We read with interest recent evaluations of SARS-CoV-2 antigen rapid diagnostic tests (Ag-RDTs), showing high sensitivity for detecting cases with higher viral loads.1 The clinical sensitivity of these tests relies on their limit of detection (LOD), and there are concerns that new SARS-CoV-2 variants may affect test performance.
New variants of SARS-CoV-2 have arisen worldwide, including the lineage B.1.1.7 (Variant of Concern (VOC) strain 202,012/01), which has become the dominant circulating SARS-CoV-2 strain in the U.K., causing >95% of new infections as of April 2021.2 Lineage B.1.1.7 has now been detected in over 114 countries in Europe, America, Australia, Asia and Africa. B.1.1.7 has 17 mutations compared with the original circulating virus, including 8 in the spike protein (S), and 2 in the nucleoprotein (N).3 Most Ag-RDTs target N protein with some targeting the S protein. Therefore, these mutations may affect antibody binding in the Ag-RDT, and consequently affect the assay sensitivity. The impact of these mutations on molecular diagnostics has been demonstrated by the failure of S gene detection probes in some nucleic acid tests.4
It is essential that Ag-RDTs are re-evaluated using new variants to determine if there is any change in test sensitivity due to mutations in the target antigen. The aim of this study was to determine the LOD of seventeen commercially available RDTs using the B.1.1.7 lineage and compare results obtained with the original dominant strain (B.1).
A clinically isolated SARS-CoV-2 strain from the B.1.1.7 lineage (Genbank accession number: MW980115), was used for the study. Frozen aliquots of the third passage of the virus were quantified via plaque assay as previously described.5 For the determination of LODs, a fresh aliquot was serially diluted from 1.0 × 106 plaque forming units (pfu)/ml to 1.0 × 102 pfu/ml. Each dilution was tested in triplicate. Two-fold dilutions were made below the ten-fold LOD dilution to confirm the lowest LOD. Culture media was used as negative control.
Viral RNA was extracted from each dilution using QIAmp Viral RNA mini kit (Qiagen, Germany) according to the manufacturer's instructions, and quantified using TaqPath COVID-19 CE-IVD RT-PCR (ThermoFisher). Genome copy number/ml (gcn/ml) were calculated as previously described.6
We evaluated 17 commercially available Ag-RDT tests (Table 1 ) following the instructions for use (IFU). The LOD was defined as the lowest dilution at which all three replicates were positive. Results were interpreted by two operators, each blinded to the result of the other. If a discrepant result was obtained, a third operator read any discrepant tests as a tie-breaker.
Table 1.
Characteristics of the Ag-RDT tested.
| In this Study | Test/Company | Target | LOD (pfu/ml) | LOD (gcn/ml) |
|---|---|---|---|---|
| ActiveXpress | ActivXpress+ COVID-19 Ag Complete Kit/ Edinburgh Genetics Ltd, UK | Nucleocapsid | 5.0 × 102 | 1.0 × 104 |
| Biocredit | Biocredit COVID-19 Ag, Rapidgen Inc., Rep.Korea | Nucleocapsid | 5.0 × 104 | 4.7 × 106 |
| Espline | ESPLINE® SARS-CoV-2/ Fujirebio Diagnostics Inc., Japan | Nucleocapsid | 2.5 × 102 | 5.1 × 103 |
| Excalibur | Rapid SARS-CoV-2 Antigen test card/ Excalibur Healthcare Services, UK | Nucleocapsid | 5.0 × 102 | 1.0 × 104 |
| Genedia | GENEDIA W COVID-19 Ag/ Green Cross Medical Sciences; Rep. Korea | Nucleocapsid | 5.0 × 102 | 1.0 × 104 |
| iChroma | iChroma COVID-19 Ag Test/ Boditech Medical Inc.,Kep Korea | Nucleocapsid | 2.5 × 102 | 5.1 × 104 |
| Innova | Innova SARS-CoV-2 Antigen Rapid/ Innova Medical Group Ltd.,UK | Nucleocapsid | 2.5 × 102 | 5.1 × 104 |
| Joysbio | SARS-CoV-2 Antigen Rapid Test Kit./ Joysbio Biotechnology Ltd. China | Nucleocapsid | 1.0 × 102 | 6.3 × 103 |
| Mologic | Mologic COVID-19 Ag Test device/ Mologic Ltd.UK | Nucleocapsid | 2.5 × 102 | 5.1 × 104 |
| NowCheck | NowCheck COVID-19 Ag test/ Bionote Inc./Mologic Ltd., Rep. Korea | Nucleocapsid | 5.0 × 102 | 1.0 × 104 |
| PanBio | Panbio™ COVID-19 Ag Rapid Test/ Abbott Rapid Diagnostics. Rep. Korea | Nucleocapsid | 5.0 × 101 | 3.2 × 103 |
| RespiStrip | Respi-Strip COVID-19 Ag/ Coris Bioconcept, Belgium | Nucleocapsid | 1.0 × 103 | 9.8 × 104 |
| Roche | SARS-CoV-2 Rapid Antigen Test/ SD Biosensor Inc., Rep. Korea / Roche Diagnostics, Switzerland | Nucleocapsid | 2.5 × 102 | 5.1 × 104 |
| Standard-F | Standard F COVID-19 Ag FIA., SD Biosensor Inc., Rep. Korea | Nucleocapsid | 1.0 × 102 | 6.3 × 102 |
| Standard-Q | Standard Q COVID-19, SD Biosensor Inc., Rep., Korea | Nucleocapsid | 2.5 × 102 | 5.1 × 103 |
| Sure-Status | Sure-Status COVID-19 Antigen Card Test, Premier Medical Corporation, India | Nucleocapsid | 5.0 × 101 |
3.2 × 103 |
| Wondfo | Wondfo 2019-nCoV Antigen Test/ Guangzhou Wondfo Biotech Co., China | Nucleocapsid | 1.0 × 103 | 2.1 × 104 |
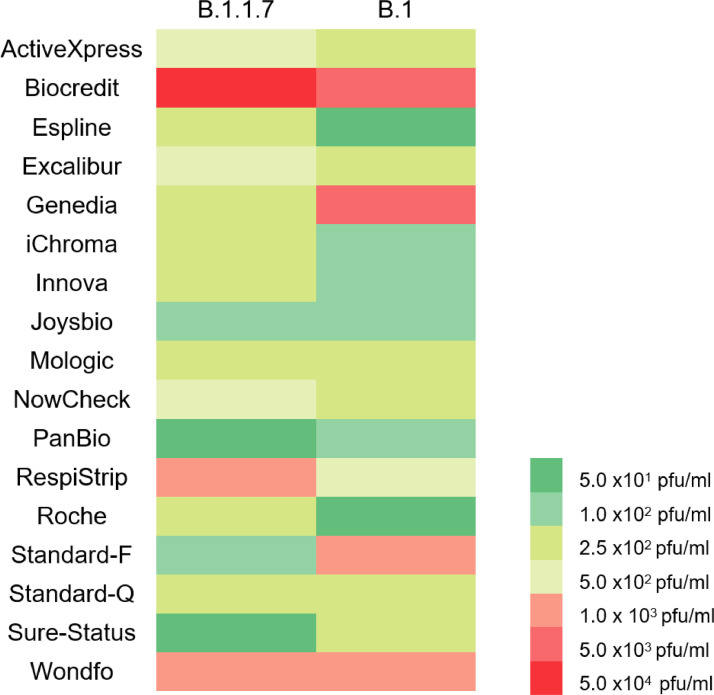
We found 14/17 Ag-RDTs had an analytical LOD of ≤ 5.0 × 102 pfu/ml (ActiveXpress, Espline, Genedia, iChroma, Innova, Mologic, NowCheck, PanBio, Excalibur, Joysbio, Roche, Standard-F, Standard-Q and Sure-Status) fulfilling the British Department of Health and Social Care (DHSC) acceptable criteria. Additionally, 16/17 (including Wondfo and RespiStrip) had an LOD of ≤ 1.0 × 106 gcn/ml, fulfilling the recommendations in the WHO Target Product Profile for SARS-CoV-2 Ag-RDT (Table 1). With an early 2020 SARS-CoV-2 strain from the B.1 lineage (GISAID accession ID EPI_ISL_464,183), 13/17 Ag-RDTs had an analytical LOD of ≤ 5.0 × 102 pfu/ml. A total of 4/17 Ag-RDTs had a lower LOD when tested with the B.1.1.7 variant, 9/17 were higher, and 4/17 were the same. However, the performance of the Ag-RDTs was not significantly different (Mann Whitney test, P 0.937) between the two strains (Fig. 1 ) and only 1/17 (RespiStript) no longer met the DHSC acceptance criteria.
Fig. 1.
Heatmap comparing the LODs of 17 Ag-RDT using the B.1.1.7 strain and the B.1. 2020 strain taken from a previously published work.8 Ag-RDT coloured green fulfilled WHO and UK DHSC.
Our results build on previous studies showing equivalent performance with the B.1.1.7 variant for a small panel of Ag-RDTs.7 The B.1.1.7 variant has two mutations in the nucleoprotein, which may be insufficient to impact antibody binding for detection of the antigen. All Ag-RDTs evaluated in this study targeted the N protein. Ag-RDTs targeting the S protein may be expected to have more difficulties in detecting B.1.1.7 due to the larger number of mutations, however, further evaluations are required to investigate this.
Other VOCs have different N gene mutations with the potential to affect Ag-RDT performance. For example, the P.1 lineage that has emerged in Manaus, Brazil, carries a single N gene mutation (P80R) 8 and B.1.243.1 carries mutation S194L, and further evaluations with these lineages are needed. New lineages and mutations in SARS-CoV-2 are constantly appearing and re-evaluation of SARS-COV-2 diagnostic tests including Ag-RDTs will be required once certain strains become more prevalent.
This study demonstrates equivalent performance of 18 Ag-RDTs with SARS-CoV-2 B1.1.7 variant 6 as with the original dominant strain from the beginning of the pandemic, and that 16 tests meet WHO limits for analytical sensitivity. This supports the continued usage of these tests in areas where B.1.1.7 is dominant.
Conflicts of Competing Interest
The authors have no conflicts of interest to declare.
Sources of funding: The study was supported by the Foundation for Innovative New Diagnostics (FIND). The funders of the study had no role in data collection and data analysis.
References
- 1.Alemanya A., Baróc B., Ouchia D. Analytical and clinical performance of the panbio COVID-19 antigen-detecting rapid diagnostic test. J. Infect. 2021;82(5):186–230. doi: 10.1016/j.jinf.2020.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies N.G., Abbott S., Barnard R.C. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538) doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A. Rambaut, N. Loman, O. Pybus, W. Barclay, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations.. 2020. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563
- 4.Kidd M. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral loads in samples tested by ThermoFisher TaqPath RT-qPCR. J Infect Dis. 2021 doi: 10.1093/infdis/jiab082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards T., A Kay G., Aljayyoussi G. SARS-CoV-2 transmission risk from sports equipment (STRIKE) MedRxiv. 2021 [Google Scholar]
- 6.Cubas-Atienzar A.I. Limit of detection in different matrices of nineteen commercially available rapid antigen tests for the detection of SARS-CoV-2. MedRxiv. 2021 doi: 10.1038/s41598-021-97489-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickering S., Batra R., Snell L.B. Comparative performance of SARS CoV-2 lateral flow antigen tests demonstrates their utility for high sensitivity detection of infectious virus in clinical specimens. MedRxiv. 2021 doi: 10.1016/S2666-5247(21)00143-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabino E.C., Buss L.F., Carvalho M.P.S. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397(10273):452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]