Abstract
Introduction
The detection and monitoring of electrolyte imbalance is essential for appropriate management of many metabolic diseases; however, there is no tool that detects such imbalances reliably and noninvasively. In this study, we developed a deep learning model (DLM) using electrocardiography (ECG) for detecting electrolyte imbalance and validated its performance in a multicenter study.
Methods and Results
This retrospective cohort study included two hospitals: 92,140 patients who underwent a laboratory electrolyte examination and an ECG within 30 min were included in this study. A DLM was developed using 83,449 ECGs of 48,356 patients; the internal validation included 12,091 ECGs of 12,091 patients. We conducted an external validation with 31,693 ECGs of 31,693 patients from another hospital, and the result was electrolyte imbalance detection. During internal, the area under the receiving operating characteristic curve (AUC) of a DLM using a 12‐lead ECG for detecting hyperkalemia, hypokalemia, hypernatremia, hyponatremia, hypercalcemia, and hypocalcemia were 0.945, 0.866, 0.944, 0.885, 0.905, and 0.901, respectively. The values during external validation of the AUC of hyperkalemia, hypokalemia, hypernatremia, hyponatremia, hypercalcemia, and hypocalcemia were 0.873, 0.857, 0.839, 0.856, 0.831, and 0.813 respectively. The DLM helped to visualize the important ECG region for detecting each electrolyte imbalance, and it showed how the P wave, QRS complex, or T wave differs in importance in detecting each electrolyte imbalance.
Conclusion
The proposed DLM demonstrated high performance in detecting electrolyte imbalance. These results suggest that a DLM can be used for detecting and monitoring electrolyte imbalance using ECG on a daily basis.
Keywords: artificial intelligence, deep learning, electrocardiography, electrolytes
1. INTRODUCTION
Electrolyte balance is important for maintaining homeostasis and protecting cellular function (El‐Sharkawy et al., 2014; Rhoda et al., 2011). Electrolytes are controlled precisely between the intra‐ and extracellular compartments to sustain the normal physiological function of the muscles and nerves (Riggs, 2002). Some forms of electrolyte imbalance cause fatal arrhythmia and sudden cardiac death and require early detection (Goldberg et al., 2004; Klingkowski et al., 2019; Soar et al., 2010). As electrolyte imbalance is a common indicator for many diseases, its evaluation is a cornerstone for diagnosis and proper treatment (Kadri, 2013; Lee et al., 2000). Screening critical electrolyte imbalance is crucial for patients with diseases that impair the retention and excretion of electrolytes, such as renal failure, and those who partake medications that affect electrolyte excretion, such as diuretics (Arampatzis et al., 2013; Pun et al., 2017).
As the symptoms of electrolyte imbalance are vague, it is difficult to diagnose with only patient histories and examinations, that is, until the condition worsens and complications occur (Kadri, 2013). The gold standard for diagnosing electrolyte imbalance is a laboratory test that measures the concentration of electrolytes. Laboratory tests are invasive, costly, and require specialized equipment and infrastructure, such as trained medical staff for sampling blood and hematology analyzers for performing assessments with biochemical reagents (Stanifer et al., 2014). Evaluation of electrolytes on a daily basis is important in order to monitor health status and prevent life‐threatening events, but using laboratory tests is not optimal for this purpose.
The status of the cardiac cell membrane is dependent upon the maintenance of a normal electrolyte balance across the membrane, and it affects cardiac function and electrocardiography (ECG) (Noordam et al., 2019). Previous studies have shown that electrolyte imbalance alters the shape of the ECG (Noordam et al., 2019). It is not easy to make diagnostic tools based on conventional statistical methods using such subtle ECG changes. Deep learning has previously been used in the medical field to identify lesions and is currently used to analyze ECGs to diagnose heart failure, valvular heart disease, anemia, and coronary artery disease (Attia, Friedman, et al., 2019; Attia, Kapa, et al., 2019; Attia, Noseworthy, et al., 2019; Cho et al., 2020; Galloway et al., 2019; Jo et al., 2020; Kwon, Cho, et al., 2020; Kwon, Kim, et al., 2020; Kwon, Lee, et al., 2020). Recent studies have shown that deep learning models can detect dyskalemia using ECG (Galloway et al., 2019; Lin et al., 2020). However, these deep learning models only focused on detecting dyskalemia. In this study, we developed and validated a deep learning model (DLM) to detect electrolyte imbalance.
2. METHODS
2.1. Study design and population
We conducted a retrospective, multicenter, diagnostic study in which a DLM was developed using ECGs and then internally and externally validated. We excluded individuals with missing demographic, electrocardiographic, and electrolyte laboratory examination information. Data from Sejong General Hospital (SGH) were used for development and internal validation. In SGH, we identified patients with at least one standard digital, 10 s, 12‐lead ECG acquired in the supine position within the study period (October 1, 2016, to August 31, 2020) and at least one electrolyte laboratory panel for three electrolytes (sodium (Na), potassium (K), and calcium (Ca)) obtained within 30 min of the index ECG. The individuals who visited the general health checkup, outpatient department, and emergency department and were admitted to SGH were the study population for the development and internal validation datasets. As shown in Figure 1, patients treated at SGH were randomly split into algorithm development (80%) and internal validation (20%) datasets. Data from Mediplex Sejong Hospital (MSH) were used for external validation. The Sejong General Hospital (SGH) is a cardiovascular disease teaching hospital, and the Mediplex Sejong Hospital (MSH) is a general community hospital. We identified patients who were admitted to MSH during the study period (March 1, 2017, to August 31, 2020) and who had at least one ECG and one electrolyte laboratory examination panel obtained within 30 min of the index ECG. Because the purpose of the validation data was to assess the accuracy of the algorithm, we only used one ECG from each patient for the internal and external validation datasets, specifically the electrolyte examination closest to their most recent ECG in the study period.
FIGURE 1.

Study flowchart. ECG denotes electrocardiography
This study was approved by the institutional review boards of SGH and MSH. Clinical data, including digitally stored ECGs, electrolyte laboratory examination panel values (K, Na, and Ca), age, and sex, were obtained from both hospitals. Both institutional review boards waived the need for informed consent because of the retrospective nature of the study, which used fully anonymized ECG and health data and caused minimal harm.
2.2. Procedures
The predictor variables were ECG, age, and sex. Digitally stored 12‐lead ECG data, amounting to 5,000 values for each lead, were recorded over 10 s (500 Hz). One second each was removed at the beginning and end of each ECG because they have more artifacts than other parts. Because of this, the length of each ECG was 8 s (4,000 values). We created a dataset using the entire 12‐lead ECG data. We also used partial datasets from 12‐lead ECG data, such as limb 6‐lead and single‐lead (I) ECG data. We selected the sets of leads because they could easily be recorded by wearable and pad devices in contact with the hands and legs. Consequently, when we developed and validated the DLM using 12‐lead ECGs, a dataset of two‐dimensional (2D) data of 12 × 4,000 values was used. When we developed and validated an algorithm using 6‐lead ECGs, we used datasets that were 6 × 4,000 values, and when using single‐lead ECGs, we used datasets that were 1 × 4,000 values.
The objective of this research was to determine abnormalities of electrolytes, defined by serum electrolyte concentrations. Normal concentration range of K, Na, and Ca were 3.5–5.5 mmol/L, 130–150 mmol/L, and 8.0–11.0 mg/dl. We developed a DLM for each electrolyte imbalance independently, because the model trained using the transfer learning method was overfitted to local optima, and it showed poor performance.
The sampling rate of the ECG data was fixed at 500 Hz. We carried out a pre‐processing step for the ECG data. First, we eliminated high‐frequency noise, such as electrical line artifacts, using a low‐pass filter set at 150 Hz. Second, we removed low‐frequency noise, such as chest wall movement due to breathing, using a high‐pass filter set at 0.05 Hz. Finally, we normalized the Z‐score for the ECG data.
As shown in Figure 2, we developed a DLM based on an ensemble network. We developed each DLM to determine the presence of each electrolyte imbalance, such as hyperkalemia, hypokalemia, hypernatremia, hyponatremia, hypercalcemia, and hypocalcemia. Each DLM was developed using six residual blocks of the neural network to learn complex hierarchical non‐linear representations from the data (LeCun et al., 2015). In a residual block with four stages, there were two convolution layers and two batch normalizations layer repeated. The last layer of the sixth residual block was connected to a flattened layer, which was fully connected to the 1D layer composed of neural nodes. The values for age and sex were inputted to the input layer of a multilayer perceptron (MLP) consisting of three 1D layers. The 1D ECG, obtained after passing the 2D ECG data through the flattened layer, and the 1D age and sex data were concatenated and fully connected to the 1st ensemble layer. The second fully connected 1D layer was connected to the output layer. The output layer was composed of two nodes. The value of the output node of each DLM represented the probability of each electrolyte imbalance, and the output node of each DLM used a softmax function as an activation function because the output of the softmax function was between 0 and 1. As a comparative model, we also developed classification models using conventional statistical model (logistic regression) and conventional machine learning model (random fores). We used glm and randomForest of R to develop prediction models.
FIGURE 2.

Architecture of deep learning‐based model for detecting electrolyte imbalance. Conv denotes convolutional neural network and ECG electrocardiography
As most of the medical test results are normal, medical data often are imbalanced between normal and positive cases. In this study, the electrolyte imbalanced data were also imbalanced. We used oversampling and undersampling simultaneously in the training process. When we trained the DLM, we oversampled the positive case, allowing duplication by 3–5 times, and undersampled the normal case by half. We determined the ratios for oversampling and undersampling using a grid search.
2.3. Statistical analysis
Continuous variables were represented as mean values (standard deviation, SD) and compared using the unpaired Student's t‐test or Mann‐Whitney U‐test. Categorical variables were expressed as frequencies and percentages and compared using the chi‐square test.
At each input (ECG) of validation data, the DLM calculated the probability of electrolyte imbalance in the range from 0 (non‐electrolyte imbalance) to 1 (electrolyte imbalance). To confirm the DLM performance, we compared the probability calculated by the DLM with the presence of electrolyte imbalance in the internal and external validation datasets. For this purpose, we used the area under the receiver operating characteristic curve (AUC). Confidence intervals (CIs) of 95% were used for all measures of diagnostic performance, except for the AUC. We selected the cutoff point for calculating the sensitivity, specificity, PPV, and NPV of the validation dataset when the sensitivity was 90% in the development dataset. The purpose of the DLM was to screen for electrolyte imbalance and to refer the patient for confirmatory laboratory tests if needed. We selected a high‐sensitivity point as the cutoff point. The CIs for the AUC were determined based on the Sun and Su optimization of the De‐long method using the pROC package in R (The R Foundation for Statistical Computing, Vienna, Austria). A significant difference in patient characteristics was defined as a two‐sided p‐value of <.001. Statistical analyses were computed using R software, version 3.4.2. In addition, we used PyTorch's open‐source software library at the backend and Python (version 3.6.11) for the analyses.
2.4. Visualizing the developed XDM for interpretation
To understand the model and draw a comparison with existing medical knowledge, it was necessary to identify a region that had a significant effect on the decision of the developed DLM. We employed a sensitivity map using a saliency method. The map was computed using the first‐order gradients of the classifier probabilities with respect to the input signals; if the probability of a classifier is sensitive to a specific region of the signal, the region would be considered significant in the model. We used a gradient class activation map as a sensitivity map with the gradient backpropagation method (Selvaraju et al., 2017, 2020).
3. RESULTS
The eligible population included 60,479 and 31,704 patients at SGH and MSH, respectively. We excluded 32 and 11 patients (from SGH and MSH, respectively) because of missing age and sex data, laboratory evaluation information, or ECG data (Figure 1). The study included 92,140 patients, of whom 4,638 had electrolyte imbalance. There were 163, 83, and 241 patients who had electrolyte imbalances “potassium and sodium,” “calcium and sodium,” and “potassium and calcium,” respectively. The DLM was developed using a development dataset of 83,449 12‐lead ECGs for 48,356 patients. Then, the performance of the algorithm was examined using 12,091 ECGs from the 12,091 patients in the internal validation dataset from SGH and 31,693 ECGs from the 31,693 patients in the external validation dataset from MSH (Figure 1 and Table 1).
TABLE 1.
Study population characteristics
| Characteristic | Sejong General Hospital (development and internal validation data) | Mediplex Sejong Hospital (external validation data) | p |
|---|---|---|---|
| Study population | 60,447 | 31,693 | |
| Age, year, mean (SD) | 59.76 (16.22) | 54.57 (16.50) | <.001 |
| Male, n (%) | 31,634 (52.3) | 15,844 (50.0) | <.001 |
| Heart rate, bpm, mean (SD) | 72.89 (18.54) | 69.83 (14.06) | <.001 |
| Atrial fibrillation, n (%) | 6,483 (10.7) | 1,491 (4.7) | <.001 |
| PR interval, ms, mean (SD) | 171.03 (30.01) | 167.13 (26.39) | <.001 |
| QRS duration, ms, mean (SD) | 96.65 (18.01) | 94.97 (14.84) | <.001 |
| QT interval, ms, mean (SD) | 404.53 (42.35) | 404.70 (36.14) | .559 |
| QTc, ms, mean (SD) | 438.62 (34.92) | 431.98 (30.89) | <.001 |
| P‐wave axis, mean (SD) | 43.91 (30.44) | 44.04 (27.44) | .544 |
| R‐wave axis, mean (SD) | 39.17 (44.74) | 40.74 (39.62) | <.001 |
| T‐wave axis, mean (SD) | 45.60 (49.34) | 39.68 (35.50) | <.001 |
| Potassium, mmol/L, mean (SD) | 4.22 (0.47) | 4.08 (0.44) | <.001 |
| Sodium, mmol/L, mean (SD) | 140.10 (3.07) | 141.29 (3.10) | <.001 |
| Calcium, mg/dl, mean (SD) | 9.37 (0.46) | 9.11 (0.45) | <.001 |
| Potassium abnormalities | <.001 | ||
| Hypokalemia (<3.5) | 2,082 (3.4) | 1,052 (3.3) | |
| Normokalemia (3.5–5.5) | 57,766 (95.6) | 30,449 (96.1) | |
| Hyperkalemia (>5.5) | 599 (1.0) | 192 (0.6) | |
| Sodium abnormalities | <.001 | ||
| Hyponatremia (<130) | 605 (1.0) | 217 (0.7) | |
| Normonatremia (130–150) | 59,793 (98.9) | 31,448 (99.2) | |
| Hypernatremia (>150) | 49 (0.1) | 28 (0.1) | |
| Calcium abnormalities | <.001 | ||
| Hypocalcemia (<8.0) | 503 (0.8) | 397 (1.3) | |
| Normocalcemia (8.0–11.0) | 59,859 (99.0) | 31,268 (98.7) | |
| Hypercalcemia (>11.0) | 85 (0.1) | 28 (0.1) |
In hyperkalemia patients, the ECGs had prolonged QRS duration, prolonged QTc, rightward T‐wave axis, prolonged PR interval, and tachycardia (Table 2). Sodium imbalance had correlation with heart rate, presence of atrial fibrillation, PR interval, QRS duration, QT interval, QTc, and T‐wave axis. Calcium abnormality had correlation with heart rate, presence of atrial fibrillation, QRS duration, QT interval, QTc, R‐wave axis, and T‐wave axis (Table 2).
TABLE 2.
Study population characteristics stratified by electrolyte abnormalities
| Potassium abnormality | ||||
|---|---|---|---|---|
| Characteristics | Hypokalemia | Normokalemia | Hyperkalemia | p |
| Study population, n | 3,134 | 88,215 | 791 | |
| Age, year, mean (SD) | 60.75 (18.02) | 57.75 (16.42) | 71.58 (13.13) | <.001 |
| Male, n (%) | 1,260 (40.2) | 45,811 (51.9) | 407 (51.5) | <.001 |
| Heart rate, bpm, mean (SD) | 82.16 (21.31) | 71.42 (16.81) | 77.02 (25.91) | <.001 |
| Atrial fibrillation, n (%) | 310 (9.9) | 7,478 (8.5) | 186 (23.5) | <.001 |
| PR interval, ms, mean (SD) | 171.70 (31.19) | 169.43 (28.46) | 186.21 (51.41) | <.001 |
| QRS duration, ms, mean (SD) | 97.51 (18.27) | 95.94 (16.82) | 104.63 (26.81) | <.001 |
| QT interval, ms, mean (SD) | 401.14 (50.74) | 404.64 (39.66) | 412.46 (60.33) | <.001 |
| QTc, ms, mean (SD) | 459.94 (39.34) | 435.36 (33.03) | 452.34 (47.21) | <.001 |
| P‐wave axis, mean (SD) | 46.03 (31.65) | 43.89 (29.21) | 43.50 (39.96) | .001 |
| R‐wave axis, mean (SD) | 37.69 (47.01) | 39.83 (42.75) | 34.67 (57.45) | <.001 |
| T‐wave axis, mean (SD) | 46.54 (63.76) | 43.22 (44.08) | 69.44 (62.04) | <.001 |
| Sodium abnormality | ||||
|---|---|---|---|---|
| Characteristics | Hyponatremia | Normonatremia | Hypernatremia | p |
| Study population, n | 822 | 91,241 | 77 | |
| Age, year, mean (SD) | 73.18 (12.86) | 57.82 (16.46) | 73.16 (16.81) | <.001 |
| Male, n (%) | 347 (42.2) | 47,092 (51.6) | 39 (50.6) | <.001 |
| Heart rate, bpm, mean (SD) | 83.68 (23.31) | 71.71 (17.07) | 94.92 (26.55) | <.001 |
| Atrial fibrillation, n (%) | 134 (16.3) | 7,827 (8.6) | 13 (16.9) | <.001 |
| PR interval, ms, mean (SD) | 179.83 (37.80) | 169.56 (28.71) | 157.17 (43.76) | <.001 |
| QRS duration, ms, mean (SD) | 101.51 (24.73) | 96.02 (16.90) | 101.31 (26.48) | <.001 |
| QT interval, ms, mean (SD) | 401.86 (58.11) | 404.63 (40.09) | 383.43 (67.75) | <.001 |
| QTc, ms, mean (SD) | 461.66 (46.02) | 436.08 (33.49) | 468.35 (50.71) | <.001 |
| P‐wave axis, mean (SD) | 45.20 (39.27) | 43.95 (29.29) | 40.28 (35.84) | .370 |
| R‐wave axis, mean (SD) | 36.74 (54.23) | 39.73 (42.92) | 44.49 (59.59) | .089 |
| T‐wave axis, mean (SD) | 59.76 (67.55) | 43.38 (44.81) | 83.08 (85.74) | <.001 |
| Calcium abnormality | ||||
|---|---|---|---|---|
| Characteristics | Hypocalcemia | Normocalcemia | Hypercalcemia | p |
| Study population, n | 900 | 91,127 | 113 | |
| Age, year, mean (SD) | 68.37 (17.10) | 57.86 (16.47) | 64.86 (14.11) | <.001 |
| Male, n (%) | 470 (52.2) | 46,965 (51.5) | 43 (38.1) | .015 |
| Heart rate, bpm, mean (SD) | 84.96 (28.76) | 71.69 (16.98) | 83.80 (23.70) | <.001 |
| Atrial fibrillation, n (%) | 135 (15.0) | 7,822 (8.6) | 17 (15.0) | <.001 |
| PR interval, ms, mean (SD) | 169.98 (36.29) | 169.62 (28.75) | 173.83 (30.34) | .340 |
| QRS duration, ms, mean (SD) | 99.07 (22.95) | 96.04 (16.93) | 100.00 (21.82) | <.001 |
| QT interval, ms, mean (SD) | 410.59 (58.95) | 404.56 (40.06) | 382.00 (54.44) | <.001 |
| QTc, ms, mean (SD) | 474.84 (50.48) | 435.95 (33.30) | 440.17 (40.41) | <.001 |
| P‐wave axis, mean (SD) | 43.45 (36.32) | 43.96 (29.30) | 43.88 (41.66) | .899 |
| R‐wave axis, mean (SD) | 34.45 (52.85) | 39.77 (42.93) | 31.10 (49.77) | <.001 |
| T‐wave axis, mean (SD) | 64.54 (70.93) | 43.35 (44.75) | 52.04 (60.04) | <.001 |
During internal validation, the AUC of the DLM using 12‐lead ECG for detecting hyperkalemia, hypokalemia, hypernatremia, hyponatremia, hypercalcemia, and hypocalcemia was 0.945 (95% confidence interval, 0.931–0.959), 0.866 (0.854–0.878), 0.944 (0.895–0.993), 0.885 (0.869–0.900), 0.905 (0.806–1.000), and 0.901 (0.880–0.922), respectively (Figure 3 and Table 3). During external validation, the AUC of the DLM using 12‐lead ECG for detecting hyperkalemia, hypokalemia, hypernatremia, hyponatremia, hypercalcemia, and hypocalcemia was 0.873 (0.843–0.902), 0.857 (0.846–0.867), 0.839 (0.727–0.951), 0.856 (0.831–0.880), 0.831 (0.723–0.939), and 0.813 (0.793–0.834), respectively (Figure 3 and Table 3). There were no significant differences in DLM performance between patients with multiple electrolyte imbalances simultaneously and patients with one electrolyte imbalance. As shown in Figure 3, the DLM outperformed the logistic regression and random forest models for internal and external validation datasets.
FIGURE 3.
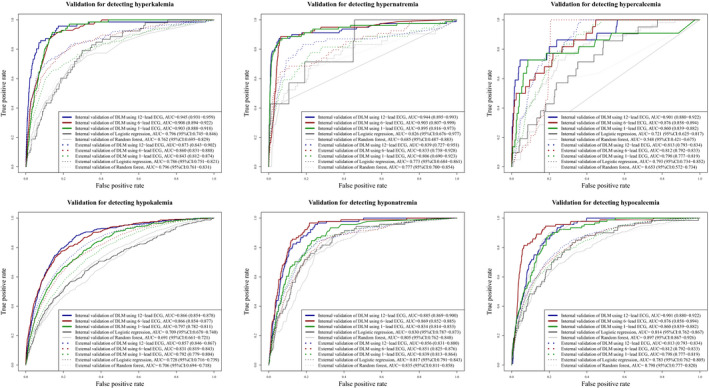
Performances of deep learning‐based model for detecting electrolyte abnormalities. AUC denotes area under the receiver operating characteristic curve, DLM deep learning‐based model, and ECG electrocardiography
TABLE 3.
Performances of deep learning‐based model for detecting electrolyte imbalance using electrocardiography
| Deep learning‐based models (DLMs) | Internal validation (95% confidence interval) | External validation (95% confidence interval) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AUC | SEN | SPE | PPV | NPV | AUC | SEN | SPE | PPV | NPV | |
| Hyperkalemia | ||||||||||
| DLM using 12‐lead ECG | 0.945 (0.931–0.959) | 0.901 (0.807–0.959) | 0.850 (0.843–0.856) | 0.038 (0.030– 0.049) | 0.999 (0.998–1.000) | 0.873 (0.843–0.902) | 0.896 (0.848–0.934) | 0.599 (0.594– 0.604) | 0.014 (0.012– 0.016) | 0.999 (0.998–1.000) |
| DLM using 6‐lead ECG | 0.908 (0.894–0.922) | 0.915 (0.825– 0.968) | 0.829 (0.822–0.836) | 0.034 (0.027–0.044) | 0.999 (0.998–1.000) | 0.860 (0.831–0.888) | 0.892 (0.842–0.930) | 0.568 (0.560–0.570) | 0.012 (0.011–0.014) | 0.999 (0.998–1.000) |
| DLM using 1‐lead ECG | 0.903 (0.888–0.918) | 0.887 (0.790– 0.950) | 0.866 (0.859–0.872) | 0.042 (0.033–0.054) | 0.999 (0.998–1.000) | 0.843 (0.812–0.874) | 0.897 (0.848–0.934) | 0.413 (0.407–0.418) | 0.009 (0.008–0.011) | 0.998 (0.998–0.999) |
| Hypokalemia | ||||||||||
| DLM using 12‐lead ECG | 0.866 (0.854–0.878) | 0.893 (0.858–0.922) | 0.704 (0.695–0.713) | 0.100 (0.091–0.111) | 0.994 (0.992–0.996) | 0.857 (0.846–0.867) | 0.896 (0.882–0.908) | 0.560 (0.554–0.565) | 0.120 (0.115–0.125) | 0.988 (0.986–0.989) |
| DLM using 6‐lead ECG | 0.866 (0.854–0.877) | 0.896 (0.861–0.924) | 0.647 (0.638–0.656) | 0.086 (0.077–0.095) | 0.994 (0.992–0.996) | 0.831 (0.819–0.843) | 0.914 (0.901–0.925) | 0.435 (0.430–0.440) | 0.098 (0.094–0.102) | 0.987 (0.985–0.989) |
| DLM using 1‐lead ECG | 0.797 (0.782–0.811) | 0.930 (0.899–0.953) | 0.465 (0.455–0.475) | 0.060 (0.054–0.067) | 0.994 (0.992–0.996) | 0.792 (0.779–0.804) | 0.888 (0.874–0.901) | 0.437 (0.432–0.443) | 0.096 (0.092–0.100) | 0.983 (0.981–0.985) |
| Hypernatremia | ||||||||||
| DLM using 12‐lead ECG | 0.944 (0.895–0.993) | 0.923 (0.640–0.998) | 0.634 (0.625–0.643) | 0.003 (0.002–0.005) | 1.000 (0.999–1.000) | 0.839 (0.727–0.951) | 0.870 (0.751–0.946) | 0.649 (0.644–0.654) | 0.004 (0.003–0.005) | 1.000 (0.999–1.000) |
| DLM using 6‐lead ECG | 0.903 (0.807–0.999) | 0.923 (0.640–0.998) | 0.488 (0.478–0.497) | 0.002 (0.001–0.004) | 1.000 (0.999–1.000) | 0.833 (0.738–0.928) | 0.889 (0.774–0.958) | 0.456 (0.451–0.461) | 0.003 (0.002–0.003) | 1.000 (0.999–1.000) |
| DLM using 1‐lead ECG | 0.895 (0.816–0.973) | 0.846 (0.546–0.981) | 0.347 (0.338–0.357) | 0.002 (0.001–0.003) | 0.999 (0.998–1.000) | 0.806 (0.690–0.923) | 0.907 (0.797–0.969) | 0.253 (0.249–0.258) | 0.002 (0.001–0.002) | 0.999 (0.999–1.000) |
| Hyponatremia | ||||||||||
| DLM using 12‐lead ECG | 0.885 (0.869–0.900) | 0.901 (0.821–0.954) | 0.820 (0.812–0.827) | 0.041 (0.033–0.051) | 0.999 (0.998–1.000) | 0.856 (0.831–0.880) | 0.887 (0.845–0.921) | 0.629 (0.624–0.634) | 0.020 (0.017–0.022) | 0.998 (0.998–0.999) |
| DLM using 6‐lead ECG | 0.869 (0.852–0.885) | 0.890 (0.807–0.946) | 0.797 (0.789–0.805) | 0.036 (0.029–0.045) | 0.999 (0.998–0.999) | 0.851 (0.825–0.876) | 0.887 (0.845–0.921) | 0.599 (0.594–0.604) | 0.018 (0.016–0.021) | 0.998 (0.998–0.999) |
| DLM using 1‐lead ECG | 0.834 (0.814–0.853) | 0.912 (0.834–0.961) | 0.686 (0.677–0.694) | 0.024 (0.019–0.030) | 0.999 (0.998–1.000) | 0.839 (0.813–0.864) | 0.915 (0.877–0.944) | 0.477 (0.472–0.483) | 0.015 (0.013–0.016) | 0.998 (0.998–0.999) |
| Hypercalcemia | ||||||||||
| DLM using 12‐lead ECG | 0.905 (0.806–1.000) | 0.909 (0.708–0.989) | 0.521 (0.511–0.530) | 0.004 (0.002–0.006) | 1.000 (0.999–1.000) | 0.831 (0.723–0.939) | 0.852 (0.738–0.930) | 0.794 (0.790–0.798) | 0.007 (0.005–0.009) | 1.000 (0.999–1.000) |
| DLM using 6‐lead ECG | 0.878 (0.791–0.966) | 0.864 (0.651–0.971) | 0.605 (0.596–0.615) | 0.004 (0.003–0.007) | 1.000 (0.999–1.000) | 0.813 (0.726–0.900) | 0.885 (0.778–0.953) | 0.690 (0.685–0.695) | 0.005 (0.004–0.006) | 1.000 (0.999–1.000) |
| DLM using 1‐lead ECG | 0.875 (0.786–0.965) | 0.909 (0.708–0.989) | 0.352 (0.343–0.361) | 0.003 (0.002–0.004) | 0.999 (0.998–1.000) | 0.634 (0.522–0.746) | 0.918 (0.819–0.973) | 0.592 (0.587–0.597) | 0.004 (0.003–0.005) | 1.000 (0.999–1.000) |
| Hypocalcemia | ||||||||||
| DLM using 12‐lead ECG | 0.901 (0.880–0.922) | 0.891 (0.809–0.947) | 0.847 (0.84–0.854) | 0.048 (0.038–0.059) | 0.999 (0.998–0.999) | 0.813 (0.793–0.834) | 0.905 (0.877–0.929) | 0.551 (0.546–0.557) | 0.031 (0.028–0.033) | 0.997 (0.996–0.998) |
| DLM using 6‐lead ECG | 0.876 (0.858–0.894) | 0.902 (0.822–0.954) | 0.777 (0.769–0.785) | 0.034 (0.027–0.042) | 0.999 (0.998–1.000) | 0.812 (0.792–0.833) | 0.928 (0.902–0.948) | 0.473 (0.467–0.478) | 0.027 (0.024–0.029) | 0.998 (0.997–0.998) |
| DLM using 1‐lead ECG | 0.860 (0.839–0.882) | 0.913 (0.836–0.962) | 0.752 (0.744–0.760) | 0.031 (0.025–0.038) | 0.999 (0.998–1.000) | 0.798 (0.777–0.819) | 0.883 (0.853–0.909) | 0.547 (0.541–0.552) | 0.030 (0.027–0.032) | 0.997 (0.996–0.997) |
During external validation, the AUC of the DLM using 6‐lead ECG for detecting hyperkalemia, hypokalemia, hypernatremia, hyponatremia, hypercalcemia, and hypocalcemia was 0.860 (0.831–0.888), 0.831 (0.819–0.843), 0.833 (0.738–0.928), 0.851 (0.825–0.876), 0.813 (0.726–0.900), and 0.812 (0.792–0.833), respectively (Figure 3 and Table 3). During external validation, the AUC of the DLM using single‐lead ECG for detecting hyperkalemia, hypokalemia, hypernatremia, hyponatremia, hypercalcemia, and hypocalcemia was 0.843 (0.812–0.874), 0.792 (0.779–0.804), 0.806 (0.690–0.923), 0.839 (0.813–0.864), 0.634 (0.522–0.746), and 0.798 (0.777–0.819), respectively (Figure 3 and Table 3).
The DLM described the important ECG region to detect each electrolyte imbalance. As shown in Figure 4, the DLM focused on the QRS complex for detecting hyperkalemia, hypokalemia, and hyponatremia. The DLM focused on the T wave for detecting hyperkalemia and on the S wave for detecting hypernatremia and hypercalcemia. We provided a full‐size sensitivity map in the supplemental material (Figures S1‐S6).
FIGURE 4.
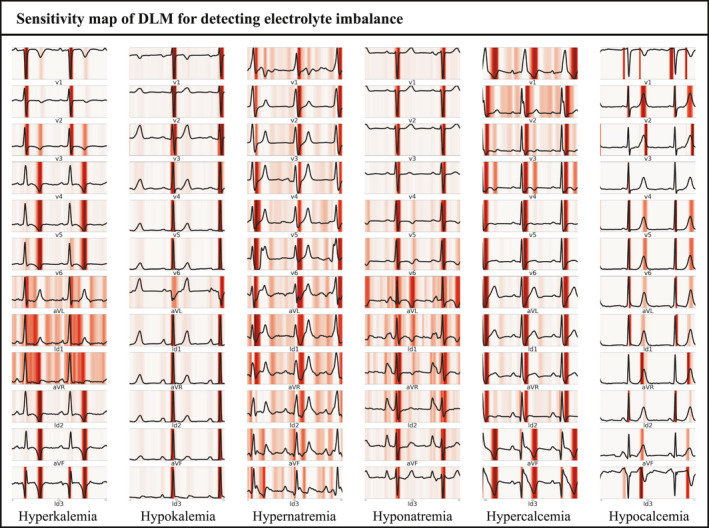
Sensitivity map of DLM for detecting electrolyte imbalance. DLM denoted deep learning‐based model
4. DISCUSSION
We developed and validated a DLM for electrolyte imbalance detection using a 12‐lead, 6‐lead, and single‐lead ECG. In addition, we showed the ECG region that had a significant effect on the decision of the developed DLM. To the best of our knowledge, this study is the first to develop an artificial intelligence algorithm for detecting electrolyte imbalance and to show the interpretable patterns of decision making using artificial intelligence in the biosignal domain. The purpose of a DLM is to screen the electrolyte imbalance using ECG, which is noninvasive, economical, and obtained using wearable devices; the DLM refers the patients to conduct confirmative laboratory examinations if electrolyte imbalance is suspected. For example, renal failure patients can be screened for critical electrolyte imbalance using wearable ECG devices and can then visit the hospital for confirmative laboratory tests; this is important to prevent deterioration of patients' condition and irreversible disease progression.
Electrolyte balance is a cornerstone to evaluate the general condition of patients and conduct proper management of many metabolic disorders (Kadri, 2013; Lee et al., 2000). Detecting electrolyte imbalance is important for the diagnosis of new metabolic diseases and management of patients with diseases that impair electrolyte homeostasis, such as renal failure, diabetes insipidus, severe diarrhea, hyperparathyroidism, and diabetes ketoacidosis (Dhondup & Qian, 2017; Liamis, 2014; Papi et al., 2014; Priyamvada et al., 2015). Monitoring electrolyte imbalance is important for managing patients who have medication, which could alter the homeostasis of electrolytes such as diuretics (Lim et al., 2016). The symptoms of electrolyte imbalance are vague and nonspecific (Kadri, 2013). Diagnostic examination is a laboratory examination that requires invasive blood sampling and cannot be conducted daily. Because of this, a new technology is required for detecting electrolyte imbalance using simple and noninvasive methods and for daily use. As ECG is a noninvasive test and changes with electrolyte imbalance, we developed a DLM for detecting electrolyte imbalance using ECG.
In many previous studies, electrolyte imbalance is shown to affect ECG. A progressive change from peaked T waves to wide QRS can correlate with hyperkalemia (Littmann & Gibbs, 2018). Increased amplitude and width of P wave, T‐wave flattening and inversion, prominent U waves, and apparent long QR intervals can correlate with hypokalemia (Levis, 2012). The most common ECG finding associated with hypercalcemia is shortening of the QR interval, and the most common ECG finding associated with hypocalcemia is a prolonged QT interval (Chorin et al., 2016). Although the ECG pattern is associated with electrolyte imbalance, diagnostic criteria and methods could not be made based on conventional statistical methods.
The most important aspect of deep learning is its ability to extract features and develop an algorithm using various types of data, such as images, 2D data, and waveforms. In previous studies, Attia and colleagues and our study group developed a deep learning‐based model to screen for heart failure, arrhythmia, valvular heart disease, left ventricular hypertrophy, and anemia (Attia, Friedman, et al., 2019; Attia, Kapa, et al., 2019; Attia, Noseworthy, et al., 2019; Cho et al., 2020; Galloway et al., 2019; Jo et al., 2020; Kwon, Cho, et al., 2020; Kwon, Kim, et al., 2020; Kwon, Lee, et al., 2020). In recent studies, Attia and colleagues showed that hyperkalemia and hypokalemia could be detected using ECG based on a deep learning model (Galloway et al., 2019). However, the studies focused only on the imbalance of potassium and could not detect other electrolyte imbalances. Therefore, we developed a DLM for detecting electrolyte imbalance, including potassium, sodium, and calcium.
We adopted a sensitivity map to describe the abnormal finding that affects the decision of DLM for detecting electrolyte imbalance. Using this methodology, we could confirm an ECG region that was associated with each electrolyte imbalance. In conventional methods, the research process was started based on the hypothesis of researchers. For example, in the association between hyperkalemia and ECG, researchers made a hypothesis based on researchers' experience of reading the ECG of hyperkalemia patients. This methodology limited the opportunity to discover knowledge in human perception. In deep learning methods, such as DLM and sensitivity map in this study, the findings were not based on previous medical knowledge of humans but data itself. Because of this, we could have the opportunity to discover new knowledge from the data itself without human prejudice. Deep learning could discover the complex hierarchical non‐linear representation that could not be discovered using conventional statistical methods, such as logistic regression. In this study, we could confirm the important ECG region for detecting each electrolyte imbalance from waveform data. For example, the DLM focused on T wave and QRS complex for detecting hyperkalemia and hypokalemia. These findings were in agreement with those in previous studies (Levis, 2012; Littmann & Gibbs, 2018).
In a previous multicenter study, Galloway et al. showed the performance of DLM‐enabled ECG for detecting hyperkalemia (Galloway et al., 2019). Following this, there were several studies conducted for detecting potassium imbalance using ECG based on a DLM. In the present study, we developed a DLM to detect not only potassium imbalance, but also sodium and calcium imbalances. As sodium and calcium imbalances are also important in the diagnosis of hidden disease and the management of patients, the results of the present study will be the basis for further studies. This is the first study to use a sensitivity map for electrolyte imbalance analysis. Using the proposed method, we clarified the important ECG lesion for a DLM to detect electrolyte imbalance. We also compared our results with previous medical knowledge regarding the correlation between electrolyte imbalance and ECG. As deep learning models suffer from a black box limitation, the proposed method would prove to be helpful for other researchers to conduct their medical research using artificial intelligence.
For detecting hypokalemia, the performance of the DLM using 12‐lead ECG and 6‐lead ECG was almost the same. We had two hypotheses. First, in some tasks, the information about precordial 6‐lead ECG is not needed to detect the disease. As a result, adding the information about the precordial 6‐lead to limb 6‐lead did not enhance the performance of the DLM. The second hypothesis is that limb 6‐lead ECG already had information on precordial 6‐lead ECG. In previous studies, Cho et al. have already developed a model to generate precordial 6‐lead ECG from limb 6‐lead ECG (Cho et al., 2020). In some tasks, information about the specific disease of precordial 6‐lead is already reflected in limb 6‐lead ECG. As the exact decision process of deep learning has not been discovered, we could explore this topic in the near future.
There were several limitations to this study. First, we developed a DLM for detecting electrolyte imbalances, including Na, K, and Ca. Although we selected three electrolytes because the four electrolytes are most commonly used in the clinical field, we need to develop DLM for including other electrolytes such as magnesium and phosphorus. Second, we validated DLM using retrospective data; therefore, we need to validate DLM with prospective studies and daily data. In this study, the performance of the DLM using single‐lead ECG was evaluated using partial Lead I data from 12‐lead ECG data. As a result of this, further studies are needed to apply the DLM to diverse wearable life‐style devices and to confirm the performance of the DLM using ECG devices. Studies related to the clinical significance of the new technology are required to apply it in clinical practice. In our next study, we will verify DLM performance and significance with a prospective study in daily clinical practice. Third, this study was only conducted in two hospitals in Korea, and it is necessary to validate the DLM with patients in other countries. As shown in Figure 3, the performance differences of the DLM between internal and external validation for hypernatremia and hypocalcemia were significant. As pitfall of deep learning is overfitting, we need a diverse dataset for enhanced performance of the DLM in robust situations. Therefore, we plan to conduct a large multicenter study involving multiple hospitals in other countries. Fourth, as the characteristics and comorbidities of patients could affect the performance of the DLM, further studies are needed to develop a robust DLM for preserving the performance for a more diverse population. As the performance in specific populations could not be determined in this study, further studies are needed to determine the performance by patient's characteristics and comorbidities. Fifth, we experiment with only three combinations of ECG leads, namely, 12‐lead, limb 6‐lead, and single‐lead (Lead I). As some ECG information can be calculated using the other ECG lead, we need to experiment with diverse combinations of lead information of ECG. In the near future, we will conduct the research on this topic and gain new insights for clinical use.
CONFLICT OF INTEREST
KHK, KHJ, SYL, and BHO declare that they have no competing interests. JK and JP are co‐founder, and YYJ, MSJ, YJL, YHC, and JHS are researchers of Medical AI Co., a medical artificial intelligence company. JK and JHB are researchers of Bodyfriend Co. There are no products in development or marketed products to declare. This does not alter our adherence to Journal.
AUTHOR CONTRIBUTIONS
MSJ and KHK performed data analysis and verified the clinical coding. YYJ, YHC, HJS, YJL, and JHB contributed to the study idea and design as well as data collection, performed data analysis, and contributed to subsequent drafts. KHJ, SYL, JP, and BHO contributed to data collection and revised the manuscript. JK is the principal investigator and contributed to the study idea and design, data analysis, verified the clinical coding, and contributed to subsequent drafts.
ETHICAL APPROVAL
This study was approved by the institutional review boards of SGH and MSH. Both institutional review boards waived the need for informed consent because of the retrospective nature of the study, which used fully anonymized ECG and health data and caused minimal harm.
Supporting information
Fig S1‐6
ACKNOWLEDGMENTS
This research was results of a study on the “AI voucher” Project, supported by the Ministry of Science and ICT and National IT Industry Promotion Agency of South Korea.
Kwon J‐M, Jung M‐S, Kim K‐H, et al. Artificial intelligence for detecting electrolyte imbalance using electrocardiography. Ann Noninvasive Electrocardiol. 2021;26:e12839. 10.1111/anec.12839
Joon‐myoung Kwon, Min‐Seung Jung, Kyung‐Hee Kim contributed equally to this work.
Funding information
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea Government (MSIT) (No. 2020R1F1A1073791).
Contributor Information
Joon‐myoung Kwon, Email: kwonjm@sejongh.co.kr.
Kyung‐Hee Kim, Email: kwonjm@sejongh.co.kr, Email: learnbyliving9@gmail.com.
DATA AVAILABILITY STATEMENT
The data underlying this article will be shared on reasonable request to the corresponding author.
REFERENCES
- Arampatzis, S. , Funk, G.‐C. , Leichtle, A. B. , Fiedler, G.‐M. , Schwarz, C. , Zimmermann, H. , Exadaktylos, A. K. , & Lindner, G. (2013). Impact of diuretic therapy‐associated electrolyte disorders present on admission to the emergency department: A cross‐sectional analysis. BMC Medicine, 11(1), 83. 10.1186/1741-7015-11-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attia, Z. I. , Friedman, P. A. , Noseworthy, P. A. , Lopez‐Jimenez, F. , Ladewig, D. J. , Satam, G. , Pellikka, P. A. , Munger, T. M. , Asirvatham, S. J. , Scott, C. G. , Carter, R. E. , & Kapa, S. (2019). Age and sex estimation using artificial intelligence from standard 12‐lead ECGs. Circulation: Arrhythmia and Electrophysiology, 12(9), e00728410. 10.1161/CIRCEP.119.007284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attia, Z. I. , Kapa, S. , Lopez‐Jimenez, F. , McKie, P. M. , Ladewig, D. J. , Satam, G. , Pellikka, P. A. , Enriquez‐Sarano, M. , Noseworthy, P. A. , Munger, T. M. , Asirvatham, S. J. , Scott, C. G. , Carter, R. E. , & Friedman, P. A. (2019). Screening for cardiac contractile dysfunction using an artificial intelligence–enabled electrocardiogram. Nature Medicine, 25(1), 70–74. 10.1038/s41591-018-0240-2 [DOI] [PubMed] [Google Scholar]
- Attia, Z. I. , Noseworthy, P. A. , Lopez‐Jimenez, F. , Asirvatham, S. J. , Deshmukh, A. J. , Gersh, B. J. , Carter, R. E. , Yao, X. , Rabinstein, A. A. , Erickson, B. J. , Kapa, S. , & Friedman, P. A. (2019). An artificial intelligence‐enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: A retrospective analysis of outcome prediction. The Lancet, 394(10201), 861–867. 10.1016/S0140-6736(19)31721-0 [DOI] [PubMed] [Google Scholar]
- Balcı, A. K. , Koksal, O. , Kose, A. , Armagan, E. , Ozdemir, F. , Inal, T. , & Oner, N. (2013). General characteristics of patients with electrolyte imbalance admitted to emergency department. World Journal of Emergency Medicine, 4(2), 113–116. 10.5847/wjem.j.issn.1920-8642.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y. , Kwon, J.‐M. , Kim, K.‐H. , Medina‐Inojosa, J. R. , Jeon, K.‐H. , Cho, S. , Lee, S. Y. , Park, J. , & Oh, B.‐H. (2020). Artificial intelligence algorithm for detecting myocardial infarction using six‐lead electrocardiography. Scientific Reports, 10(1), 20495. 10.1038/s41598-020-77599-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorin, E. , Rosso, R. , & Viskin, S. (2016). Electrocardiographic manifestations of calcium abnormalities. Annals of Noninvasive Electrocardiology, 21(1), 7–9. 10.1111/anec.12316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondup, T. , & Qian, Q. (2017). Electrolyte and acid‐base disorders in chronic kidney disease and end‐stage kidney failure. Blood Purification, 43(1–3), 179–188. 10.1159/000452725 [DOI] [PubMed] [Google Scholar]
- El‐Sharkawy, A. M. , Sahota, O. , Maughan, R. J. , & Lobo, D. N. (2014). The pathophysiology of fluid and electrolyte balance in the older adult surgical patient. Clinical Nutrition, 33(1), 6–13. 10.1016/j.clnu.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Galloway, C. D. , Valys, A. V. , Shreibati, J. B. , Treiman, D. L. , Petterson, F. L. , Gundotra, V. P. , Albert, D. E. , Attia, Z. I. , Carter, R. E. , Asirvatham, S. J. , Ackerman, M. J. , Noseworthy, P. A. , Dillon, J. J. , & Friedman, P. A. (2019). Development and validation of a deep‐learning model to screen for hyperkalemia from the electrocardiogram. JAMA Cardiology, 4(5), 428–436. 10.1001/jamacardio.2019.0640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, A. , Hammerman, H. , Petcherski, S. , Zdorovyak, A. , Yalonetsky, S. , Kapeliovich, M. , Agmon, Y. , Markiewicz, W. , & Aronson, D. (2004). Prognostic importance of hyponatremia in acute ST‐elevation myocardial infarction. American Journal of Medicine, 117(4), 242–248. 10.1016/j.amjmed.2004.03.022 [DOI] [PubMed] [Google Scholar]
- Jo, Y.‐Y. , Cho, Y. , Lee, S. Y. , Kwon, J.‐M. , Kim, K.‐H. , Jeon, K.‐H. , Cho, S. , Park, J. , & Oh, B.‐H. (2021). Explainable artificial intelligence to detect atrial fibrillation using electrocardiogram. International Journal of Cardiology, 328, 104–110. [DOI] [PubMed] [Google Scholar]
- Klingkowski, U. , Kropshofer, G. , Crazzolara, R. , Schachner, T. , & Cortina, G. (2019). Refractory hyperkalaemic cardiac arrest – What to do first: Treat the reversible cause or initiate E‐CPR? Resuscitation, 142, 81. 10.1016/j.resuscitation.2019.07.014 [DOI] [PubMed] [Google Scholar]
- Kwon, J.‐M. , Cho, Y. , Jeon, K.‐H. , Cho, S. , Kim, K.‐H. , Baek, S. D. , Jeung, S. , Park, J. , & Oh, B.‐H. (2020). A deep learning algorithm to detect anaemia with ECGs: A retrospective, multicentre study. The Lancet Digital Health, 2(7), e358–e367. 10.1016/S2589-7500(20)30108-4 [DOI] [PubMed] [Google Scholar]
- Kwon, J.‐M. , Kim, K.‐H. , Medina‐Inojosa, J. , Jeon, K.‐H. , Park, J. , & Oh, B.‐H. (2020). Artificial intelligence for early prediction of pulmonary hypertension using electrocardiography. Journal of Heart and Lung Transplantation, 39(8), 805–814. 10.1016/j.healun.2020.04.009 [DOI] [PubMed] [Google Scholar]
- Kwon, J.‐M. , Lee, S. Y. , Jeon, K.‐H. , Lee, Y. , Kim, K.‐H. , Park, J. , Oh, B.‐H. , & Lee, M.‐M. (2020). Deep learning‐based algorithm for detecting aortic stenosis using electrocardiography. Journal of the American Heart Association, 9(7), e014717. 10.1161/JAHA.119.014717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun, Y. , Bengio, Y. , & Hinton, G. (2015). Deep learning. Nature, 521(7553), 436–444. 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- Lee, C.‐T. , Guo, H.‐R. , & Chen, J.‐B. (2000). Hyponatremia in the emergency department. American Journal of Emergency Medicine, 18(3), 264–268. 10.1016/S0735-6757(00)90118-9 [DOI] [PubMed] [Google Scholar]
- Levis, J. (2012). ECG diagnosis: Hypokalemia. The Permanente Journal, 16(2), 57. 10.7812/TPP/12-015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liamis, G. (2014). Diabetes mellitus and electrolyte disorders. World Journal of Clinical Cases, 2(10), 488. 10.12998/wjcc.v2.i10.488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, L. M. , Tsai, N.‐C. , Lin, M.‐Y. , Hwang, D.‐Y. , Lin, H.‐Y.‐H. , Lee, J.‐J. , Hwang, S.‐J. , Hung, C.‐C. , & Chen, H.‐C. (2016). Hyponatremia is associated with fluid imbalance and adverse renal outcome in chronic kidney disease patients treated with diuretics. Scientific Reports, 6(1), 36817. 10.1038/srep36817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.‐S. , Lin, C. , Fang, W.‐H. , Hsu, C.‐J. , Chen, S.‐J. , Huang, K.‐H. , Lin, W.‐S. , Tsai, C.‐S. , Kuo, C.‐C. , Chau, T. , Yang, S. J. H. , & Lin, S.‐H. (2020). A deep‐learning algorithm (ECG12Net) for detecting hypokalemia and hyperkalemia by electrocardiography: Algorithm development. JMIR Medical Informatics, 8 (3), 1–12. 10.2196/15931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littmann, L. , & Gibbs, M. A. (2018). Electrocardiographic manifestations of severe hyperkalemia. Journal of Electrocardiology, 51(5), 814–817. 10.1016/j.jelectrocard.2018.06.018 [DOI] [PubMed] [Google Scholar]
- Noordam, R. , Young, W. J. , Salman, R. , Kanters, J. K. , van den Berg, M. E. , van Heemst, D. , Lin, H. J. , Barreto, S. M. , Biggs, M. L. , Biino, G. , Catamo, E. , Concas, M. P. , Ding, J. , Evans, D. S. , Foco, L. , Grarup, N. , Lyytikäinen, L.‐P. , Mangino, M. , Mei, H. , … Warren, H. R. (2019). Effects of calcium, magnesium, and potassium concentrations on ventricular repolarization in unselected individuals. Journal of the American College of Cardiology, 73(24), 3118–3131. 10.1016/j.jacc.2019.03.519 [DOI] [PubMed] [Google Scholar]
- Papi, G. , Corsello, S. M. , & Pontecorvi, A. (2014). Clinical concepts on thyroid emergencies. Frontiers in Endocrinology, 5, 102. 10.3389/fendo.2014.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyamvada, S. , Gomes, R. , Gill, R. K. , Saksena, S. , Alrefai, W. A. , & Dudeja, P. K. (2015). Mechanisms underlying dysregulation of electrolyte absorption in inflammatory bowel disease‐associated diarrhea. Inflammatory Bowel Diseases, 21(12), 2926–2935. 10.1097/MIB.0000000000000504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pun, P. H. , Goldstein, B. A. , Gallis, J. A. , Middleton, J. P. , & Svetkey, L. P. (2017). Serum potassium levels and risk of sudden cardiac death among patients with chronic kidney disease and significant coronary artery disease. Kidney International Reports, 2(6), 1122–1131. 10.1016/j.ekir.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoda, K. M. , Porter, M. J. , & Quintini, C. (2011). Fluid and electrolyte management. Journal of Parenteral and Enteral Nutrition, 35(6), 675–685. 10.1177/0148607111421913 [DOI] [PubMed] [Google Scholar]
- Riggs, J. E. (2002). Neurologic manifestations of electrolyte disturbances. Neurologic Clinics, 20(1), 227–239. 10.1016/S0733-8619(03)00060-4 [DOI] [PubMed] [Google Scholar]
- Selvaraju, R. R. , Cogswell, M. , Das, A. , Vedantam, R. , Parikh, D. , & Batra, D. (2017). Grad‐CAM: Visual explanations from deep networks via gradient‐based localization. In Proceedings of the IEEE international conference on computer vision (Vol. 1, pp. 618–626). 10.1109/ICCV.2017.74 [DOI] [Google Scholar]
- Selvaraju, R. R. , Cogswell, M. , Das, A. , Vedantam, R. , Parikh, D. , & Batra, D. (2020). Grad‐CAM: Visual explanations from deep networks via gradient‐based localization. International Journal of Computer Vision, 128(2), 336–359. 10.1007/s11263-019-01228-7 [DOI] [Google Scholar]
- Soar, J. , Perkins, G. D. , Abbas, G. , Alfonzo, A. , Barelli, A. , Bierens, J. J. , Brugger, H. , Deakin, C. D. , Dunning, J. , Georgiou, M. , Handley, A. J. , Lockey, D. J. , Paal, P. , Sandroni, C. , Thies, K. C. , Zideman, D. A. , & Nolan, J. P. (2010). European resuscitation council guidelines for resuscitation 2010 section 8. Cardiac arrest in special circumstances: Electrolyte abnormalities, poisoning, drowning, accidental hypothermia, hyperthermia, asthma, anaphylaxis, cardiac surgery, trauma, pregnancy, electrocution. Resuscitation, 81(10), 1400–1433. 10.1016/j.resuscitation.2010.08.015 [DOI] [PubMed] [Google Scholar]
- Stanifer, J. W. , Jing, B. , Tolan, S. , Helmke, N. , Mukerjee, R. , Naicker, S. , & Patel, U. (2014). The epidemiology of chronic kidney disease in sub‐Saharan Africa: A systematic review and meta‐analysis. The Lancet Global Health, 2(3), e174–e181. 10.1016/S2214-109X(14)70002-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐6
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.


