Abstract
Background
Chemoradiotherapy (CRT) is the standard of care for patients diagnosed with locally advanced cervical cancer (LACC), a human papillomavirus (HPV)-related cancer that relapses in 30%-60% of patients. This study aimed to (i) design HPV droplet digital PCR (ddPCR) assays for blood detection (including rare genotypes) and (ii) monitor blood HPV circulating tumor DNA (HPV ctDNA) levels during CRT in patients with LACC.
Methods
We analyzed blood and tumor samples from 55 patients with HPV-positive LACC treated by CRT in a retrospective cohort (n = 41) and a prospective cohort (n = 14). HPV-ctDNA detection was carried out by genotype-specific ddPCR.
Results
HPV ctDNA was successfully detected in 69% of patients (n = 38/55) before CRT for LACC, including nine patients with a rare genotype. HPV-ctDNA level was correlated with HPV copy number in the tumor (r = 0.41, P < 0.001). HPV-ctDNA positivity for HPV18 (20%, n = 2/10) was significantly lower than for HPV16 (77%, n = 27/35) or other types (90%, n = 9/10, P = 0.002). HPV-ctDNA detection (positive versus negative) before CRT was associated with tumor stage (P = 0.037) and lymph node status (P = 0.02). Taking into account all samples from the end of CRT and during follow-up in the prospective cohort, positive HPV-ctDNA detection was associated with lower disease-free survival (DFS) (P = 0.048) and overall survival (OS) (P = 0.0013).
Conclusion
This is one of the largest studies to report HPV-ctDNA detection before CRT and showed clearance of HPV ctDNA at the end of treatment in most patients. Residual HPV ctDNA at the end of CRT or during follow-up could help to identify patients more likely to experience subsequent relapse.
Key words: circulating tumor DNA, human papillomavirus, cervical cancer, chemoradiotherapy, prognostic marker
Highlights
-
•
HPV ctDNA can be detected before CRT in patients with LACC (69%).
-
•
HPV-ctDNA detection was associated with tumor stage and lymph node status.
-
•
A lower detection rate of HPV ctDNA was observed for HPV18 genotype.
-
•
Residual ctDNA levels after CRT and during follow-up had a prognostic impact.
Introduction
Cervical cancer is a commonly diagnosed cancer in women, especially in less developed countries where cervical cancer is the leading cause of cancer death among women.1 Definitive chemoradiotherapy (CRT) is the current standard of care for patients diagnosed with locally advanced cervical cancer (LACC) [International Federation of Gynecology and Obstetrics (FIGO) stage IB2-IIIC/IVA], possibly in combination with surgery and/or neoadjuvant chemotherapy in some cases.1 However, 30%-60% of these patients develop local and/or distant relapse during follow-up.2 Interpretation of conventional imaging can be challenging after CRT, as cancer regression after CRT can take >3 months.1 No sensitive blood biomarkers are currently available to predict relapse in patients, as squamous cell carcinoma (SCC) antigen demonstrated low sensitivity and specificity3 for the monitoring of patients with LACC during CRT.4
In many tumor types, circulating tumor DNA (ctDNA) has demonstrated a good correlation with tumor response or progression and is a useful complement to standard tumor imaging,5, 6, 7, 8, 9 able to detect early relapse after treatment in the non-metastatic setting.10, 11, 12, 13, 14
Cervical cancer is due to human papillomavirus (HPV) infection in the vast majority of cases, and HPV DNA sequences can be found in tumors.15, 16, 17 However, hundreds of HPV types have been described, and <20 HPV genotypes are classified as being associated with a high risk of cancer.17,18 HPV16 and HPV18 are the genotypes most commonly detected in cervical cancer, while around 20% of invasive cancers can be attributed to the less common HPV31, HPV33, HPV35, HPV45, HPV52, HPV58 and HPV73 types.19
In HPV-related cancer, HPV viral genomes are usually integrated into the tumor cell genome or episomal DNA.20 While real-time PCR was previously used,21, 22, 23 several more sensitive detection techniques are now available to detect circulating HPV in blood, such as ctDNA,24, 25, 26, 27 including digital-PCR based techniques or next-generation sequencing (NGS)-based techniques. We have previously validated that a droplet digital PCR (ddPCR) based technique is able to detect and quantify tumor-derived HPV DNA sequences in patient blood (HPV ctDNA) with high sensitivity and specificity for HPV16 and HPV1826,28 and could be used in anal carcinoma (another HPV-related cancer) to monitor the efficacy of immunotherapy25 or chemotherapy in the metastatic setting.29 Other studies have demonstrated that HPV-ctDNA detection by ddPCR can be used to monitor tumor response in cervical cancer,30 or head and neck cancer.31,32 Importantly for cervical cancer, no HPV ctDNA was detected in healthy controls or in women treated for HPV16-associated high-grade cervical intraepithelial neoplasia.26,28 Several studies in non-cervical cancers have found that the HPV-ctDNA detection after CRT was a strong predictive factor for relapse in anal carcinoma33 or head and neck cancer,27,31,34 but no study is available for LACC. As HPV types are more diverse in cervical cancer than in anal carcinoma (mostly HPV16-18) and head and neck cancer (mainly HPV16), ddPCR assays designed for each HPV type are also required for monitoring of HPV ctDNA.
The objectives of this study were: (i) to design HPV ddPCR assays for blood detection of HPV types other than HPV16-18 and (ii) to monitor blood HPV ctDNA levels during CRT in patients with LACC.
Material and methods
This study was conducted in accordance with the Declaration of Helsinki. This study was conducted according to national requirements and in accordance with the guidelines of the Declaration of Helsinki (CPP 13381). A waiver of patient informed consent was obtained for samples stored at the Institut Curie biobank. The PAIR-HPV prospective study obtained number NCT02554565.
Patients and samples
The retrospective study included patients treated at Institut Curie (France) with at least radiotherapy for LACC from 2011 to 2015 with serum or plasma samples stored at the Institut Curie biobank. Main eligibility criteria were: at least radiotherapy treatment, histologically proven HPV-related tumor (HPV subtype was determined by routine PCR diagnostic procedures on tumor samples) and serum or plasma samples banked at the time of diagnosis (before any treatment) plus at least one serum/plasma sample thereafter. Patients included in the study received standard treatment. A waiver of patient informed consent was obtained for samples stored at the Institut Curie biobank. In the retrospective cohort, samples were considered as ‘end of treatment samples’ if they were collected between D(−)21 and D(+)90 after the end of the therapeutic sequence.
The PAIR-HPV prospective study (NCT02554565) included patients with cervical cancer at any stage before treatment. We selected patients receiving first-line treatment with CRT in this cohort. Blood samples were collected at baseline (before treatment), days 7, 21 and 35 during CRT and then at 2, 6, 12, 18 and 24 months.
The 2018 FIGO classification was used for staging.
Analytic methods
All patients from both cohorts had confirmation of HPV positivity from their tumor tissue.
We used ddPCR assays to detect HPV16 or HPV18 in tumor or blood samples, as previously described in Jeannot et al.26
We developed specific ddPCR assays using TaqMan probes to detect other high-risk HPV: HPV31, HPV33, HPV35, HPV45, HPV52, HPV58 and HPV73 types (Supplementary Methods, available at https://doi.org/10.1016/j.esmoop.2021.100154). To ensure compatibility with ctDNA detection, primers were designed to generate amplicons smaller than 150 bp. To avoid cross-reactivity between the various HPV types, primers were designed to target the E7 gene, a region of the HPV genome with a lesser degree of similarity between HPV types. Primer and probe sequences, amplicon sizes and annealing temperatures are shown in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100154.
HPV-ctDNA quantification
Serum or plasma samples were stored at −80°C until DNA isolation. Cell-free circulating DNA was isolated in duplicate from 1 to 2 ml of serum or plasma using the QIAsymphony SP/AS workflow (Qiagen®, Courtaboeuf, France), according to the manufacturer's instructions. Elution was carried out in 60 μl of the supplied elution buffer; eluates were stored at −20°C until HPV-ctDNA analysis. Three replicates were carried out to detect HPV E7 gene with HPV type-specific primers, TaqMan probes, and the maximal volume of ctDNA template (8 μl). PCR reaction was multiplexed with a commercial human ddPCR assay targeting the RPP30 gene, used as human DNA reference, to quantify cell-free circulating DNA. For each patient, genomic DNA from the frozen or formalin-fixed paraffin-embedded tumor was tested as positive control and a triplicate of no DNA was tested for each plate as negative control.26
According to our previous findings, serum/plasma samples were considered to be positive in the presence of at least three positive droplets.26 A sample was considered to be negative when fewer than three droplets from all replicates were detected, whereas more than 500 copies/ml were detected for RPP30. Finally, HPV-ctDNA concentration was expressed in copies/ml of serum.
Tumour HPV copy number
HPV copy number in tumor cells extracted from the tumor sample was determined by ddPCR, using the same HPV type-specific and RPP30 assays as in the HPV-ctDNA quantification experiment. Experiments were carried out with 10 ng of tumor DNA, in duplicates for each sample. RRP30 gene was used as a reference for two copies of a DNA sequence per cell. The value used to express HPV copy number was twice that of the HPV E7/RPP30 ratio provided by the ddPCR experiment. Finally, tumor HPV copy number was expressed in copies/tumor cell.
Statistical analysis
Retrospective and prospective cohorts were analyzed together for baseline samples, and separately for following timepoints.
This report was written in accordance with REMARK criteria.35 No prespecified power was calculated for this study. Fisher's exact test was used for categorical variables and Spearman's correlation test, Mann–Whitney test and Kruskal–Wallis test were used for continuous variables. Survival curves were compared by an unstratified log-rank test, and hazard ratio with 95% confidence interval (CI) were calculated using a Cox model.
Disease-free survival (DFS) was defined as the time from the start of treatment to recurrence of tumor or death. Overall survival (OS) was defined as the time from the start of treatment to death.
Statistical analyses and figures were carried out using GraphPad Prism version v-6-07 (GraphPad Software, San Diego, CA) and R version 4.0. All statistical tests were two-tailed, and P values < 0.05 were considered statistically significant.
Results
Patient characteristics
Forty-one patients with HPV-related LACC and serum or plasma samples available before, during and after treatment were included in the retrospective cohort and 14 patients were included in the prospective cohort (Figure 1 flowchart), corresponding to a total of 55 patients. The clinical and laboratory characteristics are presented in Table 1. The median age of the patients of the whole cohort was 50 years (range: 29-78); the most common histological tumor type was SCC (87%). Tumor HPV typing detected HPV16, HPV18 or other types (31, 33, 35, 45, 52, 58, 73) in 64%, 18% and 18% of LACC, respectively. The majority of patients (60%) had stage III disease (FIGO classification, 2018). Sixty percent of patients had invaded lymph nodes at imaging and/or initial surgical staging (Table 1). Median duration of CRT in the prospective cohort was 33 days (range 21-50).
Figure 1.
Study flow chart.
PAIR-HPV, clinical study (NCT02554565); pts, patients.
Table 1.
Patient characteristics and HPV circulating tumor DNA (HPV-ctDNA) detection
| Characteristics | All patients | HPV-ctDNA detectable at diagnosis | P |
|---|---|---|---|
| Total | 55 | 38 (69%) | |
| Age (years) | |||
| Median (range) | 50 (29-78) | 50 (29-78) | |
| FIGO stage | |||
| I | 5 (9%) | 1 (20%) | |
| II | 13 (24%) | 8 (62%) | 0.04 |
| III | 33 (60%) | 25 (76%) | |
| Iva | 4 (7%) | 4 (100%) | |
| Lymph node involvement | |||
| Yes | 33 (60%) | 27 (82%) | 0.02 |
| No | 22 (40%) | 11 (50%) | |
| Histology | |||
| SCC | 48 (87%) | 36 (75%) | 0.056 (SCCvsAdk) |
| Adenocarcinoma | 6 (11%) | 2 (33%) | |
| Other | 1 (2%) | 0 | |
| HPV type | |||
| HPV16 | 35 (64%) | 27 (77%) | 0.002 |
| HPV18 | 10 (18%) | 2 (20%) | |
| Other HPV (31, 33, 35, 45, 52, 58, 73) | 10 (18%) | 9 (90%) | |
| Tumor HPV copy | |||
| ≤5 copies/tumor cell | 26 (47%) | 16 (62%) | 0.381 |
| >5 copies/tumor cell | 29 (53%) | 22 (76%) | |
| Neoadjuvant chemotherapy | |||
| No | 46 (84%) | 32 (70%) | |
| Yes | 9 (16%) | 6 (67%) | |
| Brachytherapy | |||
| No | 19 (35%) | 13 (68%) | |
| Yes | 36 (65%) | 25 (69%) | |
| Surgerya,b | |||
| No | 36 (64%) | 28 (78%) | |
| Yes | 19 (35%) | 10 (53%) |
Adk, adenocarcinoma; FIGO, International Federation of Gynecology and Obstetrics; HPV, human papilloma virus; SCC, squamous cell carcinoma.
No surgery in the prospective cohort.
Five patients had surgery before chemoradiotherapy and 14 patients after.
All patients had serum/plasma collected at baseline (before treatment). Serum/plasma samples were also available at the end of treatment of 39 patients (71%) (Figure 1). D7, D21 and D35 samples in the prospective cohort were obtained in 9, 13 and 13 patients, with a range of 4-11 days, 14-23 days and 30-43 days, respectively.
Development of new HPV ddPCR assays
We developed specific ddPCR assays for the seven additional HPV types (beside HPV16/18) that were identified in the 55 patients: HPV31, HPV33, HPV35, HPV45, HPV52, HPV58 and HPV73.
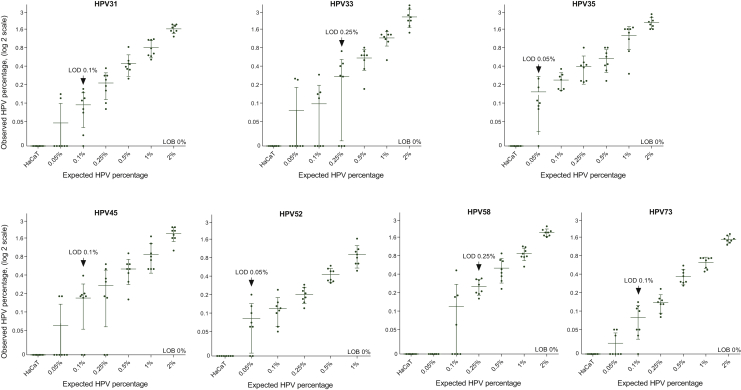
The seven HPV assays showed high specificity, as no false-positive event was observed in reactions carried out with HPV-negative DNA (Figure 2). Therefore, the limit of blank (LOB), defined as the 95% CI of the mean false-positive calls, was estimated to be 0% for the seven HPV types tested. Sensitivity was assessed by using the limit of detection (LOD), identifying the lowest HPV fraction with all replicates presenting values above the LOB. The LOD was estimated to be 0.05% for HPV35 and HPV52, 0.1% for HPV31, HPV45 and HPV73 and 0.25% for HPV33 and HPV58 (Figure 2).
Figure 2.
Limit of detection (LOD) estimation for HPV31, HPV33, HPV35, HPV45, HPV52, HPV58 and HPV73 ddPCR assays.
The limit of blank (LOB), defined as the 95% CI of the mean false-positive calls, was estimated to be 0% for the seven HPV types tested (no false-positive events observed from the HPV-negative cell line HaCaT). The LOD, defined as the lowest HPV fraction with all replicates presenting values above the LOB, are displayed by an arrow.
CI, confidence interval; HaCaT, X; HPV, human papillomavirus.
HPV-ctDNA detection
At baseline, serum/plasma HPV ctDNA was successfully detected in 38 of the 55 patients (69%) (Figure 3A). The median HPV-ctDNA level was 33 copies/ml (range: 0-56 400), with an interquartile range (IQR) in positive patients of 30-493. The median level of the human reference gene, RPP30, was 7771 copies/ml of serum (IQR 2838-28 675) (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2021.100154). Three patients (5%) of the whole cohort had higher levels of HPV-ctDNA (copies/ml) than the human reference gene (RPP30).
Figure 3.
HPV circulating tumour DNA (HPV ctDNA) by droplet digital PCR.
Group median is represented by a horizontal bar. A circle, square and triangle correspond to patients with HPV16-related tumors, HPV18-related tumors and other HPV-related tumors, respectively. (A) HPV-ctDNA levels before treatment in the whole cohort (n = 55). HPV-ctDNA levels are displayed by HPV type, P = Mann–Whitney test. (B) Positive correlation between HPV-ctDNA level and tumor HPV copy number. Spearman correlation r = 0.41 (P < 0.001). For undetectable HPV-ctDNA cases, point ‘a’ refers to three HPV16-related tumors, five HPV18-related tumors and one HPV35-related tumor, point ‘b’ refers to one HPV16-related tumor and three HPV18-related tumors. P = Kruskal–Wallis test. (C) HPV-ctDNA levels according to FIGO stage and lymph node status.
Baseline HPV-ctDNA level (copies/ml) was correlated with tumor HPV copy number (R = 0.41, P < 0.001, Spearman's) (Figure 3B), while the median tumor HPV copy number was six copies/tumor cell (range: 1-787), and HPV copy number was lower in HPV18-related tumor than in HPV16-related tumor (1.5 copies/tumor cell versus 18, P = 0.002, Wilcoxon). HPV-ctDNA positivity was significantly lower for HPV18 (20%, n = 2/10) than for HPV16-related tumor (77%, n = 27/35) or other HPV types (90%, n = 9/10) (P = 0.002), with a median of 52 copies/ml for HPV16 versus 0 for HPV 18 (Mann–Whitney P = 0.003) (Table 1). HPV-ctDNA detection (positive versus negative) before CRT was associated with tumor stage (stage I: 20%, n = 1/5; II: 62%, n = 8/13; III: 76%, n = 25/33; IV: 100%, n = 4/4; P = 0.037 for categorical variables, P = 0.004 for continuous variables) and lymph node status (N0: 50%, n = 11/22 versus N+: 82%, n = 27/33; P = 0.02 for categorical variables, P = 0.06 for continuous variables) (Figure 3C).
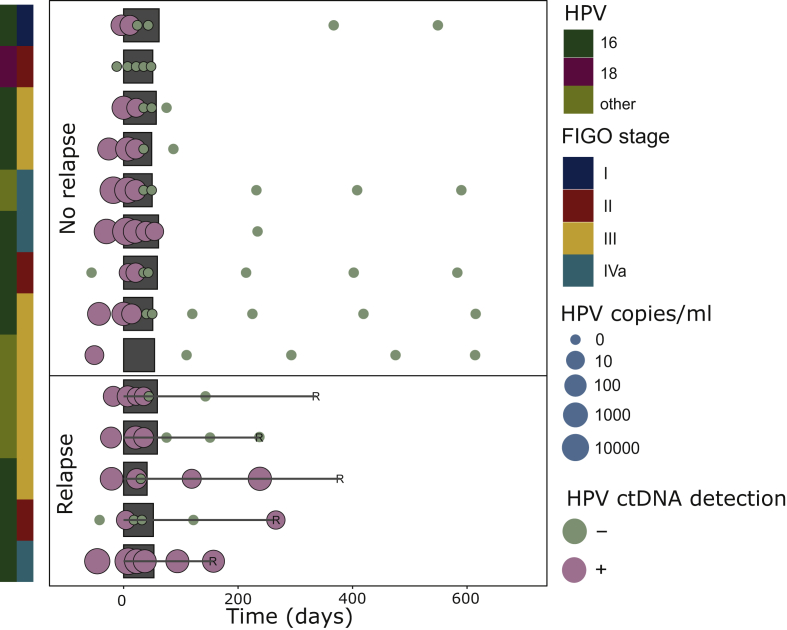
Dynamics of HPV-ctDNA level during CRT in the prospective cohort
The dynamics of HPV-ctDNA level during CRT are shown for the 14 patients of the prospective cohort in Figure 4 (the retrospective cohort was not analyzed due to the very small number of blood samples available during treatment). Interestingly, clearance of HPV ctDNA was not obtained in all patients without relapse before the end of CRT, as 6/8 (75%) of patients had positive HPV ctDNA at mid-treatment. Forty-three percent (n = 6/14) of patients had a transitory rise of HPV-ctDNA level between D7 and D21, but half of these patients did not subsequently experience relapse.
Figure 4.
HPV-ctDNA dynamics during treatment and follow-up in the prospective cohort.
Each line corresponds to a patient (N = 14). Details are provided for HPV subtype, FIGO stage of cervical cancer and detection of HPV ctDNA (positive versus negative and level in copies/ml). In the no-relapse group (upper part), all patients had a follow-up >600 days. Grey squares indicate the duration of radiotherapy.
HPV-ctDNA, human papillomavirus circulating tumor DNA.
HPV-ctDNA detection and outcome
With a median follow-up of 49.9 months (range: 10-130 months) for the retrospective cohort and 37 months (range: 15-52 months) for the prospective cohort, disease relapses were observed in 12 (29%) and 5 patients (36%), and deaths were observed in 7 (17%) and 4 patients (29%), respectively.
At baseline in the whole cohort (n = 55), neither HPV-ctDNA detection status (positive versus negative) or level (> versus ≤ median HPV-ctDNA copies/ml) had a significant prognostic impact on OS [hazard ratio (HR) = 1.4 95% CI (0.4-4.4), P = 0.57 and HR = 1.9 95% CI (0.7-5.3), P = 0.24, respectively], or DFS [HR = 0.85 95% CI (0.4-2), P = 0.71 and HR = 1.2 95% CI (0.5-2.7), P = 0.67, respectively] (Supplementary Figure S2, available at https://doi.org/10.1016/j.esmoop.2021.100154).
In the retrospective cohort, among patients with blood collected at the end of curative treatment (n = 25), 2 (8%) displayed residual detectable HPV ctDNA. These two patients experienced relapse (at 6 and 42 months, respectively).
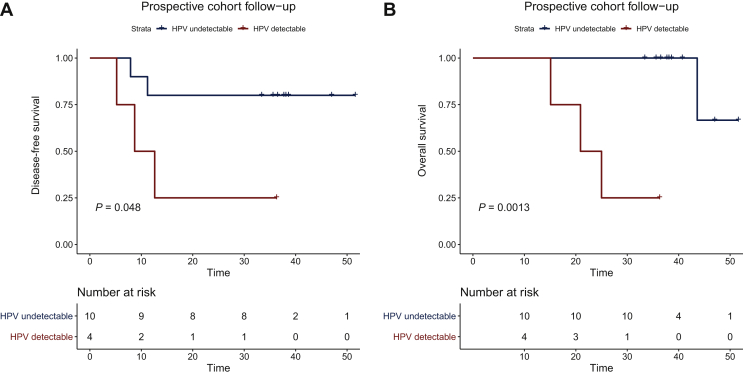
In the prospective cohort, two patients displayed residual detectable HPV ctDNA after CRT, but only one patient experienced metastatic relapse 2 months later (Figure 4). Interestingly, the patient with positive HPV-ctDNA detection without relapse had a low HPV-ctDNA level at the end of CRT (10 copies/ml), with a high baseline HPV ctDNA (1457 copies/ml) and negative HPV ctDNA at the following determination at 6 months. Taking into account all samples in the prospective cohort, positive HPV-ctDNA detection at the end of CRT and/or during follow-up was associated with lower DFS [HR = 5.1 95% CI (0.85-31), P = 0.048] and OS [HR = 25.4 95% CI (2.4 to >100), P = 0.0013] (no multivariate analysis was carried out due to the small number of patients) (Figure 5). Interestingly, all follow-up samples from patients who did not relapse were HPV-ctDNA negative.
Figure 5.
Disease-free survival (A) and overall survival (B) of patients in the prospective cohort (n = 14) according to the positivity of HPV-ctDNA detection at the end of chemoradiotherapy and/or during follow-up.
HPV-ctDNA, human papillomavirus circulating tumor DNA.
Discussion
This study showed that (i) HPV-ctDNA levels can be monitored during CRT and follow-up of patients with cervical cancer by using ddPCR assays specific to nine different HPV types, covering 90% of the women diagnosed with cervical cancer, (ii) monitoring of HPV ctDNA during follow-up could be more effective than simply assessing HPV ctDNA at the end of CRT.
The theoretical advantage of HPV-ctDNA detection over ctDNA detection of point mutations in cancer genes is that multiple HPV DNA sequences are present in a tumor cell (one to several thousand), and this can contribute to improving the sensitivity of detection, as these numerous copies are released into the blood following cancer cell death.20 This hypothesis was confirmed in this study by the positive correlation between HPV copy number in tumor cells and HPV-ctDNA detection in blood, and by the fact that the HPV-ctDNA copy number was higher than the actual total number of circulating human genome equivalent in 5% of patients. The 70% detection rate before therapy appears to be higher than that reported in other non-metastatic cancer types with ctDNA detection of point mutations.36,37 For example, in a previous study in non-metastatic gastric cancers with ddPCR, we found a ctDNA detection rate of only 30%.37 Interestingly, we found that the HPV-ctDNA detection rate differed according to the HPV type, with the lowest detection rate (20%) observed for HPV18, which was associated with a lower HPV copy number in tumor cells. This lower detection rate for HPV18 ctDNA has been reported by Cheung et al., with a detection rate of 41% for HPV18 versus 61% for HPV 16.38 Other studies have previously reported higher HPV-ctDNA detection rates (>80%) in other HPV-induced head and neck27 or gynecologic cancers,26,28,39 but with no or less HPV18 subtype, and NGS technique for the head and neck study.27 The volume of serum/plasma used in this study (1-2 ml) could be increased in future to improve sensitivity.
One of the main results of this study is that HPV-ctDNA levels drop markedly in most patients during CRT and that residual detectable ctDNA levels after CRT are associated with cancer relapse, as found in another study in LACC with 19 patients.39 However, the sensitivity of HPV-ctDNA detection at the end of CRT to predict relapse is not perfect, and we have highlighted that repeated timepoints may be required to predict relapse. Interestingly, positive HPV-ctDNA detection during CRT did not appear to be predictive of relapse, as several patients obtained complete clearance of HPV ctDNA only at the end of CRT and did not subsequently experience relapse. In terms of the specificity of HPV-ctDNA positivity at the end of CRT to predict outcome, it may be preferable to repeat HPV-ctDNA detection over the following weeks or months in patients with low levels of HPV ctDNA, especially in the case of high baseline HPV ctDNA. A large study in head and neck cancer showed that HPV-ctDNA detection on two consecutive plasma samples during post-treatment surveillance was required to achieve very high positive (94%) and negative (100%) predictive values.34 Assessing HPV ctDNA every 3-6 months may therefore be a reasonable follow-up option to predict relapse before the onset of symptoms or radiological signs.
For implementation in clinical practice, we have shown that several HPV types can be monitored, representing the majority of HPV types involved in cervical cancer, although the sensitivity of HPV-ctDNA detection was lower for HPV18-related tumors. However, this approach requires HPV genotyping and dedicated ddPCR for each genotype. NGS-based techniques27 could avoid this issue, but when repeated measures are required, the cost would rapidly increase and become prohibitive, while the cost of ddPCR is more acceptable for repeated measures and long-term follow-up.
The main limitations of our proof-of-concept study are the small number of patients analyzed, the retrospective data collection for the majority of patients with missing samples, heterogeneity in treatments and storage that could degrade DNA and impair sensitivity.
In conclusion, we showed that HPV-ctDNA monitoring by ddPCR during CRT is feasible for nine HPV types involved in at least 90% of all cervical cancers and can be used to predict relapses. A large prospective trial using the same assays, Circa HPV (NCT03739775), will analyze the clinical validity of HPV-ctDNA detection in cervical cancer during follow-up, and especially the impact of HPV-ctDNA detection on outcome.
Acknowledgements
We thank the Institut Curie biobank for providing serum and plasma samples and Michèle Cauchois for her help with clinical data.
Funding
This work was supported by Fondation ARC pour la recherche sur le cancer, Ligue contre le cancer, INCa Integrated Research Action Programme (PAIR-HPV, No. 2012-030), SIRIC 2 Curie [grant number INCa-DGOS-Inserm_12554].
Disclosure
The authors have declared no conflicts of interest.
Data sharing
The data that support the findings of this study are available from Institut Curie but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Institut Curie.
Supplementary data
HPV-ctDNA detection and concomitant RPP30 detection (human gene for quality control of cell-free circulating DNA) (copies/ml).
Overall survival (A,C) and disease-free survival (B,D) according to baseline HPV-ctDNA detection (detectable/undetectable A,B or above/below median C,D).
References
- 1.Marth C., Landoni F., Mahner S. Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28:iv72–iv83. doi: 10.1093/annonc/mdx220. [DOI] [PubMed] [Google Scholar]
- 2.Kim J.Y., Byun S.J., Kim Y.S., Nam J.-H. Disease courses in patients with residual tumor following concurrent chemoradiotherapy for locally advanced cervical cancer. Gynecol Oncol. 2017;144:34–39. doi: 10.1016/j.ygyno.2016.10.032. [DOI] [PubMed] [Google Scholar]
- 3.Kawaguchi R., Furukawa N., Kobayashi H., Asakawa I. Posttreatment cut-off levels of squamous cell carcinoma antigen as a prognostic factor in patients with locally advanced cervical cancer treated with radiotherapy. J Gynecol Oncol. 2013;24:313–320. doi: 10.3802/jgo.2013.24.4.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu J., Wang W., Wang Y., Liu C., Wang P. The role of squamous cell carcinoma antigen (SCC Ag) in outcome prediction after concurrent chemoradiotherapy and treatment decisions for patients with cervical cancer. Radiat Oncol. 2019;14:146. doi: 10.1186/s13014-019-1355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alix-Panabières C., Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 6.Cabel L., Proudhon C., Mariani P. Circulating tumor cells and circulating tumor DNA: what surgical oncologists need to know? Eur J Surg Oncol. 2017;43:949–962. doi: 10.1016/j.ejso.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Bidard F.-C., Weigelt B., Reis-Filho J.S. Going with the flow: from circulating tumor cells to DNA. Sci Transl Med. 2013;5:207ps14. doi: 10.1126/scitranslmed.3006305. [DOI] [PubMed] [Google Scholar]
- 8.Kilgour E., Rothwell D.G., Brady G., Dive C. Liquid biopsy-based biomarkers of treatment response and resistance. Cancer Cell. 2020;37:485–495. doi: 10.1016/j.ccell.2020.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Boonstra P.A., Wind T.T., van Kruchten M. Clinical utility of circulating tumor DNA as a response and follow-up marker in cancer therapy. Cancer Metastasis Rev. 2020;39(3):999–1013. doi: 10.1007/s10555-020-09876-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehl F., Schmidt K., Choti M.A. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tie J., Wang Y., Tomasetti C. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Murillas I., Schiavon G., Weigelt B. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7:302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 13.Olsson E., Winter C., George A. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med. 2015;7:1034–1047. doi: 10.15252/emmm.201404913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riva F., Bidard F.-C., Houy A. Patient-specific circulating tumor DNA detection during neoadjuvant chemotherapy in triple-negative breast cancer. Clin Chem. 2017;63:691–699. doi: 10.1373/clinchem.2016.262337. [DOI] [PubMed] [Google Scholar]
- 15.de Martel C., Plummer M., Vignat J., Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 2017;141:664–670. doi: 10.1002/ijc.30716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention (CDC) Human papillomavirus-associated cancers – United States, 2004-2008. MMWR Morb Mortal Wkly Rep. 2012;61:258–261. [PubMed] [Google Scholar]
- 17.Muñoz N., Bosch F.X., de Sanjosé S. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 18.de Villiers E.-M., Fauquet C., Broker T.R., Bernard H.-U., zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 19.de Sanjose S., Quint W.G., Alemany L. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–1056. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 20.Valmary-Degano S., Jacquin E., Prétet J.-L. Signature patterns of human papillomavirus type 16 in invasive anal carcinoma. Hum Pathol. 2013;44:992–1002. doi: 10.1016/j.humpath.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 21.Cocuzza C.E., Martinelli M., Sina F. Human papillomavirus DNA detection in plasma and cervical samples of women with a recent history of low grade or precancerous cervical dysplasia. PLoS One. 2017;12:e0188592. doi: 10.1371/journal.pone.0188592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu Y., Wan C., Qiu J., Cui Y., Jiang T., Zhuang Z. Circulating HPV cDNA in the blood as a reliable biomarker for cervical cancer: a meta-analysis. PLoS One. 2020;15:e0224001. doi: 10.1371/journal.pone.0224001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimada T., Yamaguchi N., Nishida N. Human papillomavirus DNA in plasma of patients with HPV16 DNA-positive uterine cervical cancer. Jpn J Clin Oncol. 2010;40:420–424. doi: 10.1093/jjco/hyp193. [DOI] [PubMed] [Google Scholar]
- 24.Cabel L., Proudhon C., Romano E. Clinical potential of circulating tumour DNA in patients receiving anticancer immunotherapy. Nat Rev Clin Oncol. 2018;15:639–650. doi: 10.1038/s41571-018-0074-3. [DOI] [PubMed] [Google Scholar]
- 25.Cabel L., Bidard F.-C., Servois V. HPV circulating tumor DNA to monitor the efficacy of anti-PD-1 therapy in metastatic squamous cell carcinoma of the anal canal: a case report. Int J Cancer. 2017;141:1667–1670. doi: 10.1002/ijc.30863. [DOI] [PubMed] [Google Scholar]
- 26.Jeannot E., Becette V., Campitelli M. Circulating human papillomavirus DNA detected using droplet digital PCR in the serum of patients diagnosed with early stage human papillomavirus-associated invasive carcinoma. J Pathol Clin Res. 2016;2:201–229. doi: 10.1002/cjp2.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J.Y., Garcia-Murillas I., Cutts R.J. Predicting response to radical (chemo)radiotherapy with circulating HPV DNA in locally advanced head and neck squamous carcinoma. Br J Cancer. 2017;117:876–883. doi: 10.1038/bjc.2017.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campitelli M., Jeannot E., Peter M. Human papillomavirus mutational insertion: specific marker of circulating tumor DNA in cervical cancer patients. PLoS One. 2012;7:e43393. doi: 10.1371/journal.pone.0043393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernard-Tessier A., Jeannot E., Guenat D. Clinical validity of HPV circulating tumor DNA in advanced anal carcinoma: an ancillary study to the epitopes-HPV02 trial. Clin Cancer Res. 2019;25:2109–2115. doi: 10.1158/1078-0432.CCR-18-2984. [DOI] [PubMed] [Google Scholar]
- 30.Kang Z., Stevanović S., Hinrichs C.S., Cao L. Circulating cell-free DNA for metastatic cervical cancer detection, genotyping, and monitoring. Clin Cancer Res. 2017;23:6856–6862. doi: 10.1158/1078-0432.CCR-17-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damerla R.R., Lee N.Y., You D. Detection of early human papillomavirus-associated cancers by liquid biopsy. JCO Precis Oncol. 2019;3 doi: 10.1200/PO.18.00276. PO.18.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veyer D., Wack M., Mandavit M. HPV circulating tumoral DNA quantification by droplet-based digital PCR: a promising predictive and prognostic biomarker for HPV-associated oropharyngeal cancers. Int J Cancer. 2020;147(4):1222–1227. doi: 10.1002/ijc.32804. [DOI] [PubMed] [Google Scholar]
- 33.Cabel L., Jeannot E., Bieche I. Prognostic impact of residual HPV ctDNA detection after chemoradiotherapy for anal squamous cell carcinoma. Clin Cancer Res. 2018;24(22):5767–5771. doi: 10.1158/1078-0432.CCR-18-0922. [DOI] [PubMed] [Google Scholar]
- 34.Chera B.S., Kumar S., Shen C. Plasma circulating tumor HPV DNA for the surveillance of cancer recurrence in HPV-associated oropharyngeal cancer. J Clin Oncol. 2020;38:1050–1058. doi: 10.1200/JCO.19.02444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McShane L.M., Altman D.G., Sauerbrei W., Taube S.E., Gion M., Clark G.M. REporting recommendations for tumour MARKer prognostic studies (REMARK) Br J Cancer. 2005;93:387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen J.D., Li L., Wang Y. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cabel L., Decraene C., Bieche I. Limited sensitivity of circulating tumor DNA detection by droplet digital PCR in non-metastatic operable gastric cancer patients. Cancers. 2019;11(3):396. doi: 10.3390/cancers11030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheung T.H., Yim S.F., Yu M.Y. Liquid biopsy of HPV DNA in cervical cancer. J Clin Virol. 2019;114:32–36. doi: 10.1016/j.jcv.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 39.Han K., Leung E., Barbera L. Circulating human papillomavirus DNA as a biomarker of response in patients with locally advanced cervical cancer treated with definitive chemoradiation. JCO Precis Oncol. 2018;2:1–8. doi: 10.1200/PO.18.00152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HPV-ctDNA detection and concomitant RPP30 detection (human gene for quality control of cell-free circulating DNA) (copies/ml).
Overall survival (A,C) and disease-free survival (B,D) according to baseline HPV-ctDNA detection (detectable/undetectable A,B or above/below median C,D).