Abstract
During early neural development, changes in signalling inform the expression of transcription factors that in turn instruct changes in cell identity. At the same time, switches in adhesion molecule expression result in cellular rearrangements that define the morphology of the emerging neural tube. It is becoming increasingly clear that these two processes influence each other; adhesion molecules do not simply operate downstream of or in parallel with changes in cell identity but rather actively feed into cell fate decisions. Why are differentiation and adhesion so tightly linked? It is now over 60 years since Conrad Waddington noted the remarkable "Constancy of the Wild Type” (Waddington in Nature 183: 1654–1655, 1959) yet we still do not fully understand the mechanisms that make development so reproducible. Conversely, we do not understand why directed differentiation of cells in a dish is sometimes unpredictable and difficult to control. It has long been suggested that cells make decisions as 'local cooperatives' rather than as individuals (Gurdon in Nature 336: 772–774, 1988; Lander in Cell 144: 955–969, 2011). Given that the cadherin family of adhesion molecules can simultaneously influence morphogenesis and signalling, it is tempting to speculate that they may help coordinate cell fate decisions between neighbouring cells in the embryo to ensure fidelity of patterning, and that the uncoupling of these processes in a culture dish might underlie some of the problems with controlling cell fate decisions ex-vivo. Here we review the expression and function of cadherins during early neural development and discuss how and why they might modulate signalling and differentiation as neural tissues are formed.
Keywords: Adhesion, Pluripotency, Neuroectoderm, Differentiation, Signalling
A brief introduction to the cadherin superfamily
In the late 1970s, Masatoshi Takeichi proposed that cell–cell adhesion in Chinese hamster cells was mediated by two processes: one calcium-independent and the other calcium-dependent. The activity of the calcium-dependent process correlated with the presence of a 150 kDa molecule [4]. Several groups independently found that inhibition of the activity of this molecule with antisera and antibodies resulted in the inhibition of calcium-dependent adhesion, with evident changes in the morphology of antiserum/antibody-treated cells and embryos, and in the ability of these cells to reaggregate following disaggregation [5–13]. The identified glycoprotein was thus named “cadherin” (now E-cadherin, Cdh1) after the process of calcium-dependent adhesion it was found to mediate [12]. Similar experimental strategies were used to identify close family members N-cadherin (Cdh2) [14–16] and P-cadherin (Cdh3) [17].
The cadherin superfamily of proteins comprises more than 100 members in humans [18], with roles in several physiological processes including signalling, mechanotransduction, self-recognition and tumour suppression [19–22]. Proteins of this superfamily share an extracellular cadherin (CA) domain of approximately 110 amino acids in size (Interpro IPR039808). The CA domain is formed by seven β-sheets which arrange to form an Ig domain-like β-sandwich (Fig. 1a). It is found in tandem repeats and mediates both calcium binding and interaction with other cadherin molecules (Fig. 1b, c) [23].
Fig. 1.
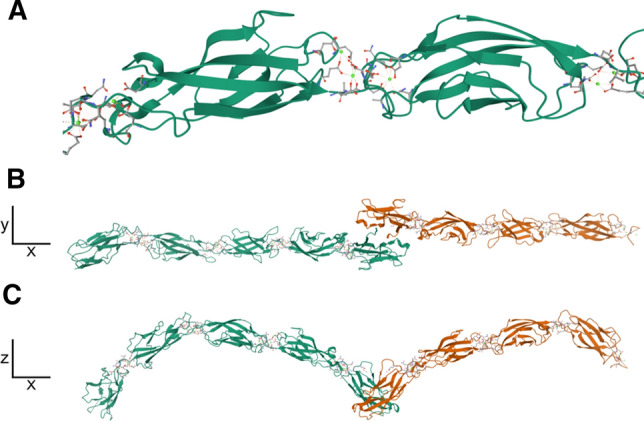
Structure of the cadherin domains of mouse Cdh1. a Two cadherin domains from mouse E-cadherin (EC2, EC3), forming seven-stranded β-sandwiches. Binding of three calcium ions (displayed in emerald green) is mediated at the aspartate and glutamate-rich interface of the two domains. Further calcium ions are visible at the interface of these domains with neighbouring cadherin domains. b–c Trans-interaction between two E-cadherin ectodomains (EC1-5 for green chain, EC1-4 for orange chain). A conserved residue (Trp2) in EC1 interacts with a hydrophobic pocket on the opposite EC1 domain. Crystal structure from [23] (PDB: 3Q2V) visualised with Mol*
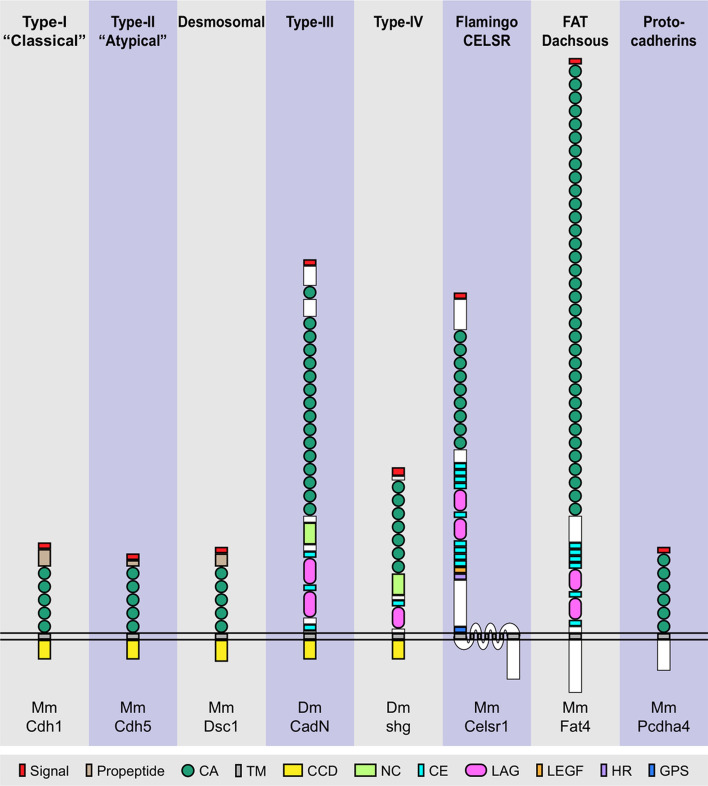
The number of CA repeats and the composition of the remainder of the molecule vary greatly between different superfamily members and across species, and can be used to classify cadherin molecules into subfamilies. The following is a list of the main cadherin subfamilies and their properties, as comprehensively reviewed in [18, 20, 24–28] (Fig. 2).
Fig. 2.
Cadherin subfamilies and representative members. Representative members of the eight cadherin subfamilies described in the text. The plasma membrane is illustrated as two parallel horizontal lines. Mm Mus musculus, Dm Drosophila melanogaster, CA cadherin domain, TM transmembrane domain, CCD cadherin cytoplasmic domain, NC non-chordate domain, CE cysteine-rich EGF-like domain, LAG laminin globular domain, LEGF laminin-type EGF-like domain, HR hormone receptor motif, GPS GPCR proteolytic site, white: other regions
Type-I or “classical” cadherins are located at adherens junctions, and are characterised by five extracellular CA (abbreviated to EC) domains, a transmembrane region, and an intracellular classical cadherin cytoplasmic domain (CCD, Interpro IPR000233), which binds armadillo family proteins β-catenin (Ctnnb1) and p120ctn (Ctnnd1). The interaction with β-catenin links cadherins to α-catenin and the actin cytoskeleton, whereas p120ctn is involved in cadherin turnover. Trans-interactions with other cadherins are mediated by conserved residues: a Trp in position 2 interacts with a hydrophobic pocket on the opposite N-terminal EC domain, which contains the conserved His-Ala-Val (HAV) motif [18, 26, 28, 29].
Type-II or “atypical” cadherins are structurally similar to type-I cadherins, but lack the HAV motif and have two Trp residues mediating trans-interactions (Trp2, Trp4) [24–26].
Desmosomal cadherins (desmogleins and desmocollins) are located at desmosomes. They have five EC domains, a transmembrane domain, and an intracellular CCD that interacts with armadillo family members plakoglobin and plakophilin, which link the cadherins to intermediate filaments via the protein desmoplakin [25, 26].
Type-III cadherins are not found in mammals other than the platypus. They have a variable number of EC domains followed by a conserved primitive classic cadherin domain (PCCD), composed of a “non-chordate” domain (NC), a cysteine-rich EGF-like (CE) and a LAG (laminin globular/LamG) domain. They have a transmembrane region and a CCD. Their EC1 domains lack conserved Trp residues but share a conserved Tyr in position 5 that may mediate trans-interactions [18, 24, 25].
Type-IV cadherins are found in arthropods and comprise seven EC domains followed by LAG and CE domains, a transmembrane region and a CCD [18, 25].
The Flamingo/CELSR (cadherin EGF LAG seven-pass G-type receptor) subfamily members have an extracellular region composed of nine EC domains followed by several LAG and CE domains, a laminin-type EGF-like domain, a hormone receptor domain and a GPS (GPCR proteolytic site) motif. This structure is reminiscent of the extracellular region of type-III cadherins, with which they also share the Tyr5 residue in EC1. They are highly unusual cadherins in that they have a 7-pass transmembrane (7TM) domain, which is why these cadherins are also referred to as the 7TM subfamily [18, 24, 25, 28].
The FAT, FAT-like and Dachsous group comprises proteins with large extracellular domains of up to 34 EC repeats, followed by LAG and CE domains for FAT and FAT-like cadherins, a transmembrane domain and a conserved C-terminal domain [18, 25].
Protocadherins are a large cadherin subfamily. They are subdivided into non-clustered protocadherins (NC-Pcdh) and clustered protocadherins (C-Pcdh). In human and mouse, C-Pcdh genes are transcribed from three adjacent gene clusters (Pcdh α, β and γ), with alternative splicing generating over 50 proteins with different N-termini and constant α, β and γ C-termini. The extracellular domains of both NC- and C-Pcdh contain 6/7 EC repeats with sequence similarity to the FAT/Dachsous group. They lack a conserved Trp2 residue, and are reported to interact in trans by means of anti-parallel interfacing of several EC domains. They share a transmembrane domain and conserved intracellular motifs. They are expressed in the mammalian brain, where they act as a cell-surface neuronal barcode to avoid self-synapsing [18, 20, 24, 27].
An evolutionary perspective
Cadherins have classically been described as molecules that regulate calcium-dependent cell adhesion in metazoans [30–33]. However, cadherins containing EC domains, EGF-like domains and a single pass transmembrane domain have been identified in the genome of the unicellular filasterean Capsaspora owczarzaki and of several choanoflagellates, implying cadherins are a shared feature of Filozoa [34, 35]. A classical-like cadherin similar to Type-IV superfamily members, containing both EC repeats and a CCD was identified in the sponge Oscarella carmela, suggesting that interaction with armadillo family proteins evolved in an ancestral metazoan [35, 36].
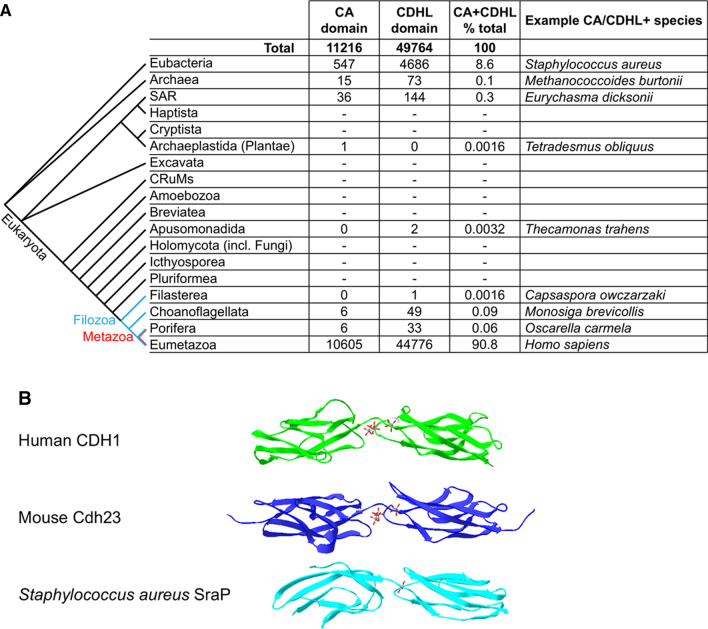
Pinpointing the evolutionary origin of cadherins, however, is not straightforward: proteins containing cadherin and cadherin-like domains have also been identified in bacteria and archaea [37–41]. The InterPro protein family database has over 60,000 combined entries for “Cadherin” (IPR039808, CA) and “Cadherin-like” (IPR002126, CDHL) domains. 90.8% of entries are specific to Eumetazoa, 8.6% to Eubacteria, 0.1% to Archaea, with the remainder spread amongst other eukaryotic clades (Fig. 3a).
Fig. 3.
Phylogeny and structure of cadherin and cadherin-like domains. a Number of InterPro database entries for “Cadherin” and “Cadherin-like” protein domains, sorted taxonomically. Phylogenetic relationships of the described clades are based on [43–45]. InterPro data retrieved in May 2020. b Structural comparison of two CA domains of human CDH1 with two CDHL domains of mouse Cdh23 and Staphylococcus aureus SraP. Calcium ions are displayed as wireframe structures. Crystal structures from [46] (PDB: 3MVS), [47] (PDB: 2O72), [42] (PDB: 4M03), visualised with Swiss-PdbViewer [48] and aligned with the CA/CDHL domains on the right of the image in similar orientation
The crystal structure of a Staphylococcus aureus protein containing CDHL domains [42] reveals that CA/CDHL domains have remained broadly structurally conserved throughout evolution, with calcium atoms bound at the interface of two Ig domain-like β-sandwiches (Fig. 3b). Variations in amino acid composition and in rotation angle between adjacent CA/CDHL domains may account for differences in the number of calcium ions bound at the interface of these domains (three for mammalian cadherins, one for S. aureus SraP).
Despite their presence in bacteria, archaea and Filozoa, CA/CDHL domain-containing proteins are not ubiquitous throughout eukaryotes. Aside from Filozoa, they are found almost exclusively in SAR, a large group of diverse species which diverged from Filozoa early in eukaryotic evolution (Fig. 3a). Two CDHL InterPro entries are recorded for the apusomonadid Thecamonas trahens [36], and one CA entry for plants (the green alga Tetradesmus obliquus). The T. obliquus entry is unreviewed, so it is unclear whether this protein truly contains CA domains.
This seemingly desultory phylogenetic distribution of CA/CDHL domain-containing proteins may be explained by horizontal gene transfer events. Ancient Filozoa were likely to be bacterivorous [49]. Similarly, T. trahens feeds on bacteria and on other flagellates [50]. These organisms could have therefore incorporated prokaryotic CA/CDHL-encoding DNA into their genomes. King and colleagues [35] suggest this is how bacterial cohesin domains may have been acquired in the choanoflagellate and Porifera coherin subfamily of cadherins.
The reverse process is also conceivable, with commensal, parasitic or pathogenic prokaryotic and SAR species potentially acquiring CA/CDHL domains from Filozoa. Proteins containing these domains could in turn support interaction with host cell surfaces, as suggested by Gachon and colleagues for the nonagonal family of SAR cadherins [51]. There are several examples of pathogenic and parasitic prokaryotic and SAR species with CA/CDHL domains (e.g., Pythium insidiosum, S. aureus, Vibrio cholerae) [39, 42, 51], but many prokaryotes and SAR species which are non-symbiotic and non-pathogenic to Filozoa also contain CA/CDHL domains (e.g., Aureococcus anophagefferens, Methanococcoides burtonii, Rhodopirellula baltica) [41, 51, 52].
Despite the uncertainty underlying the evolutionary origin of the cadherin domain, its ability to bind calcium appears to remain a defining feature throughout the tree of life [37, 40, 42, 53]. Aside from regulating adhesion in Metazoa and in some prokaryotes [33, 40], it has been suggested that non-metazoan proteins containing CA/CDHL domains may regulate cell–cell interactions in feeding processes, pathogen-host interaction and cell aggregation events. When considering the role of cadherins during neural development, we should bear in mind that they may have acquired multiple functions over evolutionary time and may be acting as more than just adhesion molecules.
Early neural development
During early embryonic development, the fertilized egg undergoes several rounds of cell division, lineage segregation, symmetry breaking, and axis specification, generating the pluripotent epiblast and culminating in gastrulation and the formation of three germ layers that will go on to generate all embryonic tissues. These processes have been extensively reviewed elsewhere [54, 55], and here we will focus on subsequent neural development.
Neurulation
From gastrulation onwards, cells of the anterior epiblast that do not ingress through the primitive streak go on to form ectodermal tissues, giving rise to neurectoderm, which generates the nervous system, and surface ectoderm [56, 57]. Neurulation refers to the process of generating the neural tube, a tubular epithelial structure, out of the flat epithelial sheet of the neural ectoderm (Fig. 4). While anatomical differences exist between various species, modes of neurulation are generally conserved across amniotes, Xenopus, zebrafish and Amphioxus (reviewed in [58]).
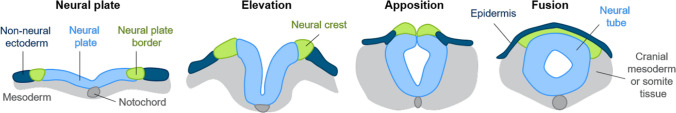
Fig. 4.
Generalised morphological events of neurulation leading to neural tube closure. Transverse view. Neurulation begins in the flat neural plate, which generates lineages of the central nervous system. It is flanked on either side by the neural plate border and non-neural ectoderm. As neurulation progresses, the neural plate thickens, stratifies and begins to fold, while a ventral hinge point is formed at the notochord, a mesodermal tissue. The neural plate borders elevate, becoming the neural crest. As the neural tube fuses dorsally, neural crest cells migrate out of it, going on to form lineages of the peripheral nervous system. Closure of the nascent neural tube disconnects it from the overlying epidermis
Neurulation begins with the formation of the neural plate, which initially consists of a layer of neuroepithelial cells. As development progresses, these cells multiply, causing the neuroepithelium to thicken and stratify [59, 60]. The neural plate is flanked by a population of cells called the neural plate border, which distinguishes the neural plate from the rest of the ectoderm. The neural plate border will go on to form the neural crest, giving rise to the peripheral nervous system as well as several non-neural cell types including melanocytes, smooth muscle cells and, in the anterior, bone and cartilage [61]. Ectodermal tissues lying outside the neural plate and neural plate border will give rise to epidermis.
As the neural plate increases in size, it begins to fold, initiating the formation of the neural tube. Ventral to the neural plate lies the notochord, a mesoderm-derived embryonic tissue that serves as the precursor to the nucleus pulposus within the vertebral column. The neural plate forms a single ventral hingepoint directly above the notochord. Meanwhile, two dorsal hinge points are formed at the neural crest, forming a central ‘valley’ called the neural groove. The neural crest hinge points then converge towards each other, bringing the contralateral dorsal ends of the neural groove together, allowing them to fuse to form a closed tube. The closure of the neural tube detaches the neural ectoderm from the epidermis, which now dorsally overlies the neural crest and the nascent neural tube. Depending on the organism and the location along the anterior–posterior axis, the neural crest cells undergo an epithelial to mesenchymal transition (EMT) and migrate dorsally from either the converging neural folds or from the roof of the nascent neural tube [59, 60].
The potencies of the epiblast regions that give rise to neurectoderm have been investigated using explant culture studies. In gastrulating mouse embryos at E6.5, the anterior epiblast can give rise to all three germ layers [62, 63]. Within half a day however, cells in this region become restricted to ectoderm but retain potency to become either epidermal or neural lineages, determined by either the presence or absence of bone morphogenetic protein 4 (BMP4), respectively [64, 65]. At E7.5, the positional identity within the ectoderm defines cellular potency: the proximal region of the ectoderm is restricted to a surface ectodermal fate, while more distal regions of the anterior neurectoderm will give rise to the central nervous system [65–67].
Neuroepithelial precursor cells can be derived by directed differentiation from mouse embryonic stem cells (ESCs) [68], human ESCs [69, 70], and induced pluripotent stem cells (iPSCs) [71] by culture in basal medium with no exogenous growth factors [72]. Transcription factors that drive the formation of the anterior neuroectoderm include Sox2 [73, 74], Zfp521 [75], Pou3f1/Oct6 [76], Sip1[77], FoxD4 [78], and the homodimerised form of E2A [79].
Signalling pathways controlling anterior neural specification
In vivo, neural fate specification depends on the inhibition of anti-neural signals BMP, Nodal, and WNT (reviewed in [80]). This has led researchers to propose a “neural default model”, which argues that pluripotent cells adopt a neuroectodermal fate unless specified otherwise. This is supported by the observations that early postimplantation embryos lacking Nodal [81] or BMP signalling [82] undergo premature and ectopic neural specification.
Pluripotent cells in culture, like their in vivo counterparts, progress to a neural identity when deprived of signalling inputs from the BMP [70, 83], Nodal [70], and Wnt [84, 85] pathways. Experiments using inhibitors of the fibroblast growth factor (FGF) pathway led to the proposal that autocrine FGF may be required for the acquisition of neural identity by mouse embryonic stem cells in culture [68], in keeping with findings in chick embryos [86, 87]. Later studies clarified that FGF can facilitate exit from naive pluripotency and subsequent induction and/or maintenance of formative [88] and primed [89–92] pluripotent states representative of post-implantation epiblast, but FGF must then be suppressed in order for primed pluripotent cells to efficiently acquire a neural identity [89, 93, 94]. Furthermore, embryos lacking BMP signalling default to a neural identity even when exposed to inhibitors of FGF signalling soon after implantation [82]. FGF signalling therefore appears to play a part in the exit from naive pluripotency and maintenance of the primed pluripotent state, but is then dispensable for neural specification. Additional factors, for example Notch signalling, can come into play to help coordinate the timing of neural fate specification between neighbouring cells possibly by dampening anti-neural signalling pathways [95], and unknown signals from adjacent tissues can refine the position of the neural plate boundary [96, 97].
In summary, the transition of primed pluripotent cells into anterior neuroectoderm is facilitated by inhibiting BMP, Nodal, Wnt, and FGF signalling pathways, while poorly understood local cell interactions signals seem to tune the response to these signals to refine the timing and position of neural fate specification.
Neuro-mesodermal progenitors
In 2009, a single-cell clonal analysis of the mouse embryo revealed a distinct population of cells arising during early gastrulation that could give rise to both neural and mesodermal lineages [98]. These neuro-mesodermal progenitors (NMPs) are a transient population of cells that emerges in the mouse at head-fold stage (around E7.5) and persists until E13.5 [99, 100], long after other organ systems become specified. They largely drive the elongation of the anteroposterior axis during development and give rise to the neural tube and paraxial mesoderm [98].
In several vertebrates, including mouse, chick, and zebrafish, NMPs can be identified by the co-expression of the transcription factors T (Brachyury) and Sox2 [100–103]. Cells that subsequently differentiate into neural lineages downregulate T but maintain Sox2 expression [104–106], while those committing to mesoderm downregulate Sox2 and upregulate Msgn1 and Tbx6 [99, 107]. T and Sox2 play antagonistic roles in NMP lineage specification, with T being essential for NMP maintenance and promoting mesodermal fate, while Sox2 promotes neural fate acquisition. Tbx6 steers cells towards a paraxial mesoderm fate [108].
The induction, maintenance and differentiation of NMPs is governed by WNT and FGF signalling, and embryonic regions containing NMPs express ligands for both of these pathways [109–112]. In the absence of components of the WNT or FGF signalling cascades, the embryo becomes truncated and ectopic neural tissue forms in place of posterior paraxial mesoderm, demonstrating that both of these signalling inputs are required for mesoderm specification and axis elongation, but are dispensable for neural commitment [100, 109, 112–115]. For a more detailed review of signalling events during specification and differentiation of neuromesodermal progenitors, see [105].
Cadherin expression patterns in neural development
Changes in cell identity are frequently accompanied by switches in expression of cadherins. For example, a switch from E-cadherin to N-cadherin is associated with the EMTs that accompany the formation of mesoderm or neural crest and with the dysregulation of tissue structure in tumorigenesis [116]. The emergence of the neural plate from the epiblast is not a classical EMT event but does bear a subset of EMT hallmarks, including loss of E-cadherin, gain of N-cadherin and upregulation of vimentin [117, 118]. Here we summarise changes in cadherin expression in the early stages of vertebrate development.
E-cadherin and N-cadherin
E-cadherin is the predominant cadherin expressed in pluripotent cells immediately prior to neural specification. In mice, it is present in the unfertilized egg as both mRNA and protein [5, 119]. At early stages of development, there is additionally significant maternal contribution of the protein to facilitate blastomere adhesion and embryonic compaction [120, 121]. E-cadherin knockout results in embryonic lethality at the morula stage (E2.5 in mouse) due to defects in morula compaction and a failure to segregate the trophectoderm and inner cell mass [121–126]. In the chick, E-cadherin is widely expressed in the epiblast of the early conceptus [61, 127]. In Xenopus, the protein first becomes expressed in ectoderm following the blastula stage [128–130].
At gastrulation, the mesodermal cells ingressing through the primitive streak are the first to lose E-cadherin expression as they begin to undergo EMT [131–133]. Strong E-cadherin expression is maintained in the anterior neurectoderm and neural plate until the later stages of neurulation. Early studies of the avian embryo indicated a lack of E-cadherin in the neural tube at HH10 [127, 134], though more recent studies have shown that the protein remains expressed at this stage, albeit at lower levels than in the overlying ectoderm [61, 135].
As pluripotent cells transit to a neural fate, they also upregulate N-cadherin. In mice, N-cadherin can first be observed in the neural plate at E7.5 [136], and by E8.5, it is widely expressed in the neural tube [137]. In the avian embryo, the earliest expression of N-cadherin in the ectoderm is in the neural plate at HH7, and expression persists in the neural plate and the closed neural tube [16, 127, 135].
The expression patterns of E- an N-cadherin in the neural crest appear more dynamic. In the mouse embryo, as neural crest cells delaminate they undergo EMT, downregulating E-cadherin and upregulating N-cadherin in its place, to acquire a migratory phenotype [61, 135]. In the chick embryo, E-cadherin expression is maintained in the neural crest, while the downregulation of N-cadherin is required for the delamination of cells in this structure [61, 135]. However, this functional patterning appears species-dependent, as it differs from that observed in Xenopus [138] or zebrafish [139].
In Xenopus, ectoderm is specified into dorsal neural ectoderm at the end of gastrulation, and this is where N-cadherin is first expressed, concomitantly with the downregulation of E-cadherin. The non-neural ectoderm, fated for epidermis, retains E-cadherin expression [130].
E- to N-cadherin switching has also been observed during the maturation of neuromesodermal progenitors. During axial elongation in the mouse embryo, NMPs mature while maintaining bipotency to generate both neural and mesodermal daughter tissues in the posterior regions of the trunk. This maturation is accompanied by a switch from epithelial to mesenchymal gene expression similar to a partial EMT, including a switch from E- to N-cadherin expression [140].
Other cadherin family members
In mice, P-cadherin (Cdh3) is first detected in the E4.0 trophectoderm but not in any embryonic tissues until later in development [141, 142]; it is not expressed in the neural ectoderm, but is instead found in non-neural ectoderm and other tissues [143]. In the chick, however, it has been suggested that P-cadherin is the predominantly expressed cadherin in the epiblast, reminiscent of E-cadherin expression in the early mouse embryo. This raises the possibility of an evolutionary exchange of E-cadherin for P-cadherin between mammals and birds, similar to the functional differences between the transcription factors Snail and Slug between the two species in EMT-like morphological events in early development [144, 145].
In the chick, K-cadherin (Cadherin-6B) is first expressed in the neural plate border at HH6, and subsequently plays a critical role in the specification and delamination of the neural crest [61]. In mice, K-cadherin (Cdh6) becomes detectable from E7.5 in neuroepithelial cells in the prospective hindbrain region, with expression in the forebrain by E8.0. At E8.5, K-cadherin is downregulated in the neural plate and neural tube, with expression persisting in the neural crest and in a band of the hindbrain at the level of rhombomere 6 (r6), and in the migrating neural crest cells emerging from r6; these neural crest cells go on to contribute to the peripheral nervous system. At E12.5, K-cadherin is found in the developing brain (expressed in a complementary pattern with R-cadherin) and in the spinal cord [146, 147]. Inoue et al. [146] suggest that K-cadherin facilitates cell sorting through differential adhesion of specific neural structures. Similarly, in zebrafish, combinatorial codes of cdh2, cdh11, and pcdh19 define different dorsoventral domains in the developing spinal cord and help to confer robust patterning by preventing the intermingling of cells from adjacent domains [148].
In mice, R-cadherin (Cdh4) is present in the neurectoderm of the developing midbrain at E8.5 [149]. Later, at E12.5, this cadherin is found in the lateral cortex of the brain in a complementary pattern with K-cadherin [146, 147]. In the chick, it is also detected in the neuroepithelium of the forebrain at E5 during later neural patterning [150].
Expression of cadherins 5 (VE-cadherin), 8, 9 (T1-cadherin), and 10 (T2-cadherin) has not been reported in the early mouse embryo [143], while cadherin 11 (OB-cadherin) is detectable in small amounts in the roof plate of the mouse neural tube [136]. Low levels of cadherin 7 transcript have been reported in the mouse neural tube at E8.5 [151]. In the chick, CDH4, 7, 8, 9, 11, 12 18 and 20 are expressed at later stages of neural patterning but not during early neurulation [152].
In Xenopus, C-cadherin (also called EP-cadherin) is the primary cadherin expressed from fertilized oocyte stage through to gastrula stages [130, 153, 154], and is critical for cell adhesion in the blastula [155]. C-cadherin continues to be expressed throughout neurulation in both neural and non-neural tissues [130]. U-cadherin is also expressed in all Xenopus cells during early development up to late neurula stage [156].
Neural developmental phenotypes of cadherins
Cadherins play important roles at multiple stages of brain development [157, 158], spinal cord neurogenesis [159, 160], and neural crest formation [61] (reviewed in [161]). Classical cadherins regulate the migration of different neuronal subtypes to the correct cortical layers in the developing brain (reviewed in [162]) and help position motor neurons within the hindbrain [163] and ocular system [164]. In addition, cadherins are located at synapses [165] where they regulate both synaptic formation [166–168] and maturation [167]. Cadherins can facilitate contact inhibition of cell proliferation in some contexts [168] although it is not known whether this occurs during neural development. It is, however, clear that cadherins influence proliferation in the nervous system through other mechanisms, for example as a consequence of their ability to modulate the activity of various signalling pathways [169]. In this review, we focus on the roles of cadherins during early stages of neural development.
In mice, knockout of E-cadherin results in embryonic lethality due to a failure in compaction of the morula and formation of trophectoderm [124, 170]. This phenotype has been attributed to the loss of adhesion and signalling functions of E-cadherin during the earliest stages of embryogenesis. Epiblast-specific replacement of E-cadherin with N-cadherin in mice leads to embryonic death at E8.5 due to improper growth and degeneration of the epiblast. Gastrulation is initiated in these embryos and all germ layers maintain their differentiation capacity, but mesoderm formation is compromised due to impaired BMP signalling [171]. These findings again implicate a specific role for E-cadherin in modulating signalling during embryonic development that may be independent of adhesion.
N-cadherin knockout is embryonic lethal in mice at around E10 due to heart defects; embryos also have malformed somites and a “wavy” neural tube, defects which appear to be caused by impaired cell–cell adhesion [137]. Work in ascidian embryos suggests that N-cadherin facilitates the directed forces that drive neural tube closure [172]. However, N-cadherin-null cells can form neural rosettes and adopt a neural tube-like organisation in teratoma assays, suggesting that the protein is not essential for the formation of simple neural structures. Cells lacking both N-cadherin and P-cadherin can give rise to rosettes but not neural tube-like structures, suggesting a certain level of functional redundancy between these two cadherins in the mouse, since embryos with knockout of P-cadherin alone are viable and have normal neural organisation [173, 174].
In Xenopus, depletion of N-cadherin in the neural plate results in abnormal invagination of the plate during neurulation (similar to spina bifida), caused by failure in actin assembly resulting in insufficient forces to direct folding of the neural plate [130]. While some bending in the neural plate can be observed in these embryos, the force for this bending is supplied exclusively by pushing forces from the non-neural ectoderm. Depletion of E-cadherin in the same structure in Xenopus embryos causes impaired spreading of the non-neural ectoderm and results in failure of neural fold closure, likely due to defects in E-cadherin-dependent cortical actin assembly and resulting in impaired movement of epidermal cells [130]. Neither of these phenotypes can be rescued by overexpressing the other cadherin, highlighting cadherin-specific roles in mechanical function during neurulation. In zebrafish, deletion of N-cadherin gives rise to a characteristic T-shaped neural tube due to a defect in convergent cell movements during neurulation [175, 176].
Overexpression of E-cadherin or N-cadherin in xenopus embryos results in failure to form the neural plate, with embryos instead developing large cysts. At later stages, these embryos do not develop a visible anteroposterior or dorsal–ventral axis. The same phenotype is seen upon depletion of β-catenin, and appears to involve a failure of neural and dorsal mesodermal differentiation [155].
In cultured neural stem cells (NSCs), overexpression of E-cadherin causes a downregulation of N-cadherin, and conversely, RNAi-mediated knockdown of E-cadherin causes N-cadherin to be upregulated, suggesting that these two cadherins may repress each other’s expression. The same study found that overexpression of E-cadherin inhibits the migration of NSCs; effects on differentiation were not examined [177].
It is clear that cadherins are crucial for early neural development to proceed correctly, raising the questions of whether this requirement for cadherins is explained predominantly by their ability to modulate cell–cell adhesion to drive morphological changes [60], or whether they may also perform other functions that directly influence the differentiation process.
Cadherins modulate neural differentiation
There is plentiful evidence that cadherins can protect or promote pluripotency in a number of contexts [178–185]. Here we will review the evidence that cadherins are also able to regulate the transition from pluripotency to neural identity.
Downregulation of E-cadherin is tightly correlated with a transition from pluripotency to neural identity in culture [186]. This timely loss of E-cadherin seems to be a limiting factor for cells to enter the neural lineage: experimental suppression of this adhesion molecule results in faster and more uniform neural differentiation [186].
What regulates the changes in cadherin expression that enable efficient differentiation? As discussed above, inhibition of BMP signalling is a key event that triggers neural differentiation [79, 82, 83, 87]. BMP also maintains high levels of E-cadherin in pluripotent cells [186]. The ability of BMP to maintain high E-cadherin seems to explain its ability to block neural differentiation; experimental suppression of E-cadherin is sufficient to overcome the inhibitory effects of BMP on neural differentiation if no other anti-neural signals are present, and if BMP levels remain below a particular threshold [186]. So, in situations where BMP levels are not too high, E-cadherin is the primary mediator of its anti-neural effects.
Higher doses of BMP do, however, have the additional ability to impose a posterior identity that favours mesodermal rather than neural priming of pluripotent cells. This second anti-neural effect of BMP seems to be independent of E-cadherin. This might explain why downregulation of E-cadherin does not result in neural differentiation in the posterior of the embryo, where several BMP family members (together with other anti-neural signals) are expressed at high levels [54].
It seems likely that gain of N-cadherin also influences the acquisition or stability of neural identity. Forcing premature N-cadherin expression can enhance neural differentiation in culture even in situations where E-cadherin initially remains present [187]. Manipulations of N-cadherin have also been used to demonstrate that cadherins can influence differentiation independently of any changes in adhesion. For example, N-cadherin can fully rescue the adhesion phenotype of E-cadherin mutant pre-implantation embryos, but does not rescue all defects in signalling and differentiation phenotypes [122, 187]. Similarly, premature activation of N-cadherin can result in pro-neural phenotypes in culture without any apparent effect on the spatial organisation or cohesion of cells [187].
These findings confirm that both E- and N-cadherin may contribute to the efficiency of neural differentiation. What might be the mechanisms by which cadherins influence cell fate? Developmental transitions are sensitive to changes in the forces that cells impose on their neighbours [188, 189] and by movements of cell populations relative to each other [190]. Both of these processes could be modulated by changes in cadherin expression. The work discussed above [122, 187] does, however, hint that at least part of the mechanism by which cadherins control differentiation may not be explained only by changes in the cell–cell adhesion. This might instead be explained by the ability of cadherins to directly interact with components of signalling pathways.
Cadherins modulate signalling
Cadherins do more than just stick cells together, something that should perhaps not come as a surprise given that cadherins first appeared in unicellular organisms. As multicellular organisms evolved, it seems that cadherins may have acquired a diverse range of functions. For example, it is now well established that cadherins can bind to various components of signal transduction pathways to modulate signal responsiveness. This could provide an opportunity for cells to coordinate changes in morphology (influenced by changes in adhesion) with changes in cell identity (influenced by changes in signalling pathway activity). There have been a number of excellent reviews [161, 191, 192] covering ways in which cadherins can modulate signalling pathways. Here we outline a few examples that may be of particular relevance to early neural development.
Perhaps the most obvious candidate mechanistic link between cadherins and neural differentiation is the Wnt signalling pathway. Wnt signalling has multiple stage-specific effects on development of the nervous system, but its earliest effects seem to be anti-neural; Wnt activity needs to be suppressed if either pluripotent or neuromesodermal progenitor cells are to transit to a neural identity [84, 85, 100, 109, 112]. β-catenin is a component of adherens junctions and also a component of the Wnt signal transduction pathway, and so provides one possible mechanistic link between cadherins and Wnt signalling. In at least some contexts [155, 193] cadherins can titrate the amount of β-catenin that is available for mediating Wnt signalling, effectively acting as a ‘dampener' of the Wnt response. Moreover, it has been suggested that E-cadherin does not merely dampen Wnt activity but can also prime it for future activation [194], for example during mesoderm formation from pluripotent human cells [195]. A positive influence of E-cadherin on Wnt responsiveness is in keeping with the observation that loss of E-cadherin can decrease rather than increase Wnt responsiveness in a mammary epithelial cell line [196].
These findings raise the possibility that the switch from E- to N-cadherin may result in a switch in Wnt responsiveness. In support of this idea, E-cadherin and N-cadherin differ in their ability to influence Wnt activity during mesoderm formation in drosophila: E-cadherin, but not N-cadherin, can effectively suppress Wnt activity. Importantly, particular point mutations in E-cadherin can abolish effects on Wnt activity without affecting cell adhesion [197]. It therefore seems likely that the switch from E- to N-cadherin during neural differentiation might modulate receptiveness to Wnt independently from effects on cell adhesion.
When pluripotent cells are forced to experience a premature cadherin switch by replacing the coding sequence for E-cadherin with that for N-cadherin in mouse ES cells [123, 171, 187], this results in a strong reduction in β-catenin levels accompanied by an increase in the efficiency of neural differentiation. Furthermore, this forced cadherin switch enables cells to resist the anti-neural effects of exogenous Wnt. Surprisingly, however, this apparent “Wnt resistance” seems to operate via an indirect mechanism, because the transcriptional response to Wnt remained intact in cells that had undergone an enforced cadherin switch. Therefore, during neural differentiation of pluripotent cells in culture, the ability of cadherins to influence neural differentiation does not seem to be explained by their ability to modulate transcriptional activity via Wnt signalling. What other signalling pathways might be regulated by cadherins during early neural development? One promising candidate is the FGF signalling pathway.
There are a number of ways in which cadherins modulate FGF activity. FGF receptors (FGFRs) interact with N-cadherin, for example during neurite outgrowth, and may even mediate ligand-independent activation [52]. Similarly, N-cadherin can bind FGF receptors and potentiate FGF activity in breast cancer cell lines [198]. Perhaps most strikingly, mice engineered to express N-cadherin in place of E-cadherin in mammary epithelia exhibit constitutive FGF activity and pre-malignant growth [199].
FGF can sustain primed pluripotency [90, 91] and suppress neural differentiation of epiblast cells [89, 94], at least in culture. N-cadherin has been reported to sustain FGF responsiveness to maintain pluripotency in EpiSCs [185], although the in vivo relevance of this is unclear given that N-cadherin is not readily detectable in the epiblast [187]. N-cadherin is, however, readily detectable at the onset of neural differentiation in culture and, as discussed above, is able to enhance the efficiency of the differentiation process. By measuring the activity of a panel of signalling pathways, Punovuori et al. [187] found that N-cadherin dampens the activity of pathways downstream of FGF receptors during early neural differentiation. Restoring FGF activity can reverse the pro-neural effects of N-cadherin. It seems therefore that N-cadherin has context-specific effects on FGF signalling, and in the context of neural differentiation it acts to dampen FGF activity and consequently enhances the transition from pluripotency to a stable neural identity (Fig. 5).
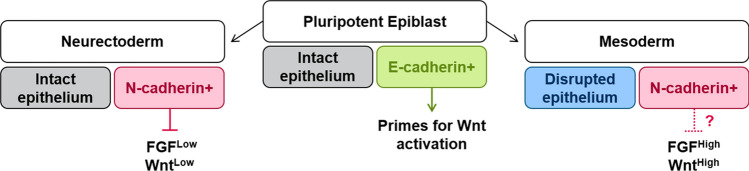
Fig. 5.
E-cadherin is exchanged for N-cadherin as the pluripotent epiblast forms neuroectoderm or mesoderm. E-cadherin in the epiblast may prime cells for future activation of Wnt [189, 197]. N-cadherin in the neuroectoderm dampens low levels of FGF that would otherwise have anti-neural activity [189]. It remains an open question whether the high levels of FGF or other differences in context in the emerging mesoderm override the FGF-dampening effects of N-cadherin
Much remains to be explored about the links between signalling pathways and neural development. For example, E-cadherin binds insulin-like growth factor 1 (IGF1) receptor and this interaction is critical for survival of cells in the preimplantation embryo [122]. IGF is also important for survival of neural progenitor cells, so it would be interesting to explore whether cadherins also influence IGF activity in the nervous system. In endothelial cells, VE-cadherin can bind and stabilise BMP receptors [200], making it tempting to speculate that other cadherins expressed before and during neural specification may also influence BMP signalling.
In summary, it seems clear that effects of cadherins on signalling are diverse and context specific. In the context of neural differentiation one might speculate that cadherins act as ‘dampeners’ of multiple signalling pathways and so help shield cells from anti-neural influences. Furthermore, a loosening of cell–cell contacts caused by changes in cadherin expression may interfere with juxtacrine signalling from surface-bound ligands, for example through the Notch receptor. It would be interesting to explore these ideas in future work.
Perspectives
Why might it be useful for the cells of the embryo to use cadherins to help inform their differentiation decisions during early neural development? Neural tissue forms within an embryo that is growing and changing shape rapidly. Signalling molecules appear and disappear from different regions of the embryo over a short space of time as they are used and then re-used for successive differentiation decisions. In this constantly changing environment, it might be difficult to ensure that there are never any ectopic residual or premature signals that might disrupt neural specification.
Cadherin switching can dampen signal responsiveness. We propose that the embryo exploits this property of cadherins in a ‘belt and braces’ approach to guard against any ectopic signals that might otherwise mislead cells into the ‘wrong’ fate. One important open question is whether cadherins influence differentiation only cell-autonomously or whether they can also share this information with neighbouring cells, and so help coordinate neural differentiation across a local neighbourhood, perhaps propagating information from cell to cell as has been described during mesoderm formation [201].
Given that cadherins first appeared in unicellular organisms, they will have had ample time to acquire multiple functions, and therefore would be well placed to coordinate multiple events during the development of multicellular organisms. In this review, we have focused on the ability of cadherins to coordinate changes in adhesion with changes in signal response, but it remains possible that cadherins influence and coordinate other biological processes. It would be interesting to explore these ideas and test their relevance to neural development.
Can our understanding of cadherins in early neural development help us to gain better control over differentiation of cells in culture? For reasons still unknown, the timing of cadherin switching relative to neural specification becomes partially dysregulated in cultured cells; in the embryo, N-cadherin only becomes readily detectable in cells that have already switched on early neural marker Sox1, whereas in culture N-cadherin can be found in cells that retain pluripotency markers and lack neural markers [187]. Does this variability in cadherins explain variability in the differentiation response? There is some evidence that this is indeed the case: forcing a more uniform cadherin switch does seem to impose a more homogenous differentiation response, at least in monolayer culture [186, 187]. It would be interesting to explore whether manipulation of cadherins can also give better control over differentiation of cells in 3D or organoid culture in which cells have a more in vivo-like spatial organisation.
Our understanding of differentiation is gradually shifting from a view dominated primarily by signalling pathways, transcription factors, and chromatin structure to a broader understanding incorporating information from adhesion and morphology. As John Gurdon wrote, “most aspects of animal development seem to proceed by the cooperation of several contributory processes, no one of which is individually indispensable" [2]. Cadherins are unlikely to be essential instructors of neural differentiation decisions, but do seem likely to improve coordination of cell decisions across fields of cells, thus contributing to the fidelity of patterning in the early embryo. Cells, it seems, do better when they stick together.
Acknowledgments
We acknowledge support from the Wellcome Trust (Senior Fellowship WT103789AIA to SL) and the Medical Research Council (MRC PhD studentship to KP).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Waddington CH. Canalization of development and genetic assimilation of acquired characters. Nature. 1959;183:1654–1655. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 2.Gurdon JB. A community effect in animal development. Nature. 1988;336:772–774. doi: 10.1038/336772a0. [DOI] [PubMed] [Google Scholar]
- 3.Lander AD. Pattern, growth, and control. Cell. 2011;144:955–969. doi: 10.1016/j.cell.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeichi M. Functional correlation between cell adhesive properties and some cell surface proteins. J Cell Biol. 1977;75:464–474. doi: 10.1083/jcb.75.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kemler R, Babinet C, Eisen H, Jacob F. Surface antigen in early differentiation. Proc Natl Acad Sci U S A. 1977;74:4449–4452. doi: 10.1073/pnas.74.10.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertolotti R, Rutishauser U, Edelman GM. A cell surface molecule involved in aggregation of embryonic liver cells. Proc Natl Acad Sci U S A. 1980;77:4831–4835. doi: 10.1073/pnas.77.8.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hyafil F, Morello D, Babinet C, Jacob F. A cell surface glycoprotein involved in the compaction of embryonal carcinoma cells and cleavage stage embryos. Cell. 1980;21:927–934. doi: 10.1016/0092-8674(80)90456-0. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida C, Takeichi M. Teratocarcinoma cell adhesion: Identification of a cell-surface protein involved in calcium-dependent cell aggregation. Cell. 1982;28:217–224. doi: 10.1016/0092-8674(82)90339-7. [DOI] [PubMed] [Google Scholar]
- 9.Damsky CH, Richa J, Solter D, Knudsen K, Buck CA. Identification and purification of a cell surface glycoprotein mediating intercellular adhesion in embryonic and adult tissue. Cell. 1983;34:455–466. doi: 10.1016/0092-8674(83)90379-3. [DOI] [PubMed] [Google Scholar]
- 10.Gallin WJ, Edelman GM, Cunningham BA. Characterization of L-CAM, a major cell adhesion molecule from embryonic liver cells. Proc Natl Acad Sci U S A. 1983;80:1038–1042. doi: 10.1073/pnas.80.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peyriéras N, Hyafil F, Louvard D, Ploegh HL, Jacob F. Uvomorulin: a nonintegral membrane protein of early mouse embryo. Proc Natl Acad Sci U S A. 1983;80:6274–6277. doi: 10.1073/pnas.80.20.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida-Noro C, Suzuki N, Takeichi M. Molecular nature of the calcium-dependent cell-cell adhesion system in mouse teratocarcinoma and embryonic cells studied with a monoclonal antibody. Dev Biol. 1984;101:19–27. doi: 10.1016/0012-1606(84)90112-x. [DOI] [PubMed] [Google Scholar]
- 13.Behrens J, Birchmeier W, Goodman SL, Imhof BA. Dissociation of Madin-Darby Canine Kidney epithelial Cells by the monoclonal antibody anti-arc-l: mechanistic aspects and identification of the antigen as a component related to uvomorulin. J Cell Biol. 1985;101:1307–1315. doi: 10.1083/jcb.101.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volk T, Geiger B. A 135-kd membrane protein of intercellular adherens junctions. EMBO J. 1984;3:2249–2260. doi: 10.1002/j.1460-2075.1984.tb02123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatta K, Okada TS, Takeichi M. A monoclonal antibody disrupting calcium-dependent cell-cell adhesion of brain tissues: possible role of its target antigen in animal pattern formation. Proc Natl Acad Sci U S A. 1985;82:2789–2793. doi: 10.1073/pnas.82.9.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatta K, Takeichi M. Expression of N-cadherin adhesion molecules associated with early morphogenetic events in chick development. Nature. 1986;320:447–449. doi: 10.1038/320447a0. [DOI] [PubMed] [Google Scholar]
- 17.Nose A, Takeichi M. A novel cadherin cell adhesion molecule: its expression patterns associated with implantation and organogenesis of mouse embryos. J Cell Biol. 1986;103:2649–2658. doi: 10.1083/jcb.103.6.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulpiau P, van Roy F. New insights into the evolution of metazoan cadherins. Mol Biol Evol. 2011;28:647–657. doi: 10.1093/molbev/msq233. [DOI] [PubMed] [Google Scholar]
- 19.Angulo-Urarte A, van der Wal T, Huveneers S. Cell-cell junctions as sensors and transducers of mechanical forces. Biochim Biophys Acta Biomembr. 2020;1862:183316. doi: 10.1016/j.bbamem.2020.183316. [DOI] [PubMed] [Google Scholar]
- 20.Canzio D, Maniatis T. The generation of a protocadherin cell-surface recognition code for neural circuit assembly. Curr Opin Neurobiol. 2019;59:213–220. doi: 10.1016/j.conb.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janiszewska M, Primi MC, Izard T. Cell adhesion in cancer: Beyond the migration of single cells. J Biol Chem. 2020;295:2495–2505. doi: 10.1074/jbc.REV119.007759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yulis M, Kusters DHM, Nusrat A. Cadherins: cellular adhesive molecules serving as signalling mediators. J Physiol. 2018;596:3883–3898. doi: 10.1113/JP275328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison OJ, Jin X, Hong S, Bahna F, Ahlsen G, Brasch J, et al. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 2011;19:244–256. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol. 2009;41:349–369. doi: 10.1016/j.biocel.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 25.Hulpiau P, Gul IS, van Roy F. New insights into the evolution of metazoan cadherins and catenins. Progress Mol Biol Transl Sci. 2013;116:71–94. doi: 10.1016/b978-0-12-394311-8.00004-2. [DOI] [PubMed] [Google Scholar]
- 26.Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- 27.Peek SL, Mah KM, Weiner JA. Regulation of neural circuit formation by protocadherins. Cell Mol Life Sci. 2017;74:4133–4157. doi: 10.1007/s00018-017-2572-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stemmler MP. Cadherins in development and cancer. Mol Biosyst. 2008;4:835–850. doi: 10.1039/b719215k. [DOI] [PubMed] [Google Scholar]
- 29.van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65:3756–3788. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallin WJ. Evolution of the“ classical” cadherin family of cell adhesion molecules in vertebrates. Mol Biol Evol. 1998;15:1099–1107. doi: 10.1093/oxfordjournals.molbev.a026017. [DOI] [PubMed] [Google Scholar]
- 31.Takeichi M. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development. 1988;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- 32.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 33.Tepass U. Genetic analysis of cadherin function in animal morphogenesis. Curr Opin Cell Biol. 1999;11:540–548. doi: 10.1016/s0955-0674(99)00025-3. [DOI] [PubMed] [Google Scholar]
- 34.Abedin M, King N. The premetazoan ancestry of cadherins. Science. 2008;319:946–948. doi: 10.1126/science.1151084. [DOI] [PubMed] [Google Scholar]
- 35.Nichols SA, Roberts BW, Richter DJ, Fairclough SR, King N. Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/β-catenin complex. Proc Natl Acad Sci U S A. 2012;109:13046–13051. doi: 10.1073/pnas.1120685109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray PS, Zaidel-Bar R. Pre-metazoan origins and evolution of the cadherin adhesome. Biol Open. 2014;3:1183–1195. doi: 10.1242/bio.20149761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdian PL, Caramelo JJ, Ausmees N, Zorreguieta A (2013). RapA2 is a calcium-binding lectin composed of two highly conserved cadherin-like domains that specifically recognize Rhizobium leguminosarum acidic exopolysaccharides. J Biol Chem 288(4):2893–2904. [DOI] [PMC free article] [PubMed]
- 38.Cao L, Yan X, Borysenko CW, Blair HC, Wu C, Yu L. CHDL: a cadherin-like domain in Proteobacteria and Cyanobacteria. FEMS Microbiol Lett. 2005;251:203–209. doi: 10.1016/j.femsle.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Chatterjee R, Nag S, Chaudhuri K. Identification of a new RTX-like gene cluster in Vibrio cholerae. FEMS Microbiol Lett. 2008;284:165–171. doi: 10.1111/j.1574-6968.2008.01199.x. [DOI] [PubMed] [Google Scholar]
- 40.Fraiberg M, Borovok I, Weiner RM, Lamed R. Discovery and characterization of cadherin domains in Saccharophagus degradans 2–40. J Bacteriol. 2010;192:1066–1074. doi: 10.1128/JB.01236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Studholme DJ, Fuerst JA, Bateman A. Novel protein domains and motifs in the marine planctomycete Rhodopirellula baltica. FEMS Microbiol Lett. 2004;236:333–340. doi: 10.1016/j.femsle.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 42.Yang Y-H, Jiang Y-L, Zhang J, Wang L, Bai X-H, Zhang S-J, et al. Structural insights into SraP-mediated Staphylococcus aureus adhesion to host cells. PLoS Pathog. 2014;10:e1004169. doi: 10.1371/journal.ppat.1004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adl SM, Simpson AGB, Farmer MA, Andersen RA, Anderson OR, Barta JR, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 44.Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, et al. Revisions to the classification, nomenclature, and diversity of eukaryotes. J Eukaryot Microbiol. 2019;66:4–119. doi: 10.1111/jeu.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elledge HM, Kazmierczak P, Clark P, Joseph JS, Kolatkar A, Kuhn P, et al. Structure of the N terminus of cadherin 23 reveals a new adhesion mechanism for a subset of cadherin superfamily members. Proc Natl Acad Sci U S A. 2010;107:10708–10712. doi: 10.1073/pnas.1006284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parisini E, Higgins JMG, Liu J-H, Brenner MB, Wang J-H. The crystal structure of human E-cadherin domains 1 and 2, and comparison with other cadherins in the context of adhesion mechanism. J Mol Biol. 2007;373:401–411. doi: 10.1016/j.jmb.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guex N, Peitsch MC (1997) SWISS‐MODEL and the Swiss‐Pdb viewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2713 [DOI] [PubMed]
- 49.Sebé-Pedrós A, Degnan BM, Ruiz-Trillo I. The origin of Metazoa: a unicellular perspective. Nat Rev Genet. 2017;18:498–512. doi: 10.1038/nrg.2017.21. [DOI] [PubMed] [Google Scholar]
- 50.Larsen J, Patterson DJ. Some flagellates (Protista) from tropical marine sediments. J Nat Hist. 1990;24:801–937. [Google Scholar]
- 51.Fletcher KIG, van West P, Gachon CMM. Nonagonal cadherins: a new protein family found within the Stramenopiles. Gene. 2016;593:64–75. doi: 10.1016/j.gene.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 52.Williams TJ, Lauro FM, Ertan H, Burg DW, Poljak A, Raftery MJ, et al. Defining the response of a microorganism to temperatures that span its complete growth temperature range (−2°C to 28°C) using multiplex quantitative proteomics. Environ Microbiol. 2011;13:2186–2203. doi: 10.1111/j.1462-2920.2011.02467.x. [DOI] [PubMed] [Google Scholar]
- 53.Gul IS, Hulpiau P, Saeys Y, van Roy F. Evolution and diversity of cadherins and catenins. Exp Cell Res. 2017;358:3–9. doi: 10.1016/j.yexcr.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 54.Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- 55.Morgani SM, Hadjantonakis A-K. Signaling regulation during gastrulation: Insights from mouse embryos and in vitro systems. Curr Top Dev Biol. 2020;137:391–431. doi: 10.1016/bs.ctdb.2019.11.011. [DOI] [PubMed] [Google Scholar]
- 56.Lawson KA, Meneses JJ, Pedersen RA. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- 57.Quinlan GA, Williams EA, Tan SS, Tam PP. Neuroectodermal fate of epiblast cells in the distal region of the mouse egg cylinder: implication for body plan organization during early embryogenesis. Development. 1995;121:87–98. doi: 10.1242/dev.121.1.87. [DOI] [PubMed] [Google Scholar]
- 58.Harrington MJ, Hong E, Brewster R. Comparative analysis of neurulation: first impressions do not count. Mol Reprod Dev. 2009;76:954–965. doi: 10.1002/mrd.21085. [DOI] [PubMed] [Google Scholar]
- 59.Smith JL, Schoenwolf GC. Neurulation: coming to closure. Trends Neurosci. 1997;20:510–517. doi: 10.1016/s0166-2236(97)01121-1. [DOI] [PubMed] [Google Scholar]
- 60.Sutherland AE. Tissue morphodynamics shaping the early mouse embryo. Semin Cell Dev Biol. 2016;55:89–98. doi: 10.1016/j.semcdb.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 61.Dady A, Duband J-L. Cadherin interplay during neural crest segregation from the non-neural ectoderm and neural tube in the early chick embryo. Dev Dyn. 2017;246:550–565. doi: 10.1002/dvdy.24517. [DOI] [PubMed] [Google Scholar]
- 62.Parameswaran M, Tam PP. Regionalisation of cell fate and morphogenetic movement of the mesoderm during mouse gastrulation. Dev Genet. 1995;17:16–28. doi: 10.1002/dvg.1020170104. [DOI] [PubMed] [Google Scholar]
- 63.Tam PP, Zhou SX. The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev Biol. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- 64.Beddington RSP. An autoradio graphic analysis of tissue potency in different regions of the embryonic ectoderm during gastrulation in the mouse. Plan Perspect. 1982;265:285. [PubMed] [Google Scholar]
- 65.Li L, Liu C, Biechele S, Zhu Q, Song L, Lanner F, et al. Location of transient ectodermal progenitor potential in mouse development. Development. 2013;140:4533–4543. doi: 10.1242/dev.092866. [DOI] [PubMed] [Google Scholar]
- 66.Cajal M, Lawson KA, Hill B, Moreau A, Rao J, Ross A, et al. Clonal and molecular analysis of the prospective anterior neural boundary in the mouse embryo. Development. 2012;139:423–436. doi: 10.1242/dev.075499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tam PP. Regionalisation of the mouse embryonic ectoderm: allocation of prospective ectodermal tissues during gastrulation. Development. 1989;107:55–67. doi: 10.1242/dev.107.1.55. [DOI] [PubMed] [Google Scholar]
- 68.Ying Q-L, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 69.Zhang S-C, Wernig M, Duncan ID, Brüstle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nature Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 70.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu B-Y, Weick JP, Yu J, Ma L-X, Zhang X-Q, Thomson JA, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pollard SM, Benchoua A, Lowell S. Neural stem cells, neurons, and glia. Methods Enzymol. 2006;418:151–169. doi: 10.1016/s0076-6879(06)18010-6. [DOI] [PubMed] [Google Scholar]
- 73.Thomson M, Liu SJ, Zou L-N, Smith Z, Meissner A, Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 75.Kamiya D, Banno S, Sasai N, Ohgushi M, Inomata H, Watanabe K, et al. Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature. 2011;470:503–509. doi: 10.1038/nature09726. [DOI] [PubMed] [Google Scholar]
- 76.Zhu Q, Song L, Peng G, Sun N, Chen J, Zhang T, et al. The transcription factor Pou3f1 promotes neural fate commitment via activation of neural lineage genes and inhibition of external signaling pathways. Elife. 2014 doi: 10.7554/eLife.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chng Z, Teo A, Pedersen RA, Vallier L. SIP1 mediates cell-fate decisions between neuroectoderm and mesendoderm in human pluripotent stem cells. Cell Stem Cell. 2010;6:59–70. doi: 10.1016/j.stem.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 78.Sherman JH, Karpinski BA, Fralish MS, Cappuzzo JM, Dhindsa DS, Thal AG, et al. Foxd4 is essential for establishing neural cell fate and for neuronal differentiation. Genesis. 2017;55:e23031. doi: 10.1002/dvg.23031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rao C, Malaguti M, Mason JO, Lowell S. The transcription factor E2A drives neural differentiation in pluripotent cells. Development. 2020 doi: 10.1242/dev.184093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stern CD. Neural induction: 10 years on since the “default model”. Curr Opin Cell Biol. 2006;18:692–697. doi: 10.1016/j.ceb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 81.Camus A, Perea-Gomez A, Moreau A, Collignon J. Absence of Nodal signaling promotes precocious neural differentiation in the mouse embryo. Dev Biol. 2006;295:743–755. doi: 10.1016/j.ydbio.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 82.Di-Gregorio A, Sancho M, Stuckey DW, Crompton LA, Godwin J, Mishina Y, et al. BMP signalling inhibits premature neural differentiation in the mouse embryo. Development. 2007;134:3359–3369. doi: 10.1242/dev.005967. [DOI] [PubMed] [Google Scholar]
- 83.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 84.Aubert J, Dunstan H, Chambers I, Smith A. Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat Biotechnol. 2002;20:1240–1245. doi: 10.1038/nbt763. [DOI] [PubMed] [Google Scholar]
- 85.Osteil P, Studdert JB, Goh HN, Wilkie EE, Fan X, Khoo P-L, et al. Dynamics of Wnt activity on the acquisition of ectoderm potency in epiblast stem cells. Development. 2019;146:dev172858. doi: 10.1242/dev.172858. [DOI] [PubMed] [Google Scholar]
- 86.Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD. Initiation of neural induction by FGF signalling before gastrulation. Nature. 2000;406:74–78. doi: 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- 87.Linker C, Stern CD. Neural induction requires BMP inhibition only as a late step, and involves signals other than FGF and Wnt antagonists. Development. 2004;131:5671–5681. doi: 10.1242/dev.01445. [DOI] [PubMed] [Google Scholar]
- 88.Sterneckert J, Stehling M, Bernemann C, Araúzo-Bravo MJ, Greber B, Gentile L et al (2010) Neural induction intermediates exhibit distinct roles of Fgf signaling. Stem Cells 28(10):1772–1781. [DOI] [PubMed]
- 89.Sterneckert J, Stehling M, Bernemann C, Araúzo-Bravo MJ, Greber B, Gentile L, et al. Neural induction intermediates exhibit distinct roles of Fgf signaling. Stem Cells. 2010;28:1772–1781. doi: 10.1002/stem.498. [DOI] [PubMed] [Google Scholar]
- 90.Brons IGM, Smithers LE, Trotter MWB, Rugg-Gunn P, Sun B, de Sousa C, Lopes SM, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 91.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 92.Lanner F, Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- 93.Stavridis MP, Lunn JS, Collins BJ, Storey KG. A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development. 2007;134:2889–2894. doi: 10.1242/dev.02858. [DOI] [PubMed] [Google Scholar]
- 94.Greber B, Coulon P, Zhang M, Moritz S, Frank S, Müller-Molina AJ, et al. FGF signalling inhibits neural induction in human embryonic stem cells. EMBO J. 2011;30:4874–4884. doi: 10.1038/emboj.2011.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lowell S, Benchoua A, Heavey B, Smith AG. Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLoS Biol. 2006;4:e121. doi: 10.1371/journal.pbio.0040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Linker C, De Almeida I, Papanayotou C, Stower M, Sabado V, Ghorani E, et al. Cell communication with the neural plate is required for induction of neural markers by BMP inhibition: evidence for homeogenetic induction and implications for Xenopus animal cap and chick explant assays. Dev Biol. 2009;327:478–486. doi: 10.1016/j.ydbio.2008.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cajal M, Creuzet SE, Papanayotou C, Saberan-Djoneidi D, de Sousa Lopes SMC, Zwijsen A, et al. A conserved role for non-neural ectoderm cells in early neural development. Development. 2014;141:4127–4138. doi: 10.1242/dev.107425. [DOI] [PubMed] [Google Scholar]
- 98.Tzouanacou E, Wegener A, Wymeersch FJ, Wilson V, Nicolas J-F. Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev Cell. 2009;17:365–376. doi: 10.1016/j.devcel.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 99.Gouti M, Delile J, Stamataki D, Wymeersch FJ, Huang Y, Kleinjung J, et al. A gene regulatory network balances neural and mesoderm specification during vertebrate trunk development. Dev Cell. 2017;41:243–261.e7. doi: 10.1016/j.devcel.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wymeersch FJ, Huang Y, Blin G, Cambray N, Wilkie R, Wong FCK, et al. Position-dependent plasticity of distinct progenitor types in the primitive streak. Elife. 2016;5:e10042. doi: 10.7554/eLife.10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martin BL, Kimelman D. Canonical Wnt signaling dynamically controls multiple stem cell fate decisions during vertebrate body formation. Dev Cell. 2012;22:223–232. doi: 10.1016/j.devcel.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Olivera-Martinez I, Harada H, Halley PA, Storey KG. Loss of FGF-dependent mesoderm identity and rise of endogenous retinoid signalling determine cessation of body axis elongation. PLoS Biol. 2012;10:e1001415. doi: 10.1371/journal.pbio.1001415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tsakiridis A, Huang Y, Blin G, Skylaki S, Wymeersch F, Osorno R, et al. Distinct Wnt-driven primitive streak-like populations reflect in vivo lineage precursors. Development. 2014;141:1209–1221. doi: 10.1242/dev.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gouti M, Tsakiridis A, Wymeersch FJ, Huang Y, Kleinjung J, Wilson V, et al. In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol. 2014;12:e1001937. doi: 10.1371/journal.pbio.1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gouti M, Metzis V, Briscoe J. The route to spinal cord cell types: a tale of signals and switches. Trends Genet. 2015;31:282–289. doi: 10.1016/j.tig.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 106.Tsakiridis A, Wilson V. Assessing the bipotency of in vitro-derived neuromesodermal progenitors. F1000Res. 2015;4:100. doi: 10.12688/f1000research.6345.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chalamalasetty RB, Garriock RJ, Dunty WC, Jr, Kennedy MW, Jailwala P, Si H, et al. Mesogenin 1 is a master regulator of paraxial presomitic mesoderm differentiation. Development. 2014;141:4285–4297. doi: 10.1242/dev.110908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Koch F, Scholze M, Wittler L, Schifferl D, Sudheer S, Grote P, et al. Antagonistic activities of Sox2 and brachyury control the fate choice of neuro-mesodermal progenitors. Dev Cell. 2017;42:514–526.e7. doi: 10.1016/j.devcel.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 109.Takada S, Stark KL, Shea MJ, Vassileva G, McMahon JA, McMahon AP. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8:174–189. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- 110.van de Ven C, Bialecka M, Neijts R, Young T, Rowland JE, Stringer EJ, et al. Concerted involvement of Cdx/Hox genes and Wnt signaling in morphogenesis of the caudal neural tube and cloacal derivatives from the posterior growth zone. Development. 2011;138:3859–3859. doi: 10.1242/dev.066118. [DOI] [PubMed] [Google Scholar]
- 111.Wilson V, Olivera-Martinez I, Storey KG. Stem cells, signals and vertebrate body axis extension. Development. 2009;136:2133–2133. doi: 10.1242/dev.021246. [DOI] [PubMed] [Google Scholar]
- 112.Yoshikawa Y, Fujimori T, McMahon AP, Takada S. Evidence that absence of Wnt-3a signaling promotes neuralization instead of paraxial mesoderm development in the mouse. Dev Biol. 1997;183:234–242. doi: 10.1006/dbio.1997.8502. [DOI] [PubMed] [Google Scholar]
- 113.Boulet AM, Capecchi MR. Signaling by FGF4 and FGF8 is required for axial elongation of the mouse embryo. Dev Biol. 2012;371:235–245. doi: 10.1016/j.ydbio.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chal J, Pourquié O. Making muscle: skeletal myogenesis in vivo and in vitro. Development. 2017;144:2104–2122. doi: 10.1242/dev.151035. [DOI] [PubMed] [Google Scholar]
- 115.Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- 116.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 117.Kohei Hatta MT. Expression of N-cadherin adhesion molecules associated with early morphogenetic events in chick development. Nature. 1986;320:447–449. doi: 10.1038/320447a0. [DOI] [PubMed] [Google Scholar]
- 118.Aaku-Saraste E, Hellwig A, Huttner WB. Loss of occludin and functional tight junctions, but not ZO-1, during neural tube closure–remodeling of the neuroepithelium prior to neurogenesis. Dev Biol. 1996;180:664–679. doi: 10.1006/dbio.1996.0336. [DOI] [PubMed] [Google Scholar]
- 119.Ohsugi M, Larue L, Schwarz H, Kemler R. Cell-junctional and cytoskeletal organization in mouse blastocysts lacking E-cadherin. Dev Biol. 1997;185:261–271. doi: 10.1006/dbio.1997.8560. [DOI] [PubMed] [Google Scholar]
- 120.Ohsugi M, Hwang SY, Butz S, Knowles BB, Solter D, Kemler R. Expression and cell membrane localization of catenins during mouse preimplantation development. Dev Dyn. 1996;206:391–402. doi: 10.1002/(SICI)1097-0177(199608)206:4<391::AID-AJA5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 121.De Vries WN, Evsikov AV, Haac BE, Fancher KS, Holbrook AE, Kemler R, et al. Maternal beta-catenin and E-cadherin in mouse development. Development. 2004;131:4435–4445. doi: 10.1242/dev.01316. [DOI] [PubMed] [Google Scholar]
- 122.Bedzhov I, Liszewska E, Kanzler B, Stemmler MP. Igf1r signaling is indispensable for preimplantation development and is activated via a novel function of E-cadherin. PLoS Genet. 2012;8:e1002609. doi: 10.1371/journal.pgen.1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kan NG, Stemmler MP, Junghans D, Kanzler B, de Vries WN, Dominis M, et al. Gene replacement reveals a specific role for E-cadherin in the formation of a functional trophectoderm. Development. 2007;134:31–41. doi: 10.1242/dev.02722. [DOI] [PubMed] [Google Scholar]
- 124.Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci U S A. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fierro-González JC, White MD, Silva JC, Plachta N. Cadherin-dependent filopodia control preimplantation embryo compaction. Nat Cell Biol. 2013;15:1424–1433. doi: 10.1038/ncb2875. [DOI] [PubMed] [Google Scholar]
- 126.Riethmacher D, Brinkmann V, Birchmeier C. A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc Natl Acad Sci U S A. 1995;92:855–859. doi: 10.1073/pnas.92.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Edelman GM, Gallin WJ, Delouvée A, Cunningham BA, Thiery JP. Early epochal maps of two different cell adhesion molecules. Proc Natl Acad Sci U S A. 1983;80:4384–4388. doi: 10.1073/pnas.80.14.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Choi YS, Gumbiner B. Expression of cell adhesion molecule E-cadherin in Xenopus embryos begins at gastrulation and predominates in the ectoderm. J Cell Biol. 1989;108:2449–2458. doi: 10.1083/jcb.108.6.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Levi G, Gumbiner B, Thiery JP. The distribution of E-cadherin during Xenopus laevis development. Development. 1991;111:159–169. doi: 10.1242/dev.111.1.159. [DOI] [PubMed] [Google Scholar]
- 130.Nandadasa S, Tao Q, Menon NR, Heasman J, Wylie C. N- and E-cadherins in Xenopus are specifically required in the neural and non-neural ectoderm, respectively, for F-actin assembly and morphogenetic movements. Development. 2009;136:1327–1338. doi: 10.1242/dev.031203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vestweber D, Kemler R. Rabbit antiserum against a purified surface glycoprotein decompacts mouse preimplantation embryos and reacts with specific adult tissues. Exp Cell Res. 1984;152:169–178. doi: 10.1016/0014-4827(84)90241-6. [DOI] [PubMed] [Google Scholar]
- 132.Butz S, Larue L. Expression of catenins during mouse embryonic development and in adult tissues. Cell Adhes Commun. 1995;3:337–352. doi: 10.3109/15419069509081018. [DOI] [PubMed] [Google Scholar]
- 133.Damjanov I, Damjanov A, Damsky CH. Developmentally regulated expression of the cell-cell adhesion glycoprotein cell-CAM 120/80 in peri-implantation mouse embryos and extraembryonic membranes. Dev Biol. 1986;116:194–202. doi: 10.1016/0012-1606(86)90056-4. [DOI] [PubMed] [Google Scholar]
- 134.Thiery JP, Delouvée A, Gallin WJ, Cunningham BA, Edelman GM. Ontogenetic expression of cell adhesion molecules: L-CAM is found in epithelia derived from the three primary germ layers. Dev Biol. 1984;102:61–78. doi: 10.1016/0012-1606(84)90175-1. [DOI] [PubMed] [Google Scholar]
- 135.Rogers CD, Sorrells LK, Bronner ME. A catenin-dependent balance between N-cadherin and E-cadherin controls neuroectodermal cell fate choices. Mech Dev. 2018;152:44–56. doi: 10.1016/j.mod.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kimura Y, Matsunami H, Inoue T, Shimamura K, Uchida N, Ueno T, et al. Cadherin-11 expressed in association with mesenchymal morphogenesis in the head, somite, and limb bud of early mouse embryos. Dev Biol. 1995;169:347–358. doi: 10.1006/dbio.1995.1149. [DOI] [PubMed] [Google Scholar]
- 137.Radice GL, Rayburn H, Matsunami H, Knudsen KA, Takeichi M, Hynes RO. Developmental defects in mouse embryos lacking N-cadherin. Dev Biol. 1997;181:64–78. doi: 10.1006/dbio.1996.8443. [DOI] [PubMed] [Google Scholar]
- 138.Kuriyama S, Theveneau E, Benedetto A, Parsons M, Tanaka M, Charras G, et al. In vivo collective cell migration requires an LPAR2-dependent increase in tissue fluidity. J Cell Biol. 2014;206:113–127. doi: 10.1083/jcb.201402093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tuttle AM, Hoffman TL, Schilling TF. Rabconnectin-3a regulates vesicle endocytosis and canonical Wnt signaling in zebrafish neural crest migration. PLoS Biol. 2014;12:e1001852. doi: 10.1371/journal.pbio.1001852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dias A, Lozovska A, Wymeersch FJ, Nóvoa A, Binagui-Casas A, Sobral D, et al. A Tgfbr1/Snai1-dependent developmental module at the core of vertebrate axial elongation. Elife. 2020 doi: 10.7554/eLife.56615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, et al. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 142.Kadokawa Y, Fuketa I, Nose A, Takeichi M, Nakatsuji N (1989) Expression pattern of E‐ and P‐Cadherin in mouse embryos and uteri during the periimplantation period. Dev Growth Diff 31(1):23–30. [DOI] [PubMed]
- 143.Richardson L, Venkataraman S, Stevenson P, Yang Y, Moss J, Graham L, et al. EMAGE mouse embryo spatial gene expression database: 2014 update. Nucleic Acids Res. 2014;42:D835–D844. doi: 10.1093/nar/gkt1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Acloque H, Ocaña OH, Abad D, Stern CD, Nieto MA. Snail2 and Zeb2 repress P-cadherin to define embryonic territories in the chick embryo. Development. 2017;144:649–656. doi: 10.1242/dev.142562. [DOI] [PubMed] [Google Scholar]
- 145.Locascio A, Manzanares M, Blanco MJ, Nieto MA. Modularity and reshuffling of Snail and Slug expression during vertebrate evolution. Proc Natl Acad Sci U S A. 2002;99:16841–16846. doi: 10.1073/pnas.262525399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Inoue T, Chisaka O, Matsunami H, Takeichi M. Cadherin-6 expression transiently delineates specific rhombomeres, other neural tube subdivisions, and neural crest subpopulations in mouse embryos. Dev Biol. 1997;183:183–194. doi: 10.1006/dbio.1996.8501. [DOI] [PubMed] [Google Scholar]
- 147.Matsunami H, Takeichi M. Fetal brain subdivisions defined by R- and E-cadherin expressions: evidence for the role of cadherin activity in region-specific, cell-cell adhesion. Dev Biol. 1995;172:466–478. doi: 10.1006/dbio.1995.8029. [DOI] [PubMed] [Google Scholar]
- 148.Tsai TY-C, Sikora M, Xia P, Colak-Champollion T, Knaut H, Heisenberg C-P, et al. An adhesion code ensures robust pattern formation during tissue morphogenesis. Science. 2020;370:113–116. doi: 10.1126/science.aba6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rhinn M, Dierich A, Le Meur M, Ang S. Cell autonomous and non-cell autonomous functions of Otx2 in patterning the rostral brain. Development. 1999;126:4295–4304. doi: 10.1242/dev.126.19.4295. [DOI] [PubMed] [Google Scholar]
- 150.Gänzler SI, Redies C.(1995) R-cadherin expression during nucleus formation in chicken forebrain neuromeres. J Neurosci 15(6):4157–4172 [DOI] [PMC free article] [PubMed]
- 151.Brunskill EW, Potter AS, Distasio A, Dexheimer P, Plassard A, Aronow BJ, et al. A gene expression atlas of early craniofacial development. Dev Biol. 2014;391:133–146. doi: 10.1016/j.ydbio.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lin J, Wang C, Redies C. Restricted expression of classic cadherins in the spinal cord of the chicken embryo. Front Neuroanat. 2014;8:18. doi: 10.3389/fnana.2014.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Choi YS, Sehgal R, McCrea P, Gumbiner B. A cadherin-like protein in eggs and cleaving embryos of Xenopus laevis is expressed in oocytes in response to progesterone. J Cell Biol. 1990;110:1575–1582. doi: 10.1083/jcb.110.5.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ginsberg D, DeSimone D, Geiger B. Expression of a novel cadherin (EP-cadherin) in unfertilized eggs and early Xenopus embryos. Development. 1991;111:315–325. doi: 10.1242/dev.111.2.315. [DOI] [PubMed] [Google Scholar]
- 155.Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, et al. Overexpression of cadherins and underexpression of beta-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell. 1994;79:791–803. doi: 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 156.Angres B, Müller AH, Kellermann J, Hausen P. Differential expression of two cadherins in Xenopus laevis. Development. 1991;111:829–844. doi: 10.1242/dev.111.3.829. [DOI] [PubMed] [Google Scholar]
- 157.Redies C, Neudert F, Lin J. Cadherins in cerebellar development: translation of embryonic patterning into mature functional compartmentalization. Cerebellum. 2011;10:393–408. doi: 10.1007/s12311-010-0207-4. [DOI] [PubMed] [Google Scholar]
- 158.Veeraval L, O’Leary CJ, Cooper HM. Adherens junctions: guardians of cortical development. Front Cell Dev Biol. 2020;8:6. doi: 10.3389/fcell.2020.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rousso DL, Pearson CA, Gaber ZB, Miquelajauregui A, Li S, Portera-Cailliau C, et al. Foxp-mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron. 2012;74:314–330. doi: 10.1016/j.neuron.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kasioulis I, Storey KG. Cell biological mechanisms regulating chick neurogenesis. Int J Dev Biol. 2018;62:167–175. doi: 10.1387/ijdb.170268ks. [DOI] [PubMed] [Google Scholar]
- 161.Stepniak E, Radice GL, Vasioukhin V. Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harb Perspect Biol. 2009;1:a002949. doi: 10.1101/cshperspect.a002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Martinez-Garay I. Molecular mechanisms of cadherin function during cortical migration. Front Cell Dev Biol. 2020;8:588152. doi: 10.3389/fcell.2020.588152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Astick M, Tubby K, Mubarak WM, Guthrie S, Price SR. Central topography of cranial motor nuclei controlled by differential cadherin expression. Curr Biol. 2014;24:2541–2547. doi: 10.1016/j.cub.2014.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Knüfer A, Diana G, Walsh GS, Clarke JD, Guthrie S. Cadherins regulate nuclear topography and function of developing ocular motor circuitry. Elife. 2020 doi: 10.7554/eLife.56725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yamagata M, Herman JP, Sanes JR. Lamina-specific expression of adhesion molecules in developing chick optic tectum. J Neurosci. 1995;15:4556–4571. doi: 10.1523/JNEUROSCI.15-06-04556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- 167.Stan A, Pielarski KN, Brigadski T, Wittenmayer N, Fedorchenko O, Gohla A, et al. Essential cooperation of N-cadherin and neuroligin-1 in the transsynaptic control of vesicle accumulation. Proc Natl Acad Sci U S A. 2010;107:11116–11121. doi: 10.1073/pnas.0914233107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mendonsa AM, Na T-Y, Gumbiner BM. E-cadherin in contact inhibition and cancer. Oncogene. 2018;37:4769–4780. doi: 10.1038/s41388-018-0304-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Miyamoto Y, Sakane F, Hashimoto K. N-cadherin-based adherens junction regulates the maintenance, proliferation, and differentiation of neural progenitor cells during development. Cell Adh Migr. 2015;9:183–192. doi: 10.1080/19336918.2015.1005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Larue L, Antos C, Butz S, Huber O, Delmas V, Dominis M, et al. A role for cadherins in tissue formation. Development. 1996;122:3185–3194. doi: 10.1242/dev.122.10.3185. [DOI] [PubMed] [Google Scholar]
- 171.Basilicata MF, Frank M, Solter D, Brabletz T, Stemmler MP. Inappropriate cadherin switching in the mouse epiblast compromises proper signaling between the epiblast and the extraembryonic ectoderm during gastrulation. Sci Rep. 2016;6:26562. doi: 10.1038/srep26562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hashimoto H, Munro E. Differential expression of a classic cadherin directs tissue-level contractile asymmetry during neural tube closure. Dev Cell. 2019;51:158–172.e4. doi: 10.1016/j.devcel.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Moore R, Radice GL, Dominis M, Kemler R. The generation and in vivo differentiation of murine embryonal stem cells genetically null for either N-cadherin or N- and P-cadherin. Int J Dev Biol. 1999;43:831–834. [PubMed] [Google Scholar]
- 174.Radice GL, Ferreira-Cornwell MC, Robinson SD, Rayburn H, Chodosh LA, Takeichi M, et al. Precocious mammary gland development in P-cadherin–deficient mice. J Cell Biol. 1997;139:1025–1032. doi: 10.1083/jcb.139.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lele Z, Folchert A, Concha M, Rauch G-J, Geisler R, Rosa F, et al. parachute/n-cadherin is required for morphogenesis and maintained integrity of the zebrafish neural tube. Development. 2002;129:3281–3294. doi: 10.1242/dev.129.14.3281. [DOI] [PubMed] [Google Scholar]
- 176.Hong E, Brewster R. N-cadherin is required for the polarized cell behaviors that drive neurulation in the zebrafish. Development. 2006;133:3895–3905. doi: 10.1242/dev.02560. [DOI] [PubMed] [Google Scholar]
- 177.Chen D, Wu Z, Luo L-J, Huang X, Qian W-Q, Wang H, et al. E-cadherin maintains the activity of neural stem cells and inhibits the migration. Int J Clin Exp Pathol. 2015;8:14247–14251. [PMC free article] [PubMed] [Google Scholar]
- 178.Chou Y-F, Chen H-H, Eijpe M, Yabuuchi A, Chenoweth JG, Tesar P, et al. The growth factor environment defines distinct pluripotent ground states in novel blastocyst-derived stem cells. Cell. 2008;135:449–461. doi: 10.1016/j.cell.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Soncin F, Mohamet L, Eckardt D, Ritson S, Eastham AM, Bobola N, et al. Abrogation of E-cadherin-mediated cell-cell contact in mouse embryonic stem cells results in reversible LIF-independent self-renewal. Stem Cells. 2009;27:2069–2080. doi: 10.1002/stem.134. [DOI] [PubMed] [Google Scholar]
- 180.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 181.Redmer T, Diecke S, Grigoryan T, Quiroga-Negreira A, Birchmeier W, Besser D. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011;12:720–726. doi: 10.1038/embor.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.del Valle I, Rudloff S, Carles A, Li Y, Liszewska E, Vogt R, et al. E-cadherin is required for the proper activation of the Lifr/Gp130 signaling pathway in mouse embryonic stem cells. Development. 2013;140:1684–1692. doi: 10.1242/dev.088690. [DOI] [PubMed] [Google Scholar]
- 183.Faunes F, Hayward P, Descalzo SM, Chatterjee SS, Balayo T, Trott J, et al. A membrane-associated-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development. 2013;140:1171–1183. doi: 10.1242/dev.085654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Livigni A, Peradziryi H, Sharov AA, Chia G, Hammachi F, Migueles RP, et al. A conserved Oct4/POUV-dependent network links adhesion and migration to progenitor maintenance. Curr Biol. 2013;23:2233–2244. doi: 10.1016/j.cub.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Takehara T, Teramura T, Onodera Y, Frampton J, Fukuda K. Cdh2 stabilizes FGFR1 and contributes to primed-state pluripotency in mouse epiblast stem cells. Sci Rep. 2015;5:14722. doi: 10.1038/srep14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Malaguti M, Nistor PA, Blin G, Pegg A, Zhou X, Lowell S. Bone morphogenic protein signalling suppresses differentiation of pluripotent cells by maintaining expression of E-Cadherin. Elife. 2013 doi: 10.7554/elife.01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Punovuori K, Migueles RP, Malaguti M, Blin G, Macleod KG, Carragher NO, et al. N-cadherin stabilises neural identity by dampening anti-neural signals. Development. 2019;146:dev183269. doi: 10.1242/dev.183269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chan CJ, Heisenberg C-P, Hiiragi T. Coordination of morphogenesis and cell-fate specification in development. Curr Biol. 2017;27:R1024–R1035. doi: 10.1016/j.cub.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 189.Gilmour D, Rembold M, Leptin M. From morphogen to morphogenesis and back. Nature. 2017;541:311–320. doi: 10.1038/nature21348. [DOI] [PubMed] [Google Scholar]
- 190.Busby L, Steventon B. Tissue tectonics and the multi-scale regulation of developmental timing. 2020. Available: https://www.preprints.org/manuscript/202006.0168. Accessed June 2020 [DOI] [PMC free article] [PubMed]
- 191.Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J Cell Sci. 2008;121:727–735. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 192.Pieters T, van Roy F. Role of cell–cell adhesion complexes in embryonic stem cell biology. J Cell Sci. 2014;127:2603–2613. doi: 10.1242/jcs.146720. [DOI] [PubMed] [Google Scholar]
- 193.Sanson B, White P, Vincent J-P. Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature. 1996;383:627–630. doi: 10.1038/383627a0. [DOI] [PubMed] [Google Scholar]
- 194.Howard S, Deroo T, Fujita Y, Itasaki N. A positive role of cadherin in Wnt/β-catenin signalling during epithelial-mesenchymal transition. PLoS ONE. 2011;6:e23899. doi: 10.1371/journal.pone.0023899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Przybyla L, Lakins JN, Weaver VM. Tissue mechanics orchestrate Wnt-dependent human embryonic stem cell differentiation. Cell Stem Cell. 2016;19:462–475. doi: 10.1016/j.stem.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hendriksen J, Jansen M, Brown CM, van der Velde H, van Ham M, Galjart N, et al. Plasma membrane recruitment of dephosphorylated beta-catenin upon activation of the Wnt pathway. J Cell Sci. 2008;121:1793–1802. doi: 10.1242/jcs.025536. [DOI] [PubMed] [Google Scholar]
- 197.Schafer G, Narasimha M, Vogelsang E, Leptin M. Cadherin switching during the formation and differentiation of the Drosophila mesoderm—implications for epithelial-to-mesenchymal transitions. J Cell Sci. 2014 doi: 10.1242/jcs.139485. [DOI] [PubMed] [Google Scholar]
- 198.Suyama K, Shapiro I, Guttman M, Hazan RB. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell. 2002;2:301–314. doi: 10.1016/s1535-6108(02)00150-2. [DOI] [PubMed] [Google Scholar]
- 199.Kotb AM, Hierholzer A, Kemler R. Replacement of E-cadherin by N-cadherin in the mammary gland leads to fibrocystic changes and tumor formation. Breast Cancer Res. 2011;13:R104. doi: 10.1186/bcr3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Benn A, Bredow C, Casanova I, Vukičević S, Knaus P. VE-cadherin facilitates BMP-induced endothelial cell permeability and signaling. J Cell Sci. 2016;129:206–218. doi: 10.1242/jcs.179960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Martyn I, Kanno TY, Ruzo A, Siggia ED, Brivanlou AH. Author Correction: self-organization of a human organizer by combined Wnt and Nodal signalling. Nature. 2018;564:E10. doi: 10.1038/s41586-018-0583-3. [DOI] [PubMed] [Google Scholar]