Abstract
Objective
This study aimed to implement and evaluate machine learning based-models to predict COVID-19’ diagnosis and disease severity.
Methods
COVID-19 test samples (positive or negative results) from patients who attended a single hospital were evaluated. Patients diagnosed with COVID-19 were categorised according to the severity of the disease. Data were submitted to exploratory analysis (principal component analysis, PCA) to detect outlier samples, recognise patterns, and identify important variables. Based on patients’ laboratory tests results, machine learning models were implemented to predict disease positivity and severity. Artificial neural networks (ANN), decision trees (DT), partial least squares discriminant analysis (PLS-DA), and K nearest neighbour algorithm (KNN) models were used. The four models were validated based on the accuracy (area under the ROC curve).
Results
The first subset of data had 5,643 patient samples (5,086 negatives and 557 positives for COVID-19). The second subset included 557 COVID-19 positive patients. The ANN, DT, PLS-DA, and KNN models allowed the classification of negative and positive samples with >84% accuracy. It was also possible to classify patients with severe and non-severe disease with an accuracy >86%. The following were associated with the prediction of COVID-19 diagnosis and severity: hyperferritinaemia, hypocalcaemia, pulmonary hypoxia, hypoxemia, metabolic and respiratory acidosis, low urinary pH, and high levels of lactate dehydrogenase.
Conclusion
Our analysis shows that all the models could assist in the diagnosis and prediction of COVID-19 severity.
Keywords: COVID-19, Diagnosis, Severity, Blood test, Urine test, Machine learning model
1. Introduction
Coronavirus disease (COVID-19) remains an emergency of global interest; up to 21 May 2021, a total of 164.52 million confirmed cases and 3.42 million deaths had accumulated from the disease [1]. Social disparity and the scarcity of hospital resources for the treatment of patients in hospital units have been identified among the main factors associated with an increased number of deaths from this disease [[2], [3], [4], [5], [6], [7]]. Thus, it is essential to identify potential prognostic biomarkers towards earlier and more targeted care, especially considering that some patients with COVID-19 develop severe disease, which is associated with a higher risk of hospitalisation. Biomarkers provide a dynamic and powerful approach to understanding the spectrum of disease with applications in observational and analytic epidemiology, randomised clinical trials, screening and diagnosis, and prognosis [8]. Recently, studies investigating biomarkers to diagnose COVID-19 in early stages have been encouraged worldwide, aiming to provide a faster referral to treatment and reducing health-related problems associated with the disease [17,18].
Machine learning (ML) is an effective and innovative tool able to assist healthcare professionals, policymakers, and other stakeholders during decision-making processes. These computer system models learn and adapt information by using algorithms and statistical networks to analyse and draw inferences from patterns in data. In the field of clinical diagnosis of COVID-19, these predictive analyses grounded on biomarkers can help optimise the screening of patients with severe disease, minimising mortality and hospitalisation, and reducing care delays. Previous machine learning studies highlight that some demographic variables, patients' comorbidities, and laboratory findings can be predictive factors for COVID-19 mortality [[9], [10], [11], [12]]. However, most of these studies included a small sample size, which may impact the model's robustness and reliability of findings (e.g., low sensitivity) [[14], [15], [16]] and prevent its use in practice. Moreover, to date, specific biomarkers associated with the disease severity and patients' hospitalisation in intensive care units are still unknown, which may hamper the development of further targeted treatments [13].
Thus, this study aims to implement and evaluate machine learning-based algorithm models for the diagnosis and prediction of the severity of COVID-19 using data from biochemical, haematological, and urine tests from a large sample size.
2. Material and methods
2.1. Data set
Data from the public Kaggle platform [19] on individuals that had an reverse transcription polymerase chain reaction (RT-PCR) exam to detect severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2) infection in the Israelita Albert Einstein hospital (São Paulo, Brazil) were collected. Regardless of the RT-PCR result (positive or negative for COVID-19), patients were included for analysis when presenting data on biochemical, haematological, and urinary parameters (Table 1 ). Two subgroups of samples were created according to RT-PCR results: (i) 5,643 patients’ samples accounting for both negative (n = 5,086) and positive (n = 557) results; (ii) 557 positive samples of asymptomatic outpatients and patients with severe COVID-19, hospitalised in intensive care units. As the SARS-Cov-2 infection resembles other respiratory diseases, to minimise the chance of obtaining false-positive samples, patients who tested positive for at least one other virus or respiratory bacteria (see Table 1) were excluded from analyses.
Table 1.
Biochemical, urinalysis, haematological, virological, and bacteriological tests performed on the patients included in the study.
| Biochemical tests |
|---|
| Glucose serum, urea, C-reactive protein, creatinine, potassium, sodium, alanine transaminase, aspartate transaminase, gamma-glutamyltransferase, total bilirubin, direct bilirubin, indirect bilirubin, alkaline phosphatase, ionised pH, blood, magnesium analysis, HCO3 (venous blood gas analysis), lactate dehydrogenase, creatine phosphokinase, ferritin, arterial lactic acid, lipase dosage, HCO3 (arterial blood gas analysis), phosphorus, pCO2 (venous blood gas analysis), Hb saturation (venous blood gas analysis), base excess (venous blood gas analysis), pO2 (venous blood gas analysis), total CO2 (venous blood gas analysis), Hb saturation (arterial blood gases), pCO2 (arterial blood gas analysis), base excess (arterial blood gas analysis), pH (arterial blood gas analysis), total CO2 (arterial blood gas analysis), pO2 (arterial blood gas analysis), arterial FiO2, and ctO2 (arterial blood gas analysis). |
| Haematological tests |
| Hematocrit, Hemoglobin, Platelets, Mean platelet volume, Red blood Cells, Lymphocytes, Mean corpuscular hemoglobin concentration, Leukocytes, Basophils, Mean corpuscular hemoglobin, Eosinophils, Mean corpuscular volume, Monocytes, Red blood cell distribution width |
| Urine tests |
| Urine pH, segmented neutrophil, promyelocytes, metamyelocytes and myeloblasts, and the international normalised ratio (INR). |
| Virological tests |
| Respiratory syncytial virus, influenza A, influenza B, parainfluenza 1, coronavirus NL63, rhinovirus/enterovirus, coronavirus HKU1, parainfluenza 3, adenovirus, parainfluenza 4, coronavirus 229E, coronavirus OC43, influenza A H1N1, influenza H1N1 test, and influenza A rapid test. |
| Bacteriological tests |
| Mycoplasma pneumoniae, Chlamydophila pneumoniae, and Streptococcus A. |
2.2. Machine learning models
The first step for implementing any ML model is to perform exploratory analysis. The exploratory analysis intends to: (i) identify the presence of possible outliers, (ii) recognise patterns of data distribution in the multidimensional space, and (iii) identify relationships between variables [20]. In this study, both data used to build the COVID-19 diagnostic model and the data used to build the severity prediction model were previously subjected to two methods of exploratory analysis: principal component analysis (PCA) and k-means cluster analysis (KMCA). Additionally, the outliers were detected and eliminated from the dataset using the graphical method of leverage versus student residuals [21].
A total of four algorithms were used: (i) artificial neural networks (ANN), (ii) decision trees (DT), (iii) discriminant analysis by partial least squares (PLS-DA), and (iv) the method of k-nearest neighbours (KNN). For implementing these models with algorithms for the diagnosis and prediction of COVID-19 severity, 70% of the samples were used for the training set and 30% for the test set. For both the diagnostic model and the severity model, the Kennard-Stone method was employed to select samples from the training set and samples from the test set [22]. The samples used for implementing the algorithms for COVID-19 diagnosis were divided into class 1 (negative samples for COVID-19) and class 2 (positive samples for COVID-19). For the severity prediction models, samples were classified into class 1 (non-severe disease; i.e., outpatients) and class 2 (severe disease; i.e., hospitalised patients).
The number of latent variables (LVs) selected for the ML models was performed using the leave-one-out cross-validation method. The number of LVs presented the lowest square root of mean cross-validation error (RMSECV). The predictive capacity of the model was evaluated using the square root of mean error of prediction (RMSEP), where the classification models were optimised considering the lower RMSEP.
The analytical validation of the models based on the machine learning algorithms was performed using the following metrics: sensitivity, specificity, and accuracy. These figures of merit were calculated using the parameters true positive (TP), true negative (TN), false positive (FP), and false-negative (FN) [20,[23], [24], [25]]. In ML, a sample is called a true positive when it belongs to class one (1) and is correctly classified by the ML algorithm as belonging to class one (1). A sample is considered a false positive when it belongs to class zero (0) and is incorrectly classified by the ML algorithm as being class one (1). A true negative sample belongs to class zero (0) and is correctly classified as class zero (0). Finally, a sample is false negative when it belongs to class one (1) and is wrongly classified by the ML algorithm as class zero (0). Sensitivity and specificity are defined as the ability of the ML-based model to correctly classify negative and positive samples, respectively. Accuracy is the ability of an ML-based model to correctly classify both negative and positive samples. The values of sensitivity, specificity, and accuracy vary from zero (0) to one (1), and the closer to 1, the more sensitive, specific, and accurate the model, respectively. These parameters were calculated according to equations (1), (2), (3), respectively:
| (1) |
| (2) |
| (3) |
where FN: false negative; FP: false positive; TN: true negative; and TP: true positive.
The accuracy of the models was calculated by integrating the area under the receiver operating characteristic (ROC) curve.
3. Results
The variation of all biomarkers (biochemical, haematological, and urinary) for the COVID-19 patients are summarised in Table 2 .
Table 2.
Levels of biochemical, haematological, and urine biomarkers variation in positive patients with severe disease on a normalised scale of patients.
| Biomarker | COVID-19 positive patients' samplesa | COVID-19 severe patients' samplesb |
|---|---|---|
| Hematocrit | Low | Low |
| Haemoglobin | Low | Low |
| Platelets | Low | Low |
| Mean platelet volume | Low | Low |
| Red blood Cells | Low | Low |
| Lymphocytes | Low | Low |
| Mean corpuscular haemoglobin concentration (MCHC) | Low | Low |
| Leukocytes | High | High |
| Basophils | Normal | Normal |
| Mean corpuscular haemoglobin (MCH) | Normal | Low |
| Eosinophils | Low | Low |
| Mean corpuscular volume (MCV) | Low | Low |
| Monocytes | High | Normal |
| Red blood cell distribution width (RDW) | Low | Normal |
| Serum glucose | High | High |
| Neutrophils | Low | Low |
| Urea | Low | Low |
| C-reactive protein | High | High |
| Creatinine | High | High |
| Potassium | Low | Low |
| Sodium | Low | Low |
| Alanine transaminase | High | High |
| Aspartate transaminase | High | High |
| Gamma-glutamyltransferase | High | High |
| Total bilirubin | High | High |
| Direct bilirubin | High | High |
| Indirect bilirubin | High | High |
| Alkaline phosphatase | High | High |
| Ionised calcium | Low | Low |
| pCO2 (venous blood gas analysis) | High | High |
| Magnesium | Low | Low |
| Hb saturation (venous blood gas analysis) | Low | Low |
| Base excess (venous blood gas analysis) | Low | Low |
| pO2 (venous blood gas analysis) | Low | Low |
| Total CO2 (venous blood gas analysis) | High | High |
| pH (venous blood gas analysis) | Low | Low |
| HCO3 (venous blood gas analysis) | High | High |
| Rods | High | High |
| Segmented | Low | Low |
| Promyelocytes | Normal | – |
| Metamyelocytes | Normal | – |
| Myelocytes | Normal | – |
| Urine pH | Low | Low |
| Urine density | Normal | Low |
| Urine red blood cells | Normal | Normal |
| International normalised ratio (INR) | High | High |
| Lactate dehydrogenase | High | High |
| Creatine phosphokinase (CPK) | Normal | Low |
| Ferritin | High | High |
| Arterial lactic acid | High | High |
| Hb saturation (arterial blood gases) | Low | Low |
| pCO2 (arterial blood gas analysis) | High | High |
| Base excess (arterial blood gas analysis) | Low | Low |
| pH (arterial blood gas analysis) | Low | Low |
| Total CO2 (arterial blood gas analysis) | High | High |
| HCO3 (arterial blood gas analysis) | High | High |
| pO2 (arterial blood gas analysis) | Low | Low |
| Arterial FiO2 | Low | Low |
| Phosphorous | Low | – |
Diagnostic data.
Disease severity data.
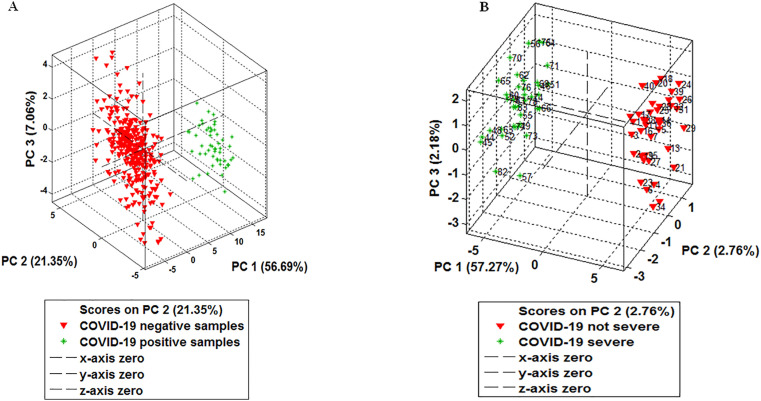
The results of the exploratory analyses of the diagnostic and severity disease data using the PCA model are depicted in Fig. 1 . Additional results with the KMCA model are available in Figures ESM 1 and ESM 2 in the Online Resource. These models were able to differentiate between positive and negative patients for COVID-19 (diagnostic data) and between patients with non-severe disease and vs severe disease (disease severity data).
Fig. 1.
Exploratory analysis. Principal component analysis (PCA) model for the discrimination of negative and positive samples (A) and samples from patients with severe and non-severe disease (B).
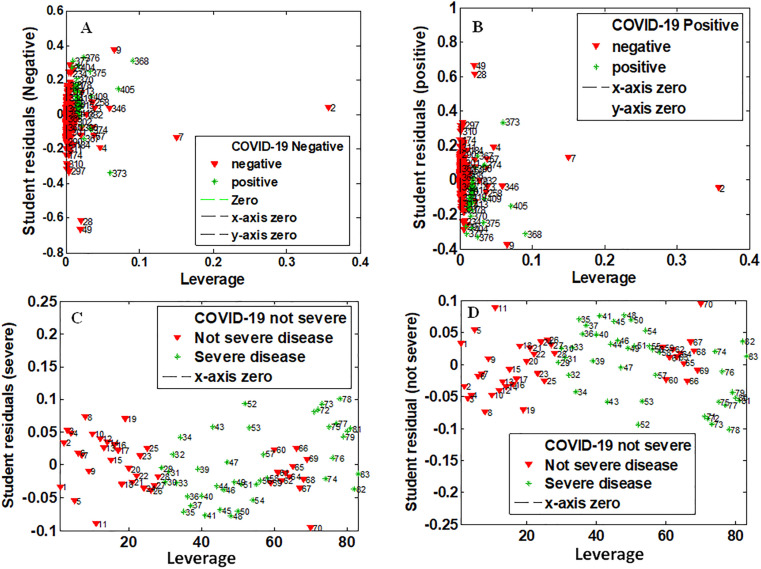
Outlier samples were analysed for both diagnostic and disease severity data using the graph of leverage versus student residuals with 95% of confidence interval (Fig. 2 ). No sample was detected as an outlier.
Fig. 2.
Graph of leverage versus student residuals for detecting outlier samples. For diagnostic data: outlier analysis of negative samples (A) and positive samples (B). For severity data: outlier analysis for samples from patients without severity (C) and with severity (D).
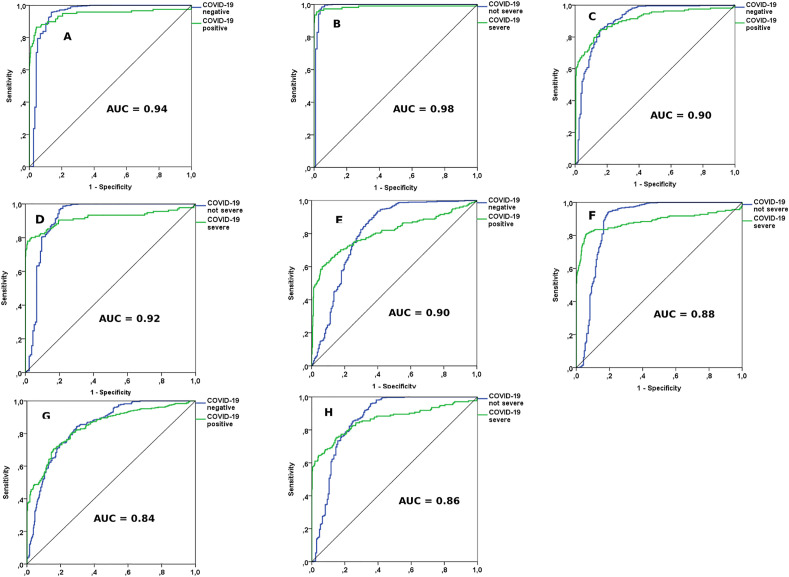
The results of the four ML-based models for the data subsets are presented in Table 3 . All models for COVID-19 diagnosis and prediction of disease severity were compared with each other using the following metrics: training time, model training error, cross-validation error, sensibility, specificity, and accuracy (area under the ROC curve). The ANN model performed better as it presented less training time and forecasting errors, and greater overall accuracy. The ROC curves of the models are depicted in Fig. 3 .
Table 3.
Performance comparison of the machine learning models for COVID-19.
| Metric | Diagnostic model |
Disease severity model |
||||||
|---|---|---|---|---|---|---|---|---|
| RNA | DT | PLS-DA | K-NN | RNA | DT | PLS-DA | K-NN | |
| Training time | 21 min. 43 s | 27 min. 11 s | 31 min. 19 s | 22 min. 15 s | 7 min. 1 s | 10 min. 19 s | 18 min. 3 s | 09 min. 53 s |
| Calibration error | 1.0% | 0.5% | 1.2% | 0.5% | 1.0% | 8.4% | 6.0% | 0.4% |
| Cross validation error | 0.8% | 1.0% | 0.9% | 0.6% | 0.5% | 1.8% | 4.0% | 0.7% |
| Sensibility | 0.93 | 0.89 | 0.88 | 0.84 | 0.99 | 0.90 | 0.87 | 0.82 |
| Specificity | 0.94 | 0.89 | 0.90 | 0.83 | 0.97 | 0.94 | 0.88 | 0.88 |
| Accuracya | 0.94 | 0.90 | 0.90 | 0.84 | 0.98 | 0.92 | 0.88 | 0.86 |
Area under the ROC curve.
Fig. 3.
ROC curves of the accuracy of the machine learning models. Artificial neural network (ANN): diagnosis (A) and severity (B). Decision tree (DT): diagnosis (C) and severity (D). Discriminant analysis by partial least squares (PLS-DA): diagnosis (E) and severity (F). K-nearest neighbours (KNN): diagnosis (G) and severity (H).
According to the ML-based models, some biomarkers were judged as critical or important for predicting COVID-19 and disease severity (see Table 4 ). Ferritin was ranked as the most important variable in all models. Further information is available on Online Resource.
Table 4.
Important biomarkers in machine learning models for the diagnosis and classification of COVID-19 severity.
| Biomarkers | Diagnostic model |
Disease severity model |
||||||
|---|---|---|---|---|---|---|---|---|
| ANN | DT | PLSDA | KNN | ANN | DT | PLSDA | KNN | |
| Ferritin | + + | + + | + + | + + | + + | + + | + + | + + |
| Gamma-glutamyltransferase | + | - | - | - | - | - | - | - |
| HCO3 (arterial) | + | - | - | + | - | - | + | + |
| Base excess (arterial) | + | - | - | - | - | - | + | + |
| Base excess (venous) | - | - | - | - | - | - | + | - |
| Sodium | + | - | - | - | - | - | - | - |
| Total O2 (arterial) | - | - | - | - | + | - | - | - |
| pO2 (arterial) | - | - | - | + | - | - | - | - |
| Total CO2 | - | + | + | + | - | - | - | + |
| pCO2 (arterial) | + | - | - | - | - | - | + | + |
| pCO2 (venous) | + | - | + | - | - | - | + | - |
| Indirect bilirubin | + | - | - | - | - | - | - | - |
| Alkaline phosphatase | + | - | - | - | + | - | - | - |
| Urine pH | - | - | + | + | + | - | + | - |
| pH (venous) | - | - | - | - | - | - | + | - |
| pH (arterial) | - | - | - | - | - | - | - | + |
| FiO2 (arterial) | - | - | - | - | + | - | - | |
| ctO2 (arterial) | - | - | - | - | - | - | - | + |
| Total bilirubin | - | - | - | - | + | - | - | - |
| Red blood cell distribution width | - | - | - | - | + | - | - | - |
| Platelets | - | - | - | - | + | - | - | - |
| C-reactive protein | - | - | - | - | + | - | + | - |
| Calcium ionised | - | - | + | + | - | - | - | - |
| Urine-density | - | - | + | + | - | - | - | - |
| Lactate dehydrogenase | - | - | - | - | - | - | + | - |
| Arterial lactic acid | - | - | - | + | - | - | - | + |
| Haemoglobin saturation (arterial) | - | - | - | - | - | - | - | + |
| Phosphorous | - | - | + | + | - | - | - | - |
| Lipase dosage | - | - | - | - | - | - | + | - |
| Rods | - | - | - | + | - | - | + | - |
Less important variable (−); Important variable (+); Critical variable (++).
4. Discussion
In the present study, we used four ML-based models (ANN, DT, PLS-DA, and KNN) with over 5,000 RT-PCR samples and data on patients’ biochemical, haematological, and urinary parameters that effectively predicted COVID-19 diagnosis and disease severity in Brazil. Considering the high likelihood of existing SARS-Cov-2 mutations (B.1.1.7, P.1, and P.2) in the sample, the complexity of these models is even higher [[38], [39], [40]].
Several ML-based models, including unsupervised approaches (e.g., PCA and hierarchical cluster analysis) and supervised models (e.g., artificial neural network, PLS-DA, DT, KMC, and KNN), are available in the scientific literature [26,27]. The performance of these models relies on several factors, including sample size and the type of data. ML-based models built with larger samples are usually more accurate and efficient for series forecasting; for instance, the deep neural networks that require a great amount of training data [[28], [29], [30]]. The larger the network architecture, the more data is needed to obtain more robust models [[31], [32], [33]].
Regarding COVID-19, previous models have been implemented aiming at predicting disease behaviour and severity. However, most of these studies used a small sample size, which may directly impact the performance of the model [[34], [35], [36]]. Banerjee (2020) developed an ML algorithm to forecast COVID-19 diagnosis using a public database with 598 patients, of which only 39 were positive for SARS-CoV-2. The authors obtained a model with good specificity (91%) but low sensitivity (43%), which can prevent the use of the model in practice for early diagnosis of the disease [14]. Similarly, Joshi (2020) implemented a logistic regression model previously trained with 390 samples, of which only 33 were positive for COVID-19, proving sensitivity and specificity values of 93% and 43%, respectively [37]. Additionally, most studies implemented ML-based models using only routine blood tests [41,42]. In our study, besides the complete blood count test, data from patients’ biochemical, urinary, bacteriological, and virological tests aiming at identifying further biomarkers associated with COVID-19 were also included.
The models reached 84%–98% accuracy. The biomarkers that most contributed to this result and predicting the diagnosis and severity of COVID-19 included: hyperferritinaemia, hypocalcemia (low levels of ionised calcium), hypoxemia (low arterial oxygen pressure, pO2), pulmonary hypoxia (low inspired fraction of arterial oxygen, FiO2), respiratory acidosis (high levels of total CO2 and pCO2), metabolic acidosis (high levels of lactic acid and low venous pH), low urinary pH, and high levels of lactate dehydrogenase (LDH).
Similar precision values were recently reported by Zhou (2021) (94%) and Wu (2021) (90%) after the implementation of ML-based models for COVID-19 diagnosis and disease severity, respectively [15,35]. However, different variables were highlighted by the authors as important for data classification. According to Zhou (2021), these were the rates of circulating lymphocytes, while Wu (2021) reported the rates of neutrophils and lymphocytes, the neutrophil/lymphocyte ratio, and platelet/lymphocyte ratio [15,35]. This variation may be associated with the different sample sizes (n = 357 vs n = 51) and type of models (decision tree model vs support vector machine model).
Recently, most patients with severe COVID-19 who require hospitalisation in intensive care units develop an atypical form of acute distress syndrome, which is usually accompanied by a preserved volume of pulmonary gas [43]. This suggests hypoxia, which results from the difficulty in performing gas exchanges at the level of the pulmonary alveoli. It has been observed that this pulmonary dysfunction can also compromise iron metabolism [43]. Imbalances in both haemoglobin and ferritin levels were reported in patients with severe disease or deaths caused by COVID-19. In a systematic review with a meta-analysis conducted by Taneri (2020), which included 189 studies (n = 57,563 patients), high-risk patients with severe disease had significantly high levels of ferritin (weighted mean difference (WMD), 473.25 ng/mL (95% CI 382.52; 563.98)) and low haemoglobin levels (WMD, 4.08 g/L (95% CI 5.12; −3.05)) when compared to patients with moderate or low-risk disease [44]. In our study, both patient groups with the disease (diagnostic model) and patients with severe disease (disease severity model) presented low levels of haemoglobin and high levels of ferritin. All developed ML-based models (ANN, PLS-DA, KNN, and DT) highlighted that the differences in the levels of these two biomarkers were critical, both for the prediction of COVID-19 diagnosis and disease severity. Recent studies have indicated anaemia and hyperferritinaemia to be strong biomarkers for the prognosis of mortality due to SARS-CoV-2, in addition to other serious respiratory diseases [[45], [46], [47], [48], [49], [50]].
Ferritin is a protein found mainly in the liver, bone marrow, and spleen and is the body's main source of iron storage. It is considered a key biomarker of immune dysregulation, mainly in a situation of hyperferritinaemia, through the direct route of proinflammatory and immunosuppressive effects contributing to a cytokine storm [[51], [52], [53]]. Some studies show that fatal outcomes caused by COVID-19 are accompanied by cytokine storm syndrome involving high levels of inflammatory markers, such as ferritin [54,55]. In the present study, ferritin was associated with predicting the severity of COVID-19, and it was the most important biomarker in predicting the diagnosis of the disease by all the ML-based models developed, corroborating the literature data.
Acid and base disorders are important indicators in the pathogenesis and severity of several diseases, especially respiratory diseases of infectious origin, such as pneumonia [[56], [57], [58]]. Acidosis can occur as a result of a significant increase in arterial carbon dioxide pressure (respiratory acidosis) or a variety of inorganic or organic compounds (metabolic acidosis), such as bicarbonate, lactic acid arterial, ketones, or as a result of renal failure or hyperchloremic acidosis; all of these factors act simultaneously in the increase of hydrogen protons and, consequently, the reduction of blood and respiratory pH levels [[59], [60], [61], [62]]. Researchers suggest that the metabolic acidosis caused by lactic acid in COVID-19 is probably due to anaerobic glycolysis, which is favoured in consequence of hypoxemia. In this condition, pyruvate, a product of the glycolytic pathway, is not translocated to mitochondria to follow the oxidative process [63,64]; instead, it is converted into lactate in the cytosol by the LDH enzyme. As the hypoxemia impairs the tissue oxygenation and the oxidative phosphorylation, the cells obtain ATP by anaerobic glycolysis. This flow relies on the conversion of pyruvate to lactate which results in high levels of this metabolite that comes out of the cells. The excessive consumption of lactate during the process of gluconeogenesis culminates in lactic acidosis [65]. It has been reported that on the 18th day of COVID-19 disease, the levels of lactic acid begin to increase significantly, triggering metabolic acidosis, although the carbon dioxide pressure is acceptable [66].
In our study, patients positively diagnosed with COVID-19 (diagnostic model) and with severe disease (disease severity model) had high levels of carbon dioxide pressure (arterial and venous gas analysis), total carbon dioxide (arterial and venous gas analysis), arterial lactic acid and bicarbonate (arterial gas analysis), and exceptionally low venous pH (pH = 0.3), which also suggests respiratory and metabolic acidosis. These results are similar to other countries, indicating that these metabolic imbalances are prevalent in COVID-19 patients [64,67,68]. In general, all the available data show that most patients with severe disease have accompanying comorbidities, such as diabetes. Recent studies indicate that metabolic acidosis is influenced by the use of metformin for the treatment of diabetes mellitus [69,70].
The present analysis detected levels of five function biomarkers, including bilirubin, direct bilirubin, indirect bilirubin, alanine transaminase, and aspartate transaminase, increased in patients with positive (diagnostic model) and severe (severity model) disease compared to patients with non-severe disease. A systematic review and meta-analysis performed by Parohan (2020) revealed similar findings, where all 1,455 patients with severe disease had extremely high levels of total bilirubin (WMD 2.30 mmol/l; 95% CI, 1.24; 3.36; p < 0.001), alanine aminotransferase (WMD 7.35 U/L; 95% CI, 4.77; 9.93; p < 0.001) and aspartate aminotransferase (WMD 8.84 U/L; 95% CI 5.97; 11.71; p < 0.001), compared to 1,973 patients with non-severe disease [71]. Liver damage has also been reported in other viral pneumonia (e.g., MERS and SARS) and is directly associated with disease severity and mortality [[72], [73], [74], [75]]. However, the biochemical/pathophysiological mechanisms that explain the liver dysfunction caused by COVID-19 are still unknown. It is unclear if the liver dysfunction is due to SARS-Cov-2 or is a consequence of multiple organ failure caused by the virus [71].
Additionally, high levels of the C-reactive protein (CRP) in the samples were found. An increase in this important biomarker in COVID-19 patients’ was previously reported by Chen (2020) (up to 86%) [50]. Recently, a systematic review with meta-analysis concluded that extremely high levels of CRP were statistically associated with COVID-19 severity [77,78]. CRP is an inflammatory protein in the acute phase of inflammatory and infectious processes that is synthesised mainly in liver cells but also smooth muscle cells, macrophages, lymphocytes, and adipocytes. High levels of CRP (increasing up to 100 times) are commonly found during infections (plasma CRP levels increase around 1–500 μg/mL within 24–72 h) [76]. However, the role of CRP isoforms and their involvement in the progression of infectious diseases is still unknown [77,78].
Finally, in addition to the biochemical parameters already mentioned, low calcium levels were also detected in our analysis as a predictor for COVID-19 diagnosis and disease severity. Calcium is essential for a wide variety of processes in the body, ranging from normal muscle contraction to enzymatic activities [79]. It is known that alteration in Ca2+ homeostasis can contribute to cell death by necrosis and apoptosis. Evidence shows that calcium metabolism disorders are associated with cardiovascular disease and early cell death [80,81]. Low calcium levels have already been associated with increased in-hospital mortality in patients with severe coronary artery disease [82], septic patients [79], bacterial pneumonia [83], and patients with dengue [84]. More recent studies have also associated hypocalcemia as an important predictor of hospitalisation and mortality risk by COVID-19 [67,[85], [86], [87]].
Although the present study has shown consistent results, it has some limitations. Cross-sectional studies, with no follow-up analysis of patients' data, are prone to selection bias, information bias, and confounding bias. In addition, biomarkers’ levels may change during the disease.
5. Conclusion
All the ML-based models (ANN, DT, PLS-DA, and KNN) were able to effectively predict COVID-19 diagnosis and disease severity with an accuracy above 84%, which is similar to the results obtained by RT-PCR and the minimum recommended threshold for diagnostic tests. The ANN was the model with the best performance (94% and 98%) and, thus, could be used as a supporting decision tool for healthcare professionals in practice. Hyperferritinaemia, hypocalcemia, hypoxemia, pulmonary hypoxia, respiratory acidosis, metabolic acidosis, low urinary pH, and high levels of lactate dehydrogenase were associated with COVID-19 diagnosis and disease severity. These biomarkers are potential therapeutic targets that should be more effectively investigated in further clinical trials.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authors’ contributions
Study concepts: AFC, RP, FST, AW, GRN.
Study design: AFC, FST, RP, GRN.
Data acquisition: AFC, MMF, DPS.
Statistical analysis: AFC, DPS, RP, MS, GRN.
Manuscript preparation: AFC, AW, MMF, MS, GRN.
Manuscript editing: AFC, AW, DPS, MMF, MS.
Manuscript review: AFC, DPS, MMF, MS, FST, RP, AW, GRN.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors express their gratitude to the CAPES (Brazilian Federal Agency for Support and Evaluation of Graduate Education within the Ministry of Education of Brazil) for research funding - Finance Code 001.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104531.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.World Health Organization, WHO WHO coronavirus disease (COVID-19) Dashboard 2021. 2021. https://covid19.who.int/ Available from:
- 2.Cobre A.F., Böger B., Fachi M.M., Vilhena R. de O., Domingos E.L., Tonin F.S., Pontarolo R. Risk factors associated with delay in diagnosis and mortality in patients with Covid-19 in the city of Rio de Janeiro, Brazil, Cienc. Saúde Coletiva. 2020;25:4131–4140. doi: 10.1590/1413-812320202510.2.26882020. [DOI] [PubMed] [Google Scholar]
- 3.Cobre A.F., Surek M., Vilhena R. de O., Böger B., Fachi M.M., Momade D.R.O., Tonin F.S., Sarti F.M., Pontarolo R. Influence of foods and nutrients on COVID-19 recovery: a multivariate analysis of data from 170 countries using a generalized linear model. Clin. Nutr. 2021 doi: 10.1016/j.clnu.2021.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Supady A., Curtis J.R., Abrams D., Lorusso R., Bein T., Boldt J., Brown C.E., Duerschmied D., Metaxa V., Brodie D. Allocating scarce intensive care resources during the COVID-19 pandemic: practical challenges to theoretical frameworks. Lancet Respir. Med. 2021;9:430–434. doi: 10.1016/S2213-2600(20)30580-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ribeiro A.L., Alves-Sousa N.W., Martins-Filho P.R., Carvalho V.O. Social disparity in magnifying glass: the inequality among the vulnerable people during COVID-19 pandemic. Int. J. Clin. Pract. 2021;75:2–3. doi: 10.1111/ijcp.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobre A.F., Böger B., Vilhena R. de O., Fachi M.M., dos Santos J.M., Tonin F.S. A multivariate analysis of risk factors associated with death by Covid-19 in the USA, Italy, Spain, and Germany. J. Public Health. 2020 doi: 10.1007/s10389-020-01397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emanuel E.J., Persad G., Upshur R., Thome B., Parker M., Glickman A., Zhang C., Boyle C., Smith M., Phillips J.P. Fair allocation of scarce medical resources in the time of Covid-19. N. Engl. J. Med. 2020;382:2049–2055. doi: 10.1056/NEJMsb2005114. [DOI] [PubMed] [Google Scholar]
- 8.Weiss P., Murdoch D.R. Clinical course and mortality risk of severe COVID-19. Lancet. 2020;395:1014–1015. doi: 10.1016/S0140-6736(20)30633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolourani S., Brenner M., Wang P., McGinn T., Hirsch J.S., Barnaby D., Zanos T.P., Barish M., Cohen S.L., Coppa K., Davidson K.W., Debnath S., Lau L., Levy T.J., Makhnevich A., Paradis M.D., Tóth V. A machine learning prediction model of respiratory failure within 48 hours of patient admission for COVID-19: model development and validation. J. Med. Internet Res. 2021;23:1–15. doi: 10.2196/24246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang W., Liang H., Ou L., Chen B., Chen A., Li C., Li Y., Guan W., Sang L., Lu J., Xu Y., Chen G., Guo H., Guo J., Chen Z., Zhao Y., Li S., Zhang N., Zhong N., He J. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern. Med. 2020;180:1081–1089. doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan X., Zhang B., Fu M., Li M., Yuan X., Zhu Y., Peng J., Guo H., Lu Y. Clinical and inflammatory features based machine learning model for fatal risk prediction of hospitalized COVID-19 patients: results from a retrospective cohort study. Ann. Med. 2021;53:257–266. doi: 10.1080/07853890.2020.1868564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pourhomayoun M., Shakibi M. Predicting mortality risk in patients with COVID-19 using machine learning to help medical decision-making. Smart Health (Amsterdam, Netherlands) 2021;20 doi: 10.1016/j.smhl.2020.100178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heffernan A.J. Host diagnostic biomarkers of infection in the ICU : where are we and where are we going ? Curr. Infect. Dis. Rep. 2021;23:4. doi: 10.1007/s11908-021-00747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee A., Ray S., Vorselaars B., Kitson J., Mamalakis M., Weeks S., Baker M., Mackenzie L.S. Use of machine learning and artificial intelligence to predict SARS-CoV-2 infection from full blood counts in a population. Int. Immunopharmacol. 2020;86:106705. doi: 10.1016/j.intimp.2020.106705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X., Wang Z., Li S., Liu T., Wang X., Xia J., Zhao Y. Machine learning-based decision model to distinguish between covid-19 and influenza: a retrospective, two-centered, diagnostic study, Risk Manag. Healthc. Policy. 2021;14:595–604. doi: 10.2147/RMHP.S291498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schöning V., Liakoni E., Baumgartner C., Exadaktylos A.K., Hautz W.E., Atkinson A., Hammann F. Development and validation of a prognostic COVID-19 severity assessment (COSA) score and machine learning models for patient triage at a tertiary hospital. J. Transl. Med. 2021;19:1–11. doi: 10.1186/s12967-021-02720-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee A., Ray S., Vorselaars B., Kitson J., Mamalakis M., Weeks S., Baker M., Mackenzie L.S. Use of machine learning and artificial intelligence to predict SARS-CoV-2 infection from full blood counts in a population. Int. Immunopharmacol. 2020;86:106705. doi: 10.1016/j.intimp.2020.106705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J., Zhang P., Zhang L., Meng W., Li J., Tong C., Li Y., Cai J., Yang Z., Zhu J., Zhao M., Huang H., Xie X., Li S. Rapid and accurate identification of COVID-19 infection through machine learning based on clinical available blood test results. MedRxiv. 2020 doi: 10.1101/2020.04.02.20051136. [DOI] [Google Scholar]
- 19.Kaagle 2020. https://www.kaggle.com/allen-institute-for-ai/CORD-19-research-challenge/discussion/139347
- 20.Ballabio D., Consonni V. Classification tools in chemistry. Part 1: linear models. PLS-DA. Anal. Methods. 2013;5:3790–3798. doi: 10.1039/c3ay40582f. [DOI] [Google Scholar]
- 21.Walczak B., Massart D.L. Multiple outlier detection revisited. Chemometr. Intell. Lab. Syst. 1998;41:1–15. doi: 10.1016/S0169-7439(98)00034-3. [DOI] [Google Scholar]
- 22.Taylor P., Kennard R.W., Stone L.A. Technometrics computer aided design of experiments. Technometric. 1969;11:137–148. [Google Scholar]
- 23.Ruiz-Perez D., Guan H., Madhivanan P., Mathee K., Narasimhan G. So you think you can PLS-DA? BMC Bioinf. 2020;21:1–10. doi: 10.1186/s12859-019-3310-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. BBA - Protein Struct. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 25.Wise R.S.K., M B., Gallagher N.B., Bro R., Shaver J.M., Windig W. Eigenvector Research Inc.; Manson, WA: 2006. PLS Toolbox 4.0 for Use with MATLAB. [Google Scholar]
- 26.Suthaharan S. Pringer; New York: 2016. Machine Learning Models and Algorithms for Big Data: Thinking with Examples for Effective Learning. [DOI] [Google Scholar]
- 27.Arthur D., Vassilvitskii S., K-means++ Proc. Annu. ACM-SIAM Symp. Discret. Algorithms. 07-09-January-2007. 2007. The advantages of careful seeding; pp. 1027–1035. [Google Scholar]
- 28.Réda C., Kaufmann E., Delahaye-Duriez A. Machine learning applications in drug development. Comput. Struct. Biotechnol. J. 2020;18:241–252. doi: 10.1016/j.csbj.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin S.J., Chiang L.H. Advances and opportunities in machine learning for process data analytics. Comput. Chem. Eng. 2019;126:465–473. doi: 10.1016/j.compchemeng.2019.04.003. [DOI] [Google Scholar]
- 30.Butler K.T., Davies D.W., Cartwright H., Isayev O., Walsh A. Machine learning for molecular and materials science. Nature. 2018;559:547–555. doi: 10.1038/s41586-018-0337-2. [DOI] [PubMed] [Google Scholar]
- 31.Shallue C.J., Lee J., Antogniniy J., Sohl-Dickstein J., Frostig R., Dahl G.E. Measuring the effects of data parallelism on neural network training. ArXiv. 2018;20:1–49. [Google Scholar]
- 32.Alwosheel A., van Cranenburgh S., Chorus C.G. Is your dataset big enough? Sample size requirements when using artificial neural networks for discrete choice analysis. J. Choice Model. 2018;28:167–182. doi: 10.1016/j.jocm.2018.07.002. [DOI] [Google Scholar]
- 33.Liu B., Wei Y., Zhang Y., Yang Q. Deep neural networks for high dimension, low sample size data. IJCAI Int. Jt. Conf. Artif. Intell. 2017:2287–2293. doi: 10.24963/ijcai.2017/318. [DOI] [Google Scholar]
- 34.Dai W., Ke P.F., Li Z.Z., Zhuang Q.Z., Huang W., Wang Y., Xiong Y., Huang X.Z. Establishing classifiers with clinical laboratory indicators to distinguish COVID-19 from community-acquired pneumonia: retrospective cohort study. J. Med. Internet Res. 2021;23 doi: 10.2196/23390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu P., Ye H., Cai X., Li C., Li S., Chen M., Wang M., Heidari A.A., Chen M., Li J., Chen H., Huang X., Wang L. An effective machine learning approach for identifying non-severe and severe coronavirus disease 2019 patients in a rural Chinese population: the wenzhou retrospective study. IEEE Access. 2021;9:45486–45503. doi: 10.1109/ACCESS.2021.3067311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laing A.G., Lorenc A., del Molino del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I., Kamdar S., Joseph M., Davies D., Davis R., Jennings A., Zlatareva I., Vantourout P., Wu Y., Sofra V., Cano F., Greco M., Theodoridis E., Freedman J., Gee S., Chan J.N.E., Ryan S., Bugallo-Blanco E., Peterson P., Kisand K., Haljasmägi L., Chadli L., Moingeon P., Martinez L., Merrick B., Bisnauthsing K., Brooks K., Ibrahim M.A.A., Mason J., Lopez Gomez F., Babalola K., Abdul-Jawad S., Cason J., Mant C., Seow J., Graham C., Doores K.J., Di Rosa F., Edgeworth J., Shankar-Hari M., Hayday A.C. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 37.Joshi R.P., Pejaver V., Hammarlund N.E., Sung H., Lee S.K., Furmanchuk A., Lee H.Y., Scott G., Gombar S., Shah N., Shen S., Nassiri A., Schneider D., Ahmad F.S., Liebovitz D., Kho A., Mooney S., Pinsky B.A., Banaei N. A predictive tool for identification of SARS-CoV-2 PCR-negative emergency department patients using routine test results. J. Clin. Virol. 2020;129:104502. doi: 10.1016/j.jcv.2020.104502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kymie C., Nonaka V., Franco M.M., Gräf T., Verena A., Mendes A., Santana De Aguiar R., Giovanetti M., Solano B., Souza F., Souza B.S.F. Genomic evidence of a Sars-Cov-2 reinfection case with E484K spike mutation in Brazil. Preprints.Org. 2021;55:6–11. https://pangolin.cog-uk.io/ [Google Scholar]
- 39.Faria N.R., Claro I.M., Candido D., Franco L.A.M., Andrade P.S., Thais M., Silva C.A.M., Sales F.C., Erika R., Aguiar R.S., Gaburo N., Cecília C., Fraiji N.A., Crispim M.A.E., Carvalho P.S.S., Rambaut A. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virol. Org. 2021;2020:1–9. [Google Scholar]
- 40.Voloch C.M., da Silva Francisco R., de Almeida L.G.P., Cardoso C.C., Brustolini O.J., Gerber A.L., Guimarães A.P. de C., Mariani D., da Costa R.M., Ferreira O.C., Frauches T.S., de Mello C.M.B., Leitão I. de C., Galliez R.M., Faffe D.S., Castiñeiras T.M.P.P., Tanuri A., de Vasconcelos A.T.R. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021 doi: 10.1128/jvi.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Formica V., Minieri M., Bernardini S., Ciotti M., D'Agostini C., Roselli M., Andreoni M., Morelli C., Parisi G., Federici M., Paganelli C., Legramante J.M. Complete blood count might help to identify subjects with high probability of testing positive to SARS-CoV-2. Clin. Med. J. R. Coll. Phys. Lond. 2020;20:114–119. doi: 10.7861/CLINMED.2020-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Federico C., Andrea C., Davide F., Chiara D.R., Daniele C., Alessandra C., Elena D.V., Giuseppe B., Massimo L., Galeazzi V.R., Carobene A. Development, evaluation, and validation of machine learning models for COVID-19 detection based on routine blood tests. MedRxiv. 2021 doi: 10.1101/2020.10.02.20205070. [DOI] [Google Scholar]
- 43.Lang M., Som A., Mendoza D.P., Flores E.J., Reid N., Carey D., Li M.D., Witkin A., Rodriguez-Lopez J.M., Shepard J.A.O., Little B.P. Hypoxaemia related to COVID-19: vascular and perfusion abnormalities on dual-energy CT. Lancet Infect. Dis. 2020;20:1365–1366. doi: 10.1016/S1473-3099(20)30367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taneri P.E., Gómez-Ochoa S.A., Llanaj E., Raguindin P.F., Rojas L.Z., Roa-Díaz Z.M., Salvador D., Groothof D., Minder B., Kopp-Heim D., Hautz W.E., Eisenga M.F., Franco O.H., Glisic M., Muka T. Anemia and iron metabolism in COVID-19: a systematic review and meta-analysis. Eur. J. Epidemiol. 2020;35:763–773. doi: 10.1007/s10654-020-00678-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasmussen L., Christensen S., Lenler-Petersen P., Johnsen S.P. Anemia and 90-day mortality in COPD patients requiring invasive mechanical ventilation. Clin. Epidemiol. 2011;3:1–5. doi: 10.2147/CLEP.S12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lomholt F.K., Laulund A.S., Bjarnason N.H., Jørgensen H.L., Godtfredsen N.S. Meta-analysis of routine blood tests as predictors of mortality in COPD. Eur. Clin. Respir. J. 2014;1:24110. doi: 10.3402/ecrj.v1.24110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Z., Sun R., Li J., Cheng W., Li L. Relations of anemia with the all-cause mortality and cardiovascular mortality in general population: a meta-analysis. Am. J. Med. Sci. 2019;358:191–199. doi: 10.1016/j.amjms.2019.05.016. [DOI] [PubMed] [Google Scholar]
- 48.Bennett T.D., Hayward K.N., Farris R.W.D., Ringold S., Wallace C.A., Brogan T.V. Very high serum ferritin levels are associated with increased mortality and critical care in pediatric patients. Pediatr. Crit. Care Med. 2011;12:233–236. doi: 10.1097/PCC.0b013e31820abca8. [DOI] [PubMed] [Google Scholar]
- 49.Fox S.E., Akmatbekov A., Harbert J.L., Li G., Quincy Brown J., Vander Heide R.S. Pulmonary and cardiac pathology in African American patients with COVID-19: an autopsy series from New Orleans. Lancet Respir. Med. 2020;8:681–686. doi: 10.1016/S2213-2600(20)30243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nadadur S.S., Srirama K., Mudipalli A. Iron transport & homeostasis mechanisms: their role in health & disease. Indian J. Med. Res. 2008;128:533–544. [PubMed] [Google Scholar]
- 52.Hunt J.R. How important is dietary iron bioavailability? Am. J. Clin. Nutr. 2001;73:3–4. doi: 10.1093/ajcn/73.1.3. [DOI] [PubMed] [Google Scholar]
- 53.Abbaspour N., Hurrell R., Kelishadi R. Review on iron and its importance for human health. J. Res. Med. Sci. Off. J. Isfahan Univ. Med. Sci. 2014;19:164–174. [PMC free article] [PubMed] [Google Scholar]
- 54.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vargas-vargas M., Cortés-rojo C. Letter to the editor ferritin levels and COVID-19. Ther. Adv. Vaccines. 2020;9:2019–2020. [Google Scholar]
- 56.Goraya N., Wesson D.E. Acid-base status and progression of chronic kidney disease. Curr. Opin. Nephrol. Hypertens. 2012;21:552–556. doi: 10.1097/MNH.0b013e328356233b. [DOI] [PubMed] [Google Scholar]
- 57.Alper S.L. Genetic diseases of acid-base transporters. Annu. Rev. Physiol. 2002;64:899–923. doi: 10.1146/annurev.physiol.64.092801.141759. [DOI] [PubMed] [Google Scholar]
- 58.Bruno C.M., Valenti M. Acid-base disorders in patients with chronic obstructive pulmonary disease: a pathophysiological review. J. Biomed. Biotechnol. 2012;2012 doi: 10.1155/2012/915150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Modi R.G.M. Assessment of acid base disorders in critically ill medical patients. Int. J. Med. Sci. Educ. 2019;6:27–31. [Google Scholar]
- 60.Gunnerson K.J. Clinical review: the meaning of acid-base abnormalities in the intensive care unit - Epidemiology. Crit. Care. 2005;9:508–516. doi: 10.1186/cc3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J., Hoskote A., Hickey C., Stephens D., Bohn D., Holtby H., Van Arsdell G., Redington A.N., Adatia I. Effect of carbon dioxide on systemic oxygenation, oxygen consumption, and blood lactate levels after bidirectional superior cavopulmonary anastomosis. Crit. Care Med. 2005;33:984–989. doi: 10.1097/01.CCM.0000162665.08685.E2. [DOI] [PubMed] [Google Scholar]
- 62.Kellum J.A., Song M., Subramanian S. Acidemia: good, bad or inconsequential? Yearb. Intens. Care Emerg. Med. 2002;2002:510–516. doi: 10.1007/978-3-642-56011-8_47. [DOI] [Google Scholar]
- 63.De Backer D. Lactic acidosis. Intensive Care Med. 2003;29:699–702. doi: 10.1007/s00134-003-1746-7. [DOI] [PubMed] [Google Scholar]
- 64.Li J., Wang X., Chen J., Zuo X., Zhang H., Deng A. COVID-19 infection may cause ketosis and ketoacidosis. Diabetes Obes. Metabol. 2020;22:1935–1941. doi: 10.1111/dom.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamel K.S., Oh M.S., Halperin M.L. L-lactic acidosis: pathophysiology, classification, and causes; emphasis on biochemical and metabolic basis. Kidney Int. 2020;97:75–88. doi: 10.1016/j.kint.2019.08.023. [DOI] [PubMed] [Google Scholar]
- 66.Kogelmann K., Jarczak D., Scheller M., Drüner M. Hemoadsorption by CytoSorb in septic patients: a case series. Crit. Care. 2017;21:1–10. doi: 10.1186/s13054-017-1662-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chhetri S., Khamis F., Pandak N., Al Khalili H., Said E., Petersen E. A fatal case of COVID-19 due to metabolic acidosis following dysregulate inflammatory response (cytokine storm) IDCases. 2020;21 doi: 10.1016/j.idcr.2020.e00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goldman N., Fink D., Cai J., Lee Y.N., Davies Z. High prevalence of COVID-19-associated diabetic ketoacidosis in UK secondary care. Diabetes Res. Clin. Pract. 2020;166:108291. doi: 10.1016/j.diabres.2020.108291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chan K.H., Thimmareddygari D., Ramahi A., Atallah L., Baranetsky N.G., Slim J. Clinical characteristics and outcome in patients with combined diabetic ketoacidosis and hyperosmolar hyperglycemic state associated with COVID-19: a retrospective, hospital-based observational case series. Diabetes Res. Clin. Pract. 2020;166 doi: 10.1016/j.diabres.2020.108279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng X., Liu Y.M., Li H., Zhang X., Lei F., Qin J.J., Chen Z., Deng K.Q., Lin L., Chen M.M., Song X., Xia M., Huang X., Liu W., Cai J., Zhang X.J., Zhou F., Zhang P., Wang Y., Ma X., Xu Q., Yang J., Ye P., Mao W., Huang X., Xia J., Zhang B.H., Guo J., Zhu L., Lu Z., Yuan Y., Wei X., She Z.G., Ji Y.X., Li H. Metformin is associated with higher incidence of acidosis, but not mortality, in individuals with COVID-19 and pre-existing type 2 diabetes. Cell Metabol. 2020;32:537–547. doi: 10.1016/j.cmet.2020.08.013. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parohan M., Yaghoubi S., Seraji A. Liver injury is associated with severe coronavirus disease 2019 (COVID-19) infection: a systematic review and meta-analysis of retrospective studies. Hepatol. Res. 2020;50:924–935. doi: 10.1111/hepr.13510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., Flemban H., Al-Nassir W.N., Balkhy H.H., Al-Hakeem R.F., Makhdoom H.Q., Zumla A.I., Memish Z.A. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Al-Hameed F., Wahla A.S., Siddiqui S., Ghabashi A., Al-Shomrani M., Al-Thaqafi A., Tashkandi Y. Characteristics and outcomes of middle east respiratory syndrome coronavirus patients admitted to an intensive care unit in Jeddah, Saudi Arabia. J. Intensive Care Med. 2016;31:344–348. doi: 10.1177/0885066615579858. [DOI] [PubMed] [Google Scholar]
- 74.Saad M., Omrani A.S., Baig K., Bahloul A., Elzein F., Matin M.A., Selim M.A.A., Al Mutairi M., Al Nakhli D., Aidaroos A.Y.A., Al Sherbeeni N., Al-Khashan H.I., Memish Z.A., Albarrak A.M. Clinical aspects and outcomes of 70 patients with Middle East respiratory syndrome coronavirus infection: a single-center experience in Saudi Arabia. Int. J. Infect. Dis. 2014;29:301–306. doi: 10.1016/j.ijid.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang H.L., Chen K.T., Lai S.K., Kuo H.W., Su I.J., Lin R.S., Sung F.C. Hematological and biochemical factors predicting SARS fatality in Taiwan. J. Formos. Med. Assoc. 2006;105:439–450. doi: 10.1016/S0929-6646(09)60183-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sproston N.R., Ashworth J.J. Role of C-reactive protein at sites of inflammation and infection. Front. Immunol. 2018;9:1–11. doi: 10.3389/fimmu.2018.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thiele J.R., Habersberger J., Braig D., Schmidt Y., Goerendt K., Maurer V., Bannasch H., Scheichl A., Woollard K.J., Von Dobschütz E., Kolodgie F., Virmani R., Stark G.B., Peter K., Eisenhardt S.U. Dissociation of pentameric to monomeric C-reactive protein localizes and aggravates inflammation: in vivo proof of a powerful proinflammatory mechanism and a new anti-inflammatory strategy. Circulation. 2014;130:35–50. doi: 10.1161/CIRCULATIONAHA.113.007124. [DOI] [PubMed] [Google Scholar]
- 78.Huang I., Pranata R., Lim M.A., Oehadian A., Alisjahbana B. C-reactive protein, procalcitonin, D-dimer, and ferritin in severe coronavirus disease-2019: a meta-analysis. Ther. Adv. Respir. Dis. 2020;14:1–14. doi: 10.1177/1753466620937175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holowaychuk M.K., Martin L.G. Review of hypocalcemia in septic patients: state-of-the-art review. J. Vet. Emerg. Crit. Care. 2007;17:348–358. doi: 10.1111/j.1476-4431.2007.00246.x. [DOI] [Google Scholar]
- 80.Appel S.A., Molshatzki N., Schwammenthal Y., Merzeliak O., Toashi M., Sela B.A., Tanne D. Serum calcium levels and long-term mortality in patients with acute stroke. Cerebrovasc. Dis. 2010;31:93–99. doi: 10.1159/000321335. [DOI] [PubMed] [Google Scholar]
- 81.Chung J.W., Ryu W.S., Kim B.J., Yoon B.W. Elevated calcium after acute ischemic stroke: association with a poor short-term outcome and long-term mortality. J. Stroke. 2015;17:54–59. doi: 10.5853/jos.2015.17.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Di Yan S., Liu X.J., Peng Y., Xia T.L., Liu W., Tsauo J.Y., Xu Y.N., Chai H., Huang F.Y., Chen M., Huang D.J. Admission serum calcium levels improve the GRACE risk score prediction of hospital mortality in patients with acute coronary syndrome. Clin. Cardiol. 2016;39:516–523. doi: 10.1002/clc.22557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sankaran R.T., Mattana J., Pollack S., Bhat P., Ahuja T., Patel A., Singhal P.C. Laboratory abnormalities in patients with bacterial pneumonia. Chest. 1997;111:595–600. doi: 10.1378/chest.111.3.595. [DOI] [PubMed] [Google Scholar]
- 84.Constantine G.R., Rajapakse S., Ranasinghe P., Parththipan B., Jayawardana P., Wijewickrama A. Hypocalcemia is associated with disease severity in patients with dengue. J. Infect. Dev. Ctries. 2014;8:1205–1209. doi: 10.3855/jidc.4974. [DOI] [PubMed] [Google Scholar]
- 85.Liu J., Han P., Wu J., Gong J., Tian D. Prevalence and predictive value of hypocalcemia in severe COVID-19 patients. J. Infect. Publ. Health. 2020;13:1224–1228. doi: 10.1016/j.jiph.2020.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Akirov A., Gorshtein A., Shraga-Slutzky I., Shimon I. Calcium levels on admission and before discharge are associated with mortality risk in hospitalized patients. Endocrine. 2017;57:344–351. doi: 10.1007/s12020-017-1353-y. [DOI] [PubMed] [Google Scholar]
- 87.di Filippo L., Formenti A.M., Doga M., Frara S., Rovere-Querini P., Bosi E., Carlucci M., Giustina A. Hypocalcemia is a distinctive biochemical feature of hospitalized COVID-19 patients. Endocrine. 2021;71:9–13. doi: 10.1007/s12020-020-02541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.