Abstract
N6-methyladenosine (m6A) is a modification that plays pivotal roles in RNA metabolism and function, although its functions in spliceosomal U6 snRNA remain unknown. To elucidate its role, we conduct a large-scale transcriptome analysis of a Schizosaccharomyces pombe strain lacking this modification and found a global change of pre-mRNA splicing. The most significantly impacted introns are enriched for adenosine at the fourth position pairing the m6A in U6 snRNA, and exon sequences weakly recognized by U5 snRNA. This suggests cooperative recognition of 5’ splice site by U6 and U5 snRNPs, and also a role of m6A facilitating efficient recognition of the splice sites weakly interacting with U5 snRNA, indicating that U6 snRNA m6A relaxes the 5’ exon constraint and allows protein sequence diversity along with explosively increasing number of introns over the course of eukaryotic evolution.
Subject terms: RNA, RNA modification, RNA splicing
Spliceosomal U6 snRNA is modified by m6A. Here the authors show that m6A modification on U6 snRNA promotes efficient recognition of the splice site through cooperating with U5 snRNA, indicating that U6 snRNA m6A allows the 5’ exon sequence variation.
Introduction
Post-transcriptional modification is a characteristic structural feature of RNAs. About 150 types of chemical modifications have been found in various RNAs from all domains of life1–3. Recent studies using deep-sequencing technologies successfully mapped several species of RNA modifications in eukaryotic mRNAs and non-coding RNAs in a transcriptome-wide manner4–6. These findings raise the concept of the “epitranscriptome” and highlight the importance of RNA modification as a previously unrecognized layer of regulatory gene expression.
N6-methyladenosine (m6A), an abundant modification in eukaryotic mRNAs and long non-coding RNAs, plays a critical role in various biological processes, including meiosis7,8, circadian rhythm9, sex determination10,11, cell proliferation12, differentiation13, reprogramming14, and stress responses15. The biogenesis and dynamics of m6A have been studied extensively. The METTL3/METTL14 writer complex introduces m6A in the RRACH motif (R = A or G; H = except G)16–23. Internal m6As are abundant in the last exon of mRNAs, near the stop codon20,21,24,25, and are decoded by several reader proteins, including YTH proteins, thereby contributing to the diverse fates of mRNAs26–33. m6A also has the ability to destabilize double-stranded regions of structured RNA, thereby facilitating recognition by hnRNP C and hnRNP G34,35. It has been proposed that m6A is a reversible modification that can be modified back to adenosine by erasers FTO and ALKBH536,37.
In addition to internal m6A, N6, 2′-O-dimethyladenosine (m6Am) is present at the transcription start site of mRNAs in vertebrates38,39. Recently, our group reported a cap-specific m6A writer, CAPAM/PCIF1, which catalyzes N6-methylation of m6Am40. Structural studies of CAPAM revealed the molecular basis of cap-specific m6Am formation, and ribosome profiling analysis revealed that CAPAM-mediated N6-methylation promotes translation of mRNAs starting from m6Am.
METTL16 is an m6A writer for U6 snRNA and MAT2A mRNA41–43, which encodes S-adenosylmethionine (SAM) synthetase. METTL16 targets a specific nonamer sequence in structured RNA (UACAGAGAA) for m6A formation41,44,45. At high SAM concentrations, METTL16 introduces m6A into six hairpin structures in the 3′ UTR of MAT2A mRNA, leading to degradation of the mRNA mediated by YTHDC1. At low SAM concentrations, METTL16 is bound to the hairpin structures and acts as a splicing enhancer for the 3′ terminal intron, thereby increasing production of mature MAT2A mRNA. Thus, METTL16 mediates a feedback mechanism that controls SAM homeostasis. This regulatory mechanism is critical for cell viability, as shown by the essentiality of METTL16 gene in human culture cells46 and Mettl16 in mouse embryo44. Mettl16 null mutations are embryonically lethal, and homozygotes die around the implantation stage (E3.5). Early embryos lacking Mettl16 contain reduced amounts of Mat2a mRNA, highlighting the importance of Mettl16 for production of this message.
U snRNAs are essential components of the spliceosome, contributing to recognition of substrate pre-mRNAs and serving as ribozyme catalysts of two consecutive transesterifications to ligate two exons concomitant with removal of an intron47–54. U snRNAs contain a number of modified internal nucleosides, including pseudouridines, ribose 2′-O-methylations, and several base methylations55–58. 2′-O-methylation and pseudouridylation of U2 snRNA are essential for snRNP biogenesis and efficient splicing59–65. In U6 snRNA from mammals56,66, S. pombe57, and broad bean67, an m6A is present in the middle position of the ACAGA box that recognizes the 5′ splice site (5′SS)56,57,66,67 (Fig. 1a and Supplementary Fig. 1). In human, the m6A at position 43 in the ACAGA box is introduced by METTL1641,42,68, whereas in Schizosaccharomyces pombe, the ortholog Mtl16 is responsible for the m6A modification at position 37 in the ACAGA box41. This m6A faces the fourth position of the intron when the ACAGA box base-pairs with the 5′SS in the spliceosome B complexes, implying a functional role of m6A in mRNA splicing42. However, because little change in global splicing is observed in null mutant mice embryos44, and because alteration in splicing has not been studied in METTL16-deficient strains from other organisms, the functional significance of m6A in U6 snRNA remains elusive.
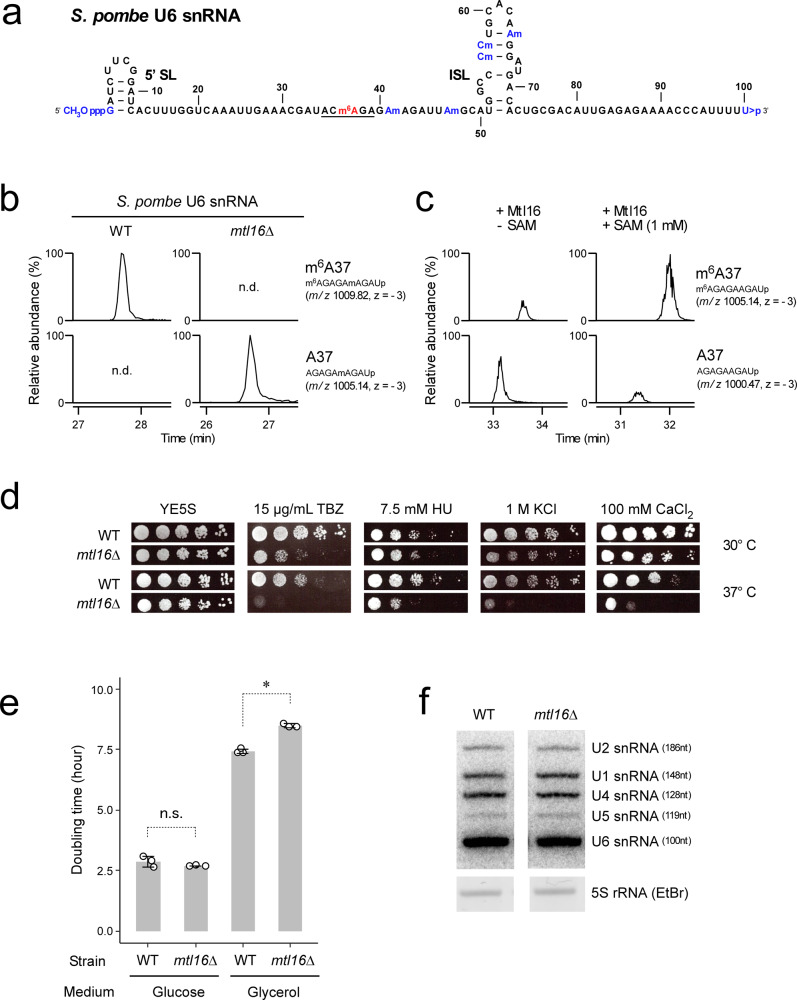
Fig. 1. Phenotypes of cells lacking the m6A modification in U6 snRNA.
a Primary and secondary structures of U6 snRNA from S. pombe with the following post-transcriptional modifications: CH3OpppG (γ-methyl triphosphate cap), N6-methyladenosine (m6A), 2′O-methylations (Nm), and 2′,3′ cyclic phosphate (>p). 5′SL and ISL represent the 5′ stem loop and internal stem loop, respectively. The underlined sequence represents the ACAGA box. b Loss of m6A in U6 snRNA isolated from SPAC27D7.08c knockout (mtl16Δ) cells. Mass chromatograms of the RNase A-digested fragments of U6 snRNA containing m6A37 or A37 isolated from S. pombe WT and mtl16Δ cells. n.d., not detected. c In vitro reconstitution of m6A in U6 snRNA catalyzed by S. pombe Mtl16 in the presence or absence of SAM. Mass chromatograms of the RNase A-digested fragments of U6 snRNA containing m6A37 or A37. d Growth phenotypes of S. pombe WT and mtl16Δ strains cultured at 30 °C or 37 °C on YE5S plates supplemented with the indicated inducers of cell stress. The plates were photographed 3–4 days after inoculation. TBZ, thiabendazole; HU, hydroxyurea. e Doubling times of WT and mtl16Δ cells were measured in glucose (YE5S) and glycerol media. OD600 was measured every hour for 12 h. The asterisk indicates a statistically significant difference in the doubling time of the two strains, as determined by two-sided Student’s t-test. *p = 7.7e-5; n = 3 biologically independent samples. Data are presented as mean values +/− SD. Source data are provided as a Source Data file. f Northern blotting of S. pombe U snRNAs in total RNA of WT and mtl16Δ cells. 5 S rRNA in total RNA was stained by EtBr as a loading control. Length of each U snRNA is indicated. This result was repeated once with a similar result. Source data are provided as a Source Data file.
To elucidate the functional role of m6A in U6 snRNA, we performed RNA-seq analyses of an S. pombe mtl16 knockout strain. We found that a subset of introns was retained in mRNAs, indicating a splicing defect caused by loss of m6A in U6 snRNA. The most significantly impacted introns were enriched for adenosine at the fourth position from the 5′SS, which base-pairs with m6A of U6 snRNA. In addition, the retained introns tended to have sequences that are weakly recognized by U5 snRNA at the 3′ termini of their 5′ exons, suggesting that U6 and U5 snRNPs cooperatively recognize the 5′SS, and that m6A in U6 snRNA facilitates efficient splicing of a subset of pre-mRNAs.
Results
Physiological phenotypes of S. pombe mtl16 knockout strains
To elucidate the functional roles of m6A in U6 snRNA (Fig. 1a and Supplementary Fig. 1), we chose S. pombe as a model organism because m6A was undetectable in the poly(A)+ RNA fraction (Supplementary Fig. 2) and the S. pombe MAT2a homolog sam1 contains no hairpin structures harboring the specific nonamer sequence targeted by METTL1669. Consequently, U6 snRNA m6A is the sole target for S. pombe Mtl16. First, we verified that U6 snRNA m6A is introduced by S. pombe Mtl16. To this end, we isolated U6 snRNAs from wild-type (WT) and mtl16 knockout (mtl16Δ) strains by reciprocal circulating chromatography (RCC)70. The isolated U6 snRNA was digested by RNase T1 and subjected to capillary LC-nano-ESI-MS to analyze the RNA fragment containing m6A37. m6A was clearly present in U6 snRNA isolated from the WT strain (Fig. 1b and Supplementary Fig. 3a), and no fragment bearing A37 was detected, indicating that U6 snRNA was fully modified with m6A37. By contrast, m6A37 was completely absent in the U6 snRNA isolated from the mtl16Δ strain (Fig. 1b and Supplementary Fig. 3b). Next, we conducted an in vitro reconstitution of m6A in U6 snRNA using recombinantly expressed S. pombe Mtl16 in the presence or absence of SAM. m6A was efficiently introduced at position 37 of U6 snRNA in the presence of SAM (Fig. 1c), whereas m6A was slightly detected even in the absence of SAM (Fig. 1c), indicating that Mtl16 contains endogenous SAM in its active site. In addition, we probed the modified RNA fragment by collision-induced dissociation (CID) analysis to confirm the presence of m6A at position 37 (Supplementary Figs. 3c, d). These results demonstrated that S. pombe Mtl16 is an m6A writer for U6 snRNA. In a previous study43, we reconstituted m6A formation in 3′ UTR hairpins of MAT2A mRNA using human METTL16. Similarly, in this study, we confirmed m6A formation in U6 snRNA by human METTL16 (Supplementary Figs. 4a, b).
We searched for physiological phenotypes of the mtl16Δ strain on media containing some inducers for cellular stress. As reported previously71, the mtl16Δ strain was sensitive to reagents that induce DNA damage, e.g., hydroxyurea (HU) and thiabendazole (TBZ) (Fig. 1d). In addition, we found that the mtl16Δ strain was sensitive to salt stress (KCl and CaCl2); these sensitivities were exacerbated when the cells were cultured at 37 °C (Fig. 1d). The mtl16Δ strain grew more slowly than the WT strain in medium containing a nonfermentable carbon source (Fig. 1e), implying slight reduction of mitochondrial activity.
To determine whether m6A loss affects stability of U6 snRNA, we analyzed steady-state levels of U snRNAs by Northern blotting (Fig. 1f). We detected no significant difference in the levels of U6 and other U snRNAs between the WT and mtl16Δ strains, implying that the phenotype of the mtl16Δ strain is due to a functional consequence of the m6A loss from U6 snRNA.
Global alteration of mRNA splicing caused by m6A loss in U6 snRNA
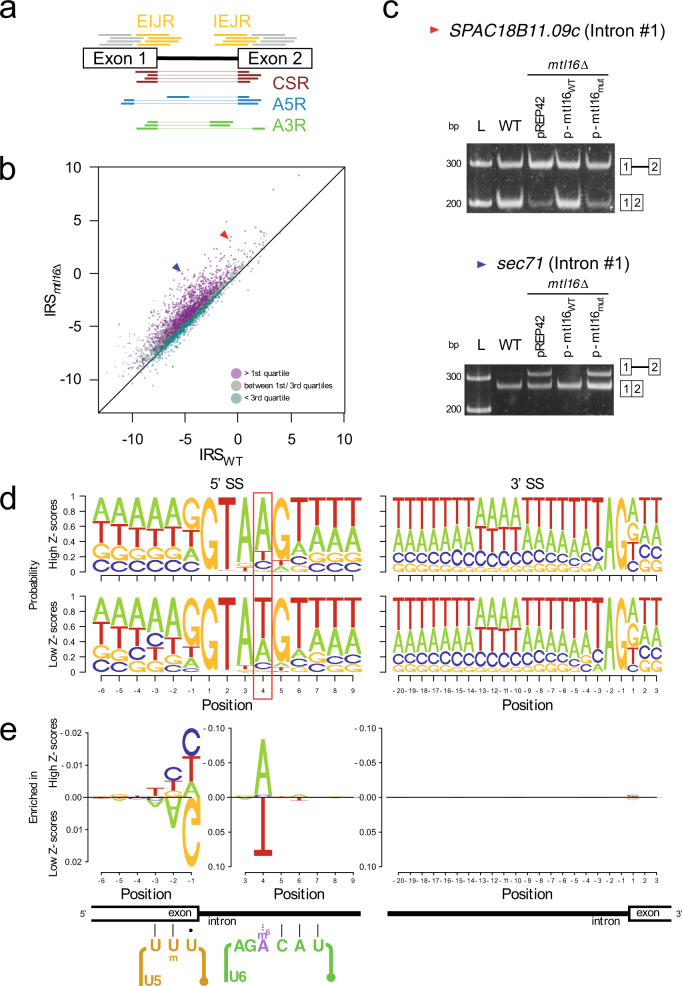
Given that m6A37 in U6 snRNA is present in the ACAGA box, which pairs with the 5′SS of the intron in the spliceosome, we next asked whether mRNA splicing would be affected globally by loss of m6A in U6 snRNA. To this end, we performed RNA-seq of poly(A)+ RNAs from WT and mtl16Δ strains and compared the splicing properties of the two samples. Based on S. pombe genome annotations, we first took reads that mapped to each intron and its neighboring exons and classified them into five groups (Fig. 2a): 5′ exon–intron junction reads (EIJR), intron-3′ exon junction reads (IEJR), exon–exon canonical splicing junction reads (CSR), 5′ alternative splice site selection reads (A5R), and 3′ alternative splice site selection reads (A3R). We then developed algorithms to detect alteration in splicing efficiency and transcript abundance. For each of five groups (Fig. 2a), the number of reads (coverage) is returned by function cov(x). Intron Retention Score (IRS) and Proportion of Canonical Splicing (PCS) are calculated by Eqs. (1) to (3) against each intron in each sample (Supplementary Data 1). IRS is used for measuring the difference in intron retention level and PCS is used for measuring the fraction of canonically spliced transcripts.
| 1 |
| 2 |
| 3 |
A plot of IRS for mtl16Δ versus WT revealed a notable bias in the distribution; specifically, a number of introns in the mtl16Δ strain had higher IRS values (Fig. 2b). Therefore, in the mtl16Δ strain, we suggest that the pre-mRNA is dissociated from the spliceosome during the steps from the association of tri-snRNP until the first transesterification, which is at the state of B, Bact or B* complexes, leaving the intron retained. For introns with large differences in IRS, we confirmed intron retention in the mtl16Δ strain by semi-quantitative RT-PCR (Fig. 2c, Supplementary Fig. 5). Complementation of the mtl16Δ strain by ectopic expression of WT or the active-site mutant (P169A/P170A) of mtl1641,44 confirmed that the alterations in splicing were dependent on the methyltransferase activity of Mtl16 (Fig. 2c).
Fig. 2. Global alteration of mRNA splicing in mtl16Δ cells.
a Five classifications of RNA-seq reads mapped to each intron with its neighboring exons annotated in the S. pombe genome: 5′ exon–intron junction (EIJR), intron-3′ exon junction (IEJR), exon–exon canonical splicing junction (CSR), 5′ alternative splice site selection (A5R), and 3′ alternative splice site selection (A3R). b Scatter-plot of IRS in mtl16Δ versus WT. Each plot represents each annotated intron with sufficient number of reads. The plots are classified based on quartiles of the Z-score of each intron. The red and blue arrows represent plots of SPAC18B11.09c intron #1 and sec71 intron #1, respectively. The black line represents an equal value of IRS. c Semi-quantitative RT-PCR analyses of two introns with large IRS difference. The upper and lower bands on the gel represent retained and spliced introns, respectively. Intron retention in the mtl16Δ strain was rescued by ectopic expression of plasmid-encoded WT mtl16 (p-mtl16WT), but not by its active-site mutant (P169A/P170A) (p-mtl16mut). This result was repeated once with a similar result. Source data are provided as a Source Data file. d Sequence logos of 5′ and 3′ splice sites with Z-scores over the top quartile (upper panels) and under the 3rd quartile (lower panels). The adenine base at the fourth position of the intron is highlighted and is associated a higher probability of intron retention in the mtl16Δ strain. e DiffLogo analysis was used to compare sequence enrichment of the 5′SS between introns with high Z-scores versus low Z-scores. The 5′SS is recognized by a part of the loop I sequence of U5 snRNA and the portion of U6 snRNA that contains m6A. Enriched and depleted nucleotides at the 5′SS are shown above and below the axis, respectively. The enrichment of A at position 4 against other nucleotides was significant by a p-value under 1.0 × 10−98 by two-sided Fisher’s exact test. The depletion of A, A, and G at positions −3, −2, and −1 against other nucleotides was significant by p-values under 0.01, 5.0 × 10−14, and 5.0 × 10−30 by two-sided Fisher’s exact test, respectively. The loop I sequence of U5 snRNA and a portion of U6 snRNA pair with the 5′ exon and intron at the 5′SS, respectively. m6A pairs with the fourth nucleotide of introns.
To analyze the effect of mtl16 knockout on gene expression, we estimated the global alteration of transcriptome (Supplementary Fig. 6a and Supplementary Data 2). Since pre-mRNA splicing is significantly affected by the mtl16 knockout, we corrected the read counts of each transcript by multiplying PCS of introns inside the CDS, and applied them for differential expression analysis. As a result, 54 and 32 genes were up- and down-regulated, respectively, significantly over 2-fold (Supplementary Fig. 6a and Supplementary Data 2). In the up-regulated gene list, a set of genes upregulated under some stress conditions, such as heat, osmotic, oxidation, and nutrient starvation, were enriched according to the gene ontology enrichment analysis, implying that the mtl16Δ strain is suffering from several cellular stresses (Supplementary Fig. 6b). We speculate that the phenotypic features of this strain might be caused by accumulation of aberrant transcripts and proteins upon mtl16 knockout. Within the down-regulated genes in the mtl16Δ strain, mrp17 (mitochondrial ribosomal protein) and icp55 (intermediate cleaving peptidase 55) are required for mitochondrial function72–74. The slow-growth phenotype of the mtl16Δ strain under nonfermentable condition (Fig. 1e) might be explained by downregulation of these proteins. In addition, downregulation of apl5 (AP-3 adaptor complex subunit)75 and pot1 (shelterin complex subunit)76 is associated with sensitivity against hydroxyurea (HU) and/or thiabendazole (TBZ) (Fig. 1d).
Introns bearing A at the 4th nucleotide were sensitive to m6A loss from U6 snRNA
To characterize the 5′ and 3′SSs of the introns that exhibited a splicing defect, we chose a subset of introns showing large differences in IRS. To this end, we gave each intron a Z-score of a two-sample Z-test with a null hypothesis of an equal IRS, as IRS values follow normal distribution (Supplementary Fig. 7). Then, their Z-scores were sorted in descending order and split by quartiles (Fig. 2b). Sequence logos around the 5′ and 3′SSs of introns with Z-scores above the 1st quartile and below the 3rd quartile were generated by SeqLogo77 (Fig. 2d), then the nucleotides enriched and depleted in the selected introns were shown by DiffLogo78 (Fig. 2e). Notably, the fourth nucleotide of the 5′SS was significantly more likely to be A and less likely to be U (genomic T) in the group of introns with the highest Z-scores (Fig. 2e) than in the set of introns with the lowest Z-scores, in which T and A are equally likely at this position (Fig. 2e). By contrast, we observed no large difference in the sequence logo around the 3′SS (Fig. 2e). Given that m6A37 in U6 snRNA faces the fourth position of the intron during the catalytic steps of splicing79, m6A loss might affect 5′SS recognition of a subset of introns bearing A at the fourth position (A4 introns).
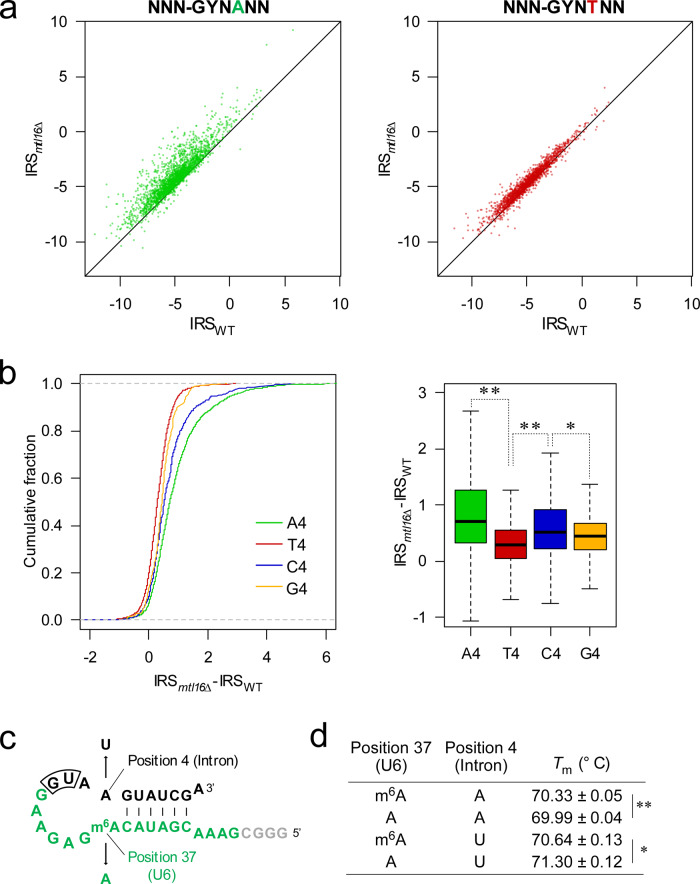
To compare the splicing errors of introns with different nucleotides at the fourth position, we classified all introns with a sufficient read depth into four groups based on their fourth nucleotide: A4 (n = 2539), T4 (n = 1846), C4 (n = 514), or G4 (n = 157). We then plotted the IRS values for each group of introns in mtl16Δ versus WT strains (Fig. 3a and Supplementary Fig. 8). A large number of A4 introns exhibited a large difference in IRS implicating splicing error in the absence of m6A, whereas T4 introns were only slightly over the line of equal IRS, implicating little effect by m6A loss (Fig. 3a). Next, we calculated the difference in IRS for each group of introns upon mtl16 knockout and generated cumulative curves (Fig. 3b). As expected, the A4 and T4 introns had the largest and smallest IRS differences, respectively. G4 introns were less strongly affected than T4 introns, whereas the IRS differences of C4 introns were significantly higher than T4 and G4 introns, but lower than those of A4 introns. As observed for the A4 introns, the C4 introns are also spliced less efficiently than T4 and G4 introns in the absence of m6A in U6 snRNA. Thus, m6A of U6 snRNA plays a functional role in splicing of most introns improving their efficiencies, but to a greater extent against A4 and C4 introns.
Fig. 3. The fourth nucleotide of the intron is the cis-element for sensitivity against m6A loss.
a Scatter-plots of IRS of A4 and T4 introns in mtl16Δ versus WT. The black lines represent an equal value of IRS. b The left panel shows a cumulative plot of IRS differences for each group of classified introns in mtl16Δ versus WT. All introns are classified into four groups based on their fourth nucleotide. The right panel shows a box plot of IRS differences for each group of introns. The first quartile, median, and third quartile are shown, and the whiskers represent 1.5 × interquartile ranges. Sample numbers are n = 2539, n = 1846, n = 514, and n = 157 for A4, T4, C4, and G4, respectively. *p = 6.6 × 10−3 and **p < 2.2e-16 (two-sided Wilcoxon’s rank-sum test). c Primary and secondary structure of the model RNA substrate and its mutation and modification introduced to the position 4 of intron and position 37 of U6 snRNA, respectively, used for measuring melting temperatures (Tm). Black and green characters represent the sequence derived from intron and U6 snRNA, respectively. The first dinucleotide at the 5′SS in the intron is boxed. d Tm values of the model RNA substrates. Asterisks indicate statistically significant differences, as determined by two-sided Student’s t-test. **p = 7.6 × 10−4, *p = 3.0 × 10−3; n = 3 independent experiments. Source data are provided as a Source Data file.
In a previous study, it was shown that an m6A protruding from a double strand stabilizes the terminal base pair through base-stacking effect enforced by the hydrophobicity of the methyl group80. To demonstrate this phenomenon in the spliceosome context, we synthesized an in vitro transcript that mimics the interaction between the ACAGA box of U6 snRNA and the 5′SS of A4 or T4 introns, and efficiently methylated it enzymatically by recombinant human METTL16 methyltransferase domain (Fig. 3c and Supplementary Fig. 9). Then, we measured the melting temperatures of the RNA hairpins with A4 or U4 in the presence or absence of m6A. As a result, consistent with previous studies80, methylation of the A–A pair to m6A–A increased the thermostability of the double strand, and an opposite effect was given to the A–U pair when it was methylated to m6A–U (Fig. 3d). This result indicates that efficient recognition of A4 introns by m6A is due to its thermostabilizing property in the sequence context of U6 snRNA and 5′SS.
AAG consensus of 5′ exon suppresses sensitivity against m6A loss
In all annotated introns (Fig. 2d), AAG triplet is present as a weak consensus sequence at the 3′ terminus of the 5′ exon (positions −3 to −1). This trinucleotide is complementary to loop I of U5 snRNA (Fig. 2e) and facilitates interaction between pre-mRNA and U5 snRNP in the Bact complex81–86. In the Two Sample Logo (Fig. 2e), the AAG triplet was depleted in A4 introns with splicing deficiencies in the mtl16Δ strain, but enriched in the rest of the introns. Accordingly, we classified the A4 introns (n = 2539) into two groups based on 5′ exonic sequence, AAG-A4 introns (n = 289) and non-AAG-A4 introns (n = 2250); we then plotted their IRS values in mtl16Δ versus WT strains (Fig. 4a), which shows a notable bias only in the non-AAG-A4 introns. We further classified non-AAG-A4 introns into seven groups (BAG, ABG, AAH, BBG, BAH, ABH, and BBH-A4) (Supplementary Fig. 10a) and calculated the IRS difference between the mtl16∆ and WT strains for each group (Supplementary Fig. 10b). The AAG-A4 introns have the smallest ratio, whereas the BBH-A4 introns have the largest (Supplementary Fig. 10c). Thus, the farther the 5′ exon sequence is from the AAG consensus, the larger the IRS difference the intron group will bear. These results suggest that the AAG consensus of the 5′ exon alleviates the splicing deficiency of the A4 introns caused by m6A loss of U6 snRNA via a stable interaction with the loop I of U5 snRNA.
Fig. 4. Splicing defect of A4 introns was suppressed by the AAG sequence of the 5′ exon.
a Scatter-plots of IRS of AAG-A4 (left panel) introns and non-AAG-A4 introns (right panels) in mtl16Δ versus WT. The black line represents an equal value of IRS. b Mutation study with a minigene construct encoding intron #1 of SPAC18B11.09c. Splicing efficiency was measured by RT-PCR using minigene-specific primers, followed by non-denaturing PAGE analysis. The IRS was quantified from the signal intensity ratio of the PCR products with and without introns. Asterisks indicate statistically significant differences in IRS between the two strains, as determined by two-sided Student’s t-test. *p = 6.4 × 10−4, **p = 8.7 × 10−5, ***p = 1.5 × 10−8; n = 4 independent PCR amplifications. Source data are provided as a Source Data file. c IRS of several A4 introns measured by RT-qPCR for WT and the mtl16Δ strains transformed with plasmids. The 5′ exon triplet for each intron and its base pairing with AGA in loop I of the mutant U5AGA is depicted below. Asterisks indicate statistically significant differences in IRS between the two strains, as determined by two-sided Student’s t-test. Exact p-values are written in the figure. Data represent average values of technical quadruplicates for nhm1 #2 or technical triplicates for other introns, with s.d. Source data are provided as a Source Data file.
To confirm this observation, we carried out a mutation study of an A4 intron using a minigene construct. We chose intron 1 of the SPAC18B11.09c gene, whose IRS values obtained by RNA-seq are −0.733 and 3.076 in the WT and mtl16Δ strains, respectively (Supplementary Data 1). The IRS values of the minigene construct obtained from semi-quantitative RT-PCR were 0.058 and 3.375 in the WT and mtl16Δ strains, respectively (Fig. 4b and Supplementary Fig. 11). When the GU at the 5′SS was mutated to GA, no splicing took place, as expected. When we mutated A at the fourth position to U, the IRS value in the WT strain increased to the same level as that in the mtl16Δ strain, indicating that the fourth nucleotide is the key cis-element for erroneous splicing in the absence of m6A in U6 snRNA. In addition, when the exonic ACC triplet was mutated to AAC or ACG, the splicing efficiency in both strains increased (i.e., IRS decreased). Furthermore, when the ACC was mutated to the AAG consensus sequence, splicing of this construct reached maximum efficiency even in the absence of m6A in U6 snRNA (Fig. 4b and Supplementary Fig. 11). This reporter experiment nicely supported our finding that m6A in U6 snRNA facilitates efficient splicing of A4 introns, especially when U5 snRNA is weakly associated with the 3′ terminus of the 5′ exon.
As an orthogonal approach to examine whether U6 and U5 snRNPs cooperatively recognize the 5′SS, we constructed a mutant of U5 snRNA in which UUU sequence of the loop I was replaced with AGA sequence (U5AGA) (Fig. 4c). Then, a plasmid harboring the mutant U5AGA was introduced in the WT and mtl16Δ strains. We measured the splicing efficiencies of several A4 introns harboring non-consensus triplet sequences at the 3′ terminus of the 5′ exon (Fig. 4c). The A4 intron of nhm1 mRNA has UCU triplet at the 5′ exonic sequence, which is weakly associated with loop I of U5 snRNA. The IRS of this intron was kept low in the WT strain, but significantly increased in the mtl16Δ strain as expected (Fig. 4c). When U5AGA was introduced, the IRS of this intron was reduced significantly, whereas WT U5 snRNA did not change its IRS (Fig. 4c), because the UCU triplet is complementary to the AGA sequence of U5AGA. The A4 intron of ysh1 has a UCC triplet that is recognized by the AGA in loop I of U5AGA with two basepairs and one mismatch. The increased IRS in the mtl16Δ strain was reduced significantly upon U5AGA expression (Fig. 4c). The A4 introns of trk2 and sec71 have CUG and CUC triplets at the 5′ exonic sequence, respectively. The AGA sequence of U5AGA weakly recognizes these triplets with single-wobble pairing at the middle of the sequence. Curiously, but as expected, the increased IRS of these A4 introns in the mtl16Δ strain was slightly but significantly reduced by U5AGA expression (Fig. 4c). As a negative control, the A4 intron of atg1803 has a canonical AAG triplet that is stably recognized by the endogenous U5 snRNA, but not recognized by the U5AGA mutant (Fig. 4c). The IRS increased by U5AGA expression (Fig. 4c), because U5AGA competes with the endogenous U5 snRNA. These results clearly show that stable recognition of the 5′ exon triplet by loop I of U5 snRNA suppresses intron retention caused by m6A loss in U6 snRNA.
Discussion
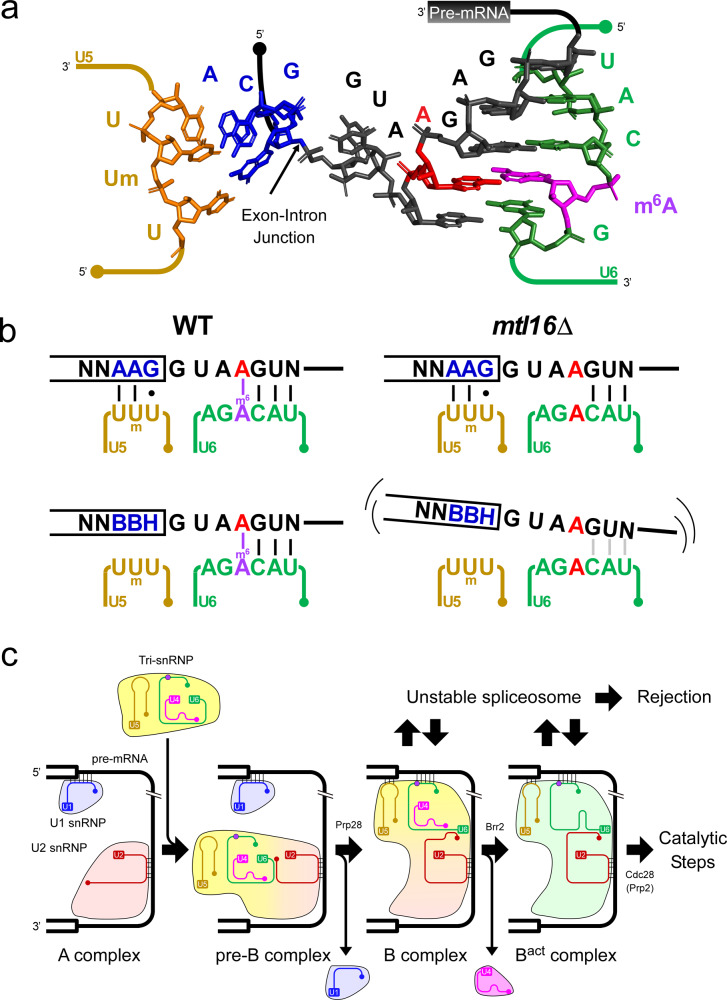
Biochemical79,81,82, genetic87–90, and structural studies83–86,91–94 reveal that the ACAGA box of U6 snRNA and loop I of U5 snRNA recognize 5′SS at B or Bact complexes, and maintain this interaction until the intron lariat–spliceosome dissociation (Fig. 5a and Supplementary Fig. 12). Recognition of 5′SS by U6/U5 snRNAs contributes to definition and proofreading of pre-mRNA splicing88,95–98, but little is known about the functional interaction between U5 snRNA–exon pairing and U6 snRNA–intron pairing. Here, we showed that in the WT S. pombe strain, the AAG-A4 introns are robustly recognized by U5 snRNA and U6 snRNA harboring the m6A modification (Fig. 5b). Even when the AAG consensus sequence is not present in the 5′ exon, the BBH-A4 introns are stably recognized by U6 snRNA with m6A modification (Fig. 5b) because m6A stabilizes the interaction between U6 snRNA and the intron via m6A–A pairing. In the mtl16Δ strain, the AAG-A4 introns are still recognized by U5 and U6 snRNPs in B or Bact complex, even when the m6A is not present in U6 snRNA (Fig. 5b). In this case, interaction between the 5′ exon and U5 snRNA plays a predominant role in efficient splicing. However, if the 5′ exon sequence deviates from the AAG consensus, the BBH-A4 introns are not stably recognized by U6 snRNA without the m6A modification (Fig. 5b). Collectively, these observations indicate that m6A in U6 snRNA facilitates efficient splicing of A4 introns only when U5 snRNA weakly recognizes the exon side of the 5′SS (Fig. 5b), indicating that U6 and U5 snRNPs interact cooperatively with pre-mRNA and stabilize the spliceosome during the splicing reaction. The cryo-EM structures of spliceosomes indicate that the interaction between pre-mRNA and U5/U6 snRNAs juxtaposes the 5′SS to the catalytic active site formed by the U snRNA backbones in the B complexes. Then, the Prp2 (S. pombe Cdc28) helicase pulls the intron to bring the branch site to the active site to form the B* complex. During these dynamic conformational changes, the 5′SS needs to be anchored to the active site, which enables the branch site 2′-OH to attack the splice site. We speculate that the stability of this anchoring interaction between pre-mRNA and U5/U6 snRNA decides the overall efficiency of the first transesterification (Fig. 5c).
Fig. 5. Mechanistic insight into the functional role of U6 snRNA m6A modification on pre-mRNA splicing.
a Cryo-EM structure of 5′SS in the B complex of the human spliceosome (PDB ID:5O9Z)92, as depicted by PyMol 2.1.0 (http://www.pymol.org/2/support.html). The model consists of U5 snRNA (orange), U6 snRNA (green), 5′ exon (blue), intron (black), A4 (red), and m6A (magenta). The coordinate of the methyl group of m6A was set to its syn conformation. b Interaction between the 5′SS of A4 intron and the U5 and U6 snRNAs in WT and mtl16Δ strains. m6A–A pairing stabilizes the complex, especially when U5 snRNA recognizes the 5′ exon weakly. c Schematic depiction of spliceosomal assembly from A to Bact complex based on cryo-EM structures of S. cerevisiae and human spliceosomes83–86,91,92. The m6A base in U6 snRNA is depicted by a dark-purple circle. The blue, red, yellow, purple, and green figures represent U1 snRNP, U2 snRNP, U4/U6.U5 tri-snRNP, U4 snRNP, and Bact complex, respectively.
As shown by the cryo-EM structures of the B and Bact complexes (Fig. 5a and Supplementary Fig. 12a), the 3′ terminal nucleotides of the 5′ exon are recognized by the UUmU sequence of loop I in U5 snRNA, whereas the intron sequence of 5′SS is base-paired with the ACm6A sequence of the ACAGA box in U6 snRNA. The m6A of U6 snRNA faces the fourth position of the intron. In the ILS complex (Supplementary Fig. 12b), the U6 snRNA–intron pairing is slightly different from that in the B complexes. Especially, m6A37 shifts out of the pairing with U4 in the intron lariat. It is possibly due to the conformational alterations of the spliceosome after the first transesterification. Tight recognition of 5′SS by U6 snRNA might not be necessary after the lariat formation. From a chemical viewpoint, the N6-methyl group of m6A has two conformations, syn and anti. Energetically, the syn conformation is favored over the anti conformation by 1.5 kcal/mol80. However, structural studies revealed that m6A–U pairing in the RNA duplex assumes the high-energy anti conformation. In support of this idea, thermodynamic measurement of RNA duplexes revealed that m6A destabilizes A–U pairs by 0.5–1.7 kcal/mol, but stabilizes A–A pairs by 0.7 kcal/mol. In addition, when m6A is present at the 5′ or 3′ overhang position, it stacks with the terminal base pair and stabilizes the RNA duplex by 0.9–1.2 kcal/mol, compared to unmodified A. Also, these thermodynamic properties were shown to be consistent with the results obtained from the sequence context of the spliceosome, by measuring the melting temperatures of modified and unmodified transcripts that mimic the base pairing between U6 snRNA and the 5′ end of an intron (Figs. 3c, d). These chemical properties of m6A neatly explain how m6A in U6 snRNA facilitates 5′SS recognition of A4 introns.
The evolutionary conservation of METTL16 implies that m6A in U6 snRNA is functionally important. METTL16 orthologs are highly conserved in metazoans, fungi, plants, and protists, implying that this modification is a fundamental component of the splicing machinery. However, m6A is not present in some organisms with small numbers of introns, including several species of budding yeasts such as Saccharomyces cerevisiae and Candida albicans and the red algae Cyanidioschyzon merolae (Supplementary Fig. 13). Presumably, METTL16 and m6A in U6 snRNA have been lost independently from these organisms over the course of evolution. In vertebrates, including humans, more than 200,000 introns are present in the set of all genes99, and the consensus sequence of 5′SS is GUAAGU (A4 intron). Among fungi, S. pombe has over 5000 introns and S. cerevisiae has 296; the consensus sequences of the 5′SS are GUAAGU (A4 intron) and GUAUGU (T4 intron), respectively69,100. Given that m6A in U6 snRNA facilitates efficient splicing of A4 introns, S. cerevisiae may preferentially use T4 introns rather than A4 introns to achieve efficient splicing in the absence of this modification. C. albicans also has a 5′SS consensus of T4100. On the other hand, C. merolae has only 27 introns, and its 5′SS consensus sequence is GUAAGU (A4 intron)101. Curiously, in C. merolae U6 snRNA, the ACAGA box is ACUGA, and the central U base pairs with the A4 position of introns with Watson–Crick geometry102. This is an alternative means of stabilizing 5′SS recognition by U6 snRNA without having m6A in the ACAGA box. Based on these cases, m6A in U6 snRNA has likely contributed to expansion of 5′SS sequence variation in organisms with large numbers of introns as it facilitates efficient splicing of non-AAG-A4 introns.
METTL16 is a key regulator of SAM synthetase (MAT2A), which maintains cellular SAM homeostasis41,43. METTL16 introduces m6A into the hairpin structures in the 3′ UTR of MAT2A mRNA by sensing cellular SAM concentrations, thereby controlling mRNA stability. In addition, METTL16 acts as a splicing enhancer to recognize the same hairpins and produce functional MAT2A protein. In fact, Mettl16 null mice die during early embryogenesis with a reduced steady-state level of Mat2a mRNA44, highlighting the importance of Mettl16 function in regulation of Mat2a levels. The 3′ UTR hairpins of MAT2A mRNA are conserved in vertebrates, but not in other organisms103. In fact, S. pombe sam1 mRNA, an ortholog of MAT2A, is not affected in the mtl16Δ strain (0.96-fold change; no significant alteration). Thus, the observed phenotypes of this strain can be explained as a function of the splicing defect caused by loss of m6A in U6 snRNA. Nematode and plant homologs of METTL16 also have physiological functions, and strains harboring knockouts of these genes exhibit severe phenotypes. mett-10, a Caenorhabditis elegans ortholog, is required for embryonic development and germ cell differentiation104. FIONA1, an Arabidopsis thaliana homolog, plays an essential role in regulating the circadian clock105. These phenotypes might be explained by splicing alterations caused by loss of m6A from U6 snRNA. Because little change in global splicing is observed in early embryos (E3.5) of Mettl16 null mice44, the functional significance of m6A in U6 snRNA in mammals remains elusive. Presumably, however, maternal U6 snRNA bearing m6A is still present at this stage. Further studies will be necessary to determine whether m6A loss of U6 snRNA affects global splicing in vertebrate systems.
Given that the frequency of m6A in U6 snRNA is nearly 100% (Fig. 1b), this modification might be a structural component of spliceosome, relaxing the 5′ exon constraint and diversifying the protein sequence along with the number of introns increases over the course of eukaryotic evolution. However, because vertebrate METTL16 is a SAM-sensitive methyltransferase45, and the m6A modifications in the 3′UTR of MAT2A mRNA are regulated by availability of cellular SAM43, m6A in U6 snRNA might be regulated by intracellular SAM concentrations. U6 snRNA m6A might also be targeted by “m6A eraser” demethylases in higher eukaryotes. This idea raises the possibility that U6 snRNA m6A is not only a structural component of the spliceosome, but also a regulator of the splicing efficiency of a subset of mRNAs under SAM-depleted conditions.
Methods
Yeast strains and cultivation
The ED668 (h+ ade6-M216 ura4-D18 leu1–32) (WT) and SPAC27D7.08cΔ (mtl16Δ) strains were obtained from the Bioneer deletion collection74. The deletion mutation was verified by PCR. Strains were grown at 30 °C in YE5S media [0.5% yeast extract and 3% glucose, supplemented with adenine, uracil, lysine, histidine, and leucine (225 mg/mL each)] or EMM media [EMM without dextrose (MP Biochemicals) and 2% glucose, supplemented with adenine and leucine (225 mg/mL each)]. For spot analyses, yeast cells were first grown to mid-log phase, serially diluted (1:5) from a starting sample of 104 cells, and spotted onto YE5S agar plates supplemented with inducers of cellular stress: 15 μg/ml thiabendazole (TBZ), 7.5 mM hydroxyurea (HU), 1 M KCl, or 100 mM CaCl2. The plates were photographed after 3–4 days of incubation at 30 °C or 37 °C. To measure non-fermentable growth, the yeast strains were cultured in medium containing 0.5% yeast extract, 0.1% glucose, and 3% glycerol supplemented with adenine, uracil, lysine, histidine, and leucine (225 mg/mL each). OD600 was measured every hour for 12 h.
RNA preparation
Total RNA from yeast cells for snRNA isolation was prepared by hot phenol extraction106. Cell pellets were resuspended in extraction buffer [50 mM NaOAc (pH 5.2), 10 mM EDTA (pH 8.0), and 10% SDS], mixed with an equal volume of neutral phenol, incubated at 95 °C for 15 min, and centrifuged at 8000 × g for 30 min at 4 °C. The supernatant was collected and mixed with 1/4th volume of chloroform, followed by centrifugation at 8000 × g for 15 min at 4 °C. The supernatant was subjected to isopropanol precipitation to obtain an RNA pellet. The RNA pellet was dissolved in 10 mL buffer consisting of 20 mM HEPES-KOH (pH 7.5) and 80 mM NaCl, and then mixed with 4 mL of 2-butoxyethanol. The mixture was placed on ice for 30 min. The precipitate containing polysaccharide was removed by centrifugation at 8000 × g for 30 min at 4 °C. The supernatant was subjected to ethanol precipitation to obtain total RNA.
Nucleoside analysis of poly(A)+ RNA
Total RNA extracted from exponentially growing S. pombe WT cells underwent four rounds of enrichment of poly(A)+ RNA using Dynabeads oligo [dT]25 (Invitrogen). Eluate RNAs from each round of enrichment (1 μg each) were digested into nucleosides with 0.07 units of nuclease P1 (Fujifilm Wako Pure Chemical Corporation), 0.127 units of phosphodiesterase I (Worthington Biochemical Corporation), and 0.08 units of bacterial alkaline phosphatase (BAP from E. coli C75, Takara). The digests were subjected to LC/MS analysis using a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific) equipped with a Dionex UltiMate 3000 LC System (Thermo Fisher Scientific) and an InertSustain C18 column (5 μm, 2.1 × 250 mm, GL Science). The mass chromatogram peak area of adenosine (A, m/z 268.10), N6-methyladenosine (m6A, m/z 282.12), N6-dimethyladenosine (m6,6A, m/z 296.14), and 2′-O-methyladenosine (Am, m/z 282.12) was measured. Then, the peak area of m6A, m6,6A, and Am was divided by the peak area of A, and these ratios at each round of enrichment were normalized by the ratio from the first eluate and then plotted for each modification. Data were analyzed on Xcalibur 4.1 and Excel 2016, and visualized by R 4.0.3 and Canvas 15.
Isolation of U6 snRNA and mass spectrometry
Isolation of yeast U6 snRNA was performed by RCC70 using a DNA probe (Supplementary Data 3). Mass spectrometric analysis of RNase T1- or A-digested fragments of the isolated U6 snRNA was performed by capillary LC-nano-ESI-MS107. Mongo Oligo Mass Calculator v2.08 (https://mods.rna.albany.edu/masspec/Mongo-Oligo) was used for assignment of the product ions in CID spectra108.
Preparation of transcripts for in vitro reconstitution
DNA templates for in vitro transcription were prepared using a series of DNA oligos (described in Supplementary Data 3). U6 snRNAs were transcribed by T7 RNA polymerase in vitro essentially as described109. Briefly, DNA template was mixed with in-house made T7 RNA polymerase, NTPs, and a transcription buffer consisting of 40 mM Tris-HCl (pH 7.5), 24 mM MgCl2, 5 mM DTT, 2.5 mM Spermidine, and 0.01% Triton X-100, and was incubated at 37 °C overnight. The reaction was stopped by addition of an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, v/v/v). The supernatant was desalted by PD-10. Desalted RNA was collected by ethanol precipitation. The transcript was resolved by 10% denaturing PAGE and purified by excision from the gel.
Expression and purification of recombinant proteins
To obtain recombinant S. pombe Mtl16, the mtl16 cDNA was amplified by RT-PCR of total RNA from WT S. pombe cells using the primers listed in Supplementary Data 3, which were designed based on genomic sequence retrieved from PomBase110. Because the original initiation codon (genomic position I:4,522,948-4,522,950) of mtl16 ORF in the database was misannotated inside an unannotated intron (genomic position I:4,522,922–4,522,964), we constructed a recombinant Mtl16 starting from an AUG codon (genomic position I:4,523,030–4,523,032) upstream of the unannotated intron. The PCR product was cloned into the NheI and XhoI sites of pET-28a(+) (Novagen) to yield pET-Spmtl16, which produces N-terminal hexahistidine-tagged Mtl16 (His6-Mtl16). The E. coli Rosetta2 (DE3) strain was transformed with pET-Spmtl16 and cultured in LB medium containing 50 μg/mL kanamycin and 30 μg/mL chloramphenicol. When the bacteria reached OD600 of 0.5, protein expression was induced by addition of 10 μM IPTG and the cells were grown for an additional 3 h at 37 °C. Cells were harvested and suspended in buffer A [50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, and 7 mM β-mercaptoethanol] supplied with 0.5 mM PMSF, followed by sonication on ice. Cell lysates were cleared by ultracentrifugation at 100,000 × g for 60 min. The supernatant was loaded onto a nickel-charged HiTrap chelating column (GE Healthcare) and unbound proteins were washed out with buffer A. The recombinant proteins were eluted with a 50-mL linear gradient comprising 0–500 mM imidazole in buffer A. A fraction containing the recombinant protein was dialyzed overnight against dialysis buffer [50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 10% glycerol, and 7 mM β-mercaptoethanol]. The concentration of the purified protein was determined in a Bradford assay (Bio-Rad, Hercules, CA, USA) using bovine serum albumin as a standard. To obtain recombinant human METTL16, the METTL16 cDNA was amplified using the primers listed in Supplementary Data 3, which were designed based on human genomic sequence retrieved from Ensembl111 and cloned into pET-28a(+) to yield pET-hMETTL16, which produces C-terminal hexahistidine-tagged METTL16 (METTL16-His6). Expression and purification were carried out as described for S. pombe Mtl16. To obtain recombinant human METTL16 MTD, cDNA coding the methyltransferase domain of METTL16 was amplified using the primers listed in Supplementary Data 3 and cloned into pE-SUMO-TEV by SLiCE to yield pE-SUMO-hMETTL16MTD. Expression was carried out as described for S. pombe Mtl16. Following expression, cells were harvested and suspended in buffer A [50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, and 7 mM β-mercaptoethanol] supplied with 0.5 mM PMSF, followed by sonication on ice. Cell lysates were cleared by ultracentrifugation at 100,000 × g for 60 min. The supernatant was loaded onto a nickel-charged HiTrap chelating column (GE Healthcare) and unbound proteins were washed out with buffer A. The recombinant proteins were eluted with a 50-mL linear gradient comprising 0–500 mM imidazole in buffer A. A fraction containing the recombinant protein was dialyzed overnight against dialysis buffer [50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 10% glycerol, and 7 mM β-mercaptoethanol] containing SUMO protease Ulp1. Dialyzed protein was applied to Ni Sepharose 6 Fast Flow (GE Healthcare) and the flow-through was collected. The cleaved protein was further purified on a HiScreen Capto SP ImpRes (GE Healthcare) column. The concentration of the purified protein was determined in a Pierce BCA Protein Assay (Thermo Fisher Scientific) using bovine serum albumin as a standard.
In vitro reconstitution using recombinant proteins
In vitro methylation of U6 snRNA was performed at 37 °C for 1 h in 50 µL of reaction mixture consisting of 10 mM HEPES-KOH (pH 7.5), 100 mM NaCl, 1 mM DTT, 2 μM U6 snRNA transcript, 1 μM His6-Mtl16, and 1 mM SAM. The reaction was stopped by addition of an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, v/v/v). RNA was collected by ethanol precipitation, digested with RNase T1, and subjected to RNA-MS analysis as described above.
Northern blotting
Total RNA (1 μg) from yeast cells was resolved by 10% denaturing PAGE and either stained with SYBR Gold (Invitrogen) or blotted onto a nylon membrane (Amersham Hybond N+, GE Healthcare) in 1 × TBE using a Transblot Turbo (Bio-Rad). Fluorescence was visualized on an FLA-7000 imaging analyzer (Fujifilm). The membrane was air-dried and irradiated twice with UV light (254 nm, 120 mJ/cm2; CL-1000, UVP) to cross-link the blotted RNA. Hybridization was performed using PerfectHyb (TOYOBO) at 52 °C with 4 pmol of 5′-32P-labeled oligonucleotides specific to each U snRNA (Supplementary Data 3). The membrane was washed four times with 2 × SSC [300 mM NaCl, 30 mM trisodium citrate (pH 7.0)] containing 0.1% SDS, dried, and exposed on an imaging plate (BAS-MS2040, Fujifilm). Radioactivity was visualized on the FLA-7000. Uncropped scans are available in the Source Data file.
RNA-seq sample preparation
Yeast were harvested, frozen in liquid nitrogen, and homogenized in TriPure Isolation Reagent (Roche Life Science) using SK-100 (Funakoshi). Total RNA was extracted with chloroform, recovered from the aqueous phase by isopropanol precipitation, ethanol precipitated, and dissolved in water. The total RNAs underwent two rounds of enrichment of poly(A)+ RNA using Dynabeads oligo [dT]25 (Invitrogen) and were sequenced on an Illumina NovaSeq sequencer (150-bp, paired-end).
RNA-seq read alignment
Raw Illumina sequencing reads were trimmed using fastp software112 to remove adaptor sequences and low-quality bases. The processed reads were first aligned to S. pombe rRNA and tRNA sequences, then the unmapped reads were aligned to S. pombe genome assembly ASM294v2.34 using STAR 2.7.3a113. The following parameters were used–twopassMode Basic–outSJfilterOverhangMin 30 8 8 8–alignIntronMax 2000–outFilterMultimapNmax 30–outFilterScoreMinOverLread 0.4–outFilterMatchNminOverLread 0.44–outFilterScoreMin 10. The statistics of trimming and mapping is listed in Supplementary Data 4. Read alignment files (.bam) were processed with custom Python scripts114 using the libraries Biopython115 and Pysam116, and the plots were written by R 4.0.3. For each segment comprising a pair of consecutive annotated exons (exon1 and exon2), reads that mapped to the annotated SS junction (CSR), exon 1–intron junction (EIJR), intron–exon 2 junction (IEJR), any alternative 5′SS to exon 2 (A5R), and exon 1 to any alternative 3′SS (A3R), were counted.
Calculation of erroneous splicing
For each annotated segment described above, we obtained IRS by Eq. (2) to assess retention level for each annotated intron. PCS was calculated by Eq. (3) to assess the proportion of canonically spliced mRNA within a mixture of mature, immature, and aberrant mRNA. To compare the difference in IRS between WT and mtl16Δ strains, as the IRS values mostly follow normal distribution (Supplementary Fig. 7), Z-scores with a null hypothesis of zero difference in IRS for each intron were obtained by Eq. (4), where and represent the mean and standard deviation, respectively, of the IRS values among the quadruplicates of WT and mtl16Δ strains.
| 4 |
All annotated GY-AG introns with a sufficient arbitrary number of reads in all four replicates [cov(Total) > 10] and an average CSR over 10 in either strain were applied for this calculation so that the number of introns picked up is maximized and the variance in read counts among the replicates is minimized. This process picked up 5056 GY-AG introns with sufficient read counts out of 5364 annotated introns. The 5′ and 3′SS sequences of the introns with splicing deficiencies were characterized by choosing a subset of introns with large and small IRS differences. Upon sorting the introns by Z-scores in a descending order, those over the 1st quartile were treated as introns with large IRS differences, and those under the 3rd quartile were treated as introns with small IRS differences. Then, sequence logos around the 5′ and 3′ SSs of introns with large or small IRS differences with sufficient read depths were generated by SeqLogo77. Then, two logos were compared by DiffLogo78. The raw count and result of the calculation is provided in Supplementary Data 1.
Differential expression analysis
Raw counts for each annotated transcript were determined using the htseq-count command of HTSeq version 0.6.1117. The splicing-adjusted read counts of each annotated gene in each sample were calculated by Eq. (5), where is the intron number and is the value of intron number . The product of each ratio of canonically spliced reads for each intron inside the CDS of the gene was multiplied against the overall read count that falls inside each genomic locus of the transcript.
| 5 |
Differential expression of adjusted counts was evaluated by edgeR version 3.12118,119, applying a statistical cutoff of FDR < 0.05 and fold-change > 2 to obtain differentially expressed genes (Supplementary Data 2). Enriched features of gene lists were searched by the AnGeLi120 tool with protein-coding genes as a background (Supplementary Data 5).
Rescue experiment and intron-specific semi-quantitative RT-PCR
The S. pombe mtl16 gene, including untranslated regions, was PCR-amplified from the genome and cloned into pREP42-mcs+ (Addgene plasmid 52691) digested by NdeI and BamHI using the primers listed in Supplementary Data 3 and the SLiCE method121 to yield the WT plasmid p-mtl16WT. To construct an active-site mutant, both P169 and P170 were mutated to Ala residues by site-directed mutagenesis using the primers listed in Supplementary Data 3 to yield p-mtl16mut. S. pombe mtl16Δ strain was transformed with the WT or mutant plasmid by the lithium acetate method122, cultivated in EMM medium devoid of thiamine until the cell density reached OD600 = 1.0, and harvested. Total RNA was prepared using the TriPure Isolation Reagent (Roche Life Science). One microgram of total RNA from WT, KO, and transformed strains were treated with RQ1 RNase-free DNase (Promega) and subjected to reverse transcription with random hexamer and oligo dT primers using the Transcriptor First Strand cDNA Synthesis Kit (Roche), followed by PCR amplification with intron-specific primers (Supplementary Data 3) under the following conditions: 30 cycles at 95 °C for 30 s, 53 °C for 45 s, and 68 °C for 30 s. PCR products were resolved on 8% non-denaturing PAGE, stained with EtBr, and visualized on FLA-7000.
Minigene analysis
The flanking sequence of the first intron of SPAC18B11.09c was amplified using primers bearing NdeI and BamHI sites (Supplementary Data 3). The PCR product was cloned into the corresponding sites of pREP42-mcs+ to yield the WT minigene plasmid pREP-AC18B11.09c. Mutations were introduced by site-directed mutagenesis using the primers listed in Supplementary Data 3. S. pombe WT and mtl16Δ strains were transformed with the WT or mutated plasmid using the lithium acetate method, cultivated in EMM medium devoid of thiamine, until the cell density reached OD600 = 1.0, and harvested. Total RNA was prepared using the TriPure Isolation Reagent. One microgram of total RNA was treated with RQ1 RNase-free DNase and then subjected to reverse transcription with nmt1 promoter-specific RT primer (Supplementary Data 3) using the Transcriptor First Strand cDNA Synthesis Kit, followed by PCR amplification with minigene-specific primers (Supplementary Data 3) under the following conditions: 16–23 cycles at 95 °C for 30 s, 53 °C for 45 s, and 68 °C for 30 s. The number of cycles was adjusted so that the signals of the PCR products were not saturated. The PCR products were resolved on 8% non-denaturing PAGE, stained with EtBr, and visualized on FLA-7000. Signals corresponding to spliced or retained introns were quantified using the Multi Gauge V3.0 software (Fujifilm). Signal intensities were converted to levels of PCR product using the signals of Loading Quick 100-bp DNA Ladder (Toyobo) as a standard. IRS is the binary logarithm of the value obtained by dividing the abundance of the retained isoform by the abundance of the spliced isoform.
Intron retention-specific RT-qPCR
The LEU2+ marker of pTN69 was replaced by ura4+ using the primers listed in Supplementary Data 3 and the SLiCE method121 to yield pDblet. The S. pombe snu5 gene and its flanking region was PCR-amplified from the genome and cloned into pDblet digested by HindIII using the primers listed in Supplementary Data 3 and the SLiCE method121. Then, site-directed mutagenesis using the primers listed in Supplementary Data 3 was used to introduce an additional 15-nt stem-loop in the 3′ end of U5 snRNA to yield the plasmid pU5WT. Site-directed mutagenesis using the primers listed in Supplementary Data 3 was utilized to introduce a TTT to AGA mutation in the snu5 gene of pU5WT to yield pU5AGA. Total RNA (2 μg) was extracted from WT or the mtl16Δ strains transformed with pDblet, pU5WT or pU5AGA grown to mid-log phase in EMM media without uracil. cDNA was synthesized by Superscript III (Thermo Fisher Scientific) using random N6 primers, according to the manufacturer’s instructions. PCR was performed in a 10 μL mixture containing a 0.2-μL aliquot of the cDNA solution, 0.2 μM of each PCR primers, and 1 × KAPA SYBR FAST Master Mix optimized for LightCycler480 (Kapa biosystems). The thermal cycling conditions included 45 cycles of 95 °C for 10 s, 58 °C for 20 s, and 72 °C for 1 s. Amplification of DNA was monitored by a LightCycler480 (Roche), according to the manufacturer’s instructions. To write a standard curve of Cp (crossing point) value against the amount of the template, depending on each set of primers, a dilution series of the S. pombe WT genomic DNA was amplified by the primers. To measure the IRS value of a single intron, primers spanning intron and exon were used to obtain the Cp value of the retained isoform, and primers constructed inside an adjacent single exon was used to obtain the Cp value of the total of both retained and spliced isoforms. Then, the abundance of cDNA derived from each isoform was calculated based on the standard curve. IRS is the binary logarithm of the value obtained by dividing the abundance of the retained isoform by the abundance of the spliced isoform.
Melting temperature observation of the transcript
In vitro transcripts U6A4, U6U4, and U6guide were synthesized from templates U6modelA4_T7R, U6modelU4_T7R, and U6modelGuide_T7R, respectively (Supplementary Data 3) as described. U6A4 and U6U4 were annealed with U6guide by incubating at 95 °C for 5 min and ramped at −0.1 °C/sec to 10 °C in a mixture consisting of 100 mM NaCl, 20 mM HEPES-KOH (pH 7.5), 75 μM U6A4 or U6U4 RNA, and 75 μM U6guide RNA. The U6guide RNA was designed to form a secondary structure with U6A4 or U6U4 RNAs that emulates human U6 snRNA secondary structure recognized by METTL1668. The annealed mixture was applied for methylation by incubating at 37 °C overnight in a mixture consisting of 200 mM NaCl, 50 mM HEPES-KOH (pH 7.5), 1 mM DTT, 2.5 mM SAM, 0.2 units/uL SUPERase-In RNase inhibitor (Thermo Fisher Scientific), 30 μM human recombinant METTL16 MTD, and 30 μM RNA. The reaction was stopped by mixing with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, v/v/v). The RNA was collected by ethanol precipitation, then U6A4 RNA or U6U4 RNA was resolved by 10% denaturing PAGE and excised from the gel piece. The methylation efficiency was verified by LC/MS as described. The methylated or unmodified transcripts were dissolved at a final concentration of 3 μM in degassed buffer consisting of 20 mM sodium phosphate (pH 7.0) and 1 M NaCl, followed by incubation at 95 °C for 5 min and ramped at −0.1 °C/sec to 10 °C for annealing. A UV–Vis spectrophotometer (V-630, JASCO) equipped with an 8 multiquartz microcell array (path length: 10 mm, JASCO) was used to monitor the hyperchromicity by monitoring absorbance at 260 nm. The temperature gradient was as follows: 25 °C for 30 sec, ramped at 5 °C/min to 40 °C and held for 5 min, and after that ramped at 0.5 °C/min to 105 °C. The measurement was performed in triplicate for each sample.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank members of Tom Suzuki’s laboratory, especially Yuriko Sakaguchi, for RNA-MS analysis, Kenjyo Miyauchi for RNA isolation with RCC, and Shunpei Okada for NGS data analysis. We thank Terumi Horiuchi and Kiyomi Imamura of Yutaka Suzuki laboratory for sample preparation, data generation, and data registration of NGS. Some computations were performed on the NIG supercomputer at the ROIS National Institute of Genetics. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), Japan Society for the Promotion of Science (JSPS) [20292782 and 18H05272 to T.S., 16H06279 (PAGS) to Y.S., and 18J13582 to Y.Ish.], and Exploratory Research for Advanced Technology (ERATO, JPMJER2002 to T.S.) from Japan Science and Technology Agency (JST).
Source data
Author contributions
Y.Ish. performed biochemical, genetic, and bioinformatic works assisted by T.O. Y.Iso performed the poly(A)+ purification-nucleoside analysis experiment. Y.Ish. and T.S. wrote this paper. Y.S. performed NGS. All authors discussed the results and revised this paper. T.S. designed and supervised all work.
Data availability
The sequence data from this study have been submitted to the DDBJ Sequence Read Archive under accession number DRA009909. Structural data was retrieved from PDB ID: 5O9Z [10.2210/pdb5o9z/pdb], 5GM6 [10.2210/pdb5gm6/pdb], and 3JB9 [10.2210/pdb3jb9/pdb]. Source data are provided with this paper.
Code availability
Python scripts written for RNA-seq data analysis are available from a public GitHub repository https://github.com/YumaIshigami/irscalc114.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Jeffrey Pleiss and other, anonymous, reviewers for their contributions to the peer review of this work. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-23457-6.
References
- 1.Boccaletto P, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki, T. The expanding world of tRNA modifications and disease relevance. Nat. Rev. Mol. Cell Biol.10.1038/s41580-021-00342-0 (2020). [DOI] [PubMed]
- 4.Harcourt EM, Kietrys AM, Kool ET. Chemical and structural effects of base modifications in messenger RNA. Nature. 2017;541:339–346. doi: 10.1038/nature21351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T. RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet. 2016;17:365–372. doi: 10.1038/nrg.2016.47. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz S, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30:4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fustin JM, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Lence T, et al. m(6)A modulates neuronal functions and sex determination in Drosophila. Nature. 2016;540:242–247. doi: 10.1038/nature20568. [DOI] [PubMed] [Google Scholar]
- 11.Haussmann IU, et al. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batista PJ, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geula S, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schibler U, Kelley DE, Perry RP. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- 17.Csepany T, Lin A, Baldick CJ, Jr., Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. J. Biol. Chem. 1990;265:20117–20122. doi: 10.1016/S0021-9258(17)30477-5. [DOI] [PubMed] [Google Scholar]
- 18.Harper JE, Miceli SM, Roberts RJ, Manley JL. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res. 1990;18:5735–5741. doi: 10.1093/nar/18.19.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rottman FM, Bokar JA, Narayan P, Shambaugh ME, Ludwiczak R. N6-adenosine methylation in mRNA: substrate specificity and enzyme complexity. Biochimie. 1994;76:1109–1114. doi: 10.1016/0300-9084(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 20.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 21.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linder B, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ke S, et al. A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 2015;29:2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang H, et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature. 2019;567:414–419. doi: 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alarcon CR, et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao W, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Patil DP, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roundtree IA, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife. 2017;6:e31311. doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi H, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li A, et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–447. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu N, et al. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keith JM, Ensinger MJ, Moss B. HeLa cell RNA (2’-O-methyladenosine-N6-)-methyltransferase specific for the capped 5’-end of messenger RNA. J. Biol. Chem. 1978;253:5033–5039. doi: 10.1016/S0021-9258(17)34652-5. [DOI] [PubMed] [Google Scholar]
- 39.Banerjee AK. 5’-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol. Rev. 1980;44:175–205. doi: 10.1128/MR.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akichika S, et al. Cap-specific terminal N(6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science. 2019;363:aav0080. doi: 10.1126/science.aav0080. [DOI] [PubMed] [Google Scholar]
- 41.Pendleton KE, et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–835 e14. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warda AS, et al. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–2014. doi: 10.15252/embr.201744940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shima H, et al. S-Adenosylmethionine synthesis is regulated by selective N(6)-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 2017;21:3354–3363. doi: 10.1016/j.celrep.2017.11.092. [DOI] [PubMed] [Google Scholar]
- 44.Mendel M, et al. Methylation of structured RNA by the m(6)A writer METTL16 Is essential for mouse embryonic development. Mol. Cell. 2018;71:986–1000 e11. doi: 10.1016/j.molcel.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doxtader KA, et al. Structural basis for regulation of METTL16, an S-adenosylmethionine homeostasis factor. Mol. Cell. 2018;71:1001–1011 e4. doi: 10.1016/j.molcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blomen VA, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 47.Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. Are snRNPs involved in splicing? Nature. 1980;283:220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- 48.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yean SL, Wuenschell G, Termini J, Lin RJ. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fica SM, et al. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503:229–234. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/S0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 52.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 53.Fica SM, Nagai K. Cryo-electron microscopy snapshots of the spliceosome: structural insights into a dynamic ribonucleoprotein machine. Nat. Struct. Mol. Biol. 2017;24:791–799. doi: 10.1038/nsmb.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan C, Wan R, Shi Y. Molecular Mechanisms of pre-mRNA Splicing through Structural Biology of the Spliceosome. Cold Spring Harb. Perspect. Biol. 2019;11:a032409. doi: 10.1101/cshperspect.a032409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karijolich J, Yu YT. Spliceosomal snRNA modifications and their function. RNA Biol. 2010;7:192–204. doi: 10.4161/rna.7.2.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Epstein P, Reddy R, Henning D, Busch H. The nucleotide sequence of nuclear U6 (4.7 S) RNA. J. Biol. Chem. 1980;255:8901–8906. doi: 10.1016/S0021-9258(18)43587-9. [DOI] [PubMed] [Google Scholar]
- 57.Gu J, Patton JR, Shimba S, Reddy R. Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: modified nucleotides are clustered in functionally important regions. RNA. 1996;2:909–918. [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou H, Qu L, Du Y, Zhou W. Identification of a novel methylated nucleoside inSchizosac-charomyces pombe U6 snRNA. Chin. Sci. Bull. 2000;45:1304–1308. doi: 10.1007/BF03182908. [DOI] [Google Scholar]
- 59.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Newby MI, Greenbaum NL. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat. Struct. Biol. 2002;9:958–965. doi: 10.1038/nsb873. [DOI] [PubMed] [Google Scholar]
- 61.Ma X, Zhao X, Yu YT. Pseudouridylation (Psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 2003;22:1889–1897. doi: 10.1093/emboj/cdg191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Donmez G, Hartmuth K, Luhrmann R. Modified nucleotides at the 5’ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004;10:1925–1933. doi: 10.1261/rna.7186504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma X, et al. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J. 2005;24:2403–2413. doi: 10.1038/sj.emboj.7600718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu G, Xiao M, Yang C, Yu YT. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 2011;30:79–89. doi: 10.1038/emboj.2010.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu G, et al. Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. EMBO J. 2016;35:654–667. doi: 10.15252/embj.201593113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shimba S, Bokar JA, Rottman F, Reddy R. Accurate and efficient N-6-adenosine methylation in spliceosomal U6 small nuclear RNA by HeLa cell extract in vitro. Nucleic Acids Res. 1995;23:2421–2426. doi: 10.1093/nar/23.13.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kiss T, Antal M, Solymosy F. Plant small nuclear RNAs. II. U6 RNA and a 4.5SI-like RNA are present in plant nuclei. Nucleic Acids Res. 1987;15:543–560. doi: 10.1093/nar/15.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aoyama T, Yamashita S, Tomita K. Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nucleic Acids Res. 2020;48:5157–5168. doi: 10.1093/nar/gkaa227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 70.Miyauchi K, Ohara T, Suzuki T. Automated parallel isolation of multiple species of non-coding RNAs by the reciprocal circulating chromatography method. Nucleic Acids Res. 2007;35:e24. doi: 10.1093/nar/gkl1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pan X, et al. Identification of novel genes involved in DNA damage response by screening a genome-wide Schizosaccharomyces pombe deletion library. BMC Genomics. 2012;13:662. doi: 10.1186/1471-2164-13-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naamati A, Regev-Rudzki N, Galperin S, Lill R, Pines O. Dual targeting of Nfs1 and discovery of its novel processing enzyme, Icp55. J. Biol. Chem. 2009;284:30200–30208. doi: 10.1074/jbc.M109.034694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vogtle FN, et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 74.Kim DU, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nguyen TT, et al. Predicting chemotherapeutic drug combinations through gene network profiling. Sci. Rep. 2016;6:18658. doi: 10.1038/srep18658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tadeo X, et al. Elimination of shelterin components bypasses RNAi for pericentric heterochromatin assembly. Genes Dev. 2013;27:2489–2499. doi: 10.1101/gad.226118.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bembom, O. & Ivanek, R. seqLogo: Sequence logos for DNA sequence alignments. R package version 1.56.0. (2020).
- 78.Nettling M, et al. DiffLogo: a comparative visualization of sequence motifs. BMC Bioinforma. 2015;16:387. doi: 10.1186/s12859-015-0767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wassarman DA, Steitz JA. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- 80.Roost C, et al. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 2015;137:2107–2115. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sontheimer EJ, Steitz JA. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 82.Newman AJ, Teigelkamp S, Beggs JD. snRNA interactions at 5’ and 3’ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- 83.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast activated spliceosome at 3.5 A resolution. Science. 2016;353:904–911. doi: 10.1126/science.aag0291. [DOI] [PubMed] [Google Scholar]
- 84.Rauhut R, et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science. 2016;353:1399–1405. doi: 10.1126/science.aag1906. [DOI] [PubMed] [Google Scholar]
- 85.Haselbach D, et al. Structure and conformational dynamics of the human Spliceosomal B(act) complex. Cell. 2018;172:454–464 e11. doi: 10.1016/j.cell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 86.Zhang X, et al. Structure of the human activated spliceosome in three conformational states. Cell Res. 2018;28:307–322. doi: 10.1038/cr.2018.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Madhani HD, Bordonne R, Guthrie C. Multiple roles for U6 snRNA in the splicing pathway. Genes Dev. 1990;4:2264–2277. doi: 10.1101/gad.4.12b.2264. [DOI] [PubMed] [Google Scholar]
- 88.Newman A, Norman C. Mutations in yeast U5 snRNA alter the specificity of 5’ splice-site cleavage. Cell. 1991;65:115–123. doi: 10.1016/0092-8674(91)90413-S. [DOI] [PubMed] [Google Scholar]
- 89.Newman AJ, Norman C. U5 snRNA interacts with exon sequences at 5’ and 3’ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 90.Lesser CF, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 91.Plaschka C, Lin PC, Nagai K. Structure of a pre-catalytic spliceosome. Nature. 2017;546:617–621. doi: 10.1038/nature22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bertram K, et al. Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell. 2017;170:701–713 e11. doi: 10.1016/j.cell.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 93.Yan C, et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 94.Wan R, Yan C, Bai R, Lei J, Shi Y. Structure of an intron lariat spliceosome from Saccharomyces cerevisiae. Cell. 2017;171:120–132 e12. doi: 10.1016/j.cell.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 95.Koodathingal P, Novak T, Piccirilli JA, Staley JP. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5’ splice site cleavage during pre-mRNA splicing. Mol. Cell. 2010;39:385–395. doi: 10.1016/j.molcel.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang F, et al. Splicing proofreading at 5’ splice sites by ATPase Prp28p. Nucleic Acids Res. 2013;41:4660–4670. doi: 10.1093/nar/gkt149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.O’Keefe RT. Mutations in U5 snRNA loop 1 influence the splicing of different genes in vivo. Nucleic Acids Res. 2002;30:5476–5484. doi: 10.1093/nar/gkf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kershaw CJ, Barrass JD, Beggs JD, O’Keefe RT. Mutations in the U5 snRNA result in altered splicing of subsets of pre-mRNAs and reduced stability of Prp8. RNA. 2009;15:1292–1304. doi: 10.1261/rna.1347409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen W, Moore MJ. Spliceosomes. Curr. Biol. 2015;25:R181–R183. doi: 10.1016/j.cub.2014.11.059. [DOI] [PubMed] [Google Scholar]
- 100.Neuveglise C, Marck C, Gaillardin C. The intronome of budding yeasts. C. R. Biol. 2011;334:662–670. doi: 10.1016/j.crvi.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 101.Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 102.Stark MR, et al. Dramatically reduced spliceosome in Cyanidioschyzon merolae. Proc. Natl Acad. Sci. USA. 2015;112:E1191–E1200. doi: 10.1073/pnas.1416879112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Parker BJ, et al. New families of human regulatory RNA structures identified by comparative analysis of vertebrate genomes. Genome Res. 2011;21:1929–1943. doi: 10.1101/gr.112516.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dorsett M, Westlund B, Schedl T. METT-10, a putative methyltransferase, inhibits germ cell proliferative fate in Caenorhabditis elegans. Genetics. 2009;183:233–247. doi: 10.1534/genetics.109.105270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim J, Kim Y, Yeom M, Kim JH, Nam HG. FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell. 2008;20:307–319. doi: 10.1105/tpc.107.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Collart MA, Oliviero S. Preparation of yeast RNA. Curr. Protoc. Mol. Biol. 2001;Chapter 13:Unit13 12. doi: 10.1002/0471142727.mb1312s23. [DOI] [PubMed] [Google Scholar]
- 107.Suzuki T, Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014;42:7346–7357. doi: 10.1093/nar/gku390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao Y, Yang J, Cancilla MT, Meng F, McLuckey SA. Top-down interrogation of chemically modified oligonucleotides by negative electron transfer and collision induced dissociation. Anal. Chem. 2013;85:4713–4720. doi: 10.1021/ac400448t. [DOI] [PubMed] [Google Scholar]
- 109.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lock A, et al. PomBase 2018: user-driven reimplementation of the fission yeast database provides rapid and intuitive access to diverse, interconnected information. Nucleic Acids Res. 2019;47:D821–D827. doi: 10.1093/nar/gky961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aken BL, et al. Ensembl 2017. Nucleic Acids Res. 2017;45:D635–D642. doi: 10.1093/nar/gkw1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ishigami, Y. YumaIshigami/irscalc: First Release. 10.5281/zenodo.4673906] (2021).
- 115.Cock PJ, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bitton DA, et al. AnGeLi: a tool for the analysis of gene lists from fission yeast. Front. Genet. 2015;6:330. doi: 10.3389/fgene.2015.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang Y, Werling U, Edelmann W. Seamless ligation cloning extract (SLiCE) cloning method. Methods Mol. Biol. 2014;1116:235–244. doi: 10.1007/978-1-62703-764-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Suga M, Hatakeyama T. A rapid and simple procedure for high-efficiency lithium acetate transformation of cryopreserved Schizosaccharomyces pombe cells. Yeast. 2005;22:799–804. doi: 10.1002/yea.1247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The sequence data from this study have been submitted to the DDBJ Sequence Read Archive under accession number DRA009909. Structural data was retrieved from PDB ID: 5O9Z [10.2210/pdb5o9z/pdb], 5GM6 [10.2210/pdb5gm6/pdb], and 3JB9 [10.2210/pdb3jb9/pdb]. Source data are provided with this paper.
Python scripts written for RNA-seq data analysis are available from a public GitHub repository https://github.com/YumaIshigami/irscalc114.