bacteria, destabilize gut microbiota [12] and provide opportunities for competitive potential pathogens to contribute to ASD [13,14]. Bacteroidetes, Clostridia, and Desulfovibrio are common bacteria that may promote GI symptoms and autistic behaviors. In addition to altering the intestinal immune system, these bacteria also produce certain potent neurotoxins that directly contribute to the pathologies of ASD.
Several findings support a microbiota–gut–brain connection in a mouse model of ASD. However, direct clinical evidence for the role of the microbiota–gut–brain axis in ASD is relatively limited, and consensus across studies has not yet been established. The sample size of previous studies has been relatively small since recruitment is difficult. To better understand the microbial profiles associated with ASD in the context of the Chinese diet, we conducted this case-control study including 71 ASD children and 18 neurotypical controls, representing the largest sample size of the gut microbiota of children with autism in China.
Methods and materials
Participants
This study was reviewed and approved by the Institutional Review Board of China-Japan Friendship Hospital. All written informed consent was obtained from the parents. The parents and the enrolled participants visited the Department of Pediatrics, China-Japan Friendship Hospital, and were provided the questionnaire data sheets. The fecal samples were obtained within three days after the visit.
According to the results of the Autism Behavior Checklist (ABC) screening tool of ASD and the Diagnostic and Statistical Manual, Fifth Edition (DSM-V) diagnostic criteria [15], the neuropsychiatric status of the affected subjects was established by two experienced experts in the Department of Pediatrics of China-Japan Friendship Hospital. The controls were evaluated in the same manner as the ASD subjects to exclude those with any developmental disturbances, including ASD.
Enrollment criteria
Autism group
-
1)
Age 3 to 6.
-
2)
Male.
-
3)
ASD diagnosed according to the ABC screening tool and DSM-V criteria.
-
4)
No usage of any type of antibiotic medications or probiotics within at least one month prior to the sample collection.
Control group
-
1)
Age and sex-matched neurotypical healthy subjects.
Exclusion criteria
-
1)
Treated with anti-inflammatory or antioxidant drugs.
-
2)
Diagnosed with neurological diseases, severe head injuries, and gastrointestinal diseases such as celiac disease, chronic diarrhea, and chronic constipation, etc.
Sample collection, DNA extraction, and sequencing
Fecal specimens from participants were collected with sterilized 2-ml tubes containing pure ethanol, aliquoted, and frozen at 80 °C until DNA extraction. Total DNA extraction from fecal samples (250 mg, wet weight) was performed using a PSP Spin StooL DNA Kit/PSP Spin StooL DNA PLus Kit according to the manufacturer's instructions. Bacterial 16S rRNA amplicon sequencing of the V1–V2 gene region [16,17] was performed on the Illumina MiSeq platform for identification and relative quantification of bacterial taxa.
Metagenomic sequencing was also performed in this study [18], which can analyze the genetic composition and function of the microbial population in a specific environment and identify which strains are contained in the mixed environment. Metagenomic sequencing and technical advances have enabled culture-free, high-resolution strain and subspecies analyses at high throughput and in complex environments [19]. During metagenomic sequencing, 20 μg of microbial metagenomic DNA was taken, and a sequencing library was constructed using the NEXTflex® Rapid DNA Sequencing Kit (Bioo Scientific, USA, NOVA-5144-02) and following the manufacturer's instructions. After the library was constructed, Qubit 2.0 was used for preliminary quantification, the library was diluted to 2 ng/μL, Agilent 2100 was used to detect the insert size of the library, and the qPCR method [20] was used to accurately quantify the effective concentration of the library (the library was effective concentration> 3 mmol/L) to ensure the quality of the library. The constructed library was sequenced on the Illumina HiSeq 2500 platform. The study utilized MOCAT [21] preprocessing to filter low-quality sequences, sequences that were too short, and error sequences to obtain clean data. Starting from the clean data after quality control of each sample, metagenomic assembly was carried out [22]. Starting from the gene catalog, bowtie was used to compare the sequence with the MicroNR library to obtain the species annotation information of unigenes, and the gene abundance table was combined to obtain the species abundance table of different classification levels.
Statistical analysis
All statistical analyses were performed using SPSS 20.0. Several types of statistical analyses were carried out, depending on the research question being addressed. Continuous data among the groups were analyzed by using ANOVA (generalized linear model). Frequencies were calculated, and a chi-square test was performed to examine the frequency differences among the groups. The calculation of P values was performed with Kruskal–Wallis H-tests and Welch's t-tests. The P values were corrected by false discovery rates (FDRs) to control for multiple hypothesis testing. Principal coordinates were computed for the unweighted distance matrices and used to generate principal coordinate analysis (PCoA) plots using evenly sampled OTU abundances. According to the QIIME (Quantitative Insights into Microbial Ecology) tutorial (http://qiime.org/), high-throughput sequencing analysis of bacterial rRNA genes was processed using the QIIME (version 1.9.1) software suite [23].
Alpha diversity metrics from the final OTU table without singletons were obtained within the QIIME pipeline. Linear discriminant effect size (LEfSe) analysis was used to explore potential bacterial biomarkers associated with different groups. LEfSe combines the Kruskal-Wallis test pairwise Wilcoxon rank-sum test with linear discriminant analysis (LDA). It ranks features by effect size, which places features that explain most of the biological differences at the top. Relative operating characteristic (ROC) curves and area under the curve (AUC) values were used to evaluate the performance of the predictive model.
To further distinguish between ASD and their comparisons, we used a machine learning method based on the random forest algorithm to build a classifier from the same input feature set. Random forest classifier is a kind of ensemble learning algorithm belonging to bagging. It can establish an unbiased estimate of the error during the generation of the tree. Random forest classifier was found to be more accurate than both the alternative cross-validation-based estimator of the underlying algorithm's error [24].
Species relative abundances and patient profiles were analyzed using the random forest package in R [25,26]. A forest was trained by supervised learning. In the forest, each tree finds an ideal split for a set of randomly chosen features that predict the outcome of each sample as the expected outcome. The data partition split by every tree in a forest is used to vote on a predicted overall outcome of each sample. Every tree was used to vote on an outcome to prevent any single tree that may have memorized the data from having a dominant prediction. In this study, outcomes were children with ASD or neurotypical children. For all analyses, differences were considered statistically significant when the FDR corrected P value was <0.05.
Results
Demographics of the children with ASD and the controls
This study enrolled a total of 71 children with ASD and 18 controls. The demographic characteristics of the study participants are listed in Table 1. Age and sex were not significantly different between the two groups (P > 0.05).
Table 1.
Demographics of the children with ASD and the controls.
| ASD group (n = 71) | Control group (n = 18) | P-value | |
|---|---|---|---|
| Sex (Male/Female) | 71/0 | 18/0 | 1.000 |
| Age (years, mean ± SD) | 4.28 ± 1.52 | 4.62 ± 1.29 | P = 0.792 |
| ABC score | 79.82 ± 13.10 | 8.77 ± 7.29 | P<0.001 |
Altered gut microbiome profile
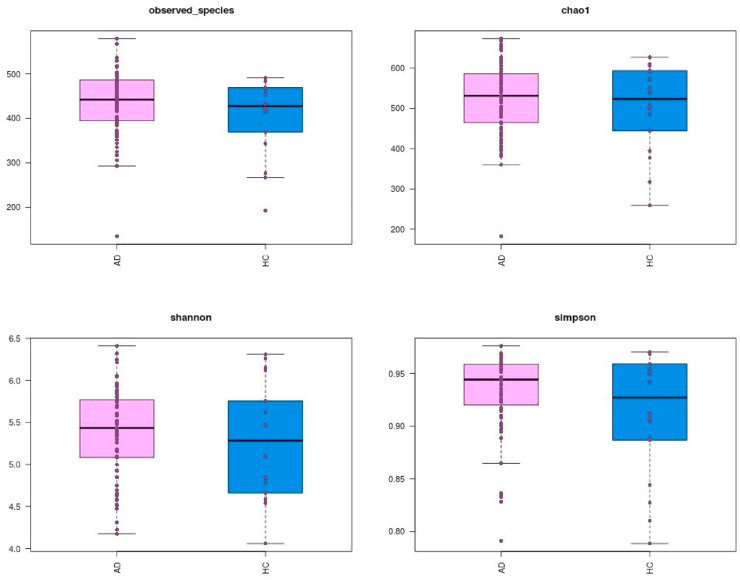
We compared the alpha diversity of the gut microbiota in the two groups. As shown in Fig. 1, significant increases in bacterial richness, including observed species and the Shannon and Simpson diversity indices (all P values < 0.05), were observed in the ASD group; no significant variations between the two groups were observed for the Chao1 diversity index.
Fig. 1.
Comparison of diversity index of two groups on the OTU profile.
The total distribution of bacteria demonstrated that the taxonomy was significantly different at the phylum level, with increases in Firmicutes, Proteobacteria, and Actinomycetes and decreases in Bacteroidetes in the ASD group (all P values < 0.05). Notably, the Bacteroidetes/Firmicutes ratio was significantly lower in the ASD group (P value < 0.001).
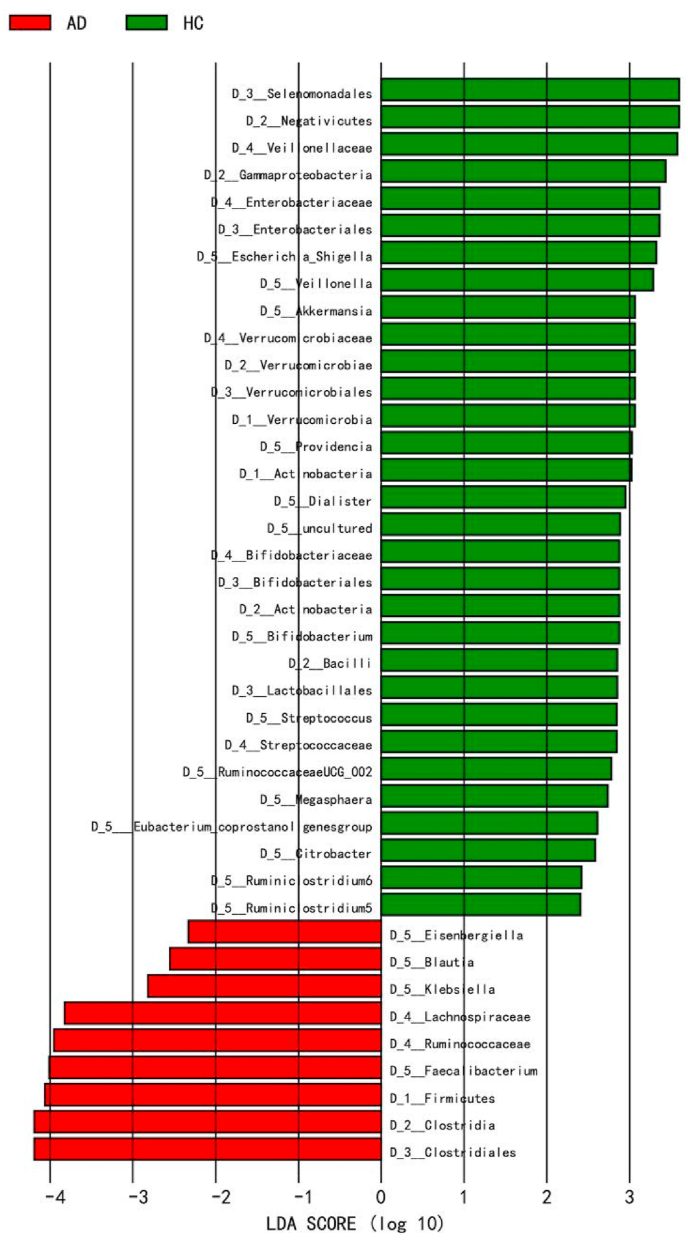
LEfSe analysis showed significantly different microbiome profiles in the two groups. A histogram of the LDA scores was computed, which indicated the effect size of each differentially abundant taxon. At the genus level, significantly higher Escherichia, Shigella, Veillonella, Akkermansia, Provindencia, Dialister, Bifidobacterium, Streptococcus, Ruminococcaceae UCG_002, Megasphaera, Eubacterium_coprostanol, Citrobacter, Ruminiclostridium_5, and Ruminiclostridium_6 were observed in the control group, while Eisenbergiella, Klebsiella, Faecalibacterium, and Blautia were higher in the ASD group (all P value < 0.05) as shown in Fig. 2.
Fig. 2.
LEfSe analysis between two groups.
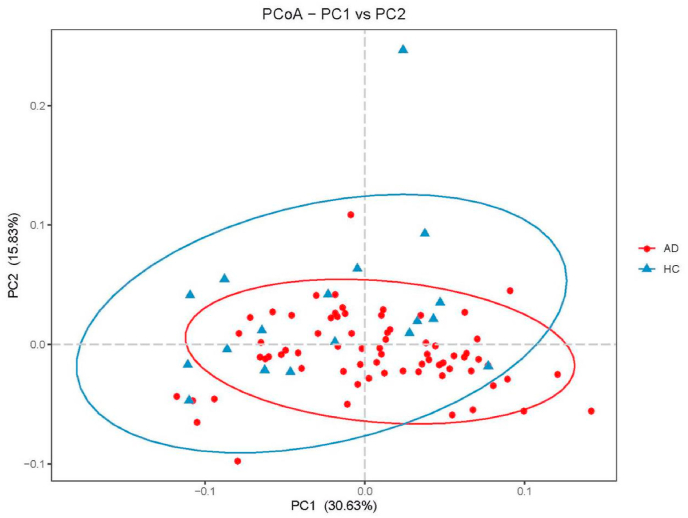
We further performed PCoA analysis of the bacterial beta diversity. As shown in Fig. 3, based on weighted UniFrac distances, the microbiome of the ASD group was distinct from that of the control group.
Fig. 3.
PCoA of bacterial diversity based on weighted distance.
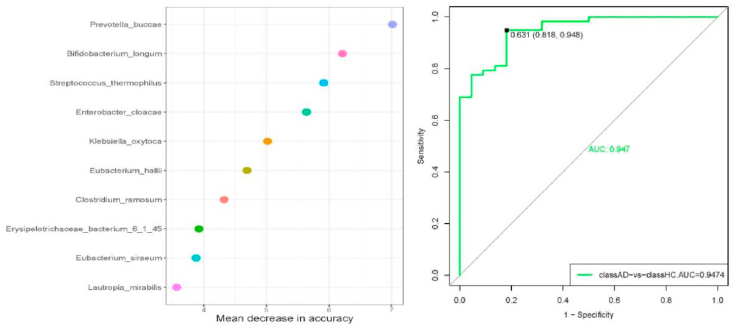
Consistent with the 16S rRNA analysis at the genus level, most of the identified differential species belonged to the genus Clostridium. We constructed a stochastic forest machine algorithm to build a prediction model of the identified bacterial strains for clinical discrimination (ASD vs. comparison group). Ten bacterial strains, namely, Prevotella buccae, Bifidobacterium longum, Streptococcus thermophilus, Enterobacter cloacae, Klebsiella oxytoca, Eubacterium hallii, Clostridium ramosum, Erysipelotrichaceae bacterium 6_1_45, Eubacterium siraeum, and Lautropia mirabilis, were selected to predict the risk of ASD in children (Fig. 4). The ROC curves are presented in Fig. 4 (Right). The highest AUC value was 0.947, with a sensitivity of 0.818 and specificity of 0.948.
Fig. 4.
Prediction model constructed by stochastic forest machine algorithm (Left) and ROC curves for clinical discrimination (Right).
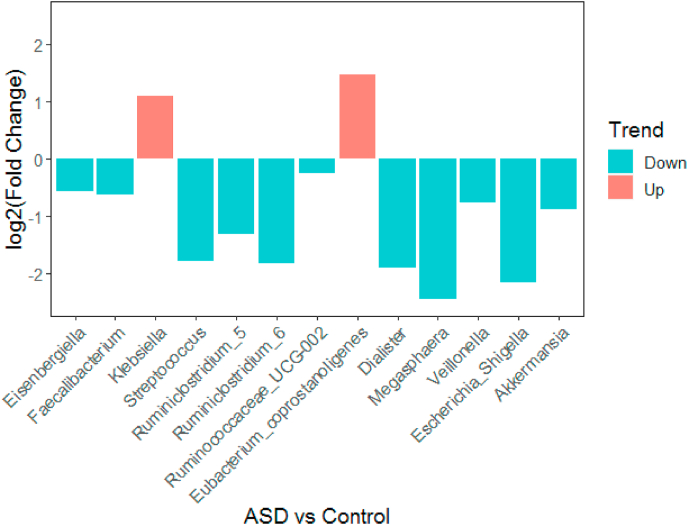
We utilized sample from an independent cohort to verify the random forest classifier derived from this study. The independent cohort include 15 asd children, with a mean age 4.17 ± 0.89 (range 3–6). Results showed that the Streptococcus,Ruminiclostridium_5,Dialister,Escherichia_Shigella was significantly lower in affected group than those in control group, which was consistent with the results of this study population, as shown in Fig. 5.
Fig. 5.
Fold change of mean relative abundance for ASD versus HC.
Discussion
ASD is a complex neurodevelopmental disorder. The microbiota plays a key role in regulating normal host metabolism, physiology, nutrition, and even brain function. Emerging evidence has revealed that the gut microbiota profile [[6], [7], [8], [9]] in individuals with ASD is different from that in neurotypical populations. To explore the profile of gut microbiota in ASD children, the current study enrolled 71 ASD children and 18 neurotypical controls. The sample size of this study is the largest of its kind in China.
Existing studies have revealed alterations in gut microbiota in ASD individuals compared with neurotypical controls. Consistent with previous clinical studies [[27], [28], [29]], the results of this study demonstrated a higher abundance of OTUs in fecal bacterial profiling in ASD patients.
The alterations in gut microbiota were related to the intensity of autistic symptomatology, indicating that bacterial metabolites may be involved in the development and severity of ASD [6]. To some extent, these findings verified the “gut–brain microbiome axis” concept, which assumes that there is a bidirectional interaction between gut microbiota and the brain. However, it is considered that this interaction is caused by hormones and neurotransmitters from the gut endocrine system. However, the specific underlying mechanisms have not yet been elucidated.
Consistent with the most recently published articles on the gut microbiome of Chinese ASD children [30], ASD patients exhibited a decreased Bacteroidetes/Firmicutes ratio.
Consistent with a previous study, Ruminococcus and Faecalibacterium were more abundant in ASD children than in the controls [31]. Studies have revealed that Faecalibacterium is related to the up- and downregulation of some genes involved in the expression of interferon (IFN) gamma. IFN is a cytokine related to ASD that is exposed during fetal development [32]. As an underlying mechanism, IFN-gamma may play an indirect role in brain plasticity and synapse formation [33]. However, its connection with the pathogenesis of autism remains to be further investigated.
The percentage of Dialister was higher in neurotypical children, in line with previous studies. However, the evidence from a previous study was not enough to observe a statistically significant difference [34]. This study reported a lower abundance of protective bacteria, such as Bifidobacterium. A number of studies have shown that some species of Bifidobacterium produce GABA (gamma aminobutyric acid) [35], the concentrations of which have been found to be low in ASD children. GABA is highly related to glutamate metabolism, which is the main excitatory neurotransmitter in the central nervous system [36]. Some studies have revealed that lower glutamate concentrations correlate with the severity of social and behavioral symptoms of ASD [37].
Consistent with recent studies, higher Veillonella, Akkermansia, and Provindencia were observed in neurological children than in the ASD group. Veillonella colonizing the intestine may supplement the Cori cycle by providing another lactic acid treatment method, thereby converting systemic lactic acid into SCFAs and re-entering the circulation [38]. Akkermansia induces immunoglobulin G1 (IgG1) antibodies and antigen-specific T cell responses, metabolic modulation, immune regulation and gut health protection [39]. Studies have shown that Akkermansia muciniphila can be a next-generation probiotic [40]. Members of the Provindencia family have been considered pathobionts, with the Providencia stuartii species being the dominant taxon. These strains swarm on semisolid (viscous) surfaces and adhere to and invade host cells, determining the specificity of disease pathogenesis and therapy [41].
The current study found an increase in Faecalibacterium and Klebsiella. Faecalibacterium prausnitzii is a late colonizer of the healthy human gut and a major butyrate producer [42,43]. Klebsiella is gaining recognition as a cause of several human infections. Recent studies of carbapenemase-producing and colistin-resistant strains demonstrate a potential reservoir of multidrug-resistant genes [44].
To our knowledge, this is the first study using the random forest algorithm to build a classifier to identify the characteristics of the gut microbiome of children with ASD, which is important for the early assessment of ASD risk. In addition, the new approach of diagnosis classifiers based on the AUC value makes it possible to plan personalized treatment and prevention strategies for ASD via microbiota modulation. Restoring the balance of the microbiota–gut–brain axis offers promising beneficial therapeutic effects on autistic deficits; however, direct clinical evidence for a role of the microbiota–gut–brain axis in ASD is relatively limited. In the future, more randomized double-blind clinical studies are required to determine the role of the microbiota–gut–brain axis in the etiology of ASD. As a potential mediator of risk factors in ASD, the microbiota is positioned at the intersection between the environment and genes. The composition and function of the microbiome are dependent on genetic background and influenced by environmental factors, including feeding patterns, age, infection, antibiotic treatment, etc. Further study could focus on the mediating effect of the microbiome on the development of ASD.
Conclusions
Significant differences were observed in the composition of the gut microbiome between boys with ASD and neurotypical controls. These findings contribute to knowledge of the alteration of the gut microbiome in ASD patients, which opens the possibility for early identification of this disease.
Availability of data and materials
The datasets used and/or analyzed during the current study are available as supplemental materials.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgments
The authors wish to thank all of the boys with ASD and their caregivers for their participation and involvement in our ASD gut microbiome Program including permission to use their clinical information. We thank all the staff who were involved with the recruitment and data collection processes. We extend sincere thanks to Mr. Liu Fei from Institute of Microbiology, Chinese Academy of Sciences for providing great support in bioinformatics analysis for the manuscript.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Liming Wang, Email: wanglm@im.ac.cn.
Pengfei Xu, Email: xpf@263.net.
Qi Zhang, Email: zhangqi0355@sina.com.
Abbreviations
- ABC
Autism Behavior Checklist
- ANOVA
Analysis of Variance
- ASD
Autism spectrum disorders
- AUC
Area Under Curve
- DNA
DeoxyriboNucleic Acid
- DSM-V
Diagnostic and Statistical Manual, Fifth Edition
- FDR
False Discovery Rates
- GABA
Gamma aminobutyric acid
- GI
gastrointestinal
- IFN
interferon
- LDA
Linear Discriminant Analysis
- LEfSe
Linear discriminant effect size
- OTUs
Operational Taxonomic Units
- PCoA
Principal coordinate Analysis
- QIIME
Quantitative Insights into Microbial Ecology
- RNA
Ribonucleic Acid
- ROC
Relative Operating Characteristic
- SPSS
Statistical Product and Service Solutions
Ethical statement and consent to participate
The study protocol and data collection instruments were approved by the ethical review boards of China-Japan Friendship Hospital. Informed consent was obtained prior to interviews.
Consent for publication
Not applicable.
Funding
The study was supported by grants from Chinese Academy of Medical Sciences medical and Health Science and Technology Innovation Project (No. 2018–12M-1–003) and China-Japan Friendship Hospital Excellence Project (No. 2018-2-QN-33).
Authors’ contributions
FY, XYG, PFX, DNH, and QZ coordinated the study, participated in the study design, supervision, and data collection. ZYW, SMC, GCL, ZTL, and LMW conducted the experiment and data analysis. FY, XYG, and ZYW participated in the drafting of the manuscript. PFX, QZ and LMW coordinated the study, participated in study design, provided supervision, and contributed to the drafting of the final manuscript. All the authors read and approved the final manuscript.
References
- 1.World Health Organization . World Health Organization; Geneva: 2004. International statistical classification of diseases and health related problems, Tenth Revision (ICD-10) [Google Scholar]
- 2.Cdc What is autism spectrum disorder? https://www.cdc.gov/ncbddd/autism/facts.html Available online:
- 3.Hsiao E.Y. Gastrointestinal issues in autism spectrum disorder. Harv. Rev. Psychiatr. 2014;22(2):104–111. doi: 10.1097/HRP.0000000000000029. [DOI] [PubMed] [Google Scholar]
- 4.McElhanon B.O., McCracken C., Karpen S., Sharp W.G. Gastrointestinal symptoms in autism spectrum disorder: a meta-analysis. Pediatrics. 2014;133(5):872–883. doi: 10.1542/peds.2013-3995. [DOI] [PubMed] [Google Scholar]
- 5.Martin C.R., Osadchiy V., Kalani A., Mayer E.A. The brain-gut-microbiome Axis. Cell Mol. Gastroenterol. Hepatol. 2018;6:133–148. doi: 10.1016/j.jcmgh.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu M., Xu X., Li J., Li F. Association between gut microbiota and autism spectrum disorder: a systematic review and meta-analysis. Front Psychiatr. 2019;10:473. doi: 10.3389/fpsyt.2019.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Son J.S., Zheng L.J., Rowehl L.M., Tian X., Zhang Y., Zhu W. Comparison of fecal microbiota in children with autism spectrum disorders and neurotypical siblings in the simons simplex collection. PloS One. 2015;10:E0137725. doi: 10.1371/journal.pone.0137725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fattorusso A., Di Genova L., Dell’isola G.B., Mencaroni E., Esposito S. Autism spectrum disorders and the gut microbiota. Nutrients. 2019;11:521. doi: 10.3390/nu11030521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bermon S., Petriz B., Kajeniene A., Prestes J., Castell L., Franco O.L. The microbiota: an exercise immunology perspective. Exerc Immunol Rev. 2015;21:70–79. [PubMed] [Google Scholar]
- 10.Di Mauro A., Neu J., Riezzo G., Raimondi F., Martinelli D., Francavilla R. Gastrointestinal function development and microbiota. Ital J Pediatr. 2013;39:15–22. doi: 10.1186/1824-7288-39-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez C.A., Kingsbury D.D., Velaz-quez E.M., Baumler A.J. Collateral damage: microbiota-derived metabolites and immune function in the antibiotic era. Cell Host Microbe. 2014;16:156–163. doi: 10.1016/j.chom.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niehus R., Lord C. Early medical history of children with autism spectrum disorders. J Dev Behav Pediatr. 2006;27:S120–S127. doi: 10.1097/00004703-200604002-00010. [DOI] [PubMed] [Google Scholar]
- 13.Finegold S.M., Dowd S.E., Gontcharova V., Liu C., Henley K.E., Wolcott R.D. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Williams B.L., Hornig M., Parekh T., Lipkin W.I. Application of novel PCR-based methods for detection, quantification, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. mBio. 2012;3:e00261. doi: 10.1128/mBio.00261-11. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudziak J.J., Novins D.K. Proposed criteria for autism spectrum disorder in the DSM-5. J Am Acad Child Adolesc Psychiatry. 2012;51:343. doi: 10.1016/j.jaac.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 16.Edwards U., Rogall T., Blöcker H., Emde M., Böttger E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fierer N., Hamady M., Lauber C.L., Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franzosa E.A., Morgan X.C., Segata N., Waldron L., Reyes J., Earl A.M., Giannoukos G., Boylan M.R., Ciulla D., Gevers D., Izard J., Garrett W.S., Chan A.T., Huttenhower C. Relating the metatranscriptome and metagenome of the human gut[J] Proc Natl Acad Sci U S A. 2014;111(22):E2329–E2338. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Rossum T., Ferretti P., Maistrenko O.M., Bork P. Diversity within species: interpreting strains in microbiomes. Nat Rev Microbiol. 2020 Sep;18(9):491–506. doi: 10.1038/s41579-020-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehta B., Venables S., Roffey P. Comparison between magnetic bead and qPCR library normalisation methods for forensic MPS genotyping[J] Int J Leg Med. 2017;132(3):1–8. doi: 10.1007/s00414-017-1591-9. [DOI] [PubMed] [Google Scholar]
- 21.Kultima J.R., Sunagawa S., Li J. MOCAT: a metagenomics assembly and gene prediction toolkit[J] PloS One. 2012;7(10) doi: 10.1371/journal.pone.0047656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo R., Liu B., Xie Y. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler[J] GigaScience. 2012;1(1):18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolpert D.H., Macready W.G. An efficient method to estimate bagging's generalization error[J] Mach Learn. 1999;35(1):41–55. [Google Scholar]
- 25.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 26.Liaw A. Classification and regression by random forest. R News. 2002;2:18–22. [Google Scholar]
- 27.Finegold S.M., Molitoris D., Song Y., Liu C., Vaisanen M.L., Bolte E. vol. 35. 2002. Gastrointestinal microflora studies in late-onset autism. Clinical infectious diseases : an official publication of the Infectious Diseases. Society of America; pp. S6–S16. [DOI] [PubMed] [Google Scholar]
- 28.Song Y., Liu C., Finegold S.M. Real-time PCR quantitation of clostridia in feces of autistic children. Appl Environ Microbiol. 2004;70:6459–6465. doi: 10.1128/AEM.70.11.6459-6465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parracho H.M., Bingham M.O., Gibson G.R., McCartney A.L. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J Med Microbiol. 2005;54:987–991. doi: 10.1099/jmm.0.46101-0. [DOI] [PubMed] [Google Scholar]
- 30.Kang D.W., Park J.G., Ilhan Z.E., Wallstrom G., Labaer J., Adams J.B. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PloS One. 2013;8 doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue R., Sakaue Y., Sawai C., Sawai T., Ozeki M., Romero-Pérez G.A. A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders. Biosci Biotechnol Biochem. 2016;80:2450–2458. doi: 10.1080/09168451.2016.1222267. [DOI] [PubMed] [Google Scholar]
- 32.Goines P.E., Croen L.A., Braunschweig D., Yoshida C.K., Grether J., Hansen R. Increased mid gestational IFN-, IL-4 and IL-5 in women bearing a child with autism:A case-control study. Mol Autism. 2011;2:13. doi: 10.1186/2040-2392-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shatz C.J. MHC class I: an unexpected role in neuronal plasticity. Neuron. 2009;64:40–45. doi: 10.1016/j.neuron.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang D.W., Adams J.B., Gregory A.C., Borody T., Chittick L., Fasano A. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome. 2017;5:10. doi: 10.1186/s40168-016-0225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srikantha P., Hasan Mohajeri M. The possible role of the microbiota-gut-brain-axis in autism spectrum disorder. Int J Mol Sci. 2019;20:2115. doi: 10.3390/ijms20092115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen J. Modeling the glutamate-glutamine neurotransmitter cycle. Front Neuroenergetics. 2013;5:1. doi: 10.3389/fnene.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horder J., Petrinovic M.M., Mendez M.A., Bruns A., Takumi T., Spooren W. Glutamate and GABA in autism spectrum disorder-a translational magnetic resonance spectroscopy study in man and rodent models. Transl Psychiatry. 2018;8:106. doi: 10.1038/s41398-018-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheiman J., Luber J.M., Chavkin T.A., MacDonald T., Tung A., Pham L.D., Wibowo M.C., Wurth R.C., Punthambaker S., Tierney B.T., Yang Z., Hattab M.W., Avila-Pacheco J., Clish C.B., Lessard S., Church G.M., Kostic A.D. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat Med. 2019 Jul;25(7):1104–1109. doi: 10.1038/s41591-019-0485-4. Epub 2019 Jun 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ansaldo E., Slayden L.C., Ching K.L., Koch M.A., Wolf N.K., Plichta D.R., Brown E.M., Graham D.B., Xavier R.J., Moon J.J., Barton G.M. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science. 2019;364(6446):1179–1184. doi: 10.1126/science.aaw7479. Jun. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhai Q., Feng S., Arjan N., Chen W. A next generation probiotic, Akkermansia muciniphila. Crit Rev Food Sci Nutr. 2019;59(19):3227–3236. doi: 10.1080/10408398.2018.1517725. [DOI] [PubMed] [Google Scholar]
- 41.Kurmasheva N., Vorobiev V., Sharipova M., Efremova T., Mardanova A. The potential virulence factors of Providencia stuartii: motility, adherence, and invasion. BioMed Res Int. 2018:3589135. doi: 10.1155/2018/3589135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coretti L., Paparo L., Riccio M.P., Amato F., Cuomo M. Gut microbiota features in young children with autism spectrum disorders. Front Microbiol. 2018;9:3146. doi: 10.3389/fmicb.2018.03146. Dec. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu M., Xu X., Li J., Li F. Association between gut microbiota and autism spectrum disorder: a systematic review and meta-analysis. Front Psychiatr. 2019;10:473. doi: 10.3389/fpsyt.2019.00473. Jul. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodríguez-Medina N., Barrios-Camacho H., Duran-Bedolla J., Garza-Ramos U. Klebsiella variicola: an emerging pathogen in humans. Emerg Microb Infect. 2019;8(1):973–988. doi: 10.1080/22221751.2019.1634981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available as supplemental materials.