Abstract
Background
XRCC2, a homologous recombination-related gene, has been reported to be associated with a variety of cancers. However, its role in glioma has not been reported. This study aimed to find out the role of XRCC2 in glioma and reveal in which glioma-specific biological processes is XRCC2 involved based on thousands of glioma samples, thereby, providing a new perspective in the treatment and prognostic evaluation of glioma.
Methods
The expression characteristics of XRCC2 in thousands of glioma samples from CGGA and TCGA databases were comprehensively analyzed. Wilcox or Kruskal test was used to analyze the expression pattern of XRCC2 in gliomas with different clinical and molecular features. The effect of XRCC2 on the prognosis of glioma patients was explored by Kaplan–Meier and Cox regression. Gene set enrichment analysis (GSEA) revealed the possible cellular mechanisms involved in XRCC2 in glioma. Connectivity map (CMap) was used to screen small molecule drugs targeting XRCC2 and the expression levels of XRCC2 were verified in glioma cells and tissues by RT-qPCR and immunohistochemical staining.
Results
We found the overexpression of XRCC2 in glioma. Moreover, the overexpressed XRCC2 was associated with a variety of clinical features related to prognosis. Cox and meta-analyses showed that XRCC2 is an independent risk factor for the poor prognosis of glioma. Furthermore, the results of GSEA indicated that overexpressed XRCC2 could promote malignant progression through involved signaling pathways, such as in the cell cycle. Finally, doxazosin, quinostatin, canavanine, and chrysin were identified to exert anti-glioma effects by targeting XRCC2.
Conclusions
This study analyzed the expression pattern of XRCC2 in gliomas and its relationship with prognosis using multiple datasets. This is the first study to show that XRCC2, a novel oncogene, is significantly overexpressed in glioma and can lead to poor prognosis in glioma patients. XRCC2 could serve as a new biomarker for glioma diagnosis, treatment, and prognosis evaluation, thus bringing new insight into the management of glioma.
Supplementary Information
The online version contains supplementary material available at 10.1186/s10020-021-00316-0.
Keywords: Glioma, XRCC2, Oncogene, Prognosis, Biomarker
Background
Glioma is the most common primary tumor among adult brain tumors and has high mortality and disability rates (Tan 2020). Currently, the treatment of glioma is mainly surgical intervention and postoperative adjuvant chemoradiotherapy (Lim et al. 2018). Although there are many new methods for treating glioma (Pulkkanen and Yla-Herttuala 2005; Kamran 2018; Mahmoudi 2019), the prognosis of glioma patients remains unsatisfactory. Targeted therapy attacking specific genes within the tumorous cells may provide new ways to kill glioma cells and improve the prognosis of patients. Therefore, it is vital to find new and effective molecular targets for the diagnosis and treatment of glioma.
In recent years, research on molecular targeted therapy has proliferated greatly, and genes related to glioma diagnosis, treatment, and prognosis have been screened and identified. For instance, the B-RAF proto-oncogene encoded protein has a regulatory role in the MAPK/ERK signaling pathway, which regulates glioma cell division and differentiation (Lassaletta 2017); KRAS mutations are associated with malignant progression of glioma (Fioravanzo 2019); the zinc finger E-box binding homeobox 1 can inhibit the transcription of interleukin-2 and inhibit the invasion and epithelial-mesenchymal transition of glioma cells (Siebzehnrubl 2013). Although these studies have enriched the molecular pool related to glioma diagnosis and treatment, the prognosis of glioma patients is yet to be significantly improved, which may be due to the fact that the malignant progression of gliomas is triggered by a variety of complex factors rather than by a single oncogene. Therefore, more genes need to be screened and identified to enrich the molecular pool for glioma diagnosis and treatment, thus bringing hope for improving the prognosis of glioma patients.
The malignant transformation of a normal cell is the result of multiple factors, among which DNA double-strand breaks (DSBs) are the most lethal form of cell damage (Ceccaldi et al. 2016; Hoppe et al. 2018; Frappart 2009). As one of the repair methods for DSBs, homologous recombination plays an important role in maintaining the stability of chromosomes and inhibiting the infinite replication of cells; the dysfunction of this repair method can increase the risk of cancer (Hoppe et al. 2018; Chen 2018; Hallam et al. 2015). It is well known that RAD51, a DNA repair protein, plays a central role in regulating the homologous recombination repair process of DSBs (Shen 2018; Cruz 2018). Moreover, studies have shown that after ionizing radiation exposure, RAD51 participates in cell cycle changes in glioblastoma stem cells (Tachon 2018). Furthermore, Ohba et al. found that RAD51-mediated homologous recombination showed an increasing trend in isocitrate dehydrogenase 1(IDH1) -mutant gliomas (Ohba et al. 2014). XRCC2, a homologous gene of RAD51, can form a complex with three RAD51 paralogs (RAD51B, RAD51C, and RAD51D), and it participates in homologous recombination by encoding members of the RECA/RAD51-related protein family (Baldock 2019; Andreassen and Hanenberg 2019). Most studies consider XRCC2 to be a breast cancer susceptibility gene and have demonstrated that it plays an important role in its pathophysiology (Kleibl and Kristensen 2016; Lin 2011). Previous studies have also shown that XRCC2 is abnormally expressed in a variety of tumors, including rectal and breast cancers, and that it participates in many cell signaling pathways (Chen 2018; Andreassen and Hanenberg 2019; Xu 2014; Bashir et al. 2014). In addition, XRCC1 has been reported as one of the possible genes involved in glioma prognosis (Jiang 2013). However, the role and expression pattern of XRCC2 in glioma have not been elucidated so far.
Currently, computational research has opened new avenues for advanced research on cancer management. Many recent reports have used in silico approaches to predict the potential role of numerous genes and proteins in the development of different types of cancer (Andreassen and Hanenberg 2019; Khan et al. 2017; Li 2020). Therefore, we attempted to clarify the expression pattern of XRCC2 in glioma through a joint analysis of multiple databases, to explore the correlation between the expression pattern of XRCC2 and the clinical and molecular characteristics of glioma, and to reveal the value of XRCC2 in the prognostic evaluation of glioma. Overall, we aimed to identify a novel therapeutic target for glioma and to contribute to the enlargement of the molecular pool for glioma diagnosis and treatment.
Materials and methods
Data collection
Gene Expression Profiling Interactive Analysis 2 (GEPIA2, http://gepia2.cancer-pku.cn/) integrates RNA-seq data from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases (Tang et al. 2019; Consortium GT 2015). We used GEPIA2 to preliminarily explore the expression of XRCC2 in different cancers and their corresponding normal tissues, including 163 glioblastoma samples and 207 normal brain tissue samples. Since the database is an online analysis platform, the expression level of XRCC2 in different tumors can be obtained by inputting XRCC2 in the search page.
The Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn/) database contains thousands of glioma gene expression data and the corresponding clinical information. We downloaded 1018 mRNA sequencing and 301 mRNA microarray data from the CGGA database, of which the mRNA sequencing and mRNA microarray data with complete clinical information were 748 and 268, respectively. The original data in the database has been standardized, so after downloading the original data, the expression of XRCC2 in samples may be extracted for further analysis.
TCGA (https://portal.gdc.cancer.gov/) is currently the most significant public database for cancer research. We downloaded 698 glioma transcriptome data from the TCGA database (Workflow Type: HTSeq-FPKM), and the remaining 653 data after the removal of incomplete clinical information were used as validation data sets. As the downloaded data types were standardized, there was no need for further processing, only the expression level changes of XRCC2 in various tissue samples were extracted for further analysis.
Further, we downloaded five glioma datasets from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database, including 279 glioma samples for meta-analysis to validate the effect of XRCC2 on glioma prognosis. They were GSE4412 containing 85 (platform: GPL96), GSE43378 containing 50 (platform: GPL570), GSE74187 containing 60 (platform: GPL6480), GSE50025 containing 34 (platform: GPL13938), and GSE83300 containing 50 samples of glioma patients (platform: GPL6480).
Firstly, the platform information corresponding to the five different datasets was replaced under the command of Perl software. Subsequently, all datasets were normalized one by one using the program in the limma package of R software. Finally, XRCC2 expression level and survival time and status of corresponding glioma patients were selected for further meta-analysis.
Cell culture and preparation of glioma samples
The glioma cell lines LN229, A172, U251, and T98, and the human astrocyte (HA) cell line were purchased from Procell Life Science & Technology Co. Ltd (Wuhan, China). Cells were cultured in DMEM high-sugar medium (Procell, PM150210) containing 10% fetal bovine serum (Gibco, lot: 10099-141c) and 1% penicillin–streptomycin mixture in a 37 °C humidified incubator with 5% carbon dioxide.
Twenty-three glioma samples from 11 women and 12 men were included in this study, with an average age of 50.35 years. Of these, six were world health organization (WHO) grade II, five cases were WHO grade III, and 12 were WHO grade IV Table 1. Nine non-glioma tissues served as controls. All tissues were rapidly frozen in liquid nitrogen within 15 min after surgical resection, and all glioma patients were diagnosed by professional pathologists. All patients provided written informed consent in accordance with the Declaration of Helsinki. The study protocol was approved by the Ethics Committee of Zhengzhou University.
Table 1.
Characteristics of patients with glioma based on clinical samples
| Characteristic | Number (%) | Low expression | High expression | P value |
|---|---|---|---|---|
| Gender | ||||
| Female | 11 (47.83) | 3 | 8 | NS |
| Male | 12 (52.17) | 7 | 5 | |
| Age (year) | ||||
| > 50.35 | 14 (60.87) | 7 | 7 | NS |
| ≤ 50.35 | 9 (39.13) | 3 | 6 | |
| Grade | ||||
| WHO II | 6 (26.09) | 5 | 1 | 0.019* |
| WHO III | 5 (21.74) | 3 | 2 | |
| WHO IV | 12 (52.17) | 2 | 10 | |
| Primary recurrent state | ||||
| Primary | 19 (0.83) | 8 | 11 | NS |
| Recurrent | 4 (0.17) | 2 | 2 | |
The high and low XRCC2 expression groups were divided according to the mean expression level of XRCC2. NS not significant. *Chi-Square Test
RNA isolation and real-time quantitative polymerase chain reaction (RT-qPCR)
According to the manufacturer’s instructions, total RNA was extracted using Trizol (Invitrogen, lot: 262306), cDNA was obtained using NovoScript Plus All-in-one 1st Strand cDNA Synthesis SuperMix (gDNA Purge) (Novoprotein, E047), and RT-qPCR was performed using 2x RealStar Green Fast Mixture with ROX (GenStar, A303-05). RNA-specific primer sequences for the internal reference gene GAPDH were forward: 5′-AAGAAGGTGGTGAAGCAGG-3′ and reverse: 5′-GTCAAAGGTGGAGGAGTGG-3′. The specific primer sequences of the target gene XRCC2 were forward: 5′-GAGCACAGACTATCCCAAAG-3′ and reverse: 5′-CAGGCTATCCAATCAAAA-3'.
Immunohistochemical staining
After deparaffinization and hydration in xylene, graded alcohols, paraffin tissue sections with a thickness of 3 µm were subjected to antigen retrieval using microwave retrieval for 15 min (EDTA, pH 8.0). Then 100 µL of primary antibodies against XRCC2 (ab180752, 1:100 dilution) was dropped onto the tissue sections and incubated overnight at 4 °C. Horseradish peroxidase-labeled IgG polymer (PV6000, Zhongshan Jinqiao Biotechnology, China) as a secondary antibody incubated the tissues for 40 min at room temperature condition. Finally, 3,3′-diaminobenzidine Kit (ZLI-9017, Zhongshan Jinqiao Biotechnology, China) was added as a chromogen, and the sections were evaluated using a light microscope.
Meta-analysis
We first determined the relationship between XRCC2 and glioma prognosis by systematically searching the studies of XRCC2 in glioma in PubMed and Web of Science databases. Since this is the first study to investigate the prognostic role of XRCC2 in glioma, no published studies relating XRCC2 with glioma prognosis were obtained from public databases. Therefore, we utilized the meta-analysis to evaluate the overall prognostic significance of XRCC2 in glioma patients based on eight datasets using R software (version 3.6.3), and data were extracted and selected according to Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) guidelines (Keerthana and Kumar 2020). The association of XRCC2 expression with the prognosis of glioma patients was evaluated by calculating the combined hazard ration (HR) and 95% confidence interval (CI). The heterogeneity of the eight datasets was assessed by the Q test (I2 statistics). I2 < 50% was considered as moderate heterogeneity and a fixed effects model was chosen for combination. Otherwise, random effects model was applied. The Begg and Egger funnel chart was used to assess the possibility of publication bias. However, this part of the content has not been presented separately, because when there are fewer than 10 studies in the Meta-analysis, the results of the publication bias test are considered unreliable (Cumpston 2019; Dalton et al. 2016; Egger et al. 1997). This study was registered with PROSPERO (ID: CRD42021245507).
Gene set enrichment analysis of XRCC2
GSEA is a very convenient tool for bioinformatics analysis, and it can be used to identify the disease-related signaling pathways in which specific genes participate. We used SVA and limma software packages to normalize and correct data downloaded from the TCGA and CCGA databases. The high and low XRCC2 expression groups were defined according to the expression level of XRCC2 in glioma. GSEA 4.0.jar software was used to evaluate the enrichment of signaling pathways. The number of permutations was 1000, and the gene database was associated with Kyoto Encyclopedia of Genes and Genomes (KEGG) cell signaling pathways.
CMap analysis
Connectivity map (CMap, https://portals.broadinstitute.org/cmap/) was used to explore small molecule drugs targeting XRCC2 in glioma. We first identified genes that were co-expressed with XRCC2 based on CGGA RNA-seq data using Pearson correlation analysis. Genes positively and negatively correlated with XRCC2 were then uploaded to the CMaP online tool, and P < 0.01 and enrichment < 0.8 were used as criteria to identify the small molecule drugs targeting XRCC2.
Statistical analysis
Statistical analysis was performed using R software (version 3.6.3). Wilcoxon test or Kruskal–Wallis test was used to analyze differences in XRCC2 expression in gliomas with different clinical and molecular characteristics. Kaplan–Meier and Cox regression were used to analyze the impact of XRCC2 on the prognosis of glioma patients and its value in prognostic diagnosis. Pearson correlation was used to analyze genes co-expressed with XRCC2 in gliomas. The expression level of XRCC2 in glioma cells and tissues was evaluated using a relative quantitative method based on the results of three independent experiments. GraphPad Prism 8.0 was used to analyze the differences between groups, and P < 0.05 was defined as statistically significant.
Results
Data characteristics
Glioma transcriptome data and their corresponding clinical information files were downloaded from the CGGA and TCGA databases, and, after deleting incomplete clinical information, 748 CGGA RNA-seq data Additional file 1: Table S1), 268 CGGA microarray data Additional file 2: Table S2), and 653 TCGA RNA-seq data Additional file 3: Table S3) were included. Each dataset contained at least three detailed data on the clinical characteristics of gliomas. The transcriptome data of thousands of glioma samples with complete clinical information obtained from these three data sets guaranteed the feasibility and reliability of subsequent studies.
Overexpression of XRCC2 in glioblastoma multiforme
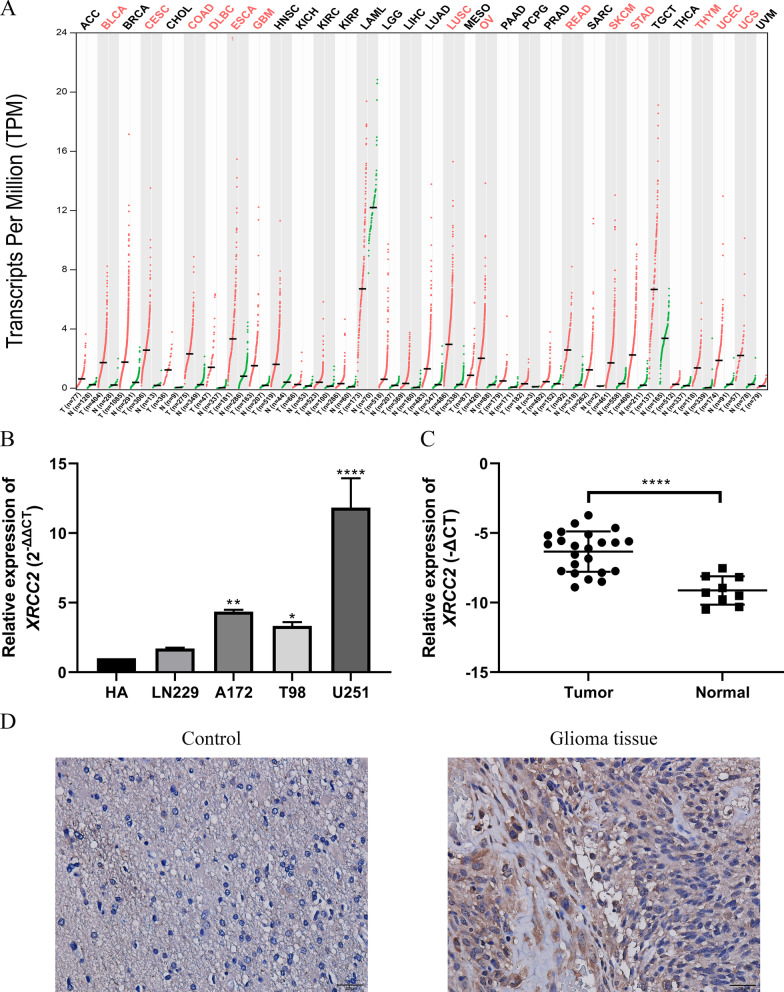
We analyzed the expression of XRCC2 in various cancers and their corresponding normal tissues using the GEPIA2 online platform. As shown in Fig. 1A, we found that the expression level of XRCC2 was significantly increased in various cancers, including colon adenocarcinoma, rectum adenocarcinoma, and glioblastoma multiforme (GBM). In order to clarify the expression pattern of XRCC2 in cancer and its impact on prognosis, we chose GBM as the research object for follow-up study. RT-qPCR results confirmed that the expression level of XRCC2 in glioma cell lines and glioma tissues was significantly higher than that in the corresponding control group Fig. 1B, C. Consistent with the mRNA expression pattern, immunohistochemical staining results indicated XRCC2 expression to be higher in glioma tissues than in benign tissues Fig. 1D.
Fig. 1.
Expression of XRCC2 in cancers. A The expression level of XRCC2 in cancers based on GEPIA2. B The expression level of XRCC2 in glioma cell lines based on RT-qPCR. C The expression level of XRCC2 in glioma tissues based on RT-qPCR. D Immunohistochemical staining results for XRCC2 in non-glioma and glioma tissues. Brown color represents positive XRCC2 staining. *P = 0.0108, **P = 0.001, ****P < 0.0001. GEPIA2 gene expression profiling interactive analysis 2, RT-qPCR real-time quantitative polymerase chain reaction
Overexpressed XRCC2 predicts poor prognosis in glioma
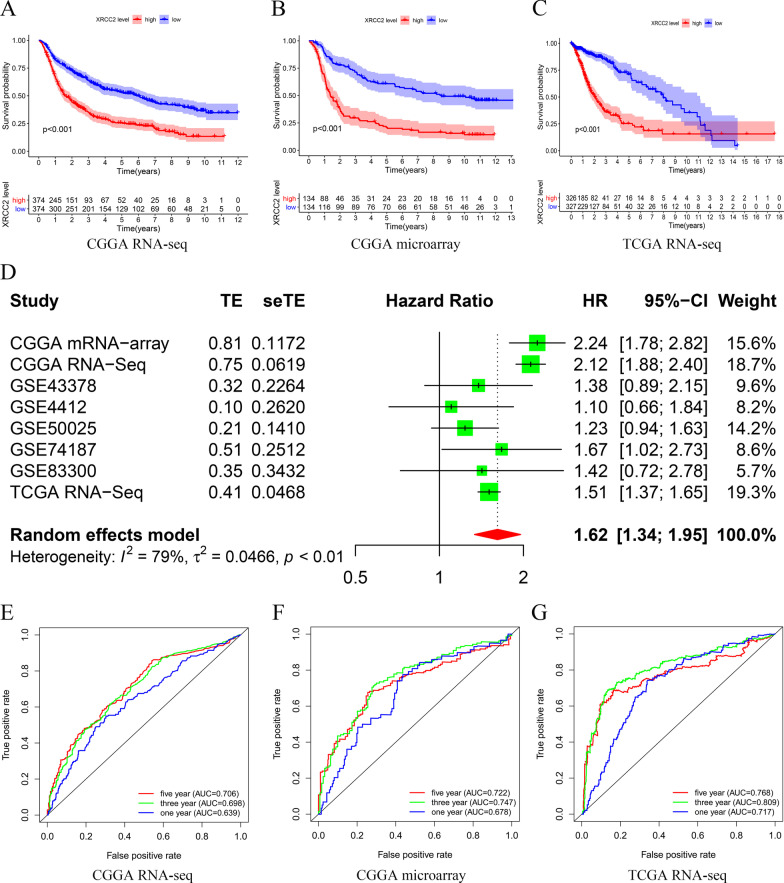
To understand the impact of XRCC2 on the prognosis of glioma, we first used Kaplan–Meier analysis to analyze the relationship between the expression level of XRCC2 and the prognosis of glioma patients based on three different data sets. The results showed that overexpressed XRCC2 could significantly shorten the survival time of glioma patients (P < 0.001, Fig. 2A–C. In the TCGA RNA-seq data set, the survival curves of glioma patients crossed each other, which may be due to the small number of patients in the late follow-up period. Nevertheless, the 5-year and 10-year survival rates of patients with low XRCC2 expression were significantly higher than those of patients with overexpression of XRCC2.
Fig. 2.
Overexpression of XRCC2 predicts poor prognosis. A–C Kaplan–Meier survival curves based on CGGA RNA-seq, CGGA microarray, and TCGA RNA-seq, respectively; D Forest plot of high XRCC2 expression with better overall survival in glioma patients from eight datasets; E–G: ROC curves based on CGGA RNA-seq, CGGA microarray, and TCGA RNA-seq, respectively. CGGA Chinese Glioma Genome Atlas, TCGA The Cancer Genome Atlas, ROC receiver operating characteristic
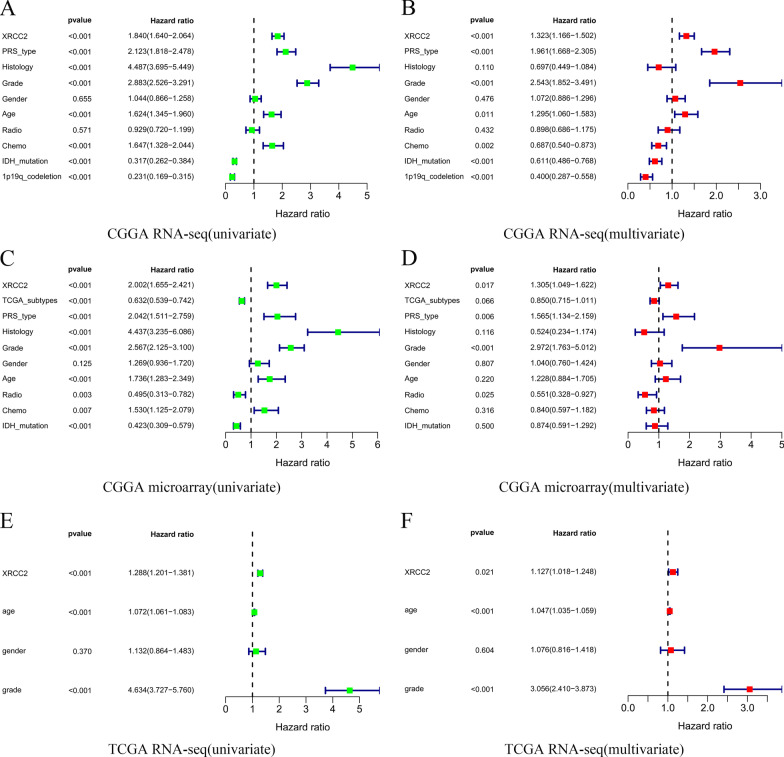
Since XRCC2 has not been previously reported in glioma, to observe the pooled effect of high expression of XRCC2 on patient prognosis, we integrated XRCC2 gene expression and patient survival information from eight datasets in the GEO, TCGA, CGGA databases, and the results of meta-analysis showed that the pooled HR along with 95% CI for the correlation between highly expressed XRCC2 and prognosis was 1.62 (1.34–1.95) Fig. 2D, which indicated that XRCC2 is a risk factor for glioma prognosis. Furthermore, the results of the Cox analysis determined by the area under the receiver operating characteristic (ROC) curve (AUC) values suggested that XRCC2 is potentially useful for the prognostic assessment of glioma, especially for 5-year outcome (P > 0.7, Fig. 2E–G. To further clarify whether XRCC2 is an independent risk factor for the prognosis of glioma patients, we conducted univariate and multivariate analyses. The results indicated that XRCC2 acts as an independent factor in the prediction of prognosis of glioma patients, and this result was mutually validated among three different data sets (P < 0.05, HR > 1, Fig. 3. Taken together, we believe that the overexpression of XRCC2 may be considered an independent risk factor of poor prognosis in glioma patients.
Fig. 3.
XRCC2 is an independent risk factor for poor glioma prognosis. A, C, E Results of the univariate analysis based on CGGA RNA-seq, CGGA microarray, and TCGA RNA-seq, respectively; B, D, F Results of the multivariate analysis based on CGGA RNA-seq, CGGA microarray, and TCGA RNA-seq, respectively. CGGA Chinese Glioma Genome Atlas; TCGA The Cancer Genome Atlas
Relationship between XRCC2 expression and clinical features of glioma
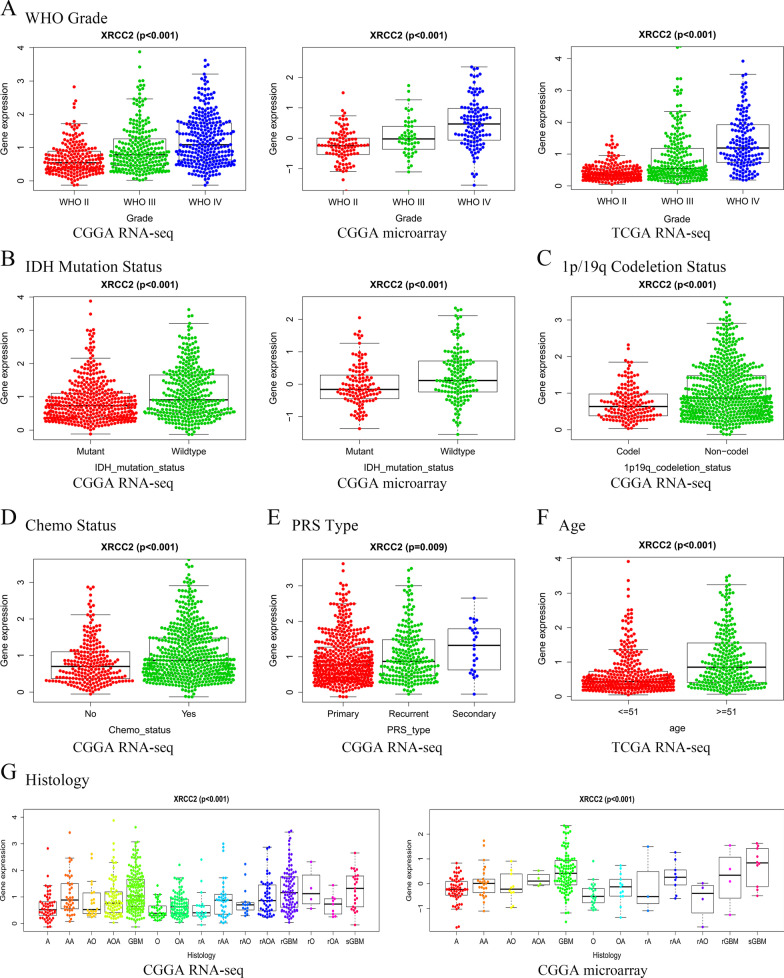
It is well known that the prognosis of glioma patients is associated with a variety of clinical features, such as glioma grade, primary recurrence status of tumors, and mutations in specific molecules. Since the results of this study suggest that overexpression of XRCC2 can lead to poor prognosis, we speculated that different clinical features of gliomas may predict differences in XRCC2 expression. Therefore, we analyzed the expression levels of XRCC2 in gliomas with different characteristics based on the three different data sets. As shown in Fig. 4A, XRCC2 has different expression levels in different grades of glioma. Moreover, the expression level of XRCC2 increased with malignity (P < 0.001). Consistently, the expression level of XRCC2 in recurrent gliomas was higher than that in primary gliomas, and this was maintained for several histological subtypes Fig. 4E, G. Interestingly, the presence of IDH mutation and 1p/19q co-deletion negatively correlated with XRCC2 expression. As shown in Fig. 4B, C, the expression of XRCC2 was reduced in gliomas with IDH mutation and 1p/19q co-deletion (P < 0.001). Since IDH mutation and 1p/19q co-deletion are currently recognized as predictors of better prognosis in glioma patients, this result indirectly supports the adverse effect of XRCC2 on the prognosis of glioma. In addition, XRCC2 has higher expression levels in patients receiving chemotherapy and in elderly patients Fig. 4D, F, P < 0.001). In addition, the samples were divided into high expression and low expression groups according to the average value of XRCC2 expression in the 23 glioma samples. The Chi-Square test showed that the expression level of XRCC2 in glioma tissues was significantly correlated with WHO grade (P = 0.019, Table 1. Simultaneously, we also found that the expression level of XRCC2 was not significant between different ages and different primary-recurrence status based on the results of clinical sample analysis (P > 0.05, Table 1, which might have been because of the relatively small sample size. These results suggest that the expression of XRCC2 is correlated with clinical features associated with the prognosis of gliomas and that highly expressed XRCC2 may be involved in the malignant progression of gliomas through different mechanisms.
Fig. 4.
Correlation between high XRCC2 expression and different clinical features of glioma. A Grade; B IDH mutation presence; C 1p/19q co-deletion presence; D Chemo status; E PRS Type; F Age; G Histology. PRS primary recurrence and secondary
GSEA-revealed XRCC2 signaling pathways involved in glioma progression
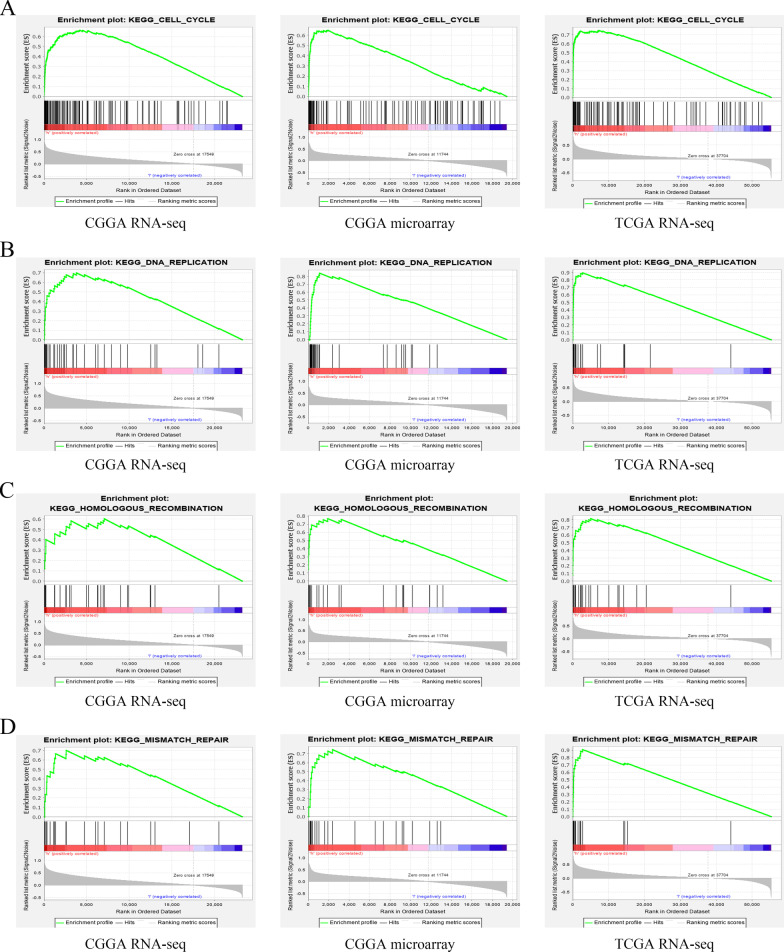
To elucidate the cellular mechanisms by which the overexpressed XRCC2 leads to tumoral progression in glioma, we used GSEA to analyze the data from the CGGA and TCGA databases. The signal pathways of cell cycle, DNA replication, homologous recombination, and mismatch repair obtained higher enrichment scores than the rest. Moreover, this result was mutually validated in the three datasets Fig. 5, Table 2). These results suggest that XRCC2 plays a cancer-promoting role in glioma by participating in different signaling pathways.
Fig. 5.
Enrichment results of GSEA based on different datasets. A Cell-cycle, B DNA-replication; C Homologous-recombination; D Mismatch-repair. GSEA gene set enrichment analysis
Table 2.
Signal pathways involved in XRCC2 in glioma based on three datasets
| Pathways | CGGA RNA-seq | CGGA microarray | TCGA RNA-seq | ||||||
|---|---|---|---|---|---|---|---|---|---|
| NES | NOM P -value | FDR q-value | NES | NOM P -value | FDR q-value | NES | NOM P -value | FDR q-value | |
| Cell cycle | 1.950 | 0.000 | 0.123 | 1.983 | 0.004 | 0.021 | 2.239 | 0.000 | 0.001 |
| DNA replication | 1.715 | 0.029 | 0.146 | 1.966 | 0.000 | 0.017 | 2.048 | 0.000 | 0.005 |
| Homologous recombination | 1.572 | 0.046 | 0.178 | 2.046 | 0.000 | 0.015 | 2.024 | 0.002 | 0.004 |
| Mismatch repair | 1.715 | 0.008 | 0.159 | 1.797 | 0.015 | 0.105 | 2.076 | 0.000 | 0.004 |
NES normalized enrichment score, NOM nominal, FDR false discovery rate. Gene sets with NOM P-value < 0.05 and FDR q-value < 0.25 were considered as significantly enriched, NOM nominal, FDR false discovery rate.
Co-expression analysis
Since XRCC2 was not involved in the malignant progression of glioma through a single cellular mechanism, we speculated that other genes co-express with XRCC2 in glioma. To test our hypothesis, we used a Pearson analysis to explore the genes co-expressed with XRCC2 in gliomas and used a circle map to show the top ten and the last ten genes co-expressed with XRCC2 Additional file 4: Figure S1, Table 3. Combined with the above results, co-expression analysis further demonstrated that XRCC2 is not involved in the malignant progression of glioma on its own, although it can independently affect the prognosis of glioma.
Table 3.
Genes co-expressed with XRCC2
| No. | Gene | Correlation index | P-value |
|---|---|---|---|
| 1 | LDHD | − 0.498 | 7.72E-65 |
| 2 | FBXW4 | − 0.49 | 1.13E-62 |
| 3 | MATK | − 0.483 | 1.06E-60 |
| 4 | CTD-2210P24.4 | − 0.475 | 1.85E-58 |
| 5 | SNCG | − 0.459 | 4.07E-54 |
| 6 | RP11-227B21.2 | − 0.457 | 1.19E-53 |
| 7 | CYP46A1 | − 0.452 | 1.75E-52 |
| 8 | ASPDH | − 0.446 | 4.97E-51 |
| 9 | SNAI3-AS1 | − 0.442 | 4.84E-50 |
| 10 | NEBL-AS1 | − 0.439 | 4.32E-49 |
| 11 | CENPI | 0.853 | 1.14E-288 |
| 12 | KIF14 | 0.852 | 4.75E-288 |
| 13 | MCM8 | 0.848 | 7.06E-282 |
| 14 | CKAP2L | 0.844 | 5.98E-277 |
| 15 | BUB1 | 0.842 | 1.97E-274 |
| 16 | FAM111B | 0.84 | 1.21E-272 |
| 17 | SGOL2 | 0.837 | 1.89E-268 |
| 18 | NCAPG2 | 0.831 | 2.95E-261 |
| 19 | ATAD5 | 0.83 | 4.20E-260 |
| 20 | RAD51AP1 | 0.822 | 3.46E-251 |
Identification of anti-glioma micromolecules targeting XRCC2
Based on the role of XRCC2 in glioma progression, it was necessary to screen for anti-glioma drugs that target XRCC2. Therefore, we used the CMap to explore anti-glioma micromolecular drugs that can target XRCC2. The results of CMap analysis suggested that doxazosin, quinostatin, canavanine, and chrysin have a high potential for clinical application against glioma by targeting XRCC2 Table 4. The two-and three-dimensional structures of these four small molecules are shown in Fig. 6.
Table 4.
Small molecules screened from CMap
| No. | CMap name | Enrichment index | P-value |
|---|---|---|---|
| 1 | Doxazosin | − 0.847 | 0.00101 |
| 2 | Quinostatin | − 0.963 | 0.00308 |
| 3 | Canavanine | − 0.859 | 0.00563 |
| 4 | Chrysin | − 0.809 | 0.0138 |
CMap connectivity map
Fig. 6.
Information regarding small molecules that target XRCC2. A Doxazosin; B Quinostatin; C Canavanine; and D Chrysin
Discussion
XRCC2 is a DNA repair gene that has been found to play an essential role in the development of many cancers (Curtin 2009; Hilbers 2016). Previous reports have suggested that XRCC2 is a target of microsatellite instability in medulloblastoma (Viana-Pereira 2009); however, no studies have revealed the expression pattern of XRCC2 in glioma, analyzed the correlation between XRCC2 and clinical features of glioma, or elucidated the impact of XRCC2 on the prognosis of glioma patients. This study comprehensively analyzed the expression pattern of XRCC2 in glioma and found that overexpression of XRCC2 can reduce the survival rate of glioma patients. Based on the mutual validation of multiple public databases, we identified the possible XRCC2 signaling pathways involved in the malignant progression of glioma and screened four small-molecule drugs with potential clinical application value.
GEPIA2 integrates the tumor data from TCGA database and the data of normal tissue samples from the GTEx database, and currently, it is the best online analysis tool to explore the levels of gene expression in cancer and normal tissue. In this study, based on the GEPIA2 analysis, we found that the expression level of XRCC2 in various cancers, including GBM, was significantly higher than that in the corresponding normal tissues. Moreover, this result was validated in a variety of glioma cell lines and tissues based on transcriptome and protein levels Fig. 1. Studies have reported that XRCC2 affects the efficacy of cancers and participates in the malignant progression of different tumors. For example, silencing XRCC2 can block the cell cycle and increase the sensitivity of rectal cancer cells to radiotherapy (Qin 2015), while the overexpression of XRCC2 in colon cancer promotes the proliferation of cancer cells (Xu 2014). In addition, studies have shown that KIF14, a co-expressed gene positively associated with XRCC2, plays an oncogenic role in numerous cancers (Corson et al. 2005), FBXW4, a gene negatively associated with XRCC2, acts as a protective factor in metastatic colorectal cancer (Zhang 2020). Therefore, we speculate that XRCC2 is involved in the malignant progression of glioma as an oncogene and, hence, its overexpression affects the prognosis of patients.
To determine the effect of XRCC2 overexpression on the prognosis of glioma patients, we performed survival analysis on different data sets, and, as predicted, we found that overexpression of XRCC2 reduced patient survival. As the occurrence of cancer is dependent on many factors, we used univariate and multivariate analyses to test whether this effect of XRCC2 on the prognosis of glioma is an independent factor. As we predicted, XRCC2 was an independent risk factor for poor prognosis in glioma, demonstrated by meta-analysis of multiple datasets. The histological characteristics of glioma play an important role in tumor classification, diagnosis, and treatment (Lapointe et al. 2018; Weller 2015). The results of multivariate analysis in this study suggest that histology is not an independent factor affecting the prognosis of gliomas (P > 0.05). This may be because the interaction between XRCC2 and histology reduces the effect of histology on the prognosis in multivariate analysis, which further reveals that XRCC2 can affect other factors to reduce the prognosis of glioma. In addition, the WHO in 2016 included IDH mutations and 1p/19q combined deletions into the classification and diagnosis of gliomas (Louis 2016), which indicates that molecular characteristics play an increasingly important role in the diagnosis, treatment, and prognosis evaluation of gliomas. Therefore, we have reason to believe that XRCC2, which is highly expressed in gliomas, is an independent risk factor affecting the prognosis of gliomas. Subsequently, it lays the foundation for studying the relationship between XRCC2 and glioma histology. Moreover, multiple data sets mutually verified and revealed that XRCC2 has a high diagnostic value in gliomas Figs. 2, 3, and it also increases the credibility of our research results. In addition, previous studies support our findings, reports suggest that overexpression of XRCC2 in rectal cancer predicts higher malignancy and more lymphatic metastasis and that abnormal overexpression of XRCC2 predicts poor prognosis in breast cancer patients (Lin 2011; Qin 2015).
It is well known that gliomas with high WHO grade have a poor prognosis and that patients with IDH mutation and 1p/19q co-deletion have a higher survival rate. Therefore, we speculated that the expression level of XRCC2 in glioma may also be correlated with multiple clinical and molecular characteristics associated with its prognosis. As expected, the results showed that XRCC2 has a higher expression level in gliomas with higher malignancy and that the expression level of XRCC2 is lower in patients with IDH mutation and 1p/19q co-deletion than in the corresponding control group. These results indirectly reveal that XRCC2 participates in the malignant progression of glioma and affects the prognosis of glioma patients.
Moreover, to understand how XRCC2 participates in the malignant progression of glioma and it actually reduces the survival rate of patients, we performed GSEA analysis on three different datasets and found that the signaling pathways of cell cycle, DNA replication, homologous recombination, and mismatch repair were significantly enriched in all three data sets. These signaling pathways play an essential role in the occurrence and development of cancer. For instance, the cell cycle signaling pathway plays a vital role in the malignant progression of multiple tumors. Studies have found that a variety of anti-glioma drugs act by targeting the cell cycle pathway (Song 2018; Lu 2018; Huang 2016). The dysregulation of DNA replication, one of the fundamental biological processes of the cell, can lead to genomic instability, which in turn promotes the occurrence of cancer (Kitao 2018; Macheret and Halazonetis 2015). The dysfunction of homologous recombination, a mechanism involved in the repair of DSBs, which mainly occurs in the late S-G2 phase of the cell cycle, can result in the process of tumorigenesis (Hoppe et al. 2018); mismatch repair plays a crucial role in maintaining gene stability and provides a new perspective for glioma immunotherapy (Hutchinson 2016; Baretti and Le 2018). These signaling pathways targeted by XRCC2 indicate that XRCC2, as an oncogene, is involved in the occurrence and malignant progression of glioma through multiple pathways, ultimately affecting the prognosis of patients. These results provide a valuable reference for further exploration of molecular therapy for glioma.
To identify small-molecule drugs that can be used to target XRCC2 against glioma, we explored four small-molecule compounds with potential clinical applications using CMap online tools based on the CGGA RNA-seq dataset: doxazosin, quinostatin, canavanine, and chrysin. Previous studies have shown that doxazosin can inhibit the growth of GBM cells through the PI3K/AKT pathway (Gaelzer 2016). Chrysin can inhibit the invasion and migration characteristics of glioma cells through ERK/NRF2 and inhibit the growth of glioma cells (Wang 2018). Quinostatin and canavanine also exhibit antitumor potential in a variety of tumors (Zhang et al. 2019; Nurcahyanti and Wink 2015). In recent years, the new application of traditional medicine has gradually gained attention and achieved satisfactory results in basic research and clinical applications. For example, atorvastatin, a previously considered anti-lipid drug, can be used to treat chronic subdural hemorrhage (Jiang 2018). Sildenafil was initially used in the treatment of cardiovascular and cerebrovascular diseases and has achieved satisfactory results in the treatment of male sexual dysfunction (Hatzimouratidis 2010). Therefore, we believe that the four small molecule compounds identified by us may soon play an essential role in anti-glioma therapy targeting XRCC2.
However, there are still some shortcomings in this study. First, because not all patients from public databases had complete clinical information, some of the glioma clinical characteristics in this study were not mutually validated between different databases. Nevertheless, each dataset contains hundreds of samples, which can reduce the bias to some extent. Second, due to the lack of clinical information, the sample sizes at different time points differed when performing survival analysis, especially in TCGA RNA-seq data. However, when we examined 5-year and 10-year survival rates, patients with high XRCC2 expression showed lower survival rates than those with low XRCC2 expression. Therefore, multicenter studies with complete clinical information still need to be continuously conducted and their data need to be incorporated into public databases, providing a complete research platform for glioma researchers worldwide.
Conclusion
This study pioneered in exploring the expression pattern of XRCC2 in glioma, clarifying that XRCC2 can act as an oncogene and reduce the survival rate of glioma patients. We found that XRCC2 is an independent risk factor for poor prognosis in glioma patients and has a high diagnostic value for glioma prognosis. Furthermore, we found that XRCC2 can participate in tumorigenesis by affecting cell cycle, DNA replication, homologous recombination, and mismatch repair signaling pathways. In order to reduce the harm caused by XRCC2 overexpression, based on the CGGA RNA-seq dataset we identified small molecules that target XRCC2 that have the potential clinical application of improving the status of glioma treatment and the prognosis of glioma patients.
Supplementary Information
Additional file 1: Table S1. Characteristics of patients with glioma based on CGGA RNA-seq data.
Additional file 2: Table S2. Characteristics of patients with glioma based on CGGA microarray data.
Additional file 3: Table S3. Characteristics of patients with glioma based on TCGA RNA-seq data.
Additional file 4: Figure S1. Circle diagram of co-expression analysis results.
Acknowledgements
Thanks the laboratory of Microbiology of Henan Provincial People’s Hospital for providing the experimental platform for this study and all patients who agreed to participate in this study.
Abbreviations
- AUC
Area under the receiver operating characteristic
- CGGA
Chinese Glioma Genome Atlas
- CI
Confidence interval
- CMap
Connectivity map
- DSBs
DNA double-strand breaks
- GBM
Glioblastoma multiforme
- GEO
Gene expression omnibus
- GEPIA2
Gene expression profiling interactive analysis 2
- GSEA
Gene set enrichment analysis
- GTEx
Genotype-tissue expression
- HA
Human astrocyte
- HR
Hazard ration
- IDH1
Isocitrate dehydrogenase 1
- KEGG
Kyoto encyclopedia of genes and genomes
- PRISMA
Preferred Reporting Items for Systematic reviews and Meta-Analyses
- ROC
Receiver operating characteristic
- RT-qPCR
Real-time quantitative polymerase chain reaction
- TCGA
The Cancer Genome Atlas
- WHO
World health organization
Authors’ contributions
WZ, ZL, JL W, ZF J, YZ G, and ZG L conceived and designed the project; HB W, LB, ZS R, and BZ downloaded and collated the data; YB W, XB C, ZB H, XY L, and BF L analyzed the data. All authors read and approved the final manuscript.
Funding
This work was supported by Henan Province Zhongyuan Thousand Talent Program (Yucai Series, No. ZYQR201912122) and the National Natural Science Foundation of China (No. U20A20383, 81772678).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Informed consent was obtained from all subjects. This study was approved by the Ethics Committee of Zhengzhou University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhendong Liu, Wang Zhang and Xingbo Cheng contributed equally to this work
Contributor Information
Zhiguo Lin, Email: linzg_neurosurgeon@163.com.
Yanzheng Gao, Email: yanzhenggaohn@gs.zzu.edu.cn.
References
- Andreassen PR, Hanenberg H. XRCC2 (X-ray repair cross complementing 2) Atlas Genet Cytogenet Oncol Haematol. 2019;23:1–7. doi: 10.4267/2042/69759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldock RA, et al. RAD51D splice variants and cancer-associated mutations reveal XRCC2 interaction to be critical for homologous recombination. DNA Repair (amst) 2019;76:99–107. doi: 10.1016/j.dnarep.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baretti M, Le DT. DNA mismatch repair in cancer. Pharmacol Ther. 2018;189:45–62. doi: 10.1016/j.pharmthera.2018.04.004. [DOI] [PubMed] [Google Scholar]
- Bashir N, Sana S, Mahjabeen I, Kayani MA. Association of reduced XRCC2 expression with lymph node metastasis in breast cancer tissues. Fam Cancer. 2014;13:611–617. doi: 10.1007/s10689-014-9745-0. [DOI] [PubMed] [Google Scholar]
- Ceccaldi R, Rondinelli B, D'Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, et al. Use of the XRCC2 promoter for in vivo cancer diagnosis and therapy. Cell Death Dis. 2018;9:420. doi: 10.1038/s41419-018-0453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GT Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corson TW, Huang A, Tsao MS, Gallie BL. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene. 2005;24:4741–4753. doi: 10.1038/sj.onc.1208641. [DOI] [PubMed] [Google Scholar]
- Cruz C, et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol. 2018;29:1203–1210. doi: 10.1093/annonc/mdy099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumpston M, et al. Updated guidance for trusted systematic reviews: a new edition of the Cochrane Handbook for Systematic Reviews of Interventions. Cochrane Database Syst Rev. 2019;10:ED000142. doi: 10.1002/14651858.ED000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin K, et al. Genetic variants in XRCC2: new insights into colorectal cancer tumorigenesis. Cancer Epidemiol Biomarkers Prev. 2009;18:2476–2484. doi: 10.1158/1055-9965.EPI-09-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton JE, Bolen SD, Mascha EJ. Publication bias: the elephant in the review. Anesth Analg. 2016;123:812–813. doi: 10.1213/ANE.0000000000001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioravanzo A, et al. Glioblastoma with tumor-to-tumor metastasis from lung adenocarcinoma. Neuropathology. 2019;39:474–478. doi: 10.1111/neup.12601. [DOI] [PubMed] [Google Scholar]
- Frappart PO, et al. Recurrent genomic alterations characterize medulloblastoma arising from DNA double-strand break repair deficiency. Proc Natl Acad Sci U S A. 2009;106:1880–1885. doi: 10.1073/pnas.0806882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaelzer MM, et al. Phosphatidylinositol 3-Kinase/AKT pathway inhibition by doxazosin promotes glioblastoma cells death, upregulation of p53 and Triggers low neurotoxicity. PLoS ONE. 2016;11:0154612. doi: 10.1371/journal.pone.0154612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam S, Govindarajulu S, Huckett B, Bahl A. BRCA1/2 mutation-associated breast cancer, wide local excision and radiotherapy or unilateral mastectomy: a systematic review. Clin Oncol (r Coll Radiol) 2015;27:527–535. doi: 10.1016/j.clon.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Hatzimouratidis K, et al. Guidelines on male sexual dysfunction: erectile dysfunction and premature ejaculation. Eur Urol. 2010;57:804–814. doi: 10.1016/j.eururo.2010.02.020. [DOI] [PubMed] [Google Scholar]
- Hilbers FS, et al. Functional analysis of missense variants in the putative breast cancer susceptibility gene XRCC2. Hum Mutat. 2016;37:914–925. doi: 10.1002/humu.23019. [DOI] [PubMed] [Google Scholar]
- Hoppe MM, Sundar R, Tan DSP, Jeyasekharan AD. Biomarkers for homologous recombination deficiency in cancer. J Natl Cancer Inst. 2018;110:704–713. doi: 10.1093/jnci/djy085. [DOI] [PubMed] [Google Scholar]
- Huang HC, et al. alpha-Carboline derivative TJY-16 inhibits tumor growth by inducing G2/M cell cycle arrest in glioma cells. J Biomed Sci. 2016;23:10. doi: 10.1186/s12929-016-0222-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L. Immunotherapy: exploiting mismatch repair in GBM. Nat Rev Clin Oncol. 2016;13:264. doi: 10.1038/nrclinonc.2016.56. [DOI] [PubMed] [Google Scholar]
- Jiang L, et al. Association between the XRCC1 polymorphisms and glioma risk: a meta-analysis of case-control studies. PLoS ONE. 2013;8:55597. doi: 10.1371/journal.pone.0055597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, et al. Safety and efficacy of atorvastatin for chronic subdural hematoma in Chinese patients: a randomized clinical trial. JAMA Neurol. 2018;75:1338–1346. doi: 10.1001/jamaneurol.2018.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamran N, et al. Current state and future prospects of immunotherapy for glioma. Immunotherapy. 2018;10:317–339. doi: 10.2217/imt-2017-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerthana S, Kumar A. Potential risks and benefits of zinc oxide nanoparticles: a systematic review. Crit Rev Toxicol. 2020;50:47–71. doi: 10.1080/10408444.2020.1726282. [DOI] [PubMed] [Google Scholar]
- Khan S, Zakariah M, Rolfo C, Robrecht L, Palaniappan S. Prediction of mycoplasma hominis proteins targeting in mitochondria and cytoplasm of host cells and their implication in prostate cancer etiology. Oncotarget. 2017;8:30830–30843. doi: 10.18632/oncotarget.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao H, et al. DNA replication stress and cancer chemotherapy. Cancer Sci. 2018;109:264–271. doi: 10.1111/cas.13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleibl Z, Kristensen VN. Women at high risk of breast cancer: molecular characteristics, clinical presentation and management. Breast. 2016;28:136–144. doi: 10.1016/j.breast.2016.05.006. [DOI] [PubMed] [Google Scholar]
- Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392:432–446. doi: 10.1016/S0140-6736(18)30990-5. [DOI] [PubMed] [Google Scholar]
- Lassaletta A, et al. Therapeutic and prognostic implications of BRAF V600E in pediatric low-grade gliomas. J Clin Oncol. 2017;35:2934–2941. doi: 10.1200/JCO.2016.71.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, et al. Analysis of salmonella typhimurium protein-targeting in the nucleus of host cells and the implications in colon cancer: an in-silico approach. Infect Drug Resist. 2020;13:2433–2442. doi: 10.2147/IDR.S258037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15:422–442. doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- Lin WY, et al. A role for XRCC2 gene polymorphisms in breast cancer risk and survival. J Med Genet. 2011;48:477–484. doi: 10.1136/jmedgenet-2011-100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- Lu WJ, et al. Licochalcone A attenuates glioma cell growth in vitro and in vivo through cell cycle arrest. Food Funct. 2018;9:4500–4507. doi: 10.1039/c8fo00728d. [DOI] [PubMed] [Google Scholar]
- Macheret M, Halazonetis TD. DNA replication stress as a hallmark of cancer. Annu Rev Pathol. 2015;10:425–448. doi: 10.1146/annurev-pathol-012414-040424. [DOI] [PubMed] [Google Scholar]
- Mahmoudi K, et al. 5-aminolevulinic acid photodynamic therapy for the treatment of high-grade gliomas. J Neurooncol. 2019;141:595–607. doi: 10.1007/s11060-019-03103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurcahyanti AD, Wink M. Cytotoxic potentiation of vinblastine and paclitaxel by L-canavanine in human cervical cancer and hepatocellular carcinoma cells. Phytomedicine. 2015;22:1232–1237. doi: 10.1016/j.phymed.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Ohba S, Mukherjee J, See WL, Pieper RO. Mutant IDH1-driven cellular transformation increases RAD51-mediated homologous recombination and temozolomide resistance. Cancer Res. 2014;74:4836–4844. doi: 10.1158/0008-5472.CAN-14-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkkanen KJ, Yla-Herttuala S. Gene therapy for malignant glioma: current clinical status. Mol Ther. 2005;12:585–598. doi: 10.1016/j.ymthe.2005.07.357. [DOI] [PubMed] [Google Scholar]
- Qin CJ, et al. XRCC2 as a predictive biomarker for radioresistance in locally advanced rectal cancer patients undergoing preoperative radiotherapy. Oncotarget. 2015;6:32193–32204. doi: 10.18632/oncotarget.4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, et al. LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res. 2018;46:717–729. doi: 10.1093/nar/gkx1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebzehnrubl FA, et al. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med. 2013;5:1196–1212. doi: 10.1002/emmm.201302827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, et al. Moxidectin inhibits glioma cell viability by inducing G0/G1 cell cycle arrest and apoptosis. Oncol Rep. 2018;40:1348–1358. doi: 10.3892/or.2018.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachon G, et al. Cell cycle changes after glioblastoma stem cell irradiation: the major role of RAD51. Int J Mol Sci. 2018;19:3018. doi: 10.3390/ijms19103018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AC. et al. Management of glioblastoma: state of the art and future directions. CA Cancer J Clin. 2020. [DOI] [PubMed]
- Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana-Pereira M, et al. Analysis of microsatellite instability in medulloblastoma. Neuro Oncol. 2009;11:458–467. doi: 10.1215/15228517-2008-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. Chrysin suppresses proliferation, migration, and invasion in glioblastoma cell lines via mediating the ERK/Nrf2 signaling pathway. Drug Des Devel Ther. 2018;12:721–733. doi: 10.2147/DDDT.S160020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller M, et al. Glioma. Nat Rev Dis Primers. 2015;1:15017. doi: 10.1038/nrdp.2015.17. [DOI] [PubMed] [Google Scholar]
- Xu K, et al. XRCC2 promotes colorectal cancer cell growth, regulates cell cycle progression, and apoptosis. Medicine (baltimore) 2014;93:294. doi: 10.1097/MD.0000000000000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang Y, Ling J, Xiang Y, Zhuo W. Screening of key genes and prediction of therapeutic agents in Arsenic-induced lung carcinoma. Cancer Biomark. 2019;25:351–360. doi: 10.3233/CBM-182333. [DOI] [PubMed] [Google Scholar]
- Zhang Y, et al. FBXW4 acts as a protector of FOLFOX-based chemotherapy in metastatic colorectal cancer identified by co-expression network analysis. Front Genet. 2020;11:113. doi: 10.3389/fgene.2020.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Characteristics of patients with glioma based on CGGA RNA-seq data.
Additional file 2: Table S2. Characteristics of patients with glioma based on CGGA microarray data.
Additional file 3: Table S3. Characteristics of patients with glioma based on TCGA RNA-seq data.
Additional file 4: Figure S1. Circle diagram of co-expression analysis results.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.