Abstract
Clinically, pain has an uneven incidence throughout lifespan and impacts more on the elderly. In contrast, preclinical models of pathological pain have typically used juvenile or young adult animals to highlight the involvement of glial populations, proinflammatory cytokines, and chemokines in the onset and maintenance of pathological signalling in the spinal dorsal horn. The potential impact of this mismatch is also complicated by the growing appreciation that the aged central nervous system exists in a state of chronic inflammation because of enhanced proinflammatory cytokine/chemokine signalling and glial activation. To address this issue, we investigated the impact of aging on the expression of genes that have been associated with neuropathic pain, glial signalling, neurotransmission and neuroinflammation. We used qRT-PCR to quantify gene expression and focussed on the dorsal horn of the spinal cord as this is an important perturbation site in neuropathic pain. To control for global vs region-specific age-related changes in gene expression, the ventral half of the spinal cord was examined. Our results show that expression of proinflammatory chemokines, pattern recognition receptors, and neurotransmitter system components was significantly altered in aged (24–32 months) versus young mice (2–4 months). Notably, the magnitude and direction of these changes were spinal-cord region dependent. For example, expression of the chemokine, Cxcl13, increased 119-fold in dorsal spinal cord, but only 2-fold in the ventral spinal cord of old versus young mice. Therefore, we propose the dorsal spinal cord of old animals is subject to region-specific alterations that prime circuits for the development of pathological pain, potentially in the absence of the peripheral triggers normally associated with these conditions.
Keywords: Dorsal horn, ageing, aging, inflammation, chemokines, Cxcl13, microglia, astrocytes, glia
Introduction
Chronic pain affects 20–30% of the population1 and considerably impacts quality of life. The prevalence of chronic pain increases with advancing age reaching 50% in later life.2 Furthermore, pain more severely interferes with daily activities in the elderly.3 The dorsal horn (DH) of the spinal cord has been implicated as a source of dysfunction in chronic pain states. For example, after injury to peripheral neurosensory elements the release of proinflammatory molecules from damaged primary afferents into the DH activates spinal glia, particularly astrocytes and microglia.4–6 These activated glia in turn modulate excitatory and inhibitory neurotransmission in the DH7,8 to promote altered sensory processing.7 Thus, the DH’s ability to discriminate sensory modalities changes following central sensitization and the development of chronic pain.9
Glial cells are strongly implicated in the generation of aberrant sensory experience in neuropathic pain. For example, microglia rapidly adopt a reactive phenotype following nerve injury. This coincides with increased release of proinflammatory cytokines, including interleukin (IL)-1β, IL-6 and tumour necrosis factor (TNF)-α,10 and increased expression of a number of proteins involved in local microenvironment sampling.10–13 Inhibition of microglia with minocycline prior to nerve injury decreases mechanical hyperalgesia and allodynia.10 In contrast, astrocytes have been suggested to participate in the maintenance of neuropathic disturbance. They undergo a delayed but prolonged activation following peripheral nerve injury,4 and evidence shows activated astrocytes are necessary and sufficient to induce hyperalgesia.14 Together the above work shows that glial-derived neuroinflammatory signalling is a key process in the establishment and maintenance of chronic pain, particularly of neuropathic origin.
Despite the burden of chronic pain falling overwhelmingly on the elderly,2,3 the above mechanisms have been largely uncovered in preclinical studies on juvenile and young-adult animals. Therefore, it remains unclear how reliably our current understanding of neuropathic pain mechanisms applies to the elderly. Despite this, work has provided clear evidence of altered excitability in central nociceptive pathways in naïve aged animals. For example, extracellular in vivo recordings show that DH neurons in aged rats exhibit significantly higher background activity and higher discharge rates during noxious stimulation.15 After-discharge is also enhanced following noxious stimulation and the receptive field of putative nociceptors is larger in aged versus young adult animals.16 Our group has also shown DH processing circuits are altered in aged mice. Specifically, DH neurons exhibit a more excitable phenotype along with decreased spontaneous excitatory synaptic input, and an increased contribution of GABAergic signalling to inhibition.17 While the ultimate outcome of these combined changes for sensory processing under normal and pathological conditions remains to be determined, collectively they confirm that the excitability and signalling in the aged dorsal horn differs from its younger counterpart.
What exactly initiates and maintains the differences in dorsal horn signalling and function with advanced age is yet to be determined. Many studies have demonstrated aging in the central nervous system is associated with chronic low-grade inflammation, or inflammaging.18 How inflammaging impacts spinal dorsal horn circuits normally and during chronic pain signalling is not known. In fact, the effects of aging on the spinal cord in general is relatively understudied, and work that focuses on the dorsal horn, where sensory information is processed, is scant. Nevertheless, the literature on inflammaging in the brain highlights many of the same neuroinflammatory pathways and cell signalling events that are implicated in neuropathic dysfunction of the dorsal horn. This raises the possibility that inflammaging in the spinal dorsal horn influences nociceptive signalling at baseline as well as the propensity of this region for dysfunctional signalling under neuropathic conditions. In line with this view, metabolite levels such as N-Acetyl-aspartate, glutamate and glutamine are disrupted in the aged spinal cord, suggesting neuronal dysfunction and neurodegeneration.19 Furthermore, age-related functional changes in the spinal cord have been described, along with greater morbidity and mortality, following spinal cord injury in older humans20–23 and rats.24–26
In summary, the available evidence suggests the mechanisms regulating neuronal excitability and neuroinflammatory signalling are altered in the aged dorsal horn. To determine the extent of any such disruption we quantified expression of a range of genes with known roles in glial function, neurotransmission and inflammation in young, middle aged, and old mouse spinal cords.
Methods
Healthy C57Bl/6 male mice were used for all experiments and were maintained under standard housing conditions, on a 12-hour light-dark cycle, with food and water available ad libitum. Eight mice were used for each of the following age groups: young (2–4 months), middle-aged (12–14 months) and old (>24 months). All animal work was undertaken in strict accordance with the University of Newcastle Animal Ethics Committee guidelines and New South Wales and Australian animal research guidelines.
Tissue processing and RNA isolation
Mice were euthanized with 1mL intraperitoneal injection of Lethabarb, and transcardially perfused with 50mL ice-cold, diethyl pyrocarbonate-treated, phosphate buffered saline (DEPC-PBS). Spinal cords were removed, and snap frozen in dry ice-cooled isopentane and stored at −80°C. Lumbar regions were separated from whole spinal cords using a clean razor blade on a cutting block cooled on dry ice. Tissues were embedded in Tissue-Tek (ProSciTech, Thuringowa, QLD, Aust), frozen on dry ice and cryosectioned at 100 µm thickness (Leica CM 1950). Cryosections were thaw mounted on RNase-free glass microscope slides and then stored at −80°C.
Between 15 and 18 transverse sections were taken from the spinal cord lumbar enlargement of each animal and mounted on slides for subsequent macro-dissection and RNA extraction. Slides were washed briefly in DEPC-PBS on ice, fixed and dehydrated for 3–5 minutes in 100% ethanol. The ethanol fixation/dehydration step was used to maintain RNA stability during the macro-dissection step. With the aid of a dissecting microscope a single horizontal cut was made on each spinal cord section at the level of the central canal. This produced a dorsal and ventral half of each spinal cord section that contained the dorsal (DH) and ventral horns (VH), respectively. Isolated dorsal and ventral halves of the spinal cord were pooled for each animal, placed into RNA later (Sigma-Aldrich) and stored at -80°C for subsequent RNA extraction. Samples were homogenised using a motorised pestle (Sigma-Aldrich) in 350 µL Buffer RL from the Norgen Single Cell RNA Purification Kit, to which β-mercaptoethanol was added (1:100). RNA was extracted as per the manufacturer’s instructions, eluted in RNase free water, quantified using a Nanodrop Spectrophotometer (Thermo Scientific), and taken for DNase treatment. To remove any residual contaminating genomic DNA, RNA was DNase treated for 15 mins with DNase I (Invitrogen) according to the manufacturer’s protocol. The DNase was subsequently inactivated by the addition of EDTA (2.5mM) and heated to 65°C for 10 minutes. To generate the first strand of cDNA, reverse transcription (RT) was carried out using the Sensifast cDNA synthesis kit (Bioline) according to the manufacturers’ instructions. Two RT reactions were run, one including the reverse transcriptase enzyme (RT+) and the other without the enzyme (RT-). RNA integrity was determined using the relative transcript abundances of each gene’s 3- and 5-prime ends. This step not only detects RNA degradation, but also serves as measure of cDNA completeness. Samples were diluted in preparation for qPCR analysis.
qPCR and statistics
qPCR gene targets were selected to assess a number of functional groups including: pattern recognition receptors; proinflammatory signalling molecules; astrocyte-specific proteins/activation markers; microglia/macrophage-specific proteins/activation markers; growth factors; glycinergic, GABAergic, glutamatergic, and purinergic neurotransmitter signalling systems, and transporters. qPCR primers were designed using the NCBI Nucleotide Primer BLAST program.27 Primers were designed across exon-exon junctions, and/or towards the 3' end of the transcript where possible. Primer sequences for all genes investigated in the study are listed in Tables 1 to 3. All qPCR reactions used the SensiFast Low Rox Sybr Green kit (Bioline). Primer pair specificity was confirmed by melt curve analysis. qPCR annealing temperatures were adjusted to ensure a single product for each primer pair. All primer pairs were found to have single peak melt curve. qPCR reaction volumes were 12 µl, which contained 5 µL of sample cDNA and 7 µL of master mix. Master mix contained 6 µL of Sybr Green, 0.25 µL primer stock (forward and reverse at 10 µM each; final reaction concentration was 210 nM for each primer), and 0.75 µL nuclease free water. RT+ samples were run in triplicate for each gene. All RT− samples were run in triplicate for primer work up, and then as singles for experimental samples, on a subset of the genes assessed. The subset included nominal “housekeeping” genes, and genes where the primer did not span over an exon junction. This allowed for determination of gDNA contamination. qPCR reactions were run either on an Applied Biosystems 7500, or Viia7 Real-Time PCR System. Ct cut off in these reactions was set at 35, where Ct averages for all age groups had to be above 35 for a particular gene to be excluded. In some cases, expression of the gene was undetectable or poorly detectable (Ct > 35) in young but expressed in old (Ct < 30), and therefore that gene remained in the analysis. The reference/housekeeping genes were detected in all samples with the highest Ct (lowest transcript abundance) being 26 for reference genes.
Table 1.
Housekeeping gene details.
| Gene name/group | Abbreviation | Forward primer | Reverse primer |
|---|---|---|---|
| Actin beta | Actb | GCAGGAGTACGATGAGTCCG | ACGCAGCTCAGTAACAGTCC |
| Glyceraldehyde-3-phosphate dehydrogenase | Gapdh | TAGGGCCTCTCTTGCTCAGT | GGTTGTCTCCTGCGACTTCA |
| Succinate dehydrogenase 3′ | Sdh3’ | CTGTGTGAAGTAGGGCAGGTC | GCACTGGCTCGATACTTACCA |
| Succinate dehydrogenase 5′ | Sdh5’ | TAACTGGGGCGTGGCCAGGA | ATGCAGCTCGCAAGCCTGCC |
Table 2.
Glial activation and signalling gene details.
| Gene name/group | Abbreviation | Forward primer | Reverse primer |
|---|---|---|---|
| Cd86 molecule | Cd86 | TCTGCCGTGCCCATTTACAA | TGTGCCCAAATAGTGCTCGT |
| C-type lectin domain containing 7a | Clec7a | CAGTACACCAGACACAGGGAG | AGGAAGTAGCTTGGGTCACG |
| NLR family pyrin domain containing 3 | Nlrp3 | TACGAAGCAATGCCCTTGGA | TTACAGTCCGGGTGCAGAAG |
| Toll-like receptor 2 | Tlr2 | CCTGAGAATGATGTGGGCGT | CATTTGCCCGGAACGAAGTC |
| Triggering receptor expressed on myeloid cells 2 | Trem2 | GCCAGTCCTTGAGGGTGTC | TCCCATTCCGCTTCTTCAGG |
| C-c motif chemokine ligand 3 | Ccl3 | TCCCAGCCAGGTGT CATTTT | ATGCAGGTGGCAGG AATGTT |
| Cd13 molecule | Cd13 | CGCTGGACAACACC CTCTTC | ACGGTTGACCCAGTTGTTGG |
| C-x-c motif chemokine ligand 13 | Cxcl13 | ACCCAACCCACATC CTTGTT | TGAAGTCCATCTCGC AAACCT |
| Interferon γ | Ifng | GGCAAAAGGATGGT GACATGA | TTTCGCCTTGCTGTTGCTGA |
| Pannexin-1 | Panx1 | GCTGCACAAGTTCTTCCCCT | ATCTCGAGCACTCTTGGCAG |
| Glial fibrillary acidic protein | Gfap | ACAGAGGAGTGGTATCGGTCT | CGTTAGCTTCGTGCTTGGCT |
| S100 calcium-binding protein B | S100b | CACCCGAAGAGGTTGCTCAT | GGAAGGGTGTAGGCGATCAG |
| Cd4 molecule | Cd4 | CACTCAAGGGAAGACGCTGG | CGATCAAACTGCGAAGGCG |
| Colony stimulating factor 1 | Csf1 | AAAGGCAATCTGGCATGAAGTC | GCCACATGATTGGGAATGGA |
| C-x3-c motif chemokine receptor 1 | Cx3cr1 | ACGCTGACTCCTCATTCCAC | CTCCCCAATGTGCAAGCAAC |
| Integrin alpha subunit M | Itgam | GATTCAGCAAGCCAGAACCC | AGGAGGCATGAGAGTCCACA |
| Brain derived neurotrophic factor | Bdnf | AGCCTGAATGAATG GACCCAA | GGAACCCAGGAGAG TAACCAC |
| Neurotrophic tyrosine receptor kinase 2 | Ntrk2 | CAGGAACTTCACCC GTCCAA | AGGACTGTAGGGAC AACCCA |
| Transforming growth factor beta-1 | Tgfb1 | AGGGCTACCATGCC AACTTC | CCACGTAGTAGACGATGGGC |
| Transforming growth factor beta-3 | Tgfb3 | TTGGTTAGGGGAAG GCACAC | TGTCCACTCGCTATCCGTTC |
Table 3.
Neurotransmitter signalling gene details.
| Gene name/group | Abbreviation | Forward primer | Reverse primer |
|---|---|---|---|
| Glycine receptor alpha1 subunit transcript variant 1 | Glra1 tv1 | TTGCCTGCTCTTCGTGTTCT | GCATGGGGCTCTTGTGATGT |
| Glycine receptor alpha1 subunit transcript variant 2 | Glra1 tv2 | TGTTTGCCTGCTCTTCGTGT | CCACCCTCATCATCC TTGTGA |
| Glycine receptor alpha2 subunit | Glra2 | GCTGGAGAGTTTTG GGTACA | GCGCTCCAGGTGAAACTTG |
| Glycine receptor alpha3 subunit | Glra3 | GGCCTCCTTACCAA AGGTGT | CCAGTGCAAAAGCTTCCGTC |
| GABA(B) receptor 1 | Gabbr1 | TGGCTCGGCATTTTCTATGG | TGGTGACAGGAGCG GTAATG |
| GABA(A) receptor alpha1 subunit | Gabra1 | AATAGGGCAAGGTG GGGCTA | ACAGCAGTGTAGCCCATTGA |
| GABA(A) receptor rho1 subunit | Gabrr1 | AGTCCCCTTAGGCA TCACCA | AGTTGACAGCTGCGTACTCC |
| Gephyrin | Gphn | GCACTACCAGGGAA TCCTGT | GGCGAGGGTCCAGTTTTACA |
| Glutamate ionotropic receptor AMPA subunit 1 | Gria1 | AAAGGGGAATGTGG AAGCAAG | AACAGAAACCCTTCA TCCGCT |
| Glutamate ionotropic receptor AMPA subunit 2 | Gria2 | ACTCATCAGGGATT CTGGAGGT | TGGAAGAGTAGGCC CAGAGG |
| Glutamate ionotropic receptor NMDA subunit 1 | Grin1 | GCCTACAAGCGACA CAAGGA | GCTTTCTTTTTAGGG TCGGGC |
| Glutamate ionotropic receptor NMDA subunit 2a | Grin2a | TCTGCTCCAGTTTGT TGGTGA | GGCTGCTCATCACCTCATTC |
| Glutamate ionotropic receptor NMDA subunit 2b | Grin2b | AACTTGGCTCCCAT TCGCTT | TGAGGCCCGTTCTATCCTCT |
| Solute carrier family 1 member 3/Excitatory amino acid transporter 1 | Slc1a3/Eaat1 | CGAACCACCACCAA CGTACT | ACGGGTTTCTCCGGTTCATT |
| Solute carrier family 1 member 2/Excitatory amino acid transporter 2 | Slc1a2/Eaat2 | TATGTCGGTTGCCG TTTGGA | TCCTCCTCGGGGCTGTATTT |
| Solute carrier family 12 member 5/Potassium chloride cotransporter 2 | Slc12a5/Kcc2 | GAAGCCGGAGTGGG AAAACT | CGGGCATGTTGAGCAAAACT |
| Solute carrier family 32 member 1/Vesicular gaba transporter | Slc32a1/Vgat | GAATCTACAGCGTC CAGCCA | CAGGCCGCAAGGTTGAAATG |
| Solute carrier family 17 member 7/Vesicular glutamate transporter 1 | Slc17a7/Vglut1 | TCCACTACCAACGT GCGAAA | GAGTGCGAGTATCCGACCAC |
| Adenosine receptor a1 | Adora1 | GACATCAGAGAAAA GCCTCGC | GGAAAGCCACTCAG GTCTCA |
| Adenosine receptor a2a | Adora2a | ACAGGGCTATCTCC CGCTAA | GCTCGGGTCCATGACTTGAT |
| 5'-nucelotidase ecto | Nt5e | GTCCTGTGACCAAG TGAGCA | ACGGTTTGGGTCAAGAGTCC |
| Purinergic Receptor P2X1 | P2rx1 | CTACCATCGGCTCT GGGATTG | GTCACGTTCACCCTCCCCAG |
| Purinergic Receptor P2X4 | P2rx4 | CATTTGCGATTCAG ACGCCA | ACCAAGAGGGTGAA GTTTTCTG |
| Purinergic Receptor P2X7 | P2rx7 | ACACAGCAATAGGC AACTGG | GAACCATAGGAGAG CAAGGCA |
| Purinergic Receptor P2Y1 | P2ry1 | CCAGGACACTAACC CATCGT | AAGGCCCACAAACCTCTTCA |
| Purinergic Receptor P2Y12 | P2ry12 | TACAGAAACACTCA AGGCTGC | TGTTGACACCAGGCACATCC |
RNA integrity was determined using the 3′: 5′ ratio for the succinate dehydrogenase (SDH). Raw qPCR CT values for each gene were normalised to the geomean of beta actin, SDH, and Glyceraldehyde 3-phosphate dehydrogenase (gapdh), to generate a ΔCT. The ΔCT was transformed to its linear form using the formula ΔΔCT = 2−ΔCT for statistical analyses and fold change calculations. SEMs were calculated and adjusted relative to fold change. Heat maps summarise gene expression comparisons. For each transcript, the group average ΔΔCT (young, middle age, and old age) was calculated and then the difference of each sample’s ΔΔCT from this group average was determined, expressed in standard deviations. Values were transformed to single colour gradients (dorsal spinal cord = white to red, ventral spinal cord = white to blue) to produce finalised heatmaps. IBM SPSS was used for statistical analyses comparing ΔΔCT values. Browne-Forsythe ANOVA was used to determine group differences, with Games-Howell post-hoc tests performed to determine specific between-group differences. All values reported are mean ± SEM, unless otherwise stated, and significance was set at α = 0.05.
Results
Our results report on 46 genes expressed in the dorsal and ventral spinal cord of young, middle-aged, and old aged mice (n=7, 7, 7, respectively). The outcome of these comparisons identified that age related upregulation (12/46 genes) was more common than downregulation (6/46 genes) in the transcripts assessed, while the remainder did not show statistically significant changes across ages (28/46 genes). Regarding region specificity, although some gene changes were conserved across the dorsal and ventral halves of the spinal cord, some genes showed selective gene expression changes in one region only.
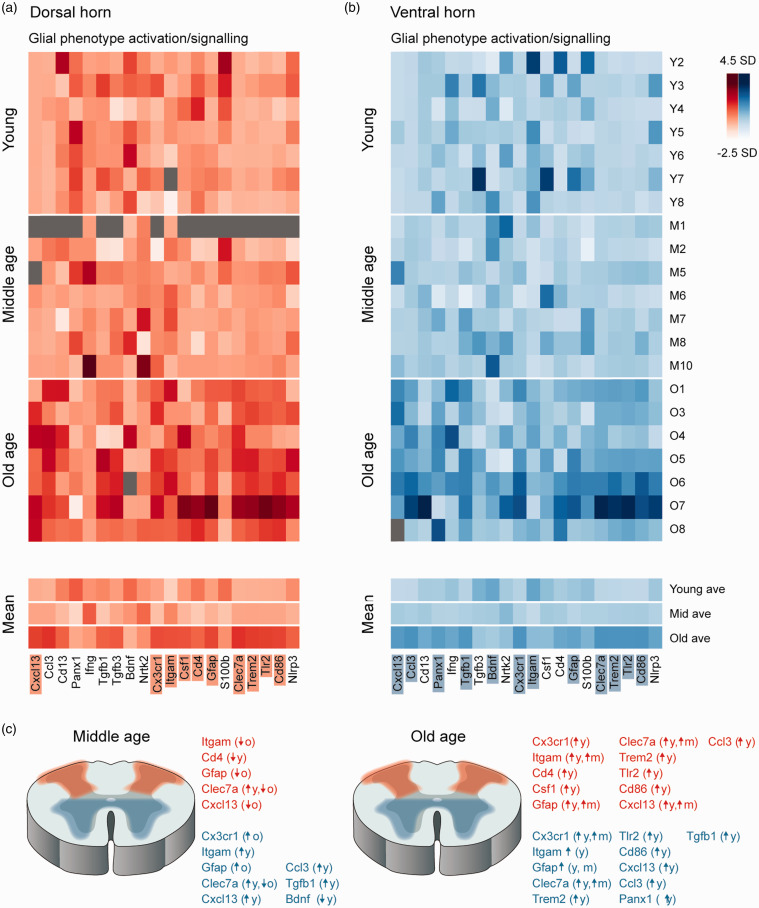
Microglia and astrocytes have been heavily implicated in the mechanisms underlying pathological dorsal horn signalling and age-related neuroinflammation, therefore a number of genes known to be enriched in these cell types were assessed in young, mid- and old-aged tissue (Figure 1). First, related to microglia, Cx3cr1 expression was affected by age in the dorsal spinal cord (F (2, 18) = 7.47, p = 0.004), undergoing upregulation at old age (1.3-fold, p = 0.006) when compared to young samples. Cx3cr1 expression was also affected by age in the ventral spinal cord (F (2, 18) = 11.42, p = 0.001), upregulated between young and old (3.8-fold, p = 0.017) and middle and old age animals (1.8-fold, p = 0.022, Figure 1). Itgam expression, also enriched in microglia, was affected by age in the dorsal spinal cord (F (2, 19) = 10.67, p = 0.001), with upregulation in old age (1.5-fold, p = 0.004) compared to young, and between middle age and old age (1.2-fold, p = 0.022). These differences were mirrored in the ventral spinal cord (F (2, 21) = 7.58, p = 0.003) with Itgam expression increased at middle age (1.3-fold, p = 0.003) and old age (1.6-fold, p = 0.023) when compared to young samples.
Figure 1.
Aging alters expression of glial activation and signalling genes in the dorsal and ventral spinal cord. Heatmaps show gene expression data presented as standard deviations (SD) from mean expression for each gene across all ages (genes listed at bottom). Samples are grouped by age: young (top), middle age (middle), and old age (lower), with mean gene expression for each age group presented at the bottom. Dark values denote elevated expression (max = 4.5 SD above mean) and light values indicate reduced gene expression (min = −2.5 SD below mean). Grey cells denote sample failed QC. Genes with significantly altered expression across age are shaded. (a) Glial activation and signalling related genes are stable in the dorsal spinal cord of the spinal cord between young and middle age, but several of these genes are increased at old age as highlighted by the generally darked expression values at the bottom of the heatmaps. (b) Expression of many glial activation and signalling related genes are also elevated in the ventral spinal cord of the spinal cord with age, with gene expression in this heatmap also appearing generally darked for aged samples, versus young and middle age. (c) Schematics summarise genes that exhibited altered expression in middle (left) and old-age (right) versus young samples in the dorsal (red) and ventral spinal cord (blue).
Two other microglia-related genes (Csf1 and Cd4) showed less consistent results, with Cd4 expression exhibiting a biphasic aging effect in the dorsal spinal cord (F (2, 18) = 5.63, p = 0.013), initially being downregulated at middle age (0.69-fold, p = 0.018), before rebounding in old age and returning to marginally higher levels than in young samples (1.1-fold, p = 0.019, Figure 1). While this same trend was apparent in the ventral spinal cord sample, Cd4 expression levels did not differ statistically across age groups (F (2, 18) = 2.118, p = 0.149). Moreover, age-related expression of Csf1 was static in the ventral spinal cord but changed in the dorsal spinal cord (dorsal: F (2, 18) = 4.008, p = 0.036; ventral: F (2, 18) = 0.238, p = 0.79).
Our analysis also assessed expression of two astrocyte-related genes. Gfap expression in the dorsal spinal cord was significantly affected by age (F (2, 18) =9.179, p = 0.002), being upregulated at old age (1.9-fold, p = 0.027) when compared to young and middle age (p = 0.013). Gfap was similarly altered by age in the ventral spinal cord (F (2, 18) =9.179, p = 0.002), being upregulated at old age (1.9-fold, p = 0.027) versus young and middle age (p = 0.013, Figure 1). In contrast S100b was unchanged across all age groups in the dorsal and ventral spinal cord (dorsal: F (2, 18) = 0.701, p = 0.509; ventral: F (2, 18) = 1.124, p = 0.347). Together, these results are consistent with an age-related change in the functional status of both microglia and astrocytes in the dorsal and ventral halves of the spinal cord.
Given several indicators of age-related disruption to microglia (above), expression of a panel of genes related to microglial phenotype and pattern recognition receptors were assessed. Of the candidates assessed in this group, Clec7a expression was significantly upregulated in the dorsal spinal cord by age (F (2, 18) = 47.227, p < 0.001) at both middle (2.3-fold, p = 0.01) and old ages (11.5-fold, p < 0.001), as well as significantly increasing between middle and old aged mice (p < 0.001). Clec7a was similarly affected in the ventral spinal cord (F (2, 18) = 13.802, p < 0.001), being upregulated at both middle (2.1-fold, p = 0.001) and old ages (28.7-fold, p = 0.018), and increased between middle and old aged mice (p = 0.03). Trem2 expression was also increased in the dorsal spinal cord with age (F (2, 18) = 22.348, p < 0.001), although upregulation was only apparent at old age (3.8-fold, p = 0.001). Upregulated Trem2 expression was also detected in the ventral spinal cord (F (2, 18) = 12.544, p < 0.001) in old age samples (5.0-fold, p = 0.018). This pattern of upregulation, limited to the later time point (old age), was also observed in Tlr2 expression in the dorsal (F (2, 18) = 16.688, p < 0.001) and ventral spinal cord (F (2, 18) = 13.594, p < 0.001), with significant increases in both regions (dorsal: 2.7-fold, p = 0.002; ventral: 1.83-fold, p = 0.021). Likewise, Cd86 expression also differed only in the old age samples in the dorsal (F (2, 18) = 13.763, p < 0.001) and ventral spinal cord (F (2, 18) = 15.303, p < 0.001), increasing in both regions (dorsal: 2.2-fold, p = 0.003; ventral: 3.3-fold, p = 0.006). Finally, expression of NLRP3 contrasted these consistent age-related changes showing relatively static expression across all age groups (dorsal: F (2, 18) = 2.209, p = 0.139; ventral: F (2, 18) = 2.902, p = 0.081).
Cytokine and growth factor signalling play key roles in both microglial and astrocyte activation as well as interactions between glia and neurons, therefore the expression of several of these molecules was assessed across our young, middle, and old age samples. Of these, the most pronounced difference was in Cxcl13 expression in the dorsal spinal cord (F (2, 17) = 10.626, p = 0.001) with a nearly 200-fold upregulation at old age (191-fold, p = 0.012), and a significant increase also between middle and old age (23-fold, p = 0.022). In the ventral spinal cord Cxcl13 expression was upregulated (F (2, 18) = 16.989, p < 0.001) at middle age (3.1-fold, p < 0.001) and old age (1.9-fold, p = 0.009), though these differences were far less dramatic than in the dorsal spinal cord. Ccl3 expression was affected in the dorsal spinal cord by age (F (2, 18) = 27.457, p < 0.001), increasing with old age (9.3-fold, p = 0.001) and was also affected in the ventral spinal cord (F (2, 18) = 16.989, p < 0.001), increasing at middle (3.0-fold, p < 0.001) and old age (23.3-fold, p = 0.009). Contrasting the conserved changes in cytokine expression across dorsal and ventral spinal cord regions described above, Panx-1 expression did not change with age in the dorsal spinal cord (F (2, 18) = 3.488, p = 0.052), but was altered with age in the ventral spinal cord (F (2, 18) = 5.926, p = 0.011) increasing at old age (1.1-fold, p = 0.037). Similarly, the growth factors assessed were stable in the dorsal spinal cord, with Tgfb1 (F (2, 18) = 3.365, p = 0.057), Tgfb3 (F (2, 18) = 2.364, p = 0.123), Bdnf (F (2, 18) = 0.850, p = 0.443), and Ntk2 (F (2, 20) = 0.532, p = 0.595) all stable across age groups. In contrast, ventral spinal cord expression of Tgfb1 was altered (F (2, 18) = 9.99, p = 0.00241), with upregulation at middle age (2.8-fold, p = 0.026) and old age (2.0-fold, p = 0.001), whereas Bdnf expression changed in the opposite direction (F (2, 21) = 3.82, p = 0.038), downregulated at middle age (0.8-fold) and remaining decreased at old age, though post-hoc tests did not resolve a significant difference in the old age sample.
The remaining category of genes assessed in these experiments included neurotransmitter receptor and transporter proteins (Figure 2). This was motivated by our previous work showing age related changes in both excitatory and inhibitory synaptic drive in dorsal spinal cord neurons. Somewhat surprisingly, the majority of genes assessed remained stable over the age, with only a few exceptions. For example, among the AMPA and NMDA receptor subunits assessed, only Gria1 expression was affected by age in the dorsal spinal cord (F (2, 20) = 4.172, p = 0.031), with middle and old age values ∼1.2 fold higher than in the young dorsal spinal cord. No other differences were detected in the dorsal (Gria2 F (2, 20) = 1.084, p = 0.357; Grin1 F (2, 20) = 0.454, p = 0.642; Grin2a F (2, 20) = 0.548, p = 0.586; Grin2b F (2, 18) = 0.359, p = 0.703) or ventral spinal cord (Gria1 F (2, 21) = 1.055, p = 0.366; Gria2 F (2, 21) = 1.903, p = 0.174; Grin1 F (2, 21) = 1.493, p = 0.248; Grin2a F (2, 21) = 1.453, p = 0.257; Grin2b F (2, 18) = 0.269, p = 0.767) expression values.
Figure 2.
Aging does not dramatically alter expression of excitatory, inhibitory, and purinergic signalling genes. Heatmaps show gene expression data presented as SD from mean expression for each gene across all ages (genes listed at bottom). Samples are grouped by age: young (top), middle age (middle), and old age (lower), with mean gene expression for each age group presented below. Dark values denote elevated expression (max = 4.5 SD above mean) and light values indicate reduced gene expression (min = −2.5 SD below mean). Grey cells denote sample failed QC. Genes with significantly altered expression across age highlighted by shading. (a) excitatory, inhibitory, and purinergic signalling genes are relatively stable in the dorsal spinal cord of the spinal cord across age, with the exception of the a2 glycine receptor subunit gene and two ATP sensitive ionotropic receptor genes (P2x1r and P2x4r). (b) expression of excitatory, inhibitory, and purinergic signalling genes were also generally stable in the ventral spinal cord of the spinal cord across age. Altered transcripts in this region all related to inhibitory signalling with the a2 glycine receptor subunit gene, α1 GABAA receptor subunit gene, and vesicular GABA transporter gene all reduced, along with a small decrease in expression of the gephyrin gene. (c) Schematics summarise genes that exhibited altered expression in middle (left) and old-age (right) versus young samples in the dorsal (red) and ventral spinal cord (blue).
As fast synaptic inhibition in the spinal cord can be mediated by glycine and GABA, both receptor types were assessed as well as the receptor stabilising protein gephyrin, and a range of transporter proteins. For glycine receptors, Glra2 differed in the dorsal spinal cord with age (F (2, 20) = 14.33, p < 0.001), downregulated at old age (0.7-fold, p = 0.005). Glra2 expression was also affected in the ventral spinal cord (F (2, 21) = 7.05, p = 0.005), with a downregulation at middle (0.6-fold, p = 0.034) and old age (0.6-fold, p = 0.018). Expression of the remining glycine receptor subunit genes assessed were stable across age in the dorsal (Glra1 tv1 F (2, 20) = 0.193, p = 0.826; Glra1 tv2 F (2, 20) = 2.495, p = 0.108; Glyra3 F (2, 20) = 1.406, p = 0.268) and ventral spinal cord (Glra1 tv1 F (2, 20) = 1.805, p = 0.190; Glra1 tv2 F (2, 21) = 1.915, p = 0.172; Glyra3 F (2, 21) = 1.065, p = 0.363). For GABA, all receptor genes assessed were stable in the dorsal spinal cord across age (Gabrr1 F (2, 18) = 0.695, p = 0.512; Gabbr1 F (2, 18) = 0.021, p = 0.979; and Gabra1 F (2, 18) = 1.449, p = 0.261). In the ventral spinal cord, Gabra1 expression did change (F (2, 18) = 8.313, p = 0.003) undergoing downregulation in old age (0.4-fold, p = 0.011), which also differed significantly from middle aged (0.7-fold, p = 0.019). The remaining GABA receptor genes were stable in the ventral spinal cord across age (Gabbr1 F (2, 18) = 0.415, p = 0.667; and Gabrr1 F (2, 18) = 0.265, p = 0.770;). Gphn expression was stable in the dorsal spinal cord (F (2, 18) = 0.717, p = 0.501), but differed in the ventral spinal cord (F (2, 17) = 6.90, p = 0.006), showing a modest downregulation with old age (0.8-fold, p = 0.004). The expression of vesicular GABA transporter, Vgat, was not affected in the dorsal spinal cord by age (F (2, 18) = 3.541, p = 0.050), but did differ in the ventral spinal cord (F (2, 18) = 11.21, p = 0.001), with a significant downregulation in old age (0.5-fold, p = 0.004), as well as downregulation between middle age (0.7-fold) and old age (p = 0.019). Also critical to effective inhibition in the dorsal spinal cord, KCC2 (a potassium-chloride co-transporter) expression has been shown to be downregulated under neuropathic conditions, in a microglial dependent manner. Despite these associations, our data suggest KCC2 expression was unchanged by age in the dorsal spinal cord (F (2, 20) = 0.696, p = 0.510) though expression in the ventral spinal cord changed with age (F (2, 21) = 3.808, p = 0.039).
The final neurotransmitter system assessed in our data was purinergic signalling proteins including ATP receptors, adenosine receptors, and associated enzymes. Among ATP receptors, expression of both P2rx1 and P2rx4 ATP-gated cation channels were altered by age (F (2, 16) = 15.42, p < 0.001; and F (2, 16) = 7.20, p = 0.006, respectively), each showing upregulation in the dorsal spinal cord with old age (2.1-fold, p = 0.007, and 1.3-fold, p = 0.007, respectively), and P2rx1 expression also increased in middle age (2.5-fold, p = 0.003). In contrast P2rx7 was unchanged in the dorsal spinal cord across all age groups (F (2, 18) = 0.813, p = 0.459). None of the remaining purinergic related proteins showed age dependent differences in the dorsal spinal cord including the ATP-sensitive G protein-coupled receptors P2ry1 (F (2, 18) = 0.986, p = 0.392) and P2ry12 (F (2, 18) = 2.516, p = 0.109), adenosine receptors Adora1 (F (2, 18) = 1.642, p = 0.221) and Adora2a (F (2, 18) = 1.315 p = 0.293), or the adenosine producing enzyme Nt5e (F (2, 18) = 0.359, p = 0.703). In the ventral spinal cord purinergic signalling gene expression did not change with age for ATP-gated cation channels, ATP-sensitive G protein-coupled receptors, or adenosine receptor and enzyme proteins. Thus, our data shows some age-related changes in glycine, GABA, and purinergic receptor gene expression, though far less pronounced than for cytokine and microglial activation genes.
Discussion
This study was motivated by the need to establish how aging affects signalling pathways in the spinal cord, with a particular emphasis on the phenotype of glial populations and the expression of neuroinflammatory signalling molecules. Comparison across three ages highlighted changes in gene expression that suggest the functional phenotypes of microglia and astrocytes are altered with age and expression of a number of neuroinflammatory molecules and receptors are elevated in naïve, uninjured animals. Altered expression of many of the molecules assessed here has also been reported in the spinal cord of young animals under neuropathic conditions This raises the prospect that these gene expression differences may contribute to the increased incidence of chronic pain in the elderly. Together, these findings provide valuable baseline data for future work assessing pathological pain mechanisms in advanced age.
There are some obvious caveats associated with our study. First, within-group variability was high for many of the genes we assessed, as shown in the intensity ranges within the columns of Figures 1 and 2. With such variability it is possible that this analysis may yield some false negatives and therefore our conclusions provide a conservative overview of these aging effects. This said, previous aging studies have documented that large variability is feature of aging and essentially explains the phenomena of healthy and unhealthy aging.28,29 The second caveat is our assessment of only male mice, necessitated by the aging colony used to source animals for the study. This limits translation of our findings as there are now well-established gender differences in chronic pain mechanisms, and chronic pain states are more common in women.30,31 Nevertheless, our data provides a necessary first step, with future work needed in aged female mice to extend these insights. Finally, our analysis examined a limited gene panel spanning glial activation and signalling molecules, as well as a number of neurotransmitter systems. (eg. the GABA receptor comes in >20 types - we only examined three). This data still provides insight into the diverse range signalling pathways that are likely altered in the dorsal and ventral spinal cord by age, as well as providing a basis for future work that interrogates these signalling pathways in greater detail.
The most dramatic age-related upregulation of gene expression observed in our work were in cytokine/chemokine and innate immune signalling pathways. For example, C-X-C motif chemokine 13 (Cxcl13) rose in the dorsal spinal cord of middle-aged mice by 23-fold, and increased further to 191-fold at old age. This observation was regionally specific, with far less dramatic expression increases in the ventral spinal cord – 3 fold in middle-age (3-fold) and ∼ 2-fold by 24 months of age. It should be noted that the magnitude of the relative increase in expression of Cxcl13 in the DH partly reflects its near undetectable expression in young animals. As to the cellular origin of spinal Cxcl13 expression, microglia and neurons have been identified as potential sources under pathological conditions in a number of studies.14,32,33 Furthermore, Cxcl13 expression has been implicated in the development of neuropathic pain following peripheral spinal- and cranial nerve injury.14 Strikingly, following spinal nerve ligation (SNL), microarray analysis revealed that Cxcl13 was the most upregulated chemokine gene in the spinal cord, showing a 47-fold increase.14 This work also confirmed a corresponding increase in Cxcl13 protein in the DH. Furthermore, Cxcl13 signalling was essential for activation of astrocytes and pain behaviours. Conversely, intrathecal injection of Cxcl13, and of Cxcl13 -stimulated astrocytes from wild type mice, but not Cxcl13 -stimulated astrocytes from Cxcr5 KO mice, induced mechanical allodynia.14 This nerve injury-induced increase in Cxcl13/Cxcr5 signalling was also demonstrated in the trigeminal ganglion (TG) following partial infraorbital nerve ligation. Intra-TG injection of Cxcl13 also increased TNF-α and IL-1β mRNA.34 Together these findings strongly implicate Cxcl13/Cxcr5 signalling, likely arising from microglia and/or neurons and activating astrocytes, in neuropathic pain and suggest the dorsal spinal cord specific increased expression in aged animals has implications for spinal pain signalling in the elderly.
Another inflammatory chemokine35 exhibiting substantially increased expression in the dorsal and ventral spinal cord (9-fold and 23-fold, respectively) was C-C motif chemokine ligand 3 (Ccl3). The source of Ccl3 is unclear, but IL-1β and TNF-α have been shown to induce Ccl3 production in astrocytes,36 while P2X7 receptor activation can induce the production of Ccl3 in microglia.37 These findings highlight neuroglia as the likely candidate for the enhanced expression of Ccl3 in aging. With respect to its role in pathological pain, expression of Ccl3 was increased following partial sciatic nerve ligation (PSNL), and neutralization of Ccl3 with an antibody ameliorated PSNL-induced neuropathic pain.38 Moreover, the direct peripheral or spinal administration of Ccl338,39 has been shown to induce pain behaviours. Thus, the dramatic increase to Ccl3 and Cxcl13 expression (above), along with both the role of both in neuropathic pain, establishes an altered baseline environment for neuropathic pain mechanisms to develop in the aged spinal cord. Further, regional differences in the degree of upregulation of Cxcl13 and Ccl3, being more pronounced in aged dorsal and ventral cord, carry implications for not just chronic pain but also motor dysfunction in the elderly.
A number of genes showing increased age-related expression relate to activation and switching in microglial and astrocyte phenotype, which would be expected to produce the altered cytokine signalling detected above. For example, Colony stimulating factor 1 (Csf1), also known as macrophage colony-stimulating factor, is a cytokine that increased in the aged dorsal spinal cord (1.3-fold). This molecule is capable of promoting the proliferation, differentiation, and survival of macrophage-derived cells, including microglia.40 The importance of this signalling pathway has been confirmed as genetic removal of Csf1r,41,42 or Csf1,43–45 produces a severe depletion in microglial numbers. Concerning its involvement in pathological pain states, the expression of the receptor for Csf1 (Csf1r) is upregulated following peripheral nerve injury, though Csf1 itself was not increased in this work.46 Regardless, this study showed a nerve injury-dependent induction of Csf1 expression in sensory neurons, which was transported centrally into the DH to activate microglia via the Csf1r and subsequently drive morphological and phenotypic change.46 Thus, elevated Csf1 in the aged dorsal spinal cord could be expected to enhance stimulation of these processes at baseline.
Also contributing to glial activation, expression of C-X3-C motif receptor 1 (Cx3cr1), also known as the fractalkine receptor, increased in both the dorsal and ventral cord (1.3-fold and 3.8-fold, respectively). Fractalkine is constitutively expressed in peripheral and central neurons, while Cx3cr1 is expressed on microglia and astrocytes.47 Accordingly, fractalkine is thought to mediate neuron-glial crosstalk in both the normal and pathological CNS.48 In a rodent prion disease model of chronic CNS inflammation both the fractalkine ligand and receptor upregulated.49 Likewise, soluble fractalkine in the neuropathic dorsal spinal cord is capable of activating microglia, and pharmacological or genetic interventions directed against fractalkine abolish hypersensitivity and the accompanying microglial activation.50–53 Moreover, direct intrathecal administration of fractalkine can induce mechanical allodynia and thermal hyperalgesia, whereas single injection intrathecal Cx3cr1 antagonist can suppress neuropathic pain induced by chronic constriction injury (CCI) and sciatic inflammatory neuropathy (SIN).54–56
The above findings support the existence of an age-related change in the activation state of microglia. However, an expanding view of microglial activation has now begun to consider these cells as a continuum of distinct, overlapping phenotypes.57–59 Importantly, a group of microglia referred to as disease-associated microglia (DAM) are now described among these phenotypes. These DAM have been associated with neurodegenerative diseases, such as Alzheimer’s and ALS, as well as in ‘aging induced’ brain damage.57,60 DAM can be identified by specific gene expression patterns with fully competent DAM requiring Triggering receptor expressed by myeloid cells (Trem2) expression. Expression of Trem2 in our data increased 3.8-fold in DH and 5-fold in the ventral cord of old mice, suggesting the presence of the DAM phenotype in both regions. Furthermore, the DAM gene signature also includes elevated expression of C-type lectin domain family 7 member A (Clec7a) and Toll Like Receptor 2 (Tlr2), which were both expressed at elevated levels in the dorsal and ventral spinal cord in aged samples. Together, these findings suggest that DAM feature in both the dorsal and ventral spinal cord of aged mice, raising the likelihood that these cells modify the vulnerability of both regions to insult and injury with advanced age. Reinforcing this point, Trem2 has been implicated in neuropathic pain, where it increases in the spinal cord following nerve injury.57,60 Nerve injury-induced increase in Trem2 expression paralleled the increase in microglial numbers in the ipsilateral DH, and intrathecal administration of a Trem2 agonist antibody in the absence of injury, reduced paw-withdrawal thresholds. In addition, evidence supports a role of TLR-mediated immune responses with neuropathic pain, as nerve transection upregulates TLR2 expression in the spinal cord.61 Furthermore, nerve injury induced pain behaviours, microglial activation, and the induction of proinflammatory cytokines are all diminished in TLR2-KO mice.62 Finally, the ability of Clec7a to induce the production of proinflammatory cytokines,63 and its responsiveness to damage-associated molecular patterns (DAMPs), may further contribute to a heightened responsiveness of the aged spinal cord and nerve-injury responses.
Consistent with the above findings a number of other genes were also increased in the spinal cord including: Glial fibrillary acidic protein (Gfap), an astrocyte-specific cytoskeletal protein increased by activation and a range pathological states64; cluster of differentiation 86 (Cd86), a transmembrane glycoprotein expressed by activated microglia and a recently identified populations of resident brain dendritic cells (bDC)65; and the P2X purinoceptor (P2rx4), an ionotropic receptor expressed in microglia and neurons and increased in spinal microglia following nerve-injury.66 Collectively these findings add to the above interpretation that the spinal cord, just like the brain, exhibits age-related priming of glial populations and neuroinflammatory pathways.66,67 Accordingly, these alterations may be due to adaptations in response to, or involvement in chronic inflammatory processes. In either case they will dramatically alter the responses of the aged dorsal and ventral spinal cord to peripheral and central insults.
As noted above, interpretation of our data also comes with a number of caveats. As is the case with any gene expression study, it cannot be assumed that these differences are expressed as changes, at a protein level, required to influence function.68 Thus, future work in young, middle and old age spinal cords prepared to preserve protein for analysis will be required to verify the age-related dorsal and ventral spinal cord phenotypes we describe. When such cross-checks have been performed in the brain, assessing age-related gene expression that overlaps with our study, there has been good correspondence between mRNA and protein levels.57,58 In addition, although we have used the literature describing the expression of genes in different cells to attribute gene expression changes to cell types such as microglia and astrocytes, the cellular origin of these signals was not directly assessed in our experiments. Immunolabelling of the related proteins in spinal cord sections, RNAscope analysis, qPCR in identified cells, or single cell RNASeq will be required to confirm these associations. Future work using the single cell RNASeq approach would also dramatically increase the coverage of gene expression from the relatively small and targeted panel of genes we assessed. This may unmask expression changes with the capacity to compensate for the largely proinflammatory network of disrupted genes we detected. In support of this premise, transforming growth factor-beta 1 (Tgf-β1) expression was increased ∼ 2-fold in the aged ventral spinal cord. Given Tgf-β1is a potent anti-inflammatory cytokine,69 it is tempting to suggest this indicates motor circuits may be somewhat protected from proinflammatory signalling. Finally, the expression of some of the genes we assessed did not show any age-related change in our analysis, including neuroinflammatory and glial related proteins, and the majority of synaptic proteins. This was surprising for the dorsal spinal cord, as our previous work has identified alterations in synaptic function in these circuits with age17 that predict altered gene expression. This discrepancy could be explained by incomplete translation of gene expression through to protein as suggested above. Alternatively, age-dependent posttranslational modification of synaptic proteins, altered trafficking or distribution of these proteins at synaptic versus extrasynaptic sites may help to explain these differences.
Conclusions and future directions
The present findings clearly indicate aging causes changes in the expression of a range of transcripts involved in inflammatory responses, and glial function with some regional specificity in the spinal cord (dorsal versus ventral spinal cord). The expression changes in the dorsal spinal cord relating to glial activation and proinflammatory cytokine signalling bear striking similarities with those seen in neuropathic pain. Of particular interest, the most upregulated chemokine following spinal nerve ligation, Cxcl13,14 underwent a more-pronounced increase in gene expression with age alone. It remains to be determined whether the neuroinflammatory remodelling underlies a heightened susceptibility or response to neuropathology. Moreover, given the identified roles in the pathophysiology of neuropathic pain, it will be important to establish the magnitude of neuroinflammatory responses to nerve injury in aged animals. For example, the altered neuroinflammatory status of the aged spinal cord may limit its capacity to increase inflammatory and glial gene expression, or further potentiate expression when a neuropathic perturbation is superimposed on an elevated baseline. Furthermore, the altered neuroinflammatory baseline may precipitate the recruitment of alternative or additional pathways that could exacerbate or prolong pathology. Future work in aged animals will be required to resolve these issues, but the ability of age to drastically alter key neuroinflammatory cytokines and glial activation, replicating aspects of the pathogenesis of neuropathic pain is undoubtedly relevant. Together, these new insights provide a potential mechanism to alter sensory signalling in dorsal horn circuits and appear of relevance to the heightened incidence of chronic pain states in the elderly.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Hunter Medical Research Institute (Glenn Moss RHD Scholarship) and National Health and Medical Research Council (1043933, 1144638, 1184974, and 631000).
ORCID iD: Brett A Graham https://orcid.org/0000-0002-8070-0503
References
- 1.Blyth FM, March LM, Brnabic AJ, Jorm LR, Williamson M, Cousins MJ. Chronic pain in Australia: a prevalence study. Pain 2001; 89: 127–134. [DOI] [PubMed] [Google Scholar]
- 2.Gibson S. Pain and aging: the pain experience over the adult life span. Proc 10th World Congr Pain 2003; 24: 767–790. [Google Scholar]
- 3.Thomas E, Peat G, Harris L, Wilkie R, Croft PR. The prevalence of pain and pain interference in a general population of older adults: cross-sectional findings from the North Staffordshire Osteoarthritis Project (NorStOP). Pain 2004; 110: 361–368. [DOI] [PubMed] [Google Scholar]
- 4.Coyle DE. Partial peripheral nerve injury leads to activation of astroglia and microglia which parallels the development of allodynic behavior. Glia 1998; 23: 75–83. [PubMed] [Google Scholar]
- 5.Scholz J, Woolf CJ. The neuropathic pain triad: neurons, immune cells and glia. Nat Neurosci 2007; 10: 1361–1368. 2007/10/30. [DOI] [PubMed] [Google Scholar]
- 6.Watkins LR, Milligan ED, Maier SF. Glial activation: a driving force for pathological pain. Trends Neurosci 2001; 24: 450–455. [DOI] [PubMed] [Google Scholar]
- 7.Gao YJ, Ji RR. Chemokines, neuronal-glial interactions, and central processing of neuropathic pain. Pharmacol Ther 2010; 126: 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao YJ, Zhang L, Samad OA, Suter MR, Yasuhiko K, Xu ZZ, Park JY, Lind AL, Ma Q, Ji RR. JNK-induced MCP-1 production in spinal cord astrocytes contributes to central sensitization and neuropathic pain. J Neurosci 2009; 29: 4096–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simone DA, Sorkin LS, Oh U, Chung JM, Owens C, LaMotte RH, Willis WD. Neurogenic hyperalgesia: central neural correlates in responses of spinothalamic tract neurons. J Neurophysiol 1991; 66: 228–246. [DOI] [PubMed] [Google Scholar]
- 10.Raghavendra V, Tanga F, DeLeo JA. Inhibition of microglial activation attenuates the development but not existing hypersensitivity in a rat model of neuropathy. J Pharmacol Exp Ther 2003; 306: 624–630. [DOI] [PubMed] [Google Scholar]
- 11.Colburn RW, DeLeo JA, Rickman AJ, Yeager MP, Kwon P, Hickey WF. Dissociation of microglial activation and neuropathic pain behaviors following peripheral nerve injury in the rat. J Neuroimmunol 1997; 79: 163–175. [DOI] [PubMed] [Google Scholar]
- 12.Sweitzer SM, White KA, Dutta C, DeLeo JA. The differential role of spinal MHC class II and cellular adhesion molecules in peripheral inflammatory versus neuropathic pain in rodents. J Neuroimmunol 2002; 125: 82–93. [DOI] [PubMed] [Google Scholar]
- 13.Tanga FY, Raghavendra V, DeLeo JA. Quantitative real-time RT-PCR assessment of spinal microglial and astrocytic activation markers in a rat model of neuropathic pain. Neurochem Int 2004; 45: 397–407. [DOI] [PubMed] [Google Scholar]
- 14.Jiang BC, Cao DL, Zhang X, Zhang ZJ, He LN, Li CH, Zhang WW, Wu XB, Berta T, Ji RR, Gao YJ. CXCL13 drives spinal astrocyte activation and neuropathic pain via CXCR5. J Clin Invest 2016; 126: 745–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwata K, Fukuoka T, Kondo E, Tsuboi Y, Tashiro A, Noguchi K, Masuda Y, Morimoto T, Kanda K. Plastic changes in nociceptive transmission of the rat spinal cord with advancing age. J Neurophysiol 2002; 87: 1086–1093. [DOI] [PubMed] [Google Scholar]
- 16.Kitagawa J, Kanda K, Sugiura M, Tsuboi Y, Ogawa A, Shimizu K, Koyama N, Kamo H, Watanabe T, Ren K, Iwata K. Effect of chronic inflammation on dorsal horn nociceptive neurons in aged rats. J Neurophysiol 2005; 93: 3594–3604. [DOI] [PubMed] [Google Scholar]
- 17.Mayhew JA, Callister RJ, Walker FR, Smith DW, Graham BA. Aging alters signaling properties in the mouse spinal dorsal horn. Mol Pain 2019; 15: 1744806919839860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci 2014; 69: S4–S9. [DOI] [PubMed] [Google Scholar]
- 19.Abdel-Aziz K, Solanky BS, Yiannakas MC, Altmann DR, Wheeler-Kingshott CA, Thompson AJ, Ciccarelli O. Age related changes in metabolite concentrations in the normal spinal cord. PLoS One 2014; 9: e105774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furlan JC, Bracken MB, Fehlings MG. Is age a key determinant of mortality and neurological outcome after acute traumatic spinal cord injury? Neurobiol Aging 2010; 31: 434–446. [DOI] [PubMed] [Google Scholar]
- 21.Rodakowski J, Skidmore ER, Anderson SJ, Begley A, Jensen MP, Buhule OD, Boninger ML. Additive effect of age on disability for individuals with spinal cord injuries. Arch Phys Med Rehabil 2014; 95: 1076–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scivoletto G, Morganti B, Ditunno P, Ditunno JF, Molinari M. Effects on age on spinal cord lesion patients’ rehabilitation. Spinal Cord 2003; 41: 457–464. [DOI] [PubMed] [Google Scholar]
- 23.Wilson JR, Davis AM, Kulkarni AV, Kiss A, Frankowski RF, Grossman RG, Fehlings MG. Defining age-related differences in outcome after traumatic spinal cord injury: analysis of a combined, multicenter dataset. Spine J 2014; 14: 1192–1198. [DOI] [PubMed] [Google Scholar]
- 24.Gwak YS, Hains BC, Johnson KM, Hulsebosch CE. Effect of age at time of spinal cord injury on behavioral outcomes in rat. J Neurotrauma 2004; 21: 983–993. [DOI] [PubMed] [Google Scholar]
- 25.Siegenthaler MM, Ammon DL, Keirstead HS. Myelin pathogenesis and functional deficits following SCI are age-associated. Exp Neurol 2008; 213: 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegenthaler MM, Berchtold NC, Cotman CW, Keirstead HS. Voluntary running attenuates age-related deficits following SCI. Exp Neurol 2008; 210: 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinf 2009; 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linssen AM, van Boxtel MP, Joore MA, Anteunis LJ. Predictors of hearing acuity: cross-sectional and longitudinal analysis. J Gerontol A Biol Sci Med Sci 2014; 69: 759–765. [DOI] [PubMed] [Google Scholar]
- 29.Olivieri F, Capri M, Bonafè M, Morsiani C, Jung HJ, Spazzafumo L, Viña J, Suh Y. Circulating miRNAs and miRNA shuttles as biomarkers: perspective trajectories of healthy and unhealthy aging. Mech Ageing Dev 2017; 165: 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mills SEE, Nicolson KP, Smith BH. Chronic pain: a review of its epidemiology and associated factors in population-based studies. Br J Anaesth 2019; 123: e273–e283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mogil JS. Qualitative sex differences in pain processing: emerging evidence of a biased literature. Nat Rev Neurosci 2020; 21: 353–365. [DOI] [PubMed] [Google Scholar]
- 32.Irani DN. Regulated production of CXCL13 within the central nervous system. J Clin Cell Immunol 2016; 7(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu XB, Cao DL, Zhang X, Jiang BC, Zhao LX, Qian B, Gao YJ. CXCL13/CXCR5 enhances sodium channel Nav1.8 current density via p38 MAP kinase in primary sensory neurons following inflammatory pain. Sci Rep 2016; 6: 34836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Q, Cao DL, Zhang ZJ, Jiang BC, Gao YJ. Chemokine CXCL13 mediates orofacial neuropathic pain via CXCR5/ERK pathway in the trigeminal ganglion of mice. J Neuroinflammation 2016; 13: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev 2002; 13: 455–481. [DOI] [PubMed] [Google Scholar]
- 36.Choi SS, Lee HJ, Lim I, Satoh J, Kim SU. Human astrocytes: secretome profiles of cytokines and chemokines. PLoS One 2014; 9: e92325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kataoka A, Tozaki-Saitoh H, Koga Y, Tsuda M, Inoue K. Activation of P2X7 receptors induces CCL3 production in microglial cells through transcription factor NFAT. J Neurochem 2009; 108: 115–125. [DOI] [PubMed] [Google Scholar]
- 38.Kiguchi N, Kobayashi Y, Maeda T, Saika F, Kishioka S. CC-chemokine MIP-1alpha in the spinal cord contributes to nerve injury-induced neuropathic pain. Neurosci Lett 2010; 484: 17–21. [DOI] [PubMed] [Google Scholar]
- 39.Oh SB, Tran PB, Gillard SE, Hurley RW, Hammond DL, Miller RJ. Chemokines and glycoprotein120 produce pain hypersensitivity by directly exciting primary nociceptive neurons. J Neurosci 2001; 21: 5027–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hume DA, MacDonald KP. Therapeutic applications of macrophage colony-stimulating factor-1 (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood 2012; 119: 1810–1820. [DOI] [PubMed] [Google Scholar]
- 41.Erblich B, Zhu L, Etgen AM, Dobrenis K, Pollard JW. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One 2011; 6: e26317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010; 330: 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kondo Y, Duncan ID. Selective reduction in microglia density and function in the white matter of colony-stimulating factor-1-deficient mice. J Neurosci Res 2009; 87: 2686–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasaki A, Yokoo H, Naito M, Kaizu C, Shultz LD, Nakazato Y. Effects of macrophage-colony-stimulating factor deficiency on the maturation of microglia and brain macrophages and on their expression of scavenger receptor. Neuropathology 2000; 20: 134–142. [DOI] [PubMed] [Google Scholar]
- 45.Wegiel J, Wisniewski HM, Dziewiatkowski J, Tarnawski M, Kozielski R, Trenkner E, Wiktor-Jedrzejczak W. Reduced number and altered morphology of microglial cells in colony stimulating factor-1-deficient osteopetrotic op/op mice. Brain Res 1998; 804: 135–139. [DOI] [PubMed] [Google Scholar]
- 46.Guan Z, Kuhn JA, Wang X, Colquitt B, Solorzano C, Vaman S, Guan AK, Evans-Reinsch Z, Braz J, Devor M, Abboud-Werner SL, Lanier LL, Lomvardas S, Basbaum AI. Injured sensory neuron-derived CSF1 induces microglial proliferation and DAP12-dependent pain. Nat Neurosci 2016; 19: 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorf ME, Berman MA, Tanabe S, Heesen M, Luo Y. Astrocytes express functional chemokine receptors. J Neuroimmunol 2000; 111: 109–121. [DOI] [PubMed] [Google Scholar]
- 48.Harrison JK, Jiang Y, Chen S, Xia Y, Maciejewski D, McNamara RK, Streit WJ, Salafranca MN, Adhikari S, Thompson DA, Botti P, Bacon KB, Feng L. Role for neuronally derived fractalkine in mediating interactions between neurons and CX3CR1-expressing microglia. Proc Natl Acad Sci U S A 1998; 95: 10896–10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hughes PM, Botham MS, Frentzel S, Mir A, Perry VH. Expression of fractalkine (CX3CL1) and its receptor, CX3CR1, during acute and chronic inflammation in the rodent CNS. Glia 2002; 37: 314–327. [PubMed] [Google Scholar]
- 50.Clark AK, Yip PK, Grist J, Gentry C, Staniland AA, Marchand F, Dehvari M, Wotherspoon G, Winter J, Ullah J, Bevan S, Malcangio M. Inhibition of spinal microglial cathepsin S for the reversal of neuropathic pain. Proc Natl Acad Sci U S A 2007; 104: 10655–10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staniland AA, Clark AK, Wodarski R, Sasso O, Maione F, D'Acquisto F, Malcangio M. Reduced inflammatory and neuropathic pain and decreased spinal microglial response in fractalkine receptor (CX3CR1) knockout mice. J Neurochem 2010; 114: 1143–1157. [DOI] [PubMed] [Google Scholar]
- 52.Verge GM, Milligan ED, Maier SF, Watkins LR, Naeve GS, Foster AC. Fractalkine (CX3CL1) and fractalkine receptor (CX3CR1) distribution in spinal cord and dorsal root ganglia under basal and neuropathic pain conditions. Eur J Neurosci 2004; 20: 1150–1160. [DOI] [PubMed] [Google Scholar]
- 53.White FA, Jung H, Miller RJ. Chemokines and the pathophysiology of neuropathic pain. Proc Natl Acad Sci U S A 2007; 104: 20151–20158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milligan E, Zapata V, Schoeniger D, Chacur M, Green P, Poole S, Martin D, Maier SF, Watkins LR. An initial investigation of spinal mechanisms underlying pain enhancement induced by fractalkine, a neuronally released chemokine. Eur J Neurosci 2005; 22: 2775–2782. [DOI] [PubMed] [Google Scholar]
- 55.Milligan ED, Sloane EM, Watkins LR. Glia in pathological pain: a role for fractalkine. J Neuroimmunol 2008; 198: 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milligan ED, Zapata V, Chacur M, Schoeniger D, Biedenkapp J, O'Connor KA, Verge GM, Chapman G, Green P, Foster AC, Naeve GS, Maier SF, Watkins LR. Evidence that exogenous and endogenous fractalkine can induce spinal nociceptive facilitation in rats. Eur J Neurosci 2004; 20: 2294–2302. [DOI] [PubMed] [Google Scholar]
- 57.Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, Itzkovitz S, Colonna M, Schwartz M, Amit I. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 2017; 169: 1276.e1217–1290.e1217. [DOI] [PubMed] [Google Scholar]
- 58.Mathys H, Adaikkan C, Gao F, Young JZ, Manet E, Hemberg M, De Jager PL, Ransohoff RM, Regev A, Tsai LH. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep 2017; 21: 366–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rangaraju S, Dammer EB, Raza SA, Rathakrishnan P, Xiao H, Gao T, Duong DM, Pennington MW, Lah JJ, Seyfried NT, Levey AI. Identification and therapeutic modulation of a pro-inflammatory subset of disease-associated-microglia in Alzheimer’s disease. Mol Neurodegener 2018; 13: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yerbury JJ, Ooi L, Dillin A, Saunders DN, Hatters DM, Beart PM, Cashman NR, Wilson MR, Ecroyd H. Walking the tightrope: proteostasis and neurodegenerative disease. J Neurochem 2016; 137: 489–505. [DOI] [PubMed] [Google Scholar]
- 61.Lim H, Kim D, Lee SJ. Toll-like receptor 2 mediates peripheral nerve injury-induced NADPH oxidase 2 expression in spinal cord microglia. J Biol Chem 2013; 288: 7572–7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim D, Kim MA, Cho IH, Kim MS, Lee S, Jo EK, Choi SY, Park K, Kim JS, Akira S, Na HS, Oh SB, Lee SJ. A critical role of toll-like receptor 2 in nerve injury-induced spinal cord glial cell activation and pain hypersensitivity. J Biol Chem 2007; 282: 14975–14983. [DOI] [PubMed] [Google Scholar]
- 63.Kerrigan AM, Brown GD. Syk-coupled C-type lectin receptors that mediate cellular activation via single tyrosine based activation motifs. Immunol Rev 2010; 234: 335–352. [DOI] [PubMed] [Google Scholar]
- 64.Seifert G, Schilling K, Steinhauser C. Astrocyte dysfunction in neurological disorders: a molecular perspective. Nat Rev Neurosci 2006; 7: 194–206. [DOI] [PubMed] [Google Scholar]
- 65.Kaunzner UW, Miller MM, Gottfried-Blackmore A, Gal-Toth J, Felger JC, McEwen BS, Bulloch K. Accumulation of resident and peripheral dendritic cells in the aging CNS. Neurobiol Aging 2012; 33: 681.e681–693.e681. [DOI] [PubMed] [Google Scholar]
- 66.Tsuda M, Shigemoto-Mogami Y, Koizumi S, Mizokoshi A, Kohsaka S, Salter MW, Inoue K. P2X4 receptors induced in spinal microglia gate tactile allodynia after nerve injury. Nature 2003; 424: 778–783. [DOI] [PubMed] [Google Scholar]
- 67.Perry VH, Holmes C. Microglial priming in neurodegenerative disease. Nat Rev Neurol 2014; 10: 217–224. [DOI] [PubMed] [Google Scholar]
- 68.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012; 13: 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spittau B, Wullkopf L, Zhou X, Rilka J, Pfeifer D, Krieglstein K. Endogenous transforming growth factor-beta promotes quiescence of primary microglia in vitro. Glia 2013; 61: 287–300. [DOI] [PubMed] [Google Scholar]