Abstract
miRNAs are important regulators of eukaryotic gene expression. The post-transcriptional maturation of miRNAs is controlled by the Drosha-DiGeorge syndrome critical region gene 8 (DGCR8) microprocessor. Dysregulation of miRNA biogenesis has been implicated in the pathogenesis of human diseases, including cancers. C-terminal–binding protein–interacting protein (CtIP) is a well-known DNA repair factor that promotes the processing of DNA double-strand break (DSB) to initiate homologous recombination–mediated DSB repair. However, it was unclear whether CtIP has other unknown cellular functions. Here, we aimed to uncover the roles of CtIP in miRNA maturation and cancer cell metastasis. We found that CtIP is a potential regulatory factor that suppresses the processing of miRNA primary transcripts (pri-miRNA). CtIP directly bound to both DGCR8 and pri-miRNAs through a conserved Sae2-like domain, reduced the binding of Drosha to DGCR8 and pri-miRNA substrate, and inhibited processing activity of Drosha complex. CtIP depletion significantly increased the expression levels of a subset of mature miRNAs, including miR-302 family members that are associated with tumor progression and metastasis in several cancer types. We also found that CtIP-inhibited miRNAs, such as miR-302 family members, are not crucial for DSB repair. However, increase of miR-302b levels or loss of CtIP function severely suppressed human colon cancer cell line tumor cell metastasis in a mouse xenograft model. These studies reveal a previously unrecognized mechanism of CtIP in miRNA processing and tumor metastasis that represents a new function of CtIP in cancer.
Keywords: CtIP, microRNA maturation, microprocessor, Drosha, DGCR8, metastasis
Abbreviations: BiFC, bimolecular fluorescence complementation; BRCA1, breast cancer 1; cDNA, complementary DNA; ChIP, chromatin immunoprecipitation; CtBP, C-terminal–binding protein; CtIP, C-terminal–binding protein–interacting protein; DDX5, DEAD-box helicase 5; DGCR8, DiGeorge syndrome critical region gene 8; DSB, double-strand break; EGFP, enhanced GFP; EMSA, electrophoretic mobility shift assay; GST, glutathione-S-transferase; HR, homologous recombination; MRE11, meiotic recombination 11; MRN, MRE11–RAD50–NBS1; NBS1, Nijmegen breakage syndrome protein 1; NETN, NaCl, EDTA, Tris–HCl, and NP-40 buffer; PIC, protease inhibitor cocktail; PLA, proximity ligation assay; pre-miRNAs, precursor miRNAs; pri-miRNA, miRNA primary transcripts; RAD50, ATP-binding cassette—ATPase; Rhed, RNA-binding heme domain; RNase, ribonuclease; RPA, replication protein A; VN, Venus N-terminal fragment; VC, Venus C-terminal fragment; qPCR, quantitative PCR; U2OS, human osteosarcoma cell line
miRNAs are a major class of short noncoding RNA molecules that post-transcriptionally modulate gene expression by repressing the translation and/or promoting the degradation of target messenger RNAs (1, 2, 3). MiRNAs are involved in a wide range of physiological and pathological processes. Generally, miRNAs are encoded as individual genes or within clusters comprising several different miRNAs. The biogenesis of miRNAs starts through the transcription of miRNA genes by RNA polymerase II, which produces miRNA primary transcripts (pri-miRNA) containing a stem–loop hairpin structure (4, 5). In the nucleus, pri-miRNAs are cleaved by the microprocessor complex containing the ribonuclease (RNase) III enzyme Drosha and its cofactor DiGeorge syndrome critical region gene 8 (DGCR8) into precursor miRNAs (pre-miRNAs) with 70 to 100 nucleotide-long hairpin structures (6, 7). The pre-miRNAs are then exported to the cytoplasm by exportin-5, where they are further processed into mature miRNAs by cytoplasmic Dicer RNase III to yield an approximately 22-nucleotide miRNA duplex (8, 9, 10, 11). One strand of the miRNA duplex is subsequently incorporated into the RNA-induced silencing complex. Base pairing between the miRNA and target mRNA transcripts at their 3′ UTRs guides the RNA-induced silencing complex to induce the mRNA degradation or translation inhibition (12, 13).
The production and maturation of miRNA must be strictly regulated, as any disruption of the control mechanisms may lead to the development of various kinds of diseases, including cancer. As noted previously, miRNA expression involves the transcription of miRNA genes and the maturation of primary transcripts. Thus, miRNA levels may be regulated in both a transcription-dependent and transcription-independent manner. However, in many cases, the levels of mature miRNAs are determined by post-transcriptional maturation rather than transcriptional regulation. The level of expression and activity of core components of the miRNA biogenesis machinery are often found to be dysregulated in cancer. For instance, the expression levels of Drosha and Dicer are either increased or decreased in various types of cancers, and they are inversely correlated with advanced tumor stages and poor clinical outcomes (14). Somatic and germline mutations of Drosha or DGCR8 have also been frequently found in some cancers (15, 16). In particular, the activity of the Drosha microprocessor is modulated by different nuclear proteins in a manner that usually affects the processing of only a small subset of miRNAs. For example, the RNA-editing enzyme adenosine deaminase acting on RNA 1 interacts with DGCR8 and suppresses microprocessor activity by reducing the availability of DGCR8 to Drosha. The expression of a proportion of miRNAs was upregulated in adenosine deaminase acting on RNA 1-defective cancer cells, which may have facilitated the malignant activity of metastatic melanoma (17). Smad proteins, the signal transducers of transforming growth factor-β, also modulate Drosha activity in the nucleus. Smads are recruited to the Drosha complex by DEAD-box helicase 5 (DDX5) and promote the pri-miRNA processing of about 20 miRNAs. Among them, miR-21 promotes the metastatic and invasive potential of cancer cells through the inhibition of a large group of tumor suppressor genes (18, 19). In addition, the central tumor suppressor p53 and several RNA-binding proteins, including KH-type splicing regulatory protein, TAR DNA-binding protein-43, DEAD-box 1, heterogeneous nuclear ribonucleoprotein A1, and breast cancer 1 (BRCA1), have also been identified as regulatory proteins that interact with Drosha complexes and modulate the maturation of specific miRNAs (20, 21, 22, 23, 24, 25, 26). It is likely that regulatory components in the core microprocessor complex select specific miRNAs by controlling miRNA processing.
Although initially identified as a transcription repressor, C-terminal–binding protein (CtBP)–interacting protein (CtIP) is better known for its functions within DNA double-strand break (DSB) processing. Together with the meiotic recombination 11 (MRE11)–ATP-binding cassette—ATPase (RAD50)–Nijmegen breakage syndrome protein 1 (NBS1) (MRN) complex, CtIP efficiently promotes end resection, which generates 3′-end ssDNA filaments to promote homologous recombination (HR)–mediated DSB repair (27, 28). CtIP and its yeast functional homolog, Sae2, harbor structure-dependent endonuclease activity, which is required for cleaning “dirty” DSB ends, R-loop processing, and the stabilization of stalled replication forks, but it is dispensable for end resection of regular DSBs (29, 30, 31, 32, 33). CtIP is a large protein containing several functional domains. The N-terminal region is required for the oligomerization of the protein and interaction with Nbs1 (34, 35, 36). The middle part of CtIP is important for its endonuclease activity and interactions with multiple proteins, including BRCA1, CtBP transcriptional repressor, retinoblastoma-associated protein, and proliferating cell nuclear antigen. Phosphorylations at multiple sites of this region facilitate the capacity of CtIP to promote MRN- and DNA2-mediated DSB end resection and end resection–dependent DSB repair (37, 38, 39, 40). The C terminus of CtIP shares the most similarity with Sae2 and thus was named the Sae2-like domain (27, 41). This region is critical for the regulation of MRN nuclease activity in vitro and used for end resection of some DSBs in vivo (27, 38). However, the mechanisms by which conserved Sae2-like domains contribute to the function of CtIP are still largely unknown.
In this study, we aimed to investigate the functions of CtIP in miRNA post-transcription processing, its association with the Drosha-DGCR8 microprocessor, and its effects in cancer cells. Using immunoprecipitation experiments, we assessed the associations CtIP with Drosha-DGCR8 microprocessor complex proteins and pri-miRNA. We then investigated the effects of knocking out the CtIP gene on colorectal cancer cell miRNA expression and metastasis as well as osteosarcoma cell DSB end processing and repair.
We found that CtIP is involved in Drosha-DGCR8 microprocessor–mediated pri-miRNA processing through its Sae2-like domain. This domain was shown to directly interact with DGCR8 and pri-miRNAs and suppress the Drosha complex processing of a subset of pri-miRNA substrates, including miR-302 family members. The depletion of CtIP or overexpression of miR-302b strongly inhibited the metastasis of human colon cancer cell line (HCT116) colorectal cancer cells in vitro and in an orthotopic xenograft mouse model. The findings suggest that the classical DSB repair factor CtIP promotes the metastatic capacity of tumor cells through the regulation of miRNA maturation.
Results
CtIP is associated with the Drosha-DGCR8 microprocessor complex
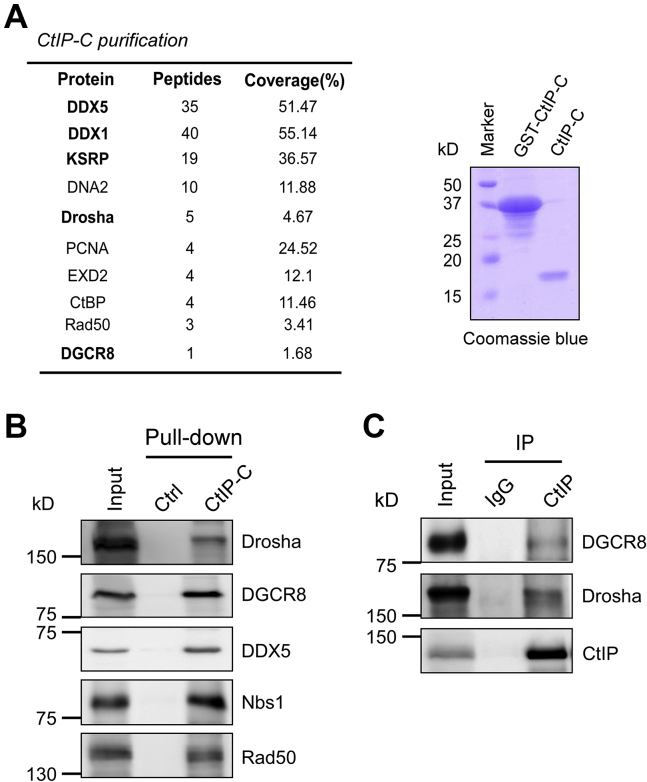
In order to explore the CtIP-associated proteins that specifically interact with its C-terminal conserved Sae2-like domain, high-purity CtIP-C (Sae2-like domain) protein (Fig. 1A) was obtained and coupled with cyanogen bromide–activated Sepharose 4B, and CtIP-C affinity purification was performed using 293T cell lysate. CtIP-C–associated proteins were then subjected to mass spectrometry analysis (Fig. 1A). We found several known CtIP-associated proteins, including DNA2, proliferating cell nuclear antigen, 3'-5' exonuclease domain-containing protein 2, and CtBP. Surprisingly, several Drosha-DGCR8 microprocessor components, such as DDX5, DEAD-box 1, and KH-type splicing regulatory protein, also were copurified (Fig. 1A).
Figure 1.
CtIP interacts with Drosha complex.A, left, mass spectrometry identification of CtIP-C–associated protein. Right, Coomassie blue staining of SDS-PAGE gel indicating purified proteins. B, pull-down experiments were performed by incubating 293T cell lysates with purified GST-CtIP-C fusion protein, followed by immunoblotting using indicated antibodies. C, anti-CtIP immunoprecipitation (IP) was performed, followed by Western blot analysis using indicated antibodies. CtIP, C-terminal-binding protein–interacting protein.
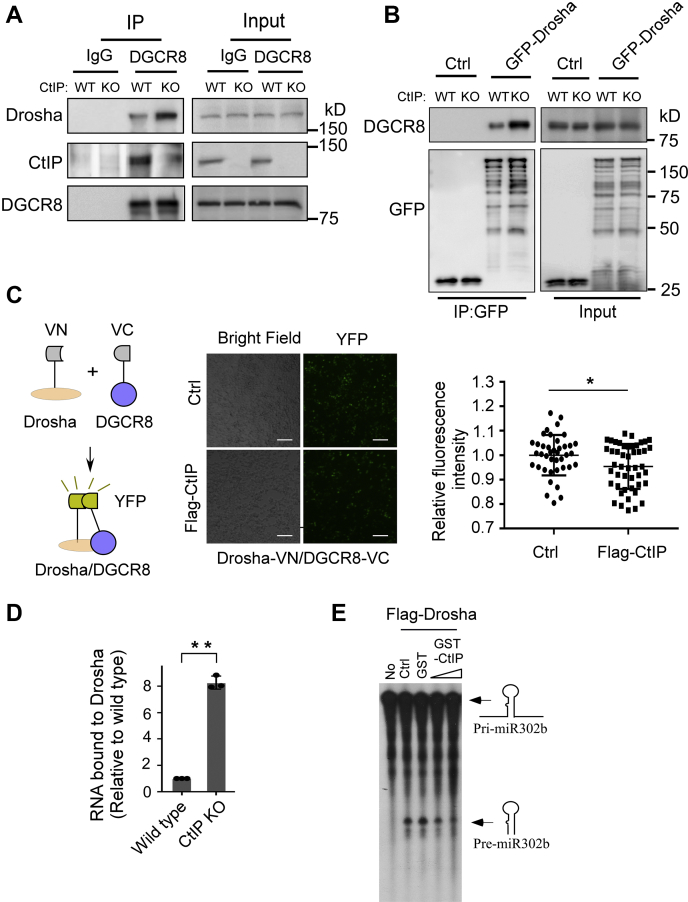
To confirm the association of CtIP with the DGCR8 microprocessor, we performed a regular glutathione-S-transferase (GST) pull-down assay using GST-CtIP-C fusion protein as bait. We again found that CtIP-C interacts with the DSB repair factors Nbs1 and Rad50 and the microprocessor components DGCR8 and DDX5 (Fig. 1B). Furthermore, we performed coimmunoprecipitation experiments to confirm the interactions and found that endogenous Drosha and DGCR8 were present in the CtIP immunocomplex (Fig. 1C). These data indicate that CtIP is a new Drosha-DGCR8–associated protein, and CtIP may have a role in miRNA biogenesis.
CtIP suppresses the expression of a subset of miRNAs
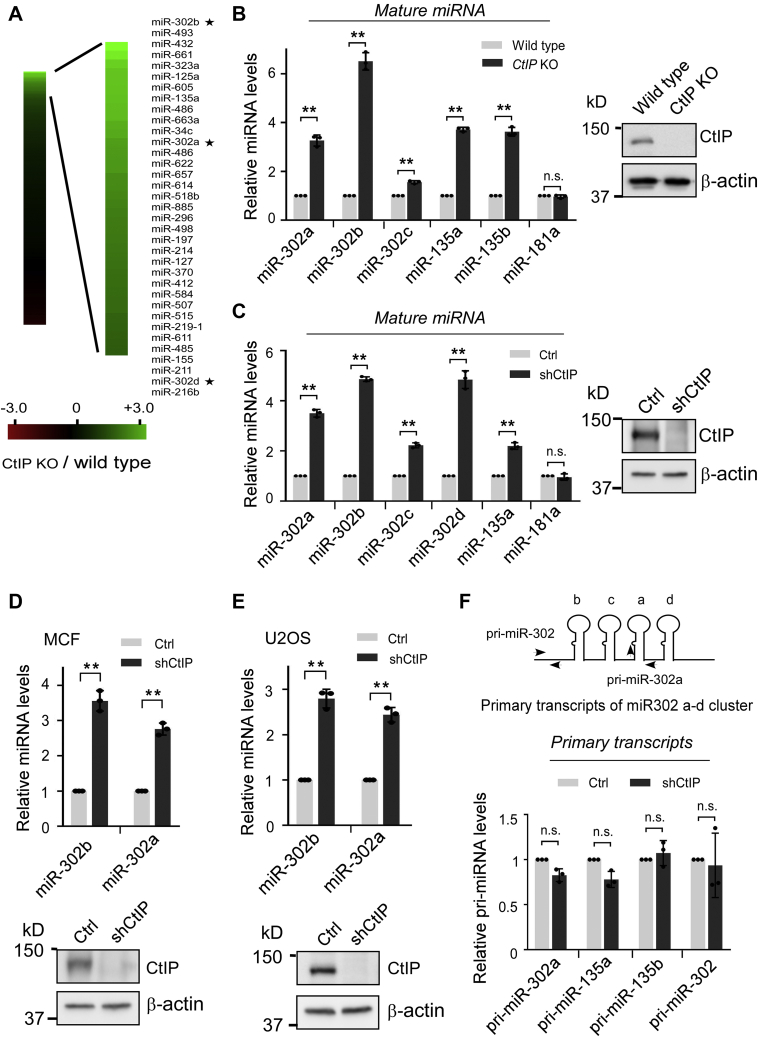
To determine the effect of CtIP on cancer miRNA biogenesis, we analyzed the miRNA expression profiles in wildtype HCT116 and CtIP-deficient HCT116 cells (CtIP-KO) using a human cancer miRNA quantitative PCR (qPCR) array. The depletion of CtIP in HCT116 cells significantly increased the expression of a subset of 49 miRNAs (cutoff greater than 3-fold; Fig. 2A), which suggests CtIP inhibited the expression of these miRNAs.
Figure 2.
CtIP suppresses Drosha-mediated pri-miRNA processing.A, depletion of CtIP promotes the expression of a subset of miRNAs. Total RNAs from control and CtIP-KO cells were subjected to miRNA-profiling analysis using Cancer miRNA quantitative PCR array. Green and red on the heat map indicate a decrease and increase in miRNA levels, respectively, and the color intensity corresponds to relative signal levels. A portion of upregulated miRNAs in CtIP-KO cells is indicated. The stars indicated miR-302 family members. B, expression levels of mature miRNAs in wildtype and CtIP-KO cells were verified by quantitative PCR. C–E, expression levels of indicated miRNAs were examined in HCT116 (C), MCF7 (D), or U2OS (E) cells expressing CtIP shRNA or vector control. Western blots showing CtIP expression. F, expression levels of indicated pri-miRNAs were examined in HCT116 cells expressing CtIP shRNA or vector control. The primer sets used for pri-miR-302 cluster and pri-miR302a detection are shown at the top. In the B–F panel, the data represent the means ± SD of three independent experiments. The p value is indicated as ∗∗p < 0.01. CtIP, C-terminal-binding protein–interacting protein; HCT116, human colon cancer cell line; ns, not significant; MCF7, Michigan Cancer Foundation-7; pri-miRNA, miRNA primary transcripts; U2OS, human osteosarcoma cell line.
We prioritized the miRNAs with the most significant changes and performed a regular qPCR assay to confirm the effects of CtIP on these selected miRNAs. Levels of mature miRNAs, including three miR-302 members (miR-302b, miR-302a, and miR-302d) and miR-135a, were upregulated in CtIP-KO cells (Fig. 2B). A similar effect was observed when CtIP expression was suppressed by shRNAs in HCT116 cells (Fig. 2C) and other common tumor cell lines, including Michigan Cancer Foundation-7 (Fig. 2D) and human osteosarcoma cell line (U2OS) (Fig. 2E). Furthermore, CtIP depletion did not alter the primary transcripts of selected miRNAs (Fig. 2F), suggesting that CtIP regulated the expression of specific miRNAs at the post-transcriptional level. As mentioned earlier, after transcription, the pri-miRNAs are processed by nuclear Drosha-DGCR8 and cytoplasmic DICER1. CtIP is only functional in cell nucleus (Fig. S1A). The expression level of DICER1 is not changed in CtIP-KO cells (Fig. S1B). CtIP may not directly or indirectly regulate maturation of miRNA through DICER1. In addition, pre-miR302b level was also upregulated in CtIP-KO cells (Fig. S1C). We thus thought that CtIP should suppress the levels of specific mature miRNAs through modulation of nucleus processing of these miRNAs.
Direct interaction of CtIP with DGCR8
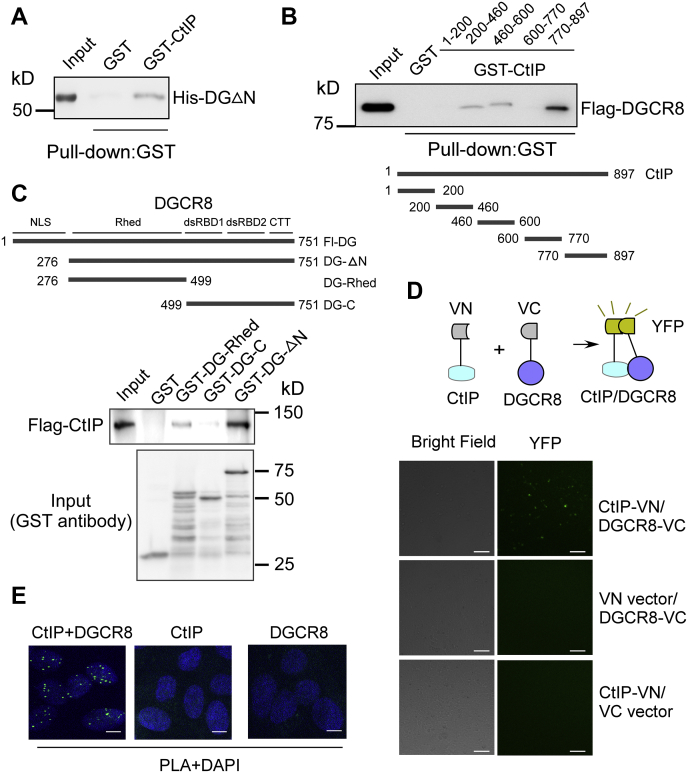
To understand how CtIP regulates miRNA processing, we performed GST pull-down assays using purified full-length CtIP from insect cells to examine the interactions between CtIP and components of the nuclear miRNA-processing complex. We found there was a direct interaction between CtIP and DGCR8 (Figs. 3A and S2). The interaction between CtIP and DGCR8 was then characterized. The central region (200–460 and 460–600 fragments) and the C-terminal 770 to 897 fragment (the Sae2-like domain) of CtIP were shown to interact with DGCR8 (Figs. 3B and S2). DGCR8 contains a nuclear localization signal at the N-terminal region, a central RNA-binding heme domain (Rhed), two double-stranded RNA-binding domains, and a C-terminal tail (42, 43, 44, 45). Using purified proteins, we found that the GST-fused Rhed domain alone (GST-DG-Rhed) and the nuclear localization signal deletion mutant (GST-DG-△N) bound to CtIP (Fig. 3C). These data suggest that the Rhed domain, but not other domains on DGCR8, is important in the interaction between DGCR8 and CtIP.
Figure 3.
Direct interaction of CtIP with GDCR8.A, the direct interaction between CtIP and DGCR8 was shown using a GST pull-down assay. B, pull-down experiments were performed by incubating lysates prepared from insect cells infected with Flag-DGCR8 baculoviruses with different purified GST-CtIP fragments. C, pull-down experiments were performed by incubating lysates prepared from insect cells infected with Flag-CtIP baculoviruses with different purified GST-DGCR8 fragments. D, 293T cells were cotransfected with CtIP-VN and DGCR8-VC. Reconstituted YFP fluorophore was visualized via total internal reflection fluorescence microscope at 24 h after transfection. Top, schematic of BiFC system. Bottom, representative fluorescence images of CtIP and DGCR8 interaction in cells. The scale bars represent 100 μm. E, detection of CtIP–DGCR8 colocalization was performed using a PLA probe in U2OS cells. Representative PLA foci (green) are shown. The scale bars represent 10 μm. CtIP, C-terminal-binding protein–interacting protein; GDCR8, DiGeorge syndrome critical region gene 8; GST, glutathione-S-transferase; U2OS, human osteosarcoma cell line; PLA, proximity ligation assay; VC, Venus C-terminal fragment; VN, Venus N-terminal fragment.
We next used a bimolecular fluorescence complementation (BiFC) reporter to examine the potential interaction of CtIP and DGCR8 in living cells. This reporter relies on protein interactions bringing together ectopically expressed Venus N-terminal (VN-) and Venus C-terminal (VC-) fragments of fluorescent protein reconstitute fluorescence of YFP protein, thus allowing direct visualization of protein interactions in their normal cellular environment. As expected, cotransfection of 293T cells with either of VN-vector and DGCR8-VC or CtIP-VN and VC-vector constructs generated no fluorescence signal. However, cotransfection of 293T cells with CtIP-VN and DGCR8-VC resulted in obvious fluorescence (Fig. 3D), suggesting a direct interaction between CtIP and DGCR8 in cells. To further confirm this finding, we performed proximity ligation assays (in situ PLAs) using antibodies directed against CtIP and DGCR8. As shown in Figure 3E, PLA foci could be readily detected in the nucleus when both CtIP and DGCR8 antibody were used. The PLA signals were not observed in the absence of one primary antibody. These results suggest that CtIP directly interacts with DGCR8 both in vitro and in the cells.
CtIP is a pri-miRNA–binding factor
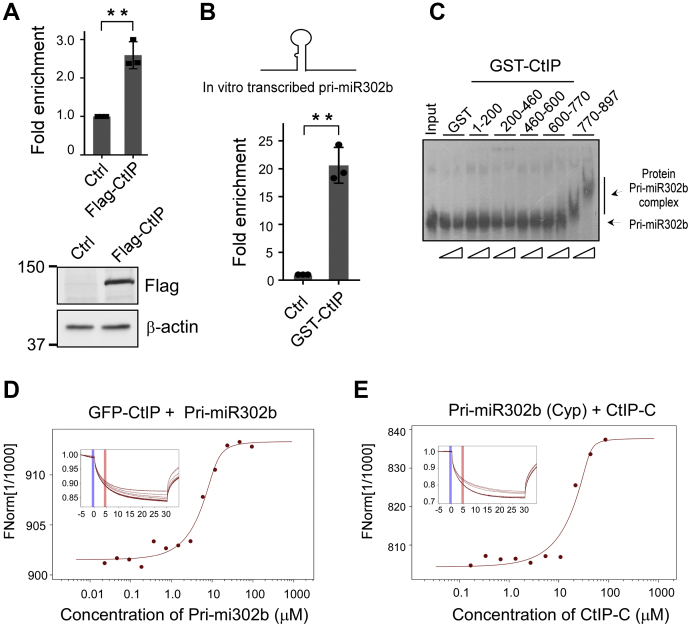
In our previous study, we found that CtIP binds to the hairpin DNA substrate (30). We thus speculated that CtIP also engages with pri-miRNA, which contains a similar secondary structure with a hairpin DNA substrate. As revealed by RNA chromatin immunoprecipitation (RNA-ChIP), overexpressed CtIP was associated with pri-miR302 transcripts in vivo (Fig. 4A). We further performed an RNA pull-down assay using a purified GST-CtIP fusion protein as bait to test whether CtIP directly interacts with pri-miRNA in vitro. We found that the in vitro–transcribed pri-miR302b was efficiently pulled down by purified GST-CtIP (full-length) in comparison with GST protein alone (Fig. 4B). These data indicate the direct binding of CtIP to pri-miR302. We next performed an electrophoretic mobility shift assay (EMSA) assay using Escherichia coli–produced CtIP fragments to map the determinants of the interaction. Intriguingly, pri-miR302 efficiently bound to a region of CtIP 770 to 897, highlighting the importance of the conserved Sae2-like domain of CtIP for pri-miRNA binding. We further confirmed the interaction between CtIP and pri-miR302 using a microscale thermophoresis (MST) assay, which can quantitatively define the interaction between two molecules. Consistent with the RNA pull-down and EMSA data, both GFP-tagged CtIP (GFP-CtIP) with pri-miR302b (Fig. 4D) and Cyp-labeled pri-miR302b with CtIP-C protein (Fig. 4E) exhibited obvious interactions in vitro. The CtIP-C protein yielded a dissociation constant (Kd) of 2.39 μM, which is comparable to the Kd value obtained for full-length CtIP (GFP-CtIP, 3.94 μM). These data support the notion that the conserved Sae2-like domain of CtIP is a secondary structure containing RNA interactor that mediates the interaction between CtIP and pri-miRNA.
Figure 4.
CtIP binds to pri-miRNA via Sae2-like domain.A, anti-Flag RNA-ChIP was performed in HCT116 cells expressing Flag-CtIP or empty vector using pri-miR302 primer sets. The ChIP value in the control was set as 1 for normalization. Data represent the means ± SD of three independent experiments. The p value is indicated as ∗∗p < 0.01. B, in vitro–transcribed pri-miR302b was mixed with bead-immobilized GST-CtIP full-length protein. RNA was eluted from beads and subjected to quantitative RT-PCR analysis using pri-miR302b primer sets. Data represent the means ± SD of three independent experiments. The p value is indicated as ∗∗p < 0.01. C, electrophoretic mobility shift assay was performed with GST alone or indicated GST-CtIP variants using an in vitro–transcribed 32P-labeled pri-miR302b probe. The band shifts resulting from specific binding to the CtIP variant are shown. D and E, microscale thermophoresis analysis showing pri-miR302b interacted with CtIP in vitro. Titration of cell lysates containing pri-miR302b in vitro–transcribed to a constant amount of GFP-CtIP or purified CtIP-C protein to Cyp-labeled pri-miR302b induced a pronounced microscale thermophoresis signal change and yielded Kd = 3.94 μM (D) and Kd = 2.39 μM (E), respectively. ChIP, chromatin immunoprecipitation; CtIP, C-terminal-binding protein–interacting protein; GST, glutathione-S-transferase; HCT116, human colon cancer cell line; pri-miRNA, miRNA primary transcripts.
CtIP reduces the binding of Drosha to DGCR8 and RNA substrate
The direct interaction of CtIP with DGCR8 and pri-miRNA substrates suggests that CtIP may interfere with the binding of DGCR8 or pri-miRNA substrate to Drosha, which is the core RNase III enzyme in the microprocessor. We compared the capacity of Drosha to bind to DGCR8 and pri-miR302 in the presence or absence of CtIP. Consistent with previous reports, we found that endogenous Drosha or overexpressed GFP-Drosha interacted with DGCR8 in HCT116 cells (Fig. 5, A and B). Interestingly, when CtIP was knocked out, the interactions increased (Fig. 5, A and B), suggesting that endogenous CtIP hinders the interaction between Drosha and its cofactors. In line with these results, ectopic expression of Flag-CtIP reduced the YFP fluorescent intensity reconstituted by cotransfection of Drosha-VN and DGCR8-VC in 293T cells (Fig. 5C). These data support a notion that CtIP may interfere with the binding of Drosha to DGCR8.
Figure 5.
CtIP weakened the binding of Drosha with DGCR8 and RNA substrates.A, coimmunoprecipitation of Drosha and DGCR8 was performed in wildtype HCT116 cells and CtIP-KO cells using anti-DGCR8 antibody. B, coimmunoprecipitation between GFP-Drosha and DGCR8 was performed in wildtype HCT116 cells and CtIP-KO cells expressing GFP-Drosha or empty vector using anti-GFP antibody. C, 293T cells cotransfected with Drosha-VN and DGCR8-VC, along with Flag-CtIP or control vector, fluorescent images were taken at 24 h after transfection. The scale bars represent 100 μm. The chart showed the cell fluorescence quantified by ImageJ. Data represent the means ± SD. The p value is indicated as ∗p < 0.05. D, RNA-ChIP analysis of association between Drosha and pri-miR302b was performed in wildtype HCT116 cells and CtIP-KO cells using anti-Drosha antibody and pri-miR302b primer sets. The ChIP value in wildtype cells was set as 1 for normalization. Data represent the means ± SD of three independent experiments. The p value is indicated as ∗∗p < 0.01. E, in vitro pri-miRNA processing assay was performed by incubating pri-miR302b substrate with immunoprecipitated Flag–Drosha complex in the presence or absence of recombinant GST-CtIP protein. ChIP, chromatin immunoprecipitation; CtIP, C-terminal-binding protein–interacting protein; DGCR8, DiGeorge syndrome critical region gene 8; HCT116, human colon cancer cell line; pri-miRNA, miRNA primary transcripts; VC, Venus C-terminal fragment; VN, Venus N-terminal fragment.
Next, we performed RNA-ChIP using Flag-Drosha to analyze the relationship between Drosha and RNA substrates. The data indicated that CtIP depletion obviously increased the association between Drosha and pri-miR302 (Fig. 5D). The attenuated binding of Drosha to a substrate might suppress its enzyme activity. Therefore, we used an in vitro pri-miRNA processing assay to assess the effect of CtIP on Drosha activity. Clearly, purified GST-CtIP but not GST alone efficiently inhibited the processing activity of Drosha on pri-miR302b (Fig. 5E). Taken together, our data suggest that CtIP reduces the binding of Drosha to its cofactor DGCR8 and pri-miRNA substrates and suppresses the processing of pri-miRNAs by Drosha.
CtIP does not promote HR by modulating biogenesis of miRNAs
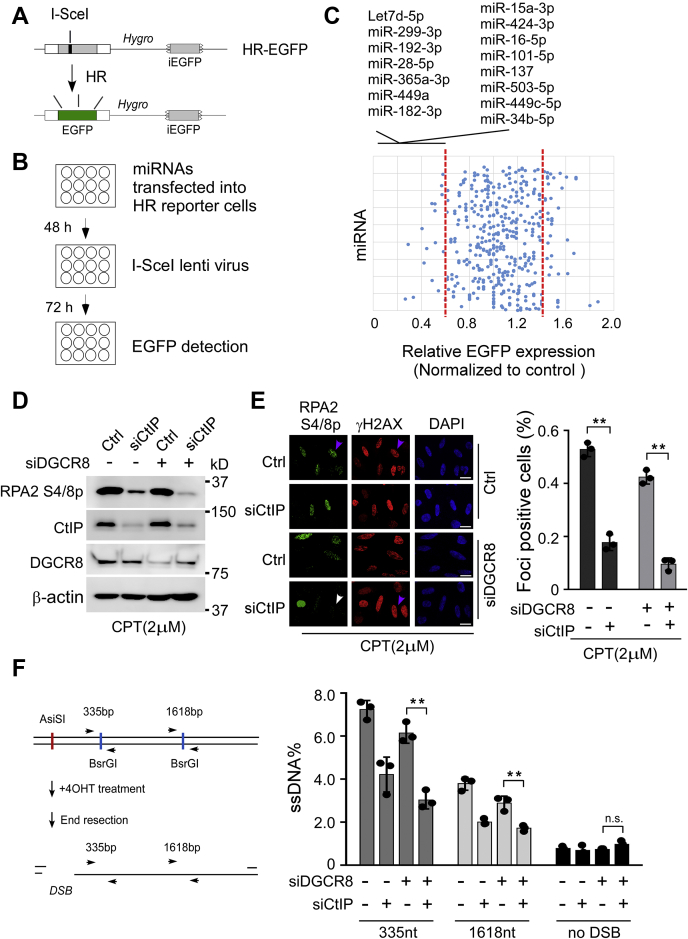
CtIP is a classical DSB repair factor. CtIP-deleted cells display severe DSB end processing defects and are hypersensitive to topoisomerase 1 poisons such as camptothecin. However, the mechanisms by which CtIP is involved in DSB end processing and repair remain unclear. We aimed to ascertain whether CtIP promotes DSB repair by modulating the processing of miRNAs. Initially, we used a cancer miRNA qPCR array containing roughly 400 cancer-related miRNAs to search for CtIP-dependent miRNAs. We synthesized all miRNA mimics on the array, transfected them into U2OS cells containing a well-characterized enhanced GFP (EGFP)-HR reporter (Fig. 6A), induced DSB by expression of I-SceI endonuclease, and examined the effects of these miRNAs on HR-mediated DSB repair (Fig. 6B). Our results show that 15 miRNAs had at least a 40% impact on HR efficiency in the cells (Fig. 6C). However, none of them belong to the CtIP-dependent miRNAs group listed in Figure 2A (Fig. 6C).
Figure 6.
CtIP-dependent miRNAs are not essential for HR.A, diagram showing the function of the EGFP-HR DSB repair reporter. B, schematic of HR assays workflow. U2OS cells carrying EGFP-HR reporter were transfected with synthesized miRNA mimics; DSB was induced by I-SceI–containing lentivirus, and EGFP-positive signals were analyzed by flow cytometry (fluorescence-activated cell sorting). C, scatter plot showing the relative levels of EGFP-HR reporter after all individual miRNAs mimicked transfection. HR value in miRNA control mimic-transfected cells was set as 1 for normalization. The dotted line represents the cutoff used to determine decreased and increased HR. MiRNAs that suppressed HR efficiency are indicated at the top. D–F, end resection function of CtIP is DGCR8 independent. D and E, U2OS cells were transfected with indicated siRNA or siRNA combination, treated with CPT (2 μM) for 1 h followed by RPA phosphorylation analysis by Western blotting (D) and RPA foci formation analysis by immunostaining (E) with indicated antibodies. E, the pink and white arrows indicate representative RPA2 and γH2AX foci-positive and foci-negative cells, respectively. The scale bars represent 10 μm. The percentage of RPA2 foci–positive cells among γH2AX-positive cells for each sample is shown. Data shown represent the means of three independent experiments, with error bars as SD. The p value is indicated as ∗∗p < 0.01. F, schematic and quantification of a quantitative PCR–based cellular resection assay of ER-AsiSI U2OS cells transfected with indicated siRNA or siRNA combination. Data represent the means ± SD of three independent experiments. The p value is indicated as ∗∗p < 0.01. CtIP, C-terminal-binding protein–interacting protein; DGCR8, DiGeorge syndrome critical region gene 8; DSB, DNA double-strand break; EGFP, enhanced GFP; HR, homologous recombination; n.s., not significant; RPA, replication protein A; U2OS, human osteosarcoma cell line.
We next sought to assess whether CtIP promotes DSB end resection by modulating DGCR8- or DGCR8-dependent miRNAs. Consistent with previous reports (27, 28), we observed end resection defect in CtIP-depleted cells, as revealed by diminished replication protein A (RPA) phosphorylation, RPA foci formation, or a more accurate assay in which resection at specific AsiSI restriction-enzyme sites can be quantified by qPCR (46) (Fig. 6, D–F). SiRNA-mediated depletion of DGCR8 also impairs end resection. This may reflect the role of Drosha in end resection (47). Significantly, depletion of CtIP strongly affects end resection in both control and DGCR8 depletion background (Fig. 6, D–F). Together, these results suggest that CtIP should not promote end resection or HR through modulation processing of miRNAs.
CtIP promotes the metastasis of colon cancer cells
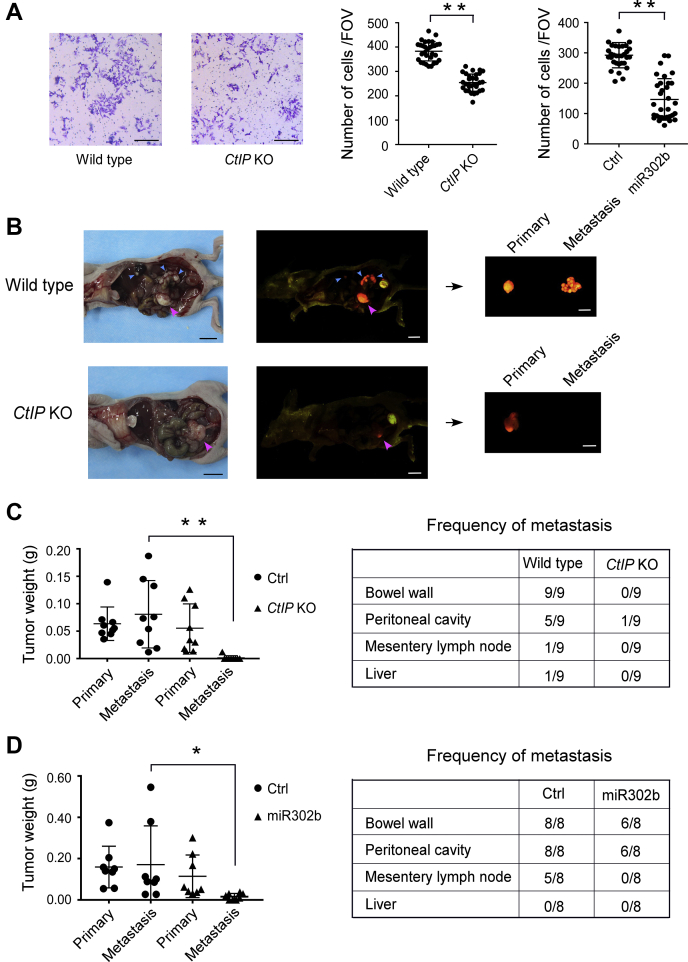
Recent reports indicated that miR-302 family members are downregulated in several cancers, and the overexpression of these miRNAs inhibits the invasion and metastasis of cancer cells (48, 49, 50, 51). We performed a Matrigel invasion assay to evaluate whether CtIP and miR-302 members are involved in the ability of HCT116 cells to migrate. We found that the ectopic expression of miR-302b or depletion of CtIP in HCT116 cells significantly reduced cell invasion activity (Fig. 7A). These in vitro data raised the possibility that CtIP promotes the metastasis of HCT116 colon cancer cells via some specific miRNAs, such as miR-302 family members, in vivo. We thus used a surgical orthotopic implantation model to examine the roles of CtIP and miR-302b in the metastasis of HCT116 colorectal cancer cells in vivo. After orthotopic implantation, in vivo fluorescence images for all mice were acquired, and fluorescence intensity over time was calculated by the in vivo fluorescence imaging system. These data roughly represent the progression of tumors in nude mice. On 42 days after orthotopic implantation, the fluorescence in the wildtype HCT116 tumor group was widespread; however, the fluorescence in the CtIP-KO tumor group was obviously less compared with the wildtype group (Fig. S3, top panel). Fluorescence intensity from wildtype tumor group was also significantly higher compared with that from CtIP-KO tumor group (Fig. S3, bottom panel). Wildtype HCT116 tumors grew faster than CtIP-KO tumors in the mice. On day 42, the mice with wildtype tumors were sacrificed, and a complete necropsy was performed to check for primary tumors and metastasis (Fig. 7B). We observed profound metastasis of wildtype HCT116 tumors, which frequently metastasized to common metastatic sites, including the bowel wall, peritoneal cavity, mesenteric lymph nodes, and liver (Fig. 7, B and C). On day 66, the mice with CtIP-KO tumors were sacrificed and inspected, and the primary tumors in these mice were comparable with those in mice with wildtype tumors. However, surprisingly, only a small number of peritoneal cavity metastatic sites were found in these mice (Fig. 7, B and C). These data suggest that depletion of CtIP strongly suppressed the metastasis of colon cancer cells in the mouse model. In the CtIP-KO tumors, the levels of miR-302 family members are also upregulated (Fig. S4A). This is consistent with the finding in the cell lines. We next overexpressed miR-302b in HCT116 cells and conducted similar assays to detect the effects of this CtIP-dependent miRNA on metastasis in the mouse model. As shown in Figure 7D, as expected, the overexpression of miR-302b resulted in an obvious decrease in metastatic tumor weight and frequency of metastasis in the mouse model. We expressed miR-302b inhibitor in CtIP-KO cells and found that miR-302b suppression weakly promoted metastasis of CtIP-KO tumors (Fig. S4B).
Figure 7.
Depletion of CtIP or overexpression of miR-302b suppresses invasion and metastasis of colon cancer cells.A, Matrigel invasion assays were performed using the indicated cells. Left, representative micrographs for the CtIP-KO and wildtype cells. The scale bars represent 100 μm. Right, quantification of the numbers of invasive cells per field of view (FOV). Data represent mean ± SD. The p values are indicated as ∗∗p < 0.01. B, left, representative photographs and corresponding fluorescence images of orthotopic RFP colon colorectal cancer nude-mouse model autopsies of the control and CtIP-KO groups. Pink and blue arrows indicate primary and metastatic tumors, respectively. Right, fluorescence images of collected primary and metastatic tumors. The scale bars represent 1 cm. C, quantification of tumor weights (left) and frequency of metastases (right) in control and CtIP-KO groups. Data represent mean ± SD. The p values are indicated as ∗∗p < 0.01. D, quantification of tumor weights (left) and frequency of metastases (right) in control and miR-302b–overexpression groups. Data represent mean ± SD. The p values are indicated as ∗p < 0.05. CtIP, C-terminal-binding protein–interacting protein.
Discussion
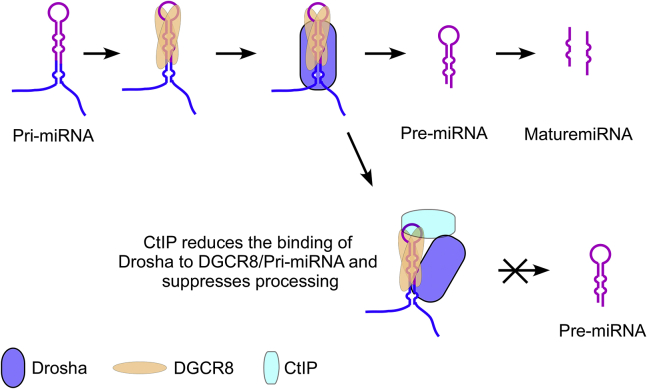
CtIP is a classic DSB repair factor. Our current study revealed a novel function for CtIP in the regulation of miRNA processing. CtIP was found to modulate the post-transcriptional maturation of a subset of miRNAs via the Drosha-DGCR8 microprocessor. The mechanisms by which the Drosha-DGCR8 microprocessor is assembled and facilitates nuclear miRNA processing remain poorly understood. Drosha alone has weak RNA-binding capacity and processing activity but is thought to work together with DGCR8 to recognize substrates and achieve high activity levels (52, 53). We have shown that CtIP directly binds to both DGCR8 and pri-miRNA substrates and reduces Drosha binding affinities to DGCR8 and specific pri-miRNA substrates. We thus propose a model to explain our findings (Fig. 8). Some of DGCR8/Drosha/pri-miRNA complexes such as DGCR8/Drosha/pri-miR-302 may form a unique structure that engages and traps CtIP. CtIP binding might hinge the tertiary structure of DGCR8/Drosha/pri-miRNA, therefore interfering Drosha binding and catalysis. It is possible that only the small percentage of DGCR8/Drosha/pri-miRNA structure can engage and trap CtIP. That could be the reason that only a subset of miRNA is modulated by CtIP. Clearly, further research is needed to clarify the mechanism details.
Figure 8.
A model description of the role of CtIP in pri-miRNA processing (seeDiscussionsection). CtIP, C-terminal-binding protein–interacting protein; pri-miRNA, miRNA primary transcripts.
BRCA1 is a close partner of CtIP and collaborate to promote DSB repair and maintain genome stability (28, 54). As was previously reported, BRCA1 also associates with the Drosha microprocessor complex and regulates the processing of specific pri-miRNAs (26). However, in contrast to CtIP, BRCA1 enhances the processing activity of Drosha (26). Therefore, the functions of CtIP and BRCA1 in miRNA processing should be mutually independent, which is distinct from the established interactions of these two factors in the repair of DSB. Recent biochemistry and structural biology studies indicated that the microprocessor is a heterotrimeric complex consisting of one Drosha and two DGCR8 molecules (52, 55, 56, 57). The Rhed domains of DGCR8 nonspecifically interact with the terminal loop and adjacent regions of pri-miRNA substrates to ensure processing accuracy and enhance efficiency (43, 52, 53, 56). BRCA1 recognizes the root of the stem–loop of pri-miRNAs, which is distant from the terminal loop (26). CtIP directly binds to both pri-miRNAs and the Rhed of DGCR8 (Figs. 3C and 4B). We therefore reasoned that the recognition regions for CtIP and BRCA1 would be different on pri-miRNA. The Rhed domain of DGCR8 and the terminal loop region of some specific pri-miRNAs might form a CtIP-favoring docking structure to facilitate the recruitment of CtIP and prevent Drosha-mediated processing and therefore has a different role than BRCA1. This is in line with the functions of BRCA1 and CtIP in stabilizing arrested replication forks. BRCA1 protects stalled forks from MRE11-dependent degradation, but CtIP defends perturbed forks from over-resection by DNA2 (31).
Our data revealed the binding capacity of CtIP to pri-miRNA, and this raises an interesting question of whether CtIP interacts with other RNA molecules and is involved in other RNA-related molecular processes, such as transcription-associated DNA damage and repair. It was recently shown that Sae2/CtIP plays important roles in R-loop processing at sites of stalled transcription. The cleavage of 5′-flaps by the endonuclease activity of Sae2/CtIP is required for the stabilization and removal of nascent R-loop–initiating structures in eukaryotic cells (32). Previous in vitro work on CtIP indicated that the CtIP N-terminal fragment, but not the C-terminal conserved Sae2-like domain, has endonuclease activity on Y-shaped DNA substrates (29, 30). It is important to determine if the RNA-binding capacity of the Sae2-like domain of CtIP indicated in this study also plays a role in R-loop recognition and processing.
One important finding of this study is that CtIP-deficient colorectal cancer cells have a reduced ability to initiate metastasis. Metastasis formation is a complex and multistep process that is probably regulated by multiple mechanisms. We have provided data to show that CtIP-dependent miRNAs, such as miR-302b, suppress the metastasis of HCT116 cancer cells. However, this does not necessarily mean all the effects CtIP has on metastasis operate via miRNAs. MiR-302b inhibitor just weakly promotes metastasis of CtIP-KO tumors (Fig. S4B), suggesting that suppression of metastasis by CtIP depletion in mice is not only mediated by miR302b. Other CtIP-dependent miRNAs or other cellular functions of CtIP may also contribute to that. Overexpression of miR-302b may cause some artificial effects, such as produce too much miR-302b in cells, which are reasons for inhibition of metastasis. Indeed, Mre11, a close partner of CtIP, is also able to promote the migration and invasion of breast cancer cells (58). In some circumstances, CtIP may work together with Mre11 to regulate metastasis, as they do in DSB repair, and further studies are required to dissect these issues. These areas have the potential to offer new opportunities for the development of novel therapeutics for tumors based on targeting these factors.
Experimental procedures
Cell culture and antibodies
Human HCT116, 293T, Michigan Cancer Foundation-7, and U2OS cells were cultured at 37 °C in a humidified atmosphere with 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics. The Sf9 insect cell line was cultured in Sf-900II serum-free medium (Gibco). CtIP-KO HCT116 cells were generated and cultured as previously described (59, 60). The antibodies used against CtIP, RPA, Nbs1, and Rad50 were used as described previously (59, 60, 61). Anti-FLAG M2 (F1804) and anti-β-actin (A5441) were purchased from Sigma. Anti-γH2AX was from Millipore, Anti-Drosha and Anti-DGCR8 were from Bethyl, and Anti-GFP was from Santa Cruz.
Plasmid construction, recombinant protein expression, and shRNA interference
Both pCK-Drosha-FLAG and Flag-DGCR8 were gifts from Dr Narry Kim (7, 45). Plasmids for BiFC assays were gifts from Dr Baohua Liu (62). Plasmids for CtIP, Drosha, or DGCR8 complementary DNA (cDNA) clones were subcloned into the mammalian expression vector pcDNA3.0 or NBLV0051 (Novo Bio) containing a 3× N-terminal Flag. EGFP-tagged Drosha was generated using the EGFP-C1 expression vector (Clontech). Bacteria expressing GST-tagged CtIP or DGCR8 variants were generated using the pGEX6T-1 system and affinity-purified with Glutathione Sepharose 4B (GE Healthcare). Recombinant baculoviruses expressing GST-tagged CtIP were generated using the Bac-to-Bac baculovirus expression system (Invitrogen) as previously described (30, 34, 37, 60). Mir-302b inhibitor expression plasmid was from GeneCopoeia.
Endogenous CtIP was silenced by lentiviral infection using pLKO vectors expressing CtIP shRNAs. The CtIP shRNA sequence was used as described previously (30, 60).
MiRNA microarray profiling and validation
Total RNA from cells was prepared using TRIzol reagent (Invitrogen) and reverse transcribed with an All-in-One miRNA First-Strand cDNA Synthesis Kit (GeneCopoeia) according to the manufacturer's protocol. The differential expression of cancer-related miRNAs in wildtype HCT116 cells and CtIP-KO HCT116 cells was analyzed using the miProfile Human Cancer miRNA qPCR Array according to the manufacturer's instructions (GeneCopoeia). Expression of the specific miRNAs was further validated using the All-in-One miRNA qRT-PCR Detection kit (GeneCopoeia), and qRT-PCR was performed on the Bio-Rad IQ5 Real-Time PCR system. U6 or GAPDH was used as the endogenous control to normalize miRNA expression. The gene-specific primers used in this study were purchased from GeneCopoeia (Table S1).
CtIP-C affinity purification
GST-fused CtIP Sae2-like domain (residues 770–897, CtIP-C) was expressed in BL21(DE3) and affinity purified with Glutathione Sepharose 4B. The GST tag was cleaved by the PreScission Protease (GenScript) on the beads. High-purity CtIP-C protein was collected, the buffer was changed, and it was coupled with cyanogen bromide–activated Sepharose 4B according to the manufacturer's instructions (GE Healthcare). The 293T cells were lysed in NETN (150 mM NaCl, 1 mM EDTA, 20 mM Tris–HCl [pH 8.0], and 0.5% Nonidet P-40) buffer containing a protease inhibitor cocktail (“PIC”; cOmplete; Roche). The cell lysate was cleared by centrifugation, and the supernatant was incubated with the homemade CtIP-C–Sepharose 4B affinity beads pre-equilibrated with the NETN buffer with PIC. After incubation at 4 °C, the beads were centrifuged and washed extensively with NETN buffer. The bound proteins were analyzed by mass spectrometry.
Immunoblotting, immunoprecipitation, and in vitro binding assay
Whole-cell lysis, coimmunoprecipitation, and Western blotting were performed as described previously (28, 34). The cells were lysed in NETN buffer containing PIC. Immunoprecipitation was performed by incubating primary antibodies with precleared cell lysates at 4 °C for 4 h, followed by the addition of protein A/G agarose for 1 h. For the in vitro binding assay, GST-fused CtIP or DGCR8 fragments were expressed in E. coli and purified using Glutathione Sepharose. In vitro binding was performed in NETN buffer.
In vitro transcription
In vitro transcription was carried out using RiboMAX Large Scale RNA Production System according to the manufacturer's instructions (Promega). A pCMV-miR vector containing the pri-miRNA coding sequence (Origene) was linearized by XhoI endonuclease digestion. Linearized DNA template was transcribed in vitro, and α-32P-UTP or Cyp-UTP was added to the transcription reaction generate internally [32P] or Cyp-labeled pri-miRNA substrate. The DNA template was removed from the transcribed product by RNase-free DNase followed by phenol extraction and ethanol precipitation. The 32P-pri-miRNA was further purified using denaturing PAGE, then extracted, denatured by heating, and renatured by gradually decreasing the temperature. The final product was ethanol precipitated, dissolved in RNase-free water, and stored at −20 °C.
RNA pull-down assay
RNA pull-down assay was performed as previously described (26). Briefly, insect cells were used to produce GST-CtIP fusion protein, which was bound to Glutathione Sepharose beads, washed, resuspended, and incubated with in vitro–transcribed pri-miRNA for 1 h at 4 °C. After extensive washing, the bound RNA was eluted in elution buffer (1% SDS and 150 mM NaCl) and purified by TRIsure reagent (Invitrogen). RNA was then reverse transcribed into cDNA, followed by qPCR to detect the relative levels of RNA binding to GST-CtIP. The primer sequences used are shown in Table S2.
EMSA and MST assay
EMSA was performed with the purified GST-CtIP fragments as described previously (30). Pri-miRNA and protein complexes were resolved on native 6% polyacrylamide gels and visualized by autoradiography.
The MST assay was performed as described previously (63, 64). Briefly, Cyp-labeled pri-miRNA302b or cell extract containing GFP-labeled CtIP or Drosha was mixed with twofold serial dilutions of the indicated proteins or pri-miRNA302b in MST buffer (100 mM NaCl, 2% glycerine, 20 mM Tris–HCl, and pH 7.5). The samples were loaded into silica capillaries (Polymicro Technologies). Measurements were performed at 25 °C using a Monolith NT.115 from NanoTemper Technologies. The data were analyzed with NanoTemper Analysis software, version 1.2.101 (NanoTemper Technologies).
RNA-ChIP
RNA-ChIP was performed as described previously with some modifications (18, 22). Briefly, cells were crosslinked with 1% formaldehyde for 10 min, and the reaction was stopped with glycine. After washing, the cell pellet was resuspended in buffer A (5 mM Pipes, pH 8.0, 85 mM KCl, and 0.5% Nonidet P-40) and sat on ice for 10 min. The crude nuclei fraction was then isolated by centrifugation, suspended in buffer B (1% SDS, 10 mM EDTA, 50 mM Tris–HCl, and pH 8.1), and disrupted by sonication. The lysates were cleared and subjected to immunoprecipitation with anti-Flag antibody (Sigma F1804). After washing and elution, the RNA was purified using TRIzol (Invitrogen), resuspended in buffer (50 mM Tris–HCl, pH 7.5, and 10 mM MgCl2) with RNase inhibitor and DNase I, and incubated for 30 min at 37 °C. After phenol/chloroform extraction, the RNA was precipitated with ethanol and dissolved in RNase-free water. An aliquot of RNA was reverse transcribed into cDNA, and qPCR analysis was performed. The primer sequences used are shown in Table S2.
In vitro pri-miRNA processing analysis
An in vitro pri-miRNA processing assay was performed as described previously with slight modifications (6, 65). Briefly, Flag-tagged Drosha was transiently transfected into 293T cells and immunoprecipitated using anti-FLAG M2-agarose (Sigma). Immunoprecipitates were extensively washed and mixed with in vitro transcribed internal 32P-labeled pri-miRNA in reaction buffer containing 32 mM MgCl2, 10 mM ATP, and 200 mM creatine phosphate. The reaction mixture was incubated at 37 °C for 90 min, and after phenol/chloroform extraction, the RNA was precipitated with ethanol and dissolved in RNase-free water. The RNA was loaded onto 8% denaturing urea polyacrylamide gels and visualized by autoradiography.
HR assay
EGFP-based HR repair substrates were used as previously described (30, 34, 60). Cell lines carrying repair substrates were induced with HA-I-SceI by lentiviral infection. After 72 h, the cells were trypsinized and collected for fluorescence-activated cell sorting analysis of EGFP-positive events using a BD Accuri C6 flow cytometer and accompanying analysis software.
Transwell assays
Transwell 24-well filters (Corning) were used for in vitro cell invasion ability analyses following the manufacturer's protocol. Transwell membranes were coated with Matrigel (BD), and serum-free medium was added to the upper chamber wells. The bottom chamber of the Transwell plate contained growth medium with 10% FBS. For the invasion assay, cells were seeded into the upper chamber and allowed to pass through the Matrigel at 37 °C with 5% CO2 for 24 h. The cells on the upper surface of the filter were removed with a cotton swab. The cells that reached the lower surface were fixed with 4% formaldehyde and stained with Giemsa.
Chromatin fractionation
Chromatin fractionation was performed as described (66). Briefly, cells were washed with cold PBS and suspended in buffer A (10 mM Pipes [pH 7.0], 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, protein inhibitor cocktail [Roche; EDTA-free], and 0.7% Triton X-100). After 30 min incubation on ice, the lysate was centrifuged at 20,000g for 10 min at 4 °C. The supernatant (soluble fraction: cytosol) was collected and kept on ice. The pellet was washed with cold PBS, suspended in buffer A, and sonicated for four pulses of 10 s at 30% amplitude with 10 s resting on ice. This sonicated solution is the chromatin fraction (Nuclear).
BiFC assay
Human embryonic kidney 293T cells cultured in 6-well plates were transfected with appropriate plasmids for each BiFC assay. Fluorescence images were captured using an invert total internal reflection fluorescence microscope at 24 h after plasmids transfected. And the fluorescence intensity was measured by ImageJ (National Institutes of Health).
In situ PLA
Cells were pre-extracted for 5 min on ice and fixed in 2% formaldehyde in PBS (w/v) for 20 min on room temperature. In situ PLA was performed using Duolink PLA technology (Sigma–Aldrich) according to the manufacturer's instructions. Briefly, coverslips were blocked for 30 min at 37 °C and incubated with the respective primary antibodies for 1 h at room temperature. Upon washing the coverslips twice in PBS for 5 min, anti-Mouse PLUS and anti-Rabbit MINUS PLA probes (Sigma–Aldrich) were coupled to the primary antibodies for 1 h at 37 °C. After three wash steps in buffer A (0.01 M Tris, 0.15 M NaCl, and 0.05% Tween-20) for 5 min, PLA probes were ligated for 30 min at 37 °C. Coverslips were then washed three times 5 min in buffer A. Amplification using the “Duolink In Situ Detection Reagents Green” (Sigma–Aldrich) was performed at 37 °C for 100 min. After amplification, coverslips were washed twice in buffer B (0.2 M Tris and 0.1 M NaCl) for 10 min and once in 0.01× buffer B for 1 min. Finally, coverslips were mounted using Vectashield Mounting Media (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole, sealed, and imaged on an Olympus IX81 FL microscope.
Orthotopic xenograft mouse models
All animal studies were conducted with the approval of the China Committee for Research and Animal Ethics in compliance with the guidance on experimental animals. All animal experiments were conducted at Anticancer Biotechnology Co, Ltd.
A modified surgical orthotopic implantation method, termed the tumor-sealing method, was used to study the metastasis of colorectal cancer in animal models, which is a more patient-like model than those previously used (67, 68). HCT116 colon cancer cells were labeled with GFP or red fluorescent protein following a well-established protocol for more accurate detection by fluorescence imaging (67). Details of the tumor-sealing method protocol were described previously (67). Briefly, GFP- or red fluorescent protein–labeled HCT116 cells were cultured, harvested, and injected into the subcutaneous layer on the right flank of 6-week-old female BALB/c-nu mice. Subcutaneous tumors were allowed to grow for several weeks until large enough for orthotopic implantation. Each subcutaneous tumor was resected aseptically and scissor minced into fragments of about 1 mm in diameter in RPMI medium. The tumor fragment was sutured to a partially torn serosa of the cecum, and the blind end of the cecum was folded over the tumor fragment and sealed with sutures. The wounds were closed using 6-0 nylon sutures. Then, in vivo fluorescence images were acquired using a FluorVivo imaging system. The mice were sacrificed at the indicated time, and a complete necropsy was performed. We recorded details of all macroscopic and microscopic tumor deposits in the liver, lung, lymph nodes, and peritoneal carcinomatosis. The primary tumors and metastatic tumors found in various organs were excised and weighed. The data are represented by the mean ± SD.
Data availability
All data generated and analyzed during this study are included in this published article and its supplementary information files.
Supporting information
This article contains supporting information (1).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Dr Narry Kim (Institute for Basic Science, Seoul) and Dr Baohua Liu (Shenzhen University School of Medicine) for kindly providing valuable reagents. We also thank Dr Xumin Zhang (Fudan University) and Dr Guohua Jiang (Beijing Normal University) for helping in mass spectrometry and MST analyses, respectively.
Author contributions
H. W., J. R., and Y. W. designed and performed experiments and analyzed the data; Y. W., Y. Z., Y. L., S. H., and S. Z. performed experiments and analyzed the data; X. X. gave valuable input to the article; and H. W. and J. R. wrote the article.
Funding and additional information
This work was supported by the National Natural Science Foundation of China (grant numbers: 31971221 and 31370841) and the Beijing Natural Science Foundation (grant number: 5182003) awarded to H. W. and the National Natural Science Foundation of China (grant numbers: 31761133012 and 31530016) and the National Basic Research Program of China (grant number: 2017YFA0503900) to X. X.
Edited by Ronald Wek
Supporting information
References
- 1.Ghildiyal M., Zamore P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel D.P. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huntzinger E., Izaurralde E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 4.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Kim V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 8.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lund E., Guttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 11.Ketting R.F., Fischer S.E., Bernstein E., Sijen T., Hannon G.J., Plasterk R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 14.Hata A., Kashima R. Dysregulation of microRNA biogenesis machinery in cancer. Crit. Rev. Biochem. Mol. Biol. 2016;51:121–134. doi: 10.3109/10409238.2015.1117054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davalos V., Esteller M. MicroRNAs and cancer epigenetics: A macrorevolution. Curr. Opin. Oncol. 2010;22:35–45. doi: 10.1097/CCO.0b013e328333dcbb. [DOI] [PubMed] [Google Scholar]
- 16.Merritt W.M., Lin Y.G., Han L.Y., Kamat A.A., Spannuth W.A., Schmandt R., Urbauer D., Pennacchio L.A., Cheng J.F., Nick A.M., Deavers M.T., Mourad-Zeidan A., Wang H., Mueller P., Lenburg M.E. Dicer, Drosha, and outcomes in patients with ovarian cancer. New Engl. J. Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nemlich Y., Greenberg E., Ortenberg R., Besser M.J., Barshack I., Jacob-Hirsch J., Jacoby E., Eyal E., Rivkin L., Prieto V.G., Chakravarti N., Duncan L.M., Kallenberg D.M., Galun E., Bennett D.C. MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth. J. Clin. Invest. 2013;123:2703–2718. doi: 10.1172/JCI62980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis B.N., Hilyard A.C., Lagna G., Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis B.N., Hilyard A.C., Nguyen P.H., Lagna G., Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol. Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guil S., Caceres J.F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 21.Michlewski G., Caceres J.F. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki H.I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 23.Trabucchi M., Briata P., Garcia-Mayoral M., Haase A.D., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M.G. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han C., Liu Y., Wan G., Choi H.J., Zhao L., Ivan C., He X., Sood A.K., Zhang X., Lu X. The RNA-binding protein DDX1 promotes primary microRNA maturation and inhibits ovarian tumor progression. Cell Rep. 2014;8:1447–1460. doi: 10.1016/j.celrep.2014.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawahara Y., Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. U. S. A. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawai S., Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J. Cell Biol. 2012;197:201–208. doi: 10.1083/jcb.201110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L., Nievera C.J., Lee A.Y., Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 29.Makharashvili N., Tubbs A.T., Yang S.H., Wang H., Barton O., Zhou Y., Deshpande R.A., Lee J.H., Lobrich M., Sleckman B.P., Wu X., Paull T.T. Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol. Cell. 2014;54:1022–1033. doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H., Li Y., Truong L.N., Shi L.Z., Hwang P.Y., He J., Do J., Cho M.J., Li H., Negrete A., Shiloach J., Berns M.W., Shen B., Chen L., Wu X. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol. Cell. 2014;54:1012–1021. doi: 10.1016/j.molcel.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Przetocka S., Porro A., Bolck H.A., Walker C., Lezaja A., Trenner A., von Aesch C., Himmels S.F., D'Andrea A.D., Ceccaldi R., Altmeyer M., Sartori A.A. CtIP-mediated fork protection synergizes with BRCA1 to suppress genomic instability upon DNA replication stress. Mol. Cell. 2018;72:568–582 e566. doi: 10.1016/j.molcel.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Makharashvili N., Arora S., Yin Y., Fu Q., Wen X., Lee J.H., Kao C.H., Leung J.W., Miller K.M., Paull T.T. Sae2/CtIP prevents R-loop accumulation in eukaryotic cells. eLife. 2018;7 doi: 10.7554/eLife.42733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lengsfeld B.M., Rattray A.J., Bhaskara V., Ghirlando R., Paull T.T. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H., Shao Z., Shi L.Z., Hwang P.Y., Truong L.N., Berns M.W., Chen D.J., Wu X. CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded breaks. J. Biol. Chem. 2012;287:21471–21480. doi: 10.1074/jbc.M112.355354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies O.R., Forment J.V., Sun M., Belotserkovskaya R., Coates J., Galanty Y., Demir M., Morton C.R., Rzechorzek N.J., Jackson S.P., Pellegrini L. CtIP tetramer assembly is required for DNA-end resection and repair. Nat. Struct. Mol. Biol. 2015;22:150–157. doi: 10.1038/nsmb.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan J., Chen J. N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J. Biol. Chem. 2009;284:31746–31752. doi: 10.1074/jbc.M109.023424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H., Shi L.Z., Wong C.C., Han X., Hwang P.Y., Truong L.N., Zhu Q., Shao Z., Chen D.J., Berns M.W., Yates J.R., 3rd, Chen L., Wu X. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anand R., Ranjha L., Cannavo E., Cejka P. Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell. 2016;64:940–950. doi: 10.1016/j.molcel.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 39.Ceppi I., Howard S.M., Kasaciunaite K., Pinto C., Anand R., Seidel R., Cejka P. CtIP promotes the motor activity of DNA2 to accelerate long-range DNA end resection. Proc. Natl. Acad. Sci. U. S. A. 2020;117:8859–8869. doi: 10.1073/pnas.2001165117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anand R., Jasrotia A., Bundschuh D., Howard S.M., Ranjha L., Stucki M., Cejka P. NBS1 promotes the endonuclease activity of the MRE11-RAD50 complex by sensing CtIP phosphorylation. EMBO J. 2019;38 doi: 10.15252/embj.2018101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Limbo O., Chahwan C., Yamada Y., de Bruin R.A., Wittenberg C., Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiohama A., Sasaki T., Noda S., Minoshima S., Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp. Cell Res. 2007;313:4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 43.Quick-Cleveland J., Jacob J.P., Weitz S.H., Shoffner G., Senturia R., Guo F. The DGCR8 RNA-binding heme domain recognizes primary microRNAs by clamping the hairpin. Cell Rep. 2014;7:1994–2005. doi: 10.1016/j.celrep.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sohn S.Y., Bae W.J., Kim J.J., Yeom K.H., Kim V.N., Cho Y. Crystal structure of human DGCR8 core. Nat. Struct. Mol. Biol. 2007;14:847–853. doi: 10.1038/nsmb1294. [DOI] [PubMed] [Google Scholar]
- 45.Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Y., Caron P., Legube G., Paull T.T. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014;42:e19. doi: 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu W.T., Hawley B.R., Skalka G.L., Baldock R.A., Smith E.M., Bader A.S., Malewicz M., Watts F.Z., Wilczynska A., Bushell M. Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nat. Commun. 2018;9:532. doi: 10.1038/s41467-018-02893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang Z., Bian X., Shim H. Inhibition of breast cancer metastasis with microRNA-302a by downregulation of CXCR4 expression. Breast Cancer Res. Treat. 2014;146:535–542. doi: 10.1007/s10549-014-3053-0. [DOI] [PubMed] [Google Scholar]
- 49.Sun L., Fang Y., Wang X., Han Y., Du F., Li C., Hu H., Liu H., Liu Q., Wang J., Liang J., Chen P., Yang H., Nie Y., Wu K. miR-302a inhibits metastasis and cetuximab resistance in colorectal cancer by targeting NFIB and CD44. Theranostics. 2019;9:8409–8425. doi: 10.7150/thno.36605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L., Min L., Wang X., Zhao J., Chen H., Qin J., Chen W., Shen Z., Tang Z., Gan Q., Ruan Y., Sun Y., Qin X., Gu J. Loss of RACK1 promotes metastasis of gastric cancer by inducing a miR-302c/IL8 signaling loop. Cancer Res. 2015;75:3832–3841. doi: 10.1158/0008-5472.CAN-14-3690. [DOI] [PubMed] [Google Scholar]
- 51.Liu J., Wang Y., Ji P., Jin X. Application of the microRNA-302/367 cluster in cancer therapy. Cancer Sci. 2020;111:1065–1075. doi: 10.1111/cas.14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen T.A., Jo M.H., Choi Y.G., Park J., Kwon S.C., Hohng S., Kim V.N., Woo J.S. Functional anatomy of the human microprocessor. Cell. 2015;161:1374–1387. doi: 10.1016/j.cell.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 53.Roth B.M., Ishimaru D., Hennig M. The core microprocessor component DiGeorge syndrome critical region 8 (DGCR8) is a nonspecific RNA-binding protein. J. Biol. Chem. 2013;288:26785–26799. doi: 10.1074/jbc.M112.446880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu X., Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwon S.C., Nguyen T.A., Choi Y.G., Jo M.H., Hohng S., Kim V.N., Woo J.S. Structure of human DROSHA. Cell. 2016;164:81–90. doi: 10.1016/j.cell.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 56.Partin A.C., Zhang K., Jeong B.C., Herrell E., Li S., Chiu W., Nam Y. Cryo-EM structures of human Drosha and DGCR8 in complex with primary MicroRNA. Mol. Cell. 2020;78:411–422 e414. doi: 10.1016/j.molcel.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin W., Wang J., Liu C.P., Wang H.W., Xu R.M. Structural basis for pri-miRNA recognition by Drosha. Mol. Cel. 2020;78:423–433 e425. doi: 10.1016/j.molcel.2020.02.024. [DOI] [PubMed] [Google Scholar]
- 58.Yuan S.S., Hou M.F., Hsieh Y.C., Huang C.Y., Lee Y.C., Chen Y.J., Lo S. Role of MRE11 in cell proliferation, tumor invasion, and DNA repair in breast cancer. J. Natl. Cancer Inst. 2012;104:1485–1502. doi: 10.1093/jnci/djs355. [DOI] [PubMed] [Google Scholar]
- 59.Liu B., Cong R., Peng B., Zhu B., Dou G., Ai H., Zhang X., Wang Z., Xu X. CtIP is required for DNA damage-dependent induction of P21. Cell Cycle. 2014;13:90–95. doi: 10.4161/cc.26810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H., Qiu Z., Liu B., Wu Y., Ren J., Liu Y., Zhao Y., Wang Y., Hao S., Li Z., Peng B., Xu X. PLK1 targets CtIP to promote microhomology-mediated end joining. Nucleic Acids Res. 2018;46:10724–10739. doi: 10.1093/nar/gky810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Z., Gong Y., Peng B., Shi R., Fan D., Zhao H., Zhu M., Zhang H., Lou Z., Zhou J., Zhu W.G., Cong Y.S., Xu X. MRE11 UFMylation promotes ATM activation. Nucleic Acids Res. 2019;47:4124–4135. doi: 10.1093/nar/gkz110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meng F., Qian M., Peng B., Peng L., Wang X., Zheng K., Liu Z., Tang X., Zhang S., Sun S., Cao X., Pang Q., Zhao B., Ma W., Songyang Z. Synergy between SIRT1 and SIRT6 helps recognize DNA breaks and potentiates the DNA damage response and repair in humans and mice. eLife. 2020;9 doi: 10.7554/eLife.55828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bai Y., Wang W., Li S., Zhan J., Li H., Zhao M., Zhou X.A., Li S., Li X., Huo Y., Shen Q., Zhou M., Zhang H., Luo J., Sung P. C1QBP promotes homologous recombination by stabilizing MRE11 and controlling the assembly and activation of MRE11/RAD50/NBS1 complex. Mol. Cel. 2019;75:1299–1314.e1296. doi: 10.1016/j.molcel.2019.06.023. [DOI] [PubMed] [Google Scholar]
- 64.Xie B., Zhang L., Zhao H., Bai Q., Fan Y., Zhu X., Yu Y., Li R., Liang X., Sun Q.Y., Li M., Qiao J. Poly(ADP-ribose) mediates asymmetric division of mouse oocyte. Cell Res. 2018;28:462–475. doi: 10.1038/s41422-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu X., Stern D.F. NFBD1/KIAA0170 is a chromatin-associated protein involved in DNA damage signaling pathways. J. Biol. Chem. 2003;278:8795–8803. doi: 10.1074/jbc.M211392200. [DOI] [PubMed] [Google Scholar]
- 67.Yoon S.N., Park J.H., Lwin T.M., Miyake K., Singh S.R., Hoffman R.M., Bouvet M. Tumor-sealing surgical orthotopic implantation of human colon cancer in nude mice induces clinically-relevant metastases without early peritoneal carcinomatosis. Anticancer Res. 2019;39:4065–4071. doi: 10.21873/anticanres.13563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puig I., Chicote I., Tenbaum S.P., Arques O., Herance J.R., Gispert J.D., Jimenez J., Landolfi S., Caci K., Allende H., Mendizabal L., Moreno D., Charco R., Espin E., Prat A. A personalized preclinical model to evaluate the metastatic potential of patient-derived colon cancer initiating cells. Clin. Cancer Res. 2013;19:6787–6801. doi: 10.1158/1078-0432.CCR-12-1740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed during this study are included in this published article and its supplementary information files.