Abstract
Objectives
In the midst of the coronavirus disease 2019 (COVID-19) outbreak, chest X-ray (CXR) imaging is playing an important role in diagnosis and monitoring of patients with COVID-19. We propose a deep learning model for detection of COVID-19 from CXRs, as well as a tool for retrieving similar patients according to the model’s results on their CXRs. For training and evaluating our model, we collected CXRs from inpatients hospitalized in four different hospitals.
Methods
In this retrospective study, 1384 frontal CXRs, of COVID-19 confirmed patients imaged between March and August 2020, and 1024 matching CXRs of non-COVID patients imaged before the pandemic, were collected and used to build a deep learning classifier for detecting patients positive for COVID-19. The classifier consists of an ensemble of pre-trained deep neural networks (DNNS), specifically, ReNet34, ReNet50¸ ReNet152, and vgg16, and is enhanced by data augmentation and lung segmentation. We further implemented a nearest-neighbors algorithm that uses DNN-based image embeddings to retrieve the images most similar to a given image.
Results
Our model achieved accuracy of 90.3%, (95% CI: 86.3–93.7%) specificity of 90% (95% CI: 84.3–94%), and sensitivity of 90.5% (95% CI: 85–94%) on a test dataset comprising 15% (350/2326) of the original images. The AUC of the ROC curve is 0.96 (95% CI: 0.93–0.97).
Conclusion
We provide deep learning models, trained and evaluated on CXRs that can assist medical efforts and reduce medical staff workload in handling COVID-19.
Key Points
• A machine learning model was able to detect chest X-ray (CXR) images of patients tested positive for COVID-19 with accuracy and detection rate above 90%.
• A tool was created for finding existing CXR images with imaging characteristics most similar to a given CXR, according to the model’s image embeddings.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00330-021-08050-1.
Keywords: COVID-19, X-rays, Machine learning, Radiography, Thoracic
Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by the SARS-CoV-2 virus, poses tremendous challenges to healthcare systems around the world, and requires physicians to make fast clinical decisions under pressure. After many months that led to exhaustion of the medical teams, hospitals are confronting renewed surges with overwhelming numbers of new patients seeking medical aid. Some patients approach the emergency departments with respiratory symptoms, and others that are being evaluated for different reasons are asymptomatic yet positive for COVID-19.
The prevalent test used for COVID-19 identification is reverse transcription polymerase chain reaction (RT-PCR) [1–3], despite its high false negative rates. The undetected fraction of active patients inevitably leads to uncontrolled viral dissemination, masking hidden essential epidemiological data [4–6]. Additionally, RT-PCR testing kits are expensive; processing them requires dedicated personnel and can take hours to days. Rapid and accurate methods of diagnosis that do not rely on medical staff are therefore becoming crucial for the control of the pandemic. CXRs of COVID-19 patients can demonstrate typical findings including peripheral opacities and ground glass patterns in the absence of pleural effusion [5, 7, 8], and therefore may be used as a triage test, for establishing and grading pulmonary manifestations, as well as for follow-up.
Deep learning models have shown impressive abilities in image-related tasks, including in many radiological contexts [9–11]. They have great potential in assisting COVID-19 management efforts but require large amounts of training data. When training neural networks for image classification, images from different classes should only differ in the task specific characteristics; it is important, therefore, that all images are taken from the same machines. Otherwise, the network could learn the differences, e.g., between machines associated with different classes rather than identifying physiological and anatomical COVID-19 characteristics.
Portable X-ray machines are predominant in COVID-19 handling [12], and most available CXRs of patients with COVID-19 in Israel come from portable X-rays. While COVID-19 is easier to detect in CT [13], CT is more expensive and exposes the patient to higher radiation, and its decontamination process is lengthy and causes severe delays between patients. The major challenge with the use of CXR in COVID-19 diagnosis is its low sensitivity and specificity in current radiological practice. A recent study found that the sensitivity of CXRs was poor for COVID-19 diagnosis [14].
This study aims to develop and evaluate machine learning tools for COVID-19 identification and management. A large dataset of images from portable X-rays collected in 4 different hospitals was used to train and evaluate a network that can detect COVID-19 in the images with high reliability and to develop a tool for retrieving CXR images that are similar to a query CXR image, based on a metric defined by the classifier. The network results in detection accuracy of 90.3%, specificity of 90%, and sensitivity of 90.5%.
Materials and methods
Data and patients
This retrospective study took place during and after the first wave of the COVID-19 pandemic in Israel, and included patients aged 18 years and older in four medical centers in Israel. The data for this study includes a total of 2427 frontal (AP/PA) CXR images from 1384 patients (63 ± 18 years, f:m = 832:552), 360 of which with a positive COVID-19 diagnosis and 1024 negative. All images came from portable X-ray machines. For COVID-19-positive patients, the standard protocol was that every symptomatic patient with positive RT PCR test for COVID 19 was admitted to the hospital, even if symptoms were mild. Routine chest X-rays were performed at the day of admission and then later for follow-up. COVID-19 positive images include a wide range of minimal to severe pulmonary damage, which, for the purpose of this work, were all read as positive COVID-19. The non-COVID-19 images were obtained from CXRs taken by the same X-ray machines from January 2017 to April 2019, before the start of the pandemic, meaning there are no false negatives in our cohort. These include normal as well as abnormal radiographs with other clinical conditions.
The test set was taken from the full CXR dataset and contains 350 CXR (15%) of which 179 (51%) are positive for COVID-19 and 171 (49%) are negative. For patients with multiple images, their images were used either for the test set or for the train set, not both. This was done to prevent the model from identifying patient-specific image features (e.g., medical implants) and associating them with the label. Both train and test sets include patients from all four hospitals.
All images were used in the highest available resolution without lossy compression; 4% (101/2426) of the images were excluded due to lateral positioning, or due to rectangular artifacts in the image, of these 98 were COVID-19 positive. No additional selection criteria were used to exclude images based on clinical radiological findings.
Image processing
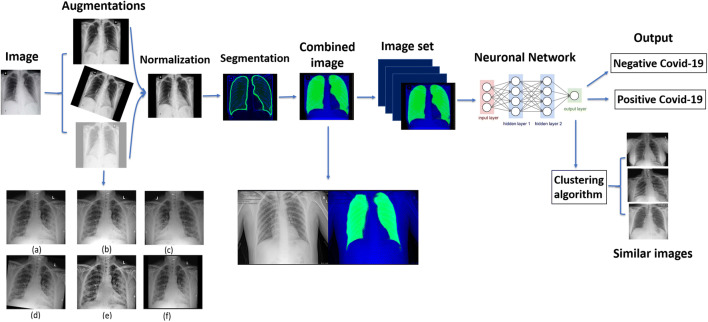
The model pipeline (Fig. 1) begins with a series of preprocessing steps, including augmentation, normalization, and segmentation of the images.
Fig. 1.
Full pipeline workflow overview. First each image undergoes processing consisting of augmentation, which is a set of visual transformations (transformations shown: (a) original image, (b) brighten, (c) horizontal flip, (d) 7 degrees rotation, (e) CLAHE transformation, (f) scale), normalization, in order to set a standard scale of image size and color, and segmentation, which emphasizes the area of the lungs and is combined to the image. The entire image set is then fed into a neural network which produces a classification outcome for each image as positive for coronavirus disease 2019 (COVID-19) or negative for COVID-19. In addition, embedded features are extracted from the last layer of the network and are used to find images with similar characteristics to a given image as learned by the network
Augmentations are transformations that change features such as image orientation and brightness. These properties are irrelevant for correct classification, but may vary during image acquisition, and can affect the training performance of the network because of its rigid registration with respect to orientation and pixel values. They serve to enlarge the dataset by creating a diverse set of images, increasing model robustness and generalizability [15, 16]. Importantly, augmentations should correspond to normal variation in CXR acquisition; to ensure this, we consulted with radiologists when defining the augmentation parameters (see Supplemental Material for details).
The normalization process aims to standardize image properties and scale. It consists of cropping black edges, standardizing the brightness and scaling the size of each image to 1024 × 1024 pixels using bilinear interpolation.
To enhance performance, we created an additional image channel using lung segmentation via a U-net pre-trained on an external dataset as detailed in [17]. This network produces a pixel-mask of the CXR indicating the probability that each pixel belongs in the lungs, allowing the network to access this information while training. Input images contain 3 channels: the original CXR, the segmentation map, and one filled with zeroes. This was done to accommodate the pre-trained models we used that use 3-channel RGB images.
Network architecture and output
We compared five network models: ResNet34, ResNet50, ResNet152 [18], VGG16 [19], and CheXpert [9]. The general approach of these architectures is to reduce images from a high-dimensional to a low-dimensional space such that a simple boundary can be used to separate image classes. The models were trained using transfer learning, i.e., loading weights pre-trained on the ImageNet database [18, 19] or on the CheXpert dataset [9] and subsequently retraining them on our data. We additionally classified the images using an ensemble model that outputs the average of the networks’ results.
In addition to classification, we propose a method for retrieving a number of CXR images that are the most similar to a given image. The activation of layers of the neural network serves as embeddings of the images into a vector space, and should capture information about clinical indications observed in the images. We used the embeddings produced by the network’s last layer to search for similarity between the resulting vectors and retrieve the nearest neighbors of each image.
Evaluation
For model evaluation we used accuracy, sensitivity, specificity, and area under the curve (AUC) for receiver operating characteristic (ROC) and precision recall (P-R) curves. Confidence intervals (CIs) were calculated for 10 different random divisions of the data into training and testing sets. For each division, the CI was obtained by taking 100 bootstrap samples out of the test set and calculating the requested metrics on each sample. The CIs are then given by the 2.5th and 97.5th percentiles for each metric. In the paper, we report the CIs for the original data split. See Supp. s7 for more detailed results from all 10 data divisions.
We evaluate the model with and without image pre-processing and examine its performance on a set of 22 CXRs determined by a radiologist as hard to diagnose. We provide additional analysis of the model’s performance by visualizing its results with t-distributed stochastic neighbor embedding (t-SNE) [20], a method that maps multi-dimensional data into a two-dimensional space to enable visualization. See Supp. s8 for more details on t-SNE.
Results
Data acquisition
The patient data included in this study are shown in Table 1. The imaging dataset consists of a total of 2426 CXRs, of which 53% (1289/2426) are positive for COVID-19 and 47% (1138/2426) are negative; 4% (101 of 2426, 98 positive) of the images were excluded due to lateral positioning or having rectangular artifacts covering parts of the image. To our knowledge, this is one of the largest datasets of original COVID-19-labeled X-ray images.
Table 1.
Demographic statistics on patients and chest images in this study
| Label | No. of patients | No. of images | Sex (men/women/unknown) | Age (years mean ± std) |
|---|---|---|---|---|
| COVID-19 positive | 360 | 1191 | 199 (55%)/132 (36%)/29 (9%) | 60 ± 18 |
| COVID-19 negative | 1024 | 1135 | 353 (34%)/323 (32%)/348 (34%) | 65 ± 19 |
Quantitative analysis of the model
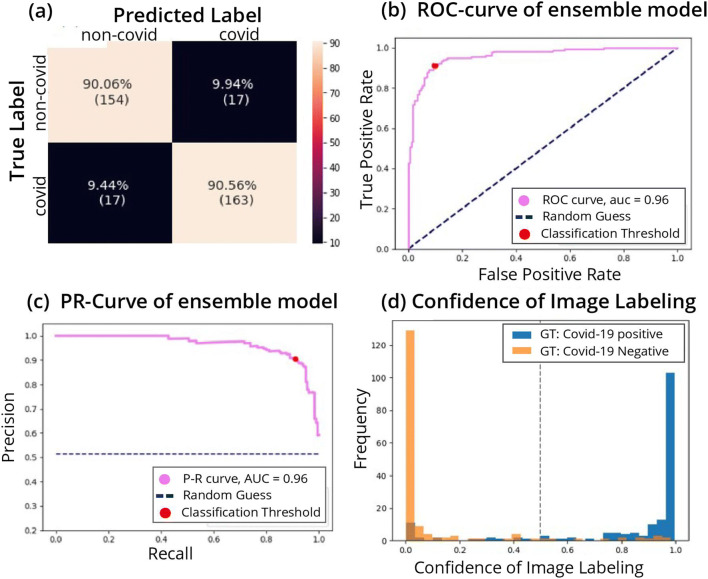
The performance of the network was tested on 15% (350 of 2426) of the images that were not used for training. The metrics we used are accuracy (proportion of successful classifications), sensitivity (also—recall, the proportion of positively labeled images that were classified correctly), and specificity (proportion of correctly classified negative images). Results for five different networks can be seen in Table 2. The ResNet50 architecture had the best performance and was used for analyses requiring network embeddings (t-SNE and KNN). The ensemble model, which averages over the output of multiple networks, achieved accuracy of 90.3% (95% CI: 86.3–93.7%), specificity 90.0% (95% CI: 84.3–94%), and sensitivity 90.5% (95% CI: 85–94%) on the test images. The AUC of the ROC curve is 0.96 (95% CI: 0.93–0.97). The ROC curve is provided in Fig. 2a, showing the relationship between the false positive rate (FPR) and the true positive rate (TPR) for different classification threshold values. Figure 2b presents the P-R curve, which shows a similar tradeoff between precision (proportion of positively classified images that were correctly classified) and recall, with AUC of 0.96 (95% CI: 0.94–0.97). Both figures show a broad range of thresholds for which both high performance metrics are attainable.
Table 2.
Comparison of accuracy, sensitivity, and specificity of various deep networks trained and tested on the same test set
| Training model | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| ResNet34 | 86.8 (305 of 351) | 83.81 (151 of 180) | 90.0 (154 of 171) |
| ResNet50 | 90.0 (316 of 351) | 90.5 (163 of 180) | 89.4 (153 of 171) |
| ResNet50 - No preprocessing | 85.1 (298 of 350) | 82.1 (147 of 179) | 88.3 (151 of 171) |
| ResNet152 | 87.1 (306 of 351) | 83.3 (150 of 180) | 91.2 (156 of 171) |
| CheXpert | 80.6 (283 of 351) | 81.1 (146 of 180) | 80.6 (137 of 171) |
| VGG16 | 85.2(299 of 351) | 81.6 (147of 180) | 88.8 (152 of 171) |
| Ensemble* | 90.3 (317 of 351) | 90.5 (163 of 180) | 90.0 (154 of 171) |
*Bold: model with best accuracy and sensitivity is the ensemble shown in bold
Fig. 2.
Performance of the model. a Confusion matrix of the classification. True positive rate (TPR) at the bottom right corner, true negative rate (TNR) at the top left corner, false positive rate (FPR) at the top right corner, and false negative rate (FNR) at the bottom left corner. b Receiver operating characteristic (ROC) curve. The curve shows the relation between true positive rate (TPR) and false positive rate (FPR) as the threshold of the separation between positive and negative classification is varied. The performance of the model is measured by the area under the curve (AUC). Ideally, the curve should cover as much area as possible up to the upper left corner (AUC score of 1), which minimizes the FPR while maximizing the TPR. The AUC is 0.95. c Precision-recall curve. Shows the relation between precision and recall. Precision and recall are affected from different classes of the data, thus can vary in scores when data is imbalanced (e.g., more observations of positive or negative compared to the other). We would like to have the AUC as large as possible up to the upper right corner, which maximizes both precision and recall. d Classification score histogram. Ground truth (GT) labels are in colors. Every image is scored on a scale between 0 and 1 with threshold of 0.5, seen as a dashed line, such that all images with a higher score will be classified as positive for COVID-19 and images below as negative. Negatively labeled images that received a score above 0.5 are, therefore, incorrectly classified images, and vice versa with respect to positively labeled images. However, the closer the image score is to one of the edges (0 or 1), the stronger the confidence in the image’s classification. The accumulation of two distinct colors on the edges point to good separation of many observations with strong confidence in the classification
We additionally evaluated the model on test sets with a COVID-19 prevalence of 5% and 10% that adhere more closely to a realistic disease prevalence in the patient population. The model maintained similar scores, as seen in Supp. s9.
We trained the ResNet50 model on the dataset with and without all preprocessing stages. As seen in Table 2, preprocessing incurs an improvement of 4% in accuracy and 5% in sensitivity. In analyzing subgroups of our patient cohort, we found that prediction accuracies are higher for females than males (Supp. Fig. 1), but there is no strong effect of age on model performance (Supp. Fig. 2).
Qualitative analysis of the model
The binary decision of whether a patient has COVID-19 is based on an activation score between 0 and 1 outputted by the network that corresponds to the probability the network assigns to the positive label. We generated a histogram of these scores (Fig. 2d), and observe that the majority of the correctly classified points are accumulated at the edges, while the wrongly classified images are more spread out along the x-axis.
We additionally visualize the distinction made by the model using t-SNE, which uses a nonlinear method to reduce high dimensional vectors into two dimensions, making it possible to visualize the data points and reveal similarities and dissimilarities between them. We used the last layer of the network to obtain an embedding of the images into a vector space. These embeddings were then inputted to the t-SNE. Figure 3 shows these image embeddings as points in a 2-dimensional space, colored by their GT labels. The figure depicts two distinct clusters, revealing a similarity between most images belonging to the same label.
Fig. 3.
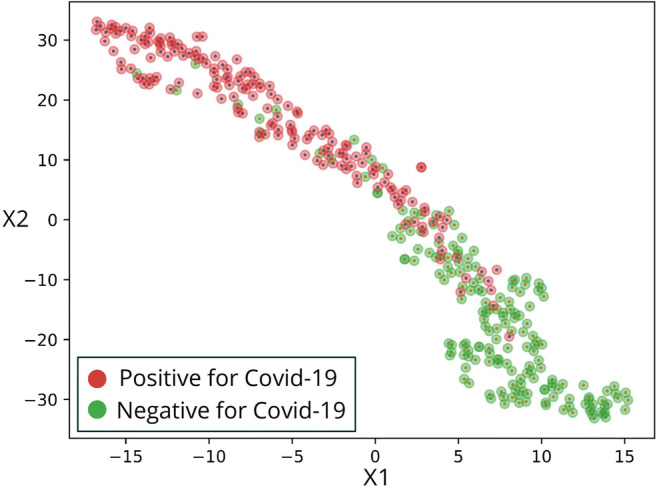
t-Distributed stochastic neighbor embedding (t-SNE). A high-dimensional feature vector was extracted for each image from the last layer before the network output, and reduced into 2 dimensions. Each point on the graph represents the features of an image after dimension reduction and arrangement in space. Next the images were colored according to their ground truth (GT), thus revealing two main clusters. The clusters are mostly in one color each, which essentially shows a strong association of the features, extracted from the decision layer and are used to arrange in space, with the GT of the images, represented by the colors
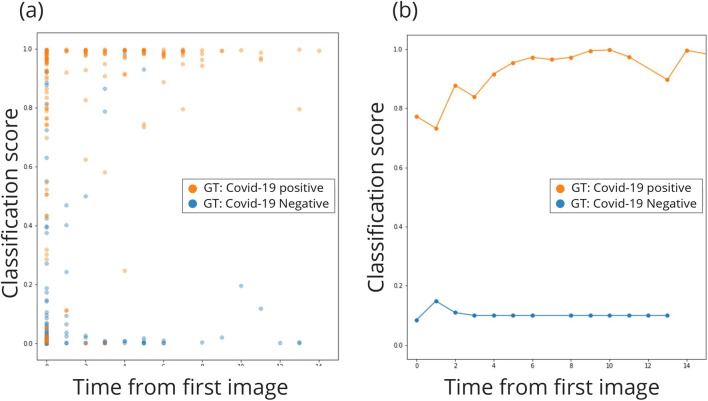
We also examine the model’s performance over time, by plotting the prediction scores according to the days from admission. As the disease progresses, lung findings tend to become more prominent. This is in line with the results, seen in Fig. 4, where the model’s performance improves over time, with most classification errors occurring on a patient’s first image, taken upon hospital admission.
Fig. 4.
Classification score as a function of time change. The first image of each patient was acquired at the same day of first admission; we note that time value as day 0. Other images of patients which were scanned more than once were noted with time value according to the number of days since the first image was acquired, thus representing the time elapsed from first admission and is ordered on the x-axis. The y-axis shows the classification score of each image between 0 (= negative for COVID-19) and 1 (= positive for COVID-19), such that a score closer to the edge indicates more confidence in the network’s classification. a The classification score with respect to change in time. The more days elapse since first admission, the more confident the classification. b Mean values of classification scores for all images of the same day value
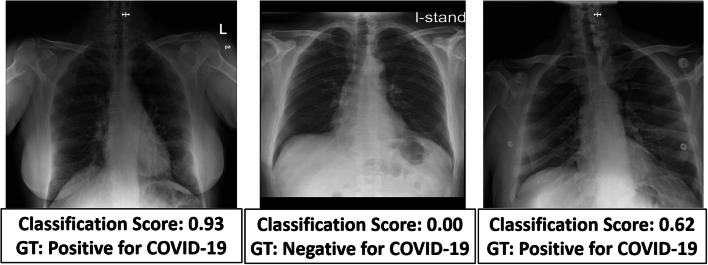
In order to consider a more difficult task, we used our model to classify 22 CXRs, 9 positive for COVID-19, and 13 negative, determined by radiologists as challenging to diagnose. Challenging images included images from patients with a positive COVID-19 PCR that either have minimal parenchymal abnormalities and look normal to the radiologist’s eye, or have pulmonary infiltrates similar to preexisting diseases other than COVID 19. The model achieved accuracy 77%, and sensitivity 77% on this task. In Fig. 5, three correctly classified images from this test are shown with the network’s classification score and the GT.
Fig. 5.
Three images labeled by a radiologist as hard to diagnose. Despite this, the model was able to classify them correctly. Each image is scored with a classification score on a scale between 0 and × 1 with threshold of 0.5 such that all images with score above the threshold will be labeled as positive for COVID-19 and images below as negative. The ground truth (GT) label of each image is also shown
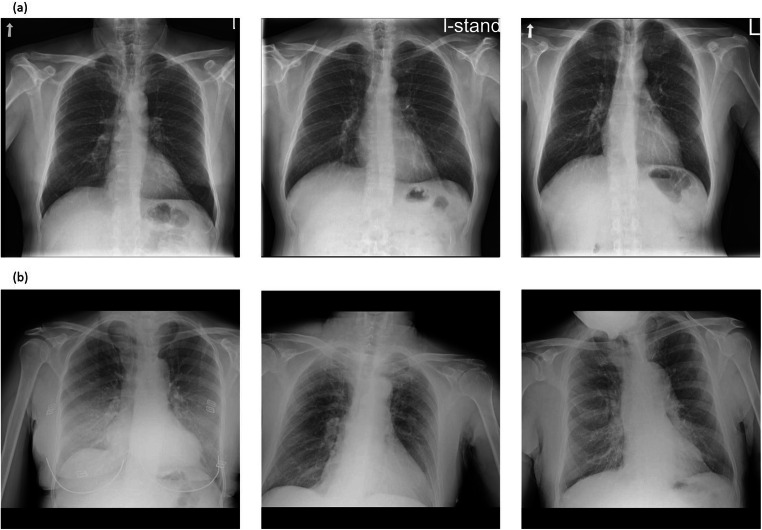
As an additional tool, we applied K-Nearest Neighbors (KNN) on the image embeddings in order to retrieve images similar to each other as shown in Fig. 6. For each image, we retrieved 4 images with the closest image embeddings; averaging over these images’ predictions achieves 87% accuracy (305/350), 91.2% specificity (156/171), and 83.2% sensitivity (149/179), meaning that the nearest images typically have the same labels.
Fig. 6.
In the figures, the left image is a CXR from the test set, and the two on the right are the two images closest to it from the training set, given the image embeddings from the network’s last layer. a All three images are COVID-19 negative. The distances between the middle and rightmost images to the left one are 0.54 and 0.56 respectively. b All three images are COVID-19 positive. The distances between the middle and rightmost images to the left one are 0.51 and 0.55 respectively. The overall mean distance between training and test images is 3.9 ± 2.5 (mean ± std). The mean distance between all positive training and positive test images is 1.4 ± 1.9, between negative training and negative test images 2.2 ± 1.3, and between images from different classes is 5.8 ± 1.9. We see that images from different classes are further away from each other, but whether a close distance truly corresponds to similar lung findings still requires verification
Discussion
In this study, we developed a deep learning pipeline to classify chest X-ray (CXR) images of patients as positive or negative for coronavirus disease 2019 (COVID-19), achieving a detection rate of above 90%. Our classifier offers instant COVID-19 detection, to allow for fast response and improved turnaround times in medical centers. In addition, we created a tool that retrieves the CXR images most similar to a given image. This tool can provide physicians with a reference to previous patients that had similar CXR findings. The internal information the hospital has about these previous patients can then be used to inform decisions for further treatment.
Early approaches to COVID-19 classification using neural networks relied on publicly available image sources, including COVID-19 image data collection [21] with 481 COVID-19-positive images and COVID-Net open-source initiative with 473 COVID-19-positive X-ray images [22–26]. Some efforts include classification into multiple lung and chest conditions including COVID-19 [27], and others attempt outcome prediction [28, 29].
Such efforts have a number of drawbacks, highlighted in the detailed review presented in [30]. The datasets they rely upon were compiled from various sources, often using one source only for COVID-19 images and another only for COVID-19-negative images and other non-COVID conditions [30]. Positive and negative images in these datasets may therefore be produced by different X-ray machines, in particular portable and fixed machines, which give rise to images with different expressions of acquisition-related features. As a result, the network’s predictions may rely on features related to the source rather than the relevant medical information [31]. Moreover, the limited number of positive COVID-19 CXR images in these datasets may cause the models to overfit [32] and impair their ability to generalize to external datasets. A dataset with more positive COVID-19 images as used in this study, containing 1191 positive CXRs, tends to produce more stable results.
In this work, we sought to address the limitations of previous studies in several ways. Most importantly, we collected CXRs from the same portable X-ray machines for both patients positive and negative to COVID-19. We used raw images without compression that may result in loss of features and introduction of source-dependent artifacts. Moreover, our dataset contains diverse data from four medical centers and is balanced between COVID-19 and non-COVID-19 images.
A recent effort has shown more reliable results based on a larger, more uniformly sourced dataset and comes closer to the goal of developing tools that can be used in clinical settings [11]. They achieved a sensitivity of 88% with a specificity of 79%. Our approach improves upon these notably solid results in terms of performance (sensitivity of 90.5% and specificity of 90.0%). As we show, this performance increase may be the result of the image pre-processing pipeline, particularly image augmentations and the addition of a segmentation channel. These steps lead to a performance increase of 8.4 percentage points in sensitivity and 1.1 percentage points in specificity (Table 2—ResNet50 vs. ResNet50 no preprocessing).
Another novelty of our work is that we introduced a content-based image retrieval tool that identifies similar CXRs based on a metric defined by using the image embeddings given by the second to last layer of ResNet50. As ResNet50 was trained for COVID-19 classification, we expect similar images under this metric to represent similar cases in terms of their clinical condition. This tool enables medical staff to search the database to identify relevant study cases for a new case under consideration. We note that the scoring process for this similarity measure still requires further investigation in a clinical setting. We would ideally like to compare the disease progression for patients that were found by our tool to have similar lung findings.
In the future, we intend to deploy our model for testing in a clinical setting within the hospitals. We also plan to work on COVID-19 severity classification. A limitation of our study is that preexisting medical illnesses and comorbidities were not integrated into the analysis of both COVID-19 and control datasets, due to a lack of access to clinical data of the patients. Our COVID-19-negative cohort comprises patients with a multitude of diseases, but with the absence of precise labels, we cannot analyze our ability to separate between COVID-19 and any specific lung morbidity. Moreover, our classifier is tailored towards portable X-rays within the four hospitals that provided the data. It requires further fine tuning to be used in other hospitals or diagnostic settings.
In summary, we developed a deep neural network which is able to reliably and rapidly detect patients with COVID-19. Even though medical imaging has not yet been approved as a standalone diagnosis tool [12], we believe it can be used as an aid to medical judgment with the advantage of immediate outcome, leading to improved turnaround times. We further created a tool for X-ray image retrieval based on lung similarities, drawing connections between patients with similar disease manifestations.
Supplementary information
(DOCX 911 kb)
Acknowledgements
We would like to acknowledge Avithal Elias, Nadav Nehmadi and the 8400 Health Network for their contribution and facilitation of the initial stages of the project.
Abbreviations
- AUC
Area under the curve
- CIs
Confidence intervals
- COVID-19
Coronavirus disease 2019
- CXR
Chest X-ray
- FPR
False positive rate
- GT
Ground truth
- KNN
K-Nearest Neighbors
- P-R curve
Precision-recall curve
- ROC
Receiver operating characteristic
- RT-PCR
Reverse transcription polymerase chain reaction
- TPR
True positive rate
- t-SNE
t-Distributed stochastic neighbor embedding
Funding
This study has received funding by Jean and Terry de Gunzburg Corona Research fund and from the Manya Igel Centre for Biomedical Engineering and Signal Processing.
Declarations
Guarantor
The scientific guarantor of this publication is Yonina C. Eldar, PhD.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• multicenter study
Footnotes
The code development and analysis was performed by six of the authors who are not radiologists (Y. E., D. K., D. Y., Y. S., E. G., A. B.). The clinical images were collected and approved by the authors (L. C., E. A., L. L, D. L., Z. N., M. M., M. H., N. E., E. S., B. N. G, S. T., Y. R., D. S., A. D., N. R. B., A. G., N. S.) which are employed as radiologists in the hospitals which provided the data.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Daphna Keidar and Daniel Yaron contributed equally to this work.
References
- 1.Vogels C, Brito A, Wyllie AL et al (2020) Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR assays. medRxiv. 10.1101/2020.03.30.20048108 Accessed September 27, 2020
- 2.Udugama B, Kadhiresan P, Kozlowski HN et al (2020) Diagnosing COVID-19: the disease and tools for detection. ACS Nano:3822–3835. 10.1021/acsnano.0c02624 Accessed September 27, 2020 [DOI] [PubMed]
- 3.Yang Y, Yang M, Shen C et al (2020) Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv. 10.1101/2020.02.11.20021493 Accessed September 27, 2020
- 4.Luo L, Liu D, Liao X et al (2020) Modes of contact and risk of transmission in COVID-19 among close contacts. medRxiv. 10.1101/2020.03.24.20042606 Accessed September 27, 2020
- 5.Arevalo-Rodriguez I, Buitrago-Garcia D, Simancas-Racines D et al (2020) False-negative results of initial rt-pcr assays for COVID-19: a systematic review. medRxiv. 10.1101/2020.04.16.20066787 Accessed September 27, 2020 [DOI] [PMC free article] [PubMed]
- 6.Fang Y, Zhang H, Xie J et al (2020) Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology:E115–E117. 10.1148/radiol.2020200432 Accessed September 27, 2020 [DOI] [PMC free article] [PubMed]
- 7.Kong W, Agarwal PP. Chest imaging appearance of COVID-19 infection. Radiol Cardiothorac Imaging. 2020;2(1):e200028. doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Y, Guan H, Zhou S, et al. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur Radiol. 2020;30(6):3306–3309. doi: 10.1007/s00330-020-06731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irvin J, Rajpurkar P, Ko M, et al. CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison. Proc AAAI Conf Artif Intell. 2019;33(01):590–597. [Google Scholar]
- 10.Van Sloun RJG, Cohen R, Eldar YC (2020) Deep learning in ultrasound imaging. Proc IEEE 108(1):11–29. 10.1109/JPROC.2019.2932116
- 11.Zhang R, Tie X, Qi Z et al (2020) Diagnosis of COVID-19 pneumonia using chest radiography: value of artificial intelligence. Radiology 298(2):E88–E97. 10.1148/radiol.2020202944 [DOI] [PMC free article] [PubMed]
- 12.ACR Recommendations for the use of Chest Radiography and Computed Tomography (CT) for Suspected COVID-19 Infection | American College of Radiology. https://www.acr.org/Advocacy-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection. Accessed September 27, 2020
- 13.Ng M-Y, Lee EY, Yang J, et al. Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiol Cardiothorac Imaging. 2020;2(1):e200034. doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon SH, Lee KH, Kim JY et al (2020) Chest radiographic and ct findings of the 2019 novel coronavirus disease (Covid-19): analysis of nine patients treated in Korea. Korean J Radiol 21(4):494–500. 10.3348/kjr.2020.0132 [DOI] [PMC free article] [PubMed]
- 15.Abdollahi B, Tomita N, Hassanpour S (2020) Data augmentation in training deep learning models for medical image analysis. Intell Syst Ref Libr:167–180 https://link.springer.com/chapter/10.1007/978-3-030-42750-4_6. Accessed September 27, 2020
- 16.Perez L, Wang J (2017) The effectiveness of data augmentation in image classification using deep learning. http://arxiv.org/abs/1712.04621. Accessed 27 Sept 2020
- 17.Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. Lect Notes Comput Sci:234–241 http://lmb.informatik.uni-freiburg.de/. Accessed September 27, 2020
- 18.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit:770–778 http://image-net.org/challenges/LSVRC/2015/. Accessed September 27, 2020
- 19.Simonyan K, Zisserman A (2015) [1409.1556] Very deep convolutional networks for large-scale image recognition. Int Conf Learn Represent. https://arxiv.org/abs/1409.1556. Accessed September 27, 2020
- 20.Van Der Maaten L, Hinton G (2008) Visualizing data using t-SNE. J Mach Learn Res 9(86)2579–2605
- 21.Cohen JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M (2020) COVID-19 image data collection: prospective predictions are the future. http://arxiv.org/abs/2006.11988. Accessed 27 Sept 2020
- 22.Shah FM, Joy SKS, Ahmed F et al (2020) A comprehensive survey of COVID-19 detection using medical images. engrXiv. https://engrxiv.org/9fdyp/. Accessed September 27, 2020 [DOI] [PMC free article] [PubMed]
- 23.Wang L, Wong A (2020) COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. http://arxiv.org/abs/2003.09871. Accessed September 27, 2020 [DOI] [PMC free article] [PubMed]
- 24.Khan AI, Shah JL, Bhat MM (2020) CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Programs Biomed 196 [DOI] [PMC free article] [PubMed]
- 25.Minaee S, Kafieh R, Sonka M, Yazdani S, Jamalipour Soufi G (2020) Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med Image Anal 65 https://pubmed.ncbi.nlm.nih.gov/32781377/. Accessed November 1, 2020 [DOI] [PMC free article] [PubMed]
- 26.Bressem KK, Adams LC, Erxleben C, Hamm B, Niehues SM, Vahldiek JL (2020) Comparing different deep learning architectures for classification of chest radiographs. Sci Rep 10(1) https://pubmed.ncbi.nlm.nih.gov/32788602/. Accessed November 1, 2020 [DOI] [PMC free article] [PubMed]
- 27.Sharma A, Rani S, Gupta D (2020) Artificial intelligence-based classification of chest X-ray images into COVID-19 and other infectious diseases. Int J Biomed Imaging. 10.1155/2020/8889023 [DOI] [PMC free article] [PubMed]
- 28.Bae J, Kapse S, Singh G et al (2020) Predicting mechanical ventilation requirement and mortality in COVID-19 using radiomics and deep learning on chest radiographs: a multi-institutional study. ArXiv. http://www.ncbi.nlm.nih.gov/pubmed/32699815. Accessed November 1, 2020 [DOI] [PMC free article] [PubMed]
- 29.Zhu J, Shen B, Abbasi A, Hoshmand-Kochi M, Li H, Duong TQ (2020) Deep transfer learning artificial intelligence accurately stages COVID-19 lung disease severity on portable chest radiographs. PLoS One 15(7 July):e0236621. https://pubmed.ncbi.nlm.nih.gov/32722697/. Accessed 1 Nov 2020 [DOI] [PMC free article] [PubMed]
- 30.DeGrave AJ, Janizek JD, Lee SI (2020) AI for radiographic COVID-19 detection selects shortcuts over signal. medRxiv. https://pubmed.ncbi.nlm.nih.gov/32995822/. Accessed November 1, 2020
- 31.Maguolo G, Nanni L (2020) A critic evaluation of methods for COVID-19 automatic detection from x-ray images. http://arxiv.org/abs/2004.12823. Accessed 27 Sept 2020 [DOI] [PMC free article] [PubMed]
- 32.Halevy A, Norvig P, Pereira F. The unreasonable effectiveness of data. IEEE Intell Syst. 2009;24(2):8–12. doi: 10.1109/MIS.2009.36. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 911 kb)