Abstract
Lipid nanoparticle (LNP) formulations of messenger RNA (mRNA) have demonstrated high efficacy as vaccines against SARS-CoV-2. The success of these nanoformulations underscores the potential of LNPs as a delivery system for next-generation biological therapies. In this article, we highlight the key considerations necessary for engineering LNPs as a vaccine delivery system and explore areas for further optimisation. There remain opportunities to improve the protection of mRNA, optimise cytosolic delivery, target specific cells, minimise adverse side-effects and control the release of RNA from the particle. The modular nature of LNP formulations and the flexibility of mRNA as a payload provide many pathways to implement these strategies. Innovation in LNP vaccines is likely to accelerate with increased enthusiasm following recent successes; however, any advances will have implications for a broad range of therapeutic applications beyond vaccination such as gene therapy.
Keywords: Lipid nanoparticles, Vaccines, Targeted delivery, Endosomal escape, mRNA
Graphical abstract
Lipid nanoparticles: from gene therapy to vaccine in a hurry
Nanoparticle delivery systems have shown significant promise for therapeutic delivery, and in 2020, they delivered on this potential. The first two vaccines approved for immunisation against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) were lipid nanoparticle (LNP) formulations of messenger RNA (mRNA) [1]. These new nanoparticle formulations proved to be more effective and faster to develop than traditional vaccination approaches. Although the first LNP formulation containing antisense RNA oligonucleotides was licenced in 2018, recent improvements in lipid formulation, nucleic acid chemistry and a global pandemic have accelerated the adoption of longer but more fragile mRNA cargoes. Unlike traditional vaccines, they contain no viral components and use technology developed for gene therapy to direct the production of antigens by cells at the inoculation site. The recent success of these formulations highlights the growing potential of nanotherapeutics, and in particular, LNP technology, as the delivery system for the next generation of biological therapies. Importantly, the accelerated approval of these nanoformulations has simplified the path for developing other LNP delivery systems for the treatment of a wide range of diseases, from cancer to diabetes and cystic fibrosis.
LNPs are typically 80–200 nm in diameter [2] and are composed of nucleic acids complexed with lipids, which protect the fragile nucleic acid cargo. LNPs spontaneously self-assemble when the appropriate ratio of lipids and nucleic acids are mixed, typically with the aid of a microfluidic micromixer device. Microfluidic devices are capable of rapid and efficient mixing of the LNP components to generate LNPs with controlled size [3]. By altering the mixing conditions, type and proportion of each lipid used in the formulation, the properties of an LNP can be tuned to meet different requirements. An overview of typical LNP compositions is discussed in Section 3. After assembly, dialysis is used to remove the solvents used to solubilise the lipid component. The size and polydispersity of LNPs is typically characterised by dynamic light scattering or nanoparticle tracking analysis. Zeta potential measurements are also used to estimate the optimal ratio of lipid to nucleic acids [4]. The dialysis step typically only removes solvent and does not remove free DNA/RNA, so it is important to characterise the encapsulation of the cargo. The encapsulation efficiency of nucleic acids is typically determined using a RiboGreen assay [5].
Despite the remarkable effectiveness of LNPs as vaccines, they are a relatively novel technology, and many gaps remain in our knowledge of these systems. Therefore, there is considerable scope to improve the effectiveness of LNPs. Presently, lipid nanoparticle-encapsulated mRNA (mRNA/LNP) vaccines require cold chain storage (<−20 °C), which has delayed the widespread distribution of the vaccines and limits their deployment in areas with less developed infrastructure. The RNA cargo of these vaccines is transcribed into protein in the cytosol; however, the efficiency of delivering RNA to the cytosol once a cell has taken up a LNP is estimated to be as low as 1% [6]. Improving this efficiency at the cellular level could reduce the number of LNPs required per dose. Maximising the potency of LNP vaccines could enable the production of a larger number of smaller doses, lowering cost, increasing access and limiting any side-effects caused by excipients in the formulation. There is also a need to investigate the tolerability of the lipids used to formulate LNPs. Cationic lipids are known to exhibit cytotoxicity and there is the potential to raise an undesirable immune response against polyethylene glycol (PEG) coatings on LNPs. The future use of LNPs as a platform for the delivery of different vaccines and other gene therapies could be compromised if repeated administration is not tolerable.
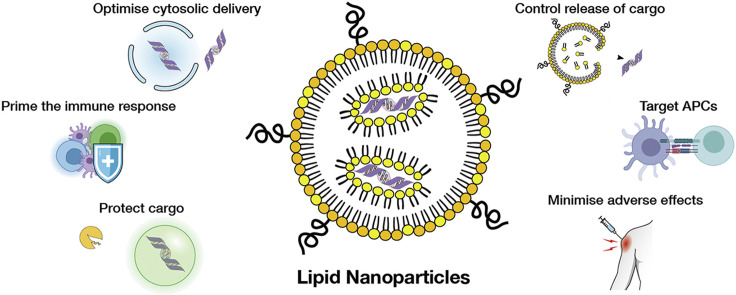
In this article, we highlight the key parameters for engineering LNP vaccine delivery systems and examine the properties of LNPs that have made them an innovative vaccine format with significant potential for further improvement and customisation (Figure 1 ). We focus on efforts to improve the immune response by incorporating adjuvating activity (Section 4) and optimising the antibody response (Section 5). From the materials delivery perspective, we explore strategies for increasing the transport of RNA to the cytosol (Section 6), controlling the release of RNA from LNPs (Section 7) and targeting LNPs to antigen-presenting cells (APCs) (Section 8). Finally, we look at efforts to improve the tolerability of LNPs (Section 9) and examine the implications for other applications of LNPs in light of their success as vaccines.
Figure 1.
Key considerations in developing mRNA/LNP vaccines.
A vaccine immunology primer for materials scientists
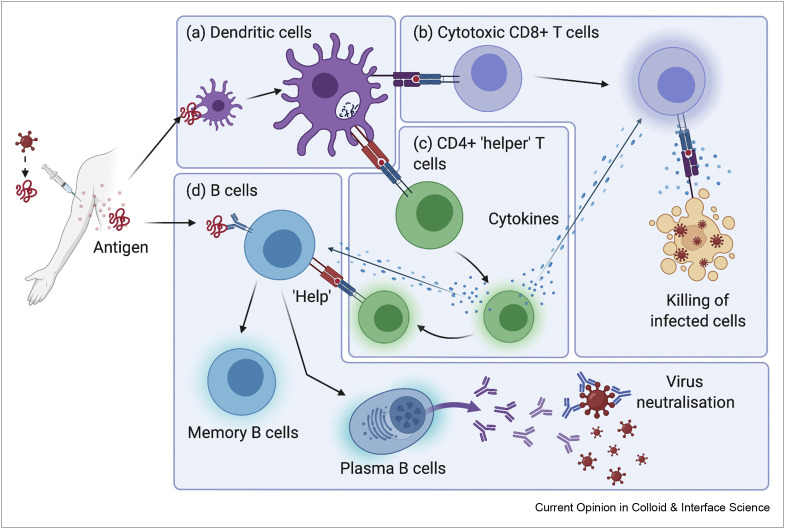
At the most basic level, the function of a vaccine is to safely expose the immune system to an infectious pathogen or facsimile [7]. This allows the pre-emptive development of a protective or prophylactic immune response against future encounters with the same pathogen without experiencing disease. An effective vaccine elicits long-lasting adaptive humoural (B cell and antibody) and cellular (cytotoxic and helper T cells) immunity, tailored to the type of pathogen and location of infection. In the context of viral immunity, antibodies help to clear the circulating virus and prevent new infection of healthy cells. Cytotoxic T cells detect and destroy infected cells to prevent the production of a new virus, whereas helper T cells amplify and direct aspects of the immune system through secreted cytokines and intercellular signalling (Figure 2 ).
Figure 2.
Antigen-presenting cells such as dendritic cells (a) are first to encounter antigens delivered by vaccination and present processed antigen to (b) cytotoxic CD8+ T cells and (c) CD4+ ‘helper’ T cells. Upon subsequent infection, these cytotoxic T cells can remove infected cells, whereas helper T cells secrete cytokines to enhance and direct the function of both T and B cells. B cells can capture antigen directly (d); however, helper T cells are required for B cell differentiation and high titre antibody production.
To initiate these adaptive responses, antigen from the site of vaccine inoculation must be processed into a format recognised by T cells. The presence of local inflammation (induction of cytokines) and pathogen-identifying molecular patterns (e.g. viral nucleic acids vs bacterial lipids) modify the type and magnitude of adaptive response required. These operations are performed by tissue-resident immune surveillance cells known as APCs such as dendritic cells (DCs), macrophages and monocytes. After capturing and recognising antigen, APCs can migrate to the local lymph node where they encounter T and B cells and direct the development of the subsequent immune response. The components of a vaccine are chosen to manipulate the development of immunity through this mechanism.
LNPs as vaccines
Antiviral vaccines vary in complexity from live attenuated viruses, to inactivated viruses, virus-like particles, through to purified recombinant proteins. Their complexity loosely reflects a balance between safety and effectiveness; however, modern developments are starting to disrupt this relationship. Live attenuated vaccines are able to infect cells and replicate, producing the strongest immunogenic signals by causing inflammation, cell death or injury, but can also result in reduced tolerability, which restricts use in individuals with weakened immunity. At the other end of the continuum are subunit vaccines, which consist of purified recombinant protein formulated with an adjuvant and represent the near-minimal immunological stimuli required. Many subunit vaccines require secondary ‘booster’ inoculations to provide additional antigen and immune stimulation and can be less effective in the elderly.
New vaccine technologies that use genetic material lie between these extremes and have features of both. These vaccines deliver nucleic acids to cells in the body rather than directly delivering protein antigens. Once inside cells, these nucleic acids are translated into protein antigens by cellular machinery, which can subsequently be presented to the immune system. Notably, nucleic acids are a consistent and biochemically defined payload, regardless of the proteins encoded in their sequence. An effective LNP formulation can therefore be adapted to deliver other genetically encoded antigens with minimal reoptimisation. This property provides a method to rapidly tailor these vaccines to address different viruses, as well as escape mutations that may arise in circulating strains by simply updating the coding sequence.
The main challenges of using nucleic acids as vaccines relate to their structure and function. Unlike protein antigens, long protein-encoding nucleic acids are highly labile and rapidly degraded by nucleases present in vivo and in the general environment. Single-stranded nucleic acids such as mRNA are particularly fragile, as cleavage of a single phosphodiester bond will interrupt the coding sequence. Furthermore, mRNA is a highly negatively charged macromolecule (>500,000 Da) unable to diffuse through cell membranes. To be active, the mRNA must be transported into the cytosol where it can access cellular machinery (e.g. ribosomes) to be translated into protein. Overcoming these issues using effective delivery systems is key to the development of nucleic acid vaccines.
The two types of nucleic acid vaccine in current use against SARS-CoV-2 deliver viral vector DNA or synthetic mRNA. Viral vectors, such as ChAdOx1 used in AstraZeneca/Oxford vaccine (AZD1222), use a replication-deficient adenovirus, where the native viral genome is replaced with the vaccine DNA. The DNA is protected from degradation by the viral capsid and native viral machinery is used to ensure efficient delivery of the genetic material. The disadvantage of this approach is that the viral vector itself can elicit an immune response, potentially clearing the vector before it has a chance to deliver its DNA cargo, and limiting the readministration of vaccines formulated in a similar vector.
Delivery of synthetic mRNA requires a different approach. To protect the mRNA from degradation and to improve cytosolic delivery, the most well-studied non-viral delivery systems are LNPs. LNPs are used in Moderna's (MRNA-1273) and Pfizer/BioNTech's (BNT162b2) SARS-CoV-2 vaccines, which are authorised for conditional/emergency use by major regulatory agencies such as the FDA and EMA [8,9]. A standard LNP formulation is composed of ionizable cationic lipids, PEGylated lipids, structural lipids and cholesterol (Figure 3 ). Cationic lipids play multiple roles in the mRNA/LNP system. During the formation of LNPs, the positively charged lipids complex with negatively charged mRNA molecules [10]. This electrostatic interaction is responsible for the high encapsulation efficiency of mRNA (and other nucleic acids) into LNPs. They also facilitate intracellular delivery of the cargo to the cytoplasm of cells by inducing endosomal escape (see Section 6) [11]. Cationic lipids can also function as adjuvants because of their immunogenicity (see Section 4) [12]. PEGylated lipids provide a hydrating ‘stealth’ layer to improve colloidal stability. Structural lipids contribute to the formation of the lipid bilayer and often resemble endogenous cell membrane lipids. Finally, cholesterol stabilises the lipid bilayer, increasing fluidity and decreasing permeability by mediating the ‘packing’ of lipid chains in the membrane [2,13].
Figure 3.
Computational model of a LNP/nucleic acid complex, with a LNP composition ratio of 4:1:4:1 cationic lipid/DSPC/cholesterol/PEG-lipid. The lipid polar moieties (cyan) of cationic lipids (yellow) surround nucleic acids (red). Structural lipids such as DSPC (grey) and cholesterol (pink) intersperse to stabilise the structure. PEG-lipid (violet) provides a hydrating ‘stealth’ layer to improve colloidal stability. Reprinted in part with permission from J. Phys. Chem. C. 2012;116(34):18440-50. Copyright 2012 American Chemical Society [14].
mRNA/LNP vaccines can be readily adapted to change the antigen breadth in response to viral mutations, much more easily than other vaccine types that contain protein antigens. The antigen content and the lipid formulation can be considered independently — changing the encoded antigens on the mRNA requires only a change in sequence and does not require reformulation to accommodate changes.
Current SARS-CoV-2 LNP vaccines already encode engineered antigens such as the prefusion stabilised spike mutant [15], but future iterations could include T helper epitopes derived from other coronavirus proteins, or additional spike mutants covering escape mutations observed in circulation. Future approaches could even implement protein engineering techniques used to produce more complex forms of antigen used in some subunit and virus-like particle vaccines. Presentation of antigenic proteins in a multimeric format can help to crosslink B cell receptors and enhance B cell activation [16].
mRNA is very susceptible to chemical degradation — it undergoes oxidation with reactive oxygen species (ROS) [17], hydrolytic cleavage of the phosphodiester bond (catalysed by metallic complexes or nucleases) [18], and internal phosphoester transfer (catalysed by RNAse A and ribozymes) [19,20]. Chemical and sequence modifications have been used to stabilise mRNAs, with artificial 5’ cap analogues, nucleobases such as 5-methylcytosine and pseudouridine, and secondary sequence structures providing protection from nuclease degradation while preserving function [21, 22, 23]. Condensation with cationic lipids can also provide some protection for mRNA, although the extent of this protection is lipid-dependent [5]. Despite both Pfizer/BioNTech and Moderna vaccines using these modifications, strict storage conditions are still required (−70 °C Pfizer/BioNTech. −20 °C Moderna vaccine). Owing to the novelty of these formulations, stability data are scarce and ongoing studies are investigating if the formulations may remain stable at higher temperatures [9]. Additional stability data have recently (as of writing) been submitted to regulatory authorities demonstrating short-term stability of the Pfizer/BioNTech vaccine at −20 °C (Pfizer; URL: https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-submit-covid-19-vaccine-stability-data). Modifying other nanoparticle components can potentially improve the protection of mRNA. The choice of ionisable lipid alters the protection afforded to mRNA, although it is not yet known which properties are required for maximum protection. Optimising each component of the carrier system can improve the stability of the system, further enabling its ability to shield mRNA from degradation. Many of these protective strategies, although discussed in the context of mRNA, are also applicable to other nucleic acids.
Adjuvants — from manufacturing impurity to molecular patterns
Vaccine development beyond initial live-attenuated and whole-pathogen formulations was driven by a need to reduce the reactogenicity and adverse effect profiles of the treatments. However, in purifying the antigen from other viral matter to improve tolerability, the resulting vaccines had notably decreased effectiveness [24]. To restore the immunogenicity of these safer vaccines, adjuvants must be added to the formulation. Adjuvants enhance the activation of APCs to amplify the subsequent T and B cell responses. This is particularly important for long-term protection as only a minor proportion of a de novo T and B cell response is directed to differentiate into specialised long-lived memory phenotypes [25,26]. Potent short-term immune responses prevent reinfection (weeks to months); however, this protection wanes and subsequent encounters require a ‘recall’ response (years to decades) mediated by memory cells. Increasing the magnitude of the initial response increases the number of memory cells and therefore the longevity (population half-life) of their protection. Adjuvants also substitute for pathogen-associated molecular patterns (PAMPs) that identify distinct pathogens (e.g. viral double-stranded RNA, bacterial lipids) [27]. The class of PAMP detected by APCs directs the type of response raised in T and B cells (Figure 4 ). For antiviral responses, the desirable outcome is a ‘T helper 1’ (TH1) bias, characterised by antibody class switching in B cells, generation of cytotoxic CD8+ T cells and CD4+ helper T cells producing cytokines such as interferon gamma. These features are important for the clearance of intracellular pathogens [28]. Lastly, adjuvants such as alum can provide a ‘depot’ for the gradual release of antigen over time [29], which allows B cell affinity maturation, a process that increases the potency of antibodies [30].
Figure 4.
The addition of adjuvants to a vaccine formulation can bias the immune response towards the most effective pathways for the target disease. (a) Viral patterns (e.g. double-stranded RNA, modified nucleobases) detected by APCs bias the system towards (b) antiviral (ideally TH1) responses. (c) Bacterial patterns (e.g. lipopolysaccharides, flagellin) bias the system towards (d) antimicrobial responses.
The presently licenced SARS-CoV-2 LNP vaccines are not formulated with conventional adjuvants. Instead, the presence of extracellular RNA and/or contaminating double-stranded RNA produced during manufacturing act as PAMPs [31] and they are often described as ‘self-adjuvanting’ [32]. However, the cationic lipids from LNPs are also speculated to provide a nonspecific adjuvating effect through inflammation caused by cellular toxicity. Necrotic cell death, cell stress or injury can cause the release of intracellular components such as histones and heat shock proteins. These normally sequestered molecules can be recognised by APCs through pattern recognition receptors in the same manner as PAMPs. These damage-associated molecular patterns can similarly activate APCs [33] and may explain the ability of LNP vaccines to function without specific adjuvants. Further developments in LNP vaccine technology are likely to lead to a reduction in toxicity, potentially resulting in a reduction in the nonspecific adjuvant effect of the system. Instead, supplementing the adjuvanting effect of LNPs with better characterised adjuvants can afford greater control over the immunostimulatory response. This could potentially enable increased control over the adverse effect profile of the vaccine.
In future iterations, the recombinant expression of transcription factors (TFs) such as interferon response factors and nuclear factor kappa-light-chain-enhancer of activated B cells (NFkB) could replace adjuvants and provide greater control over immune signalling. In DCs, these regulatory proteins control cellular programs such as immune maturation and cytokine release [34,35]. Encoding expression of these TFs in mRNA alongside vaccine antigens could result in direct modulation of the transcriptional programming of APCs, bypassing signalling through adjuvants and pattern recognition receptors altogether. Alternatively, control of these cellular pathways can be altered by overexpressing cellular receptors that signal through these TFs. The mRNA-based adjuvant TriMix, for example, encodes immunostimulatory receptors that can increase the activation of immune cells [36]. All these strategies would only be desirable in APCs, therefore such LNPs would ideally be targeted to specific cells (see Section 8). In the future, such vaccines could become in situ DC gene therapies, with no requirement of bystander cells and substantially fewer side-effects following administration.
Antibodies for protection and in dysfunction
Antibodies play a key role in removing circulating virus through a number of mechanisms. One type of antibody that can be generated is the neutralising antibody, which can directly inactivate virions by interfering with the function of the bound protein (Figure 5 a). SARS-CoV-2 virions can be neutralised by antibody binding to the receptor-binding domain of the viral spike protein, which blocks its interaction with the cellular entry receptor on human cells and prevents infection [37]. Raising a high titre, virus-specific neutralising antibody response is the primary objective for most prophylactic vaccines.
Figure 5.
Antibodies raised against different viral epitopes initiate different immune responses. (a) Neutralising antibodies block the binding site of viral particles, preventing cell entry. Non-neutralising antibodies can provide broader protection against disease by (b) opsonising virus for enhanced uptake and (c) directing natural killer cells to infected cells for apoptosis. (d) Rare, aberrant antibody responses such as ADE can increase the internalisation of viral particles.
Non-neutralising antibodies can also be beneficial against viruses and may provide broader protection against disease. Antibodies that recognise and bind to viral proteins can direct their destruction and clearance by other elements of the immune system (opsonisation — Figure 5b). Antibodies can also recognise viral proteins expressed on the surface of infected cells, directing natural killer cells to induce apoptosis of infected cells (Figure 5c). An example of this complex interplay occurs in influenza infection, where neutralising antibody epitopes occur on the haemagglutinin protein of influenza virus. Antibodies binding to the neuraminidase protein help prevent the release of virus progeny from cells, agglutinate and opsonise virus for clearance by phagocytes, and direct killing of infected cells expressing the protein [38].
Although rare, a concern regarding vaccines is the generation of aberrant antibody responses. A small number of viruses such as Dengue and Zika [39] are naturally capable of infecting and/or replicating in Fc-receptor expressing cells such as macrophages, which can lead to viral replication in these cells and immune dysfunction (Figure 5d). Antibodies to these viruses can cause increased infection of these cells by enhancing their uptake without inactivating the virus. Antibody-directed enhancement (ADE) of infection can also occur following natural infection and is understood to be caused by an antibody response unusually dominated by non-neutralising antibodies.
A concern for the current development of SARS-CoV-2 vaccines is raising non-neutralising antibodies that could contribute to ADE. Some experimental MERS-CoV vaccines encountered ADE during development, while ADE was reported in SARS-CoV-1 [40]. When dealing with a novel pandemic virus such as SARS-CoV-2, which has not been thoroughly characterised, the safest approach may be to exclude everything but the target antigen. In the case of SARS-CoV-2, this means delivering only the spike protein to avoid generating non-neutralising antibodies directed at other viral proteins. This cautious approach favours the use of vaccines with more strictly defined antigens such as subunit and mRNA/LNPs, rather than live-attenuated or inactivated virus vaccine candidates. However, as our understanding of the virus improves, increasing the breadth of coverage across multiple epitopes should be considered. This modern approach, known as ‘reverse vaccinology’ [41], is particularly suited to LNP vaccine development because of its flexibility; multiple epitopes can be incorporated simply by adding additional mRNA sequences to the formulation. This circumvents the need to recombinantly express and purify each new antigen for each reformulation and is a major strength of the platform. Outside of the current pandemic, selective incorporation of epitopes may also be useful for avoiding the generation of antibodies that cross-react with human targets. Diseases such as Guillain–Barre syndrome [42], for example, are thought to involve the activity of antibodies reactive against both influenza virus and human proteins. Although such complications are exceedingly rare, eliminating these concerns could further increase the safety and acceptability of vaccines.
Improving cytosolic delivery
For translation of therapeutic mRNA to occur, it must reach the cytosol. LNPs are taken into cells via endocytosis and are typically trafficked into endo/lysosomal compartments where biomolecules like mRNA are rapidly degraded [43]. A key requirement of LNPs is the need to promote the delivery of RNA from the endo/lysosomes in a process referred to as endosomal escape. However, our current strategies to induce endosomal escape are extremely inefficient, and LNP-mediated cytosolic delivery has been estimated to be as low as 1–2% [6]. Although it is notable that LNP vaccines and therapeutics have demonstrated efficacy despite a low percentage of mRNA reaching the cytosol, the current low efficiency of endosomal escape leaves significant potential for improvement.
Efforts to improve endosomal escape are limited by an incomplete understanding of the mechanisms involved in cytosolic delivery. Part of the challenge in this area arises from our inability to directly measure endosomal escape. Gene knockdown (when studying siRNA) or expression (when studying mRNA) are sometimes used as surrogate measurements of endosomal escape. However, these endpoint measurements are dependent on downstream processing after endosomal escape occurs. Recent advances in endosomal escape assays are helping to understand cytosolic delivery [44,45]. These cellular assays can be combined with liposomal leakage assays, which can be used to probe the membrane disruption potential of different LNP formulations [46].
There are a variety of proposed mechanisms by which endosomal escape may occur [44,47]; however, no single proposed mechanism comprehensively explains the behaviour of all endosomal escape materials. Endosomal escape for LNP cargo is thought to largely occur via destabilisation of the endosomal membrane. It has been proposed that cationic lipids in the LNP structure preferentially complex with anionic lipids that are present in the endosomal membrane. The resulting ion pair adopts a ‘cone’ shape, promoting the formation of hexagonal HII phase structures over the bilayer phase. This disrupts the lipid bilayer structure of the endosome, allowing cargo (in this case, mRNA) to ‘escape’ into the cytosol [10,11,48].
Optimising the endosomal escape ability of cationic lipids is already a major focus in the field, with both Pfizer/BioNTech and Moderna using proprietary ionisable lipid formulations in their SARS-CoV-2 vaccines (ALC-0315 and SM-102, respectively). Modifying the chemical structure (and therefore properties) of the lipid by altering the hydrophilic head group, hydrophobic lipid chain or linker region can drastically change its endosomal escape capacity [48]. For instance, the degree of saturation of the lipid chain alters the endosomal escape ability of otherwise identical lipid structures [49]. We still do not completely understand the molecular features that promote endosomal escape behaviour in certain cationic lipids. For instance, a pKa range between 6.2 and 6.5 for ionisable lipids has been observed to be ‘optimal’ to induce endosomal escape [50]. However, not all lipids within this pKa range effectively promote endosomal escape. Combinatorial libraries of lipids have been used to screen for new lipids with greater endosomal escape properties. Ramishetti et al. investigated the relationship between the structure of ionisable lipids and their endosomal escape ability [51]. By designing lipids with novel structural components, they determined that LNPs containing lipids with piperazine head groups (two ionisable amine groups) demonstrated greater transfection efficiency than the classical single tertiary amine headgroup.
Strategies to improve the endosomal escape of LNPs can also be adapted from other systems. A variety of polymers have been developed to facilitate endosomal escape [44]. Cationic polymers such as polyethylenimine (PEI), poly(2-(diisopropylamino)ethyl methacrylate) (PDPAEMA), poly(2-diethylamino)ethyl methacrylate (PDEAEMA), poly-l-histidine and poly-l-arginine have demonstrated endosomal escape ability, and can be incorporated into formulations to improve cytosolic delivery. These polymers are similar to ionisable lipids in their pH-dependent cationic properties. This enables strong electrostatic interactions between the polymers and nucleic acids, improving the stability of the nucleic acid cargo. However, this strong complexation may hinder nucleic acid dissociation from the carrier material (see Section 7). In contrast, poly(propylacrylic acid) (PPAA) is notable among pH-sensitive membrane-disruptive polymers because of its negative charge. As such, it does not complex with nucleic acid cargoes. At endosomal pH, PPAA transitions into an unionised, hydrophobic form and is thought to facilitate cargo release by partitioning into and disrupting the endosomal membrane [52,53].
Cell-penetrating peptides (CPPs) are a class of short peptides that are able to translocate across the bilayer membrane without causing significant damage. Although many CPPs have successfully delivered cargo to the cytosol, their efficiency is low [54]. Recent developments in the field have led to the design of conformationally constrained CPPs with greater binding affinity for the endosomal membrane, and a resultant improvement in endosomal escape ability [55].
Another interesting class of materials are dendrimers, polymers with branched monomer units that extend in ‘generations’ from a central core [56]. Dendrimers have been used in the cellular delivery of large nucleic acid structures such as plasmid DNA [57]. Owing to their structure, dendrimers have higher cationic charge density than their linear counterparts, particularly at higher generations. This promotes the complexation of nucleic acids and endosomal escape activity (although the exact mechanism is still to be determined). Complexation with dendrimers is also able to protect nucleic acids from enzymatic degradation, and the dendrimer surface is populated with functional groups that can be readily modified with stealth or targeting moieties.
Controlling release
An important but potentially overlooked facet of cytosolic LNP delivery is release of the mRNA (or other nucleic acids) payload from the LNP carrier. The strong electrostatic interactions between mRNA and cationic lipids are necessary to promote efficient encapsulation of mRNA during formulation. However, this interaction is detrimental to the subsequent disassembly of the LNP during delivery, where mRNA that remains complexed with cationic lipids cannot be translated into protein. It has been proposed that the release of negatively charged nucleic acids from cationic LNPs involves displacement of nucleic acids from the LNPs by anionic lipids present in the endosomal membrane [58,59]. This displacement occurs because the dual effect of electrostatic interaction of the headgroups and hydrophobic interactions of the side chains [60] is stronger than the sole electrostatic interaction between nucleic acids and cationic lipid. The efficiency of the process is likely quite low and some mRNA will remain complexed to cationic lipids. To control the association of mRNA with ionic lipids during delivery, the ionic lipids used in the LNP must be carefully designed. The pKa of the ionisable lipids in LNPs play an important role in governing release. The 6.2–6.5 pKa optimal range identified for ionisable lipids in endosomal escape screens is likely related to the strength of the electrostatic interaction and release of nucleic acids from the ionic lipids.
A potential strategy to improve nucleic acid release from LNPs is the use of charge-shifting materials. For instance, hydrolysable ester bonds have been used to conjugate side chains containing cationic tertiary amine groups to a poly(acrylamide) backbone. The initial charge on the resulting polymer is positive; however, hydrolysis of the ester bonds causes loss of the cationic amine groups and introduces anionic carboxylate functionality [61]. A similar strategy could be applied to the design of lipids for LNPs. Temporal control over lipid charge will allow effective dissociation from nucleic acids during delivery without compromising the LNP mechanisms that rely on the charge of the ionic lipids.
Targeting specific cells
Increasing the affinity of mRNA/LNPs for APCs may enable the delivery of a greater proportion of mRNA to APCs, and therefore increase the proportion of total antigen being synthesised that is actively being used to generate an immune response. There are many potential benefits to this: (1) increased potency of the immune response; (2) decreased toxicity via reduced distribution to nontargeted cells (swelling, irritation at site of administration, liver use); and (3) decreased dose required for effective protection.
Presently, LNP vaccines [62] are administered intramuscularly and result in the transfection of cells with mRNA at the injection site [2]. LNPs are thought to be taken up primarily by muscle cells and tissue-resident APCs. Transfected APCs will migrate to lymph nodes draining the injection site to present antigenic material (such as the spike protein in SARS-CoV-2) to T cells [63]. Although incompletely understood, the roles of transfected muscle and other non-APC types are also likely important. For example, cell death of transfected cells would release soluble spike antigens that would be recognised by B cells. It is yet to be established if mRNA is preferentially delivered to certain cell types or if certain cells translate the antigen more efficiently. Although the synthesis of antigen by non-APCs may contribute positively to the immune response, APCs remain the likely ideal target for mRNA/LNP vaccines because of their direct role in mediating the cellular immune response. Targeting the LNPs to APCs may increase delivery efficiency, leading to greater protectivity and lower dose vaccines. Furthermore, reducing the proportion of non-APCs transfected may alleviate local side-effects such as itching and swelling.
A number of promising targets are available on the APC surface that can be used to direct LNP delivery, including C–C chemokine receptors [64], CD80 [65], DEC205 and Clec9A [66]. Targeting can be achieved by attaching target-specific antibodies to the LNP carrier. Although antibodies are the gold standard for specificity, they are large molecules that can impact the biodistribution and colloidal stability of the LNPs. In addition, the Fc region of antibodies can elicit immunogenicity. Although some adjuvanting activity is desirable in a vaccine context, excess stimulation of the immune system can lead to an increased rate and severity of adverse events, such as anaphylaxis. Therefore, antibody fragments, such as single-chain variable fragments (scFv) or single-domain antibody fragments (sdAb or nanobodies), may be preferable. The smaller size (30 kDa for scFv and 15 kDa for nanobodies) is less likely to have a detrimental effect on the colloidal stability of the LNPs. Furthermore, the absence of the antibody Fc region curtails any concerns or risks of excess immunogenicity. However, it should be noted that the current range of scFvs and nanobodies available is significantly narrower than that of traditional antibodies.
Protein-based targeting moieties are typically attached to LNPs via reactive groups on the amino acid side chains. For instance, reacting amine residues (lysine) with N-hydroxysuccinimide esters present on the LNP will conjugate the two via an amide bond. These simple conjugation reactions are relatively uncontrolled, as modification can occur at multiple residues on the protein. The resulting random orientation of the targeting moieties impedes their ability to bind to their targets. To optimise the orientation of these attachments, unique functional groups can be introduced into the protein. This enables site-specific conjugation through the use of ‘bio-orthogonal’ reactions. Using site-specific orientation of nanobodies on nanoparticles has shown >6-fold improvement in nanoparticle binding to target cells [67,68].
Overcoming adverse responses
There are two types of adverse responses that have been noted in current LNP formulations: (1) toxicity of the cationic lipids, leading to localised cell death; and (2) immune responses to components of the LNP formulation, especially PEG.
Although cationic lipids are an integral part of mRNA/LNP formulations, they also exert cytotoxic effects. The positive charge of the lipid can destabilise the cellular membrane, leading to cell death. Furthermore, cationic lipids have been shown to induce the production of ROS, a key indicator of apoptosis [69,70]. To combat these issues, pH-dependent ionisable lipids and lipid-like materials have been developed [13]. By adjusting the pKa of these lipids, their ionisation states can be environmentally controlled. They are ionised at low pH, allowing us to leverage their charged properties during formulation and upon internalisation into the acidic environment of the endosome (pH ~6.5) [10,71]. Under systemic conditions (pH ~7.4), they are unionised, reducing the toxicity associated with surface charge and membrane disruption.
Another potentially undesirable response to the vaccine is the generation of an immune response to the LNP carrier. Despite its reputation as an ‘inert’ shielding particle, there is an established body of research showing that IgM antibodies are raised against PEG in humans. Owing to its GRAS (generally regarded as safe) designation, the use of PEG has become increasingly common in nontherapeutic products such as cosmetics and food. This has led to anti-PEG antibodies being observed in a growing proportion of the population, including those who have never been exposed to a PEG-containing therapeutic [72]. Anti-PEG IgMs promote accelerated blood clearance, leading to the rapid clearance of PEG and therefore PEG-conjugated nanoparticles upon readministration [73]. This can lead to significant changes in the biodistribution of the therapeutic, simultaneously reducing delivery to the target organ(s) and increasing unwanted accumulation of drug and carrier in others. Generation of an anti-PEG immune response would preclude the effectiveness of any PEG-conjugated therapeutics in the affected patient. This may limit the ability to deliver multiple doses of LNP-based therapeutics; an increasing concern as more LNP vaccines are developed, and LNPs are increasingly adopted as carrier systems for novel therapeutics.
To overcome undesired PEG-related immunogenicity, alternative ‘shielding’ can fill a similar role to the current linear PEG system. Shiraishi et al. introduced anionic poly(aspartic acid) blocks between PEG chains and the nanoparticle surface to reduce the binding affinity of anti-PEG IgM antibodies to PEG-conjugated nanoparticles [74]. They observed that anti-PEG IgM have much stronger affinity to PEG when conjugated to a hydrophobic surface, rather than PEG alone. Modifying the architecture of PEG to a ‘bottlebrush’ structure over the traditional linear structure can also reduce its immunogenicity [75]. Modifying the side chain length of these brushes can substantially reduce the binding of both PEG-backbone-selective and PEG-end group-selective IgMs. These strategies, where the architecture of PEG polymers is altered to reduce immunogenicity without replacing the PEG entirely, are appealing because of the existing investment and large body of research that already exists for PEG polymers.
A variety of synthetic, natural, hydrophilic and zwitterionic polymer approaches have been taken when designing novel ‘shielding’ materials. Polyoxazolines are synthetic hydrophilic lipids that have shown promising stealth properties and low immunogenicity; however, the relative youth of the field limits the available data regarding their in vivo behaviour, particularly in the context of lipid-conjugated polyoxazolines as decoration for LNPs [76].
Zwitterionic materials such as poly(carboxybetaine) are other alternative materials to PEG. Their strong hydration ability confers stealth properties that are potentially even greater than traditional PEG approaches. Furthermore, they contain abundant reactive groups that can be leveraged for specific properties (e.g. conjugation) and can be prepared as hydrolysable esters with ‘transforming’ properties upon hydrolysis [77]. As their development has mostly been focussed on use as anti-fouling surface coating materials, there is little research on their use in LNP formulations (or drug delivery systems at large). They are, however, a very promising candidate for next-generation stealth materials, as they provide strong stealth behaviour and other benefits not afforded by PEG.
Conclusion and future applications of LNPs
mRNA/LNPs have shown significant potential to address the rapid outbreak of a global pandemic like COVID-19. The ease and speed at which the mRNA cargo can be formulated with viral antigen without affecting the physical properties of the vaccine have outpaced the development of traditional vaccines. This same quality also allows LNPs to address potential viral mutations, which would decrease the efficacy of an initial vaccine formulation. The use of this cutting-edge vaccine technology to address the pandemic has rapidly advanced the field and LNP vaccines are likely to be increasingly adopted for a range of other diseases in the coming years.
Although the success of LNPs has been remarkable, there is still significant room to enhance their effectiveness and safety. We have identified a number of areas with significant potential for optimisation. Although some of these parameters are specific to the development of viral vaccines, others (protection of mRNA, optimisation of cytosolic delivery and controlling the release of cargo) can also be applied to LNP formulations in a wider context. LNP carriers have already shown promise in a wide range of therapeutic contexts, including cancer treatment [2] and gene therapy [10]. The success of LNP vaccines is likely to draw renewed interest in the field of LNP formulations. It is our hope that the knowledge gained from the development and clinical use of LNPs in SARS-CoV-2 vaccines gives new impetus to investment in and development of other LNP therapeutics.
Acknowledgements
This research was supported by a National Health and Medical Research Council Project Grant (1129672 — APRJ) and Career Development Fellowship (1141551 — APRJ) as well as the Australian Research Council through the Discovery Project Scheme (DP200100475, DP210103174 — APRJ). APRJ was also supported through the Monash University Larkin's Fellowship Scheme.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
This review comes from a themed issue on Hot Topic: COVID-19
Edited by Reinhard Miller and Libero Liggieri
References
- 1.Nanomedicine and the COVID-19 vaccines. Nat Nanotechnol. 2020;15:963. doi: 10.1038/s41565-020-00820-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines — a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive review of mRNA vaccines in development
- 3.Maeki M., Kimura N., Sato Y., Harashima H., Tokeshi M. Advances in microfluidics for lipid nanoparticles and extracellular vesicles and applications in drug delivery systems. Adv Drug Deliv Rev. 2018;128:84–100. doi: 10.1016/j.addr.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J., Shrivastava S., Cleveland R.O., Rabbitts T.H. Lipid-mRNA nanoparticle designed to enhance intracellular delivery mediated by shock waves. ACS Appl Mater Interfaces. 2019;11:10481–10491. doi: 10.1021/acsami.8b21398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blakney A.K., McKay P.F., Yus B.I., Aldon Y., Shattock R.J. Inside out: optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019;26:363–372. doi: 10.1038/s41434-019-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- Pollard A.J., Bijker E.M. A guide to vaccinology: from basic principles to new developments. Nat Rev Immunol. 2021;21:83–100. doi: 10.1038/s41577-020-00479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive introduction to the state of art in vaccinology
- 8.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]; Phase 2/3 clinical trial evaluating safety, immunogenicity and efficacy of BNT162b2 (Pfizer/BioNTech vaccine) in preventing COVID-19
- 10.Kulkarni J.A., Cullis P.R., Meel R.v.d. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Therapeut. 2018;28:146–157. doi: 10.1089/nat.2018.0721. [DOI] [PubMed] [Google Scholar]
- 11.Hafez I.M., Maurer N., Cullis P.R. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 12.Perrie Y., Crofts F., Devitt A., Griffiths H.R., Kastner E., Nadella V. Designing liposomal adjuvants for the next generation of vaccines. Adv Drug Deliv Rev. 2016;99:85–96. doi: 10.1016/j.addr.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung A.K.K., Hafez I.M., Baoukina S., Belliveau N.M., Zhigaltsev I.V., Afshinmanesh E., et al. Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J Phys Chem C. 2012;116:18440–18450. doi: 10.1021/jp303267y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh C.-L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.-C., Javanmardi K., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiller J., Lowy D. Explanations for the high potency of HPV prophylactic vaccines. Vaccine. 2018;36:4768–4773. doi: 10.1016/j.vaccine.2017.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabre A.-L., Colotte M., Luis A., Tuffet S., Bonnet J. An efficient method for long-term room temperature storage of RNA. Eur J Hum Genet. 2014;22:379–385. doi: 10.1038/ejhg.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zagórowska I., Kuusela S., Lönnberg H. Metal ion-dependent hydrolysis of RNA phosphodiester bonds within hairpin loops. A comparative kinetic study on chimeric ribo/2′-O-methylribo oligonucleotides. Nucleic Acids Res. 1998;26:3392–3396. doi: 10.1093/nar/26.14.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raines R.T., Ribonuclease A. Chem Rev. 1998;98:1045–1066. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 20.Emilsson G.M., Nakamura S., Roth A., Breaker R.R. Ribozyme speed limits. RNA. 2003;9:907–918. doi: 10.1261/rna.5680603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muttach F., Muthmann N., Rentmeister A. Synthetic mRNA capping. Beilstein J Org Chem. 2017;13:2819–2832. doi: 10.3762/bjoc.13.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohnsack K.E., Höbartner C., Bohnsack M.T. Eukaryotic 5-methylcytosine (m⁵C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes. 2019;10:102. doi: 10.3390/genes10020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Pasquale A., Preiss S., Tavares Da Silva F., Garçon N. Vaccine adjuvants: from 1920 to 2015 and beyond. Vaccines. 2015;3:320–343. doi: 10.3390/vaccines3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaech S.M., Wherry E.J., Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 26.Shinnakasu R., Kurosaki T. Regulation of memory B and plasma cell differentiation. Curr Opin Immunol. 2017;45:126–131. doi: 10.1016/j.coi.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Mogensen T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiko G.E., Horvat J.C., Beagley K.W., Hansbro P.M. Immunological decision-making: how does the immune system decide to mount a helper T-cell response? Immunology. 2008;123:326–338. doi: 10.1111/j.1365-2567.2007.02719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Awate S., Babiuk L.A., Mutwiri G. Mechanisms of action of adjuvants. Front Immunol. 2013;4:114. doi: 10.3389/fimmu.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kranich J., Krautler N.J. How follicular dendritic cells shape the B-cell antigenome. Front Immunol. 2016;7:225. doi: 10.3389/fimmu.2016.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kallen K.J., Heidenreich R., Schnee M., Petsch B., Schlake T., Thess A., et al. A novel, disruptive vaccination technology: self-adjuvanted RNActive® vaccines. Hum Vaccines Immunother. 2013;9:2263–2276. doi: 10.4161/hv.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roh J.S., Sohn D.H. Damage-associated molecular patterns in inflammatory diseases. Immune Netw. 2018;18:e27. doi: 10.4110/in.2018.18.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vander Lugt B., Riddell J., Khan A.A., Hackney J.A., Lesch J., DeVoss J., et al. Transcriptional determinants of tolerogenic and immunogenic states during dendritic cell maturation. J Cell Biol. 2017;216:779–792. doi: 10.1083/jcb.201512012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizumoto N., Hui F., Edelbaum D., Ryan Weil M., Wren J.D., Shalhevet D., et al. Differential activation profiles of multiple transcription factors during dendritic cell maturation. J Invest Dermatol. 2005;124:718–724. doi: 10.1111/j.0022-202X.2005.23616.x. [DOI] [PubMed] [Google Scholar]
- 36.Guardo A.C., Joe P.T., Miralles L., Bargalló M.E., Mothe B., Krasniqi A., et al. Preclinical evaluation of an mRNA HIV vaccine combining rationally selected antigenic sequences and adjuvant signals (HTI-TriMix) AIDS. 2017;31:321–332. doi: 10.1097/QAD.0000000000001276. [DOI] [PubMed] [Google Scholar]
- 37.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Memoli M.J., Shaw P.A., Han A., Czajkowski L., Reed S., Athota R., et al. Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. mBio. 2016;7 doi: 10.1128/mBio.00417-16. e00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martín-Acebes M.A., Saiz J.-C., Jiménez de Oya N. Antibody-dependent enhancement and Zika: real threat or phantom menace? Front Cell Infect Microbiol. 2018;8:44. doi: 10.3389/fcimb.2018.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Cheng Y., Ling R., Dai Y., Huang B., Huang W., et al. Antibody-dependent enhancement of coronavirus. Int J Infect Dis. 2020;100:483–489. doi: 10.1016/j.ijid.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Evidence of ADE in SARS-CoV-1 infection
- Burton D.R., Walker L.M. Rational vaccine design in the time of COVID-19. Cell Host Microbe. 2020;27:695–698. doi: 10.1016/j.chom.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; Applying modern immunology to mRNA/LNP vaccines
- 42.Cusick M.F., Libbey J.E., Fujinami R.S. Molecular mimicry as a mechanism of autoimmune disease. Clin Rev Allergy Immunol. 2012;42:102–111. doi: 10.1007/s12016-011-8294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rennick J.J., Johnston A.P.R., Parton R.G. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat Nanotechnol. 2021;16:266–276. doi: 10.1038/s41565-021-00858-8. [DOI] [PubMed] [Google Scholar]
- Selby L.I., Cortez-Jugo C.M., Such G.K., Johnston A.P.R. Nanoescapology: progress toward understanding the endosomal escape of polymeric nanoparticles. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2017;9:e1452. doi: 10.1002/wnan.1452. [DOI] [PubMed] [Google Scholar]; A review highlighting the challenges of engineering endosomal escape in nanoparticles
- 45.FitzGerald L.I., Aurelio L., Chen M., Yuen D., Rennick J.J., Graham B., et al. A molecular sensor to quantify the localization of proteins, DNA and nanoparticles in cells. Nat Commun. 2020;11:4482. doi: 10.1038/s41467-020-18082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erazo-Oliveras A., Najjar K., Truong D., Wang T.-Y., Brock Dakota J., Prater Austin R., et al. The late endosome and its lipid BMP act as gateways for efficient cytosolic access of the delivery agent dfTAT and its macromolecular cargos. Cell Chem Biol. 2016;23:598–607. doi: 10.1016/j.chembiol.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith S.A., Selby L.I., Johnston A.P.R., Such G.K. The endosomal escape of nanoparticles: toward more efficient cellular delivery. Bioconjugate Chem. 2019;30:263–272. doi: 10.1021/acs.bioconjchem.8b00732. [DOI] [PubMed] [Google Scholar]
- 48.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 49.Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Contr Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 50.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing In Vivo. Angew Chem Int Ed. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramishetti S., Hazan-Halevy I., Palakuri R., Chatterjee S., Naidu Gonna S., Dammes N., et al. A combinatorial library of lipid nanoparticles for RNA delivery to leukocytes. Adv Mater. 2020;32:1906128. doi: 10.1002/adma.201906128. [DOI] [PubMed] [Google Scholar]; Used a combinatorial lipid library to optimise the composition of LNP and improve gene silencing.
- 52.Kyriakides T.R., Cheung C.Y., Murthy N., Bornstein P., Stayton P.S., Hoffman A.S. pH-Sensitive polymers that enhance intracellular drug delivery in vivo. J Contr Release. 2002;78:295–303. doi: 10.1016/s0168-3659(01)00504-1. [DOI] [PubMed] [Google Scholar]
- 53.Jones R.A., Cheung C.Y., Black F.E., Zia J.K., Stayton P.S., Hoffman A.S., et al. Poly(2-alkylacrylic acid) polymers deliver molecules to the cytosol by pH-sensitive disruption of endosomal vesicles. Biochem J. 2003;372:65–75. doi: 10.1042/BJ20021945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erazo-Oliveras A., Muthukrishnan N., Baker R., Wang T.-Y., Pellois J.-P. Improving the endosomal escape of cell-penetrating peptides and their cargos: strategies and challenges. Pharmaceuticals. 2012;5:1177–1209. doi: 10.3390/ph5111177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pei D., Buyanova M. Overcoming endosomal entrapment in drug delivery. Bioconjugate Chem. 2019;30:273–283. doi: 10.1021/acs.bioconjchem.8b00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mendes L.P., Pan J., Torchilin V.P. Dendrimers as nanocarriers for nucleic acid and drug delivery in cancer therapy. Molecules. 2017;22:1401. doi: 10.3390/molecules22091401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kretzmann J.A., Ho D., Evans C.W., Plani-Lam J.H.C., Garcia-Bloj B., Mohamed A.E., et al. Synthetically controlling dendrimer flexibility improves delivery of large plasmid DNA. Chem Sci. 2017;8:2923–2930. doi: 10.1039/c7sc00097a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu Y., Szoka F.C. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 59.Zelphati O., Szoka F.C., Jr. Mechanism of oligonucleotide release from cationic liposomes. Proc Natl Acad Sci USA. 1996;93:11493–11498. doi: 10.1073/pnas.93.21.11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhattacharya S., Mandal S.S. Evidence of interlipidic ion-pairing in anion-induced DNA release from cationic Amphiphile−DNA complexes. Mechanistic implications in transfection. Biochemistry. 1998;37:7764–7777. doi: 10.1021/bi971772j. [DOI] [PubMed] [Google Scholar]
- 61.Zhang J., Lynn D.M. Ultrathin multilayered films assembled from “charge-shifting” cationic polymers: extended, long-term release of plasmid DNA from surfaces. Adv Mater. 2007;19:4218–4223. [Google Scholar]
- 62.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., et al. An mRNA vaccine against SARS-CoV-2 — preliminary report. N Engl J Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang N.-N., Li X.-F., Deng Y.-Q., Zhao H., Huang Y.-J., Yang G., et al. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182:1271–1283.e16. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grødeland G., Mjaaland S., Tunheim G., Fredriksen A.B., Bogen B. The specificity of targeted vaccines for APC surface molecules influences the immune response phenotype. PloS One. 2013;8 doi: 10.1371/journal.pone.0080008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang H.-C., Zou Z.-Z., Wang Q.-H., Li J., Jin H., Yin Q.-X., et al. Targeting and specific activation of antigen-presenting cells by endogenous antigen-loaded nanoparticles elicits tumor-specific immunity. Adv Sci. 2020;7:1900069. doi: 10.1002/advs.201900069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Macri C., Dumont C., Johnston A.P., Mintern J.D. Targeting dendritic cells: a promising strategy to improve vaccine effectiveness. Clin Transl Immunol. 2016;5:e66. doi: 10.1038/cti.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong K.W., Yuen D., Chen M.Z., Porter C.J.H., Johnston A.P.R. Pointing in the right direction: controlling the orientation of proteins on nanoparticles improves targeting efficiency. Nano Lett. 2019;19:1827–1831. doi: 10.1021/acs.nanolett.8b04916. [DOI] [PubMed] [Google Scholar]; Demonstrates that controlling the orientation of targeting groups on nanoparticles increases cell binding
- 68.Yong K.W., Yuen D., Chen M.Z., Johnston A.P.R. Engineering the orientation, density, and flexibility of single-domain antibodies on nanoparticles to improve cell targeting. ACS Appl Mater Interfaces. 2020;12:5593–5600. doi: 10.1021/acsami.9b20993. [DOI] [PubMed] [Google Scholar]
- 69.Aramaki Y., Takano S., Tsuchiya S. Induction of apoptosis in macrophages by cationic liposomes. FEBS Lett. 1999;460:472–476. doi: 10.1016/s0014-5793(99)01386-1. [DOI] [PubMed] [Google Scholar]
- 70.Yan W., Chen W., Huang L. Reactive oxygen species play a central role in the activity of cationic liposome based cancer vaccine. J Contr Release. 2008;130:22–28. doi: 10.1016/j.jconrel.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 71.Such G.K., Yan Y., Johnston A.P., Gunawan S.T., Caruso F. Interfacing materials science and biology for drug carrier design. Adv Mater. 2015;27:2278–2297. doi: 10.1002/adma.201405084. [DOI] [PubMed] [Google Scholar]
- 72.Hoang Thi T.T., Pilkington E.H., Nguyen D.H., Lee J.S., Park K.D., Truong N.P. The importance of poly(ethylene glycol) alternatives for overcoming PEG immunogenicity in drug delivery and bioconjugation. Polymers. 2020;12:298. doi: 10.3390/polym12020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shiraishi K., Yokoyama M. Toxicity and immunogenicity concerns related to PEGylated-micelle carrier systems: a review. Sci Technol Adv Mater. 2019;20:324–336. doi: 10.1080/14686996.2019.1590126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shiraishi K., Kawano K., Maitani Y., Aoshi T., Ishii K.J., Sanada Y., et al. Exploring the relationship between anti-PEG IgM behaviors and PEGylated nanoparticles and its significance for accelerated blood clearance. J Contr Release. 2016;234:59–67. doi: 10.1016/j.jconrel.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 75.Joh D.Y., Zimmers Z., Avlani M., Heggestad J.T., Aydin H.B., Ganson N., et al. Architectural modification of conformal PEG-bottlebrush coatings minimizes anti-PEG antigenicity while preserving stealth properties. Adv Healthc Mater. 2019;8:1801177. doi: 10.1002/adhm.201801177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sedlacek O., Hoogenboom R. Drug delivery systems based on poly(2-oxazoline)s and poly(2-oxazine)s. Adv Ther. 2020;3:1900168. [Google Scholar]
- 77.Jiang S., Cao Z. Ultralow-fouling, functionalizable, and hydrolyzable zwitterionic materials and their derivatives for biological applications. Adv Mater. 2010;22:920–932. doi: 10.1002/adma.200901407. [DOI] [PubMed] [Google Scholar]