Abstract
HLA haplotypes were found to be associated with increased risk for viral infections or disease severity in various diseases, including SARS. Several genetic variants are associated with COVID-19 severity. Studies have proposed associations, based on a very small sample and a large number of tested HLA alleles, but no clear association between HLA and COVID-19 incidence or severity has been reported. We conducted a large-scale HLA analysis of Israeli individuals who tested positive for SARS-CoV-2 infection by PCR. Overall, 72,912 individuals with known HLA haplotypes were included in the study, of whom 6413 (8.8%) were found to have SARS-CoV-2 by PCR. A total of 20,937 subjects were of Ashkenazi origin (at least 2/4 grandparents). One hundred eighty-one patients (2.8% of the infected) were hospitalized due to the disease. None of the 66 most common HLA loci (within the five HLA subgroups: A, B, C, DQB1, DRB1) was found to be associated with SARS-CoV-2 infection or hospitalization in the general Israeli population. Similarly, no association was detected in the Ashkenazi Jewish subset. Moreover, no association was found between heterozygosity in any of the HLA loci and either infection or hospitalization. We conclude that HLA haplotypes are not a major risk/protecting factor among the Israeli population for SARS-CoV-2 infection or severity. Our results suggest that if any HLA association exists with the disease it is very weak, and of limited effect on the pandemic.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10875-021-01071-x.
Keywords: HLA, SARS-CoV-2, association, hospitalization, Israel, Ashkenazi
Introduction
The major histocompatibility complex (MHC) molecules are cell surface protein complexes encoded in the human leukocyte antigens (HLA) locus. The HLA locus is highly polymorphic with thousands of different alleles and millions of haplotypes reported [1, 2]. HLAs are associated with several infectious diseases. Previous disease association studies showed that some HLA haplotypes are highly correlated with multiple viral infections [3–5] and have been reported to confer differential susceptibility to infection and the severity of the resulting disease [6]. In particular, HLA class I alleles are key players in the immune defense against viruses as they initiate the activity of T cells against the invading intracellular pathogens [7, 8]. Although the presentation of viral antigens relies classically on MHC class I molecules, MHC class II genes have also been associated with the outcome of many viral infections [9–11]. Studies in a range of species [12–15], including humans [16, 17], imply a heterozygous selection mechanism operating on the HLA loci as an explanation for the extensive variability of the MHC molecules [18, 19]. However, recent results on human populations suggest that such an advantage may be limited [20].
In the context of COVID-19, specific HLAs have been associated with coronaviruses in the past, including SARS coronavirus infections [21, 22]. Several efforts were made to identify HLA-related susceptibility to SARS-CoV-1 after the first SARS epidemic in East Asia [23–26] and the MERS-CoV outbreak in 2014 in Saudi Arabia [27]. Most of these case studies provided weak or conflicting results and required further validation due to the relatively small sample size. An association between the geographic distributions of COVID-19 and HLA alleles has been suggested [28]. Similarly, an association between severe disease outcome and specific HLAs was suggested by another small-scale whole-genome sequencing study [29]. In silico model raised the possibility that HLA supertypes may have a role in the severity of the disease [30]. Moreover, expression of human leukocyte antigen class DR (HLA-DR) was decreased [31] or contrarily increased [32] in peripheral blood mononuclear cells of COVID-19 patients. Nevertheless, genome-wide association study (GWAS) among 1980 Italian and Spanish patients with severe COVID-19 detected a few loci associated with that state but no association with the HLA loci at chromosome 6 or heterozygote advantage was found [33]. However, another GWAS study analyzing 2244 critically ill COVID-19 patients in the UK detected three variants in chromosome 6 but these variants were not replicated in other studies. Since population stratification in chromosome 6 (the major histocompatibility complex) is difficult to control, further studies will be required to determine whether these associations are real [34]. Two other large unpublished GWAS studies did not report a signal at the HLA locus on chromosome 6 [35, 36]. To the best of our knowledge, there are no previous studies looking specifically at HLA subgroups among individuals with COVID-19.
HLA allele and haplotype frequencies differ between populations belonging to different ethnic groups [37, 38]. For example, among Jewish populations, similarities of HLA alleles may be seen within ethnicities, allowing an estimation of an individual’s origin based on his HLA genotypes [39]. It has been estimated that decreased HLA variability may assist in detecting signals relevant to a specific disease state. Indeed, specific HLAs were associated with different disorders in the Ashkenazi population, a large genetically isolated group [40–42]. Given the diversity of HLAs among populations, testing genetically isolated and genetically homogenous populations may provide an advantage in detecting the effects of specific genetic variants, in particular in Ashkenazi Jews. Note that this is not a genome-wide associate study, where the case and controls can be matched by the PCA components of their genotypes. In HLA association studies, self-reporting is the standard [43, 44].
Our study aimed to utilize the presence of a large-scale cohort having HLA genotypes and a high rate of COVID-19 to test the possible role of specific HLA alleles in SARS-CoV-2 infection and its severity. Using a dataset of this magnitude also allowed us to analyze associations between COVID-19 infection and HLA zygosity. Understanding how variation in HLA may affect both susceptibility and severity of COVID-19 infection could help identify and stratify individuals at higher risk for the disease and may support future vaccination strategies.
Methods
Study Design
A case-control study with a test-negative design [45] was used to evaluate the infection outcome, while a retrospective cohort study was used to evaluate the hospitalization outcome. In the case-control design, patients who tested positive or negative for SARS-CoV-2 were evaluated for their HLA alleles. In the cohort design, patients who were diagnosed with COVID-19 were followed up to see who were hospitalized.
The study was based on patient data taken from the data warehouse of Clalit Health Care (CHS), a large integrated payer-provider healthcare organization operating in Israel. CHS insures over 4.6 million Israelis (~50%), which are a representative sample of the entire population.
HLA Dataset
HLA alleles were obtained from the Ezer Mizion Bone Marrow Donor Registry, which enlists the highest number of registered unrelated volunteer donors per capita in the world. The HLA resolution varies from serologic (8%) to DNA-based testing at low (32%), intermediate (22%), and allele resolution (38%) [46]. The initial analysis included HLA genotypes of 1,040,250 donors which represent all the populations composing the Ezer Mizion registry. High-resolution five-locus typing for HLA-A, -B, -C, -DRB1, and -DQB1 was imputed for all donors using GRIMM (GRaph IMputation and Matching for HLA Genotypes) [47], which produces the most likely five-locus genotype consistent with the low-resolution typing and the self-defined ethnicity. For each subject in the dataset, we chose the unphased genotype with the highest probability. We also used the low-resolution two-digit typing of each subject, as well as the homozygosity status of each allele.
Cases and Controls
The study included all patients tested for SARS-CoV-2 by PCR that were members of CHS at the date of testing and had HLA data available. Cases were those patients tested positive, while controls were patients tested negative. Covariates for adjustment included age, sex, number of children in the household, population sector (ultra-orthodox vs. general, which has had an important impact on the epidemic in Israel), socioeconomic status, and ethnic origin (Ashkenazi vs. other). The analyses for the hospitalization outcome were further adjusted for obesity and history cardiovascular disease, diabetes, chronic respiratory disease, chronic renal disease, chronic hepatic disease, chronic neurological disease, chronic hematological disease, and immunosuppression. These covariates were extracted as of the test date. The number of children in the household was calculated based on CHS’ demographic registry. The population sector was determined using the patient’s primary-care clinic address. Socioeconomic status was determined based on a patient’s home address. Patients were defined as Ashkenazi if they were Jewish and were born themselves or had at least one parent or 2 grandparents born in central or western Europe. A severe disease course was defined by hospitalization less than 2 weeks after the molecular diagnosis.
Statistical Analysis
The association between each HLA allele and the outcome, adjusted for the covariates listed above, was tested separately for each allele using logistic regression, where all the covariates and the presence or absence of the HLA were used as an input, and the outcome (infection or hospitalization) was used as the prediction. The effect of homozygosity was tested similarly, as a binary predictor, separately for each locus, and a similar logistic regression was performed on the same outputs. A Bonferroni correction was applied to the p-values and confidence intervals. The results were similar when the Benjamini-Hochberg false discovery rate was controlled [48]. Zygosity was computed based on two-digit HLA alleles at the appropriate locus. The study was performed with the approval of the Institutional Research Community (IRB) #0080-20-COM2).
Results
Overall, 72,912 individuals with HLA genotyping and at least one PCR test for COVID-19 were included in the study between March 1, 2020, and October 31, 2020. Of them, 8.8% (6413) tested positive for SARS-CoV-2 by PCR. Demographic and socioeconomic data related to the population are available in Table 1. The sample is large enough to allow for the detection of OR with a significance of 1.3 (Supplemental Figure S1).
Table 1.
Demographic characterization of study groups
| Overall | Ashkenazi | |||||
|---|---|---|---|---|---|---|
| Infected | Non-infected | Overall | Infected | Non-infected | Overall | |
| Total number (%) | 6413 (8.8) | 66,499 (91.2) | 72,912 | 1495 (7.1) | 19,442 (92.9) | 20,937 |
| Mean age (SD) | 39.47 | 40.33 | 40.26 | 40.82 | 41.12 | 41.1 |
| Females (%) | 3176 (49.5) | 39,493 (59.4) | 42,669 (58.5) | 764 (51.1) | 11,859 (61.0) | 12,623 (60.3) |
| Males (%) | 3237 (50.5) | 27,006 (40.6) | 30,243 (41.5) | 731 (48.9) | 7583 (39.0) | 8314 (39.7) |
| Mean # of children (SD) | 2.49 (2.27) | 2.04 (1.72) | 2.08 (1.78) | 2.53 (2.24) | 2.01 (1.67) | 2.05 (1.73) |
| Ultraorthodox sector (%) | 920 (14.3) | 2895 (4.4) | 3815 (5.2) | 204 (13.6) | 721 (3.7) | 925 (4.4) |
| Socioeconomic level-low (%) | 1177 (18.4) | 5704 (8.6) | 6881 (9.4) | 211 (14.1) | 1136 (5.8) | 1347 (6.4) |
| Socioeconomic level-med (%) | 2813 (43.9) | 27,009 (40.6) | 29,822 (40.9) | 601 (40.2) | 6811 (35.0) | 7412 (35.4) |
| Socioeconomic level-high (%) | 2423 (37.8) | 33,786 (50.8) | 36,209 (49.7) | 683 (45.7) | 11,495 (59.1) | 12,178 (58.2) |
| Hospitalized (%) | 181 (2.8) | |||||
| Hospitalized mean age (SD) | 39.39 (11.85) | |||||
| Hospitalized females (%) | 74 (40.9) | |||||
| Hospitalized males (%) | 107 (59.1) | |||||
Characterization of the individuals with HLA genotyping and at least one PCR test for COVID-19 that were included in the study
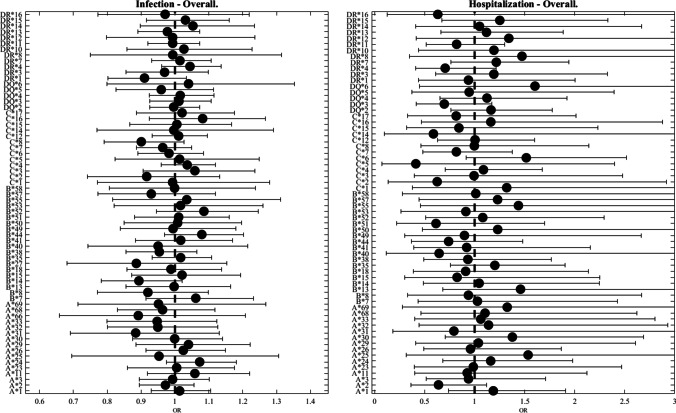
All HLA alleles were grouped by two-digit representations to minimize the number of tests, and only alleles with a frequency of at least 1 % were used. A total of 16 HLA-A, 20 HLA-B, 13 HLA-C, 5 HLA-DQB1, and 12 HLA-DRB1 (Table 2) met the criteria for analysis. No significant differences in HLA- allele frequency were detected between individuals with negative and positive SARS-CoV-2 PCR (Fig. 1, left panel).
Table 2.
HLA- subgroups (2-digit) analyzed
| HLA-A | HLA-B | HLA-C | HLA-DQB1 | HLA-DRB1 | |
|---|---|---|---|---|---|
| A*1 | B*7 | C*1 | DQ*2 | DR*1 | |
| A*2 | B*8 | C*2 | DQ*3 | DR*3 | |
| A*3 | B*13 | C*3 | DQ*4 | DR*4 | |
| A*11 | B*14 | C*4 | DQ*5 | DR*7 | |
| A*23 | B*15 | C*5 | DQ*6 | DR*8 | |
| A*24 | B*18 | C*6 | DR*10 | ||
| A*25 | B*27 | C*7 | DR*11 | ||
| A*26 | B*35 | C*8 | DR*12 | ||
| A*29 | B*38 | C*12 | DR*13 | ||
| A*30 | B*40 | C*14 | DR*14 | ||
| A*31 | B*41 | C*15 | DR*15 | ||
| A*32 | B*44 | C*16 | DR*16 | ||
| A*33 | B*49 | C*17 | |||
| A*66 | B*50 | ||||
| A*68 | B*51 | ||||
| A*69 | B*52 | ||||
| B*53 | |||||
| B*55 | |||||
| B*57 | |||||
| B*58 |
List of two-digit HLA alleles with a population frequency > 1%, analyzed in the study
Fig. 1.
Odds ratio (OR) as a function of HLA allele. In all figures, the dot represents the odds ratio, and the interval represents the 95% confidence intervals (Bonferroni correction). The left plot is for SARS-CoV-2 infection and the right plot is for hospitalization. We trim error bars at 3 for better visualization. The analysis presented here is for the full population. One can see that no HLA allele is significantly associated
We further tested whether HLA genotypes may be associated with severe COVID-19 infection. Of the 6413 infected individuals, 181 (2.6%) were hospitalized within 2 weeks. Again, no significant HLA genotypes were found to be associated with hospitalization among individuals positive for SARS-CoV-2 (Fig. 1, right panel).
Given the association between HLA and ethnic origin and the fact that the Ashkenazi Jewish population is genetically isolated [36], we tested for SARS-CoV-2 presence and severity within the Ashkenazi population. This analysis included 20,937 Ashkenazi donors, of which 1495 were positive for SARS-CoV-2 and 45 were hospitalized. No significant association was observed between either SARS-CoV-2 positive frequency (Fig. 2, left panel) or COVID-19 severity (Fig. 2, right panel) and any HLA subgroup in the Ashkenazi population (Fig. 2).
Fig. 2.
Odds ratio (OR) as a function of HLA allele in the Ashkenazi population. In all figures, the dot represents the odds ratio, and the interval represents the 95% confidence intervals (Bonferroni correction). The left plot is for SARS-CoV-2 infection and the right plot is for hospitalization. We trim error bars at 3 for better visualization. The analysis presented here is for the Ashkenazi population. One can see that no HLA allele is significantly associated
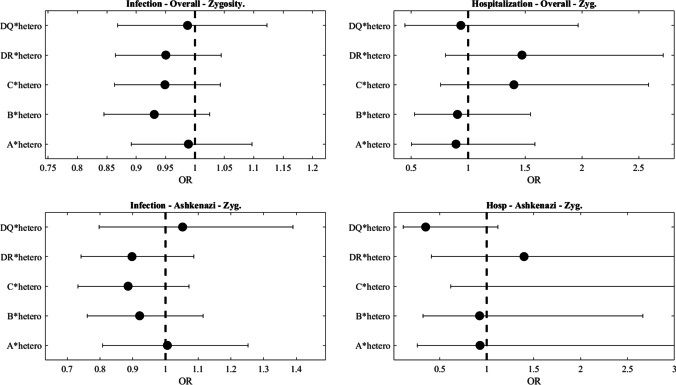
To investigate whether HLA heterozygosity is associated with the outcome of SARS-CoV-2 infection, we further tested whether an increased probability of SARS-CoV-2 infection or severe disease course is associated with a higher homozygous rate of each of the 5 HLA loci subgroup. Homozygosity was defined at the two-digit level. No such association was detected either in the general population or in the Ashkenazi population (Fig. 3).
Fig. 3.
Effect of homozygosity on either infection or hospitalization in the entire studied population or the Ashkenazi population. Again no significant association was found. Homozygosity at a given locus is defined as having the same low-resolution allele in this locus
To ensure our results were not the results of lowering the resolution of the HLA, we repeated the analysis above for both infection and hospitalization for the general population and the Ashkenazi population using the 50 most frequent HLA alleles in the population (4 digits), and none had a significant effect in any of these tests (Supplemental Table S1).
Discussion
The COVID-19 pandemic is associated with heavy medical, economic, and social costs. A few factors were associated with decreased or increased risks of SARS-CoV-2 infection, including smoking [49] and gender (male) [50]. Genetic factors were also found to be associated with COVID-19 disease severity [51], including a genomic segment of around 50 kilobases in size that is inherited from Neanderthals [52]. Similarly, a meta-analysis of GWAS studies of patients from Spain and Italy with severe COVID-19, which was defined as a hospitalization with respiratory failure, detected three significant loci with odd ratios of 1.32–1.77 per locus [33]. Given the important role of HLA in several viral infections, the above study analyzed the extended HLA region (chromosome 6, 25 through 34 Mb) but did not find significant SNP association signals at the HLA complex. Another GWAS study analyzing 2244 critically ill COVID-19 patients in the UK detected potential associations between variants in chromosome 6 and COVID-19 [34].
Our study, performed on a large-scale dataset, did not detect any specific HLA subgroups associated with increased or decreased risk for SARS-CoV-2 infection, or a severe disease course causing hospitalization. We also did not detect any associations of heterozygote advantage with COVID-19 when we analyzed the entire study population or when analyzing a sub-population. Despite clear successes in identifying novel disease susceptibility genes, GWAS approaches have not been without controversy [53]. A major limitation of this approach is the need to adopt a high level of significance to account for the multiple tests burden due to the very large number of loci compared concurrently. As such, it can miss important associations. In contrast with GWAS studies, we incorporated HLA alleles with a prevalence of >1% in the Israeli population. This approach enabled a large-scale analysis not of a few HLA subtypes but of 66 variants, and still no association was found in the HLA region
Two main caveats should be considered. First, the imputation of HLA alleles may induce errors. However, the precision of the imputation was previously established [54]. Moreover, no association was found with the A alleles that were typed in the vast majority of subjects. Finally, the analysis was performed at the two-digit level, which is practically unaffected by imputation. Another possible limitation is the age distribution. Our study included only a small proportion of hospitalized cases 181/6413 (2.6%). This may be related to the average young age of individuals for whom we have HLA data. However, this should not affect the results related to SARS-CoV-2 infection. While hospitalization alone may not be an indicator of disease severity, we expect a significant enrichment of severe cases in hospitalized patients.
The lack of HLA association reported here and in previous smaller studies, and the absence of advantage for heterozygote may suggest that heterozygote advantage in HLA may be more limited in general than often considered. This is in agreement with more recent claims that other mechanisms beyond heterozygote advantage or pathogen-driven balancing selection may be the source of the large HLA polymorphism [2, 20].
We did not detect an HLA association with the initial phases of infection and hospitalization. As in other diseases, the initial response to the disease in the first few days is mainly mediated by the innate immune response, limiting the involvement of HLA and T cells. HLA may still be involved in the later phases of the disease progression, such as long-term sequels, or complications following hospitalization. A long-term follow-up will be needed to test for those.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
YL, SM, and SB wrote the paper. NB performed the statistical analysis. SI performed the HLA imputation. SC corrected the manuscript and helped with the final version. NA, RB, and BZ supplied the data analyzed here.
Data Availability
The data is personal HLA typing and COVID-19 status and cannot be provided. All the aggregated data is explicated in the figures and tables. The frequency of each allele in each dataset can be obtained from the authors upon demand.
Declarations
Ethical Approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of the Clalit medical services (IRB) #0080–20-COM2.
Consent to Participate
No experiment was performed on humans, so there is no consent to report.
Consent to Publish
All the authors agreed to this publication.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/30/2021
The original version of this paper was updated to present the first author’s family name.
References
- 1.Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE. IPD-IMGT/HLA database. Nucleic Acids Res Oxford University Press. 2020;48:D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter I, Gragert L, Fingerson S, Maiers M, Louzoun Y. HLA class I haplotype diversity is consistent with selection for frequent existing haplotypes. PLoS Comput Biol. 2017;13:e1005693. doi: 10.1371/journal.pcbi.1005693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin GG, Scott JG. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2012;100:130–134. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loffredo JT, Sidney J, Bean AT, Beal DR, Bardet W, Wahl A, et al. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol. 2009;182:7763–7775. doi: 10.4049/jimmunol.0900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwell JM, Jamieson SE, Burgner D. HLA and infectious diseases. Clin Microbiol Rev. 2009;22:370–385. doi: 10.1128/CMR.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez-Mazas A. A review of HLA allele and SNP associations with highly prevalent infectious diseases in human populations. Swiss Med Wkly. 2020;150:w20214. doi: 10.4414/smw.2020.20214. [DOI] [PubMed] [Google Scholar]
- 7.Crux NB, Elahi S. Human Leukocyte Antigen (HLA) and immune regulation: how do classical and non-classical HLA alleles modulate immune response to human immunodeficiency virus and hepatitis C virus infections? Front Immunol. 2017;8:1–26. doi: 10.3389/fimmu.2017.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parham P. The immune system. 4. New York: Garland Science; 2014. [Google Scholar]
- 9.Tamori A, Kawada N. HLA class II associated with outcomes of hepatitis B and C infections. World J Gastroenterol. 2013;19:5395–5401. doi: 10.3748/wjg.v19.i33.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ranasinghe S, Cutler S, Davis I, Lu R, Soghoian DZ, Qi Y, et al. Association of HLA-DRB1-restricted CD4+ T cell responses with HIV immune control. Nat Med. 2013;19:930–933. doi: 10.1038/nm.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sierra B, Alegre R, Pérez AB, García G, Sturn-Ramirez K, Obasanjo O, et al. HLA-A, -B, -C, and -DRB1 allele frequencies in Cuban individuals with antecedents of dengue 2 disease: advantages of the Cuban population for HLA studies of dengue virus infection. Hum Immunol United States. 2007;68:531–540. doi: 10.1016/j.humimm.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Penn DJ, Damjanovich K, Potts WK. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc Natl Acad Sci U S A. 2002;99:11260–11264. doi: 10.1073/pnas.162006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeshima S, Matsumoto Y, Chen J, Yoshida T, Mukoyama H, Aida Y. Evidence for cattle major histocompatibility complex (BoLA) class II DQA1 gene heterozygote advantage against clinical mastitis caused by Streptococci and Escherichia species. Tissue Antigens England. 2008;72:525–531. doi: 10.1111/j.1399-0039.2008.01140.x. [DOI] [PubMed] [Google Scholar]
- 14.Evans ML, Neff BD. Major histocompatibility complex heterozygote advantage and widespread bacterial infections in populations of Chinook salmon (Oncorhynchus tshawytscha) Mol Ecol England. 2009;18:4716–4729. doi: 10.1111/j.1365-294X.2009.04374.x. [DOI] [PubMed] [Google Scholar]
- 15.Niskanen AK, Kennedy LJ, Ruokonen M, Kojola I, Lohi H, Isomursu M, et al. Balancing selection and heterozygote advantage in major histocompatibility complex loci of the bottlenecked Finnish wolf population. Mol Ecol England. 2014;23:875–889. doi: 10.1111/mec.12647. [DOI] [PubMed] [Google Scholar]
- 16.Arora J, Pierini F, McLaren PJ, Carrington M, Fellay J, Lenz TL. HLA heterozygote advantage against HIV-1 is driven by quantitative and qualitative differences in HLA allele-specific peptide presentation. Mol Biol Evol. 2020;37:639–650. doi: 10.1093/molbev/msz249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hraber P, Kuiken C, Yusim K. Evidence for human leukocyte antigen heterozygote advantage against hepatitis C virus infection. Hepatology United States. 2007;46:1713–1721. doi: 10.1002/hep.21889. [DOI] [PubMed] [Google Scholar]
- 18.Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proceedings Biol Sci. 2010;277:979–988. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sommer S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front Zool. 2005;2:16. doi: 10.1186/1742-9994-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobkovsky AE, Levi L, Wolf YI, Maiers M, Gragert L, Alter I, et al. Multiplicative fitness, rapid haplotype discovery, and fitness decay explain evolution of human MHC. Proc Natl Acad Sci U S A. 2019;116:14098–14104. doi: 10.1073/pnas.1714436116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh HLJ, Gan SKE, Bertoletti A, Tan YJ. Understanding the T cell immune response in SARS coronavirus infection. Emerg Microbes Infect [Internet]. 2012;1:0. Available from: 10.1038/emi.2012.26. [DOI] [PMC free article] [PubMed]
- 22.Sanchez-Mazas A. HLA studies in the context of coronavirus outbreaks. Swiss Med Wkly. 2020;150:w20248. doi: 10.4414/smw.2020.20248. [DOI] [PubMed] [Google Scholar]
- 23.Keicho N, Itoyama S, Kashiwase K, Phi NC, Long HT, Ha LD, et al. Association of human leukocyte antigen class II alleles with severe acute respiratory syndrome in the Vietnamese population. Hum Immunol. 2009;70:527–531. doi: 10.1016/j.humimm.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin M, Tseng HK, Trejaut JA, Lee HL, Loo JH, Chu CC, et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 2003;4:1–7. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen YMA, Liang SY, Shih YP, Chen CY, Lee YM, Chang L, et al. Epidemiological and genetic correlates of severe acute respiratory syndrome coronavirus infection in the hospital with the highest nosocomial infection rate in Taiwan in 2003. J Clin Microbiol. 2006;44:359–365. doi: 10.1128/JCM.44.2.359-365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong P, Zeng X, Song MS, Jia SW, Zhong MH, Xiao LL, et al. Lack of association between HLA-A, -B and -DRB1 alleles and the development of SARS: a cohort of 95 SARS-recovered individuals in a population of Guangdong, southern China. Int J Immunogenet. 2008;35:69–74. doi: 10.1111/j.1744-313X.2007.00741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hajeer AH, Balkhy H, Johani S, Yousef MZ, Arabi Y. Association of human leukocyte antigen class II alleles with severe Middle East respiratory syndrome-coronavirus infection. Ann Thorac Med [Internet]. Medknow Publications & Media Pvt Ltd; 2016;11:211–3. Available from: https://pubmed.ncbi.nlm.nih.gov/27512511. Accessed Feb 2021. [DOI] [PMC free article] [PubMed]
- 28.Pisanti S, Deelen J, Gallina AM, Caputo M, Citro M, Abate M, et al. Correlation of the two most frequent HLA haplotypes in the Italian population to the differential regional incidence of Covid-19. J Transl Med [Internet]. BioMed Central; 2020;18:1–16. 10.1186/s12967-020-02515-5. [DOI] [PMC free article] [PubMed]
- 29.Wang F, Huang S, Gao R, Zhou Y, Lai C, Li Z, et al. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov [Internet]. Springer US; 2020;6. 10.1038/s41421-020-00231-4. [DOI] [PMC free article] [PubMed]
- 30.Tomita Y, Ikeda T, Sato R, Sakagami T. Association between HLA gene polymorphisms and mortality of COVID-19: An in silico analysis. Immun Inflammation Dis. 2020;8:684–694. doi: 10.1002/iid3.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arunachalam PS, Wimmers F, Mok CKP, Perera RAPM, Scott M, Hagan T, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science (80- ) 2020;369:1210–20. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuri-Cervantes L, Pampena MB, Meng W, Rosenfeld AM, Ittner CAG, Weisman AR, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, et al. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, et al. Genetic mechanisms of critical illness in Covid-19. Nature England.; 2020; [DOI] [PubMed]
- 35.Roberts GHL, Park DS, Coignet M V, McCurdy SR, Knight SC, Partha R, et al. AncestryDNA COVID-19 host genetic study identifies three novel loci. medRxiv [Internet]. 2020;2020.10.06.20205864. Available from: http://medrxiv.org/content/early/2020/10/09/2020.10.06.20205864.abstract. Accessed Feb 2021.
- 36.Shelton JF, Shastri AJ, Ye C, Weldon CH, Filshtein-Somnez T, Coker D, et al. Trans-ethnic analysis reveals genetic and non-genetic associations with COVID-19 susceptibility and severity. medRxiv [Internet]. 2020;2020.09.04.20188318. Available from: http://medrxiv.org/content/early/2020/09/07/2020.09.04.20188318.abstract. Accessed Feb 2021. [DOI] [PubMed]
- 37.Oksenberg JR, Barcellos LF, Cree BAC, Baranzini SE, Bugawan TL, Khan O, et al. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am J Hum Genet. 2004;74:160–167. doi: 10.1086/380997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mignot E, Lin L, Rogers W, Honda Y, Qiu X, Lin X, et al. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am J Hum Genet. 2001;68:686–699. doi: 10.1086/318799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klitz W, Gragert L, Maiers M, Fernandez-Viña M, Ben-Naeh Y, Benedek G, et al. Genetic differentiation of Jewish populations. Tissue Antigens [Internet]. 2010 [cited 2016 Nov 20];76:442–58. Available from: http://doi.wiley.com/10.1111/j.1399-0039.2010.01549.x. Accessed Feb 2021. [DOI] [PubMed]
- 40.Khankhanian P, Matsushita T, Madireddy L, Lizée A, Din L, Moré JM, et al. Genetic contribution to multiple sclerosis risk among Ashkenazi Jews. BMC Med Genet [Internet]. BMC Medical Genetics; 2015;16:1–10. 10.1186/s12881-015-0201-2. [DOI] [PMC free article] [PubMed]
- 41.Liu X, Cheng R, Verbitsky M, Kisselev S, Browne A, Mejia-Sanatana H, et al. Genome-wide association study identifies candidate genes for Parkinson’s disease in an Ashkenazi Jewish population. BMC Med Genet [Internet]. BioMed Central Ltd; 2011;12:104. Available from: http://www.biomedcentral.com/1471-2350/12/104. Accessed Feb 2021. [DOI] [PMC free article] [PubMed]
- 42.Goes FS, McGrath J, Avramopoulos D, Wolyniec P, Pirooznia M, Ruczinski I, et al. Genome-wide association study of schizophrenia in Ashkenazi Jews. Am J Med Genet Part B, Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet United States. 2015;168:649–59. doi: 10.1002/ajmg.b.32349. [DOI] [PubMed] [Google Scholar]
- 43.Dehn J, Setterholm M, Buck K, Kempenich J, Beduhn B, Gragert L, et al. HapLogic: a predictive human leukocyte antigen–matching algorithm to enhance rapid identification of the optimal unrelated hematopoietic stem cell sources for transplantation. Biol Blood Marrow Transplant [Internet]. 2016 [cited 2016 Nov 30];22:2038–46. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1083879116302580. Accessed Feb 2021. [DOI] [PubMed]
- 44.Kollman C, Maiers M, Gragert L, Müller C, Setterholm M, Oudshoorn M, et al. Estimation of HLA-A, -B, -DRB1 Haplotype frequencies using mixed resolution data from a national registry with selective retyping of volunteers. Hum Immunol [Internet]. 2007 [cited 2016 Nov 20];68:950–8. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0198885907004612. Accessed Feb 2021. [DOI] [PubMed]
- 45.Jackson ML, Nelson JC. The test-negative design for estimating influenza vaccine effectiveness. Vaccine Netherlands. 2013;31:2165–2168. doi: 10.1016/j.vaccine.2013.02.053. [DOI] [PubMed] [Google Scholar]
- 46.Manor S, Halagan M, Shriki N, Yaniv I, Zisser B, Maiers M, et al. High-resolution HLA A∼B∼DRB1 haplotype frequencies from the Ezer Mizion Bone Marrow Donor Registry in Israel. Hum Immunol. 2016;77:114–1119. doi: 10.1016/j.humimm.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 47.Maiers M, Halagan M, Gragert L, Bashyal P, Brelsford J, Schneider J, et al. GRIMM: GRaph IMputation and matching for HLA genotypes. Bioinformatics England. 2019;35:3520–3523. doi: 10.1093/bioinformatics/btz050. [DOI] [PubMed] [Google Scholar]
- 48.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 49.Polosa R, Caci G. COVID-19: counter-intuitive data on smoking prevalence and therapeutic implications for nicotine. Intern Emerg Med [Internet]. Springer International Publishing; 2020;15:853–6. Available from: 10.1007/s11739-020-02361-9. [DOI] [PMC free article] [PubMed]
- 50.Abate BB, Kassie AM, Kassaw MW, Aragie TG, Masresha SA. Sex difference in coronavirus disease (COVID-19): a systematic review and meta-analysis. BMJ Open. 2020;10:e040129. doi: 10.1136/bmjopen-2020-040129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nasir J WB and VK. COVID-19 HGI results for data freeze 4. https://www.covid19hg.org/blog/2020-11-24-covid-19-hgi-results-for-data-freeze-4-october-2020/ [Internet]. 2020; Available from: https://www.covid19hg.org/blog/2020-11-24-covid-19-hgi-results-for-data-freeze-4-october-2020/. Accessed Feb 2021.
- 52.Zeberg H, Pääbo S. The major genetic risk factor for severe COVID-19 is inherited from Neandertals. bioRxiv. 2020;2020.07.03.186296. [DOI] [PubMed]
- 53.Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet [Internet]. Springer US; 2019;20:467–84. 10.1038/s41576-019-0127-1. [DOI] [PubMed]
- 54.Maiers M, Halagan M, Gragert L, Bashyal P, Brelsford J, Schneider J, et al. GRIMM: GRaph IMputation and matching for HLA genotypes. Bioinformatics Oxford University Press. 2019;35:3520–3523. doi: 10.1093/bioinformatics/btz050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is personal HLA typing and COVID-19 status and cannot be provided. All the aggregated data is explicated in the figures and tables. The frequency of each allele in each dataset can be obtained from the authors upon demand.