Abstract
Thanks to the unbiased exploration of genomic variants at large scale, hundreds of thousands of disease-associated loci have been uncovered. In parallel, network-based approaches have proven to be essential to understand the molecular mechanisms underlying human diseases. The use of these approaches has been boosted by the abundance of information about disease associated genes and variants, high quality human interactomics data, and the emergence of new types of omics data.
The DisGeNET Cytoscape App combines the capabilities of Cytoscape with those of DisGeNET, a knowledge platform based on a comprehensive catalogue of disease-associated genes and variants. The DisGeNET Cytoscape App contains functions to query, analyze, and visualize different network representations of the gene-disease and variant-disease associations available in DisGeNET. It supports a wide variety of applications through its query and filter functionalities, including the annotation of foreign networks generated by other apps or uploaded by the user. The new release of the DisGeNET Cytoscape App has been designed to support Cytoscape 3.x and incorporates novel distinctive features such as visualization and analysis of variant-disease networks, disease enrichment analysis for genes and variants, and analytic support through Cytoscape Automation. Moreover, the DisGeNET Cytoscape App features an API to access its core functionalities via the REST protocol fostering the development of reproducible and scalable analysis workflows based on DisGeNET data.
Keywords: Disease genomics, Variant interpretation, Network biology, Text mining, Cytoscape
1. Introduction
We are in an unprecedented moment for disease genomics. The maps of the genomic architecture of human diseases, both complex and rare, are being delineated [1]. These maps are built upon the results of large scale Genome Wide Association Studies (GWAS) and from the widespread use of next generation sequencing in clinical laboratories. Genomics has been adopted in high-quality patient management in rare diseases and in oncology for diagnosis and stratified health care [2]. It is envisioned that genomic medicine will be applied more widely to cover other types of diseases (e.g. complex diseases) and especially for early detection and risk prediction [2].
Sequencing of an individual's genome can produce several millions of genomic variants; determining which of these variants lead to a disease phenotype remains a significant challenge [3]. Variant assessment still involves manual exploration of multiple sources of data, which requires a significant amount of effort from domain experts. In this context, bioinformatic tools and resources that enable the automation of every possible step in this process are crucial. Another important challenge in variant interpretation is that a significant fraction of the genomic variants identified in a personal genome has little or no information in current databases [4]. Since 2010, more than 100,000 papers are published each year on disease genomics1. Therefore, most recent findings remain locked in the literature and are incorporated at a very low pace into databases. Text mining technologies are key to assist in finding, extracting and standardizing this information from the published literature to assist the development of databases and genomic resources.
DisGeNET is an open access platform that integrates information on genes and variants associated with human diseases from several resources with information extracted by text mining from the scientific literature [5]. DisGeNET covers the full spectrum of human diseases, including traits and phenotypes that are measured as a manifestation of diseases or as a consequence of drug treatment (drug side effects). The information integrated within DisGeNET is standardized following community driven terminologies and ontologies, according to FAIR data principles [6], and is annotated with different attributes and metrics to aid searching, filtering and visualizing the information.
Another important challenge faced in genomic medicine is that even for the so-called monogenic diseases, the manifestation of the disease results from a complex interplay among genomic variants and environmental factors. For complex diseases, the number of susceptibility loci discovered by genome wide genotyping has steadily increased, imposing challenges in the interpretation of these data and in the translation of these findings into actionable information [7]. In this context, network based data representation and analysis are key approaches to model the complexity inherent to disease physiopathology [8], [9], and for the identification of disease-relevant pathways [10], [11]. In particular, network representation of genotype-phenotype data can be useful for visualization and analytic purposes, especially for diseases with high genetic heterogeneity, and can also support the analysis of shared genes and genomic variants between different diseases and phenotypes.
To serve as a bridge between network modeling and genomic data analysis, we present the DisGeNET Cytoscape App, that combines the capabilities of Cytoscape [12] with those of DisGeNET [5]. In this contribution we present the current release of the DisGeNET database (v7.0) and a new version of the DisGeNET Cytoscape App (v7.3.0). This new release of the DisGeNET Cytoscape App has been redesigned to support Cytoscape 3.x and to provide novel distinctive features including: visualization and analysis of variant-disease networks; disease enrichment analysis for genes and variants; and analytic support through Cytoscape Automation.
The DisGeNET Cytoscape App is a new desktop application that supports interactive network analysis of gene-disease and variant-disease association networks. Due to its unique functionalities enables the analysis of user’s own data with DisGeNET information, and the incorporation of other analysis tools available in the rich ecosystem of Cytoscape Apps. More importantly, by exposing its functionalities through Cytoscape Automation, DisGeNET Cytoscape App allows the automatization of queries and analysis based on DisGeNET data, and at the same time supports the development of reproducible bioinformatic pipelines for a variety of applications in precision medicine and drug R&D. In summary, with its newly implemented capabilities, the DisGeNET Cytoscape App provides the biomedical community a tool that enables a systems-level analysis of human diseases in an easy to use and reproducible way.
2. Design and implementation
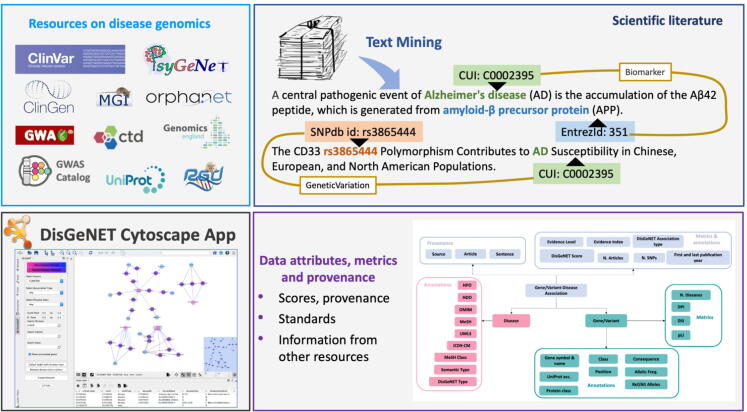
The DisGeNET Cytoscape App is designed to visualize, query and analyze a network representation of the gene-disease and the variant-disease associations contained in DisGeNET, and to analyze the data provided by the user in the context of DisGeNET’s information. Fig. 1 illustrates the main features of the DisGeNET Cytoscape App, including the data sources integrated in the current version. The DisGeNET Cytoscape App contains a set of functions to query, analyze, and visualize DisGeNET data from a network biology perspective. The gene-disease and variant-disease associations (GDAs and VDAs, respectively) are represented, queried and filtered as bipartite networks. The new version of the App includes functions to query DisGeNET for specific diseases, genes, and variants, and their combinations, and for filtering the information by a variety of data attributes and metrics, such as the original data source, the DisGeNET score and DisGeNET association type, the Evidence Index and MeSH disease class. A new feature of the App is a function for the annotation of entities from users’ provided networks with DisGeNET data, such as protein, gene or variants generated by other Cytoscape Apps or networks uploaded by the user. Finally, the App exposes its functions through Cytoscape Automation to programmatically execute different functionalities using programming languages such as R and Python.
-
1.
DisGeNET database release 7.0
Fig. 1.
The DisGeNET Cytoscape App provides access to query, analyze and visualize a network representation of DisGeNET data. The DisGeNET database integrates information on Gene-Disease and Variant-Disease Associations from multiple expert-curated resources on disease genomics and text-mined information (upper panels). Different attributes and metrics are available to customize and filter the queries, and all the provenance information is available for each association (bottom and right panel).
The DisGeNET Cytoscape App runs on the current version (7.0) of the DisGeNET database containing 1,134,942 GDAs between 21,671 genes and 30,170 diseases, and 369,554 VDAs between 194,515 variants and 14,155 diseases. GDAs and VDAs are collected from several sources (Fig. 1), that use different ontologies and terminologies to represent their data. To integrate these datasets in a harmonized database we (i) mapped gene identifiers to NCBI Entrez Gene identifiers, (ii) mapped disease vocabulary terms to the Unified Medical Language System® (UMLS®) Concept Unique Identifiers (CUIs), and (iii) integrated associations through the DisGeNET gene-disease association ontology (for more details see [5]). Fig. 2 provides a schema of the organization of the data within DisGeNET, and the different attributes, metrics and information on provenance available to the user.
Fig. 2.
The schema of the DisGeNET data and the different metrics and annotations available for the entities and associations, and the provenance. The metrics and annotations can be used both to customize the queries and to filter and prioritize the search results.
In DisGeNET, the GDAs and VDAs are grouped according to their source database. In the case of GDAs the classification encompasses the following groups: CURATED (containing GDAs from human expert curated data sources), ANIMAL MODELS (containing GDAs from animal model repositories), INFERRED (containing GDAs from HPO, GWASdb and the GWAS Catalog), and ALL (including CURATED, ANIMAL MODELS, INFERRED, and data derived from text mining the biomedical literature, named as BEFREE). In a similar manner, the VDAs are grouped in CURATED (containing VDAs from human expert curated data sources), and ALL (including CURATED data and data derived from BEFREE). For the up-to-date list and description of data sources available in DisGeNET, please visit the DisGeNET Discovery Platform Website at http://disgenet.org/dbinfo, section “Original Data Sources”.
-
2.
DisGeNET Cytoscape App
The DisGeNET Cytoscape App is an open source Cytoscape 3.x App designed to visualize, query and analyze a network representation of the gene-disease and the variant-disease associations contained in DisGeNET. The structure of the sqlite version of the DisGeNET database that supports the App is shown at the DisGeNET website2. A detailed tutorial on how to use the App is available online [13], [14]. Here we briefly describe the main functionalities of the App.
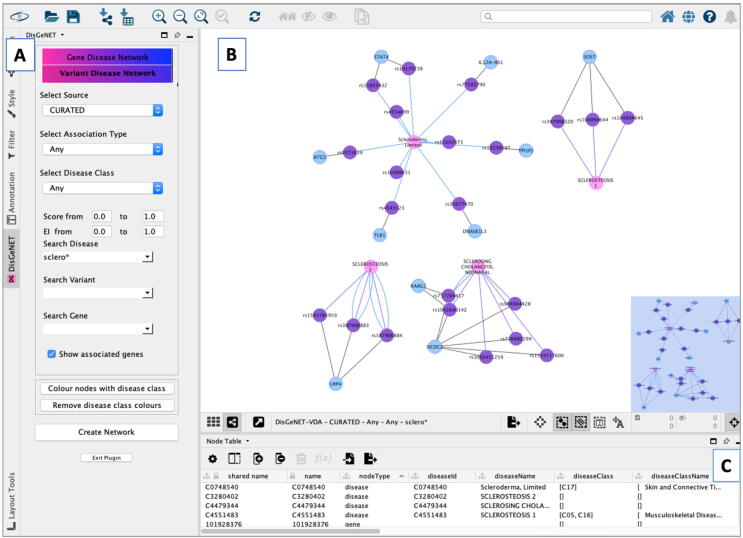
Once the DisGeNET App is installed and launched, the DisGeNET control panel (Fig. 3) contains two tabs to guide the query over different views of the information represented as networks: the Gene-Disease and the Variant-Disease networks. The Gene-Disease Network and Variant-Disease Network are bipartite graphs with two types of vertices (genes/variants and diseases) and edges connecting the vertices of different types (e.g., a gene with a disease). In these graphs, two vertices can be connected by more than one edge, representing the multiple pieces of evidence reporting the GDAs or the VDAs, such as different database source, publications, or DisGeNET association type. Fig. 4 shows a VDA network represented as a bipartite graph.
Fig. 3.
The DisGeNET Cytoscape App. A: The DisGeNET control panel with its two tabs to query GDA and VDA networks. The networks can be created customizing different parameters such as source database, association type, disease class, score and EI. B: The networks are displayed using the DisGeNET visual style and can be rendered using Cytoscape layouts. C: The lower panel shows 3 tables: Node, edge and network, displaying all the information from the network.
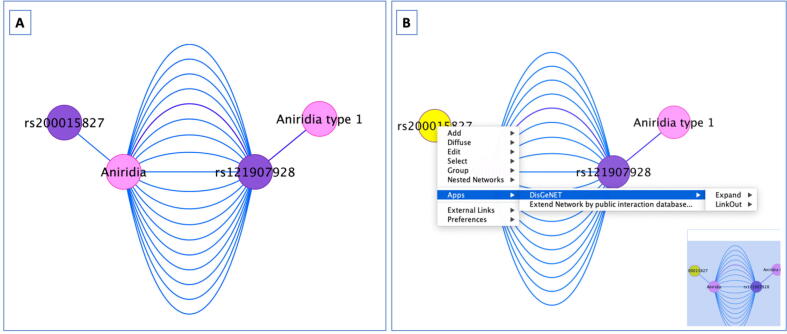
Fig. 4.
A. VDA networks are represented as bipartite graphs, with two types of vertices (variants in purple and diseases in pink) and edges connecting the vertices of different types (a variant with a disease). In these graphs, two vertices can be connected by more than one edge, representing the multiple pieces of evidence reporting the VDAs, such as different database source, publications, or different DisGeNET association types. In the example, the edge connecting Aniridia type 1 and variant rs121907928 is classified as CausalMutation in the DisGeNET association type ontology (shown in purple) whereas the edge connecting Aniridia and variant rs200015827 is classified as GeneticVariation in the DisGeNET association type ontology (shown in blue). GDA networks are represented as bipartite graphs in a similar manner (not shown in the image). B. By right-clicking on a selected node of the network, a drop-down menu is display allowing the application of the advanced Expand and Linkout functions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The Gene-Disease and Variant-Disease networks can be queried by single or multiple entities (gene, disease or variants), using free text searches or standard identifiers. The query can be restricted by the source database (e.g., UniProt, Orphanet or group of source databases, such as CURATED, ANIMAL_MODELS, etc.), the DisGeNET association type or the Evidence Level for the association, or the MeSH Disease class of the disease. In addition, all the queries can be further filtered by other DisGeNET attributes, such as the Evidence Index and the DisGeNET score. The attributes and metrics are displayed in Fig. 2. The results of the queries are displayed on the right-hand side of the Cytoscape panel as a network using the DisGeNET Visual style, and as Node and Edge tables at the bottom of the Cytoscape window displaying in a tabular format all the data visually represented in the networks. The networks can be rendered using different layouts, exported in different formats, or analyzed by other Apps available in Cytoscape.
In summary, the basic functions of the DisGeNET Cytoscape app allows the user to perform the following tasks:
-
•
retrieve and visualize the diseases associated with a gene or variant of interest: for instance, the 93 diseases associated with the variant rs7903146 in the gene TCF7L2; or the 29 diseases associated with the gene TCF7L2 reported by CURATED sources.
-
•
retrieve and visualize the list of genes or variants associated with a particular disease: for example, the 127 genes associated with Schizoaffective Disorder can be visualized as a GDA network.
-
•
generate networks for lists of genes, diseases or variants and to create GDA or VDA networks for a specific source database: for instance, to represent all the GDAs reported in a database such Genomics England as a GDA network.
-
•
retrieve those GDAs involving germline modifying mutations by selecting the “germline modifying mutations” type in the DisGeNET control panel.
-
•
create a network for a specific disease class: for instance, the GDAs for diseases classified as “Nutritional and Metabolic Diseases”
-
•
retrieve a GDA or VDA network combining different parameters: database source, DisGeNET association type, Disease Class, Evidence Level, Evidence Index and DisGeNET score.
The App also offers additional functions that allow a) coloring genes, variants, and diseases in the network according to MeSH disease classes, b) expanding the information for specific nodes in the network, c) exploring different link outs for the nodes and the edges thanks to DisGeNET data standardization, and d) disease enrichment analysis. In the following sections we describe the advanced functions available. For further details on how to use them, see section 3.b of the tutorial (https://www.disgenet.org/app/guide).
-
a.
Coloring nodes by disease class
The DisGeNET app allows coloring the nodes of a network according to the MeSH disease classification. This function can be activated by clicking on the “Colour nodes by disease class” button in the DisGeNET App control panel. In the case of diseases, the coloring is based on the disease class annotation of the disease node. Diseases belonging to more than one disease class will appear in more than one color. In the case of genes and variants, the MeSH disease class is assigned from the disease(s) associated to them. This functionality enhances the visual inspection of both the GDA and VDA networks.
-
b.
DisGeNET expand function
The DisGeNET expand function allows to retrieve all the data available in DisGeNET for a specific node in a network. The expand function is particularly useful when an initial search on a single database (e.g., UniProt) is performed, and once the network is obtained, the user wants to know if there is more information in other data sources available in DisGeNET. The function can be used to create a new DisGeNET network using the selected node(s) for the query or to expand the existing network with nodes and edges found in DisGeNET ALL. Several parameters to customize the search can be applied to the expand function, which can be accessed from the DisGeNET app menu that is displayed by right-clicking on a node in a DisGeNET network.
-
c.
DisGeNET Linkouts
In order to get more information about a specific gene, disease or variant from a DisGeNET network from reference databases, a LinkOut function is available to easily jump to a particular entry in those reference databases. In the case of edges, the LinkOuts allows the user to explore the publication supporting the GDA or VDA, by clicking on the edge connecting the pair of interest and accessing the LinkOut to NCBI PubMed. Users can access the linkout function by right clicking the node or edge of interest, in the DisGeNET app menu, as illustrated in Fig. 4, panel B.
-
d.
Annotating user’s data with DisGeNET
The DisGeNET Cytoscape App also allows annotating networks generated by other applications or uploaded by the user. Furthermore, the App enables two new functionalities to work with foreign networks of genes, variants, either uploaded by the user or generated by other apps. The first functionality allows to annotate a network of genes or variants, with disease information from DisGeNET. The annotations can be filtered by source, DisGeNET score, and by disease class. These functions can be accessed by right-clicking on an empty space on the main Network View Window of Cytoscape.
-
e.
Disease enrichment analysis
A new function is available to perform a disease enrichment analysis, also known as over-representation analysis or gene set enrichment analysis, on a gene/variant network with disease information from DisGeNET. It maps genes/variants to diseases in DisGeNET and detects statistically significantly enriched diseases by means of a Fisher’s test and p-values corrected for multiple testing using the Benjamini-Hochberg method. This functionality is useful to evaluate if the number of genes or variants associated with a particular disease in a user’s dataset is higher than expected by chance. After performing the disease enrichment analysis, a new table is created (DisGeNET Enrichment table) that contains the statistically significant diseases, the number of genes/variants from the input list that are annotated to the specific disease, the number of genes/variants associated with the disease in the selected source, and the p-value of the Fisher test. The user can choose to create a new network containing the diseases that result from the enrichment, and their associated genes/variants from the input network uploaded by the user. Additionally, the user can select a minimum number of genes or variants, and a p-value threshold to create the network. This function can also be accessed by right-clicking on an empty space on the main Network View Window of Cytoscape
-
f.
Functions exposed through Cytoscape automation
Finally, the App features a new API automation module that allows to programmatically execute different functionalities using popular programming languages such as R and Python. The API exposes four functions (Table 1) that have been documented using Swagger following the Cytoscape Automation documentation standards [15], supporting the development of reproducible and scalable analysis workflows based on DisGeNET data. A detailed tutorial describing the use of the automation module is available at [13].
Table 1.
DisGeNET Cytoscape App functions exposed through Cytoscape Automation. Documentation on the functions is available in Swagger.
| Function | Description | Protocol |
|---|---|---|
| version | Returns the version of the database, App and API | GET |
| gene-disease-net | Returns a GDA network | POST |
| variant-disease-net | Returns a VDA network | POST |
| gene-enrichment | Performs a disease enrichment analysis on genes | POST |
| variant-enrichment | Performs a disease enrichment analysis on variants | POST |
3. Application of DisGeNET to variant analysis
DisGeNET can be used to support several applications in precision medicine and drug R&D. To showcase the functionalities of the DisGeNET Cytoscape App, we illustrate how it can be used to support the annotation of genomic variants discovered by high-throughput analysis of the human genome. The purpose of this example is twofold, first to show the kind of analysis that can be performed with the DisGeNET Cytoscape App, and second to illustrate how all these analyses can be performed through scripts to enable their automation and reproducibility. In addition, we provide the detailed protocol on how to conduct this analysis using the functions of the DisGeNET App exposed through automation in the R notebook available online [13].
To showcase the application of DisGeNET in variant annotation and analysis we use data reported by Olafsdottir and colleagues [16] on a genome-wide association meta-analysis of 69,189 cases and 702,199 controls from Iceland and UK biobanks. With the purpose of identifying risk variants for asthma, they find 88 variants at 56 loci, 19 of them previously unreported. In the R notebook available at online [13], we show how to use DisGeNET to analyze the list of 88 risk variants identified in this study [16]. In particular, with the help of the DisGeNET Cytoscape App, we sought to answer the following questions:
-
•
Are the risk variants associated with asthma? What is the evidence for their association?
-
•
Are the risk variants associated with different types of asthma?
-
•
Are the risk variants associated with other diseases or phenotypes?
-
•
Are the genes to which these variants map also associated with asthma?
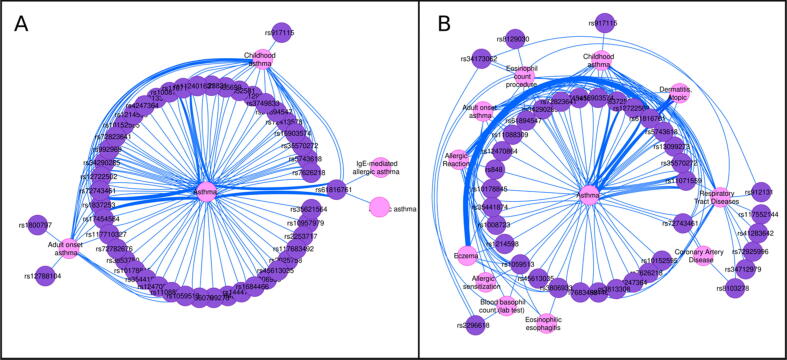
Using the “variant-disease-net” automation function endpoint available in the CyREST API (see more details in section 2.4 “Create a VDA network associated to a list of variants” [13]) we can interrogate DisGeNET with the list of 88 risk variants identified by their dbSNP identifiers (also known as rs numbers). By querying DisGeNET with the list of variants, we obtain a variant-disease association network containing 64 out of the 88 variants. The 64 variants are associated with 143 different diseases and phenotypes, among them respiratory tract diseases and disorders of the immune system. In particular, using the “variant-disease-net“ endpoint in the CyREST API of the DisGeNET Cytoscape App and setting the diseaseClass filter to ”Respiratory Tract Diseases“, we can easily retrieve a VDA network for the subset of variants associated with different types of asthma (Fig. 5, panel A). While 46 variants are associated with Asthma, 23 are associated with Childhood asthma, and 14 to Adult-onset asthma. We can also identify genomic variants, such as the one identified as rs61816761, associated with different asthma subtypes, namely Asthma, Childhood asthma, Allergic asthma, and IgE-mediated allergic asthma.
Fig. 5.
Annotating user’s data with DisGeNET. A) Variant-disease network for the genomic variants reported in [16] and associated with different types of Asthma according to DisGeNET. B: Variant-disease network representation of the disease enrichment analysis of the variants reported in [16]. We show only the diseases associated with more than one variant.
DisGeNET allows easy exploration of the supporting evidence for each of the VDA associations. By exploring the edges table, we can see that variant rs61816761 is associated with Asthma and Childhood asthma with high DisGeNET scores (0.78 and 0.71, respectively) indicating that those associations have been collated in expert-curated databases and are supported by several publications (more than 10 publications for the association with Asthma). The multiple edges connecting the variants to Asthma indicate that there are several pieces of evidence supporting the association. For example, 12 publications support the association between rs61816761 and Asthma, the oldest from 2007 [17], [18] and the more recent from 2019 [19].
Notably, by using DisGeNET to annotate the variants we find out that from 19 “unreported loci” in the original publication, 10 were already reported in DisGeNET, from which 4 are associated with different types of asthma (rs7626218 and rs3813308: Asthma, rs1800797 and rs12788104: Adult onset asthma and rs7626218: Childhood asthma) with publications dating back to 2016.
A new functionality that offers the DisGeNET Cytoscape App is the disease enrichment analysis. This kind of analysis enables discovery of the diseases and phenotypes in which the list of variants is significantly enriched. Fig. 5 (panel B) shows the network resulting from performing a disease enrichment analysis using DisGeNET (CURATED data source). The enrichment is performed by calling the automation function “variant-enrichment” endpoint in the CyREST API (See section 2.5 “Perform a variant enrichment analysis” from the R notebook [13]). The top statistically significant diseases are Asthma (p-value = 7.8e-85), Respiratory Tract Diseases (p-value 1.18e-69), Childhood Asthma (p-value 1.2e-48) and Allergic Reaction (p-value 1.9e-42). Other processes related to the immune system are also enriched in this list: Eosinophil count procedure (p-value 1.3e-18), and Blood Basophil count (p-value 5.5e-6). More information about the enrichment can be explored in the DisGeNET enrichment table in the Cytoscape panel. A complete step by step tutorial is available at [13].
In summary, the DisGeNET Cytoscape App can be used to easily annotate the results of a GWAS by interrogating multiple data sources and the scientific literature in a fast and customizable manner. Moreover, its Automation capabilities enable the automatization, sharing and reproducibility of the analysis.
4. Conclusions
The DisGeNET Cytoscape App leverages the capabilities of Cytoscape to explore genotype-phenotype associations provided by DisGeNET, including the prioritization metrics and attributes available in the latest version of DisGeNET (v7.0). As a remarkable feature it allows the exploration of variant-disease networks. The DisGeNET VDA and GDA networks are also accessible from the NDEX project [20]. The DisGeNET App can be used in a variety of applications: a) for variant analysis both in Mendelian and complex diseases; b) to analyze omics or biomedical datasets in the context of Cytoscape; c) to explore the association between diseases and traits through shared variants.
This new release of the DisGeNET Cytoscape App has been redesigned to support Cytoscape 3.x and to provide novel distinctive features including: visualization and analysis of variant-disease networks; disease enrichment analysis for genes and variants; and analytic support through Cytoscape Automation. We showcased how DisGeNET can be used to support variant analysis and interpretation, supporting different applications on precision medicine. Moreover, the example shown illustrates how the use of DisGeNET can help the user to recover information not readily available from other resources, thanks to its comprehensive database that includes findings extracted from the literature by text mining approaches.
As for the time of writing this manuscript (May 2021) the DisGeNET Cytoscape App accounts for more than 18,000 downloads from the Cytoscape App Store3, while the number of citations of DisGeNET currently exceeds 2,000.
Future improvements of the App include the incorporation of new functionalities to support the interpretation of the functional impact of genomic variants, based on the analysis of protein–protein interaction networks to discover disease-associated modules, or the incorporation of functional genomics data such as eQTL to support the analysis of non-coding disease associated variants.
In summary, with its newly implemented capabilities, the DisGeNET Cytoscape App provides the biomedical community a tool that enables a systems-level analysis of human diseases in an easy and automatic way.
Funding
IMI-JU resources of which are composed of financial contribution from the EU-FP7 [FP7/2007–2013] and EFPIA companies in kind contribution [116030 to TransQST, 777365 to eTRANSAFE], and the EU H2020 Programme 2014–2020 [676559 to Elixir-Excelerate.]; Project 001-P-001647 - Valorisation of EGA for Industry and Society funded by the European Regional Development Fund (ERDF) and Generalitat de Catalunya; Agència de Gestió d’Ajuts Universitaris i de Recerca Generalitat de Catalunya [2017SGR00519]. The Research Programme on Biomedical Informatics (GRIB) is a member of the Spanish National Bioinformatics Institute (INB), funded by ISCIII and FEDER (PRB2-ISCIII [PT13/0001/0023, of the PE I + D + i 2013–2016]). The DCEXS is a ‘Unidad de Excelencia María de Maeztu’, funded by the MINECO [MDM-2014-0370].
6. Availability
The DisGeNET App is available at the Cytoscape app store (https://apps.cytoscape.org/apps/disgenet). A detailed tutorial is provided at https://www.disgenet.org/app/guide.
CRediT authorship contribution statement
Janet Piñero: Conceptualization, Validation, Data curation, Writing - original draft, Writing - review & editing. Josep Saüch: Software. Ferran Sanz: Funding acquisition, Writing - review & editing. Laura I. Furlong: Conceptualization, Validation, Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Janet Piñero, Laura I. Furlong and Ferran Sanz are co-founders of MedBioinformatics Solutions SL.
Footnotes
The PubMed search (“Psychiatry and Psychology Category” [Mesh] AND “genetics” [Subheading]) OR (“Diseases Category” [Mesh] AND “genetics” [Subheading]) results on 1.89M citations and 102,600 citations/month since 2010 (accessed Feb 2021)
References
- 1.Claussnitzer M., Cho J.H., Collins R., Cox N.J., Dermitzakis E.T., Hurles M.E. A brief history of human disease genetics. Nature. 2020;577(7789):179–189. doi: 10.1038/s41586-019-1879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott R.H., Fowler T.A., Caulfield M. Genomic medicine: time for health-care transformation. Lancet. 2019;394(10197):454–456. doi: 10.1016/S0140-6736(19)31796-9. [DOI] [PubMed] [Google Scholar]
- 3.Eilbeck K., Quinlan A., Yandell M. Settling the score: variant prioritization and Mendelian disease. Nat Rev Genet. 2017;18(10):599–612. doi: 10.1038/nrg.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio T.A., Rowley R., Williams M.S., Roden D., Ginsburg G.S., Bult C. Opportunities, resources, and techniques for implementing genomics in clinical care. Lancet. 2019;394(10197):511–520. doi: 10.1016/S0140-6736(19)31140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., Ronzano F., Centeno E., Sanz F. update. Nucleic Acids Res. 2019;2019 doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3(1) doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeggini E, Gloyn AL, Barton AC, Wain L V. Translational genomics and precision medicine: Moving from the lab to the clinic. Science (80-) 2019;365:1409–13. https://doi.org/10.1126/science.aax4588. [DOI] [PubMed]
- 8.Caldera M., Buphamalai P., Müller F., Menche J. Interactome-based approaches to human disease. Curr Opin Syst Biol. 2017;3:88–94. doi: 10.1016/j.coisb.2017.04.015. [DOI] [Google Scholar]
- 9.Capriotti E., Ozturk K., Carter H. Integrating molecular networks with genetic variant interpretation for precision medicine. Wiley Interdiscip Rev Syst Biol Med. 2019;11(3):e1443. doi: 10.1002/wsbm.2019.11.issue-310.1002/wsbm.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donato L., Scimone C., Alibrandi S., Pitruzzella A., Scalia F., D’Angelo R. Possible A2E mutagenic effects on RPE mitochondrial DNA from innovative RNA-Seq bioinformatics pipeline. Antioxidants. 2020;9(11):1158. doi: 10.3390/antiox9111158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donato L., Scimone C., Alibrandi S., Abdalla E.M., Nabil K.M., D’Angelo R. New omics—derived perspectives on retinal dystrophies: could ion channels-encoding or related genes act as modifier of pathological phenotype? Int J Mol Sci. 2020;22(1):70. doi: 10.3390/ijms22010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piñero J., Saüch-Pitarch J. LI; DisGeNET Cytoscape App and Automation Tutorial: 2021. Furlong. https://www.disgenet.org/app/rmd-notebook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piñero J, Furlong. LI. DisGeNET Cytoscape App user’s guide 2021. https://www.disgenet.org/static/disgenet_ap1/files/current/DisGeNET_Cytoscape_v7.pdf. [DOI] [PMC free article] [PubMed]
- 15.Otasek D., Morris J.H., Bouças J., Pico A.R., Demchak B. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019;20(1) doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olafsdottir T.A., Theodors F., Bjarnadottir K., Bjornsdottir U.S., Agustsdottir A.B., Stefansson O.A. Eighty-eight variants highlight the role of T cell regulation and airway remodeling in asthma pathogenesis. Nat Commun. 2020;11(1) doi: 10.1038/s41467-019-14144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer C.N.A., Ismail T., Lee S.P., Terron-Kwiatkowski A., Zhao Y., Liao H. Filaggrin null mutations are associated with increased asthma severity in children and young adults. J Allergy Clin Immunol. 2007;120(1):64–68. doi: 10.1016/j.jaci.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Morar N., Cookson W.O.C.M., Harper J.I., Moffatt M.F. Filaggrin mutations in children with severe atopic dermatitis. J Invest Dermatol. 2007;127(7):1667–1672. doi: 10.1038/sj.jid.5700739. [DOI] [PubMed] [Google Scholar]
- 19.Pividori M., Schoettler N., Nicolae D.L., Ober C., Im H.K. Shared and distinct genetic risk factors for childhood-onset and adult-onset asthma: genome-wide and transcriptome-wide studies. Lancet Respir Med. 2019;7(6):509–522. doi: 10.1016/S2213-2600(19)30055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillich RT, Chen J, Rynkov V, Welker D, Pratt D. NDEx: A community resource for sharing and publishing of biological networks. Methods Mol. Biol., vol. 1558, Humana Press Inc.; 2017, p. 271–301. https://doi.org/10.1007/978-1-4939-6783-4_13. [DOI] [PubMed]