Abstract
Spatial organization of protein biosynthesis in the eukaryotic cell has been studied for more than fifty years, thus many facts have already been included in textbooks. According to the classical view, mRNA transcripts encoding secreted and transmembrane proteins are translated by ribosomes associated with endoplasmic reticulum membranes, while soluble cytoplasmic proteins are synthesized on free polysomes. However, in the last few years, new data has emerged, revealing selective translation of mRNA on mitochondria and plastids, in proximity to peroxisomes and endosomes, in various granules and at the cytoskeleton (actin network, vimentin intermediate filaments, microtubules and centrosomes). There are also long-standing debates about the possibility of protein synthesis in the nucleus. Localized translation can be determined by targeting signals in the synthesized protein, nucleotide sequences in the mRNA itself, or both. With RNA-binding proteins, many transcripts can be assembled into specific RNA condensates and form RNP particles, which may be transported by molecular motors to the sites of active translation, form granules and provoke liquid-liquid phase separation in the cytoplasm, both under normal conditions and during cell stress. The translation of some mRNAs occurs in specialized “translation factories,” assemblysomes, transperons and other structures necessary for the correct folding of proteins, interaction with functional partners and formation of oligomeric complexes. Intracellular localization of mRNA has a significant impact on the efficiency of its translation and presumably determines its response to cellular stress. Compartmentalization of mRNAs and the translation machinery also plays an important role in viral infections. Many viruses provoke the formation of specific intracellular structures, virus factories, for the production of their proteins. Here we review the current concepts of the molecular mechanisms of transport, selective localization and local translation of cellular and viral mRNAs, their effects on protein targeting and topogenesis, and on the regulation of protein biosynthesis in different compartments of the eukaryotic cell. Special attention is paid to new systems biology approaches, providing new cues to the study of localized translation.
Keywords: localized translation, endoplasmic reticulum, mitochondria, nuclear translation, translation factories, assemblysomes, viral factories, SARS-CoV-2 induced COVID-19, stress granules, mRNA transport
INTRODUCTION
The eukaryotic cell has a complex ultrastructure, including the nucleus, membrane organelles, cytoskeleton elements, and numerous types of protein and nucleoprotein granules. All this provides for the compartmentalization and spatial regulation of most intracellular processes. Biogenesis and the functioning of organelles require the synthesis of new proteins and each of them needs to occupy the right niche in order to function properly. Due to the huge structural and functional diversity of polypeptides and the high energy consumption of their production, diverse mechanisms for addressing proteins to the places of their localization and functioning have been developed in the eukaryotic cell. Decades of research have uncovered the bases of these processes. Depending on the stage at which the delivery of the polypeptide to the desired organelle occurs, two localization pathways are distinguished—post-translational and co-translational. The first one is undoubtedly very important, and sometimes even irreplaceable—for example, in the case of nuclear proteins that are imported from the cytoplasm into the nucleus. However, in this review, we will focus on the second pathway, when the protein is placed in the right location immediately after the end of its synthesis.
A classic example of such a pathway is the synthesis of secreted and membrane proteins, which mainly occurs on the membrane of the endoplasmic reticulum (ER). For a long time, it was considered to be the only example of localized translation. However, recent data reveal a much more complex spatial organization of protein biosynthesis in eukaryotic cells. Many alternative mechanisms have been found, including the selective translation of mRNA on the surface of other intracellular organelles: on the outer membranes of mitochondria, plastids and other membrane organelles, in specialized granules and on elements of the cytoskeleton. All these processes have been studied much less than the classical ER-coupled translation, but it is clear that they are based on the recognition of special signals either in the sequence of the mRNA, or in the structure of the synthesized product. Quite often these two mechanisms act in combination. In rare cases, particular properties of specialized ribosomes may also play a role.
Many studies indicate the widespread existence of cases when mRNA is first transported to the desired compartment in the form of inactive ribonucleoprotein (RNP) complexes, and only then the active translation phase begins, leading to the synthesis of proteins in a strictly defined area of the cell. This mode has a number of advantages over the delivery of a mature protein, since it reduces energy costs for transportation and competition with other sorting mechanisms, and also gives an advantage in the assembly of multicomponent systems and the organization of local regulatory “hubs.”
The diversity and accuracy of mechanisms for addressing proteins and localizing the process of their synthesis is ensured by the effective cooperation of various cellular systems, including the cytoskeleton, the translocation complexes built into membranes, as well as networks of specific RNA–protein and protein–protein interactions. Disruption of the normal targeting of proteins leads to their incorrect localization and inactivation, and in the worst cases, to the development of toxic effects, the formation of aggregates and other consequences harmful to the cell and the organism, including severe pathologies.
Here we review various strategies for protein targeting to eukaryotic cell compartments, which are based on mRNA localization and co-translational mechanisms. In addition to the well-known classical models, we discuss alternative or auxiliary, less studied pathways that significantly expand our understanding of the spatial organization of protein biosynthesis process in the cell.
TRANSLATION ASSOCIATED WITH ER MEMBRANES
Hypotheses concerning spatially differentiated translation pathways in eukaryotes were put forward back in the 50s of the previous century, when G. Palade, using electron microscopy, discovered several populations of cellular ribosomes (then still “Palade granules”), which were either bound to the ER membrane or freely distributed in the cytoplasm [1]. The distinct functional roles of these ribosomes were confirmed in subsequent studies on the mechanism of synthesis of secreted and cytoplasmic proteins. A question arose: if there is a distinction between ribosomes according to the type of produced protein, then how is it determined whether the synthesis occurs on the ER membrane or in the cytoplasm? Subsequent studies have revealed the existence of a special population of ER-associated mRNA [2–5], most of which encoded secreted or membrane proteins. However, it is now known that soluble cytosolic proteins can also be synthesized on the ER [6–11], and there are several ways to address mRNA to the ER membranes [12–14]. In addition, the existence of an entire cellular compartment, the TIGER domain, associated with the ER and organizing specific mRNA–protein interactions near the membrane, was recently announced [15].
Product-dependent mechanisms of ER-localized translation: the classical SRP-dependent pathway and other modes. The first attempt to explain the localized synthesis of secreted and membrane proteins was the “signal hypothesis” proposed by G. Blobel [16, 17]. According to it, a special amino acid sequence at the N-terminus of a nascent protein, a signal peptide (SP), carries information that allows establishing a connection between the translating ribosome and the membrane of the ER. Later, in the early 80s, a soluble mediator that recognizes SP was identified—it turned out to be an RNP called SRP (Signal Recognition Particle) [18]. The second component of this system was also found—the SRP receptor (SR) on the membrane of the ER [19]. As a result, a classical model of SRP-dependent membrane localization of ribosomes synthesizing membrane and secreted proteins was proposed (Fig. 1, I).
Fig. 1.
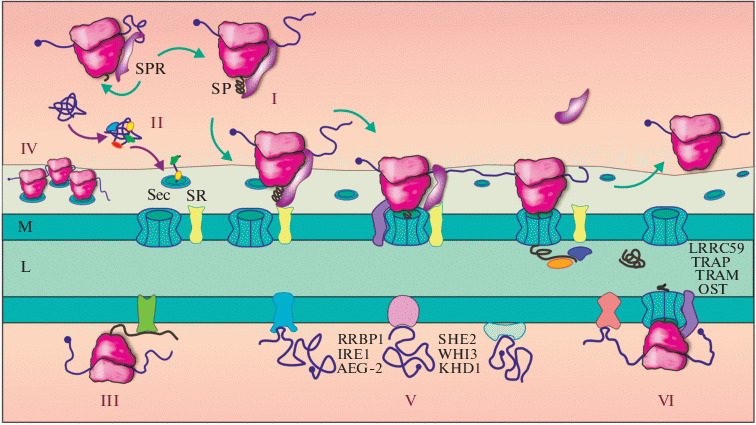
Diversity of mechanisms for protein targeting to the ER and localization of mRNA on the ER membrane. (I) The classic SRP-dependent pathway: the SRP particle, pre-associated with the ribosome, recognizes the signal peptide (SP) or transmembrane domain (TMD) emerging from the ribosomal tunnel, and arrests elongation, after which the transport of the mRNA/ribosome/nascent peptide/SRP complex onto the ER membrane (M) occurs, where SRP binds to its membrane receptor, SR. The translocon (Sec-complex) is involved in the binding of the ribosome on the ER and the translocation of the peptide across the membrane into lumen (L), with the help of proteins associated with the translocon (TRAP, TRAM, OST, etc.). (II) Post-translational mechanism of protein import into the ER mediated by chaperones and other proteins. (III) Localization through the interaction of the synthesized product with the ER resident protein. (IV) Retention of mRNA on the ER membrane in complex with the polysome. (V) Translation-independent mRNA localization mediated by 3'-untranslated region (3'-UTR). Some of the known mRNA-binding proteins involved in various mRNA localization pathways are shown. (VI) Association of the translation complex with the ER through membrane receptors of the ribosome and mRNA-binding proteins.
The mechanism of selective recognition of mRNA by the SRP particle is based on a special amino acid “message” contained in the SP and includes positively charged N-terminal amino acids, a hydrophobic core (which plays the main role in binding) and subsequent polar residues [20]. Transmembrane domains (TMDs) of membrane proteins are recognized in a similar manner [21]. Recognition and binding of SP or TMD occurs co-translationally, as the synthesized peptide is released from the ribosomal tunnel. Other proteins may be involved in this process [22]. After recognition, one of the SRP domains blocks the elongation cycle, and the mRNA/ribosome/nascent peptide/SRP complex is transported to the ER membrane, where SRP binds to its membrane receptor, SR. A special multisubunit protein complex—the translocon (Sec61-complex), which forms a channel in the ER membrane, participates in the binding of the ribosome to the ER and in the translocation of the peptide across the membrane [23, 24]. After attachment of the ribosome to the translocon, SRP dissociates, elongation resumes, and subsequent translocation of the nascent peptide into the lumen or incorporation into the ER membrane occurs [25, 26].
An interesting variation of the classic pathway is employed by an unusual mRNA coding for the animal transcription factor XBP1 (as well as its plant and yeast analogs, bZIP60 and HAC1). It is an important element of the cellular stress response to the accumulation of unfolded proteins in the ER, UPR (Unfolded Protein Response) [27]. XBP1u protein, encoded by an unspliced form of this mRNA, co-translationally recruits the translation complex to the ER membrane via the SRP-dependent pathway [28]. However, the end of the synthesis and release of XBP1u is prevented by its C-terminal region: it contains a special amino acid sequence that causes ribosome stalling [29, 30]. Complexes of ribosomes with peptide and mRNA are tightly bound on the membrane due to the highly conserved hydrophobic domain of XBP1u, which interacts with the translocon; however, full translocation does not occur in this case [28, 31, 32]. The details of this phenomenon are not fully understood and are controversial: for example, some studies have shown the release of the XBP1u protein and its active role in the stress response regulation (see discussion in [33]), while others claim the destruction of XBP1u by the ribosomal quality control (RQC) system, which recognizes stalled translational complexes [34]. In the case of UPR, non-canonical splicing of XBP1 mRNA occurs by the specific nuclease IRE1, which is localized in the ER membrane near the translocon and is activated by this type of stress. Excision of the 26-nucleotide intron leads to a shift in the open reading frame in mRNA and the production of active XBP1s protein, which is directed to the nucleus and activates the expression of genes involved in the UPR [27].
Subsequent studies have revealed additions and exceptions to the classical model. It turns out that inactivation of SRP and/or SR in yeast [35], trypanosomes [36] and human cells [37] does not lead to fatal consequences, while some proteins continue to integrate into membranes or translocate into the ER lumen, although not as efficiently as in the control. This gave rise to the idea of the existence of alternative, SRP-independent mechanisms of protein delivery to the ER [38–40]. Post-translational mechanisms play an important role (Fig. 1, II): classic GET/TRC40-dependent import and the recently discovered SND pathway (from SRP-iNDependent targeting), which description is beyond the scope of this review [41]. However, in addition, product-independent co-translational mechanisms based on the localization of mRNA have been described (discussed in detail in the next section). There are also cases when localized translation on the ER is nevertheless determined by the properties of the polypeptide, but does not depend on SP and SRP. For example, the mRNA of the mammalian and avian cytosolic protein DIAPH1/Dia1 lacking SP is localized on the membranes of the perinuclear ER during translation; however, this occurs not due to the binding of the growing protein to the translocon, but through the interaction of its N-terminal part with a membrane-associated protein of the Rho GTPase family (Fig. 1, III) [42].
Finally, we would like to call attention to a not very obvious issue. It is clear that a long mRNA encoding a large membrane or secreted protein can be translated simultaneously by several ribosomes. In this case, such mRNA will be continuously retained near the membrane due to the fact that at any given time there is at least one ribosome associated with translocon (Fig. 1, IV), even if the other components of the polysome are not associated with the membrane [8, 10]. This is a rather important circumstance, since it gives rise to the idea of the existence of a certain permanent pool of membrane-associated transcripts. Moreover, it cannot be ruled out that some ribosomes may also have a “membrane” specialization. Polysomes on the ER membrane are often circular [43], which implies repeated cycles of translation of a specific mRNA by the same ribosomes, passing from a stop codon to a start codon by CLAR (closed-loop assisted re-initiation) [44]. Thus, the concept of some specialized translational apparatus, which is engaged in the synthesis of membrane and secreted proteins, appears already at this stage.
Translation product-independent localization of mRNA and ribosomes on the ER membrane. The mechanisms of localized translation of mRNA described above are associated with the structure or function of the encoded protein products. However, in many cases, neither SP nor TMD is found in the polypeptide sequences, but the mRNAs encoding them are nevertheless found associated with the ER; sometimes they code for soluble cytosolic proteins unrelated to any membrane organelle [3–14]. The binding of such mRNAs to the membrane can occur independently of translation (Fig. 1, V) and is due to the presence of special localization signals in its nucleotide sequence (cis-elements), most often located in the 3'-UTR [14, 45]. An example is the Saccharomyces cerevisiae yeast PMP1 mRNA, which contains a UG-rich motif in the 3'-UTR, which forms a specific hairpin structure and provides interaction with the ER [46]. These interactions can be mediated by some RNA-binding proteins (see below). The yeast mRNAs USE1 and SUC2, encoding membrane-anchored ER and secreted proteins, respectively, also remain on the ER when SRP is inactivated and translation is blocked [12]. Recently, a special motif, SECReTE, was discovered in the mRNAs encoding secretory and membrane proteins (including the aforementioned SUC2 mRNA), which promotes the retention of mRNA on the ER membrane, enhances its stability, and increases the secretion of encoded proteins [47]. Many mRNAs encoding membrane-associated proteins contain AU-rich elements (ARE) in their 3'-UTRs, bound by the TIS11B protein, and aggregate into special TIS-granules on the ER membrane, then forming a separate subcellular compartment, the TIGER domain (for “TIS granule-ER”) [15, 48]. According to some reports, mRNA can also bind to ER nonspecifically due to the interaction of the poly (A)-tail with membrane receptors [9, 49].
The mRNA receptors on the ER membrane are the subject of active research. Thus, a large role in this field is assigned to the multifunctional membrane protein RRBP1/p180, which is involved in maintaining the integrity of the ER network inside the cell, interacts with microtubules, nonspecifically binds mRNA on the ER surface due to the presence of a lysine-rich domain, regulates the stability of transcripts, and promotes assembly and retention of polysomes on the membrane [49–51]. Another RNA-binding protein, MTDH/AEG-1, is known to preferentially associate with mRNAs encoding transmembrane and secreted proteins [52]. Interestingly, AEG-1 binding sites are found mainly in the coding regions of mRNA, to a lesser extent in the 5'-UTR, and they are almost absent in the 3'-UTR. Recently, the ability to bind ER-associated mRNAs, ribosomes, and tRNAs with high affinity has also been shown for IRE1 [53]. In yeast, the association of mRNA with the ER membrane is likely provided by a number of non-membrane proteins: SHE2, WHI3, KHD1, and others [54–56]. Modern high-throughput methods identify dozens of potential RNA-binding proteins among ER residents [9, 57–59], however, their role in mRNA localization requires careful verification. Many of them lack the canonical RNA-binding domains, therefore, the mechanisms of their interaction with RNA are poorly understood. Another difficulty is that these proteins often turn out to be multifunctional, which complicates their study.
In the study performed by C. Nicchitta’s group, the list of ER membrane proteins with potential RNA-binding properties has been expanded to two dozen representatives [9]. Cytosolic and secretory protein-encoding mRNAs displayed similar RNA-binding protein compositions, which were distinct from those utilized by endomembrane protein-encoding mRNAs. In another study [11], the authors analyzed a repertoire of transcripts bound to ribosomes associated with either SEC61B/Sec61β (a translocon subunit) or LRRC59 (an ER resident protein). They revealed both shared mRNA targets (in particular, mRNAs of soluble cytosolic proteins) and specific ones: e.g. transcripts encoding ER proteins were predominantly associated with SEC61B, while mRNAs of integral plasma membrane proteins—with LRRC59. This supports the above hypothesis of different binding mechanisms of mRNAs encoding membrane and secreted proteins.
A particular, extremely intriguing question concerns the membrane receptors of the ribosomes themselves. In the above-mentioned study [11] four potential ribosome receptors on the ER membrane were examined: LRRC59, RPN1/ribophorin I (a subunit of the ER oligosaccharyltransferase complex, OST) and two components of the translocon, SEC61B and SEC62. The ability of SEC61B, LRRC59, and, to a lesser extent, ribophorin I to bind ribosomes in living cells has been proven. Apart from the Sec translocon and OST, other ER-localized proteins are also the candidates for this role, e.g. the heterotetrameric TRAP complex and the membrane protein TRAM. These ER components interact with the ribosome during the stages of its binding to the translocon and subsequent translocation of the polypeptide into the lumen (Fig. 1, VI) [9, 11, 60–62]. In addition, the aforementioned membrane RNA-binding protein RRBP1/p180 was originally identified precisely as a ribosome receptor on the ER membrane [50].
Thus, in the case of the ER, both modes to ensure localized translation of mRNA are implemented: co-translational recruitment through product recognition and translation-independent mRNA targeting. Most likely, these two mechanisms act cooperatively.
TRANSLATION ASSOCIATED WITH THE OUTER MITOCHONDRIAL MEMBRANE
The processes of translation on the membranes of another important organelle—the mitochondrion, are less studied. A mitochondrion has two membranes— the outer and inner, and translation can occur in the immediate vicinity or on the surface of both, depending on where the transcript is encoded: in the nuclear or mitochondrial genome [63]. Mitochondria independently synthesize only a small number of proteins (components of the respiratory chain). For this, a bacterial type mitochondrial translational apparatus is used; biosynthesis and incorporation of such proteins into the inner membrane, most likely, occurs co-translationally. Other mitochondrial proteins, of which there are more than a thousand, are encoded in the nuclear genome, produced by cytosolic ribosomes, and need to be imported into the mitochondria [64]. The main mechanism of this import is considered to be post-translational translocation (Fig. 2, I), based on the recognition of special mitochondrial targeting peptides at the N-terminus—MTS (mitochondrial targeting sequence) by chaperones and translocations across the outer and inner membranes with the participation of mitochondrial pore complexes TOM/TIM [65]. As in the case of ER, description of the post-translational transport of proteins into mitochondria is beyond the scope of this review, thus we refer the interested readers to specialized articles [63–65] for further details.
Fig. 2.
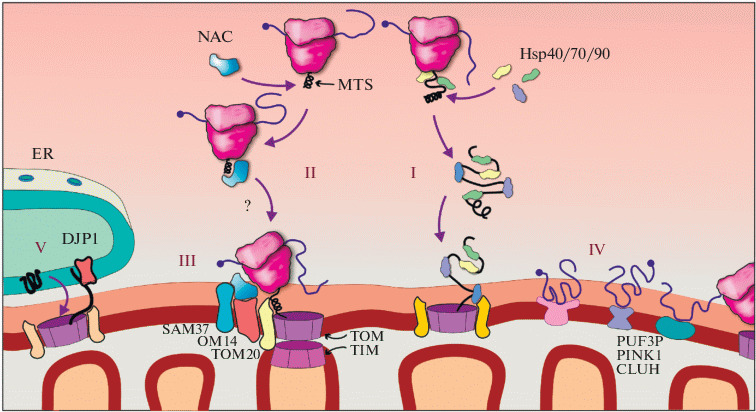
Diversity of mechanisms for proteins targeting in mitochondria. (I) Chaperone-mediated post-translational targeting. (II) Co-translational targeting of the complex consisting of mRNA, ribosome and MTS-containing nascent peptide to the TOM/TIM pore, mediated by NAC (Nascent polypeptide-Associated Complex). (III) Direct binding of NAC to proteins of the outer mitochondrial membrane. (IV) Binding of transcripts to the outer mitochondrial membrane through mRNA-binding proteins. (V) ER-SURF pathway: DJP1-mediated insertion of proteins synthesized on the ER membrane into the mitochondrial membrane. Details are in the text.
Co-translational import of proteins into mitochondria. The binding of cytoplasmic ribosomes and polysomes to the outer membrane of mitochondria was discovered back in the 70s of the previous century [66, 67]. A little later, the enrichment of these polysomes with transcripts encoding mitochondrial proteins was shown [68], as well as their ability to synthesize these proteins and integrate them into the organelle [69]. The existence of co-translational import of proteins into mitochondria, when translation is coupled with translocation across the membrane (Fig. 2, II), was proved in the 90s. In experiments with yeast, it was shown that inhibition of post-translational import is unable to completely inhibit protein translocation into the mitochondria, while inactivation of cytoplasmic ribosomes blocks the import [70]. Over time, the theory began to be supported by new experimental data. In one of the studies [71], an artificial construct encoding the fluorescent protein EGFP with two different targeting sequences was expressed in cultured human cells: the localization signal to the mitochondria was located at the N-terminus of the protein, and the ER-targeting sequence—at the C-terminus. Observations showed that the product was found exclusively in mitochondria, which means that the localization of the protein is determined even before the completion of translation. Nevertheless, for most mitochondrial proteins, the two import pathways seem to complement each other, but in some cases (for example, for the fumarase enzyme, as was shown in another study [72]), the co-translational pathway is strictly dominant. New methods, including ribosome profiling, revealed that the translocation of about 30% of mitochondrial proteins is most likely coupled with translation [73]. This is especially true for proteins of the inner mitochondrial membrane [73].
The nature of ribosome binding to mitochondria was studied by both biochemical and microscopic methods. The important role of the interaction of outer membrane proteins with components of the ribosome has been shown [74]. Later, using cryoelectron tomography, it was possible to visualize this binding [75] and reveal the decisive role of the interaction of the target peptide with the translocon of the outer membrane TOM, by analogy with the interaction of SP or TMD with the Sec complex of the ER translocon. The importance of TOM components for MTS recognition in co-translational import has been shown in another study [76]. It was also found that ribosomes on the mitochondrial membrane are located in a special way—they form clusters in specific areas where the outer and inner membranes are close together and form the contact regions of the TOM and TIM translocons [75].
The precise mechanism of the cytosolic ribosome recruitment to the outer membrane of mitochondria is not yet clear, but it is known that the NAC protein complex (Nascent polypeptide–Associated Complex) interacts with the nascent peptide on the ribosome and plays an important role in this process [77, 78]. Removal of NAC in Caenorhabditis elegans nematodes causes erroneous relocalization of ribosomes synthesizing mitochondrial proteins to the ER membrane, which leads to disruption of the proteostasis of both organelles and to a reduction in the life span of animals [79]. A complementary observation has also been made in yeast: removal of SRP from cells leads to erroneous targeting of ER proteins to mitochondria [21]. These data allow to conclude that NAC interferes with the binding of ribosomes on the ER translocon. This conclusion is consistent with early observations of competition between NAC and SRP for interaction with the growing chain of the synthesized polypeptide and/or to the ribosome [80, 81]. Later studies made additional adjustments to this idea: according to the current model, both complexes can be located on the ribosome at the same time, while the presence of NAC causes structural rearrangements in the SRP that affect the activity of the particle (see discussion in [82]).
Although NAC is usually assigned only the role of a “competitor” to SRP, there is evidence for its active function in protein targeting to mitochondria (Fig. 2, III). For example, direct participation of yeast NAC in co-translational import has been shown in an in vitro system [83]. It was also demonstrated in yeast that OM14, a protein of the outer membrane of mitochondria, functions as a NAC receptor, and the knockout of its gene leads to a decrease in the proportion of ribosomes associated with the outer membrane of mitochondria, and to a disturbance of the import of proteins into the organelle [84]. Another mitochondrial protein, SAM37, a component of the yeast membrane protein sorting and assembly complex (SAM-complex), dynamically associated with the TOM translocon, was also reported to be a NAC partner [85].
More recently, the picture has become even more complicated due to the emergence of a new player— the HEL2/ZNF598 protein, a component of the RQC system [22]. In yeast, this protein is probably capable of additional control of SP recognition by the SRP particle and delivery of the corresponding mRNA to the translocon. Thus, a full understanding of the mechanisms of co-translational import of proteins into mitochondria, the role of NAC and SRP in this process are still lacking, the data are rather limited both in terms of depth and the set of model organisms used—therefore, interest in this topic is constantly growing [64, 86, 87].
Localization of mRNA on the mitochondrial membrane. Using various methods and different model systems, it was reliably shown that many transcripts of nuclear genes encoding mitochondrial proteins are localized on the outer membrane of mitochondria [4, 5, 73, 88–92]. This points to a large contribution of local translation to the targeting of mitochondrial proteins [93]. As in the case of ER, both nucleotide sequence motifs in the encoding transcript and signals in the synthesized polypeptide can be responsible for the mRNA delivery to the mitochondrial surface (Fig. 2, IV). The former mode is exemplified by the OXA1 mRNA of both humans and yeast, containing a functionally conserved cis-acting element in its 3'-UTR, which is necessary for the localization of this mRNA on mitochondria and the correct targeting of the corresponding protein [94]. The latter, translation-dependent mode can be mediated by MTS (interacting with the components of the TOM translocon) or by other signals in the encoded polypeptide [95, 96]. For mRNA of the yeast protein ATM1, the ABC-transporter of the inner membrane of mitochondria, independent contribution of both mechanisms has been shown [97].
Recently Poulsen et al. [98] analyzed the common properties of yeast mRNAs associated with mitochondria, and came to the unexpected conclusion that such transcripts usually have features that prevent effective translation initiation and a lower ribosome coverage. As a result, they proposed a model according to which mitochondrial localization of mRNA is determined by slow initiation and greater mobility, while actively translated mRNAs are less mobile and rarely reach the mitochondrial surface. Another recent finding [99] links the special translational properties of mRNAs encoding mitochondrial and membrane proteins with the involvement of the initiation factor eIF3 in a number of first elongation cycles. The lack of eIF3 led to a defect in mitochondrial biogenesis in the muscles of mice due to the difficulty in the synthesis of mitochondrial proteins at the stage of incorporation of the first several tens of amino acids [99].
The repertoire of the characterized RNA-binding proteins of the outer membrane of mitochondria is still very limited, but their role in the localization and fate of mitochondrial protein transcripts, in biogenesis and maintenance of the organelle functions is undeniable [59, 100]. This is supported, for example, by the detection of certain nucleotide motifs in mRNAs encoding mitochondrial proteins in yeast [101]. The function of mRNA receptors on the outer membrane of mitochondria in yeast can probably be performed by some members of the Puf family of proteins (for example, PUF3), which direct the localization of such mRNAs to the organelle surface, regulate their stability and translation [102–108]. Another RNA-binding protein that can act as a mitochondrial anchor for a number of mRNAs in human cells is SYNJ2BP [109]. Knockout of the SYNJ2BP gene showed that this protein is necessary for the rapid resumption of translation of such mRNAs after stress.
In drosophila, PINK1 and Parkin proteins are implicated in binding of mRNAs encoding respiratory chain components to the mitochondrial surface, orchestrating their local translation and import of protein products. Membrane-associated mitochondrial kinase PINK1 recruits E3 ubiquitin ligase Parkin, which displaces translational repressors (in particular, Dcp1, POP2, Pum-1, and Glorund/hnRNP-F/H) from localized mRNAs and promotes recruitment of translation activators (eIFG4A and eIF4A) [110]. On the other hand, in oocytes PINK1 is predominantly localized on defective mitochondria and, through local phosphorylation of Larp, ensures their “blockade” by preventing the synthesis of new proteins on their surface [111]. This contributes to the rejection of mitochondria with defective genomes from the intracellular population so that they are not passed on to the offspring. Another conserved protein, AKAP1/MDI, which controls local translation at the outer mitochondrial membrane by attracting the mRNA-binding protein Larp, is also necessary for selective inheritance of non-mutant mtDNA in heteroplasmic flies [111, 112].
The mRNA-binding cytosolic protein CLUH, which is present in all eukaryotes from yeast to mammals, binds predominantly to transcripts encoding mitochondrial proteins, including components of oxidative phosphorylation complexes and the tricarboxylic acid cycle [113]. CLUH deficiency causes serious dysfunctions of cellular respiration and energy metabolism, which are brought on by changes in the stability and translation of associated mRNA [114]. The CLUH homologue in drosophila (Cluless) is also found in the mitochondrial fraction, where it is associated with mitochondrial membrane proteins (Tom20), as well as in a complex with ribosomes [115, 116]. Taken together, these data indicate a functional model in which CLUH of higher eukaryotes, like yeast Puf3p, participates in the binding and localization of nuclear transcripts of mitochondrial proteins on the outer mitochondrial membrane and regulates their stability and translation.
Summing up this section, we would like to once again draw attention to the similarity of the principles of localized translation on the outer membrane of the mitochondria and on the ER membranes. In fact, these organelles have much closer interactions than is commonly believed. The places of close contact between the membranes of mitochondria and the ER play a key role in maintaining lipid and calcium homeostasis, in the initiation of autophagy, mitochondrial division, and other processes [117, 118]. A significant proportion of the transcripts localized on these two organelles turns out to be associated with both of them, which suggests their translation at the sites of contact between the two organelles [5]. OSM1 is an example of a dual-localized mRNA; this is based on the use of alternative translation starts, which give different signal sequences in the protein [73]. The phenomenon of double localization of the polypeptides is not a rarity [119]. A new mechanism for the import of proteins into mitochondria has recently been discovered in yeast—ER-SURF (ER surface-mediated protein targeting): mitochondrial proteins are initially incorporated into the ER, but then are redirected to the mitochondria using the ER-localized chaperone DJP1 (Fig. 2, V) [120].
THE ROLE OF LOCALIZED TRANSLATION IN THE IMPORT OF PROTEINS INTO PEROXISOMES AND OTHER MEMBRANE ORGANELLES
Peroxisomes are single-membrane organelles responsible for the oxidation of fatty acids, detoxification of hazardous oxygen and nitrogen forms, as well as a number of other important functions and have an important structural and functional relationship with the ER and mitochondria [121, 122]. The only known way of importing proteins into peroxisomes is post-translational: unlike mitochondrial and ER proteins, most peroxisome-targeted polypeptides are imported in a completely folded form, often as oligomers [121]. Targeting is mediated by special amino acid motifs, PTS (peroxisomal targeting sequences), which are located either at the C- (PTS1) or at the N-terminus (PTS2) of the proteins. Nevertheless, a special translation-coupled import mechanism has been described for some peroxisomal proteins [123]—however, in this case, the initial incorporation of polypeptides occurs into the ER membrane, after which the proteins are delivered to the peroxisomes by directed vesicular transport. This ER-mediated topogenesis of peroxisomal proteins serves as the basis for de novo peroxisome formation and biogenesis of their membrane [121]. Another translation-related feature of some peroxisomal proteins is their dual localization, in which some isoforms are targeted to peroxisomes, while others remain in the cytosol [124]. In a number of cases, these isoforms, characterized by the presence of PTS1 at the C-terminus, are synthesized from the same mRNA due to an unusually high level of stop codon read-through [125, 126].
A significant role in the targeting of proteins to peroxisomes is assigned to the localization of the transcripts encoding them in the immediate vicinity of these organelles or directly on their surface. There are still very few studies on the mRNA localization on peroxisomes, but this line of research is rapidly developing [127]. In 2009, the localization of 50 yeast mRNAs encoding peroxisomal proteins was analyzed [128], 12 of them (including mRNA of eight peroxins and four matrix enzymes) showed a high level of co-localization with peroxisomes—from 50 to 80% of mRNA molecules. Then, a large-scale analysis of peroxisome-associated transcriptome in mice [129] revealed an enrichment in representatives encoding peroxisomal proteins, including peroxins and matrix enzymes involved in β‑oxidation of fatty acids and bile acid biosynthesis. The peroxisomal fraction contained also mRNAs encoding mitochondrial and secreted proteins.
Rare data on the relationship between translation and other membrane organelles, as well as on the specific localization of mRNA on them, can be found in the literature. Thus, in a recent screening, selective localization of mRNA of several genes at once on endosomes, the Golgi apparatus and on the outer side of the nuclear envelope was found [130]. In addition, localized translation of septin mRNA on endosomes was shown in fungal hyphae [131, 132]. In Xenopus neurons, late endosomes, often associated with mitochondria, serve as a platform for translation of many mRNAs and are necessary for efficient protein synthesis from lamin B2 mRNA [133].
TRANSLATION IN THE NUCLEUS?
Perhaps the most controversial subject in the topic of localized translation is associated with the discovery of 80S ribosomal complexes and, presumably, translating ribosomes in the cell nucleus. The history of these studies is a chain of attempts to confirm or disprove these observations [134–136]. First reports about “nuclear translation” originate in the 50s of the previous century, when in the studies of Allfrey and Mirsky [137, 138] the inclusion of labeled amino acids in the isolated nuclei of calf thymocytes was shown. However, the presence of ribosomes on the outer nuclear membrane (which directly passes into the ER membrane) is well known, and the degree of purification of nuclei from the cytoplasmic fraction at that time was clearly insufficient to draw unambiguous conclusions. Nevertheless, in the future, experiments on the incorporation of labeled amino acids into the nuclei of different cells were repeated many times using various purification methods; the energy dependence of this process, sensitivity to protein synthesis inhibitors, DNase and RNase were shown (a detailed analysis of these numerous studies of the 50-70s can be found in the review [139]). It was possible to isolate actively translating polysomes from the purified nuclei [140]. Isolated nucleoli also efficiently incorporated amino acids [134, 141], which looks especially plausible given the presence of maturing ribosomal subunits in this structure.
However, as data accumulated, it became clear that the “legitimate” place for the synthesis of both cytosolic and nuclear proteins is exclusively the cytoplasm. It turned out that maturation and nuclear export of large and small ribosomal subunits occurs separately, and the final stages of their biogenesis and “quality control” take place outside the nucleus [142], while up to this point, the subunits are associated with special inactivating factors that prevent translation initiation [143]. On the other hand, it is surprising that many tRNAs, translation factors, and auxiliary enzymes can be found in the nucleus (although some of these data are disputable) [144]. The absence of 80S ribosomes in the nucleus was also questioned: Al-Jubran and co-authors [145] applied the method of bimolecular fluorescent complementation, attaching different parts of the fluorescent protein to the components of the small and large subunits, and observed fluorescence in the nucleus. It is also intriguing that a small number of mRNA species are found exclusively in the nucleus yet produce proteins [130].
In addition, over time, evidence of various “non-canonical” translational events in the cell began to appear—and they almost immediately were linked to putative “nuclear translation” [146]. At the moment, this topic is mainly associated with the still poorly understood process of synthesis of hypothetical “defective ribosomal products” (DRiPs), or “pioneer translation products” (PTPs), which are the result of translation of unspliced pre-mRNAs and serve as one of the sources of peptides for presentation in the class I of the major histocompatibility complex (MHC I) [147–149]. Another branch of research links “nuclear translation” with some manifestations of the nonsense-mediated mRNA decay (NMD)—an important mechanism of mRNA quality control that eliminates transcripts with premature stop codons, including products of aberrant splicing [150–152]. The main argument in favor of this relationship is that many NMD factors have nuclear or perinuclear localization [153, 154]. Both of these concepts can coexist with each other, since the production of PTPs can be associated with the “pioneer round” of translation of incorrectly spliced transcripts from which some introns have not been removed—the only question is whether these events actually occur in the nucleus.
The emergence of these arguments triggered a new wave of research by proponents of “nuclear translation,” which used modern techniques of protein labeling in combination with electron and confocal microscopy. A significant contribution to the formation of the updated concept was made by the studies of P. Cook’s laboratory [150, 152, 155], in which different types of pulse-labeling of the nascent polypeptides were used followed by their microscopic detection. Another group, using a method based on the incorporation of puromycin into the growing polypeptide chain (followed by visualization of the product using puromycin-specific antibodies) showed an intense accumulation of the label in the nucleus [156]. This fact has long been considered a very strong argument in favor of “nuclear translation.” However, in two recently published studies [157, 158] the puromycin labeling approach was heavily criticized: the authors found that under the conditions that are regularly used during the reaction, the puromycilated products leave the ribosome and quickly diffuse from the site of synthesis, often ending up in the nucleus as well. It is possible that other types of labeling are not devoid of similar artifacts. Thus, today it is hardly possible to say with confidence that there are any irrefutable arguments in favor of the existence of “nuclear translation.”
CYTOSOLIC RIBONUCLEOPROTEIN GRANULES AND “TRANSLATION FACTORIES” AS A PLATFORM FOR mRNA LOCALIZATION AND TRANSLATION
Stress granules, processing bodies and other types of RNP granules. In addition to constitutively present organelles, temporary non-membrane-bound assemblies can be observed in the cytoplasm of a cell under various conditions—in particular, RNP granules, relatively large condensates, which represent multicomponent multifunctional complexes. The most well-known and studied types of such assemblies are stress granules (SGs) and processing bodies (P-bodies, PBs). [159, 160]. PBs (Fig. 3, I) present in the cell under both normal and stress conditions, and are enriched with factors involved in RNA degradation. However, according to current views, the mRNA decay is not the main purpose of PBs. Unlike PBs, SGs (Fig. 3, II) are predominantly formed under certain types of stress; they contain mRNA in a complex with small ribosomal subunits, translation initiation factors, and a variety of RNA-binding proteins [159]. The main function of both SGs and PBs is now considered to be the regulation of RNA metabolism: first of all, selective translation repression, as well as controlled degradation and other processes [159, 160]. It is generally accepted that the phenomenon of liquid–liquid phase separation (LLPS), associated with the peculiar properties of RNA and proteins with unstructured or repetitive regions, has an important role in the formation of PBs, SGs, and other large RNP-condensates and non-membrane-bound organelles [161–164]. Most likely, the “seeds” of SGs—high molecular weight RNPs (pre-SGs) and RNA–RNA condensates, which are in a dynamic equilibrium between growth and disassembly—are present in the cell under normal conditions [165, 166]. Under stress, they serve as a focus for the condensation of mRNAs and proteins to form mature SGs due to LLPS.
Fig. 3.
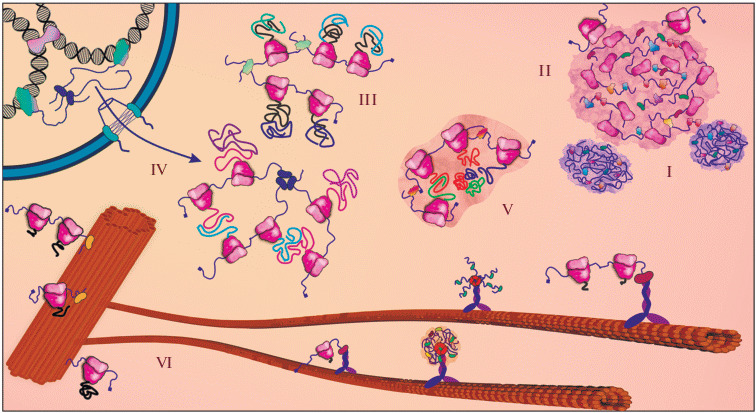
Translation associated with non-membrane cell organelles. Shown schematically: P-bodies (I); stress granule (II); “ranslation factory” (III); transperon (IV); assemblysome (V); ribosomes and nucleoprotein complexes associated with elements of the cytoskeleton (centrosome and microtubules) (VI). Details are in the text.
SGs and PBs are traditionally considered to be translationally inactive compartments (if only because ribosomes are not found in PBs, and 60S subunits in SGs). However, according to some data, both structures can sometimes also act as a “platform” for localized mRNA translation. For example, it has been shown that in drosophila oocytes, gurken mRNA is translated directly on the surface of PBs [167]. This mRNA is synthesized in trophocytes and, in complex with translational repressors, is transported into oocytes, where it binds to the PB surface and is activated. Another mRNA, bicoid, is localized in the core of PBs in the early stages of oocyte development, and as it matures, the mRNA moves to their periphery, where it is translated [168]. Localization of translation initiation factors and polysomes on the near periphery of PBs was also found in mammalian cells [169]. Probably, such proximity is important for the rapid resumption of translation of mRNA immediately after its release from a PB. Thus, this structure, which was initially considered a storage site for transcripts committed to degradation, can actually play a role in the dynamic regulation of protein biosynthesis.
The issue of translational activity associated with SGs is also highly controversial. According to classical model, mRNAs are stored in the SGs in a completely repressed state and their translation is resumed only after SG disassembly [159]. However, it was showed that translated mRNAs can temporarily interact with SGs [170]. Moreover, recent studies have revealed the possibility of active mRNA translation directly within the SGs [171], as well as in some kind of SG-like granules located in cell protrusions [172, 173]. These facts may indicate the need to revise the established model.
In addition to PBs and SGs, other types of RNA granules can be found in cells under different conditions, which can be tissue- and mRNA-specific [172, 174–177]. For example, granular high molecular weight RNA-protein complexes are widely present in nerve cells [178, 179], where they serve as a transport form of inactive polysomes during the traffic of mRNA along neurites [180, 181]. Active translation can take place directly in such granules [182]. Some neuronal granules are likely to be single polysomes “frozen” in an inactive state [183] and able to quickly resume elongation upon synapse activation [181]. In the cells of the germ line of animals, another type of RNP condensates is widely represented—the so-called germinal granules. Depending on the organism and the type of cells (gametes or their precursors, cells of surrounding tissues or embryonic germ line cells), this term can mean slightly different structures—polar granules, P-granules, sponge and chromatoid bodies, Balbiani bodies, granules in “mitochondrial clouds” or a specific perinuclear compartment, termed nuage [184]. It is believed that these granules, among their other functions, serve as a repository of untranslated parental transcripts that will be activated during meiosis, after fertilization or at a certain stage of early embryogenesis [185, 186]. In some cases, the dynamics of these granules is closely related to the unusual perturbations that mitochondria undergo during meiosis [184]. Localization of many coding and non-coding RNA, components of these granules, demonstrates complex dynamics [187, 188]. It is curious that the granules are also associated with the mitochondrial ribosomal RNA, which is found at this moment in the cytoplasm [189, 190] and can be translated [191]. The complex pattern and dynamics of mRNA distribution seem to provide regulation of translation during the maturation of germ cells and in the early stages of embryogenesis [185].
“Translation factories,” assemblysomes, transperons and other translationally active mRNA condensates. New data indicates that actively translated transcripts can also form certain condensates in the cell, sometimes referred to as “translation factories” (Fig. 3, III). Thus, in yeast, mRNAs of some glycolysis enzymes under normal conditions form translationally active granules, which, in case of stress, serve as the basis for PB formation [192, 193]. This feature is apparently also characteristic of human cells [193]. This new structures are named “CoFe-granules” (from “Core Fermentation”). The same group in a new study showed the existence of another type of translationally active mRNP condensates in yeast, in which transcripts encoding a number of translation factors are concentrated [194]. These granules are specifically inherited by daughter cells and appear to play a role in focusing of protein synthesis activity in the region of polarized growth. Co-localization of several mRNA molecules encoded by the same gene was also shown in human cells: visualization of transcripts from which the heavy chain of dynein was translated revealed that a significant proportion of them were co-localized into clusters of 3–7 copies [195]. In the recently published results of a large-scale (more than 500 mRNA) screening, it was shown that approximately 6% of the human mRNA species are clearly localized in the cell [130]. In most cases, this is granular localization or concentration of mRNA in certain cellular compartments and organelles, although condensation into a uniformly distributed network is also possible, as in the case of the already mentioned membrane-associated TIS granules [15, 48].
Thus, RNP granules can not only act as negative regulators of translation, “masking” mRNA transcripts with the help of RNA-binding proteins, but also serve as a platform for active translation. The mechanism of their formation and the principles of mRNA recruitment are still poorly understood. It is possible that mRNA condensates can form partly due to the interaction of RNAs themselves—especially their long unstructured regions [48, 161]. In addition, the effects of nuclear events on the co-localization of functionally similar mRNAs are discussed in literature. It could be that the proximity of genes in the nucleus and their cooperative transcription subsequently ensures the proximity of mRNA in the cytoplasm, mediated by nuclear-cytoplasmic RNA-binding proteins [196]—such mRNA groups are called “transperons” (Fig. 3, IV), by analogy with prokaryotic operons.
However, at least in some cases, the formation of cytoplasmic mRNA condensates is mediated by protein–protein interactions of the encoded products. Co-translational binding of the synthesized protein with its future functional partners has long been known for polycistronic mRNAs of prokaryotes—however, it also turned out to be quite common in the eukaryotic world [197]. This is true not only for the products synthesized by neighboring ribosomes from the same transcript [198], but also, more importantly, for the products encoded by different mRNAs. This was shown for the first time for several hetero-oligomeric complexes in yeast [199–201], then it was confirmed for membrane [202], nuclear [203] and cytoplasmic [204–206] protein complexes in human cells. Co-translational interaction of products cannot but influence the localization of mRNA.
In the case of such large multimeric complexes as the proteasome, co-translational condensates of mRNAs and their products can even form special compartments called assemblysomes (Fig. 3, V) [204]. The proteasome-synthesizing assemblysome is formed not only due to the interaction of translation products with each other, but also using a special transcript-specific mechanism of mRNA segregation mediated by the NOT1 protein, a component of the Ccr4-Not complex. The Ccr4-Not complex is also involved in the formation of other co-translational condensates [201, 207]. Interestingly, soon after starting the synthesis of two components of the proteasome, RPT1 and RPT2, the ribosomes synthesizing them stop. This stalling occurs at well-defined positions of the coding regions, allowing the protein subdomains emerging from the ribosomal peptide tunnel to bind to each other. Translation resumes only after the establishment of the protein-protein interaction and the transfer of mRNAs to the assemblysome [204]. This scenario strongly resembles the SRP-mediated control of the synthesis of secreted or membrane proteins described in the first chapters of the review, and once again emphasizes that nature can use the same principle in the regulation of similar processes. Moreover, the principle of “freezing” mRNA-ribosome complexes and their subsequent activation “on demand” is probably used in other cases as well, for example, when transporting inactive mRNAs in neurons, as described below.
LOCALIZATION OF TRANSLATION FACTORS IN THE CELL
Discussion of localized translation, hypothetical “translation factories” and transperons would not be complete without viewing the issue of intracellular localization of the translation machinery components themselves. It is well known that in stressed cells 40S ribosomal subunits, many initiation factors and mRNA-binding proteins are relocated into special non-membrane structures, SGs, described above. However, some data indicate that the translational machinery components are non-randomly distributed in the cytoplasm under normal conditions as well. Thus, both fractionation- and immunocytochemistry-based studies reveal a specific distribution of the translation initiation factors eIF4E and eIF4G in mouse fibroblasts: in addition to the diffuse localization in the cytoplasm, they were enriched in the region of the ER, Golgi apparatus, as well as the leading edge and protrusions of cells [208, 209]. This localization coincided with the distribution of ribosomal proteins and was observed only during active protein synthesis [209]. Poly(A)-binding protein PABP also demonstrates complex and dynamic localization [210]. The yeast initiation factor eIF2B, a guanine nucleotide exchange factor for eIF2, has the most striking distribution pattern [211]: under conditions of active protein synthesis, a significant proportion of eIF2B molecules are located in specific granules, called “eIF2B bodies”, through which eIF2 is cycled. In neurons, translation machinery can be locally concentrated in synapses or associated with receptors of stimulating ligands, which is necessary for the timely local activation of translation of certain mRNAs [212–214]. In addition, it is known that many components of the translational apparatus (for example, eIF3, eIF4G, eEF1A, eEF2, eRF3 and, possibly, the ribosomes) interact with elements of the cytoskeleton – microtubules, intermediate and actin filaments, as well as with the corresponding motors [215, 216], participating in active transport of translation complexes.
INTRACELLULAR TRANSPORT OF mRNAs AND THEIR INTERACTION WITH CYTOSKELETAL ELEMENTS
Localization of mRNAs for their subsequent compartmentalized translation is not necessarily associated with organelles or granules. Often, some zones of the cell represents the site of localization: for example, lamellipodia of fibroblasts and areas of focal contacts are enriched in mRNAs encoding β-actin and actin-binding proteins [217–219], yeast mRNA ASH1 is localized exclusively in the bud of the mother cell preparing for the division [220], and the transcripts of morphogens and homeotic genes oskar, nanos, bicoid, and gurken, after transportation from trophocytes, are located at certain poles of the drosophila oocyte, which further determines the dorso-ventral and anterior-posterior polarity of the embryo [221, 222]. These models can be called classical, they are described in textbooks and in many reviews. Another popular object of such studies is neurons: in these cells localization of mRNA largely determines the proteomes of neurites and soma [223]. In the axons of neurons, even local synthesis of ribosomal proteins has evolved for the “repair” of ribosomes in situ, since the transport of new ribosomes from the nucleus located far in the soma takes too long in these cells [224]. Systematic analysis of mRNA localization in cultured human cells [130] and drosophila embryos [225, 226] shows that a well-defined subcellular mRNA location is more often the rule, rather than the exception.
In most cases, the localization of mRNA determines the subcellular positioning of the polypeptide product, but sometimes it is needed for some intermediate stages of its maturation or functioning. For example, mammalian RAB13 GTPase is synthesized from mRNA localized in cell protrusions, and although the final product itself has a perinuclear localization, its local synthesis is important for GTPase activation, since during the production the nascent RAB13 chain interacts with its GDP/GTP exchange factor, RABIF, anchored in that specific location [205]. In mesenchymal-like human cells, LARP6-mediated localization of mRNAs encoding ribosomal proteins at the leading edge is likely to play an important role in the regulation of their translation and in the control of cell migration [227]—although, in order to perform their function, these proteins, most likely, must first enter the nucleolus; however, participation in the local “repair” of ribosomes, as in neurons [224], cannot be ruled out either.
Nevertheless, the meaning of mRNA localization in certain areas of cells is not always obvious [130]. It is possible that the spatial arrangement of translation complexes relative to other cell structures can serve as a factor that sets the desired orientation of higher-order protein complexes. Recently, such a mechanism was reconstituted in a cell lysate [228]: the authors were able to organize the structure of actin filaments in a certain way by localizing the transcripts encoding actin-binding protein using magnetic nanoparticles.
The transport of mRNA to the localization site is usually accomplished as part of RNP complexes, in which translation is inhibited by specific repressors (Fig. 3, VI). The delivery address is encoded in the mRNA sequence—in cis-acting localization elements, sometimes figuratively called “zipcodes.” Most often, these sequences are located in the 3'-UTR of mRNA, but they are also found in the coding region or 5'-UTR; they can vary significantly both in length (from a few nucleotides to several hundred) and in structure [220, 229, 230]. Various protein factors interact with the localization elements of mRNAs, forming RNP complexes. Such particles are transported along the elements of the cytoskeleton—as a rule, through directed active transport along microtubules or, less often, intermediate or actin filaments, with the participation of molecular motors [218, 231, 232].
ZBP1/IGF2BP1/IMP1 is a classic example of an mRNA-binding protein that provides targeting of transcripts with zipcodes. It recognizes a region in the 3'-UTR of β-actin mRNA (as well as in a number of other transcripts) and simultaneously forms a complex with the kinesin-like motor protein KIF11 [219, 233, 234]. This RNP particle also includes other mRNA-binding proteins, as well as 40S ribosome subunits and a number of components of “pioneer round of translation” [235]. This suggests that the mRNAs are engaged in transport immediately after export from the nucleus, even before the first round of translation occurs. Similar principles underlie the transport mechanism of the aforementioned mRNAs from drosophila oocytes (based on a complex of the RNA-binding protein Egalitarian and the dynein cargo adaptor, Bicaudal, or their functional analogs, Staufen and kinesin-1), as well as the yeast ASH1 mRNA (a complex of SHE2 RNA-binding protein and myosin motor MYO4). Numerous studies on the identification of these components can be found in the specialized reviews [221, 236]. Apparently, many different mRNA-binding proteins are involved in the transport of mRNA in vertebrate neurons, including STAU1 and STAU2 (homologues of Staufen), FMRP, TDP-43, and an important SG component, G3BP1 [236, 237]. Some RNA-binding proteins are known to combine several mRNA species into specialized transport aggregates [238, 239]. The identity of the motor protein in the case of neuronal transport is still less clear. However, recent studies have identified the key role of the APC protein and the KIF3A/B/KAP3 kinesin complex in the specific transport of mRNA along axons [173, 240].
In addition to transportation as part of an individual RNP and a frozen translation complex, mRNA can also move along the elements of the cytoskeleton as part of large structures (SGs, PBs, or other granules), as well as on membrane organelles (mitochondria, endosomes, lysosomes, etc.). In axons, the latter mRNA transport mode seems to be quite common and has been figuratively termed “hitchhiking” [133, 236, 237].
At the final location, mRNA is released from complexes with transport and repressor proteins and becomes available for the translational apparatus [241–243]. For example, in the case of the already mentioned mRNAs encoding human β-actin and yeast ASH1, this release is regulated by phosphorylation of some RNP components that interact with translation factors and prevent initiation of protein synthesis during transportation [234, 244]. For neurons, two additional unusual mechanisms of mRNA activation has been shown: by cutting the 3'-UTR [245] and by releasing inactive mRNA-ribosomal complexes from specific aggregates with transmembrane receptors of neuron-stimulating ligands [212, 213]. The rule of “silent” transportation is not always fulfilled: for example, in the case of APC-dependent transport, mRNAs can be translated during delivery [246].
Localization of mRNA can also be accomplished as a result of passive diffusion and “anchoring” onto certain cellular structures [247]. Much less often, the asymmetric distribution of mRNA in the cell can be achieved using local protection against degradation [248]. Interestingly, the localization of mRNA in the cytoplasm can be influenced by events that happened to it back in the nucleus—for example, it can depend on the type of promoter from which the transcript was synthesized [242], or on belonging of the gene to a certain transperon [196].
In addition to the transport “highway” function, elements of the cytoskeleton can act as sites of permanent residence of mRNA and non-coding transcripts [130, 249, 250], thereby providing local synthesis of transport proteins and other functions [251–254]. Thus, localized translation of cyclin B mRNA on the mitotic spindle in rapidly dividing cells of amphibian embryos helps to coordinate the production of this regulator with the cycles of cell division [255]. Localized translation of axonemal dynein mRNA at the base of the cilia (cellular antenna) is critical for spermatogenesis in drosophila [256]. The centrosomes seem to be an important translational “hub” (Fig. 3, VI): they accumulate various translational components [257–260] and specific RNAs [130, 226, 250, 255, 260, 261], although the meaning and significance of such localization are still unclear. It is only known that during cell division, some mRNAs are directed along microtubules into the pericentriole matrix, and then redistributed between daughter cells via actin transport, thus providing asymmetric segregation of transcripts [261]. Disruption of the centrosomal localization of only one mRNA (cen) leads to errors in the assembly of the division spindle and genome instability in the drosophila embryo model [262]. It is interesting that the synthesis of a number of proteins of the centrosomes also occurs directly on these organelles, which is due to the product-dependent localization of the corresponding mRNA [263]. This localization is regulated in the cell cycle and is conserved from drosophila to humans.
SPECIAL CASES OF LOCALIZATION AND TRANSLATION OF VIRAL AND TRANSPOSON mRNA
mRNA of many viruses (especially those, whose entire life cycle takes place in the cytoplasm) are translated in special structures—as part of particles, granules or in membrane compartments that viruses form during infection and where certain stages of their life cycle occur [264]. Cellular mRNAs transcribed from DNA copies of some retrotransposons can also form similar structures. Although a detailed description of this topic would require writing a separate review, we will touch on some of its aspects, illustrating them with examples.
The ER membrane, in particular, serves as a platform for the translation of mRNA of the Flaviviridae family viruses, including Zika viruses, dengue, hepatitis C and others [265–267]. Throughout most of the life cycle of these RNA viruses, their plus and minus strands are associated with ER membranes and with special membrane structures, “replication factories.” As the infection progresses, the ER and the Golgi apparatus are reorganized, and the translational properties of membranes change in such a way that they become the site of preferential translation of viral mRNAs, usurping resources from the host cell transcripts. Due to spatial isolation, all this occurs without significant activation of the interferon response, UPR, and other signaling pathways aimed at suppressing viral infection. Complex membrane structures, sometimes two-layered, necessary for replication and originating from the ER, the Golgi apparatus, lysosomes and, less often, mitochondria, are also formed by other animal and plant (+)RNA-containing viruses— picornaviruses, coronaviruses, togaviruses, caliciviruses and many others [264]. In particular, in cells infected with coronaviruses, from two to six new types of membrane structures were found, of which “convoluted membranes” and “double-membrane vesicles” (DMVs) are the best characterized [268, 269]. The transcripts of many RNA viruses contain multiple copies of the SECReTE motif [270]. For example, in the genome of the SARS-CoV-2 coronavirus, which causes COVID-19, 40 such regions are found, as well as signals of possible mitochondrial RNA localization [270, 271]. In the case of genomic mRNA of the human immunodeficiency virus HIV-1, localization on mitochondria was shown experimentally [272].
Unfortunately, little is usually known about the role of such structures in translation. In some cases (for example, during infection with the Sindbis virus), these foci have been shown to be enriched with some translation factors (eIF3, eEF2) and depleted in others (eIF2, eIF4G) [273]. Orthoreoviruses have a special mechanism for actively attracting components of the translational apparatus to the border and inside their virus factories [274]; with coronavirus infection, enrichment of membrane structures with translational components is also observed [275]. In the case of poliovirus, membrane localization of mRNA can confer additional translation stability due to CReP-mediated recruitment of eIF2 [276].
Many DNA viruses are characterized by an even more impressive reorganization of the cell membrane compartments. For example, in poxviruses, their complex reproductive cycle is carried out in special perinuclear cytoplasmic loci of the host cell (virosomes, or Guarnieri bodies), in which translation initiation factors (eIF4E, eIF4G, PABP) and other important translational components are concentrated [277]. At the same time, a special gel-like compartment (ATI bodies) containing numerous copies of the ATI protein is used to assemble virions. It is interesting that the translation of the mRNA encoding ATI occurs in these bodies, and not in the virosomes [278], and for its localization, apparently, the same principle is used as in the cellular assemblysomes: ATI peptides growing from the ribosome interact with other ATI protein molecules already localized in this structure, which provides the anchoring of the ATI mRNA and targeting of the products.
Simultaneously with the remodeling of membrane structures, viruses can have a strong effect on the formation of cellular RNP granules—SGs and PBs. Interestingly, some viruses (for example, influenza and vesicular stomatitis viruses, Zika virus or picornaviruses) actively prevent the formation of SGs, while others (like the rabies virus), on the contrary, stimulate their formation, using this mechanism to suppress cell translation or to regulate own replication cycle [279–281]. Coronavirus SARS-CoV-2, upon infection of cells, induces disassembly of PBs and interferes with the assembly of SGs, which impede productive infection [282, 283]. At the same time, the virus uses LLPS to create its own RNP condensates (see [284, 285] and references therein). The literature on the relationship of viruses with SGs and PBs is so extensive that we recommend readers to refer to specialized reviews on this topic [286, 287].
The yeast retrotransposon Ty1 uses an even more sophisticated pathway, inextricably linked with the ER, to assemble virus-like particles in which its genome is replicated [288]. During translation, Ty1 RNA is recognized by the SRP particle of the host cell and delivered to the ER membrane, while the Gag envelope protein synthesized from this RNA enters the ER lumen. In this case, Gag is found in the cytoplasm in association with the complex of Ty1 RNA and SPR, although in the absence of translocation in the ER, Gag is synthesized, but is rapidly degraded. According to the proposed model, upon translocation into the ER lumen, Gag acquires a stable conformation, then returns to the cytoplasm and binds to the RNA–SRP complex, initiating the nucleation of the virus-like particles [288].
RNAs of other known retrotransposons can also exhibit characteristic localization patterns in the cell. For example, the unique bicistronic mRNA of the autonomous retrotransposon LINE-1 (L1) is localized in special nucleoprotein particles that are enriched in proteins—the products of its translation (ORF1p and ORF2p) required for transposition, as well as markers of stress granules [289, 290]. In addition, the composition of these particles changes depending on the life cycle of LINE-1 and on the localization in the cell—cytoplasmic or nuclear [291]. Interestingly, RNAs of non-autonomous retrotransposons that use LINE-1 reverse transcriptase for transposition (Alu-repeats and SVA-elements) are found in other structures (see discussion in [292])— however, the localization of non-coding RNAs, including transcripts of these elements, are beyond the scope of this review.
PECULIARITIES OF LOCALIZED TRANSLATION REGULATION
The question of whether there are any differences in the mechanisms of regulation of protein biosynthesis depending on the localization and translation of encoding mRNAs is of great interest in the study of local translation. In particular, it is unclear whether translation by free cytoplasmic ribosomes and ribosomes associated with organelle membranes is equally regulated, whether there are any peculiarities in the functioning and circulation of ribosomes, translation factors, and mRNA within and between different compartments. The set of hypotheses assumed in this regard is quite extensive and includes contrary statements. Some authors argue that localized translation is characterized by a special, compartment-dependent mode of control, which can differ significantly from the classic intracellular mechanisms of protein biosynthesis regulation. Polysomes localized on the membrane differ from cytosolic ones in dynamics and structure: for example, it was shown that these polysomes are much less mobile [10], and the number of ribosomes in them is, on average, greater than in the composition of free polysomes [7, 10]. This suggests that such separate regulation may well take place. However, other authors insist that there are no significant differences. The truth is probably somewhere in between, but investigation is complicated by the lack of experimental data on this topic, ambiguous interpretation, as well as the presence of publications with conflicting data. Below we propose to look at the arguments of both parties.
Arguments in favor of the hypothesis of compartment-dependent translation regulation. An ardent supporter of the compartment-specific translation hypothesis is C. Nicchitta. In a number of his studies, it is argued that mRNA can go through the entire translational cycle, from initiation to termination and recycling, on the ER membrane without exchange of ribosomes and mRNA with the cytosol [293–296]. This may occur due to the fact that ribosomes associated with the ER translocon or other ribosomal receptors on the ER, after termination, are able to remain on the membrane and initiate the translation of neighboring mRNAs, which can also be associated with the membrane through various mechanisms described earlier (Fig. 1). Moreover, this mechanism encompasses mRNAs encoding not only secreted or membrane proteins, but also cytosolic proteins [7, 8, 45], released after synthesis into the cytoplasm and in no way associated with the translocon. Thus, in fact, it is argued that translation processes in the cytosol and on the ER membrane can proceed independently of each other, which, in turn, creates the possibility of their compartment-dependent regulation.
Facts demonstrating such differentiated control have been provided by Nicchitta and co-authors. For example, they showed that in the case of certain stresses (stress of the ER and viral infection), translation in the cytosol was suppressed, while on the ER membrane it remained stable or even stimulated [297, 298]. In another study [299], selective relocalization of mRNA of membrane and secreted proteins from the ER membrane to the cytoplasm during ER stress was shown, while the mRNA of cytosolic proteins on ER, as well as mRNA of proteins participating in the stress response, did not change localization. This happened in the first half hour after stress, and by the end of the first hour, the mRNA that left the ER membrane returned. This way of decreasing the rate of protein entry into the ER by selective relocalization of mRNA has been proposed as an alternative or additional mechanism to the degradation of membrane-associated mRNA during the stress response [299]. The authors of these studies put forward a new paradigm, according to which the ER membrane is assigned a primary role in the localization of translation of most mRNAs (including those encoding cytosolic proteins) and in creating special conditions for the regulation of its translation [296]. Under stress, the ER membrane can serve as a platform for the synthesis of anti-stress proteins, even if these proteins are cytosolic.
This idea finds support in the studies of other scientific groups. The study by Staudacher et al. [300] demonstrates the selective relocalization of a number of mRNAs on the ER membrane and the stimulation of its translation on the membrane under hypoxic conditions. These mRNAs included transcripts encoding elements of an adaptive response to hypoxia, glycolysis enzymes, and other targets of the HIF1A transcription factor. At the same time, conservative motifs were found in their 5'- and 3'-UTRs, which are necessary for localization on the ER and facilitate translation on ER-associated ribosomes.
The specific localization of mRNA can also determine the rapid resumption of translation after the end of stress. For example, in the absence of the “mitochondrial anchor” SYNJ2BP, some mRNAs associated with the outer membrane of mitochondria are involved in translation much later [109].
When analyzing the differentiated effects of stress on translation in different compartments, one cannot ignore their different enrichment with translation factors. Thus, in a recent study by the Gromeier group, it was stated that the key initiation factor eIF2 is retained on the ER membranes through interaction with the CReP protein. According to the authors, this provides the selective resistance of translation of some transcripts (for example, mRNA of poliovirus or mRNA of the ER resident chaperone BiP) to stresses causing inactivation of eIF2 or eIF4F [276]. In addition, one should not forget that the localization of individual components can vary greatly depending on conditions—for example, the relocalization of a number of translation factors in stress granules can lead to a change in their distribution between compartments during stress.
From a historical point of view, the ideas of spatially-mediated regulation of protein synthesis are not new: similar questions were raised by the pioneers of localized translation, including Palade. However, in modern science, these ideas find both positive responses and criticism.
Arguments against compartment-dependent regulation and conflicting data. The issue of translation of mRNA encoding cytosolic proteins on the ER membrane remains controversial, since different studies give different estimates of the proportion of such transcripts. According to ribosome profiling of yeast and mammalian cells [8], they make up no more than 7–15%, adjusted for possible contamination. However, in [7] it is stated that at least 20% of “cytosolic” (according to the official annotation) proteins are synthesized by ER-associated ribosomes. Using another methodological approach based on the visualization of translation of single mRNA molecules, the authors of study [10] counted about 7–8% of the mRNA of a reporter protein with cytoplasmic localization, translated on the ER membrane. For another, secreted reporter, 60% of the encoding mRNA was associated with ER, indicating the dynamic nature of mRNA binding to the ER and the possibility of its circulation between the membrane and the cytoplasm.
In the study [8], the model of stable association of ribosomes with ER, even after translation termination, is refuted. The authors argue that, after termination, ribosomes rapidly dissociate from the membrane and exchange with cytosolic ones. This study gave rise to a controversy between two groups of scientists [301, 302], which vividly characterized the situation in the field of studying localized translation: due to the insufficient development of the issue, there may be different interpretations of the same results. New data brought some clarity to the discussed theme [11], showing both possible scenarios—under conditions of global cellular translation inhibition, ribosomes can dissociate from the ER membrane both quickly and slowly, depending on the receptor to which the ribosome is bound.
Two interesting observations at once were made in another study [303]. The authors showed that under oxidative stress induced by sodium arsenite, ER-associated mRNAs do not go into stress granules, as “cytosolic” transcripts do. The second interesting observation is that in cells exposed to oxidative stress, ER-associated polysomes are resistant to treatment with puromycin, while polysomes synthesizing cytosolic proteins disassembled when puromycin is added. The authors saw here an analogy with the “heavy” polysomes formed during mitosis: they are translationally less active than polysomes in the active growth phase, and are also insensitive to puromycin [304]. During cell division, this is necessary to protect mRNA from degradation and rapid resumption of translation after the end of mitosis—in accordance with this analogy, the authors of another study [7] showed that translation of a chimeric transcript targeted to ER recovers faster after stress cancellation than translation of a “cytosolic” reporter.
Structural and functional peculiarities of ribosomes depending on the type of translated mRNA. To conclude this section, we will discuss the evidences of differences in the dynamics and structure of ribosome, associated with membranes of organelles and localized in the cytoplasm, as well as the functional specialization of ribosomes that translate different mRNA pools.
Dozens of different ligands and partners interact with the ribosome at different stages of the translational cycle. In the case of two populations of ribosomes—free and associated with membranes—differences are found in the composition of related proteins and other components of the translational apparatus. The SRP particle, which interacts with the ribosome at the stages of SP or TMD recognition and transport of the complex to the ER membrane, is one such component. According to classical concepts, SRP recognizes SP or TMD as they emerge from the ribosomal tunnel. However, the amount of SRP in the cell is extremely small, and the question arises—how is the effective scanning of all translating ribosomes for the presence of SP or TMD ensured? In the study [305] this paradox was elegantly resolved—SRP can be associated with the ribosome, which synthesizes the secreted protein, even before the appearance of the N‑terminal SP from the ribosomal tunnel. According to this model, the primary recruitment of SPR to the ribosome occurs independently of the signal in the growing peptide, since it preliminarily binds to special regions in the 3'-UTR of mRNAs.
As the peptide is synthesized and appears from the ribosome, the above-mentioned NAC complex can interact with it. In yeast, it has been observed that different NACs (heterodimers of αβ-NAC and αβ'-NAC) may be involved in the biogenesis of different sets of proteins through the binding of different substrates. The αβ'-NAC complex has a specific role in the biogenesis of mitochondrial proteins, since it is predominantly associated with ribosomes that actively synthesize these proteins [85, 306].
These data are consistent with the “ribosome filter” hypothesis and the concept of “specialized ribosomes,” according to which ribosomes and ribosomal complexes are heterogeneous in composition, that can provide selective translation of various mRNA pools [307–310]. In addition to the classic components mentioned above, heterogeneity can also apply to other ribosome-associated proteins. For example, the protein PKM (muscle pyruvate kinase), was found in ribosomes localized on the membrane, exhibiting the activity of a non-canonical RNA-binding protein specific for transcripts localized on the ER [311]. We cannot but mention the idea of the “ribosome code” [312, 313], according to which different paralogs of ribosomal proteins can regulate cellular processes in different ways, in particular, translation of localized mRNAs [312]. In yeast, 59 of 79 ribosomal proteins are encoded by paralogous gene pairs, and although their sequences may show a high percentage of identity, the removal of different paralogs can lead to different phenotypes. In study [314], mRNA-specific regulation of the synthesis of mitochondrial proteins, which is accomplished by a pool of ribosomes with a distinct composition of paralogous proteins, was shown. However, some authors rightly point out that there are still not enough arguments in favor of the concept of “specialized ribosomes” [315].
METHODS OF SYSTEMS BIOLOGY IN STUDYING OF LOCALIZED TRANSLATION
A large number of discoveries in the field of localized translation, made recently and resulting in explosive interest in this area, would not have been possible without the development of a number of new experimental approaches based on the methods of systems biology. These approaches allow analyzing the localization of a large amount of mRNAs simultaneously and to reveal the translation pattern as a whole. The advent of microarrays, and then high-throughput sequencing, made it possible to determine the set of mRNAs associated with certain organelles. Initially, this was done by fractionation of the cytoplasm: for example, a genome-wide analysis of the transcriptome associated with membranes in yeast and human cells [2, 3], with mitochondria in the same organisms [88, 316, 317] and with peroxisomes in mice was carried out [129], as well as a set of transcripts localized in the pseudopodia of migrating fibroblasts [173] and in neuronal processes [318–320]. In later studies, ribosome profiling (isolation and sequencing of mRNA fragments contained in translating ribosomes) was performed from the membrane fraction of cells or different parts of neurons [7, 223].
The next generation of methods is associated with the in cellulo modification of RNA located in the compartment of interest, followed by its isolation and sequencing. Such local modification (including biotinylation or base oxidation) can be performed directly in a living cell by introducing a modifying enzyme gene (for example, APEX or miniSOG), providing its appropriate localization [5, 321, 322]. RNA-binding proteins can also be subjected to local biotinylation, which, after chemical cross-linking, makes it possible to isolate RNA-protein complexes and thus obtain even more information [4, 323].
The method of proximal-specific ribosome profiling, developed in the laboratory of J. Weissman [8, 73], is also extremely interesting. It allows labeling with biotin and isolating only those ribosomes that are currently in the compartment of interest, and then sequencing the mRNA fragments that are being translated by these ribosomes. This method allowed to assess the dynamics of translation of mRNA by ribosomes localized on the membranes of the ER [8] and mitochondria [73].
A methodological pinnacle of “localized transcriptomics” was the recently developed method for sequencing RNA fragments in situ, i.e. directly on a fixed sample of cells or tissues [324–326]. This approach yields qualitative and quantitative information about the distribution of transcripts on a slice or even in 3D, although the method is limited by the resolution of the microscope.
A fundamentally different approach is associated with the development of the method of fluorescent in situ hybridization (FISH). This method, even in its classic version, made it possible to collect information on the subcellular localization of hundreds and even thousands of individual transcripts [225], although this required the production of individual fixed samples. Modern variants of FISH make use of multiplexing to allow detection of hundreds and thousands of individual transcripts simultaneously, and immediately in a large number of cells [130, 226, 327–329]. In combination with advanced methods of fluorescence imaging of single reporter mRNA molecules that have appeared recently [330], they also give the opportunity to assess the heterogeneity in the nature of the translation of individual transcripts. The disadvantage of these methods over approaches using sequencing is the “analog” principle of signal generation, but it is they that allow direct visualization of the location of transcripts in the cell.
The use of systems biology methods have provided ample evidence that a significant proportion of mRNA (depending on the organism, type of cells, stage of development, conditions and the applied method—from a few percent to ~ 3/4 of the detected transcripts) has a clearly determined subcellular localization in the cell: apical-basal, in protrusions and places of cell contacts, associated with membranes of organelles, with centrosome, microtubules, various types of granules, and so on [13, 225, 331–333]. Thus, targeted delivery of mRNA with its subsequent localized translation is an extremely important mechanism for the formation of intracellular structures, compartments, and functionally distinct poles of cells.
CONCLUSIONS
The above mentioned facts allow us to conclude that the role of intracellular localization of mRNA and local translation in maintaining the integrity of the proteome and in the normal functioning of a eukaryotic cell is undoubtedly high. The study of these aspects of protein biosynthesis is of great interest in modern science and is progressing rapidly. This is facilitated by an ever-expanding arsenal of various methods. Nevertheless, there are still quite a few unresolved issues. For example, while the processes of co-translational recruitment of ribosomes to the ER membrane and translocation of the peptide into the lumen are described in sufficient detail, similar events on mitochondria, and even more so the co-translational events in proximity to other organelles, require more in-depth study. The contribution of “translation factories” and other RNA condensates to the vital processes of the cell is unclear, their structural properties have not been studied, and there are practically no data on what happens to these elements of the cell under conditions of stress, viral infections and other perturbations. Questions about the features of the structure of translational complexes and the regulation of protein biosynthesis in different subcellular compartments remain unresolved. Nevertheless, the ever-increasing amount of studies in this area allows us to hope that in the near future many of these mysteries will be solved, and many more striking discoveries in this exciting area await us.
FUNDING
This work was supported by the Russian Foundation for Basic Research (“Expansion” program, project no. 19-14-50309). The authors are members of the Interdisciplinary Scientific and Educational School of Moscow University “Molecular Technologies of the Living Systems and Synthetic Biology.”
COMPLIANCE WITH ETHICAL STANDARDS
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
ADDITIONAL INFORMATION
The text was submitted by the authors in English.
Footnotes
Abbreviations: ER, endoplasmic reticulum; SRP, signal recognition particle; SP, signal peptide; TMD, transmembrane domain; MTS, mitochondria targeting sequences; PTS, peroxisome targeting sequences; UTR, untranslated region; NAC, nascent polypeptide-associated complex; UPR, unfolded protein response; SG, stress-granule; PB, processing body; RNP, ribonucleoprotein.
REFERENCES
- 1.Palade G.E. A small particulate component of the cytoplasm. J. Biophys. Biochem. Cytol. 1955;1:59–68. doi: 10.1083/jcb.1.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diehn M., Bhattacharya R., Botstein D., Brown P.O. Genome-scale identification of membrane-associated human mRNAs. PLoS Genet. 2006;2:e11. doi: 10.1371/journal.pgen.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diehn M., Eisen M.B., Botstein D., Brown P.O. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat. Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- 4.Kaewsapsak P., Shechner D.M., Mallard W., Rinn J.L., Ting A.Y. Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. eLife. 2017;6:e29224. doi: 10.7554/eLife.29224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fazal F.M., Han S., Parker K.R., Kaewsapsak P., Xu J., Boettiger A.N., Chang H.Y., Ting A.Y. Atlas of subcellular RNA localization revealed by APEX-Seq. Cell. 2019;178:473–490, e426. doi: 10.1016/j.cell.2019.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lerner R.S., Seiser R.M., Zheng T., Lager P.J., Reedy M.C., Keene J.D., Nicchitta C.V. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reid D.W., Nicchitta C.V. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J. Biol. Chem. 2012;287:5518–5527. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jan C.H., Williams C.C., Weissman J.S. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jagannathan S., Hsu J.C., Reid D.W., Chen Q., Thompson W.J., Moseley A.M., Nicchitta C.V. Multifunctional roles for the protein translocation machinery in RNA anchoring to the endoplasmic reticulum. J. Biol. Chem. 2014;289:25907–25924. doi: 10.1074/jbc.M114.580688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voigt F., Zhang H., Cui X.A., Triebold D., Liu A.X., Eglinger J., Lee E.S., Chao J.A., Palazzo A.F. Single-molecule quantification of translation-dependent association of mRNAs with the endoplasmic reticulum. Cell Rep. 2017;21:3740–3753. doi: 10.1016/j.celrep.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman A.M., Chen Q., Zheng T., Nicchitta C.V. Heterogeneous translational landscape of the endoplasmic reticulum revealed by ribosome proximity labeling and transcriptome analysis. J. Biol. Chem. 2019;294:8942–8958. doi: 10.1074/jbc.RA119.007996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraut-Cohen J., Afanasieva E., Haim-Vilmovsky L., Slobodin B., Yosef I., Bibi E., Gerst J.E. Translation- and SRP-independent mRNA targeting to the endoplasmic reticulum in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2013;24:3069–3084. doi: 10.1091/mbc.E13-01-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weis B.L., Schleiff E., Zerges W. Protein targeting to subcellular organelles via MRNA localization. Biochim. Biophys. Acta. 2013;1833:260–273. doi: 10.1016/j.bbamcr.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Kraut-Cohen J., Gerst J.E. Addressing mRNAs to the ER: cis sequences act up! Trends Biochem. Sci. 2010;35:459–469. doi: 10.1016/j.tibs.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Ma W., Mayr C. 2018. A membraneless organelle associated with the endoplasmic reticulum enables 3'UTR-mediated protein–protein interactions. Cell. 175, 1492‒1506, e1419. 10.1016/j.cell.2018.10.007 [DOI] [PMC free article] [PubMed]
- 16.Blobel G., Sabatini D.D. Biomembranes. Boston, MA: Springer; 1971. [Google Scholar]
- 17.Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 1975;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walter P., Ibrahimi I., Blobel G. Translocation of proteins across the endoplasmic reticulum: 1. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilmore R., Blobel G., Walter P. Protein translocation across the endoplasmic reticulum: 1. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 1982;95:463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nilsson I., Lara P., Hessa T., Johnson A.E., von Heijne G., Karamyshev A.L. The code for directing proteins for translocation across ER membrane: SRP cotranslationally recognizes specific features of a signal sequence. J. Mol. Biol. 2015;427:1191–1201. doi: 10.1016/j.jmb.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costa E.A., Subramanian K., Nunnari J., Weissman J.S. Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science. 2018;359:689–692. doi: 10.1126/science.aar3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuo Y., Inada T. The ribosome collision sensor Hel2 functions as preventive quality control in the secretory pathway. Cell Rep. 2021;34:108877. doi: 10.1016/j.celrep.2021.108877. [DOI] [PubMed] [Google Scholar]
- 23.Pfeffer S., Burbaum L., Unverdorben P., Pech M., Chen Y., Zimmermann R., Beckmann R., Forster F. Structure of the native Sec61 protein-conducting channel. Nat. Commun. 2015;6:8403. doi: 10.1038/ncomms9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voorhees R.M., Hegde R.S. Structure of the Sec61 channel opened by a signal sequence. Science. 2016;351:88–91. doi: 10.1126/science.aad4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nyathi Y., Wilkinson B.M., Pool M.R. Co-translational targeting and translocation of proteins to the endoplasmic reticulum. Biochim. Biophys. Acta. 2013;1833:2392–2402. doi: 10.1016/j.bbamcr.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 26.Aviram N., Schuldiner M. Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J. Cell Sci. 2017;130:4079–4085. doi: 10.1242/jcs.204396. [DOI] [PubMed] [Google Scholar]
- 27.Hetz C., Papa F.R. The unfolded protein response and cell fate control. Mol. Cell. 2018;69:169–181. doi: 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 28.Yanagitani K., Imagawa Y., Iwawaki T., Hosoda A., Saito M., Kimata Y., Kohno K. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol. Cell. 2009;34:191–200. doi: 10.1016/j.molcel.2009.02.033. [DOI] [PubMed] [Google Scholar]
- 29.Yanagitani K., Kimata Y., Kadokura H., Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331:586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- 30.Shanmuganathan V., Schiller N., Magoulopoulou A., Cheng J., Braunger K., Cymer F., Berninghausen O., Beatrix B., Kohno K., von Heijne G., Beckmann R. Structural and mutational analysis of the ribosome-arresting human XBP1u. eLife. 2019;8:e46267. doi: 10.7554/eLife.46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanda S., Yanagitani K., Yokota Y., Esaki Y., Kohno K. Autonomous translational pausing is required for XBP1u mRNA recruitment to the ER via the SRP pathway. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E5886–E5895. doi: 10.1073/pnas.1604435113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plumb R., Zhang Z.R., Appathurai S., Mariappan M. A functional link between the co-translational protein translocation pathway and the UPR. eLife. 2015;4:e07426. doi: 10.7554/eLife.07426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C.Y., Malchus N.S., Hehn B., Stelzer W., Avci D., Langosch D., Lemberg M.K. Signal peptide peptidase functions in ERAD to cleave the unfolded protein response regulator XBP1u. EMBO J. 2014;33:2492–2506. doi: 10.15252/embj.201488208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han P., Shichino Y., Schneider-Poetsch T., Mito M., Hashimoto S., Udagawa T., Kohno K., Yoshida M., Mishima Y., Inada T., Iwasaki S. Genome-wide survey of ribosome collision. Cell Rep. 2020;31:107610. doi: 10.1016/j.celrep.2020.107610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mutka S.C., Walter P. Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol. Biol. Cell. 2001;12:577–588. doi: 10.1091/mbc.12.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L., Liang X.H., Uliel S., Unger R., Ullu E., Michaeli S. RNA interference of signal peptide-binding protein SRP54 elicits deleterious effects and protein sorting defects in trypanosomes. J. Biol. Chem. 2002;277:47348–47357. doi: 10.1074/jbc.M207736200. [DOI] [PubMed] [Google Scholar]
- 37.Ren Y.G., Wagner K.W., Knee D.A., Aza-Blanc P., Nasoff M., Deveraux Q.L. Differential regulation of the TRAIL death receptors DR4 and DR5 by the signal recognition particle. Mol. Biol. Cell. 2004;15:5064–5074. doi: 10.1091/mbc.e04-03-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson N., Powis K., High S. Post-translational translocation into the endoplasmic reticulum. Biochim. Biophys. Acta. 2013;1833:2403–2409. doi: 10.1016/j.bbamcr.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Shao S., Hegde R.S. A calmodulin-dependent translocation pathway for small secretory proteins. Cell. 2011;147:1576–1588. doi: 10.1016/j.cell.2011.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ast T., Schuldiner M. All roads lead to Rome (but some may be harder to travel): SRP-independent translocation into the endoplasmic reticulum. Crit. Rev. Biochem. Mol. Biol. 2013;48:273–288. doi: 10.3109/10409238.2013.782999. [DOI] [PubMed] [Google Scholar]
- 41.Borgese N., Coy-Vergara J., Colombo S.F., Schwappach B. The ways of tails: The GET pathway and more. Protein J. 2019;38:289–305. doi: 10.1007/s10930-019-09845-4. [DOI] [PubMed] [Google Scholar]
- 42.Liao G., Ma X., Liu G. An RNA-zipcode-independent mechanism that localizes Dia1 mRNA to the perinuclear ER through interactions between Dia1 nascent peptide and Rho-GTP. J. Cell Sci. 2011;124:589–599. doi: 10.1242/jcs.072421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christensen A.K., Kahn L.E., Bourne C.M. Circular polysomes predominate on the rough endoplasmic reticulum of somatotropes and mammotropes in the rat anterior pituitary. Am. J. Anat. 1987;178:1–10. doi: 10.1002/aja.1001780102. [DOI] [PubMed] [Google Scholar]
- 44.Alekhina O.M., Terenin I.M., Dmitriev S.E., Vassilenko K.S. Functional cyclization of eukaryotic mRNAs. Int. J. Mol. Sci. 2020;21:1677. doi: 10.3390/ijms21051677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pyhtila B., Zheng T., Lager P.J., Keene J.D., Reedy M.C., Nicchitta C.V. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA. 2008;14:445–453. doi: 10.1261/rna.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loya A., Pnueli L., Yosefzon Y., Wexler Y., Ziv-Ukelson M., Arava Y. The 3'-UTR mediates the cellular localization of an mRNA encoding a short plasma membrane protein. RNA. 2008;14:1352–1365. doi: 10.1261/rna.867208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen-Zontag O., Baez C., Lim L.Q.J., Olender T., Schirman D., Dahary D., Pilpel Y., Gerst J.E. A secretion-enhancing cis regulatory targeting element (SECReTE. involved in mRNA localization and protein synthesis. PLoS Genet. 2019;15:e1008248. doi: 10.1371/journal.pgen.1008248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma W., Zheng G., Xie W., Mayr C. 2021. In vivo reconstitution finds multivalent RNA-RNA interactions as drivers of mesh-like condensates. Elife. 10, e64252.10.7554/eLife.64252 [DOI] [PMC free article] [PubMed]
- 49.Cui X.A., Zhang H., Palazzo A.F. p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PLoS Biol. 2012;10:e1001336. doi: 10.1371/journal.pbio.1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savitz A.J., Meyer D.I. Identification of a ribosome receptor in the rough endoplasmic reticulum. Nature. 1990;346:540–544. doi: 10.1038/346540a0. [DOI] [PubMed] [Google Scholar]
- 51.Ueno T., Kaneko K., Sata T., Hattori S., Ogawa-Goto K. Regulation of polysome assembly on the endoplasmic reticulum by a coiled-coil protein, p180. Nucleic Acids Res. 2012;40:3006–3017. doi: 10.1093/nar/gkr1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsu J.C., Reid D.W., Hoffman A.M., Sarkar D., Nicchitta C.V. Oncoprotein AEG-1 is an endoplasmic reticulum RNA-binding protein whose interactome is enriched in organelle resident protein-encoding mRNAs. RNA. 2018;24:688–703. doi: 10.1261/rna.063313.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Acosta-Alvear D., Karagoz G.E., Frohlich F., Li H., Walther T.C., Walter P. The unfolded protein response and endoplasmic reticulum protein targeting machineries converge on the stress sensor IRE1. eLife. 2018;7:e43036. doi: 10.7554/eLife.43036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Genz C., Fundakowski J., Hermesh O., Schmid M., Jansen R.P. Association of the yeast RNA-binding protein She2p with the tubular endoplasmic reticulum depends on membrane curvature. J. Biol. Chem. 2013;288:32384–32393. doi: 10.1074/jbc.M113.486431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colomina N., Ferrezuelo F., Wang H., Aldea M., Gari E. Whi3, a developmental regulator of budding yeast, binds a large set of mRNAs functionally related to the endoplasmic reticulum. J. Biol. Chem. 2008;283:28670–28679. doi: 10.1074/jbc.M804604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Syed M.I., Moorthy B.T., Jenner A., Fetka I., Jansen R.P. Signal sequence-independent targeting of MID2 mRNA to the endoplasmic reticulum by the yeast RNA-binding protein Khd1p. FEBS Lett. 2018;592:1870–1881. doi: 10.1002/1873-3468.13098. [DOI] [PubMed] [Google Scholar]
- 57.Castello A., Fischer B., Frese C.K., Horos R., Alleaume A.M., Foehr S., Curk T., Krijgsveld J., Hentze M.W. Comprehensive identification of RNA-binding domains in human cells. Mol. Cell. 2016;63:696–710. doi: 10.1016/j.molcel.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baltz A.G., Munschauer M., Schwanhausser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M., Wyler E., Bonneau R., Selbach M., Dieterich C., Landthaler M. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 59.Bethune J., Jansen R.P., Feldbrugge M., Zarnack K. Membrane-associated RNA-binding proteins orchestrate organelle-coupled translation. Trends Cell Biol. 2019;29:178–188. doi: 10.1016/j.tcb.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 60.Kalies K.U., Gorlich D., Rapoport T.A. Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J. Cell Biol. 1994;126:925–934. doi: 10.1083/jcb.126.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harada Y., Li H., Li H., Lennarz W.J. Oligosaccharyltransferase directly binds to ribosome at a location near the translocon-binding site. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6945–6949. doi: 10.1073/pnas.0812489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfeffer S., Dudek J., Zimmermann R., Forster F. Organization of the native ribosome-translocon complex at the mammalian endoplasmic reticulum membrane. Biochim. Biophys. Acta. 2016;1860:2122–2129. doi: 10.1016/j.bbagen.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 63.Hansen K.G., Herrmann J.M. Transport of proteins into mitochondria. Protein J. 2019;38:330–342. doi: 10.1007/s10930-019-09819-6. [DOI] [PubMed] [Google Scholar]
- 64.Becker T., Song J. Versatility of preprotein transfer from the cytosol to mitochondria. Trends Cell Biol. 2019;29:534–548. doi: 10.1016/j.tcb.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 65.Wiedemann N., Pfanner N. Mitochondrial machineries for protein import and assembly. Ann. Rev. Biochem. 2017;86:685–714. doi: 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- 66.Kellems R.E., Butow R.A. Cytoplasmic-type 80 S ribosomes associated with yeast mitochondria. I. Evidence for ribosome binding sites on yeast mitochondria. J. Biol. Chem. 1972;247:8043–8050. doi: 10.1016/S0021-9258(20)81806-7. [DOI] [PubMed] [Google Scholar]
- 67.Kellems R.E., Allison V.F., Butow R.A. Cytoplasmic type 80S ribosomes associated with yeast mitochondria: 2. Evidence for the association of cytoplasmic ribosomes with the outer mitochondrial membrane in situ. J. Biol. Chem. 1974;249:3297–3303. doi: 10.1016/S0021-9258(19)42672-0. [DOI] [PubMed] [Google Scholar]
- 68.Suissa M., Schatz G. Import of proteins into mitochondria. Translatable mRNAs for imported mitochondrial proteins are present in free as well as mitochondria-bound cytoplasmic polysomes. J. Biol. Chem. 1982;257:13048–13055. doi: 10.1016/S0021-9258(18)33620-2. [DOI] [PubMed] [Google Scholar]
- 69.Ades I.Z., Butow R.A. The products of mitochondria-bound cytoplasmic polysomes in yeast. J. Biol. Chem. 1980;255:9918–9924. doi: 10.1016/S0021-9258(18)43480-1. [DOI] [PubMed] [Google Scholar]
- 70.Fujiki M., Verner K. Coupling of cytosolic protein synthesis and mitochondrial protein import in yeast. Evidence for cotranslational import in vivo. J. Biol. Chem. 1993;268:1914–1920. doi: 10.1016/S0021-9258(18)53941-7. [DOI] [PubMed] [Google Scholar]
- 71.Mukhopadhyay A., Ni L., Weiner H. A co-translational model to explain the in vivo import of proteins into HeLa cell mitochondria. Biochem. J. 2004;382:385–392. doi: 10.1042/BJ20040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yogev O., Karniely S., Pines O. Translation-coupled translocation of yeast fumarase into mitochondria in vivo. J. Biol. Chem. 2007;282:29222–29229. doi: 10.1074/jbc.M704201200. [DOI] [PubMed] [Google Scholar]
- 73.Williams C.C., Jan C.H., Weissman J.S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.MacKenzie J.A., Payne R.M. Ribosomes specifically bind to mammalian mitochondria via protease-sensitive proteins on the outer membrane. J. Biol. Chem. 2004;279:9803–9810. doi: 10.1074/jbc.M307167200. [DOI] [PubMed] [Google Scholar]
- 75.Gold V.A., Chroscicki P., Bragoszewski P., Chacinska A. Visualization of cytosolic ribosomes on the surface of mitochondria by electron cryo-tomography. EMBO Rep. 2017;18:1786–1800. doi: 10.15252/embr.201744261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eliyahu E., Pnueli L., Melamed D., Scherrer T., Gerber A.P., Pines O., Rapaport D., Arava Y. Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner. Mol. Cell. Biol. 2010;30:284–294. doi: 10.1128/MCB.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.George R., Beddoe T., Landl K., Lithgow T. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2296–2301. doi: 10.1073/pnas.95.5.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.George R., Walsh P., Beddoe T., Lithgow T. The nascent polypeptide-associated complex (NAC) promotes interaction of ribosomes with the mitochondrial surface in vivo. FEBS Lett. 2002;516:213–216. doi: 10.1016/s0014-5793(02)02528-0. [DOI] [PubMed] [Google Scholar]
- 79.Gamerdinger M., Hanebuth M.A., Frickey T., Deuerling E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science. 2015;348:201–207. doi: 10.1126/science.aaa5335. [DOI] [PubMed] [Google Scholar]
- 80.Wiedmann B., Sakai H., Davis T.A., Wiedmann M. A protein complex required for signal-sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- 81.Powers T., Walter P. The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr. Biol. 1996;6:331–338. doi: 10.1016/s0960-9822(02)00484-0. [DOI] [PubMed] [Google Scholar]
- 82.Hsieh H.H., Lee J.H., Chandrasekar S., Shan S.O. A ribosome-associated chaperone enables substrate triage in a cotranslational protein targeting complex. Nat. Commun. 2020;11:5840. doi: 10.1038/s41467-020-19548-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Funfschilling U., Rospert S. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell. 1999;10:3289–3299. doi: 10.1091/mbc.10.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lesnik C., Cohen Y., Atir-Lande A., Schuldiner M., Arava Y. OM14 is a mitochondrial receptor for cytosolic ribosomes that supports co-translational import into mitochondria. Nat. Commun. 2014;5:5711. doi: 10.1038/ncomms6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ponce-Rojas J.C., Avendano-Monsalve M.C., Yanez-Falcon A.R., Jaimes-Miranda F., Garay E., Torres-Quiroz F., DeLuna A., Funes S. αβ'-NAC cooperates with Sam37 to mediate early stages of mitochondrial protein import. FEBS J. 2017;284:814–830. doi: 10.1111/febs.14024. [DOI] [PubMed] [Google Scholar]
- 86.Ahmed A.U., Fisher P.R. Import of nuclear-encoded mitochondrial proteins: a cotranslational perspective. Int. Rev. Cell Mol. Biol. 2009;273:49–68. doi: 10.1016/S1937-6448(08)01802-9. [DOI] [PubMed] [Google Scholar]
- 87.Lesnik C., Golani-Armon A., Arava Y. Localized translation near the mitochondrial outer membrane: An update. RNA Biol. 2015;12:801–809. doi: 10.1080/15476286.2015.1058686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sylvestre J., Vialette S., Corral Debrinski M., Jacq C. Long mRNAs coding for yeast mitochondrial proteins of prokaryotic origin preferentially localize to the vicinity of mitochondria. Genome Biol. 2003;4:R44. doi: 10.1186/gb-2003-4-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gadir N., Haim-Vilmovsky L., Kraut-Cohen J., Gerst J.E. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA. 2011;17:1551–1565. doi: 10.1261/rna.2621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Matsumoto S., Uchiumi T., Saito T., Yagi M., Takazaki S., Kanki T., Kang D. Localization of mRNAs encoding human mitochondrial oxidative phosphorylation proteins. Mitochondrion. 2012;12:391–398. doi: 10.1016/j.mito.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 91.Zabezhinsky D., Slobodin B., Rapaport D., Gerst J.E. An essential role for COPI in mRNA localization to mitochondria and mitochondrial function. Cell Rep. 2016;15:540–549. doi: 10.1016/j.celrep.2016.03.053. [DOI] [PubMed] [Google Scholar]
- 92.Sabharwal A., Sharma D., Vellarikkal S.K., Jayarajan R., Verma A., Senthivel V., Scaria V., Sivasubbu S. Organellar transcriptome sequencing reveals mitochondrial localization of nuclear encoded transcripts. Mitochondrion. 2019;46:59–68. doi: 10.1016/j.mito.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 93.Golani-Armon A., Arava Y. Localization of nuclear-encoded mRNAs to mitochondria outer surface. Biochemistry. 2016;81:1038–1043. doi: 10.1134/S0006297916100023. [DOI] [PubMed] [Google Scholar]
- 94.Sylvestre J., Margeot A., Jacq C., Dujardin G., Corral-Debrinski M. The role of the 3' untranslated region in mRNA sorting to the vicinity of mitochondria is conserved from yeast to human cells. Mol. Biol. Cell. 2003;14:3848–3856. doi: 10.1091/mbc.e03-02-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Garcia M., Delaveau T., Goussard S., Jacq C. Mitochondrial presequence and open reading frame mediate asymmetric localization of messenger RNA. EMBO Rep. 2010;11:285–291. doi: 10.1038/embor.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eliyahu E., Lesnik C., Arava Y. The protein chaperone Ssa1 affects mRNA localization to the mitochondria. FEBS Lett. 2012;586:64–69. doi: 10.1016/j.febslet.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 97.Corral-Debrinski M., Blugeon C., Jacq C. In yeast, the 3' untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 2000;20:7881–7892. doi: 10.1128/mcb.20.21.7881-7892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Poulsen T.M., Imai K., Frith M.C., Horton P. 2019. Hallmarks of slow translation initiation revealed in mitochondrially localizing mRNA sequences. bioRxiv. 614255. 10.1101/614255
- 99.Lin Y., Li F., Huang L., Polte C., Duan H., Fang J., Sun L., Xing X., Tian G., Cheng Y., Ignatova Z., Yang X., Wolf D.A. eIF3 associates with 80S ribosomes to promote translation elongation, mitochondrial homeostasis, and muscle health. Mol. Cell. 2020;79:575–587, e7. doi: 10.1016/j.molcel.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 100.Schatton D., Rugarli E.I. A concert of RNA-binding proteins coordinates mitochondrial function. Crit. Rev. Biochem. Mol. Biol. 2018;53:652–666. doi: 10.1080/10409238.2018.1553927. [DOI] [PubMed] [Google Scholar]
- 101.Gerber A.P., Herschlag D., Brown P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jacobs Anderson J.S., Parker R. Computational identification of cis-acting elements affecting post-transcriptional control of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 2000;28:1604–1617. doi: 10.1093/nar/28.7.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saint-Georges Y., Garcia M., Delaveau T., Jourdren L., Le Crom S., Lemoine S., Tanty V., Devaux F., Jacq C. Yeast mitochondrial biogenesis: A role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One. 2008;3:e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Quenault T., Lithgow T., Traven A. PUF proteins: Repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 105.Lee C.D., Tu B.P. Glucose-regulated phosphorylation of the PUF protein Puf3 regulates the translational fate of its bound mRNAs and association with RNA granules. Cell Rep. 2015;11:1638–1650. doi: 10.1016/j.celrep.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kershaw C.J., Costello J.L., Talavera D., Rowe W., Castelli L.M., Sims P.F., Grant C.M., Ashe M.P., Hubbard S.J., Pavitt G.D. Integrated multi-omics analyses reveal the pleiotropic nature of the control of gene expression by Puf3p. Sci. Rep. 2015;5:15518. doi: 10.1038/srep15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lapointe C.P., Stefely J.A., Jochem A., Hutchins P.D., Wilson G.M., Kwiecien N.W., Coon J.J., Wickens M., Pagliarini D.J. 2018. Multi-omics reveal specific targets of the RNA-binding protein Puf3p and its orchestration of mitochondrial biogenesis. Cell Syst. 6, 125–135, e126. 10.1016/j.cels.2017.11.012 [DOI] [PMC free article] [PubMed]
- 108.Wang Z., Sun X., Wee J., Guo X., Gu Z. Novel insights into global translational regulation through Pumilio family RNA-binding protein Puf3p revealed by ribosomal profiling. Curr. Genet. 2019;65:201–212. doi: 10.1007/s00294-018-0862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qin W., Myers S.A., Carey D.K., Carr S.A., Ting A.Y. 2020. Functional proximity mapping of RNA binding proteins uncovers a mitochondrial mRNA anchor that promotes stress recovery. bioRxiv. 2020.11.17.387209. 10.1101/2020.11.17.387209
- 110.Gehrke S., Wu Z., Klinkenberg M., Sun Y., Auburger G., Guo S., Lu B. PINK1 and Parkin control localized translation of respiratory chain component mRNAs on mitochondria outer membrane. Cell Metab. 2015;21:95–108. doi: 10.1016/j.cmet.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang Y., Wang Z. H., Liu Y., Chen Y., Sun N., Gucek M., Zhang, F., Xu H. 2019. PINK1 inhibits local protein synthesis to limit transmission of deleterious mitochondrial DNA mutations. Mol. Cell. 73, 1127–1137, e1125. 10.1016/j.molcel.2019.01.013 [DOI] [PMC free article] [PubMed]
- 112.Zhang Y., Chen Y., Gucek M., Xu H. The mitochondrial outer membrane protein MDI promotes local protein synthesis and mtDNA replication. EMBO J. 2016;35:1045–1057. doi: 10.15252/embj.201592994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gao J., Schatton D., Martinelli P., Hansen H., Pla-Martin D., Barth E., Becker C., Altmueller J., Frommolt P., Sardiello M., Rugarli E.I. CLUH regulates mitochondrial biogenesis by binding mRNAs of nuclear-encoded mitochondrial proteins. J. Cell Biol. 2014;207:213–223. doi: 10.1083/jcb.201403129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schatton D., Pla-Martin D., Marx M.C., Hansen H., Mourier A., Nemazanyy I., Pessia A., Zentis P., Corona T., Kondylis V., Barth E., Schauss A.C., Velagapudi V., Rugarli E.I. CLUH regulates mitochondrial metabolism by controlling translation and decay of target mRNAs. J. Cell Biol. 2017;216:675–693. doi: 10.1083/jcb.201607019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sen A., Kalvakuri S., Bodmer R., Cox R.T. Clueless, a protein required for mitochondrial function, interacts with the PINK1-Parkin complex in Drosophila. Dis. Model. Mech. 2015;8:577–589. doi: 10.1242/dmm.019208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sen A., Cox R.T. Clueless is a conserved ribonucleoprotein that binds the ribosome at the mitochondrial outer membrane. Biol. Open. 2016;5:195–203. doi: 10.1242/bio.015313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Giacomello M., Pellegrini L. The coming of age of the mitochondria–ER contact: A matter of thickness. Cell Death Differ. 2016;23:1417–1427. doi: 10.1038/cdd.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shimizu S. Organelle zones in mitochondria. J. Biochem. 2019;165:101–107. doi: 10.1093/jb/mvy068. [DOI] [PubMed] [Google Scholar]
- 119.Yogev O., Pines O. Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim. Biophys. Acta. 2011;1808:1012–1020. doi: 10.1016/j.bbamem.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 120.Hansen K.G., Aviram N., Laborenz J., Bibi C., Meyer M., Spang A., Schuldiner M., Herrmann J.M. An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science. 2018;361:1118–1122. doi: 10.1126/science.aar8174. [DOI] [PubMed] [Google Scholar]
- 121.Farre J.C., Mahalingam S.S., Proietto M., Subramani S. Peroxisome biogenesis, membrane contact sites, and quality control. EMBO Rep. 2019;20:e46864. doi: 10.15252/embr.201846864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Islinger M., Voelkl A., Fahimi H.D., Schrader M. The peroxisome: An update on mysteries 2.0. Histochem. Cell Biol. 2018;150:443–471. doi: 10.1007/s00418-018-1722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mayerhofer P.U., Bano-Polo M., Mingarro I., Johnson A.E. Human peroxin PEX3 is co-translationally integrated into the ER and exits the ER in budding vesicles. Traffic. 2016;17:117–130. doi: 10.1111/tra.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ast J., Stiebler A.C., Freitag J., Bolker M. Dual targeting of peroxisomal proteins. Front. Physiol. 2013;4:297. doi: 10.3389/fphys.2013.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jungreis I., Chan C.S., Waterhouse R.M., Fields G., Lin M.F., Kellis M. Evolutionary dynamics of abundant stop codon readthrough. Mol. Biol. Evol. 2016;33:3108–3132. doi: 10.1093/molbev/msw189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schueren F., Thoms S. Functional translational readthrough: a systems biology perspective. PLoS Genet. 2016;12:e1006196. doi: 10.1371/journal.pgen.1006196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Haimovich G., Cohen-Zontag O., Gerst J.E. A role for mRNA trafficking and localized translation in peroxisome biogenesis and function? Biochim. Biophys. Acta. 2016;1863:911–921. doi: 10.1016/j.bbamcr.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 128.Zipor G., Haim-Vilmovsky L., Gelin-Licht R., Gadir N., Brocard C., Gerst J. E. Localization of mRNAs coding for peroxisomal proteins in the yeast, Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19848–19853. doi: 10.1073/pnas.0910754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yarmishyn A.A., Kremenskoy M., Batagov A.O., Preuss A., Wong J.H., Kurochkin I.V. Genome-wide analysis of mRNAs associated with mouse peroxisomes. BMC Genomics. 2016;17:1028. doi: 10.1186/s12864-016-3330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chouaib R., Safieddine A., Pichon X., Imbert A., Kwon O.S., Samacoits A., Traboulsi A.M., Robert M.C., Tsanov N., Coleno E., Poser I., Zimmer C., Hyman A., Le Hir H., Zibara K. A dual protein-mRNA localization screen reveals compartmentalized translation and widespread co-translational RNA targeting. Dev. Cell. 2020;54:773–791, e5. doi: 10.1016/j.devcel.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 131.Baumann S., Konig J., Koepke J., Feldbrugge M. Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep. 2014;15:94–102. doi: 10.1002/embr.201338037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Higuchi Y., Ashwin P., Roger Y., Steinberg G. Early endosome motility spatially organizes polysome distribution. J. Cell Biol. 2014;204:343–357. doi: 10.1083/jcb.201307164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cioni J.M., Lin J.Q., Holtermann A.V., Koppers M., Jakobs M.A.H., Azizi A., Turner-Bridger B., Shigeoka T., Franze K., Harris W.A., Holt C.E. Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell. 2019;176:56–72, e15. doi: 10.1016/j.cell.2018.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.McLeod T., Abdullahi A., Li M., Brogna S. Recent studies implicate the nucleolus as the major site of nuclear translation. Biochem. Soc. Trans. 2014;42:1224–1228. doi: 10.1042/BST20140062. [DOI] [PubMed] [Google Scholar]
- 135.Reid D.W., Nicchitta C.V. The enduring enigma of nuclear translation. J. Cell Biol. 2012;197:7–9. doi: 10.1083/jcb.201202140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dahlberg J., Lund E. Nuclear translation or nuclear peptidyl transferase? Nucleus. 2012;3:320–321. doi: 10.4161/nucl.20754. [DOI] [PubMed] [Google Scholar]
- 137.Allfrey V.G. Amino acid incorporation by isolated thymus nuclei: 1. The role of desoxyribonucleic acid in protein synthesis. Proc. Natl. Acad. Sci. U. S. A. 1954;40:881–885. doi: 10.1073/pnas.40.10.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Allfrey V.G., Mirsky A.E. Protein synthesis in isolated cell nuclei. Nature. 1955;176:1042–1049. doi: 10.1038/1761042a0. [DOI] [PubMed] [Google Scholar]
- 139.Stvolinskaya N.S., Gazaryan K.G. The problem of intranuclear protein synthesis. Tsitologiya. 1976;18:125–138. [PubMed] [Google Scholar]
- 140.Goidl J.A., Canaani D., Boublik M., Weissbach H., Dickerman H. Polyanion-induced release of polyribosomes from HeLa cell nuclei. J. Biol. Chem. 1975;250:9198–9205. doi: 10.1016/S0021-9258(19)40706-0. [DOI] [PubMed] [Google Scholar]
- 141.Birnstiel M.L., Hyde B.B. Protein synthesis by isolated pea nucleoli. J. Cell Biol. 1963;18:41–50. doi: 10.1083/jcb.18.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pena C., Hurt E., Panse V.G. Eukaryotic ribosome assembly, transport and quality control. Nat. Struct. Mol. Biol. 2017;24:689–699. doi: 10.1038/nsmb.3454. [DOI] [PubMed] [Google Scholar]
- 143.Espinar-Marchena F.J., Babiano R., Cruz J. Placeholder factors in ribosome biogenesis: Please, pave my way. Microb. Cell. 2017;4:144–168. doi: 10.15698/mic2017.05.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dahlberg J.E., Lund E., Goodwin E.B. Nuclear translation: What is the evidence? RNA. 2003;9:1–8. doi: 10.1261/rna.2121703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Al-Jubran K., Wen J., Abdullahi A., Roy Chaudhury S., Li M., Ramanathan P., Matina A., De S., Piechocki K., Rugjee K.N., Brogna S. Visualization of the joining of ribosomal subunits reveals the presence of 80S ribosomes in the nucleus. RNA. 2013;19:1669–1683. doi: 10.1261/rna.038356.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Baboo S., Cook P.R. “Dark matter” worlds of unstable RNA and protein. Nucleus. 2014;5:281–286. doi: 10.4161/nucl.29577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yewdell J.W., David A. Nuclear translation for immunosurveillance. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17612–17613. doi: 10.1073/pnas.1318259110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Apcher S., Millot G., Daskalogianni C., Scherl A., Manoury B., Fahraeus R. Translation of pre-spliced RNAs in the nuclear compartment generates peptides for the MHC class I pathway. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17951–17956. doi: 10.1073/pnas.1309956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Martins R.P., Malbert-Colas L., Lista M.J., Daskalogianni C., Apcher S., Pla M., Findakly S., Blondel M., Fahraeus R. Nuclear processing of nascent transcripts determines synthesis of full-length proteins and antigenic peptides. Nucleic Acids Res. 2019;47:3086–3100. doi: 10.1093/nar/gky1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Iborra F.J., Jackson D.A., Cook P.R. Coupled transcription and translation within nuclei of mammalian cells. Science. 2001;293:1139–1142. doi: 10.1126/science.1061216. [DOI] [PubMed] [Google Scholar]
- 151.Kim S.H., Koroleva O.A., Lewandowska D., Pendle A.F., Clark G.P., Simpson C.G., Shaw P.J., Brown J.W. Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell. 2009;21:2045–2057. doi: 10.1105/tpc.109.067736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Baboo S., Bhushan B., Jiang H., Grovenor C.R., Pierre P., Davis B.G., Cook P.R. Most human proteins made in both nucleus and cytoplasm turn over within minutes. PLoS One. 2014;9:e99346. doi: 10.1371/journal.pone.0099346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lykke-Andersen J., Shu M.D., Steitz J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 154.Wilkinson M.F., Shyu A.B. RNA surveillance by nuclear scanning? Nat. Cell Biol. 2002;4:E144–E147. doi: 10.1038/ncb0602-e144. [DOI] [PubMed] [Google Scholar]
- 155.Iborra F.J., Jackson D.A., Cook P.R. The case for nuclear translation. J. Cell Sci. 2004;117:5713–5720. doi: 10.1242/jcs.01538. [DOI] [PubMed] [Google Scholar]
- 156.David A., Dolan B.P., Hickman H.D., Knowlton J.J., Clavarino G., Pierre P., Bennink J.R., Yewdell J.W. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J. Cell Biol. 2012;197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Enam S.U., Zinshteyn B., Goldman D.H., Cassani M., Livingston N.M., Seydoux G., Green R. Puromycin reactivity does not accurately localize translation at the subcellular level. eLife. 2020;9:e60303. doi: 10.7554/eLife.60303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hobson B.D., Kong L., Hartwick E.W., Gonzalez R.L., Sims P.A. Elongation inhibitors do not prevent the release of puromycylated nascent polypeptide chains from ribosomes. eLife. 2020;9:e60048. doi: 10.7554/eLife.60048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ivanov P., Kedersha N., Anderson P. Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol. 2019;11:a032813. doi: 10.1101/cshperspect.a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Corbet G.A., Parker R. RNP Granule formation: lessons from P-bodies and stress granules. Cold Spring Harb. Symp. Quant. Biol. 2019;84:203–215. doi: 10.1101/sqb.2019.84.040329. [DOI] [PubMed] [Google Scholar]
- 161.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang H., Elbaum-Garfinkle S., Langdon E.M., Taylor N., Occhipinti P., Bridges A.A., Brangwynne C.P., Gladfelter A.S. RNA controls polyQ protein phase transitions. Mol. Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Langdon E.M., Qiu Y., Ghanbari Niaki A., McLaughlin G.A., Weidmann C.A., Gerbich T.M., Smith J.A., Crutchley J.M., Termini C.M., Weeks K.M., Myong S., Gladfelter A.S. mRNA structure determines specificity of a polyQ-driven phase separation. Science. 2018;360:922–927. doi: 10.1126/science.aar7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Van Treeck B., Parker R. Emerging roles for intermolecular RNA–RNA interactions in RNP assemblies. Cell. 2018;174:791–802. doi: 10.1016/j.cell.2018.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Begovich K., Wilhelm J.E. 2020. An in vitro assembly system identifies roles for RNA nucleation and ATP in yeast stress granule formation. Mol. Cell.79, 991–1007, e1004. 10.1016/j.molcel.2020.07.017 [DOI] [PubMed]
- 166.Kershaw C.J., Nelson M.G., Lui J., Bates C.P., Jennings M.D., Hubbard S.J., Ashe M.P., Grant C.M. 2020. Integrated multi-omics reveals common properties underlying stress granule and P-body formation. bioRxiv. 2020.05.18.102517. 10.1101/2020.05.18.102517 [DOI] [PMC free article] [PubMed]
- 167.Davidson A., Parton R.M., Rabouille C., Weil T.T., Davis I. Localized translation of gurken/TGF-alpha mRNA during axis specification is controlled by access to Orb/CPEB on processing bodies. Cell Rep. 2016;14:2451–2462. doi: 10.1016/j.celrep.2016.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Weil T.T., Parton R.M., Herpers B., Soetaert J., Veenendaal T., Xanthakis D., Dobbie I.M., Halstead J.M., Hayashi R., Rabouille C., Davis I. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat. Cell Biol. 2012;14:1305–1313. doi: 10.1038/ncb2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cougot N., Molza A.E., Giudice E., Cavalier A., Thomas D., Gillet R. Structural organization of the polysomes adjacent to mammalian processing bodies (P-bodies) RNA Biol. 2013;10:314–320. doi: 10.4161/rna.23342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Moon S.L., Morisaki T., Khong A., Lyon K., Parker R., Stasevich T.J. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol. 2019;21:162–168. doi: 10.1038/s41556-018-0263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mateju D., Eichenberger B., Voigt F., Eglinger J., Roth G., Chao J.A. Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell. 2020;183:1801–1812, e13. doi: 10.1016/j.cell.2020.11.010. [DOI] [PubMed] [Google Scholar]
- 172.Yasuda K., Zhang H., Loiselle D., Haystead T., Macara I.G., Mili S. The RNA-binding protein Fus directs translation of localized mRNAs in APC-RNP granules. J. Cell Biol. 2013;203:737–746. doi: 10.1083/jcb.201306058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mili S., Moissoglu K., Macara I.G. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453:115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Buchan J.R. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014;11:1019–1030. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Moujaber O., Stochaj U. Cytoplasmic RNA granules in somatic maintenance. Gerontology. 2018;64:485–494. doi: 10.1159/000488759. [DOI] [PubMed] [Google Scholar]
- 176.van Leeuwen W., Rabouille C. Cellular stress leads to the formation of membraneless stress assemblies in eukaryotic cells. Traffic. 2019;20:623–638. doi: 10.1111/tra.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Burke J.M., Lester E.T., Tauber D., Parker R. RNase L promotes the formation of unique ribonucleoprotein granules distinct from stress granules. J. Biol. Chem. 2020;295:1426–1438. doi: 10.1074/jbc.RA119.011638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pimentel J., Boccaccio G.L. Translation and silencing in RNA granules: a tale of sand grains. Front. Mol. Neurosci. 2014;7:68. doi: 10.3389/fnmol.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Formicola N., Vijayakumar J., Besse F. Neuronal ribonucleoprotein granules: dynamic sensors of localized signals. Traffic. 2019;20:639–649. doi: 10.1111/tra.12672. [DOI] [PubMed] [Google Scholar]
- 180.Bramham C.R., Wells D.G. Dendritic mRNA: Transport, translation and function. Nat. Rev. Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 181.Graber T.E., Hebert-Seropian S., Khoutorsky A., David A., Yewdell J.W., Lacaille J.C., Sossin W.S. Reactivation of stalled polyribosomes in synaptic plasticity. Proc. Natl. Acad. Sci. U. S. A. 2013;110:16205–16210. doi: 10.1073/pnas.1307747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tatavarty V., Ifrim M.F., Levin M., Korza G., Barbarese E., Yu J., Carson J.H. Single-molecule imaging of translational output from individual RNA granules in neurons. Mol. Biol. Cell. 2012;23:918–929. doi: 10.1091/mbc.E11-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Batish M., van den Bogaard P., Kramer F.R., Tyagi S. Neuronal mRNAs travel singly into dendrites. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4645–4650. doi: 10.1073/pnas.1111226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schisa J.A. New insights into the regulation of RNP granule assembly in oocytes. Int. Rev. Cell Mol. Biol. 2012;295:233–289. doi: 10.1016/B978-0-12-394306-4.00013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kotani T., Maehata K., Takei N. Regulation of translationally repressed mRNAs in zebrafish and mouse oocytes. Results Probl. Cell Differ. 2017;63:297–324. doi: 10.1007/978-3-319-60855-6_13. [DOI] [PubMed] [Google Scholar]
- 186.Winata C.L., Korzh V. The translational regulation of maternal mRNAs in time and space. FEBS Lett. 2018;592:3007–3023. doi: 10.1002/1873-3468.13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Horie M., Kotani T. Formation of mos RNA granules in the zebrafish oocyte that differ from cyclin B1 RNA granules in distribution, density and regulation. Eur. J. Cell Biol. 2016;95:563–573. doi: 10.1016/j.ejcb.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 188.Kloc M., Dougherty M.T., Bilinski S., Chan A.P., Brey E., King M.L., Patrick C.W., Etkin L.D. Three-dimensional ultrastructural analysis of RNA distribution within germinal granules of Xenopus. Dev. Biol. 2002;241:79–93. doi: 10.1006/dbio.2001.0488. [DOI] [PubMed] [Google Scholar]
- 189.Kloc M., Bilinski S., Chan A.P., Etkin L.D. Mitochondrial ribosomal RNA in the germinal granules in Xenopus embryos revisited. Differentiation. 2001;67:80–83. doi: 10.1046/j.1432-0436.2001.067003080.x. [DOI] [PubMed] [Google Scholar]
- 190.Kobayashi S., Amikura R., Okada M. Localization of mitochondrial large rRNA in germinal granules and the consequent segregation of germ line. Int. J. Dev. Biol. 1994;38:193–199. [PubMed] [Google Scholar]
- 191.Merry T.L., Chan A., Woodhead J.S.T., Reynolds J.C., Kumaga H., Kim S.J., Lee C. Mitochondrial-derived peptides in energy metabolism. Am. J. Physiol. Endocrinol. Metab. 2020;319:E659–E666. doi: 10.1152/ajpendo.00249.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lui J., Castelli L.M., Pizzinga M., Simpson C.E., Hoyle N.P., Bailey K.L., Campbell S.G., Ashe M.P. Granules harboring translationally active mRNAs provide a platform for P-body formation following stress. Cell Rep. 2014;9:944–954. doi: 10.1016/j.celrep.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Morales-Polanco F., Bates C., Lui J., Casson J., Solari C.A., Pizzinga M., Forte G., Griffin C., Garner K.E.L., Burt H.E., Dixon H.L., Hubbard S., Portela P., Ashe M.P. 2021. Core Fermentation (CoFe) granules focus coordinated glycolytic mRNA localization and translation to fuel glucose fermentation. iScience.24, 102069. 10.1016/j.isci.2021.102069 [DOI] [PMC free article] [PubMed]
- 194.Pizzinga M., Bates C., Lui J., Forte G., Morales-Polanco F., Linney E., Knotkova B., Wilson B., Solari C.A., Berchowitz L.E., Portela P., Ashe M.P. Translation factor mRNA granules direct protein synthetic capacity to regions of polarized growth. J. Cell Biol. 2019;218:1564–1581. doi: 10.1083/jcb.201704019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pichon X., Bastide A., Safieddine A., Chouaib R., Samacoits A., Basyuk E., Peter M., Mueller F., Bertrand E. Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J. Cell Biol. 2016;214:769–781. doi: 10.1083/jcb.201605024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nair R.R., Zabezhinsky D., Gelin-Licht R., Haas B., Dyhr M.C.A., Sperber H.S., Nusbaum C., Gerst J.E. 2020. Multiplexed mRNA assembly into ribonucleoprotein particles plays an operon-like role in the control of yeast cell physiology. bioRxiv. 2020.06.28.175851. 10.1101/2020.06.28.175851 [DOI] [PMC free article] [PubMed]
- 197.Schwarz A., Beck M. The benefits of cotranslational assembly: A structural perspective. Trends Cell Biol. 2019;29:791–803. doi: 10.1016/j.tcb.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 198.Bertolini M., Fenzl K., Kats I., Wruck F., Tippmann F., Schmitt J., Auburger J.J., Tans S., Bukau B., Kramer G. Interactions between nascent proteins translated by adjacent ribosomes drive homomer assembly. Science. 2021;371:57–64. doi: 10.1126/science.abc7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Shiber A., Doring K., Friedrich U., Klann K., Merker D., Zedan M., Tippmann F., Kramer G., Bukau B. Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature. 2018;561:268–272. doi: 10.1038/s41586-018-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Duncan C.D., Mata J. Widespread cotranslational formation of protein complexes. PLoS Genet. 2011;7:e1002398. doi: 10.1371/journal.pgen.1002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kassem S., Villanyi Z., Collart M.A. Not5-dependent co-translational assembly of Ada2 and Spt20 is essential for functional integrity of SAGA. Nucleic Acids Res. 2017;45:1186–1199. doi: 10.1093/nar/gkw1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liu F., Jones D.K., de Lange W.J., Robertson G.A. Cotranslational association of mRNA encoding subunits of heteromeric ion channels. Proc. Natl. Acad. Sci. U. S. A. 2016;113:4859–4864. doi: 10.1073/pnas.1521577113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kamenova I., Mukherjee P., Conic S., Mueller F., El-Saafin F., Bardot P., Garnier J.M., Dembele D., Capponi S., Timmers H.T.M., Vincent S.D., Tora L. Co-translational assembly of mammalian nuclear multisubunit complexes. Nat. Commun. 2019;10:1740. doi: 10.1038/s41467-019-09749-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Panasenko O.O., Somasekharan S.P., Villanyi Z., Zagatti M., Bezrukov F., Rashpa R., Cornut J., Iqbal J., Longis M., Carl S.H., Pena C., Panse V.G., Collart M.A. Co-translational assembly of proteasome subunits in NOT1-containing assemblysomes. Nat. Struct. Mol. Biol. 2019;26:110–120. doi: 10.1038/s41594-018-0179-5. [DOI] [PubMed] [Google Scholar]
- 205.Moissoglu K., Stueland M., Gasparski A.N., Wang T., Jenkins L.M., Hastings M.L., and Mili S. 2020. RNA localization and co-translational interactions control RAB13 GTPase function and cell migration. EMBO J. 39, e104958. 10.15252/embj.2020104958 [DOI] [PMC free article] [PubMed]
- 206.Wagner S., Herrmannova A., Hronova V., Gunisova S., Sen N.D., Hannan R.D., Hinnebusch A.G., Shirokikh N.E., Preiss T., Valasek L.S. Selective translation complex profiling reveals staged initiation and co-translational sssembly of initiation factor complexes. Mol. Cell. 2020;79:546–560, e7. doi: 10.1016/j.molcel.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Villanyi Z., Ribaud V., Kassem S., Panasenko O.O., Pahi Z., Gupta I., Steinmetz L., Boros I., Collart M.A. The Not5 subunit of the ccr4-not complex connects transcription and translation. PLoS Genet. 2014;10:e1004569. doi: 10.1371/journal.pgen.1004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Willett M., Flint S.A., Morley S.J., Pain V.M. Compartmentalisation and localisation of the translation initiation factor (eIF) 4F complex in normally growing fibroblasts. Exp. Cell Res. 2006;312:2942–2953. doi: 10.1016/j.yexcr.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 209.Willett M., Brocard M., Davide A., Morley S.J. Translation initiation factors and active sites of protein synthesis co-localize at the leading edge of migrating fibroblasts. Biochem. J. 2011;438:217–227. doi: 10.1042/BJ20110435. [DOI] [PubMed] [Google Scholar]
- 210.Gray N.K., Hrabalkova L., Scanlon J.P., Smith R.W. Poly(A)-binding proteins and mRNA localization: who rules the roost? Biochem. Soc. Trans. 2015;43:1277–1284. doi: 10.1042/BST20150171. [DOI] [PubMed] [Google Scholar]
- 211.Campbell S.G., Hoyle N.P., Ashe M.P. Dynamic cycling of eIF2 through a large eIF2B-containing cytoplasmic body: Implications for translation control. J. Cell Biol. 2005;170:925–934. doi: 10.1083/jcb.200503162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Koppers M., Cagnetta R., Shigeoka T., Wunderlich L.C., Vallejo-Ramirez P., Qiaojin Lin J., Zhao S., Jakobs M.A., Dwivedy A., Minett M.S., Bellon A., Kaminski C.F., Harris W.A., Flanagan J.G., Holt C.E. Receptor-specific interactome as a hub for rapid cue-induced selective translation in axons. eLife. 2019;8:e48718. doi: 10.7554/eLife.48718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tcherkezian J., Brittis P.A., Thomas F., Roux P.P., Flanagan J.G. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell. 2010;141:632–644. doi: 10.1016/j.cell.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang D.O., Kim S.M., Zhao Y., Hwang H., Miura S.K., Sossin W.S., Martin K.C. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324:1536–1540. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hoyle N.P., Ashe M.P. Subcellular localization of mRNA and factors involved in translation initiation. Biochem. Soc. Trans. 2008;36:648–652. doi: 10.1042/BST0360648. [DOI] [PubMed] [Google Scholar]
- 216.Chudinova E.M., Nadezhdina E.S. Interactions between the translation machinery and microtubules. Biochemistry. 2018;83:S176–S189. doi: 10.1134/S0006297918140146. [DOI] [PubMed] [Google Scholar]
- 217.Lawrence J.B., Singer R.H. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45:407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- 218.Mingle L.A., Okuhama N.N., Shi J., Singer R.H., Condeelis J., Liu G. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci. 2005;118:2425–2433. doi: 10.1242/jcs.02371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Condeelis J., Singer R.H. How and why does beta-actin mRNA target? Biol. Cell. 2005;97:97–110. doi: 10.1042/BC20040063. [DOI] [PubMed] [Google Scholar]
- 220.Chaudhuri A., Das S., Das B. Localization elements and zip codes in the intracellular transport and localization of messenger RNAs in Saccharomyces cerevisiae. Wiley Interdiscip. Rev. RNA. 2020;11:e1591. doi: 10.1002/wrna.1591. [DOI] [PubMed] [Google Scholar]
- 221.Weil T.T. mRNA localization in the Drosophila germline. RNA Biol. 2014;11:1010–1018. doi: 10.4161/rna.36097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lasko P. mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harb. Perspect. Biol. 2012;4:a012294. doi: 10.1101/cshperspect.a012294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zappulo A., van den Bruck D., Ciolli Mattioli C., Franke V., Imami K., McShane E., Moreno-Estelles M., Calviello L., Filipchyk A., Peguero-Sanchez E., Muller T., Woehler A., Birchmeier C., Merino E., Rajewsky N. RNA localization is a key determinant of neurite-enriched proteome. Nat. Commun. 2017;8:583. doi: 10.1038/s41467-017-00690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Shigeoka T., Koppers M., Wong H.H., Lin J.Q., Cagnetta R., Dwivedy A., de Freitas Nascimento J., van Tartwijk F.W., Strohl F., Cioni J.M., Schaeffer J., Carrington M., Kaminski C.F., Jung H., Harris W.A., Holt C.E. On-site ribosome remodeling by locally synthesized ribosomal proteins in axons. Cell Rep. 2019;29:3605–3619.e10. doi: 10.1016/j.celrep.2019.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wilk R., Hu J., Blotsky D., Krause H.M. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. 2016;30:594–609. doi: 10.1101/gad.276931.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Lecuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T.R., Tomancak P., Krause H.M. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 227.Dermit M., Dodel M., Lee F.C.Y., Azman M.S., Schwenzer H., Jones J.L., Blagden S.P., Ule J., Mardakheh F.K. Subcellular mRNA localization regulates ribosome biogenesis in migrating cells. Dev. Cell. 2020;55:298–313, e210. doi: 10.1016/j.devcel.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kashida S., Wang D.O., Saito H., Gueroui Z. Nanoparticle-based local translation reveals mRNA as a translation-coupled scaffold with anchoring function. Proc. Natl Acad. Sci. U. S. A. 2019;116:13346–13351. doi: 10.1073/pnas.1900310116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Chabanon H., Mickleburgh I., Hesketh J. Zipcodes and postage stamps: mRNA localisation signals and their trans-acting binding proteins. Brief. Funct. Genomic. Proteomic. 2004;3:240–256. doi: 10.1093/bfgp/3.3.240. [DOI] [PubMed] [Google Scholar]
- 230.Andreassi C., Riccio A. To localize or not to localize: mRNA fate is in 3'UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 231.Bullock S.L. Messengers, motors and mysteries: sorting of eukaryotic mRNAs by cytoskeletal transport. Biochem. Soc. Trans. 2011;39:1161–1165. doi: 10.1042/BST0391161. [DOI] [PubMed] [Google Scholar]
- 232.Marchand V., Gaspar I., Ephrussi A. An intracellular transmission control protocol: assembly and transport of ribonucleoprotein complexes. Curr. Opin. Cell Biol. 2012;24:202–210. doi: 10.1016/j.ceb.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 233.Song T., Zheng Y., Wang Y., Katz Z., Liu X., Chen S., Singer R.H., Gu W. Specific interaction of KIF11 with ZBP1 regulates the transport of β-actin mRNA and cell motility. J. Cell Sci. 2015;128:1001–1010. doi: 10.1242/jcs.161679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Huttelmaier S., Zenklusen D., Lederer M., Dictenberg J., Lorenz M., Meng X., Bassell G.J., Condeelis J., Singer R.H. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 235.Jønson L., Vikesaa J., Krogh A., Nielsen L. K., Hansen T., Borup R., Johnsen A.H., Christiansen J., Nielsen F.C. Molecular composition of IMP1 ribonucleoprotein granules. Mol. Cell. Proteomics. 2007;6:798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- 236.Abouward R., Schiavo G. 2020. Walking the line: mechanisms underlying directional mRNA transport and localisation in neurons and beyond. Cell. Mol. Life Sci.10.1007/s00018-020-03724-3 [DOI] [PMC free article] [PubMed]
- 237.Dalla Costa I., Buchanan C.N., Zdradzinski M.D., Sahoo P.K., Smith T.P., Thames E., Kar A.N., Twiss J.L. The functional organization of axonal mRNA transport and translation. Nat. Rev. Neurosci. 2021;22:77–91. doi: 10.1038/s41583-020-00407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Cosker K.E., Fenstermacher S.J., Pazyra-Murphy M.F., Elliott H.L., Segal R.A. The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat. Neurosci. 2016;19:690–696. doi: 10.1038/nn.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kim E., Jung H. Local mRNA translation in long-term maintenance of axon health and function. Curr. Opin. Neurobiol. 2020;63:15–22. doi: 10.1016/j.conb.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 240.Baumann S., Komissarov A., Gili M., Ruprecht V., Wieser S., Maurer S.P. 2020. A reconstituted mammalian APC-kinesin complex selectively transports defined packages of axonal mRNAs. Sci. Adv. 6, eaaz1588. 10.1126/sciadv.aaz1588 [DOI] [PMC free article] [PubMed]
- 241.Bergalet J., Lecuyer E. The functions and regulatory principles of mRNA intracellular trafficking. Adv. Exp. Med. Biol. 2014;825:57–96. doi: 10.1007/978-1-4939-1221-6_2. [DOI] [PubMed] [Google Scholar]
- 242.Chin A., Lecuyer E. RNA localization: Making its way to the center stage. Biochim. Biophys. Acta Gen. Subj. 2017;1861:2956–2970. doi: 10.1016/j.bbagen.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 243.Lazzaretti D., Bono F. mRNA localization in metazoans: A structural perspective. RNA Biol. 2017;14:1473–1484. doi: 10.1080/15476286.2017.1338231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Paquin N., Menade M., Poirier G., Donato D., Drouet E., Chartrand P. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol. Cell. 2007;26:795–809. doi: 10.1016/j.molcel.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 245.Andreassi C., Luisier R., Crerar H., Darsinou M., Blokzijl-Franke S., Lenn T., Luscombe N.M., Cuda G., Gaspari M., Saiardi A., Riccio A. Cytoplasmic cleavage of IMPA1 3' UTR is necessary for maintaining axon integrity. Cell Rep. 2021;34:108778. doi: 10.1016/j.celrep.2021.108778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Moissoglu K., Yasuda K., Wang T., Chrisafis G., Mili S. Translational regulation of protrusion-localized RNAs involves silencing and clustering after transport. eLife. 2019;8:e44752. doi: 10.7554/eLife.44752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Forrest K.M., Gavis E.R. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 2003;13:1159–1168. doi: 10.1016/S0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 248.Semotok J.L., Lipshitz H.D. Regulation and function of maternal mRNA destabilization during early Drosophila development. Differentiation. 2007;75:482–506. doi: 10.1111/j.1432-0436.2007.00178.x. [DOI] [PubMed] [Google Scholar]
- 249.Blower M.D., Feric E., Weis K., Heald R. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J. Cell Biol. 2007;179:1365–1373. doi: 10.1083/jcb.200705163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Alliegro M.C. The centrosome and spindle as a ribonucleoprotein complex. Chromosome Res. 2011;19:367–376. doi: 10.1007/s10577-011-9186-7. [DOI] [PubMed] [Google Scholar]
- 251.Blower M.D., Nachury M., Heald R., Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 252.Rosic S., Kohler F., Erhardt S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol. 2014;207:335–349. doi: 10.1083/jcb.201404097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Mamon L.A., Ginanova V.R., Kliver S.F., Yakimova A.O., Atsapkina A.A., Golubkova E.V. RNA-binding proteins of the NXF (nuclear export factor) family and their connection with the cytoskeleton. Cytoskeleton. 2017;74:161–169. doi: 10.1002/cm.21362. [DOI] [PubMed] [Google Scholar]
- 254.Ito K.K., Watanabe K., Kitagawa D. The emerging role of ncRNAs and RNA-binding proteins in mitotic apparatus formation. Noncoding RNA. 2020;6:13. doi: 10.3390/ncrna6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Groisman I., Huang Y.S., Mendez R., Cao Q., Theurkauf W., Richter J.D. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103:435–447. doi: 10.1016/S0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- 256.Fingerhut J.M., Yamashita Y.M. mRNA localization mediates maturation of cytoplasmic cilia in Drosophila spermatogenesis. J. Cell Biol. 2020;219:e202003084. doi: 10.1083/jcb.202003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Amato R., Morleo M., Giaquinto L., di Bernardo D., Franco B. A network-based approach to dissect the cilia/centrosome complex interactome. BMC Genomics. 2014;15:658. doi: 10.1186/1471-2164-15-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Muller H., Schmidt D., Steinbrink S., Mirgorodskaya E., Lehmann V., Habermann K., Dreher F., Gustavsson N., Kessler T., Lehrach H., Herwig R., Gobom J., Ploubidou A., Boutros M., Lange B.M. Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 2010;29:3344–3357. doi: 10.1038/emboj.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Davies A.H., Barrett I., Pambid M.R., Hu K., Stratford A.L., Freeman S., Berquin I.M., Pelech S., Hieter P., Maxwell C., Dunn S.E. YB-1 evokes susceptibility to cancer through cytokinesis failure, mitotic dysfunction and HER2 amplification. Oncogene. 2011;30:3649–3660. doi: 10.1038/onc.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Iaconis D., Monti M., Renda M., van Koppen A., Tammaro R., Chiaravalli M., Cozzolino F., Pignata P., Crina C., Pucci P., Boletta A., Belcastro V., Giles R.H., Surace E.M., Gallo S. The centrosomal OFD1 protein interacts with the translation machinery and regulates the synthesis of specific targets. Sci. Rep. 2017;7:1224. doi: 10.1038/s41598-017-01156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lambert J.D., Nagy L.M. Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages. Nature. 2002;420:682–686. doi: 10.1038/nature01241. [DOI] [PubMed] [Google Scholar]
- 262.Ryder P.V., Fang J., Lerit D.A. centrocortin RNA localization to centrosomes is regulated by FMRP and facilitates error-free mitosis. J. Cell Biol. 2020;219:e202004101. doi: 10.1083/jcb.202004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Safieddine A., Coleno E., Salloum S., Imbert A., Traboulsi A.M., Kwon O.S., Lionneton F., Georget V., Robert M.C., Gostan T., Lecellier C.H., Chouaib R., Pichon X., Le Hir H., Zibara K. A choreo-graphy of centrosomal mRNAs reveals a conserved localization mechanism involving active polysome transport. Nat. Commun. 2021;12:1352. doi: 10.1038/s41467-021-21585-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Romero-Brey I., Bartenschlager R. Membranous replication factories induced by plus-strand RNA viruses. Viruses. 2014;6:2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Paul D., Madan V., Bartenschlager R. Hepatitis C virus RNA replication and assembly: Living on the fat of the land. Cell Host Microbe. 2014;16:569–579. doi: 10.1016/j.chom.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Reid D.W., Campos R.K., Child J.R., Zheng T., Chan K.W.K., Bradrick S.S., Vasudeva S.G., Garcia-Blanco M.A., Nicchitta C.V. Dengue virus selectively annexes endoplasmic reticulum-associated translation machinery as a strategy for co-opting host cell protein synthesi. J. Virol. 2018;92:e01766–17. doi: 10.1128/JVI.01766-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Ropidi M.I.M., Khazali A.S., Rashid N. N., Yusof R. Endoplasmic reticulum: a focal point of Zika virus infection. J. Biomed. Sci. 2020;27:27. doi: 10.1186/s12929-020-0618-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Knoops K., Kikkert M., Worm S. H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Ulasli M., Verheije M.H., de Haan C.A., Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 2010;12:844–861. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Haimovich G., Olender T., Baez C., Gerst J.E. 2020. Identification and enrichment of SECReTE cis-acting RNA elements in the Coronaviridae and other (+) single-strand RNA viruses. bioRxiv. 2020.04.20.050088. 10.1101/2020.04.20.050088
- 271.Wu K.E., Fazal F.M., Parker K.R., Zou J., Chang H.Y. RNA-GPS predicts SARS-CoV-2 RNA residency to host mitochondria and nucleolus. Cell Syst. 2020;11:102–108, e103. doi: 10.1016/j.cels.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Somasundaran M., Zapp M.L., Beattie L.K., Pang L., Byron K.S., Bassell G.J., Sullivan J.L., Singer R.H. Localization of HIV RNA in mitochondria of infected cells: Potential role in cytopathogenicity. J. Cell Biol. 1994;126:1353–1360. doi: 10.1083/jcb.126.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Sanz M.A., Castello A., Ventoso I., Berlanga J.J., Carrasco L. Dual mechanism for the translation of subgenomic mRNA from Sindbis virus in infected and uninfected cells. PLoS One. 2009;4:e4772. doi: 10.1371/journal.pone.0004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Desmet E.A., Anguish L.J., Parker J.S. Virus-mediated compartmentalization of the host translational machinery. mBio. 2014;5:e01463–01414. doi: 10.1128/mBio.01463-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.V'Kovski P. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. eLife. 2019;8:e42037. doi: 10.7554/eLife.42037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Kastan J.P., Dobrikova E.Y., Bryant J.D., Gromeier M. 2020. CReP mediates selective translation initiation at the endoplasmic reticulum. Sci. Adv.6, eaba0745. 10.1126/sciadv.aba0745 [DOI] [PMC free article] [PubMed]
- 277.Walsh D., Arias C., Perez C., Halladin D., Escandon M., Ueda T., Watanabe-Fukunaga R., Fukunaga R., Mohr I. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol. Cell. Biol. 2008;28:2648–2658. doi: 10.1128/MCB.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Katsafanas G.C., Moss B. Specific Anchoring and local translation of poxviral ATI mRNA at cytoplasmic inclusion bodies. J. Virol. 2020;94:e01671–19. doi: 10.1128/JVI.01671-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Ng C.S., Jogi M., Yoo J.S., Onomoto K., Koike S., Iwasaki T., Yoneyama M., Kato H., Fujita T. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J. Virol. 2013;87:9511–9522. doi: 10.1128/JVI.03248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Bonenfant G., Williams N., Netzband R., Schwarz M.C., Evans M.J., Pager C.T. Zika virus subverts stress granules to promote and restrict viral gene expression. J. Virol. 2019;93:e00520–19. doi: 10.1128/JVI.00520-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Nikolic J., Civas A., Lama Z., Lagaudriere-Gesbert C., Blondel D. Rabies virus infection induces the formation of stress granules closely connected to the viral factories. PLoS Pathog. 2016;12:e1005942. doi: 10.1371/journal.ppat.1005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Robinson C.-A., Kleer M., Mulloy R.P., Castle E.L., Boudreau B.Q., Corcoran J.A. 2020. Human coronaviruses disassemble processing bodies. bioRxiv. 2020.11.08.372995. 10.1101/2020.11.08.372995
- 283.Luo L., Li Z., Zhao T., Ju X., Ma P., Jin B., Zhou Y., He S., Huang J., Xu X., Zou Y., Li P., Liang A., Liu J., Chi T., Huang X., Ding Q., Jin Z., Huang C., Zhang Y. 2021. SARS-CoV-2 nucleocapsid protein phase separates with G3BPs to disassemble stress granules and facilitate viral production. Sci. Bull. (Beijing). 10.1016/j.scib.2021.01.013 [DOI] [PMC free article] [PubMed]
- 284.Savastano A., Ibanez de Opakua A., Rankovic M., Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020;11:6041. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Perdikari T.M., Murthy A.C., Ryan V.H., Watters S., Naik M.T., Fawzi N.L. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 2020;39:e106478. doi: 10.15252/embj.2020106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Reineke L.C., Lloyd R.E. Diversion of stress granules and P-bodies during viral infection. Virology. 2013;436:255–267. doi: 10.1016/j.virol.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.McCormick C., Khaperskyy D.A. Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 2017;17:647–660. doi: 10.1038/nri.2017.63. [DOI] [PubMed] [Google Scholar]
- 288.Doh J.H., Lutz S., Curcio M.J. Co-translational localization of an LTR-retrotransposon RNA to the endoplasmic reticulum nucleates virus-like particle assembly sites. PLoS Genet. 2014;10:e1004219. doi: 10.1371/journal.pgen.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Doucet A.J., Hulme A.E., Sahinovic E., Kulpa D.A., Moldovan J.B., Kopera H.C., Athanikar J.N., Hasnaoui M., Bucheton A., Moran J.V., Gilbert N. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6:e1001150. doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Goodier J.L., Zhang L., Vetter M.R., Kazazian H.H. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Mita P., Wudzinska A., Sun X., Andrade J., Nayak S., Kahler D.J., Badri S., LaCava J., Ueberheide B., Yun C.Y., Fenyo D., Boeke J.D. LINE-1 protein localization and functional dynamics during the cell cycle. eLife. 2018;7:e30058. doi: 10.7554/eLife.30058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Goodier J.L., Mandal P.K., Zhang L., Kazazian H.H. Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum. Mol. Genet. 2010;19:1712–1725. doi: 10.1093/hmg/ddq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Potter M.D., Nicchitta C.V. Regulation of ribosome detachment from the mammalian endoplasmic reticulum membrane. J. Biol. Chem. 2000;275:33828–33835. doi: 10.1074/jbc.M005294200. [DOI] [PubMed] [Google Scholar]
- 294.Nicchitta C.V., Lerner R.S., Stephens S.B., Dodd R.D., Pyhtila B. Pathways for compartmentalizing protein synthesis in eukaryotic cells: The template-partitioning model. Biochem. Cell Biol. 2005;83:687–695. doi: 10.1139/o05-147. [DOI] [PubMed] [Google Scholar]
- 295.Jagannathan S., Reid D.W., Cox A.H., Nicchitta C.V. De novo translation initiation on membrane-bound ribosomes as a mechanism for localization of cytosolic protein mRNAs to the endoplasmic reticulum. RNA. 2014;20:1489–1498. doi: 10.1261/rna.045526.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Reid D.W., Nicchitta C.V. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2015;16:221–223. doi: 10.1038/nrm3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Stephens S.B., Dodd R.D., Brewer J.W., Lager P.J., Keene J.D., Nicchitta C.V. Stable ribosome binding to the endoplasmic reticulum enables compartment-specific regulation of mRNA translation. Mol. Biol. Cell. 2005;16:5819–5831. doi: 10.1091/mbc.e05-07-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Lerner R.S., Nicchitta C.V. mRNA translation is compartmentalized to the endoplasmic reticulum following physiological inhibition of cap-dependent translation. RNA. 2006;12:775–789. doi: 10.1261/rna.2318906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Reid D.W., Chen Q., Tay A.S., Shenolikar S., Nicchitta C.V. The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell. 2014;158:1362–1374. doi: 10.1016/j.cell.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Staudacher J.J., Naarmann-de Vries I.S., Ujvari S.J., Klinger B., Kasim M., Benko E., Ostareck-Lederer A., Ostareck D.H., Bondke Persson A., Lorenzen S., Meier J.C., Bluthgen N., Persson P.B., Henrion-Caude A., Mrowka R., Fahling M. Hypoxia-induced gene expression results from selective mRNA partitioning to the endoplasmic reticulum. Nucleic Acids Res. 2015;43:3219–3236. doi: 10.1093/nar/gkv167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Reid D.W., Nicchitta C.V. Comment on “Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling”. Science. 2015;348:1217. doi: 10.1126/science.aaa7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Jan C.H., Williams C.C., Weissman J.S. Response to Comment on “Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling”. Science. 2015;348:1217. doi: 10.1126/science.aaa8299. [DOI] [PubMed] [Google Scholar]
- 303.Unsworth H., Raguz S., Edwards H.J., Higgins C.F., Yague E. mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J. 2010;24:3370–3380. doi: 10.1096/fj.09-151142. [DOI] [PubMed] [Google Scholar]
- 304.Sivan G., Kedersha N., Elroy-Stein O. Ribosomal slowdown mediates translational arrest during cellular division. Mol. Cell. Biol. 2007;27:6639–6646. doi: 10.1128/MCB.00798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Chartron J.W., Hunt K.C., Frydman J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature. 2016;536:224–228. doi: 10.1038/nature19309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.del Alamo M., Hogan D.J., Pechmann S., Albanese V., Brown P.O., Frydman J. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol. 2011;9:e1001100. doi: 10.1371/journal.pbio.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Mauro V.P., Edelman G.M. The ribosome filter redux. Cell Cycle. 2007;6:2246–2251. doi: 10.4161/cc.6.18.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Shi Z., Fujii K., Kovary K.M., Genuth N.R., Rost H.L., Teruel M.N., Barna M. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell. 2017;67:71–83, e77. doi: 10.1016/j.molcel.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Genuth N.R., Barna M. Heterogeneity and specialized functions of translation machinery: From genes to organisms. Nat. Rev. Genet. 2018;19:431–452. doi: 10.1038/s41576-018-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Emmott E., Jovanovic M., Slavov N. Ribosome stoichiometry: From form to function. Trends Biochem. Sci. 2019;44:95–109. doi: 10.1016/j.tibs.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Simsek D., Tiu G.C., Flynn R.A., Byeon G.W., Leppek K., Xu A.F., Chang H.Y., Barna M. The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell. 2017;169:1051–1065.e1018. doi: 10.1016/j.cell.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Komili S., Farny N.G., Roth F.P., Silver P.A. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Gerst J.E. Pimp my ribosome: Ribosomal protein paralogs specify translational control. Trends Genet. 2018;34:832–845. doi: 10.1016/j.tig.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 314.Segev N., Gerst J.E. Specialized ribosomes and specific ribosomal protein paralogs control translation of mitochondrial proteins. J. Cell Biol. 2018;217:117–126. doi: 10.1083/jcb.201706059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Ferretti M.B., Karbstein K. Does functional specialization of ribosomes really exist? RNA. 2019;25:521–538. doi: 10.1261/rna.069823.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Marc P., Margeot A., Devaux F., Blugeon C., Corral-Debrinski M., Jacq C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Mercer T.R., Neph S., Dinger M.E., Crawford J., Smith M.A., Shearwood A.M., Haugen E., Bracken C.P., Rackham O., Stamatoyannopoulos J.A., Filipovska A., Mattick J.S. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Minis A., Dahary D., Manor O., Leshkowitz D., Pilpel Y., Yaron A. Subcellular transcriptomics-dissection of the mRNA composition in the axonal compartment of sensory neurons. Dev. Neurobiol. 2014;74:365–381. doi: 10.1002/dneu.22140. [DOI] [PubMed] [Google Scholar]
- 319.Gumy L.F., Yeo G.S., Tung Y.C., Zivraj K.H., Willis D., Coppola G., Lam B.Y., Twiss J.L., Holt C.E., Fawcett J.W. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17:85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Taylor A.M., Berchtold N.C., Perreau V.M., Tu C.H., Li Jeon N., Cotman C.W. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J. Neurosci. 2009;29:4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Padron A., Iwasaki S., Ingolia N.T. Proximity RNA labeling by APEX-Seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol. Cell. 2019;75:875–887.e875. doi: 10.1016/j.molcel.2019.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Wang P., Tang W., Li Z., Zo Z., Zhou Y., Li R., Xiong T., Wang J., Zou P. Mapping spatial transcriptome with light-activated proximity-dependent RNA labeling. Nat. Chem. Biol. 2019;15:1110–1119. doi: 10.1038/s41589-019-0368-5. [DOI] [PubMed] [Google Scholar]
- 323.Benhalevy D., Anastasakis D.G., Hafner M. Proximity-CLIP provides a snapshot of protein-occupied RNA elements in subcellular compartments. Nat. Methods. 2018;15:1074–1082. doi: 10.1038/s41592-018-0220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Lee J.H., Daugharthy E.R., Scheiman J., Kalhor R., Yang J.L., Ferrante T.C., Terry R., Jeanty S.S., Li C., Amamoto R., Peters D.T., Turczyk B.M., Marblestone A.H., Invers S.A., Bernard A. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Wang X., Allen W.E., Wright M.A., Sylwestrak E.L., Samusik N., Vesuna S., Evans K., Liu C., Ramakrishnan C., Liu J., Nolan G.P., Bava F.A., Deisseroth K. 2018. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 361 (6400), eaat5691. 10.1126/science.aat5691 [DOI] [PMC free article] [PubMed]
- 326.Fürth D., Hatini V., Lee J.H. 2019. In situ transcriptome accessibility sequencing (INSTA-seq). bioRxiv. 722819. 10.1101/722819
- 327.Chen K.H., Boettiger A.N., Moffitt J.R., Wang S., Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Eng C.L., Lawson M., Zhu Q., Dries R., Koulena N., Takei Y., Yun J., Cronin C., Karp C., Yuan G.C., Cai L. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 2019;568:235–239. doi: 10.1038/s41586-019-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Xia C., Fan J., Emanuel G., Hao J., Zhuang X. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc. Natl. Acad. Sci. U. S. A. 2019;116:19490–19499. doi: 10.1073/pnas.1912459116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Biswas J., Liu Y., Singer R.H., Wu B. Fluorescence imaging methods to investigate translation in single cells. Cold Spring Harb. Perspect. Biol. 2019;11:a032722. doi: 10.1101/cshperspect.a032722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Koppel I., Fainzilber M. Omics approaches for subcellular translation studies. Mol. Omics. 2018;14:380–388. doi: 10.1039/c8mo00172c. [DOI] [PubMed] [Google Scholar]
- 332.Taliaferro J.M. Classical and emerging techniques to identify and quantify localized RNAs. Wiley Interdisc. Rev. RNA. 2019;10:e1542. doi: 10.1002/wrna.1542. [DOI] [PubMed] [Google Scholar]
- 333.Buxbaum A.R., Haimovich G., Singer R.H. In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 2015;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]


