Abstract
Childhood maltreatment (CM) is an established major risk factor for a number of negative health outcomes later in life. While epigenetic mechanisms, such as DNA methylation (DNAm), have been proposed as a means of embedding this environmental risk factor, little is known about its timing and trajectory, especially in very young children. It is also not clear whether additional environmental adversities, often experienced by these children, converge on similar DNAm changes.
Here, we calculated a cumulative adversity score, which additionally to CM includes socioeconomic status (SES), other life events, parental psychopathology and epigenetic biomarkers of prenatal smoking and alcohol consumption. We investigated the effects of CM alone as well as the adversity score on longitudinal DNAm trajectories in the Berlin Longitudinal Child Study. This is a cohort of 173 children aged 3–5 years at baseline of whom 86 were exposed to CM. These children were followed-up for 2 years with extensive psychometric and biological assessments as well as saliva collection at 5 time points providing genome-wide DNAm levels.
Overall, only a few DNAm patterns were stable over this timeframe, but less than 10 DNAm regions showed significant changes. At baseline, neither CM nor the adversity score associated with DNAm changes. However, in 6 differentially methylated regions (DMRs), CM and the adversity score significantly moderated DNAm trajectories over time. A number of these DMRs have previously been associated with adverse prenatal exposures. In our study, children exposed to CM also presented with epigenetic signatures indicative of increased prenatal exposure to tobacco and alcohol, as compared to non-CM exposed children. These epigenetic signatures of prenatal exposure strongly correlate with DNAm regions associated with CM and the adversity score. Finally, weighted correlation network analysis revealed a module of CpGs exclusively associated with CM.
While our study identifies DNAm loci specifically associated with CM, especially within long non-coding RNAs, the majority of associations were found with the adversity score with convergent association with indicators of adverse prenatal exposures. This study highlights the importance of mapping not only of the epigenome but also the exposome and extending the observational timeframe to well before birth.
Keywords: Epigenetics, Early-life adversity, Prenatal exposure, Risk factors, Stress
1. Introduction
Exposure to adversities in early life, as early as prenatally, is a major risk factor for a variety of negative health outcomes later in life (Guinosso et al., 2016). Adverse environmental conditions include exposure to toxins and famine but also effects of the social environment including socio-economic status (SES; Kumar et al., 2014). Early-life stress (ELS) in form of childhood maltreatment (CM) has widely been described to have sustained effects that contribute to increased risk for major psychiatric disorders later in life (Provençal and Binder, 2015). Examples of CM include exposure to neglect and/or physical, sexual or emotional abuse.
Many studies have presented evidence showing that the epigenome is responsive to external environmental conditions including the social environment (Szyf et al., 2008; Vineis et al., 2017). As a result, epigenetic mechanisms were proposed as a key factor in the biological embedding of ELS (Berens et al., 2017). DNA methylation (DNAm) is one of the most commonly studied epigenetic mechanisms in this context and describes the addition of a methyl group to a cytosine base in the DNA. Recent findings include associations of DNAm patterns with a range of environmental factors commencing as early as in utero (Provençal and Binder, 2015; Teh et al., 2014), with some of the strongest effects observed for prenatal tobacco and alcohol exposure (Portales-Casamar et al., 2016; Reynolds et al., 2011; Richmond et al., 2018; Wiklund et al., 2019). These epigenetic alterations can remain stable long after the exposure itself has taken place, supporting the role of DNAm in the embedding of environmental exposures (Heijmans et al., 2008).
To date, over 70 studies have investigated associations between ELS and DNAm changes. However, findings remain inconsistent, possibly also due to differences in methodology and sample characteristics (Cecil et al., 2020). So far, the majority of studies investigating effects of ELS in targeted (McGowan et al., 2009) or epigenome-wide association studies (EWAS) (Suderman et al., 2014) focused on adults and used retrospective assessment of ELS and measured DNAm in peripheral tissues. Targeted approaches investigated epigenetic modifications in genes related to the stress hormone system, including the genes encoding the glucocorticoid receptor (NR3C1), the serotonin system or growth factors. Findings for NR3C1 have been replicated by multiple studies across different tissues (Turecki and Meaney, 2016), while data for other candidates (e.g., SLC6A4, FKBP5, BDNF, OXTR) seem less robust (Cecil et al., 2020).
While EWAS in adults identified some significant associations of methylation profiles and childhood adversity (Suderman et al., 2014), it is not clear how robust these effects are. In a study investigating DNAm in peripheral blood at age 18 in a large cohort of over 2000 individuals with repeated assessment of victimization at ages 5, 7, 10, 12, and 18 years, no robust associations could be identified, and candidate-gene level associations could not be confirmed (Marzi et al., 2018). Another EWAS for childhood adversity in two adult cohorts measuring DNAm in peripheral blood and buccal cells, respectively, did not find any replicated associations on the level of individual CpGs but on the level of differentially methylated regions (DMRs), encompassing several CpGs. These DMRs were associated with specific forms of early adversity, mainly parental loss and illness (Houtepen et al., 2018).
Only a few studies have examined the effects of ELS in children immediately following exposure (Cecil et al., 2020). Most of these employed a candidate gene approach, extending some of the findings from adult samples to children, namely NR3C1 (Marzi et al., 2018), (Romens et al., 2015). Only a handful of studies have employed EWAS in samples from children (Cecil et al., 2020) whereby two studies by Weder et al., in 2014 and Yang et al., in 2013 are based on the same sample of 192 children with 96 substantiated cases of abuse and/or neglect over the prior 6 months and 96 demographically matched controls. Up to now, DNA methylation in two specific loci (NR3C1 and FKBP5) was investigated longitudinally in children. Parent et al. examined DNA methylation in preschoolers aged 2–5 years (n=260) within 6 months of documentation of maltreatment and one year later, showing that NR3C1 methylation is dynamic over time and the relationship with maltreatment is complex (Parent et al., 2017). Another study investigated FKBP5 methylation in preschoolers (n=231) with moderate to severe maltreatment in the past 6 months. Child maltreatment was associated with change in FKBP5 methylation over time in a six-month period, but only when children were exposed to high levels of other contextual stressors (Parade et al., 2017).
While all of these studies report significant epigenetic associations, it is not reported whether similar loci are affected across studies.
While these findings support some association between ELS and alterations in DNAm, the specificity to CM is often not clear as potential confounding by other life events, current symptoms, SES and other factors makes it difficult to attribute the findings solely to CM. In addition, the majority of past studies in children have not consistently controlled for population stratification or cell-type composition, which are important confounders of DNAm levels (Cecil et al., 2020). Furthermore, previous studies have varied in age, tissues, measures of exposure and how candidate genes were targeted, further limiting the conclusions which can be drawn. The lack of longitudinal assessments often prohibits inferences about the dynamics over time in childhood.
To the best of our knowledge, our study is the first longitudinal study to analyze the genome-wide effects of CM on DNAm in children (n = 173) within a narrow age range (from 3 to 5 years of age at baseline) over the course of 2 years. The cohort's in-depth assessment of biological data, psychiatric diagnostic assessment using the Preschool Age Psychiatric Assessment (PAPA), behavioral measures such as the Child Behavioral Check List (CBCL) and developmental measures such as the Wechsler Preschool and Primary Scale of Intelligence (WSPPI) enables the investigation of the association of CM on DNAm as well as a variety of different outcomes. In addition, the assessment of other life events, SES and contextual stressors were used to develop a cumulative adversity score, based on the fact that CM is often embedded in an environment with multiple adversities. Thus, we could assess associations between variation in DNAm and CM as well as CM in the context of other adversities. The main objectives of this study were to determine, 1) if variation in DNAm patterns are specific to CM or more accentuated in the context of additional adversity, 2) if ELS-associated variation in DNAm is associated with behavioral or biological outcomes and 3) how ELS-associated variation in DNAm changes over time.
2. Material & methods
2.1. Study population
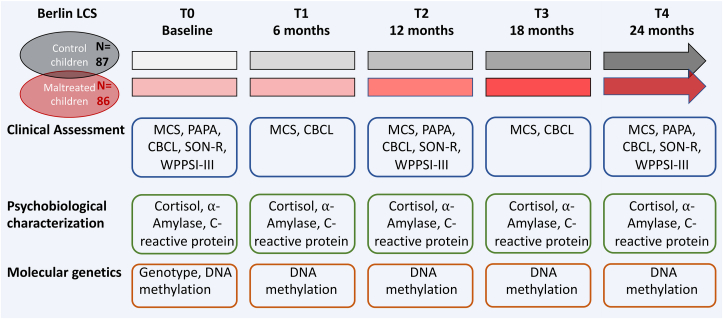
The Berlin Longitudinal Child Study (Berlin LCS) cohort consists of 173 children, who were aged 49 months on average (SD = 9.48) at the first visit (Entringer et al., 2020). Participants were recruited via child care centers, child and youth social services, child psychiatric departments or pediatricians. At baseline, 86 children were included in the maltreated group. The inclusion criterion was exposure to at least one maltreatment event with sufficient severity as described in the section “2.6 Measures of childhood adversity” (Winter et al., unpublished work). Maltreatment incidents were assessed by the MMCI and maltreatment features were recorded in the Maltreatment Classification System (Cicchetti, 1993). The control group included 87 children not exposed to maltreatment as verified using the MMCI. After the baseline visit, children were assessed every 6 months for 2 years with extensive psychometric and biological assessments (see Fig. 1, Appendix A, Appendix A). In addition, DNA from saliva samples was collected at 5 time points over the course of the study (every 6 months). Demographics of the cohort are summarized in Appendix A, Appendix A. While we did not specifically recruit for sexual abuse, as sexual abuse usually leads to removal of the child from the home, eight cases with mild sexual abuse were present in our sample. Maltreated children did receive “care as usual”, meaning that those children for whom treatment was assessed as necessary were referred to the Social Pediatric Center of the Charité or other appropriate facilities. At each visit families received feedback about the child's health and developmental status and recommendations for follow-up, where necessary (e.g., psychological consultation, dentist visit, etc.). Between 44.7% and 62.2% of the families adhered to the recommendations across the time points.
Fig. 1.
Study design. Participants were followed for 2 years with extensive biological and psychological assessments at five time points every 6 months. Some of the assessments including the structural imaging data were only collected at three time points (T0, T2, T4).
Approval for the study was obtained from the ethics committee of Charité – Universitätsmedizin Berlin. All procedures are in accordance with the Ethical Principles for Medical Research as established by the Medical Association Declaration of Helsinki. Written informed consent was obtained from all participants after the procedures were fully explained. Children gave consent by painting or signing a form that was appropriate for the children's age range. Caregivers received monetary compensation for participation. Children received a small gift. All caregivers received diagnostic results and referral for psychosocial or medical follow-up.
2.2. Saliva samples & biodata assays
Saliva samples were collected during at each clinical visit at 9, 10 and 11 am using oral swabs specially designed for small children (Salimetrics) and were immediately stored at −80 °C.
Salivary C-reactive protein levels (CRP) concentration was determined using a commercial ELISA kit (Salimetrics) with a sensitivity of 10 pg/ml. Intra-assay and inter-assay coefficients of variability were 6% and 13%, respectively. Salivary cortisol (Cort) concentration was measured using a commercial ELISA kit (Salimetrics) with a sensitivity of 0.007 μg/dL. Intra-assay and inter-assay coefficients of variability were 7% and 11%, respectively. Salivary α-amylase (AA) was analyzed using a commercially available kinetic enzyme assay kit (Salimetrics) according to the manufacturer's instructions. Intra-assay and inter-assay coefficients of variability were 4% and 10%, respectively. AA and Cort were measured at three timepoints on the day of assessment (9 a.m., 10 a.m., 11 a.m.). CRP was measured at the 11 a.m. time point. In this study we used to area under the curve (AUC) for each of the markers with respect to ground (AUCg) which was computed on flow-rate corrected levels (FR) as measured for AA and Cort. We used a log-transformation of the CRP values for downstream analyses.
From our cortisol measurements we could extract following readouts of the cortisol dynamics: baseline (9:00 a.m.), peak (10:00 a.m.), increase (peak-baseline), AUC with respect to ground, AUC with respect to increase. In our study we decided to report the AUCg measure, as it significantly correlated with all measures and best with baseline (r = 0.59, p = 6.66*10−16) and peak (r = 0.76, p < 2.2*10−26) and was thus chosen to reflect HPA-axis activity in this sample. Another reason for us to use the AUCg measure is that the other assessments of the children could not always been performed in the same order and the AUCg measure is robust against this kind of difference.
Saliva for genomic DNA extraction was collected using ORAgene DNA kits (OG500) at 9 a.m. and extracted together for all time points (T0-T4). DNA extraction was performed with a standardized and automated procedure based on magnetic beads for 2 × 400 μl saliva samples using the PerkinElmer Chemagic360 system.
2.3. DNA methylation
The Infinium Methylation EPIC BeadChip (Illumina Inc, San Diego, CA, USA) was used to measure DNAm. Samples from all timepoints were extracted, plated and run together. We randomized with regards to maltreatment, age and sex to avoid confounding between CM and batch effects. Hybridization and array processing were performed as specified by the manufacturer. Functional normalization implemented by the minfi package (Aryee et al., 2014) was used to normalize the data. Thirty-four samples with artefacts in the beta-value distribution were excluded, another three samples were removed as they presented with >5% missing values. Batch effects were identified and removed with the Empirical Bayes’ method ComBat (Müller et al., 2016) included in the R package sva (Leek et al., 2012). Chip barcode and position were the most significant batches and corrected for iteratively. Probes were filtered excluding known cross reactive as well as polymorphic probes (McCartney et al., 2016).Additionally, probes with detection p-value > 0.01 in more than 25% of the samples were also removed. CpG-sites located on the X or Y chromosome were excluded. In total 830,206 CpGs and 634 samples (by timepoint (case; control) T0: 167(84,83), T1: 128(63,65), T2: 125(57,68), T3: 104(44,60), T4: 110(46,64)) were used for downstream analysis. Cell composition of the buccal swab samples was estimated using the deconvolution method described by (Smith et al., 2015) and was corrected for in all statistical models.
2.4. Genotyping and imputation
Genotyping was performed with the Illumina GSA-24 v2.0 BeadChip. After filtering by SNP call rate (exclusion at < 95%), sample call rate (exclusion at < 98%) as well as for Hardy-Weinberg Equilibrium (HWE, p-value for HWE < 10−5) and minor allele frequency (MAF, MAF < 0.01), 469,592 SNPs remained. Samples were pre-phased with shapeit v2 (Delaneau et al., 2008) and imputed with impute 2 (Howie et al., 2011) using the 1000 genomes phase 3 reference panel (Altshuler et al., 2010). SNPs were filtered by imputation quality (info-score < 0.6), MAF > 0.01 and p-value for HWE < 10−5 with qctool v2 (Marchini et al., 2007). 9,522,926 SNPs and 173 IDs were present after quality control (QC). Remaining SNPs were pruned for linkage disequilibrium (window size = 100, step size = 5, r2 = 0.2) with plink v1.9 (Chang et al., 2015) and used to compute principal components. The first three principal components explaining 35% of the genotypic variance were used as covariates in the analysis to correct for population structure and relatedness.
2.5. Psychopathological and developmental assessments
The developmental status of the children was assessed using the Snijders-Oomen Non-verbal Intelligence Test (SON_IQ; Tellegen and Laros, 1993) and the and Wechsler Preschool and Primary Scale of Intelligence (WPPSI; Wechsler, 2002). The SON_IQ was assessed between the first (9 am) and second collection (10 am) of saliva samples. The WPPSI was assessed after the collection of the third saliva sample (11 am). Any behavioral problems were identified using the Child Behavioral Checklist (CBCL; Aschenbach et al., 2000). Psychopathology was assessed using the Preschool Age Psychiatric Assessment (PAPA; Egger and Angold, 2004). The PAPA is a structured, glossary-based psychiatric interview to assess psychiatric symptoms, symptom scale scores and diagnoses according to DSM-IV (Naftolowitz et al., 1995). In this study we used the subscales for internalizing symptoms (PAPA_int) and externalizing symptoms (PAPA_ext).
2.6. Measures of childhood adversity
Children who were exposed to maltreatment were identified by the maltreatment classification system (MCS; Cicchetti, 1993). Maltreatment experiences were coded for 7 categories: emotional maltreatment, physical neglect with insufficient care and insufficient supervision, physical abuse, sexual abuse, moral/legal/educative maltreatment as well as education-related maltreatment. The MCS provides operational definitions of maltreatment and neglect subtypes (with inclusion and exclusion criteria) as well as five different levels of severity for each of the subtypes. Further, this assessment includes the measurement of onset and for each subtype (individuals can be subjected to multiple types) chronicity, severity, developmental period and perpetrator of the incident(s). The severity of each maltreatment event was evaluated on a 5-point scale ranging from mild (1) to severe or life-threatening maltreatment (5). Cut-offs of the scores were used to include children in the maltreatment group (emotional maltreatment ≥ 2, physical abuse ≥ 1, physical neglect ≥ 1). This identified 86 children as victims of maltreatment, including cases of sexual abuse (n = 8), physical abuse (n = 36), physical neglect (n = 29) or emotional maltreatment (n = 85), with multiple types of exposure possible (Entringer et al., 2020). A cumulative severity score was calculated for each maltreatment incident by summing up the severity of the maltreatment event weighted by the duration (in months) of the events experienced.
Further, healthy controls (n = 87) were matched according to age, sex and socio-economic status (SES). The socio-economic status was assessed according to the Winkler & Stolzenberg Index (Winkler and Stolzenberg, 1999). The index score (ranging from 3 to 21) is the sum of three components including education and occupational qualification, occupational status, and net income of the household.
In addition, we assessed critical life events apart from CM using the life event list provided within the Preschool Age Psychiatric Assessment at all time points (T0-T4). The sum of the critical life events was used for our analysis and was used as exclusion criterion from the control group. This resulted in a control group exposed to few stressful events making the control group distinctly different from the maltreated group (Winter et al., unpublished work).
Maltreatment and the burden of stress in general were analyzed separately in order to identify alterations which were specific to maltreatment. Therefore, we computed a composite adversity score reflecting the general burden of stress. The adversity is a composite adversity score (ranged: 0–10) based on Adverse Childhood Experience (ACE; Felitti et al., 2019) categories: and sums up: low SES (0 or 1, Winkler & Stolzenberg Index), exposure to contextual stressors and critical life events (0 or 1, list included in the Preschool Age Psychiatric Assessment), and exposure to different maltreatment categories (range 0–7). Children were included into a high or low adversity group by using a median split of the adversity score (high: score ≥ 3, low: score ≤ 2).
2.7. Statistical analysis
Differentially methylated CpGs were identified using general linear models (glm function in R; (McCullagh, 1984). Significant changes over time for single CpGs were detected using linear mixed models (lmer function from the lme4 package; (Bates et al., 2007).
All models included age, sex and cell type composition (Buccal, CD14, CD34) as well as the first three principal components (PC1, PC2, PC3) from the genotypes in order to correct for different ethnicities and relatedness. The following models were tested: assessing significant DNAm changes occurring over time (model 1), assessing DNAm changes over time with additive effect of the environment (maltreatment or adversity in general) (model 2) and assessing interactive effects of DNAm over time and the environment (model 3).
-
1.
Model over time: (methylation changes due to the ageing of the children)
| Beta ~ age + sex + PC1 + PC2 + PC3 + Buccal + CD14 + CD34 + Time + (1|Subject) |
-
2.
Additive model (changes over time where the environment adds to the effect):
| Beta ~ age + sex + PC1 + PC2 + PC3 + Buccal + CD14 + CD34 + Time + E + (1|Subject) |
where E represents the stress measure of the environment (maltreatment or adversity score).
-
3.
Interactive model (the environment modulates the changes over time):
| Beta ~ age + sex + PC1 + PC2 + PC3 + Buccal + CD14 + CD34 + Time x E + (1|Subject) |
where E represents the stress measure of the environment (maltreatment or adversity score).
P-values were computed by comparing the models with a corresponding nested model using ANOVA (model1, model2) and ANOVA (model2, model3).
We then aggregated the results of the linear mixed morels to identify DMRs. To do so the p-values for each model of all CpGs available (n = 830,206) were combined into regions using the comb-p software (Pedersen et al., 2012) with 1 × 10−4 as seed p-value. A region was extended by a significant neighboring CpG when its distance from the region did not exceed 500bp. All results reported were corrected for multiple testing at FDR at 10% over all identified regions using the Benjamini-Hochberg Method (Benjamini and Hochberg, 1995).
Top hits (nominal p < 1 × 10−4) from the DMR analysis were used to perform pathway analysis. Enrichment for pathways was computed by mapping the CpGs to genes (EPIC array annotation) and then using FUMA, a webtool for functional mapping and annotation (Watanabe et al., 2017).Associations between DMRs and other outcome measures were reported using Pearson's correlation coefficient and nominal p-values. We investigated the robustness of our findings in multiple ways:
We tested if the DMRs remained stable when correcting for confounders such as the SES or prenatal smoking exposure (details are described in supplementary method 10).
Further, we performed a sensitivity analysis by re-running the linear mixed models described above using only the complete cases at T3 (n = 102) and T4 (n = 83). This was done in order to test the effect of missing data over time due to dropouts on the models.
Finally, we performed a post-hoc power-analysis in order to investigate if our findings are reasonable considering the sample size. We performed a power-analysis using the longpower package in R, which is especially tailored to longitudinal linear mixed models (Lu et al., 2008). Mixed effect model parameters (e.g. random intercept and/or slope) of a pilot model (we used the fitted model of the CpG with median effect size) are translated into marginal model parameters so that the formulas can be applied to investigate the power-sample size relationship for two sample longitudinal designs assuming known variance. We estimated the power for our given sample size of the smaller group (n = 81) for each of our models (parameters of interest being: time, time + adversity and time x adversity) with an effect size estimate for a 5% methylation change in the pilot model.
2.8. DNA methylation based-scores for prenatal exposures
Risk scores for prenatal smoking- and alcohol exposure were calculated based on previous epigenetic studies. We estimated prenatal smoking exposure using a DNAm score based on 15 CpG-sites identified by Richmond and colleagues (Richmond et al., 2018) and maternal alcohol intake during pregnancy using a DNAm score including 658 CpGs from a fetal alcohol syndrome (FAS) study by (Portales-Casamar et al., 2016). For the construction of the epigenetic scores for prenatal alcohol and smoking exposure, we used the CpGs identified in the previous studies: n=658 differentially methylated CpGs in FASD cases reported by Portales-Casamar et al. (2016) and n=15 that were identified as strong predictors for prenatal smoke exposure by Richmond et al. (2018). For the prenatal alcohol exposure score we weighted the beta-values measured in the Berlin_LCS cohort of the 658 CpGs with the effect size reported by Portales-Casamar et al. and then summed up the weighted CpGs to construct the epigenetic scores. For the prenatal smoking exposure, we followed a similar procedure using the weights provided by the authors in the supplement of Richmond et al. (2018).
2.9. Weighted correlation network analysis (WGCNA)
We selected the most variable 10% of all CpGs (n = 83,021) that passed QC for downstream analyses by filtering by median absolute deviation (MAD; Rousseeuw and Croux, 1993) and taking the CpGs presenting with a MAD-score > 90th percentile into further analyses.
Weighted correlation network analysis (WGCNA; Langfelder and Horvath, 2008) was conducted on the baseline DNAm levels (T0) to identify co-methylation structures. The best soft thresholding power was determined to be 5 and the tree-cut height was set to 0.25. As recommended by the authors, we constructed a signed network. For downstream analysis, we mapped the modules correlated with interesting outcomes to pathways. Stability analysis of the results were performed using a bootstrap approach (detailed description in supplementary method S11).
3. Results
3.1. Effects of early life adversity on longitudinal epigenetic trajectories
We first performed association analyses with the baseline data comparing methylation levels between the maltreated (n = 86) and the control group (n = 87) as well as the groups with high (n = 81) and low adversity score (n = 92) for each single CpG (n = 830,621). We then computed linear mixed models in order to test for associations of childhood maltreatment (CM) or adversity in general (adversity score) with longitudinal epigenetic trajectories: (1) a model over time (Model 1) estimating the general effects over time occurring due to the ageing of the children, (2) an additive model (Model 2) estimating the effects where maltreatment or adversity add to the effects over time and finally (3) an interactive model (Model 3) estimating the effects over time that are moderated by CM or adversity in general.
All of the effects on each single CpG identified by the different linear mixed models were then aggregated into DMRs as this approach has more power than the analysis of individual CpGs (Gu et al., 2010). Starting from a seed CpG with p < 1 × 10−4, a region was constructed by adding all CpGs within 500bp distance with a nominal p-value < 0.05. We found no significantly DMRs with CM nor adversity when aggregating the results of the association studies at baseline.
3.1.1. Effect of ageing
In order to assess changes over time, we applied Model 1 to the DNAm data from all 5 time points. Aggregating these results yielded 7 DMRs with significant DNAm changes over the total observation period. These DMRs mapped onto six genes (GSTM3/5, MCCC1, GSDMS, KCNQ1, AURKC, BLCAP) and one long coding RNA (LINC22001). We also applied Model 1 to the first three time points only, to assess short-term effects over time due to the low sample size at the last two time points (n(T3) = 111 and, n(T4) = 90). Here, we identified 9 DMRs mapping onto C5orf63, RUFY1, HLA response elements, NPY, RP11-73B2.6, MESTIT1, PIWIL2, CIDEB and AIRE. Due to the small sample sizes at time points 4 (n = 111) and 5 (n = 90), we only used the first three time points for the DMR analysis in Model 2 and Model 3.
3.1.2. Additive effects of time and CM or the adversity score
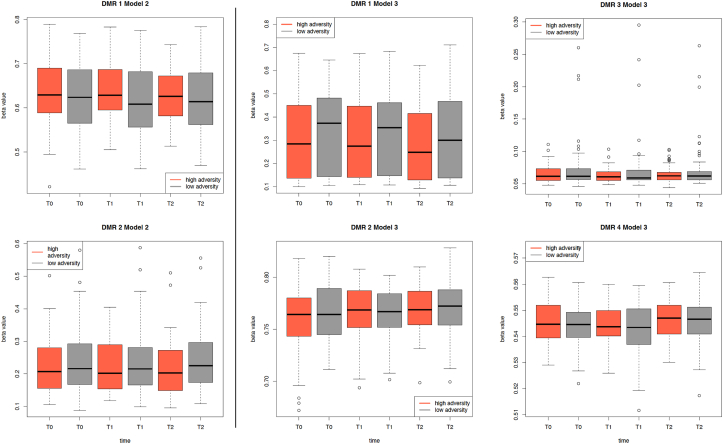
Using Model 2, which tests additive effects of time and CM or the adversity score, we identified 2 significant DMRs with additive effects of time and CM. The two DMRs mapped onto the HLA-B gene (dmr1_m2) and ZFP91 pseudogene (dmr2_m2). The same two DMRs emerged for additive effects of time and the adversity score (Appendix A, Appendix A). For these DMRs, effects of time were observed in non-exposed group, but blunted in the exposed individuals (see Fig. 2, left panel). These DMRs also showed associations with sex (Fig. 3) with lowest methylation levels over time in exposed boys and highest in non-exposed girls for both DMRs.
Fig. 2.
Differentially methylated regions (DMRs) over time. Distribution of mean methylation across the respective DMRs, split by adversity score (the high scoring group is shown in red, the low scoring group in grey) and time points. The DMRs from the time + adversity model (Model 2) are shown in the left panel, and the DMRs from the time x adversity model (Model 3) are shown in the right panel. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
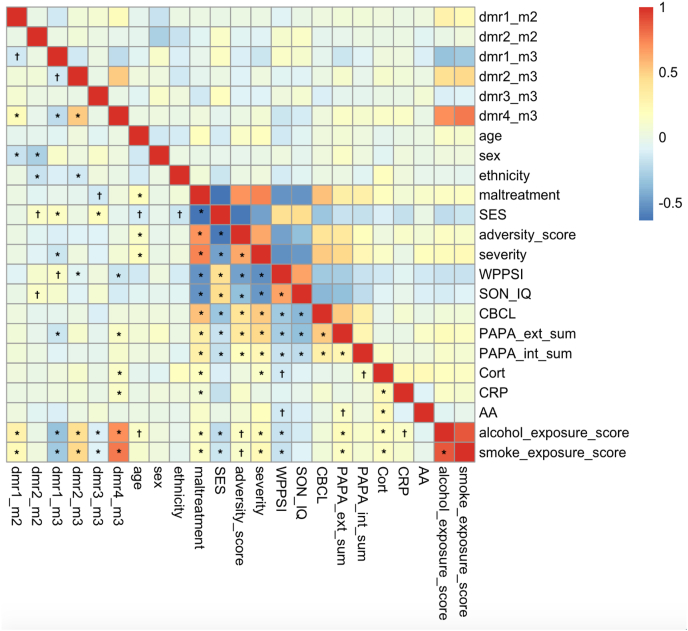
Fig. 3.
Summary of correlations within the baseline data (T0). Correlations at baseline between significant hits from differentially methylated regions (DMRs; across 3 time points), age, sex, ethnicity, maltreatment, socio-economic status (SES), composite adversity score and maltreatment severity, developmental outcomes (SON_IQ, WPPSI), psychiatric assessments (CBCL, PAPA internalizing subscale, PAPA externalizing subscale), biodata (alpha-amylase (AA), C-reactive protein (CRP), Cort (cortisol)) as well as prenatal exposure. Significant correlations are marked “* “, correlations with 0.05 ≤ nominal p ≤ 0.10 are marked " † “.
3.1.3. Interactive effects of time and CM or the adversity score
There were no significant results for Model 3 investigating the changes over time moderated by CM (time x CM). The model time x adversity score (Model 3) on the other hand yielded 4 significant DMRs mapping to the genes GAREML (dmr1_m3), P3H2 (dmr2_m3), ZNF562 (dmr3_m3) and GSTT1(dmr4_m3) (Fig. 2, right panel). Baseline methylation levels within all these regions were negatively associated with the adversity score (Appendix A, Appendix A, Fig. 2, Fig. 3). For dmr1_m3 and dmr2_m3, methylation changes over time were reduced in the exposed group (3.0% and 1.2% difference in exposed vs 2.6% and 0.4% difference in non-exposed). Dmr3_m3 showed small changes over time but different directions for the high adversity group (hypermethylation of 0.2%) and low adversity group (demethylation of 0.1%). Dmr4_m3 is demethylated over time with larger changes over time in the high adversity group (0.09%) compared to the low adversity group (0.05%).
At baseline, the DMR mapping to GSTT1 (dmr4_m3) was correlated with externalizing symptoms (Pearson correlation r = 0.14, nominal p = 0.03), cortisol- (Pearson correlation r = 0.13, nominal p = 0.04) and CRP levels (Pearson correlation r = 0.14, nominal p = 0.03).
All results from the DMR analyses are summarized in Fig. 2, Fig. 3 and Appendix A, Appendix A. Many of the DMRs described above mapped to genes previously reported to be associated with prenatal exposures (Jiang et al., 2020; Kunkle et al., 2017; Leighton et al., 2019; Roberson-Nay et al., 2020; Terasaki and Schwarz, 2016), including maternal smoking and FAS as shown in Appendix A, Appendix A.
3.2. Epigenetic signatures of prenatal exposure
Given the above described putative link to prenatal exposure, we next investigated established epigenetic markers of prenatal adversity such alcohol and tobacco exposure in our cohort. The epigenetic prenatal smoke exposure score and the epigenetic prenatal alcohol exposure were highly correlated (Pearson correlation r = 0.89, nominal p < 2.2 × 10−16). However none of the CpGs used to compute the scores (prenatal alcohol exposure score: n = 658, prenatal smoke exposure score: n = 15) overlapped, and only one pair of CpGs were located within the same locus (MYO1G), suggesting correlations of the exposures themselves.
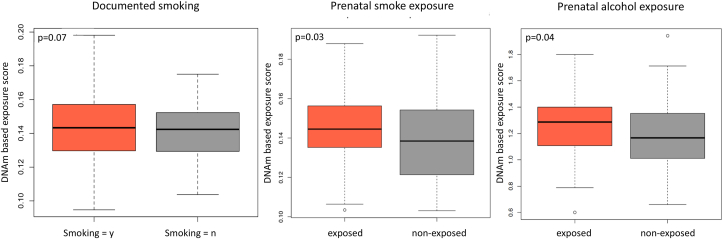
Individuals with documented prenatal smoke exposure (n = 23) and with missing information (n = 36) on average had a higher epigenetic score for smoking exposure as compared to those without reported exposure (t-test, n = 108, mean (smoking/missing) = 0.143, mean(non-smoking) = 0.139, nominal p = 0.14; documented smoking yes vs no: nominal p = 0.07). This finding validates the score in this cohort. At baseline, maltreated children had a significantly higher epigenetic score for prenatal smoke exposure (nominal p = 0.03) than non-maltreated children (Fig. 4). Epigenetic scores for prenatal smoke exposure were positively correlated with the adversity score (Pearson correlation r = 0.16, nominal p = 8.8*10−5). Similarly, maltreated children had a significantly higher epigenetic score for prenatal alcohol exposure at baseline than non-maltreated individuals (nominal p = 0.04) (see Fig. 4). Both epigenetic prenatal exposure scores correlated with the adversity score, however these correlations were not significant (smoke exposure: r = 0.12, nominal p = 0.11 and alcohol exposure: r = 0.14, nominal p = 0.05).
Fig. 4.
Prenatal exposure. DNA methylation-based scores for prenatal exposure was compared between the group exposed to maltreatment (red, n = 84) and the non-maltreated group (grey, n = 83). On average, cases had higher levels of smoke exposure (nominal p = 0.03) and alcohol exposure (nominal p = 0.04) compared to controls. The documented cases of smoke exposure (n = 23 + 36 with missing information) had a higher DNAm based score than children without exposure (n = 120). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We then investigaed the relationship between the scores, DMRs and other outcomes. The epigenetic prenatal smoke exposure score positively correlated with the baseline cortisol AUC (Pearson correlation r = 0.14, nominal p = 7.6*10−4), and psychiatric symptoms (Pearson correlation r = 0.16, p = 0.04). Both DMRs measures at baseline from Model 2 and 3 DMRs of Model 3 that reflect additive or interactive associations of time and adversity score correlated with prenatal smoke exposure (strongest correlation r = 0.78, nominal p < 2.2 × 10−16) – see Fig. 3. There was no overlap between the CpGs included in the prenatal smoke exposure epigenetic score and the CpGs within the DMRs identified.
The epigenetic prenatal alcohol exposure score also positively correlated with the AUC of cortisol at T0 (Pearson correlation r = 0.09, nominal p = 0.03), CRP levels (Pearson correlation r = 0.09, nominal p = 0.03), AA levels (Pearson correlation r = 0.15, nominal p = 0.04) and low maternal SES (Pearson correlation r = −0.12, nominal p = 0.002). Furthermore, the epigenetic prenatal alcohol exposure score was associated with cognitive impairment reflected by the WPSSI (Pearson correlation r = −0.14, nominal p = 0.05). Children with high prenatal alcohol exposure score scored lower at the WPSSI. Children with high prenatal exposure scores also presented with significantly more externalizing symptoms captured by the PAPA subscale (Pearson correlation r = 0.19, nominal p = 0.01).
One DMRs from Model 2 and all DMRs of Model 3 correlated with prenatal alcohol and smoke exposure (strongest Pearson correlation r = 0.71, nominal p < 2.2*10−16), suggesting again convergence of prenatal and postnatal exposures on DNAm. There was no overlap between the CpGs included in the prenatal alcohol exposure score and the CpGs within the DMRs identified.
In order to disentangle the effects of prenatal exposure and maltreatment on the DMRs identified, we re-ran models 2 (time + adversity score) and 3 (time x adversity score) with prenatal exposure (smoking + alcohol score) as covariates. Correcting for prenatal exposure 13 of 17 CpGs from model 2 (additive model) remained significant after correcting for prenatal exposures with some of the CpGs having even lower p-values as compared to when not correcting for prenatal exposures (lowest nominal p = 1.1*10−12). Similarly, 25 of 31 CpGs from model 3 (interaction model) remained significant after correcting for prenatal exposures (lowest nominal p = 1.4*10−38).
3.3. Weighted correlation network analysis (WGCNA) reveals maltreatment specific differential methylation
We next performed WGCNA on baseline DNAm levels using the 10% most variable CpGs (n = 83,021) to cluster changes in the variable methylome and associate obtained CpG modules with differences in environmental factors including CM, adversity score, SES, and prenatal exposure scores. This analysis identified 9 co-methylated modules (plus one module containing the unassigned CpGs), the number of CpGs in the respective modules ranged from 56 to 56,344. We performed module stability analysis by resampling 50 times and repeating network construction with 66% of the samples. Nine modules remained stable across all 50 runs and were consistent with modules identified in the complete data set (Appendix A, Appendix A and Appendix A, Appendix A). The tenth module (56 CpGs) was unstable and thus excluded from the downstream analyses.
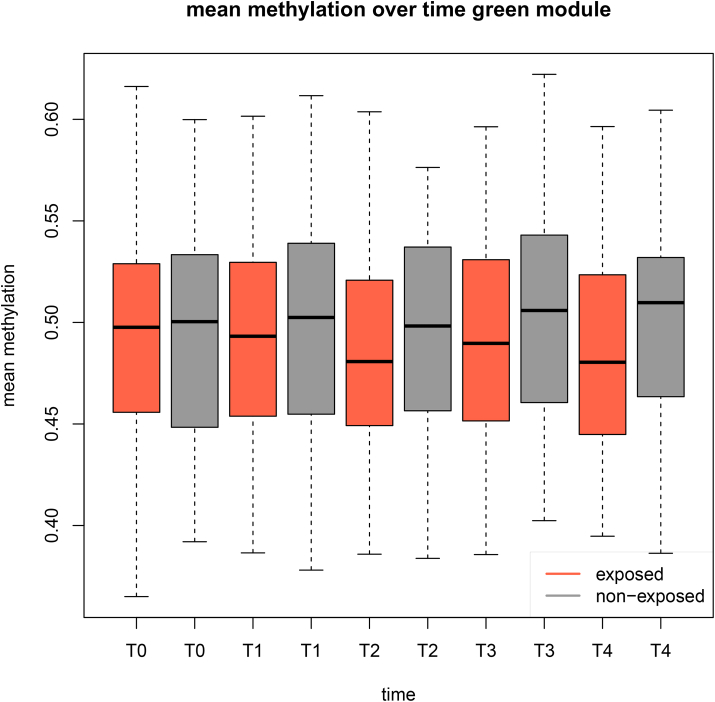
One module (green module) was associated with CM (Pearson correlation r = 0.17, nominal p = 0.04) specifically and neither adversity nor prenatal exposures (Fig. 5). This module consisted of 268 CpGs which map to 165 unique genes (listed in Appendix A, Appendix A). These genes showed no significant overlap with any specific pathway or result from genome-wide association studies (GWAS). A large portion of the genes in the module, however, mapped to non-coding RNA genes (47 lincRNA, 3 snoRNA and 3 microRNA). It also included interesting genes from the C21-steroid biosynthesis pathway (CYP1A1, CYP2A6, CYP2A7) and genes which have been reported to be important in early development (such as DNM1, FOXR1, ZNF570). The mean methylation of the green module was lower in maltreated children across all five time points as compared to non-maltreated children (Fig. 6).
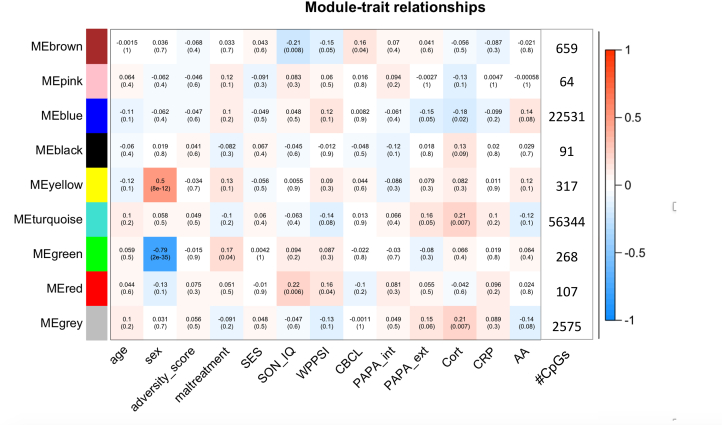
Fig. 5.
Module Trait Relationship. Associations at baseline between assigned weighted correlation network analysis (WGCNA) modules and general traits, different measures of exposure: adversity score, maltreatment, socio-economic status (SES) and prenatal exposure proxies: alcohol and smoking. Additionally, correlations between the modules and developmental outcomes (SON_IQ, WPPSI), psychiatric assessments (CBCL, PAPA internalizing subscale, PAPA externalizing subscale) and biodata (alpha-amylase (AA), C-reactive protein (CRP), Cort (cortisol)) are shown. Shown on the right are the number of CpGs included in each module. The coloring of the tiles reflects the Pearson correlation, the number within the tile is the nominal p-value.
Fig. 6.
Green module mean methylation over time. Across all time points mean methylation of CpGs in the green (n = 256) module is lower in children exposed to maltreatment (red) than in non-maltreated children (grey). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Interestingly, the green module also showed a strong negative correlation with sex (Pearson correlation r = −0.79, nominal p = 2*10−35). Methylation levels were lowest in exposed boys. Two other modules (red and brown) were correlated with developmental (SON_IQ, WPPSI) or behavioral measures (CBCL). None of the modules were associated with the adversity score or SES. The turquoise module (the largest module, consisting of 56,344 CpGs) was nominally significantly associated with the AUC of cortisol and externalizing symptoms (PAPA subscale). Associations for all modules are summarized in Fig. 5.
We functionally annotated the modules that were associated with interesting outcomes or environment variables. Small modules (black, pink, red, yellow and green) with less than 500 CpGs showed no significant enrichments. The other modules were mainly enriched for immune signaling and cytoskeleton organizational pathways (Appendix A, Appendix A). An overview of all modules is shown in Appendix A, Appendix A.
4. Discussion
To date, most studies focusing on the effect of CM on DNAm have been performed in adults using retrospective measures. Only a few studies have examined the effects of CM in children and this is the first study on the longitudinal effects of CM on DNAm at the critical time period of early childhood. In addition, CM often occurs in the context of multiple other adversities and in our study, we tried to identify whether there are DNAm changes specific to CM or whether they are more reflective of overall adversity, including prenatal exposures.
4.1. DNAm changes over time in the context of CM and adversity
We first investigated the association of DNAm levels with exposure to CM as well as a more global adversity score that included CM severity as well as SES, other life events and contextual stressors. We used linear mixed models with data from the first three time points spanning a time frame of 18 months as well as from all five timepoints (30 months) to investigate short-term and long-lasting alterations of longitudinal DNAm level trajectories. We focused our analysis on DMRs. Observed methylation changes in DMRs are more credible as neighboring sites must show similar changes because CpGs are expected to function in groups to regulate gene expression (Jiang et al., 2020; Kunkle et al., 2017).
In our analysis, we observed relatively few DMRs changing over 24 or 12 months, suggesting that overall DNA methylation pattern in saliva is rather stable over this developmental time frame. The changes over 24 months included 9 DMRs mapping to regions associated with early-onset forms of psychiatric disorders (Jiang et al., 2020; Kunkle et al., 2017) and impairments during early development (Leighton et al., 2019; Terasaki and Schwarz, 2016). The changes over 12 months included 7 DMRs, of which 5 mapped to regions that were previously associated with prenatal exposures (tobacco; Alexander et al., 2013, alcohol; Nguyen et al., 2018, lead and beta blockers; Rojas et al., 2015). Effects over time were moderated by exposure. Two DMRs showed significant additive effects of time and both adversity and maltreatment. These DMRs mapped to HLA-B an immune related gene which previously has been associated with psychopathology (Engström et al., 2015; Resendiz et al., 2013) and ZFP91 pseudogene. For time × exposure interactions, significant results were only observed for the adversity score and not for CM. Here 4 DMRs were identified, which mapped to genes which previously have been associated with prenatal alcohol exposure (Reynolds et al., 2011; J. Yang et al., 2015), childhood abuse (Fang et al., 2020; B. Z. Yang et al., 2013) and early onset MDD (Roberson-Nay et al., 2020; Zhu et al., 2003).
It should be noted that the effects sizes of the DMRs in our study are comparatively large for social epigenetics studies (Breton et al., 2017; Marzi et al., 2018) with up to 7% difference in DNAm levels between the groups with high and low adversity scores (see Appendix A, Appendix A). Interestingly, exposure to adversity or CM appeared to lead to blunted dynamics of DNAm changes over time. Such blunted responses with exposure to ELS have been described for cortisol responses as well as heart rate changes and reward-related brain activity (Agorastos et al., 2019; Hanson et al., 2016). Between the measurements of cortisol (at 9,10 and 11am), the children completed assessments, which could be perceived as stressors and therefore elicit a cortisol response.
Two of the DMRs from Model 3 (dmr1_m3 and dmr4_m3) significantly correlated with the number of externalizing symptoms (PAPA_ext). Both DMRs map onto genes (GAREML, GSTT1) that have previously been associated with psychopathology (Masliah et al., 2013; Narayanan et al., 2014; Saadat et al., 2007). Two of the DMRs (dmr2_m3 and dmr4_m3) also significantly correlated with WPPSI, a developmental measure. Interestingly, both DMRs map onto genes which have previously been associated with cognitive decline in Alzheimer's Disease (Sun et al., 2019). One of these DMRs (dmr4_m3) also correlated with cortisol and CRP levels. Dmr4_m3 maps onto GSTT1, which encodes for a protein with protective function against oxidative stress and inflammation (Tang et al., 2010).
4.2. Prenatal exposures
The fact that a number of the identified DMRs lie in regions previously associated with prenatal exposures, prompted us to investigate epigenetic proxies of such exposures in our cohort. For this we constructed two DNAm exposure scores, for prenatal tobacco exposure and prenatal alcohol exposure based on previous findings (Portales-Casamar et al., 2016) given that this information was incomplete in our study and such scores are also less biased than self-reports (McCartney et al., 2018) on smoking or alcohol consumption during pregnancy. Indeed, both the DNAm based smoking score and alcohol exposure scores were higher in CM-exposed children. In addition, of the six DMRs of Model 2 and Model 3 associated with additive and interactive effects of time and maltreatment or adversity score, five significantly correlated with both prenatal exposure measures (strongest correlation r = 0.71, p < 2.2 × 10−16). These findings suggest that maltreated children might present with a higher extent of prenatal exposures as compared to controls and that this might also influence DNAm pattern, possibly with larger effect sizes than maltreatment itself.
To disentangle effects of adversity/maltreatment from prenatal exposures, we reran models 2 and 3 correcting for prenatal exposure and found that the majority of CpGs within the DMRs remained significantly associated with adversity/maltreatment. This supports independent effects of adversity on DNA methylation, even if there seems to be a correlation of DNA methylation at these sites with prenatal exposures (see Fig. 3).
4.3. Findings from the weighted correlation network analysis (WGCNA)
We also performed WGCNA in order to identify co-methylated modules of CpG-sites and to be able to analyze correlated, functionally relevant regions together. For this analysis, we only included the 10% most variable CpGs in terms of DNAm levels, as variable CpGs are enriched for functional regions and correlate with gene expression (Allum and Grundberg, 2020; Lioznova et al., 2019). Such module-centric analyses have the advantage of not only focusing on functionally relevant DNAm regions but also of alleviating the multiple testing problem. This analysis yielded one module (green module) that was specifically associated with maltreatment (p = 0.04, r = 0.17) and did not correlate with adversity in general (p = 0.8, r = −0.2), prenatal exposure (p = 0.8, r = 0.023) nor SES (p = 0.9, r = 0.009). This finding suggests that although we did not observe strong effects on the individual CpG level, a specific set of co-methylated CpGs and regions is correlated with CM. The CpGs in this module were on average demethylated in exposed children and this remained stable across time (Fig. 6). The green module also showed a strong association with sex (p = −0.79, r = 4*10−34), with lowest DNAm seen in exposed boys. Previous studies have reported sex differences in resilience to CM as well as moderating effects of sex on the consequences of CM (Samplin et al., 2013; White and Kaffman, 2019). However, larger studies would be needed in order to investigate these effects.
While there were no strong functional enrichments in this module, a large proportion of genes (54 of 164 of the genes) mapped to long non-coding RNAs. Long non-coding RNAs can regulate gene expression by multiple mechanisms and are considered important players in developmental processes such as cell differentiation and genomic imprinting (Fatica and Bozzoni, 2014).
In our data, this cortisol response (AUC) was positively correlated with the DNAm based prenatal exposure scores for smoking (r = 0.14, p = 7.6*10−4) and alcohol (r = 0.09, p = 0.03). While the majority of studies report an attenuated cortisol response after prenatal tobacco exposure in human and in mice (Azar et al., 2010; Eiden et al., 2015), studies investigating the relationship between prenatal alcohol exposure and infant stress reactivity show an increased response following light to moderate exposure (Haley et al., 2006; Keiver et al., 2015; May and Gossage, 2011). Given previous findings of altered cortisol reactivity with adversity and CM (Agorastos et al., 2019; Cecil et al., 2020), converging effects of prenatal exposure on the stress system need to be considered. Such converging effects may be mediated by epigenetic mechanisms given the joint correlations of adversity, prenatal alcohol and smoking scores and cortisol response with dmr4_m3 for example.
We would like to point out that while the clinical visit would be perceived as a stressful event for the children our repeated cortisol measurements do not reflect the response to a standardized stress test. For a more established measure of the HPA-axis activity such as the awakening response, samples would have to be collected at the home, which could not be requested from these families. Also, the children (3–5 years at baseline) were too young to perform a Trier Social Stress Test. We acknowledge that our repeated cortisol measures are more difficult to interpret than those standard tests, especially for direction of effect, however they still allow a comparative evaluation of HPA-axis activity.
4.4. Limitations
Our study has several limitations. We measured DNAm in saliva samples. While this is more easily accessible in children than blood, several studies (Smith et al., 2015; Walton et al., 2016) have explored the correlation of DNAm in different peripheral tissues (blood, buccal swabs, saliva samples) and multiple brain regions and reported higher variability in saliva samples. This might be linked to heterogeneity in cell composition. We accounted for cell type heterogeneity using established algorithms to estimate cell type proportions. This is standard procedure in DNAm studies (Smith et al., 2015). However, only studies focusing on cell-specific DNAm patterns can show if our results are restricted to specific cell-types. Furthermore, our study presents with a relatively small sample size and a high drop-out rate of 47,97% (drop-out rate of 57,83% for maltreated and 40,22% for non-maltreated) over time. We observed that more cases dropped out than controls and found that the dropouts had a significantly higher adversity scores at baseline than the remaining individuals (mean(dropouts) = 2.92, mean(remaining) = 2.18, p = 0.02). Some of these children might have been removed from their social context and therefore were excluded from the study. Another hypothesis could be, that some of the children reached school age and attending the follow-up assessments might have been too time intensive for the families. Especially the interaction models over all time points are underpowered to detect small effects, as the last time point only included 90 samples.
We performed a sensitivity analysis in order to capture the effects of dropouts on the models and the DMRs aggregated from the results. We re-ran the additive model (time + adversity) and the interactive model (time x adversity) using only the complete samples for time point 4 (n = 102) and time point 5 (n = 83). We observed very consistent findings with completers at time point 4 but lost significance with completers at time point 5. However, we would like to point out that here only 38 cases remained in the analysis.
We also performed a power analysis tailored to longitudinal models. A pilot model of the CpG with the median effect was used to estimate power to detect a 5% change in methylation levels attributed to the fixed effect of interest (time, adversity, time:adversity). The power of the model over time was calculated to be 76,2%, for the additive model (G + E) it was 79,79% (time) and 15,9% (adversity). For the interaction model the power calculate for the interaction term was 47%. While a GxE analysis for the CpGs detected by our models would be very interesting considering recent data by (Czamara et al., 2021) that highlight the relevance of combined effects of G and childhood adversity on explaining variance in DNA methylation, the sample size and resulting power of this study seems prohibitive for such focused analyses. Although our study is underpowered for testing interaction of genotype and environment, our findings highlight the importance of the effect childhood adversity specifically on DNAm trajectories.
Larger longitudinal studies will be needed to replicate our results, especially with regards to prenatal epigenetic signatures and CpG modules. Furthermore, we also lack DNAm measures before the CM occurred, so that we cannot disentangle the sequence of events between changes in DNAm and exposure. Additionally, it would be interesting to investigate the effects of CM on other composite DNAm measures such as epigenetic aging. In particular it would be interesting to look at the interactions of CM and a recently developed pediatric clock which reliably predicts a range of child health outcomes (McEwen et al., 2020).
5. Conclusion
Overall, our data point to the fact that CM does not occur in an environmental vacuum and that prenatal exposures, SES and other life events and adversities likely contribute to DNAm changes observed with CM and possibly also the overall risk and resilience trajectories. Mainly, most of our findings related to both CM and the overall adversity score, with stronger effects in some time series model of the latter. Our findings also support the convergence of prenatal and postnatal adverse exposures on DNAm. Only a small DNAm module (green module) seemed to be more selectively associated with CM and not the other exposures. While our study does identify some interesting loci, especially long non-coding RNAs, its main message is highlighting the importance of not only mapping the epigenome but also the environ, extending the timeframe to well before birth.
Funding
This study was funded by the German Federal Ministry of Education and Research (BMBF) 01KR1301-A (to CH) and 01KR1301-B and 01GL1743-C (to EBB). The research was conducted at Charité − Universitätsmedizin Berlin, Berlin, Germany, and the Max Planck Institute of Psychiatry, Munich, Germany.
CRediT authorship contribution statement
Jade Martins: Formal analysis, Data curation, Writing – original draft, Writing – review & editing, Visualization. Darina Czamara: Formal analysis, Data curation, Writing – review & editing, Supervision. Susann Sauer: Investigation, Resources. Monika Rex-Haffner: Investigation, Resources. Katja Dittrich: Investigation, Resources. Peggy Dörr: Investigation, Resources. Karin de Punder: Formal analysis, Investigation, Resources, Data curation. Judith Overfeld: Formal analysis, Investigation, Resources, Data curation. Andrea Knop: Investigation, Resources. Felix Dammering: Investigation, Resources. Sonja Entringer: Supervision, Analysis. Sibylle M. Winter: Conceptualization (clinical part), Investigation, Resources, Supervision, Funding acquisition. Claudia Buss: Conceptualization, Investigation, Supervision, Funding Acquisition. Christine Heim: Conceptualization, Supervision, Project administration, Funding acquisition. Elisabeth B. Binder: Writing – review & editing, Conceptualization, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
None.
Acknowledgement
We would like to thank Jessica Keverne for language assistance and proof-reading the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2021.100336.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Agorastos A., Pervanidou P., Chrousos G.P., Baker D.G. Frontiers in Psychiatry. 2019. Developmental trajectories of early life stress and trauma: a narrative review on neurobiological aspects beyond stress system dysregulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M., Karmaus W., Holloway J.W., Zhang H., Roberts G., Kurukulaaratchy R.J., Arshad S.H., Ewart S. Effect of GSTM2-5 polymorphisms in relation to tobacco smoke exposures on lung function growth: a birth cohort study. BMC Pulm. Med. 2013 doi: 10.1186/1471-2466-13-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allum F., Grundberg E. Molecular Metabolism. 2020. Capturing functional epigenomes for insight into metabolic diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D.L., Durbin R.M., Abecasis G.R., Bentley D.R., Chakravarti A., Clark A.G., Collins F.S., de La Vega F.M., Donnelly P., Egholm M., Flicek P., Gabriel S.B., Gibbs R.A., Knoppers B.M., Lander E.S., Lehrach H., Mardis E.R., McVean G.A., Nickerson D.A., Peterson J.L. A map of human genome variation from population-scale sequencing. Nature. 2010 doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achenbach T.M., Rescorla L.A. Research Center for Children, Youth and Families, University of Vermont; Burlington, VT: 2000. Manual for the ASEBA Preschool Forms & Profiles: an Integrated System of Multi-Informant Assessment; Child Behavior Checklist for Ages 1 1/2-5; Language Development Survey; Caregiver-Teacher Report Form. [Google Scholar]
- Azar R., Paquette D., Stewart D.E. Prenatal tobacco exposure and cortisol levels in infants of teen mothers. J. Perinat. Med. 2010 doi: 10.1515/JPM.2010.100. [DOI] [PubMed] [Google Scholar]
- Bates D., Sarkar D., Bates M.D., Matrix L. 2007. The Lme4 Package. October. [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995 doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Berens A.E., Jensen S.K.G., Nelson C.A. BMC Medicine. 2017. Biological embedding of childhood adversity: from physiological mechanisms to clinical implications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton C.v., Marsit C.J., Faustman E., Nadeau K., Goodrich J.M., Dolinoy D.C., Herbstman J., Holland N., LaSalle J.M., Schmidt R., Yousefi P., Perera F., Joubert B.R., Wiemels J., Taylor M., Yang I.v., Chen R., Hew K.M., Hussey Freeland D.M., Murphy S.K. Environmental Health Perspectives. 2017. Small-magnitude effect sizes in epigenetic end points are important in children's environmental health studies: the children's environmental health and disease prevention research center's epigenetics working group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil C.A.M., Zhang Y., Nolte T. Neuroscience and Biobehavioral Reviews. 2020. Childhood maltreatment and DNA methylation: a systematic review. [DOI] [PubMed] [Google Scholar]
- Chang C.C., Chow C.C., Tellier L.C.A.M., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015 doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti D. Maltreatment classification system. In: Barnett D., Manly J.T., Cicchetti D., editors. Defining Child Maltreatment: the Interface between Policy and Research. Child Abuse. Child Development and Social Policy; 1993. pp. 7–73. [Google Scholar]
- Czamara D., Tissink E., Tuhkanen J., Martins J., Awaloff Y., Drake A.J., Khulan B., Palotie A., Winter S.M., Nemeroff C.B., Craighead W.E., Dunlop B.W., Mayberg H.S., Kinkead B., Mathew S.J., Iosifescu D.v., Neylan T.C., Heim C.M., Lahti J., Binder E.B. Combined effects of genotype and childhood adversity shape variability of DNA methylation across age. Transl. Psychiatry. 2021;11(1):88. doi: 10.1038/s41398-020-01147-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O., Coulonges C., Zagury J.F. Shape-IT: new rapid and accurate algorithm for haplotype inference. BMC Bioinf. 2008 doi: 10.1186/1471-2105-9-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger H.L., Angold A. Handbook of Infant and Toddler Mental Health Assessment. 2004. The Preschool Age Psychiatric Assessment (PAPA): a structured parent interview for diagnosing psychiatric disorders in preschool children. [Google Scholar]
- Eiden R.D., Molnar D.S., Granger D.A., Colder C.R., Schuetze P., Huestis M.A. Developmental Psychobiology; 2015. Prenatal Tobacco Exposure and Infant Stress Reactivity: Role of Child Sex and Maternal Behavior. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström K., Rydbeck F., Kippler M., Wojdacz T.K., Arifeen S., Vahter M., Broberg K. Prenatal lead exposure is associated with decreased cord blood DNA methylation of the glycoprotein VI gene involved in platelet activation and thrombus formation. Environ. Epigenetics. 2015 doi: 10.1093/eep/dvv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entringer S., de Punder K., Overfeld J., Karaboycheva G., Dittrich K., Buss C., Winter S.M., Binder E.B., Heim C. Immediate and longitudinal effects of maltreatment on systemic inflammation in young children. Dev. Psychopathol. 2020;32(5):1725–1731. doi: 10.1017/S0954579420001686. [DOI] [PubMed] [Google Scholar]
- Fang R., Yang H., Gao Y., Cao H., Goode E.L., Cui Y. Briefings in Bioinformatics. 2020. Gene-based mediation analysis in epigenetic studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Bozzoni I. Nature Reviews Genetics. 2014. Long non-coding RNAs: new players in cell differentiation and development. [DOI] [PubMed] [Google Scholar]
- Felitti V.J., Anda R.F., Nordenberg D., Williamson D.F., Spitz A.M., Edwards V., Koss M.P., Marks J.S. Relationship of childhood abuse and household dysfunction to many of the leading causes of death in adults: the adverse childhood experiences (ACE)study. Am. J. Prev. Med. 2019 doi: 10.1016/j.amepre.2019.04.001. [DOI] [PubMed] [Google Scholar]
- Gu H., Bock C., Mikkelsen T.S., Jäger N., Smith Z.D., Tomazou E., Gnirke A., Lander E.S., Meissner A. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat. Methods. 2010 doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinosso S.A., Johnson S.B., Riley A.W. Pediatric Research. 2016. Multiple adverse experiences and child cognitive development. [DOI] [PubMed] [Google Scholar]
- Haley D.W., Handmaker N.S., Lowe J. Infant stress reactivity and prenatal alcohol exposure. Alcohol Clin. Exp. Res. 2006 doi: 10.1111/j.1530-0277.2006.00251.x. [DOI] [PubMed] [Google Scholar]
- Hanson J.L., Albert D., Iselin A.M.R., Carré J.M., Dodge K.A., Hariri A.R. Social Cognitive and Affective Neuroscience. 2016. Cumulative stress in childhood is associated with blunted reward-related brain activity in adulthood. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans B.T., Tobi E.W., Stein A.D., Putter H., Blauw G.J., Susser E.S., Slagboom P.E., Lumey L.H. Proceedings of the National Academy of Sciences of the United States of America. 2008. Persistent epigenetic differences associated with prenatal exposure to famine in humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtepen L.C., Hardy R., Maddock J., Kuh D., Anderson E.L., Relton C.L., Suderman M.J., Howe L.D. Childhood adversity and DNA methylation in two population-based cohorts. Transl. Psychiatry. 2018 doi: 10.1038/s41398-018-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B., Marchini J., Stephens M. 2011. Genotype Imputation with Thousands of Genomes. G3: Genes, Genomes, Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Han T., Duan W., Dong Q., Hou W., Wu H., Wang Y., Jiang Z., Pei X., Chen Y., Li Y., Sun C. Prenatal famine exposure and estimated glomerular filtration rate across consecutive generations: association and epigenetic mediation in a population-based cohort study in Suihua China. Aging. 2020 doi: 10.18632/aging.103397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiver K., Bertram C.P., Orr A.P., Clarren S. Salivary cortisol levels are elevated in the afternoon and at bedtime in children with prenatal alcohol exposure. Alcohol. 2015 doi: 10.1016/j.alcohol.2014.11.004. [DOI] [PubMed] [Google Scholar]
- Kumar S., Kroon J., Lalloo R. Health and Quality of Life Outcomes. 2014. A systematic review of the impact of parental socio-economic status and home environment characteristics on children's oral health related quality of life. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle B.W., Vardarajan B.N., Naj A.C., Whitehead P.L., Rolati S., Slifer S., Carney R.M., Cuccaro M.L., Vance J.M., Gilbert J.R., Wang L.S., Farrer L.A., Reitz C., Haines J.L., Beecham G.W., Martin E.R., Schellenberg G.D., Mayeux R.P., Pericak-Vance M.A. Early-onset Alzheimer disease and candidate risk genes involved in endolysosomal transport. JAMA Neurol. 2017 doi: 10.1001/jamaneurol.2017.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008 doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton L.J., Wei W., Marshall P.R., Ratnu V.S., Li X., Zajaczkowski E.L., Spadaro P.A., Khandelwal N., Kumar A., Bredy T.W. Neurobiology of Learning and Memory. 2019. Disrupting the hippocampal Piwi pathway enhances contextual fear memory in mice. [DOI] [PubMed] [Google Scholar]
- Lioznova A.v., Khamis A.M., Artemov A.v., Besedina E., Ramensky V., Bajic V.B., Kulakovskiy I.v., Medvedeva Y.A. CpG traffic lights are markers of regulatory regions in human genome. BMC Genom. 2019 doi: 10.1186/s12864-018-5387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Luo X., Chen P.Y. Sample size estimation for repeated measures analysis in randomized clinical trials with missing data. Int. J. Biostat. 2008;4(1) doi: 10.2202/1557-4679.1098. [DOI] [PubMed] [Google Scholar]
- Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007 doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Marzi S.J., Sugden K., Arseneault L., Belsky D.W., Burrage J., Corcoran D.L., Danese A., Fisher H.L., Hannon E., Moffitt T.E., Odgers C.L., Pariante C., Poulton R., Williams B.S., Wong C.C.Y., Mill J., Caspi A. Analysis of DNA methylation in young people: limited evidence for an association between victimization stress and epigenetic variation in blood. Am. J. Psychiatr. 2018 doi: 10.1176/appi.ajp.2017.17060693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah E., Dumaop W., Galasko D., Desplats P. 2013. Distinctive Patterns of DNA Methylation Associated with Parkinson Disease: Identification of Concordant Epigenetic Changes in Brain and Peripheral Blood Leukocytes. Epigenetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P.A., Gossage J.P. Alcohol Research and Health. 2011. Maternal risk factors for fetal alcohol spectrum disorders: not as simple as it might seem. [PMC free article] [PubMed] [Google Scholar]
- McCartney D.L., Hillary R.F., Stevenson A.J., Ritchie S.J., Walker R.M., Zhang Q., Morris S.W., Bermingham M.L., Campbell A., Murray A.D., Whalley H.C., Gale C.R., Porteous D.J., Haley C.S., McRae A.F., Wray N.R., Visscher P.M., McIntosh A.M., Evans K.L., Marioni R.E. Epigenetic prediction of complex traits and death. Genome Biol. 2018 doi: 10.1186/s13059-018-1514-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney D.L., Walker R.M., Morris S.W., McIntosh A.M., Porteous D.J., Evans K.L. 2016. Identification of Polymorphic and Off-Target Probe Binding Sites on the Illumina Infinium MethylationEPIC BeadChip. Genomics Data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh P. Generalized linear models. Eur. J. Oper. Res. 1984 doi: 10.1016/0377-2217(84)90282-0. [DOI] [Google Scholar]
- McEwen L.M., O'Donnell K.J., McGill M.G., Edgar R.D., Jones M.J., MacIsaac J.L., Lin D.T.S., Ramadori K., Morin A., Gladish N., Garg E., Unternaehrer E., Pokhvisneva I., Karnani N., Kee M.Z.L., Klengel T., Adler N.E., Barr R.G., Letourneau N., Kobor M.S. The PedBE clock accurately estimates DNA methylation age in pediatric buccal cells. Proc. Natl. Acad. Sci. U.S.A. 2020;117(38):23329–23335. doi: 10.1073/pnas.1820843116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan P.O., Sasaki A., D'Alessio A.C., Dymov S., Labonté B., Szyf M., Turecki G., Meaney M.J. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009 doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C., Schillert A., Röthemeier C., Trégouët D.A., Proust C., Binder H., Pfeiffer N., Beutel M., Lackner K.J., Schnabel R.B., Tiret L., Wild P.S., Blankenberg S., Zeller T., Ziegler A. Removing batch effects from longitudinal gene expression - quantile normalization plus comBat as best approach for microarray transcriptome data. PloS One. 2016 doi: 10.1371/journal.pone.0156594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naftolowitz D.F., Donovan S., Frances A. 1995. DSM-IV. CNS Drugs. [DOI] [Google Scholar]
- Narayanan M., Huynh J.L., Wang K., Yang X., Yoo S., McElwee J., Zhang B., Zhang C., Lamb J.R., Xie T., Suver C., Molony C., Melquist S., Johnson A.D., Fan G., Stone D.J., Schadt E.E., Casaccia P., Emilsson V., Zhu J. Common dysregulation network in the human prefrontal cortex underlies two neurodegenerative diseases. Mol. Syst. Biol. 2014 doi: 10.15252/msb.20145304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Li G.E., Chen H., Cranfield C.G., McGrath K.C., Gorrie C.A. Chemical Research in Toxicology. 2018. Maternal E-cigarette exposure results in cognitive and epigenetic alterations in offspring in a mouse model. [DOI] [PubMed] [Google Scholar]
- Parade S.H., Parent J., Rabemananjara K., Seifer R., Marsit C.J., Yang B.Z., Zhang H., Tyrka A.R. Development and Psychopathology; 2017. Change in FK506 Binding Protein 5 (FKBP5) Methylation over Time Among Preschoolers with Adversity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent J., Parade S.H., Laumann L.E., Ridout K.K., Yang B.Z., Marsit C.J., Seifer R., Tyrka A.R. Development and Psychopathology; 2017. Dynamic Stress-Related Epigenetic Regulation of the Glucocorticoid Receptor Gene Promoter during Early Development: the Role of Child Maltreatment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen B.S., Schwartz D.A., Yang I.v., Kechris K.J. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E., Lussier A.A., Jones M.J., MacIsaac J.L., Edgar R.D., Mah S.M., Barhdadi A., Provost S., Lemieux-Perreault L.P., Cynader M.S., Chudley A.E., Dubé M.P., Reynolds J.N., Pavlidis P., Kobor M.S. 2016. DNA Methylation Signature of Human Fetal Alcohol Spectrum Disorder. Epigenetics and Chromatin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provençal N., Binder E.B. Experimental Neurology. 2015. The effects of early life stress on the epigenome: from the womb to adulthood and even before. [DOI] [PubMed] [Google Scholar]
- Resendiz M., Chen Y., Öztürk N.C., Zhou F.C. Epigenomics. 2013. Epigenetic medicine and fetal alcohol spectrum disorders. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J.N., Weinberg J., Clarren S., Beaulieu C., Rasmussen C., Kobor M., Dube M.P., Goldowitz D. Seminars in Pediatric Neurology. 2011. Fetal alcohol spectrum disorders: gene-environment interactions, predictive biomarkers, and the relationship between structural alterations in the brain and functional outcomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond R.C., Suderman M., Langdon R., Relton C.L., Smith G.D. DNA methylation as a marker for prenatal smoke exposure in adults. Int. J. Epidemiol. 2018 doi: 10.1093/ije/dyy091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson-Nay R., Lapato D.M., Wolen A.R., Lancaster E.E., Webb B.T., Verhulst B., Hettema J.M., York T.P. An epigenome-wide association study of early-onset major depression in monozygotic twins. Transl. Psychiatry. 2020 doi: 10.1038/s41398-020-00984-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas D., Rager J.E., Smeester L., Bailey K.A., Drobná Z., Rubio-Andrade M., Stýblo M., García-Vargas G., Fry R.C. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicol. Sci.: Off. J. Soc. Toxicol. 2015 doi: 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romens S.E., Mcdonald J., Svaren J., Pollak S.D. Child Development; 2015. Associations between Early Life Stress and Gene Methylation in Children. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw P.J., Croux C. Alternatives to the median absolute deviation. J. Am. Stat. Assoc. 1993 doi: 10.1080/01621459.1993.10476408. [DOI] [Google Scholar]
- Saadat M., Mobayen F., Farrashbandi H. Genetic polymorphism of glutathione S-transferase T1: a candidate genetic modifier of individual susceptibility to schizophrenia. Psychiatr. Res. 2007 doi: 10.1016/j.psychres.2006.03.024. [DOI] [PubMed] [Google Scholar]
- Samplin E., Ikuta T., Malhotra A.K., Szeszko P.R., DeRosse P. Sex differences in resilience to childhood maltreatment: effects of trauma history on hippocampal volume, general cognition and subclinical psychosis in healthy adults. J. Psychiatr. Res. 2013 doi: 10.1016/j.jpsychires.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.K., Kilaru V., Klengel T., Mercer K.B., Bradley B., Conneely K.N., Ressler K.J., Binder E.B. American Journal of Medical Genetics, Part B: Neuropsychiatric Genetics. 2015. DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suderman M., Borghol N., Pappas J.J., Pinto Pereira S.M., Pembrey M., Hertzman C., Power C., Szyf M. Childhood abuse is associated with methylation of multiple loci in adult DNA. BMC Med. Genom. 2014 doi: 10.1186/1755-8794-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.L., Yang S.L., Sun H., Li W. da, Duan S.R. Molecular differences in Alzheimer's disease between male and female patients determined by integrative network analysis. J. Cell Mol. Med. 2019 doi: 10.1111/jcmm.13852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M., McGowan P., Meaney M.J. Environmental and Molecular Mutagenesis. 2008. The social environment and the epigenome. [DOI] [PubMed] [Google Scholar]
- Tang J.J., Wang M.W., Jia E.Z., Yan J.J., Wang Q.M., Zhu J., Yang Z.J., Lu X., Wang L.S. Molecular Biology Reports. 2010. The common variant in the GSTM1 and GSTT1 genes is related to markers of oxidative stress and inflammation in patients with coronary artery disease: a case-only study. [DOI] [PubMed] [Google Scholar]
- Teh A.L., Pan H., Chen L., Ong M.L., Dogra S., Wong J., MacIsaac J.L., Mah S.M., McEwen L.M., Saw S.M., Godfrey K.M., Chong Y.S., Kwek K., Kwoh C.K., Soh S.E., Chong M.F.F., Barton S., Karnani N., Cheong C.Y., Holbrook J.D. The effect of genotype and in utero environment on interindividual variation in neonate DNA methylomes. Genome Res. 2014 doi: 10.1101/gr.171439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellegen P., Laros J. The construction and validation of a nonverbal test of intelligence: the revision of the Snijders-Oomen tests. Eur. J. Psychol. Assess. 1993 [Google Scholar]
- Terasaki L.S., Schwarz J.M. Effects of moderate prenatal alcohol exposure during early gestation in rats on inflammation across the maternal-fetal-immune interface and later-life immune function in the offspring. J. Neuroimmune Pharmacol. 2016 doi: 10.1007/s11481-016-9691-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecki G., Meaney M.J. Biological Psychiatry. 2016. Effects of the social environment and stress on glucocorticoid receptor gene methylation: a systematic review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vineis P., Chatziioannou A., Cunliffe V.T., Flanagan J.M., Hanson M., Kirsch-Volders M., Kyrtopoulos S. Epigenetic memory in response to environmental stressors. FASEB (Fed. Am. Soc. Exp. Biol.) J. 2017 doi: 10.1096/fj.201601059RR. [DOI] [PubMed] [Google Scholar]
- Walton E., Hass J., Liu J., Roffman J.L., Bernardoni F., Roessner V., Kirsch M., Schackert G., Calhoun V., Ehrlich S. Correspondence of DNA methylation between blood and brain tissue and its application to schizophrenia research. Schizophr. Bull. 2016 doi: 10.1093/schbul/sbv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Taskesen E., van Bochoven A., Posthuma D. 2017. FUMA: Functional Mapping and Annotation of Genetic Associations. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. The Psychological Corporation; San Antonio, TX: 2002. Wechsler Preschool and Primay Scale of Intelligence - Third Edition (WPPSI-III) Technical and Interpretive Manual. [Google Scholar]
- Weder N., Zhang H., Jensen K., Yang B.Z., Simen A., Jackowski A., Lipschitz D., Douglas-Palumberi H., Ge M., Perepletchikova F., O'Loughlin K., Hudziak J.J., Gelernter J., Kaufman J. Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. J. Am. Acad. Child Adolesc. Psychiatry. 2014 doi: 10.1016/j.jaac.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.D., Kaffman A. Frontiers in Neuroscience. 2019. The moderating effects of sex on consequences of childhood maltreatment: from clinical studies to animal models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklund P., Karhunen V., Richmond R.C., Parmar P., Rodriguez A., de Silva M., Wielscher M., Rezwan F.I., Richardson T.G., Veijola J., Herzig K.H., Holloway J.W., Relton C.L., Sebert S., Järvelin M.R. Clinical Epigenetics. 2019. DNA methylation links prenatal smoking exposure to later life health outcomes in offspring. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J., Stolzenberg H. Gesundheitswesen (Bundesverband Der Arzte Des Offentlichen Gesundheitsdienstes (Germany)); 1999. [Social Class Index in the Federal Health Survey] [PubMed] [Google Scholar]
- Winter, S. M., Dittrich, K., Dörr, P., Overfeld, J., Moebus, I., Murray, E., Zimmermann, C., Knop, A., Voelkele, M., Entringer, S., Buss, C., Haynes, J.-D., Binder, E. B., & Heim, C. (n.d.). Immediate Impact of Child Maltreatment on Mental, Developmental, and Physical Health Trajectoriesle. (In Review). [DOI] [PubMed]
- Yang B.Z., Zhang H., Ge W., Weder N., Douglas-Palumberi H., Perepletchikova F., Gelernter J., Kaufman J. Child abuse and epigenetic mechanisms of disease risk. Am. J. Prev. Med. 2013 doi: 10.1016/j.amepre.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Qiu H., Qu P., Zhang R., Zeng L., Yan H. Prenatal alcohol exposure and congenital heart defects: a meta-analysis. PloS One. 2015 doi: 10.1371/journal.pone.0130681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Pollak L., Mottagui-Tabar S., Wahlestedt C., Taubman J., Virkkunen M., Goldman D., Heilig M. NPY leu7pro and alcohol dependence in Finnish and Swedish populations. Alcohol Clin. Exp. Res. 2003 doi: 10.1111/j.1530-0277.2003.tb02715.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.