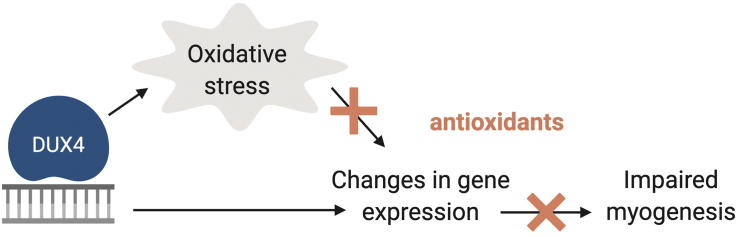
Abstract
Muscles of patients with facioscapulohumeral dystrophy (FSHD) are characterized by sporadic DUX4 expression and oxidative stress which is at least partially induced by DUX4 protein. Nevertheless, targeting oxidative stress with antioxidants has a limited impact on FSHD patients, and the exact role of oxidative stress in the pathology of FSHD, as well as its interplay with the DUX4 expression, remain unclear. Here we set up a screen for genes that are upregulated by DUX4 via oxidative stress with the aim to target these genes rather than the oxidative stress itself. Immortalized human myoblasts expressing DUX4 (MB135-DUX4) have an increased level of reactive oxygen species (ROS) and exhibit differentiation defects which can be reduced by treating the cells with classic (Tempol) or mitochondria-targeted antioxidants (SkQ1). The transcriptome analysis of antioxidant-treated MB135 and MB135-DUX4 myoblasts allowed us to identify 200 genes with expression deregulated by DUX4 but normalized upon antioxidant treatment. Several of these genes, including PITX1, have been already associated with FSHD and/or muscle differentiation. We confirmed that PITX1 was indeed deregulated in MB135-DUX4 cells and primary FSHD myoblasts and revealed a redox component in PITX1 regulation. PITX1 silencing partially reversed the differentiation defects of MB135-DUX4 myoblasts. Our approach can be used to identify and target redox-dependent genes involved in human diseases.
Keywords: DUX4, FSHD, Oxidative stress, Mitochondrial ROS, Muscle differentiation, PITX1
Graphical abstract
Highlights
-
•
Double homeobox transcription factor DUX4 misregulates hundreds of genes and induces oxidative stress in human myoblasts.
-
•
ROS, notably those of mitochondrial origin, contribute to the differentiation defects in myoblasts expressing DUX4.
-
•
A subset of genes is deregulated by DUX4 indirectly, via oxidative stress.
-
•
A strategy to identify the genes deregulated by DUX4 via oxidative stress was developed.
-
•
PITX1 is deregulated by DUX4 via oxidative stress and can be targeted to improve myogenesis in DUX4-expressing myoblasts.
1. Introduction
Facioscapulohumeral dystrophy (FSHD) is an autosomal dominant hereditary disease characterised by progressive muscle weakness and degeneration. It is caused by the combination of genetic and epigenetic factors leading to the aberrant expression of DUX4, the double homeobox protein 4, encoded within the D4Z4 repeat array on chromosome 4q35. DUX4 is involved in embryogenesis and is normally repressed in most adult tissues [1]. When abnormally expressed in adult muscles DUX4 disrupts multiple signalling pathways [2] and causes oxidative stress and DNA damage [[3], [4], [5]], but its exact function in FSHD pathology is not yet clear.
A characteristic feature of FSHD muscles is oxidative stress, which is at least partially attributed to DUX4 expression [3]. Muscle biopsies from FSHD patients have increased levels of oxidative stress markers which correlates with functional muscle impairment [5]. Patient-derived as well as healthy DUX4-transfected myoblasts are prone to DNA damage due to increased levels of reactive oxygen species [4] and exhibit reduced viability when exposed to oxidative stressors [3,6]. Multiple studies have revealed oxidative stress-related genes and proteins to be deregulated in FSHD [[6], [7], [8], [9], [10], [11]] as well as in DUX4 overexpression models [2,3]. Inversely, DUX4 expression itself may be induced by oxidative stress [12]. Despite being effective in in vitro models [4,13,14], antioxidants had a limited success in two clinical trials (reviewed in Ref. [15]): the dietary supplementation of vitamins C, vitamin E, zinc and selenium did not improve the 2-min walk test in individuals with FSHD, though the maximum voluntary contraction and endurance of the quadriceps were improved [16]. Methionine and folic acid supplement did not have any effect on DNA methylation or muscle state of FSHD patients [17], despite the antioxidant properties of these compounds. Generally, antioxidants may be effective for combating an initial acute response to oxidative stress, but are much less efficient when reversing the oxidative damage from a chronic state [18], such as FSHD. Moreover, a certain level of ROS is required for normal myogenesis (reviewed in Refs. [19,20]), thus totally removing all cellular ROS might not be a good treatment strategy for muscular dystrophies. Therefore, alternative approaches are needed to address the consequences of oxidative stress in FSHD patients. One of the strategies would be to target genes and pathways deregulated by DUX4 via oxidative stress rather than oxidative stress itself, although up to now, these targets remain largely unknown.
Here, we have successfully applied a transcriptomic approach to identify genes deregulated by DUX4 indirectly through oxidative stress and identified several genes relevant for the FSHD pathogenesis. As a proof of principle, we silenced one of these genes, PITX1; this improved the differentiation capacity of DUX4-expressing myoblasts. Our strategy may be considered as an alternative to antioxidant treatment in FSHD.
2. Results
2.1. Non-induced MB135-DUX4 myoblasts express DUX4 and exhibit differentiation defects
In this work we used immortalized MB135-DUX4 myoblasts with doxycycline-inducible DUX4 expression generated from MB135 myoblasts derived from the muscle biopsy of a healthy individual with the normal number of D4Z4 repeats [2]. When induced with doxycycline, these cells robustly express DUX4 and die within 48 h [21], but even without induction, when cultivated in the presence of the commercial non-Tet-free serum, MB135-DUX4 myoblasts express DUX4 at low levels due to promoter leakage, a known feature of doxycycline-inducible systems [22]. Low levels of DUX4 expression are very difficult to detect by qRT-PCR; therefore we monitored DUX4 expression in non-induced MB135-DUX4 myoblasts by the upregulation of its targets Zscan and Trim43 (Supplementary Fig. 1). The level of DUX4 expression observed in non-induced MB135-DUX4 is not lethal for the cells, allowing for long term studies, but is sufficient to cause visible defects in myogenic differentiation (Fig. 1C). While normal MB135 myoblasts differentiate efficiently to form thick and branched myotubes, MB135-DUX4 myoblasts form myotubes which are thin and sparse (Fig. 2A, lower left panel). A similar DUX4 expression pattern and differentiation defects are observed in primary and immortalized myoblasts derived from FSHD patients [4,7,13]; therefore here we used the MB135-DUX4 cell line without induction as a model of FSHD.
Fig. 1.
Oxidative stress and differentiation defects in MB135-DUX myoblasts. (A) ROS production in MB135 and MB135-DUX4 cells, treated or non-treated with antioxidants. Cells were incubated in the presence of 40 nM SkQ1 or 100 uM Tempol for four days and then stained with a ROS indicator CM-H2DCFD for 30 min. CM-H2DCFD fluorescence was detected by flow cytometry. The data are presented as means ± SEM, N = 5–16. Mean value for the untreated MB135-DUX4 cells is set to 100%. (B) Cardiolipin peroxidation in MB135 and MB135-DUX4 cells, treated or non-treated with antioxidants. Cells were incubated in the presence of 40 nM SkQ1 or 100 uM Tempol for four days and stained with MitoCLox for 5 h. MitoCLox fluorescence in the FITC and PE channels was then detected by flow cytometry. The data are presented as mean ± SEM, N = 4–6. The representative histograms show the fluorescence of MitoCLox-stained cells in the FITC and PE channels. The rightmost peak in the FITC channel corresponds to the cells with the oxidized MitoCLox. (C) Analysis of myotubes formed by MB135 and MB135-DUX4 myoblasts after four days of differentiation. Cells were treated with H202 for four days and then seeded on glass coverslips in 100% confluence. Myogenic differentiation was induced the next day by serum starvation and the cells were left to differentiate for four days. The myotubes were stained with monoclonal antibodies against skeletal troponin T (green) and DAPI (blue). To create a large image of the specimen, the images from adjacent fields were captured and stitched together using the Cartograph software (Microvision). Differentiation efficiency was assessed by measuring the troponin T - positive area normalized to the number of nuclei and the fusion index in 5–6 large images per sample. The fusion index is defined as the percentage of nuclei residing inside the troponin T-positive area. The data are presented as mean ± SEM, N = 3–5. (D) Apoptosis assessment in MB135 and MB135-DUX4 myoblasts treated with H202. Cells were treated with 200 or 400 uM H202 for four days, stained with Annexin V - FITC and PI and analyzed by flow cytometry. The percentage of viable (annexin V -, PI -), early apoptotic (annexin V +, PI -), late apoptotic (annexin V +, PI +) and necrotic (annexin V -, PI +) calls was calculated, N = 3. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
Antioxidant pre-treatment improves differentiation of MB135-DUX4 myoblasts. (A) Myotubes formed by MB135 and MB135-DUX4 myoblasts pre-treated with antioxidants before differentiation induction. Representative myotubes are marked with white errors. Cells were treated with 40 nM SkQ1 or 100 uM Tempol for four days and then seeded in six-well plates in 100% confluence. The next day differentiation was induced by serum starvation and the cells were left to differentiate for two days. The myotubes were stained with May-Grunwald Giemsa histological dye and 5 microscopic fields per specimen were captured. The differentiation efficiency was assessed by measuring the areas of myotubes in each of the 5 fields captured for each sample. The data is presented as mean myotube area ±SEM, N = 3. (B) Myotubes formed by MB135-DUX4 myoblasts treated with antioxidants during differentiation. The non-treated cells were seeded in 6-well plates in 100% confluence and induced to differentiate the next day. Antioxidants (40 nM SkQ1 or 100 uM Tempol) were added directly to the differentiation medium. The cells were left to differentiate for two days and then stained and analyzed as in (A). The data is presented as mean myotube area ±SEM, N = 3.
2.2. Elevated levels of ROS and mitochondrial lipid peroxidation сontribute to differentiation defects in MB135-DUX4 myoblasts
As DUX4 is known to cause oxidative stress, we assessed the levels of ROS in control and MB135-DUX4 myoblasts using the 2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA) assay (Fig. 1A). The ROS levels were increased more than twofold in MB135-DUX4 cells. Treatment with two types of antioxidants: a superoxide dismutase mimetic Tempol (100 uM) and a mitochondria-targeted antioxidant SkQ1 (40 nM), effectively reduced the ROS levels to the level of control MB135 cells. This suggests that a large proportion of ROS generated upon DUX4 expression are of mitochondrial origin. To specifically address the mitochondrial ROS (mitoROS), we used MitoCLox, a ratiometric fluorescent probe reporting cardiolipin peroxidation in living cells [23]. Cardiolipin is a diphosphatidylglycerol phospholipid, which is unique to the inner mitochondrial membrane and is also extremely oxidation-sensitive, thus it can serve as a good indicator of ROS produced specifically in mitochondria. MitoCLox was previously tested on MB135 myoblasts to discover a small subpopulation of cells with oxidized cardiolipin which increased proportionally to the density of myoblast culture [24]. Here we observed that the fraction of cells with oxidized MitoCLox is increased in MB135-DUX4 cells (46 ± 4.5%) compared to the equally dense MB135 culture (25 ± 5.7%) (Fig. 1B). Antioxidant treatment reduced the proportion of cells with oxidized MitoCLox: most of the cells treated with Tempol did not contain oxidized cardiolipin, while SkQ1 reduced the proportion of MB135-DUX4 cells with oxidized cardiolipin to the control level (29 ± 4.5%).
To understand whether the increased level of ROS alone could contribute to the differentiation defects observed in MB135-DUX4 myoblasts, we induced oxidative stress in normal MB135 cells without DUX4 expression by treating them with moderate concentrations of H202 (200 uM and 400 uM) for four days prior to differentiation induction. This significantly inhibited the differentiation of MB135 cells (Fig. 1C). Importantly, H202 concentrations used for oxidative stress induction were not lethal for the myoblasts and did not lead to significant changes in the apoptosis rate (Fig. 1D). Nevertheless, MB135-DUX4 myoblasts were slightly more susceptible to H202 -induced cell death in accordance with the previously published data that DUX4 renders the cells more sensitive to oxidative stress [3,4,6]. Myotube formation was completely inhibited in the MB135-DUX4 myoblasts after four days of H202 treatment (data not shown).
2.3. Antioxidant treatment improves the differentiation of MB135-DUX4 myoblasts
As excessive ROS production potentially contributes to the impaired myogenic differentiation of myoblasts expressing DUX4, we tested whether their differentiation capacity would be improved by antioxidants. We treated the myoblasts with a classic antioxidant Tempol (100 uM) or a mitochondria-targeted antioxidant SkQ1 (40 nM) and induced differentiation four days after. Antioxidant treatment partially restored the differentiation capacity of MB135-DUX4 myoblasts (Fig. 2A). Both antioxidants were equally effective in MB135-DUX4 cells, but in normal MB135 myoblasts, Tempol treatment slightly reduced the differentiation efficiency (Fig. 2A). This agrees with the data that a certain level of ROS is necessary for the normal myogenesis (reviewed in Refs. [19,20]) and also suggests that the ROS contributing to differentiation defects in DUX4-expressing cells are mostly of mitochondrial origin.
Importantly, the antioxidant treatment was only effective when the MB135-DUX4 myoblasts were supplemented with antioxidants four days prior to differentiation induction. Addition of antioxidants directly to the differentiation medium without the four-day pretreatment produced no effect (Fig. 2B), suggesting that ROS mostly affected the myoblasts before the differentiation began.
2.4. RNA-seq based approach to identify genes deregulated by DUX4 via oxidative stress
Gene expression changes caused by DUX4 can be attributed either to its direct transactivation activity or to the oxidative stress induced by DUX4 expression (Fig. 3A). To identify the genes which are deregulated by DUX4 through oxidative stress we compared the transcriptome profiles of MB135 and MB135-DUX4 myoblasts treated or not treated with antioxidants. We could identify the gene sets which were deregulated as a result of oxidative stress generated by DUX4 rather than by DUX4 directly by extracting genes which were differentially expressed between MB135 and MB135-DUX4 but not between MB135 and MB135-DUX4 treated with antioxidants (Fig. 3C).
Fig. 3.
Identification of genes deregulated by DUX4 via oxidative stress. (A) Volcano plot and heatmap for the genes differentially expressed between MB135-DUX4 and MB135 myoblasts. Experiments were performed in biological duplicates. The genes with padj <0.01 and |log2Fold| > 1 were considered significant. The heatmap shows 50 significant genes with the lowest padj. The color scale represents relative expression levels as row mean-centered rlog normalized counts. Hierarchical clustering was performed using Euclidean distance. The volcano plot shows all the genes differentially expressed between MB135-DUX4 and MB135, with the genes passing the significance threshold of padj < 0.01 colored blue (log2Fold < −1, downregulated) or orange (log2Fold > 1, upregulated). Darker colors correspond to lower padj values.
(B) Schematic representation of the DUX4-driven gene expression deregulation and the venn diagram showing the numbers of differentially expressed genes in different comparisons (with padj < 0.01 and |log2Fold| > 1). 200 genes that were differentially expressed between MB135 and MB135-DUX4, but not between MB135 and MB135-DUX4 treated with SkQ1 or Tempol, were considered antioxidant-sensitive. (C) Gene Ontology Slim annotation of the 200 genes falling into the antioxidant-sensitive group. (D) Gene Ontology overrepresentation analysis of the 200 genes falling into the antioxidant-sensitive group. (E) Heatmap for 10 selected differentially expressed genes from the antioxidant-sensitive group. The genes are significantly differentially expressed between MB135 and MB135-DUX4 cells (with padj < 0.01 and |log2Fold| > 1), but not between the MB135 and MB135-DUX4 cells treated with antioxidants. The color scale represents relative expression levels as row mean-centered rlog normalized counts. Hierarchical clustering was performed using Euclidean distance. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
782 genes were differentially expressed (padj < 0.01 and |Log2Fold| > 1) between MB135-DUX4 and MB135 cells (Fig. 3C). 506 genes were still deregulated in MB135-DUX4 myoblasts treated with any of the two antioxidants, and a subset of genes (242 in case of SkQ1, 234 in case of Tempol, 200 in common) were no longer differentially expressed between antioxidant-treated MB135-DUX4 cells and the control (Fig. 3E, Supplementary Table 1). Most of these antioxidant-sensitive genes (200) were similar both for the mitochondria-targeted antioxidant SkQ1 and the classical antioxidant Tempol. Gene ontology enrichment analysis of these 200 antioxidant-sensitive genes (Fig. 3D and E) revealed muscle tissue development to be the most significant overrepresented term with 10 genes falling into this category, including PITX1, the gene previously linked to FSHD [25]. Other relevant enriched categories included positive regulation of cellular protein localization (9 genes), regulation of developmental growth (8 genes), and apoptotic mitochondrial changes (5 genes). The antioxidant-sensitive genes were both protein-coding (151 genes) and non-coding (34 genes). Some of these genes, including PITX1, SORBS2 and GPX1 were previously identified as being differentially expressed in FSHD, confirming the validity of our assay. PITX1 encodes for a transcription factor involved in early tissue patterning and was shown to be specifically deregulated in muscle biopsies of FSHD patients [25]. SORBS2 (SH3 domain-containing protein 2) was misregulated in FSHD myoblasts upon telomere shortening [26]. GPX1 (glutathione peroxidase 1) was previously identified to be oxidative stress sensitive in FSHD myoblasts [14]. Among the non-protein coding genes, H19, encoding for a long noncoding RNA (lncRNA), was most robustly downregulated. H19 lncRNA has been shown to promote muscle differentiation and regeneration [27,28].
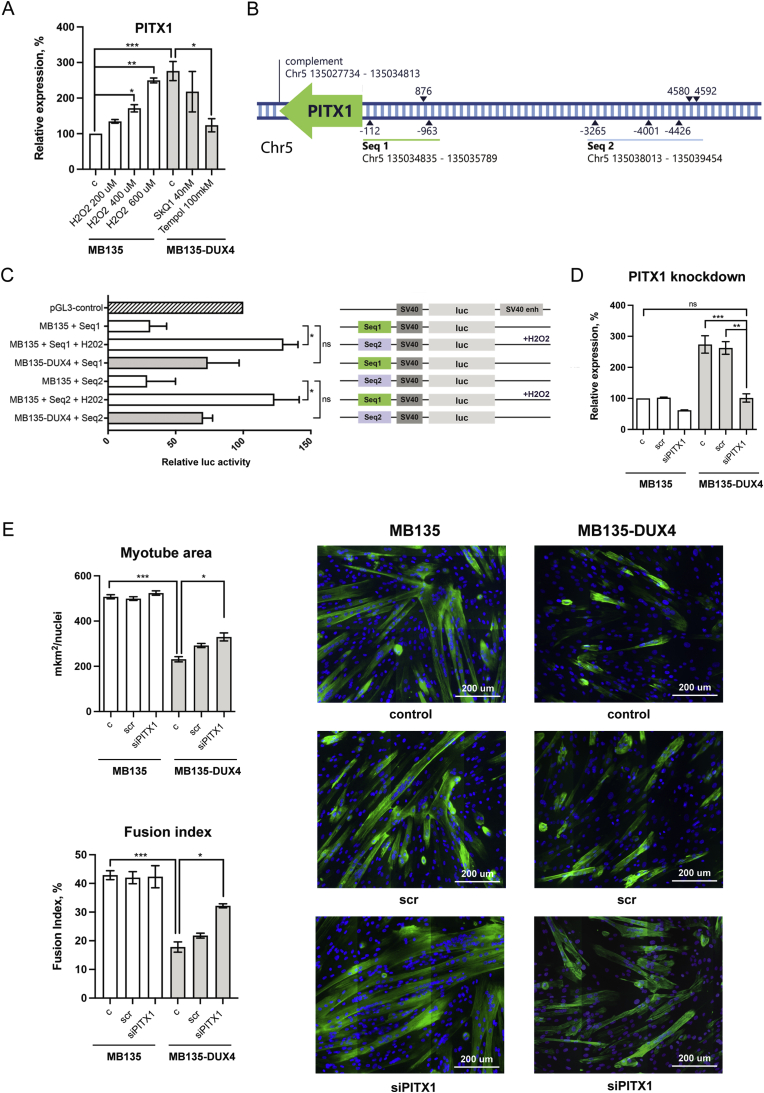
2.5. PITX1 expression in MB135-DUX4 myoblasts is ROS-sensitive and can be targeted to improve myogenesis
Transcriptomic screening revealed PITX1 as a candidate gene upregulated in DUX4-expressing cells via oxidative stress. RT-qPCR results confirmed that the expression of PITX1 was 2.5 times higher in MB135-DUX4 cells compared to the control, while Tempol treatment decreased PITX1 expression to the control level (Fig. 4A). Elevated PITX1 expression was also observed in primary myoblasts from FSHD patients (data not shown). PITX1 expression was similarly 2.5 times increased in MB135 cells treated with moderate concentrations of H202 for 24 h. As the H202 concentrations used for oxidative stress induction did not lead to significant changes in apoptosis rates in MB135 cells (Fig. 1D), PITX1 overexpression was not likely to result from the gene expression changes associated with apoptosis.
Fig. 4.
PITX1 expression is redox-sensitive and can be silenced to improve the differentiation of MB135-DUX4 myoblasts. (A)PITX1 expression assessed by RT-qPCR. Myoblasts were treated with 40 nM SkQ1 or 100 uM Tempol for four days or 200–600 uM H2O2 for 24 h. The data are represented as mean relative expression ± SEM, N = 4–10. Mean value for the MB135 cells is set to 100%. (B) Regions upstream of PITX1 contain potential ARE sequences. Blue triangles mark potential AREs, numbers indicate the distances in base pairs from the start of PITX1 gene. Seq1 and Seq2 are the sequences used for reporter assays. Genomic coordinates are for GRCh38. (C) Dual luciferase reporter assay for enhancer activity of potential ARE-containing sequences. Seq1 and Seq2 sequences were cloned into pGL3 luciferase reporter plasmid upstream of the SV40 promoter. MB135 and MB135-DUX4 cells were treated with 400 uM H202 for 24 h and then transfected with the pGL3-Seq1/Seq2 plasmids and pCMV-Red Firefly Luc plasmid, used as a as a normalization control. The data are presented as mean relative luciferase activity ± SEM, mean luciferase activity in the cells transfected with a pGL3 control vector, containing an SV40 enhancer, is set to 100%. N = 4–5. (D) PITX1 expression four days after siRNA transfection. Myoblasts were seeded on six-well plates (2 × 105 cells/well) and transfected with scr siRNA (negative control) or siRNA against PITX1 the next day. The cells were left to proliferate in siRNA-containing medium for 4 days and then PITX1 expression was assessed by RT-qPCR. The data are presented as mean relative expression ± SEM, mean value for non-transfected MB135 cells is set to 100%, N = 3. (E) Myotubes formed by MB135 and MB135-DUX4 myoblasts upon PITX1 siRNA knockdown. The cells were seeded and treated as described in (D). After 4 days of proliferation with siRNA, myoblasts were induced to differentiate by serum starvation. On the fourth day of differentiation, the cells were fixed and stained with monoclonal antibodies against skeletal troponin T (green) and DAPI (blue). To create a large image of the specimen, the images from adjacent fields were captured and stitched together using Cartograph software (Microvision). Differentiation efficiency was assessed by measuring the troponin T - positive area normalized to the number of nuclei and the fusion index in 5–6 large images per sample. The fusion index is defined as the percentage of nuclei residing inside the troponin T-positive area. The data are presented as mean ± SEM, N = 3. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Given the responsiveness of PITX1 expression to oxidative stress and antioxidant treatment we have searched the PITX1 upstream region for putative antioxidant response elements (AREs), consensus sequences recognized by NRF2, a redox-sensitive transcription factor playing an important role in redox homeostasis and cytoprotection [29]. We have identified eight potential ARE sequences [30] in the 5 kb upstream region of PITX1 (Fig. 4B, Supplementary Fig. 3). Sequences containing these putative AREs (Seq1 977 bp, Seq2 1461 bp) were cloned into the pGL3 luciferase reporter plasmid upstream of the SV40 promoter and an increased expression of the reporter gene was observed when the transfected MB135 cells were treated with H202. Elevated expression of the reporter gene was also observed in non-treated MB135-DUX4 cells (Fig. 4C). Treatment of the pGL3-Seq1/pGL3-Seq2 - transfected MB135-DUX4 myoblasts with H202 led to high cellular mortality, therefore the luminescence could not be measured. All together, these data point to the presence of redox-sensitive elements in the PITX1 promoter.
To test whether PITX1 abnormal expression could contribute to differentiation defects in MB135-DUX4 myoblasts, we have performed PITX1 knockdown by siRNA transfection (Fig. 4D). MB135-DUX4 myoblasts with the PITX1 knockdown partially restored their differentiation capacity (Fig. 4E). In normal MB135 myoblasts, PITX1 silencing did not produce any visible effect. Thus the increased expression of PITX1 contributes to the DUX4-induced impairment of myogenic differentiation in MB134-DUX4 cells.
3. Discussion
FSHD muscles are subjected to oxidative stress induced by DUX4, the protein involved in the etiology of the disease. Here we studied the involvement of ROS in the myogenesis of DUX4-expressing myoblasts and used an original strategy to identify the genes deregulated by DUX4 via oxidative stress.
We used human immortalized myoblasts MB135-DUX4 which express DUX4 at a low level and exhibit differentiation defects, similarly to myoblasts from FSHD patients [4,7,13]. MB135-DUX4 cells have elevated levels of ROS and mitochondrial lipid peroxidation, which is in line with data on impaired mitochondrial morphology and functions in patients with FSHD [5]. MB135 and MB135-DUX4 cells were treated with antioxidants, 100 uM of the SOD-mimetic Tempol or 40 nM of the mitochondria-targeted antioxidant SkQ1, that were previously used in the murine models in concentrations similar to those used in the present study [[31], [32], [33]]. Elimination of ROS with Tempol, as well as specific elimination of mitochondrial ROS (mitoROS) with SkQ1 decreased ROS and mitochondrial lipid peroxidation levels and improved the morphology of myotubes formed by MB135-DUX4 myoblasts. This is consistent with the previous works demonstrating the beneficial effect of antioxidant treatment on FSHD myoblasts [4], and suggests the involvement of ROS, specifically those produced by mitochondria, in DUX4-induced myogenesis impairment. Antioxidants could only improve myogenesis if used as pretreatment, before the differentiation induction, suggesting that oxidative stress-induced deregulation occurs at the myoblast level. Both antioxidants were.
While antioxidants are quite efficient in reversing the FSHD phenotype in primary and immortalized myoblasts derived from FSHD patients in vitro [4,14], their effect is much more limited when tested in clinics [16,17]. In search for an alternative to the antioxidant treatment, we used a transcriptomic approach to identify the genes deregulated in MB135-DUX4 myoblasts through oxidative stress rather than directly by DUX4 (Fig. 3A). We compared the transcriptomes of the control MB135 and DUX4-expressing MB135-DUX cells treated with antioxidants for four days and revealed 200 genes which were no longer differentially expressed between MB135 and MB135-DUX4 myoblasts after the antioxidant treatment (Supplementary Table 1). One of the antioxidant-sensitive genes discovered in our screen was PITX1, the gene previously shown to be specifically upregulated in the muscle biopsies of FSHD patients [25].
In mice, conditional overexpression of PITX1 causes muscle atrophy necrosis and inflammatory infiltration [34]. PITX1 has long been considered a direct DUX4 target [25], though a recent study exploring the DNA-binding specificity of DUX4 calls this into question [35]. In our screen, PITX1 emerged as a gene regulated by DUX4 indirectly. Its upregulation in DUX4-expressing myoblasts disappeared after the antioxidant treatment, while treatment with H202 induced PITX1 overexpression in the control MB135 myoblasts. Our data thus suggests the existence of a redox component in the regulation of this gene.
As a proof of principle for our approach, we tested whether PITX1 silencing could reproduce the positive effect of antioxidants on myoblasts differentiation. We could not fully restore the myotube morphology, but the number and thickness of the myotubes formed by MB135-DUX4 myoblasts treated with siRNA targeting PITX1 was significantly increased. Thus, PITX1 deregulation through oxidative stress may be one of the important factors causing differentiation defects in DUX4-expressing myoblasts.
PITX1 gene encodes for a transcription factor of bicoid-class homeodomain proteins. It is involved in the embryonic development of hind limbs musculature [[36], [37], [38], [39]], but whether it has a role in adult myogenesis is largely unknown. Some insight about PITX1 function in adults can be obtained from studies on mice. Pitx1 expression was undetectable in adult mouse muscle satellite cells, while the other genes of the family, Pitx2 and Pitx3, were expressed in a differentiation-dependent pattern, with Pitx2 and Pitx3 silencing impairing myoblast fusion into myotubes [40]. Multiple evidence suggests that Pitx2 is an important regulator of adult myogenesis [41]. In our study, PITX1 silencing did not impair the differentiation of normal MB135 myoblasts, but improved the fusion of DUX4-expressing myoblasts which had an initially elevated PITX1 level. As PITX isoforms are almost identical in their homeodomains and vary mainly in the N-terminal region [36], they can bind similar DNA sequences [36,[42], [43], [44]]. Thus PITX1 overexpression might possibly interfere with other isoforms’ signaling due to competition for binding sites. In fetal myogenesis, Pitx2 and Pitx3 control ROS levels during the transition from proliferation to differentiation, and their absence leads to irreversible DNA damage and apoptosis, mostly due to the direct downregulation of Nrf1 and its downstream genes with antioxidant function [45]. Importantly, the timeline of redox regulation by Pitx2 and Pitx3 is in accordance with our data, demonstrating that the antioxidant treatment is effective only before differentiation. Redox regulation by Pitx2 and Pitx3 happens before and during the differentiation onset, thus their signaling can only be modulated at myoblasts level. This agrees with our data demonstrating that antioxidants were only effective in pretreatment.
In conclusion, we demonstrated the involvement of mitoROS in DUX4-induced myogenic differentiation impairment. Using a transcriptomic approach, we identified a subset of genes deregulated by DUX4 indirectly, via ROS as potential targets for future FSHD treatments and validated our approach by silencing PITX1 expression. This approach can be used in other circumstances and oxidative stress-related diseases.
4. Materials and Methods
4.1. Cell cultures
Normal human immortalized myoblasts (MB135) derived from the muscle biopsy of a healthy individual with the normal number of D4Z4 repeats and MB135 myoblasts with Doxycycline-inducible DUX4 expression (MB135-DUX4) [2]were a kind gift of Dr.Stephen Tapscott. Cells were cultured at 37 °C, 5% CO2 in the growth medium composed of 60% DMEM (Sigma Aldrich, D5796), 25% 199 Medium (Sigma Aldrich, M4530) and 15% FBS (Life technology, 10270) supplemented with 0.5 ng/ml bFGF (Life technology, PHG0026), 0.2 ug/ml Dexamethasone, 100 units/mL penicillin and 100 μg/ml streptomycin (Thermo Fisher, 15140122).
4.2. Antioxidant treatment
Cells were plated onto 100 mm Petri dishes (1 × 106 cells per dish) or 6-well plates (2 × 10^5 cells per well) and left overnight for attachment. The next day the cells were treated with antioxidants (100 μM Tempol, 20 nM or 40 nM SkQ1), left to proliferate in the antioxidant-containing medium for 4 days and then used for differentiation experiments, flow cytometry, RT-qPCR or RNAseq.
4.3. Myoblast differentiation
Differentiation of MB135 cells was induced in 100% confluent cell cultures by replacing the Growth Medium with a low-serum Differentiation Medium composed of 98% DMEM (Sigma Aldrich, M4530) and 2% FBS (Life technology, 10270) supplemented with 10 ug/ml Insulin, 5.5 μg/ml Transferrin, 6.7 ng/ml Sodium Selenite (Insulin-Transferrin-Selenium Supplement 100X, Thermo Fisher, 51500056), 100 units/mL penicillin and 100 μg/ml streptomycin (Thermo Fisher, 15140122). After 2–4 days of differentiation (denoted in the figures) the cells were fixed with 4% PFA for 5 min (Euromedex) and stained either with May-Grunwald Giemsa or antibodies.
4.4. May-Grunwald Giemsa staining
The wells with PFA-fixed cells were washed with phosphate-buffered saline (PBS), and stained with 200 μL of May-Grunwald dye for 5 min. Then 1 ml of PBS was added to the wells without May-Grunwald dye removal and the cells were stained for additional 15 min in the diluted May-Grunwald solution. Afterwards the wells were washed 3 times with distilled water, stained for 1 h with 1 ml of Giemsa stain (diluted 1:10 in PBS), washed with distilled water again and let dry. All procedures were performed at room temperature. The samples were observed and photographed using the Axio Imager microscope (Zeiss). Five random fields of view were captured for each sample.
4.5. Immunostaining
The cells were fixed with 4% PFA (Euromedex) for 5 min, permeabilized with 0.25% Triton X-100 (Sigma-Aldrich, T8787) for 5 min and blocked with 0.5% BSA (Euromedex) for 1 h. To stain the myotubes, the slides were incubated with primary monoclonal anti-troponin T antibodies (mAb, Sigma-Aldrich, T6277) diluted 1:50 for 2 h and then secondary anti-mouse Alexa-488 IgG conjugated antibodies (Life Technologies, A-21200, 1:100, excitation/emission: 488/519 nm, green fluorescence) for 1 h at room temperature. The slides were then mounted with a DAPI-containing mounting medium (Vector Laboratories) and let dry for 1 h. The stained myotubes were observed and photographed with a fluorescent microscope (Microvision instruments) (Excitation/Emission: 488/519 nm). The images from adjacent fields of view were stitched together using Cartograph software (Microvision) to create a large image of the specimen. For each specimen 5–6 large images were generated.
4.6. Differentiation assessment
Image analysis for differentiation assessment was performed via Java-based image processing program Image J. For Giemsa stained slides, 5 fields of view per sample were captured and the areas of the myotubes in each field were measured. The structure with no less than four nuclei was considered a myotube. The mean myotube area for each sample was calculated. The procedure was repeated in three independent experiments. For immunostained slides, 5–6 large stitched images per sample were generated. For each large image of the specimen, the normalized troponin area (NTA) was calculated by dividing the overall troponin-positive area by the overall number of nuclei. The mean NTA was obtained for each specimen. The procedure was repeated in three independent experiments.
4.7. Flow cytometry
For flow cytometry the cells (non-treated or treated with antioxidants as described in the “Antioxidant treatment” of Materials and Methods) were incubated with 10 μM of general oxidative stress indicator CM-H2DCFD (Thermo Fisher, C6827) for 30 min or with 200 nM of ratiometric mitochondrial lipid peroxidation indicator MitoCLox [23] for 5 h. Then the cells were washed with PBS two times, lysed with Trypsin-EDTA solution (Sigma aldrich, T3924) and collected (on ice) into flow cytometry tubes with 200 μl of serum-containing Growth Medium to inactivate trypsin. The fluorescence was measured using BD FACS Aria III equipped with 5 lasers (375 nm, 405 nm, 488 nm, 561 nm, and 633 nm). For CM-H2DCFD, the mean fluorescence for each sample was calculated and the mean background fluorescence of a non-stained sample was subtracted from all the values. MitoCLox ratiometric analysis was performed using the Flowing software 2.4 (Cell Imaging Core, Turku Centre for Biotechnology). The same cell-culture density for all the samples was confirmed before running the assay as it is crucial for between-sample comparisons as reported before [24]. For the apoptosis assay, the cells were treated with 200 or 400 uM of H2O2 for four days and then stained with Annexin V with eBioscience™ Annexin V-FITC Apoptosis Detection Kit (Life Technologies #BMS500FI-20) according to the manufacturer's instructions. Stained cells were analyzed on a C6 flow cytometer (BD Biosciences).
4.8. RNA-seq
The samples were prepared in biological duplicates. The cells were grown on 100 mm Petri dishes and treated with antioxidants as described earlier. RNA was extracted using Nucleospin RNA isolation kit (Macherey–Nagel, 740955) according to the manufacturer's protocol. RNA-seq libraries were prepared using the Agilent SureSelect Automated Strand-Specific RNA Library Prep, with polyA selection. Prepared libraries were sequenced on an Illumina HiSeq2000. Raw paired-end sequence reads were mapped to the human transcriptome (Homo sapiens genome sequence GRCh38 with v99 gene annotations downloaded from Ensembl) using STAR (release 2.7.3a) [46] in the 2-pass mode. Multiple-mapping reads were excluded (-outFilterMultimappedNmax 1). Reads were assigned to genes via featureCounts (v 2.0.0) [47] including fractions. Differential expression analysis was performed using the DESeq2 R package [48]. Genes with |log2Fold change| > 1 and padj <0.01 were considered significantly differentially expressed. The genes differentially expressed between MB135-DUX4 and MB135, but not between MB135-DUX4 + Tempol/SkQ1 and MB135, were considered sensitive to antioxidant treatment. These treatment-sensitive genes are listed in the Supplementary Table 1. Supplementary Table 2 contains summary statistics for the RNA-Seq transcriptomics data. Raw fastq files, raw counts matrix and tables with differentially expressed genes are deposited to GEO (accession number GSE173762).
4.9. Gene Ontology (GO) analysis
GO analysis was performed using WebGestalt2019 [49]. Enrichment analysis was performed using over-representation analysis (ORA) method against the Gene Ontology Biological Process (noRedundant) functional database with all the genes detected in the RNA-seq experiment as a background. p-values were adjusted using Benjamini-Hochberg multiple testing correction.
4.10. RT-qPCR
Total RNA was isolated from the cultured cells using Nucleospin RNA isolation kit (Macherey–Nagel, 740955) following the manufacturer's protocols. Reverse transcription was carried out with 1000 ng of total RNA using RevertAid reverse transcriptase (Thermo Fisher, EP0441) according to manufacturer's instructions. The resulting cDNA was used to perform Real-time qPCR with PowerUp™ SYBR™ Green Master Mix (Life Technologies, A25778), following the manufacturer's protocols. The reaction was performed in a StepOnePlus thermal cycler (Life Technologies) in the following conditions: 50 °C 2’ → 95 °C 2’ → (95 °C 15″, 60 °C 1′) x 40. The relative gene expression was calculated using the 2 −ΔΔCt quantification method. Primer sequences are as follows:PITX1 F 5′-TCCACCAAGAGCTTCACCTT-3′PITX1 F 5′-TCCACCAAGAGCTTCACCTT-3′, PITX1 R 5′-CGGTGAGGTTGTTGATGTTG-3′, ZSCAN4 F: 5′-GGGAGCAGGGTGTATCTCTC-3′,
ZSCAN4 R: 5′-ACTCGGGACAAACAAATGGC-3’.
TRIM43 F: 5′-ACCCATCACTGGACTGGTGT-3′,
TRIM43 R: 5′-CACATCCTCAAAGAGCCTGA-3’.
4.11. Plasmid constructs
Putative ARE-containing sequences upstream of PITX1 gene (Seq1: chr5 135034835–135035789, 977 bp, 3 putative AREs; Seq2: chr5 135038013–135039454, 1461 bp, 5 putative AREs) were amplified from the gDNA of MB135 human myoblasts with the following primers:
Seq1 F 5′-TAAATTACGCGTGCCTGCGCTCTAATGGG-3’.
Seq1 R 5′-ATACAAAGATCTCAGCTGACCCTGG-3’.
Seq2 F 5′-ATGTATGCTAGCAGGACTGCCGCC-3’.
Seq2 R 5′-TAAGGTCTCGAGCCTTCGAGGCAGC-3′, and cloned into the pGL3-Promoter vector (Promega) upstream of the SV40 promoter and luciferase reporter gene into MluI, BglII, NheI, XhoI restriction sites.
4.12. Dual luciferase reporter assay
Dual luciferase reporter assay for assessing the enhancer activity of Seq1 and Seq2 in human myoblasts MB135 and MB135-DUX4 was performed with a Pierce Renilla-Firefly Luciferase Dual Assay Kit (Thermo Fisher, 16186) according to the manufacturer's protocol. The cells were seeded onto 48-well plates at a density of 4.5 × 104 cells per well and transfected the next day in the state of 70–80% confluence with the total of 300 ng of DNA per well. Transfection was performed with TurboFect Transfection Reagent (Thermo Fisher, R0533) according to the manufacturer's instructions. pGL3-control vector (Promega, E1741) containing SV40 enhancer, pGL3-basic vector (Promega, E1751) and pCMV-Red Firefly Luc vector (Thermo Fisher, 16156) were used as positive, negative and normalization controls respectively. The luciferase activity was assessed 72 h post-transfection.
4.13. siRNA knockdown of endogenous PITX1
The cells were seeded on 6-well plates (2 × 105 cells/well) and next day transfected with 15 μl of 10 μM PITX1 siRNA (Thermo Fisher, AM16708) using Lipofectamine RNAiMAX reagent (Thermo Fisher, 13778030) according to the manufacturer's instructions. Transfection was performed in serum-free medium for 5 h.Then the cells were supplemented with the 2X growth medium and left to proliferate for 4 days before being fixed and analyzed by microscopy. Silencing efficiency was monitored on the second and fourth days post-transfection by RT-qPCR using the primers listed in Table S2.
4.14. Statistical analysis
Statistical analyses were performed in GraphPad Prism 9 using non-parametric Kruskall-Wallis test with Dunn's correction for multiple comparisons. P-values less than 0.05 (*), 0.01 (**) and 0.001 (***) were considered significant. All data are presented as the mean ± SEM. All experiments were performed in no less than 3 biological replicates. The number of biological replicates in each experiment (N) is denoted in corresponding figure legends.
Declaration of competing interest
Declare that they have no conflict of interests.
Acknowledgements
We thank Drs. S.Tapscott and A.Evstafieva for sharing the materials. This work was supported by grants from the FSHD Society, the AFM (CTCFSHD), the Presidium of RAS, the IDB RAS government basic research program 0088-2021-0007 and the Russian Foundation for Basic Research grant 19-04-01020.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102008.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Expression of DUX4 targets ZSCAN4 and TRIM43 in MB135-DUX4 myoblasts assessed by RT-qPCR. The data are represented as mean log10fold change ± SEM, N = 7. Mean value for control MB135 cells is set to 100%.
ARE elements in the upstream region of PITX1. (A) ARE consensus sequence (adapted from Wang et al., 2007) (B) Potential ARE sequences discovered in the region 5 kb upstream from the start of PITX1. Letters matching with the most common variant in ARE consensus are colored in red, letters matching with at least one variant encountered in ARE consensus are colored in pink, mismatches are colored in black.
References
- 1.Karpukhina A., Tiukacheva E., Dib C., Vassetzky Y.S. Control of DUX4 expression in facioscapulohumeral muscular dystrophy and cancer. Trends Mol. Med. 2021 doi: 10.1016/j.molmed.2021.03.008. cited 13 Apr 2021. [DOI] [PubMed] [Google Scholar]
- 2.Jagannathan S., Shadle S.C., Resnick R., Snider L., Tawil R.N., van der Maarel S.M. Model systems of DUX4 expression recapitulate the transcriptional profile of FSHD cells. Hum. Mol. Genet. 2016;25:4419–4431. doi: 10.1093/hmg/ddw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008;27:2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dmitriev P., Bou Saada Y., Dib C., Ansseau E., Barat A., Hamade A. DUX4-induced constitutive DNA damage and oxidative stress contribute to aberrant differentiation of myoblasts from FSHD patients. Free Radic. Biol. Med. 2016;99:244–258. doi: 10.1016/j.freeradbiomed.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Turki A., Hayot M., Carnac G., Pillard F., Passerieux E., Bommart S. Functional muscle impairment in facioscapulohumeral muscular dystrophy is correlated with oxidative stress and mitochondrial dysfunction. Free Radic. Biol. Med. 2012;53:1068–1079. doi: 10.1016/j.freeradbiomed.2012.06.041. [DOI] [PubMed] [Google Scholar]
- 6.Winokur S.T., Barrett K., Martin J.H., Forrester J.R., Simon M., Tawil R. Facioscapulohumeral muscular dystrophy (FSHD) myoblasts demonstrate increased susceptibility to oxidative stress. Neuromuscul. Disord. NMD. 2003;13:322–333. doi: 10.1016/s0960-8966(02)00284-5. [DOI] [PubMed] [Google Scholar]
- 7.Barro M., Carnac G., Flavier S., Mercier J., Vassetzky Y., Laoudj-Chenivesse D. Myoblasts from affected and non-affected FSHD muscles exhibit morphological differentiation defects. J. Cell Mol. Med. 2010;14:275–289. doi: 10.1111/j.1582-4934.2008.00368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celegato B., Capitanio D., Pescatori M., Romualdi C., Pacchioni B., Cagnin S. Parallel protein and transcript profiles of FSHD patient muscles correlate to the D4Z4 arrangement and reveal a common impairment of slow to fast fibre differentiation and a general deregulation of MyoD-dependent genes. Proteomics. 2006;6:5303–5321. doi: 10.1002/pmic.200600056. [DOI] [PubMed] [Google Scholar]
- 9.Cheli S., François S., Bodega B., Ferrari F., Tenedini E., Roncaglia E. Expression profiling of FSHD-1 and FSHD-2 cells during myogenic differentiation evidences common and distinctive gene dysregulation patterns. PloS One. 2011;6 doi: 10.1371/journal.pone.0020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laoudj-Chenivesse D., Carnac G., Bisbal C., Hugon G., Bouillot S., Desnuelle C. Increased levels of adenine nucleotide translocator 1 protein and response to oxidative stress are early events in facioscapulohumeral muscular dystrophy muscle. J. Mol. Med. Berl Ger. 2005;83:216–224. doi: 10.1007/s00109-004-0583-7. [DOI] [PubMed] [Google Scholar]
- 11.Macaione V., Aguennouz M., Rodolico C., Mazzeo A., Patti A., Cannistraci E. RAGE-NF-kappaB pathway activation in response to oxidative stress in facioscapulohumeral muscular dystrophy. Acta Neurol. Scand. 2007;115:115–121. doi: 10.1111/j.1600-0404.2006.00724.x. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki-Honda M., Jonouchi T., Arai M., Hotta A., Mitsuhashi S., Nishino I. A patient-derived iPSC model revealed oxidative stress increases facioscapulohumeral muscular dystrophy-causative DUX4. Hum. Mol. Genet. 2018;27:4024–4035. doi: 10.1093/hmg/ddy293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bou Saada Y., Dib C., Dmitriev P., Hamade A., Carnac G., Laoudj-Chenivesse D. Facioscapulohumeral dystrophy myoblasts efficiently repair moderate levels of oxidative DNA damage. Histochem. Cell Biol. 2016;145:475–483. doi: 10.1007/s00418-016-1410-2. [DOI] [PubMed] [Google Scholar]
- 14.El Haddad M., Jean E., Turki A., Hugon G., Vernus B., Bonnieu A. Glutathione peroxidase 3, a new retinoid target gene, is crucial for human skeletal muscle precursor cell survival. J. Cell Sci. 2012;125:6147–6156. doi: 10.1242/jcs.115220. [DOI] [PubMed] [Google Scholar]
- 15.Denny A.P., Heather A.K. Are antioxidants a potential therapy for FSHD? A review of the literature. Oxid. Med. Cell. Longev. 2017;2017:7020295. doi: 10.1155/2017/7020295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Passerieux E., Hayot M., Jaussent A., Carnac G., Gouzi F., Pillard F. Effects of vitamin C, vitamin E, zinc gluconate, and selenomethionine supplementation on muscle function and oxidative stress biomarkers in patients with facioscapulohumeral dystrophy: a double-blind randomized controlled clinical trial. Free Radic. Biol. Med. 2015;81:158–169. doi: 10.1016/j.freeradbiomed.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 17.van der Kooi E.L., de Greef J.C., Wohlgemuth M., Frants R.R., van Asseldonk R.J.G.P., Blom H.J. No effect of folic acid and methionine supplementation on D4Z4 methylation in patients with facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. NMD. 2006;16:766–769. doi: 10.1016/j.nmd.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Steinhubl S.R. Why have antioxidants failed in clinical trials? Am. J. Cardiol. 2008;101:14D–19D. doi: 10.1016/j.amjcard.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Barbieri E., Sestili P. Reactive oxygen species in skeletal muscle signaling. J. Signal Transduct. 2012;2012:982794. doi: 10.1155/2012/982794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Moal E., Pialoux V., Juban G., Groussard C., Zouhal H., Chazaud B. Redox control of skeletal muscle regeneration. Antioxidants Redox Signal. 2017;27:276–310. doi: 10.1089/ars.2016.6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao Z., Snider L., Balog J., Lemmers R.J.L.F., Van Der Maarel S.M., Tawil R. DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet. 2014;23:5342–5352. doi: 10.1093/hmg/ddu251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer-Ficca M.L., Meyer R.G., Kaiser H., Brack A.R., Kandolf R., Küpper J.-H. Comparative analysis of inducible expression systems in transient transfection studies. Anal. Biochem. 2004;334:9–19. doi: 10.1016/j.ab.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 23.Lyamzaev K.G., Sumbatyan N.V., Nesterenko A.M., Kholina E.G., Voskoboynikova N., Steinhoff H.-J. MitoCLox: a novel mitochondria-targeted fluorescent probe for tracing lipid peroxidation. Oxid. Med. Cell. Longev. 2019;2019:9710208. doi: 10.1155/2019/9710208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyamzaev K.G., Panteleeva A.A., Karpukhina A.A., Galkin, Popova E.N., Pletjushkina O.Y. Novel fluorescent mitochondria-targeted probe MitoCLox reports lipid peroxidation in response to oxidative stress in vivo. Oxid. Med. Cell. Longev. 2020;2020:3631272. doi: 10.1155/2020/3631272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robin J.D., Ludlow A.T., Batten K., Gaillard M.-C., Stadler G., Magdinier F. SORBS2 transcription is activated by telomere position effect-over long distance upon telomere shortening in muscle cells from patients with facioscapulohumeral dystrophy. Genome Res. 2015;25:1781–1790. doi: 10.1101/gr.190660.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dey B.K., Pfeifer K., Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X., Ji S., Li W., Yi B., Li H., Zhang H. LncRNA H19 promotes the differentiation of bovine skeletal muscle satellite cells by suppressing Sirt1/FoxO1. Cell. Mol. Biol. Lett. 2017;22:10. doi: 10.1186/s11658-017-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raghunath A., Sundarraj K., Nagarajan R., Arfuso F., Bian J., Kumar A.P. Antioxidant response elements: discovery, classes, regulation and potential applications. Redox Biol. 2018;17:297–314. doi: 10.1016/j.redox.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X., Tomso D.J., Chorley B.N., Cho H.-Y., Cheung V.G., Kleeberger S.R. Identification of polymorphic antioxidant response elements in the human genome. Hum. Mol. Genet. 2007;16:1188–1200. doi: 10.1093/hmg/ddm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hermes T. de A., Mizobuti D.S., da Rocha G.L., da Silva H.N.M., Covatti C., Pereira E.C.L. Tempol improves redox status in mdx dystrophic diaphragm muscle. Int. J. Exp. Pathol. 2020;101:289–297. doi: 10.1111/iep.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zakharova V.V., Pletjushkina O.Y., Galkin, Zinovkin R.A., Chernyak B.V., Krysko D.V. Low concentration of uncouplers of oxidative phosphorylation decreases the TNF-induced endothelial permeability and lethality in mice. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2017;1863:968–977. doi: 10.1016/j.bbadis.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 33.Demyanenko I.A., Popova E.N., Zakharova V.V., Ilyinskaya O.P., Vasilieva T.V., Romashchenko V.P. Mitochondria-targeted antioxidant SkQ1 improves impaired dermal wound healing in old mice. Aging. 2015;7:475–485. doi: 10.18632/aging.100772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandey S.N., Cabotage J., Shi R., Dixit M., Sutherland M., Liu J. Conditional over-expression of PITX1 causes skeletal muscle dystrophy in mice. Biol. Open. 2012;1:629–639. doi: 10.1242/bio.20121305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Lee J.K., Toso E.A., Lee J.S., Choi S.H., Slattery M. DNA-binding sequence specificity of DUX4. Skeletal Muscle. 2016;6:8. doi: 10.1186/s13395-016-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gage P.J., Suh H., Camper S.A. The bicoid-related Pitx gene family in development. Mamm. Genome Off. J. Int. Mamm Genome Soc. 1999;10:197–200. doi: 10.1007/s003359900970. [DOI] [PubMed] [Google Scholar]
- 37.Lanctôt C., Lamolet B., Drouin J. The bicoid-related homeoprotein Ptx1 defines the most anterior domain of the embryo and differentiates posterior from anterior lateral mesoderm. Dev. Camb. Engl. 1997;124:2807–2817. doi: 10.1242/dev.124.14.2807. [DOI] [PubMed] [Google Scholar]
- 38.Logan M., Tabin C.J. Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science. 1999;283:1736–1739. doi: 10.1126/science.283.5408.1736. [DOI] [PubMed] [Google Scholar]
- 39.Szeto D.P., Rodriguez-Esteban C., Ryan A.K., O'Connell S.M., Liu F., Kioussi C. Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 1999;13:484–494. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knopp P., Figeac N., Fortier M., Moyle L., Zammit P.S. Pitx genes are redeployed in adult myogenesis where they can act to promote myogenic differentiation in muscle satellite cells. Dev. Biol. 2013;377:293–304. doi: 10.1016/j.ydbio.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 41.Hernandez-Torres F., Rodríguez-Outeiriño L., Franco D., Aranega A.E. Pitx2 in embryonic and adult myogenesis. Front. Cell. Dev. Biol. 2017;5:46. doi: 10.3389/fcell.2017.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitamura K., Miura H., Yanazawa M., Miyashita T., Kato K. Expression patterns of Brx1 (Rieg gene), Sonic hedgehog, Nkx2.2, Dlx1 and Arx during zona limitans intrathalamica and embryonic ventral lateral geniculate nuclear formation. Mech. Dev. 1997;67:83–96. doi: 10.1016/s0925-4773(97)00110-x. [DOI] [PubMed] [Google Scholar]
- 43.Lamba P., Hjalt T.A., Bernard D.J. Novel forms of Paired-like homeodomain transcription factor 2 (PITX2): generation by alternative translation initiation and mRNA splicing. BMC Mol. Biol. 2008;9:31. doi: 10.1186/1471-2199-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tremblay J.J., Goodyer C.G., Drouin J. Transcriptional properties of Ptx1 and Ptx2 isoforms. Neuroendocrinology. 2000;71:277–286. doi: 10.1159/000054547. [DOI] [PubMed] [Google Scholar]
- 45.L’honoré A., Commère P.-H., Ouimette J.-F., Montarras D., Drouin J., Buckingham M. Redox regulation by Pitx2 and Pitx3 is critical for fetal myogenesis. Dev. Cell. 2016;39:756. doi: 10.1016/j.devcel.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 46.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinforma. Oxf. Engl. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinforma. Oxf. Engl. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 48.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of DUX4 targets ZSCAN4 and TRIM43 in MB135-DUX4 myoblasts assessed by RT-qPCR. The data are represented as mean log10fold change ± SEM, N = 7. Mean value for control MB135 cells is set to 100%.
ARE elements in the upstream region of PITX1. (A) ARE consensus sequence (adapted from Wang et al., 2007) (B) Potential ARE sequences discovered in the region 5 kb upstream from the start of PITX1. Letters matching with the most common variant in ARE consensus are colored in red, letters matching with at least one variant encountered in ARE consensus are colored in pink, mismatches are colored in black.