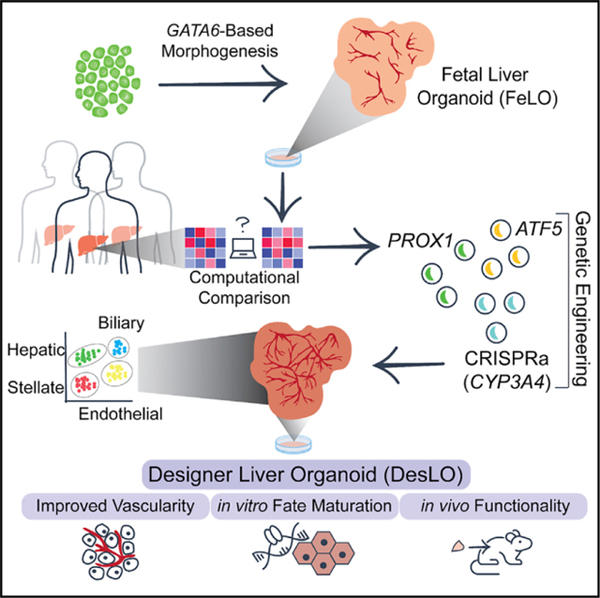
SUMMARY
Pluripotent stem cell (PSC)-derived organoids have emerged as novel multicellular models of human tissue development but display immature phenotypes, aberrant tissue fates, and a limited subset of cells. Here, we demonstrate that integrated analysis and engineering of gene regulatory networks (GRNs) in PSC-derived multilineage human liver organoids direct maturation and vascular morphogenesis in vitro. Overexpression of PROX1 and ATF5, combined with targeted CRISPR-based transcriptional activation of endogenous CYP3A4, reprograms tissue GRNs and improves native liver functions, such as FXR signaling, CYP3A4 enzymatic activity, and stromal cell reactivity. The engineered tissues possess superior liver identity when compared with other PSC-derived liver organoids and show the presence of hepatocyte, biliary, endothelial, and stellate-like cell populations in single-cell RNA-seq analysis. Finally, they show hepatic functions when studied in vivo. Collectively, our approach provides an experimental framework to direct organogenesis in vitro by systematically probing molecular pathways and transcriptional networks that promote tissue development.
Graphical Abstract
In Brief
Informed by computational analyses, Velazquez et al, exercised overexpression of ATF5, PROX1 transcription factors, and CRISPR-mediated activation of CYP3A4 to advance the maturity and vascularity of human iPSC-derived fetal liver organoids in vitro. The findings highlight the importance of integrative systems and synthetic biology to engineer multicellular systems.
INTRODUCTION
Pluripotent stem cell (PSC)-derived organoids have opened numerous opportunities to study human development and disease, which were not previously possible (Clevers, 2016). However, they often stall developmentally and exhibit aberrant tissue signatures with limited vascular systems, which can hamper their use for biomedical applications (Del Dosso et al., 2020; Holloway et al., 2019). Organoid implantation in animal hosts can promote maturation (Holloway et al., 2019; Takebe et al., 2013), but this process is expensive, lengthy, resource-intensive, and lacks scalability or control over the tissue developmental processes. Technologies that can accelerate tissue development in vitro and bypass the several months inherent to natural human development will be attractive for understanding human development and the practical application of organoids for therapeutics (Takebe and Wells, 2019; Velazquez et al., 2018).
Most organoids developed thus far have been generated through sequential administration of growth factor cocktails in a culture medium. However, current approaches usually suffer from limitations, such as lack of control over multilineage differentiations and cell states, or challenges associated with optimal media that can direct and maintain distinct cell types. Additionally, the fidelity of developed cell fates is often not tested quantitatively against their in vivo counterparts (Cahan et al., 2014; Radley et al., 2017; Tan and Cahan, 2019).
Gene regulatory networks (GRNs) within individual cells are naturally evolved to direct multicellular fates and promote collective cellular behaviors toward final tissue composition, identity, and function (Bolouri and Davidson, 2002; Peter and Davidson, 2011). Transcription factor-based fate programing has been explored before to direct cell state (Ang et al., 2018; Busskamp et al., 2014; Sugimura et al., 2017) in human induced pluripotent stem cell (hiPSC) derived mono-cell cultures. However, the manipulation of tissue GRN to override fate barriers and promote tissue development has not been examined in multicellular tissues and organoids where the presence of multiple cell types triggers cellular communication, co-differentiation, and morphogenesis.
Our previous work showed engineering self-organization of hiPSCs to a fetal liver organoid (FeLO) via heterogeneous overexpression of GATA6 (Guye et al., 2016). However, the developed FeLO presents immature hepatic characteristics (e.g., limited FXR signaling, lack of CYP3A4 mediated metabolic function), and a limited vascular network. In this study, we use FeLO as a testbed to devise a framework to rationally direct FeLO morphogenesis toward adult liver using unbiased computational analysis and reprogramming of GRNs. We examine if a small set of factors can be identified and genetically introduced into FeLO to initiate global changes at the level of chromatin and transcriptional signature of single cells and whole tissue. We ask whether the observed genetic and epigenetic changes produce a functional phenotypic response in organoids, and simultaneously can enrich our understanding of human liver development. To this end, we reverse engineer GRNs in FeLOs and compare them with adult human livers. We identify, build, and deliver genetic circuits that successfully advance hepatic fate maturation and functional characteristics, decrease aberrant GRNs, and promote the formation of vascular networks. We broadly name this rationally designed, genetically engineered tissue as a designer liver organoid (DesLO). Our studies set forth the utility of genetically guided morphogenesis in human organoids.
RESULTS
Computational Assessment of GRNs to Advance Hepatic Fate
Previously, we employed transient lentiviral expression of GATA6 to develop FeLO from hiPSCs in vitro (Guye et al., 2016). Here, to refine transgene expression with efficient and stable integration and minimal toxicity, we first generated and validated FeLO using PiggyBac transposition approach (Figures 1A and S1A–S1J). We then performed a computational analysis of FeLO transcriptomics using CellNet (Cahan et al., 2014; Radley et al., 2017). CellNet compares RNA-seq data from input samples to native cell types and tissues by both classification and GRN scores, which are measures of similarity to the target cell type or tissue and the extent to which a cell-type- or tissue-specific GRN is established, respectively. CellNet sources RNA-seq data from 97 studies to develop training sets, with no cell type or tissue training set using fewer than 4 distinct studies (Figure S1K). A total of 107 adult human livers from 10 studies were used to build the CellNet liver training set. Therefore, the platform offers an unbiased approach for defining tissue identity and classification of target and aberrant GRNs. CellNet analysis of RNA-seq data from our uninduced hiPSC line (day 0) and developing tissue at days 5, 10, and 17 (FeLO) revealed high scores for embryonic stem cells (ESC) at day 0 and day 5 that steadily dropped over time concurrently with increasing liver scores (Figures 1B and S1L). We also noticed a residual intestinal signature (Figure S1L).
Figure 1. Generation and Assessment of Fetal Liver Organoid (FeLO).
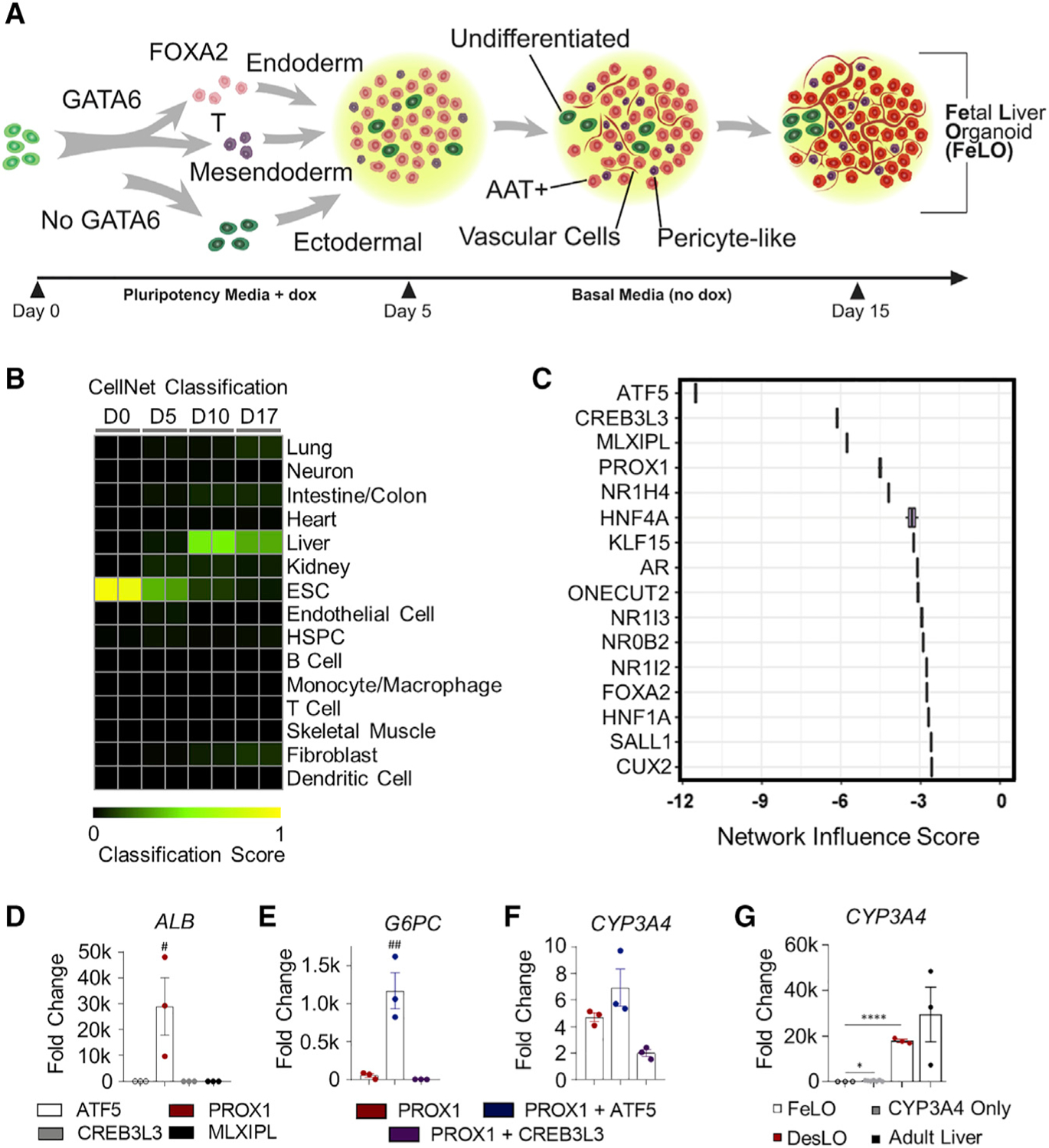
(A) Schematic of FeLO generation process from GATA6-engineered hiPSC. Full culture details are listed in the STAR Methods section.
(B) Heatmap showing the CellNet classification scores of FeLO for listed tissues or cell types before induction of GATA6 (day 0) and at day 5, 10, and 17 of culture.
(C) Liver NIS analysis using CellNet reveals transcriptional regulators that show deficiency (negative score) in day 17 FeLO compared with the human liver training set (n = 2).
(D–G) qPCR data for transcription factor screening in FeLO, increase in ALB following PROX1 induction (D), synergistic activation of G6PC with PROX1 and ATF5 (E), modest increase in CYP3A4 without SynTF(CYP3A4) (F), and significant CYP3A4 upregulation with SynTF(CYP3A4) and synergistic effect of PROX1 and ATF5 co-expression on CYP3A4 activation (G). Reference sample is untransduced. #p < 0.05, ##p < 0.01 over each other condition, *p < 0.05, ****p < 0.0001 (n = 3 except n = 6 for CYP3A4 only in L). Significance was determined by one-way ANOVA with Tukey’s multiple comparison test.
Data are represented as mean ± SEM for (D–G).
See also Figure S1.
CellNet calculates a network influence score (NIS) for transcription factors according to the degree to which their perturbation will lead to improved GRN scores. In this case, CellNet scored transcription factors according to their predicted ability to developmentally advance FeLO (Figure 1C). The top four transcription factors, whose negative score indicate under-expression relative to the liver training set (the target tissue), were activating transcription factor 5 (ATF5), cAMP-responsive element binding protein 3 like 3 (CREB3L3), MLX Interacting Protein Like (MLXIPL), and Prospero-related homeobox1 (PROX1) (Figure 1C).
Reprogramming of Gene Regulatory Networks in FeLO
Focusing on these four factors, we next examined the effect of their overexpression on a panel of hepatic and endothelial markers. We devised a method to efficiently deliver expression vectors (a.k.a. gene circuits) on day 5 by dissociation to single cells followed by lentiviral transduction (see STAR Methods). Initial analysis with a reporter showed our method yielded high transduction efficiency (Figure S1M). We transduced FeLO with each transgene individually, comparing against an untransduced control (Figure S1N). Analysis of gene expression by qPCR at day 17 revealed PROX1 to substantially outperform the other factors in upregulation of a panel of hepatic genes such as albumin (ALB), asialoglycoprotein receptor 1 (ASGR1), and farnesoid X receptor (FXR or NR1H4) or the endothelial gene CD34 (Figures 1D and S1O).
PROX1 is required for hepatocyte migration and endothelial cell development, and loss of PROX1 leads to decreased hepatocyte number (Kamiya et al., 2008; Seth et al., 2014). ATF5 is associated with the maturation of several cell types, including hepatocytes (Nakamori et al., 2016; Pascual et al., 2008; Wang et al., 2014). Overexpression of PROX1 with ATF5 and cocktails of liver-related transcription factors have been used to convert fibroblasts to hepatocyte-like cells (Du et al., 2014; Nakamori et al., 2017). However, these factors were not tested before in the context of multilineage tissues, and their downstream cellular GRNs were not fully examined. We tested whether the expression of ATF5 in tandem with PROX1 would confer any additional benefit. PROX1 and ATF5 co-expression augmented expression of several mature markers and displayed a strong synergistic effect on genes, such as G6PC (Figures 1E and S1P). However, overexpression of PROX1 and ATF5 led to a moderate induction of Cytochrome P450 3A4 (CYP3A4) in organoids (Figure 1F). CYP3A4 is a central hepatic enzyme responsible for metabolizing endogenous molecules as well as a large portion (~30%) of clinically used drugs (Zanger and Schwab, 2013). Achieving CYP3A4 levels comparable to the adult human liver is highly sought after in PSC-derived hepatocytes for applications in drug discovery, yet, previous attempts to reach this goal have been mostly ineffective (Davidson et al., 2015; Wobus and Loser, 2011). Therefore, we asked whether targeted activation of the endogenous CYP3A4 locus could meet this need and whether CYP3A4 activation in parallel with PROX1/ATF5 could further advance the developmental stage of FeLO.
We devised a synthetic transcription factor for endogenous CYP3A4 activation (SynTF(CYP3A4)) using clustered regularly interspaced short palindromic repeat (CRISPR)-based transcriptional activation (CRISPRa). To confine CYP3A4 activation to hepatocytes, we used a version of CRISPRa in which dCas9 is driven by an AAT promoter (pAAT) and two engineered hairpin aptamers in the gRNA recruit an MS2 bacteriophage coat protein (MCP)-fused transcriptional activation complex (Konermann et al., 2015) (Figures S1Q and S1R). We verified the absence of known SNPs in our hiPSC cell line that could influence CYP3A4 activity (Figure S1S). We then designed two gRNA targeting the CYP3A4 promoter and transduced FeLO at day 5 with all components of the CRISPRa complex. On day 17, we found significant upregulation of CYP3A4 expression (approximately 400-fold) (Figure 1G). When we co-delivered SynTF(CYP3A4) with the PROX1 and ATF5 circuits, we discovered a dramatic synergistic effect which resulted in an almost 20,000-fold upregulation of CYP3A4, reaching expression levels on par with adult liver tissue (Figure 1G). Combined delivery of PROX1, ATF5, and SynTF(CYP3A4) to FeLO leads to a new engineered tissue that we have dubbed as DesLO.
DesLO Demonstrates Advancement in Tissue Transcriptional Signature and Elimination of Aberrant Tissue Signatures
To investigate the effect of each genetic factor, we generated deconstructed variations of DesLO (Figure 2A) by introducing circuits encoding PROX1 only, PROX1 and ATF5, and the full combination of PROX1, ATF5, and SynTF(CYP3A4), and compared with age-matched FeLO. CellNet analyses revealed that PROX1, ATF5, and SynTF(CYP3A4) each contribute to a stepwise increase in global liver GRN and tissue classification scores (Figures 2B and 2C). Comprehensive gene set enrichment and pathway analyses using Enrichr also revealed increasing alignment to liver identity and pathways with the introduction of each circuit (Figures S2A and S2B). Whole transcriptome analysis and comparison with FeLO shows enrichment of pathways such as a nuclear receptor, complement and coagulation pathways, bile secretion, FXR pathway, and lipid metabolism similar to human liver tissue used as a positive control (clusters 1, 7, 8, 9) (Figure S2C). We also noticed the downregulation of gene clusters (clusters 5 and 6) in DesLO and human liver tissue associated with cardiac differentiation and regulation of actin cytoskeleton (Figure S2C). Additionally, we noticed the presence of cell proliferation in DesLO, which was not prominent in the human liver (cluster 4).
Figure 2. Genetic Engineering Improves Hepatic Identity and Tissue Maturation.
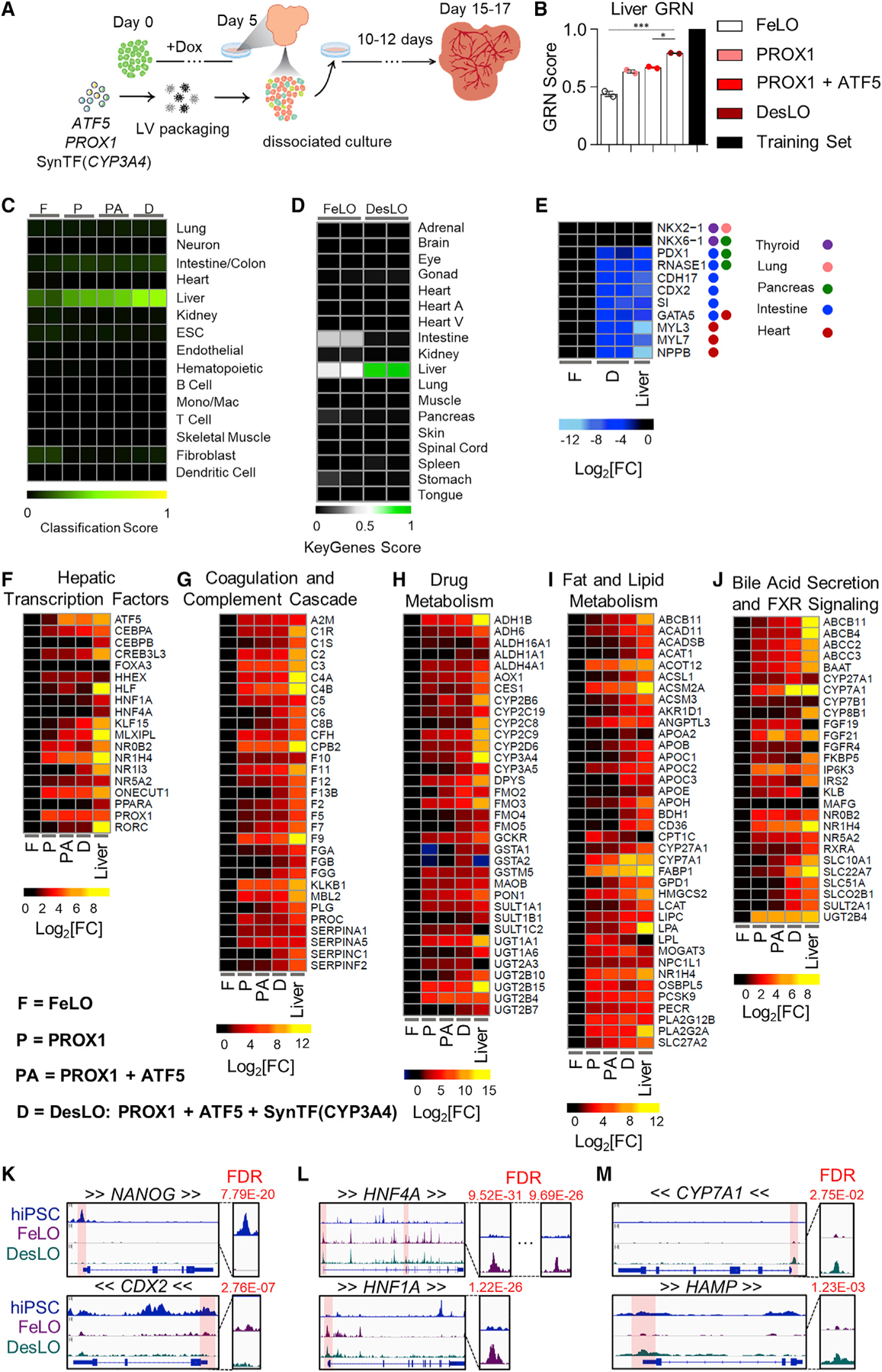
(A) Schematic of engineering strategy using dox-inducible GATA6 and lentiviral delivery of ATF5, PROX1, and SynTF(CYP3A4) gene circuits to develop DesLO. Full details in methods.
(B) Stepwise increase in CellNet liver GRN score by increasing the components transduced at day 5 from FeLO to DesLO in day 17 tissues. The training set represents the GRN score of the CellNet liver tissue library. *p < 0.05, ***p < 0.001 (n = 2). Significance determined by one-way ANOVA with Tukey’s multiple comparison test.
(C) Heatmap showing the CellNet classification scores for the listed tissues/organs or cell types for day 17 FeLO (F), PROX1 only (P), PROX1+ATF5 only (PA), and DesLO (D) samples (Mono/Mac: monocyte/Macrophage).
(D) Heatmap showing the KeyGenes scores for the listed tissues or cell types for day 17 FeLO and DesLO. (Heart A, Heart atrium; Heart V, Heart ventricle).
(E) Heatmap of gene expression in FeLO and DesLO from RNA-seq data highlighting decreased DesLO expression of aberrant lineages (values are relative to FeLO).
(F–J) Heatmaps showing enrichment in pathways relative to FeLO for hepatic transcription factors, complement and coagulation cascade, drug metabolism, cholesterol and lipid metabolism, glucose metabolism, and bile acid secretion and FXR signaling by increasing the components transduced at day 5 from FeLO to DesLO in day 17 tissues. (n = 2, mean shown, except liver n = 1).
(K) ATAC-seq results showing loss of chromatin accessibility on NANOG and CDX2 between hiPSCs, FeLO, and DesLO. FDR of differential peak comparison is indicated for the zoomed promoter regions highlighted in transparent red. Arrows around gene symbols represent the direction of transcription.
(L) ATAC-Seq results showing increases in chromatin accessibility in HNF4A and HNF1A between hiPSCs and FeLO. FDR of differential peak comparison is indicated for the zoomed promoter regions highlighted in transparent red. Arrows around gene symbols represent the direction of transcription.
(M) ATAC-seq results showing increases in chromatin accessibility in CYP7A1 and HAMP between FeLO and DesLO. FDR of differential peak comparison is indicated for the zoomed promoter regions highlighted in transparent red. Arrows around gene symbols represent the direction of transcription.
Data are represented as mean ± SEM for B, no SEM listed for the library training set.
See also Figure S2.
To cross-validate CellNet findings with an alternative methodology, we used KeyGenes (Roost et al., 2015). Using human tissue training sets, KeyGenes classified DesLO as liver, demonstrating clear improvement in liver identity and noticeable elimination of the aberrant intestinal GRN compared with FeLO (Figure 2D). These findings were in line with the repression of genes for alternative fates such as CDX2, a master regulator of intestinal epithelial differentiation, Cadherin 17 (CDH17); a CDX2 regulated gene, sucrase-isomaltase (SI) enzyme, expressed in the intestinal brush border and PDX1 important for both pancreas and duodenum (Figure 2E). We also observed a decrease in genes associated with cardiac signature (i.e., MYL3 and 7) (Figures 2E and S2C).
Further examination of specific pathways critical for adult liver function showed that each layer of engineering activated a subset of liver GRNs, successively moving the synthetic tissue closer to a native liver signature. Through our intervention, we observed a progressive enrichment in important pathways for glucose, lipid, cholesterol, and drug metabolism, complement and coagulation cascades, FXR signaling and bile secretion, and characteristic hepatic transcription factors (Figures 2F–2J, S2D, and S2E). Closer examination shows that PROX1 induces a large portion of the increases in gene expression across these pathways, including liver transcription factors NR1H4 (FXR), HNF1A, ONECUT1 (HNF6), and NR5A2 (LRH-1). The addition of the ATF5 circuit further enriched genes across pathways such as transcription factors MLXIPL and CEBPA, the bile acid transporter SLC10A1 (NTCP), regulator of glucose homeostasis G6PC, hepatokine a regulator of plasma lipids angiopoietin like 3 (ANGPTL3), and the enzyme CYP2C8, which metabolizes retinoic acid and polyunsaturated fatty acids.
Introduction of SynTF(CYP3A4) in tandem with PROX1 and ATF5 revealed pervasive upregulation across pathways and raised global liver GRN score by ~20% over PROX1/ATF5 alone (Figures 2B, 2F–2J, S2D, and S2E). The presence of CYP3A4 enriched expression of transcription factors CREB3L3, HNF4A, PPARA, and NR1I3 (CAR); coagulation and complement cascade components (e.g., FGB, F7, F9, and C6); detoxification enzymes glutathione S-transferases GSTA1 and GSTA2; apolipoprotein subunits; rate-limiting enzyme in gluconeogenesis PCK1; ATP-binding cassette transporters; ANGPTL3; and CYP7A1, which metabolizes cholesterol to synthesize bile acids.
ATAC-Seq Analysis Shows Chromatin Remodeling During Formation of FeLO and DesLO from hiPSCs
We next employed transposase-accessible chromatin sequencing (ATAC-seq) to epigenetically survey genomic loci enriched for open chromatin, which denotes sites associated with active transcription factor binding. Analysis of significant peaks indicated similarity between the FeLO and DesLO samples, which both contrasted significantly with the signature of the hiPSC samples (Figure S2F). Differentiation from hiPSC to FeLO captures the closing of chromatin at the promoter regions of pluripotency genes, such as NANOG and OCT4 (Figures 2K and S2G). Sites associated with aberrant intestinal lineages such as the CDX2 promoter were also significantly decreased in accessibility, further supporting the lineage specificity and stability of the system (Figure 2K). Conversely, promoter regions for liver identity specification such as HNF4A (at both of its well-known promoters), HNF1A, FOXA2, NR5A2, and ATF5 were opened in FeLO and DesLO (Figures 2L and S2G). This coincides with enhanced promoter-localized peaks in functional liver genes for glucose metabolism (G6PC), complement cascade (C3), and bile acid regulation (NR0B2) after differentiation to FeLO and DesLO (Figure S2G). Furthermore, the generation of DesLO significantly increases accessibility at promoter regions of loci such as CYP7A1, HAMP, and INHBE, which are important nodes for bile acid synthesis, iron homeostasis, and activin signaling in the liver, respectively (Figures 2M and S2G).
DesLO Demonstrates Advancement of Hepatic Functions In Vitro
We first confirmed the presence of the hepatic, stellate-like (SLCs), and endothelial-like cells (ELCs), via phase imaging and immunofluorescence staining of DesLO on day 17 (Figures 3A–3H, S3A, and S3B). Using several key hepatic metrics informed by past literature (Huch et al., 2015; Si-Tayeb et al., 2010b; Takebe et al., 2013; Zhu et al., 2014) we analyzed hepatocyte functions in DesLO. We also compared with FeLO, primary human hepatocytes (PHH), and commercially available hiPSC-derived hepatocytes (iHEPs). Staining for glycogen storage (Periodic acid-Schiff, PAS), lipid accumulation (Oil red O), and solute transport (Indocyanine Green, ICG) in iHEP, PHH, FeLO and DesLO revealed DesLO to perform similarly to PHH for each metric. (Figure 3I). We quantified a panel of secreted proteins in culture media known to be important for liver function (Figure 3J). In DesLO, we detected the improvement of albumin production from FeLO to levels comparable with PHH. DesLO and PHH also produced comparable levels of AAT, ANGPTL3, and complement 3 (C3), the latter of which is a central component of the complement activation cascade and innate immunity produced by the liver (Sarma and Ward, 2011). The secretion of these proteins was detected in FeLO and iHEPs for all measures but usually at lower levels than DesLO and PHH. We also showed strong upregulation of the enzymatic activity of CYP3A4 in DesLO in comparison to FeLO (Figure 3J). When testing urea synthesis and CYP2C19 activity, we noticed a slight improvement over FeLO with lower values relative to PHH (Figures 3J and S3C).
Figure 3. Immunofluorescence and Functional Comparison of DesLO, FeLO, iHEP, and PHH Cultures.
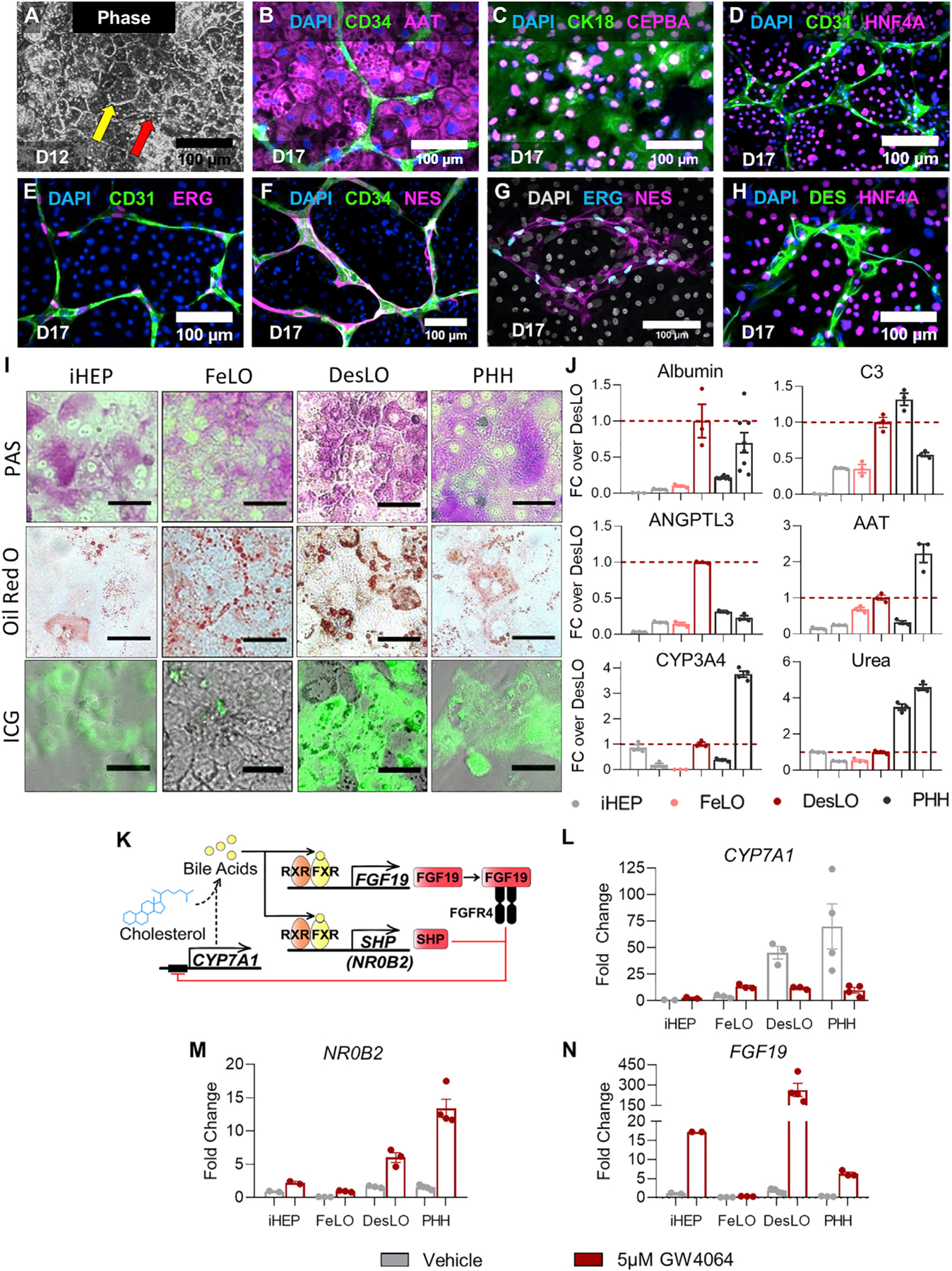
(A) Phase imaging of day 12 DesLO cultures shows the clear formation of tight junctions (indicated by the yellow arrow) characteristic of hepatic cells in culture and visible vascular structure (indicated by the red arrow) (D12: day 12). Scale bars, 100 µm.
(B–H) Immunofluorescence staining for DAPI with CD34 and AAT (B), CK18 and CEBPA (C), CD31 and HNF4A (D), CD31 and ERG (E), CD34 and NES (F), and DES and HNR4A (H) of day 17 DesLO cultures confirming the presence of HLC, SLC, and ELC populations through staining of cell-type indicative markers (D17: day 17). Scale bars, 100 µm.
(I) PAS, Oil Red O, and indocyanine green (ICG) stains indicating glycogen storage, lipid production, and solute transport activity, respectively, in iHEPs, FeLO, DesLO, and PHH cultures. Scale bar, 50 µm.
(J) ELISAs of liver-specific proteins, CYP3A4 enzymatic activity, and urea production measured in iHEP (2 lots, n = 3 each), FeLO (n = 3), DesLO, (n = 3), and PHH (2 lots, n = 3 except ANGPTL3 first lot is n = 2, and CYP3A4 second lot is n = 4). For albumin day <2 PHH samples (left PHH bar) is 2 lots, n = 3 each (n = 6 total) and day 3 PHH samples (right PHH bar) is 2 lots, n = 3 each (n = 6 total). All other PHH samples are <2 days in culture. Dotted line indicates DesLO average.
(K) Schematic of FXR regulation of CYP7A1 expression and bile acid synthesis.
(L–N) qPCR showing expression of CYP7A1 (L), NR0B2 (M), and FGF19 (N) in iHEP (n = 2), FeLO (n = 3), DesLO (n = 3), and PHH (2 lots, n = 2 each) vehicle controls and with addition of GW4064. Fold change is over iHEP vehicle.
Data are represented as mean ± SEM for (J) and (L–N).
See also Figure S3.
DesLO Shows Farnesoid X Receptor (FXR)-Mediated Response
Activation of FXR signaling in the liver maintains bile acid homeostasis and controls metabolic hemostasis (Rizzo et al., 2005). Bile acid-binding to FXR results in the secretion of fibroblast growth factor 19 (FGF19), which binds to the FGFR4-beta Klotho complex and represses CYP7A1 metabolism of cholesterol in a small heterodimer partner (SHP, NR0B2)-dependent manner (Holt et al., 2003; Inagaki et al., 2005) (Figure 3K).
We found key mediators of the FXR and bile acid related pathway upregulated in DesLO, including CYP7A1, FXR, and NR0B2 (Figure 2J). We, therefore, investigated whether the activation of this network can model the physiological dynamics of the bile acid synthesis pathway. qPCR confirmed increased expression of CYP7A1 in DesLO. Upon addition of a potent synthetic FXR agonist, GW4064, CYP7A1 was repressed 4-fold in DesLO, but was not repressed in FeLO cultures (Figure 3L). DesLO was further tested with the addition of the natural bile acid chenodeoxycholic acid (CDCA) and FGF19 to the media, which also resulted in the expected downregulation of CYP7A1 (Figure S3D). When compared with PHH and iHEPs, DesLO mirrored the response of PHH, while iHEPs and FeLO were unable to recapitulate this effect (Figures 3K–3N and S3E).
To further interrogate the dynamics of this network we surveyed NR0B2, FGF19, and FXR expression following GW4064 treatment (Figures 3K–3N and S3E). DesLO displayed a potent upregulation of both NR0B2 and FGF19 (Figures 3M and 3N). Such events, also observed in PHH, were less pronounced in FeLO and iHEP cultures, further supporting the establishment of hepatic GRNs responsive to environmental perturbations in DesLO and PHH (Figures 3K–3N). Finally, an assay of total bile acid in culture media revealed that while FeLO was unable to generate detectable amounts of bile acid from cholesterol in the medium, DesLO was capable of both producing bile acid and successfully recapitulating the role of FXR activation in the repression of bile acid synthesis upon addition of GW4064 (Figure S3F).
FXR agonists are being tested in clinical trials for controlling non-alcoholic fatty liver disease, and fibrosis (Han, 2018). Since DesLO contains a population of DES+ SLCs (Figure 3H), we asked whether SLCs can sense and respond to fibrogenic and FXR-mediated antifibrogenic stimuli. We induced fibrosis via transforming growth factor beta-1 (TGFβ1) supplementation to the culture medium (Coll et al., 2018) with and without FXR activation. Staining showed DES+ SLCs expand relative to the vehicle control (Figure S3G) with qPCR corroborating the increase in DES levels (Figure S3H). Interestingly, while FeLO and DesLO both responded modestly to TGFβ1 treatment, only DesLO could reliably capture the antifibrotic effect of FXR activation (Figure S3H). We also assessed an array of genes relevant to the antifibrotic response in DesLO. DesLO upregulated fibrogenic genes such as COL1A1 and ACTA2, which were downregulated in response to the FXR agonist (Figure S3I).
Augmented Vascular Network in DesLO
Next, we examined the development of vascular networks in DesLO. PROX1 overexpression induced many key genes associated with angiogenesis, which were maintained or enriched by the introduction of ATF5 and SynTF(CYP3A4) gene circuits (Figure 4A). CellNet analyses also revealed a significant increase in GRN score in DesLO (Figure S4A). We found upregulation of key growth factors and receptors such as VEGFA, placental growth factor (PGF), and kinase insert domain receptor (KDR), along with endothelial identity markers such as ERG, CD34, and vascular endothelial cadherin (CDH5). Image analysis of vascular network revealed that DesLO contained a vast, interconnected network of vasculature whereas FeLO was sparser (Figures 4B–4D). Analysis on days 11, 14, and 17 revealed that the total vessel length, vessel percentage area, and the number of vascular junctions were increased in DesLO relative to control FeLO (Figures 4C and S4B). Additionally, vessel metrics showed decreasing trends in FeLO from day 14 to 17, indicating instability of the vascular network, while the same measurements for DesLO cultures remained stable (Figures 4C and 4D).
Figure 4. Vascular Development in DesLO.
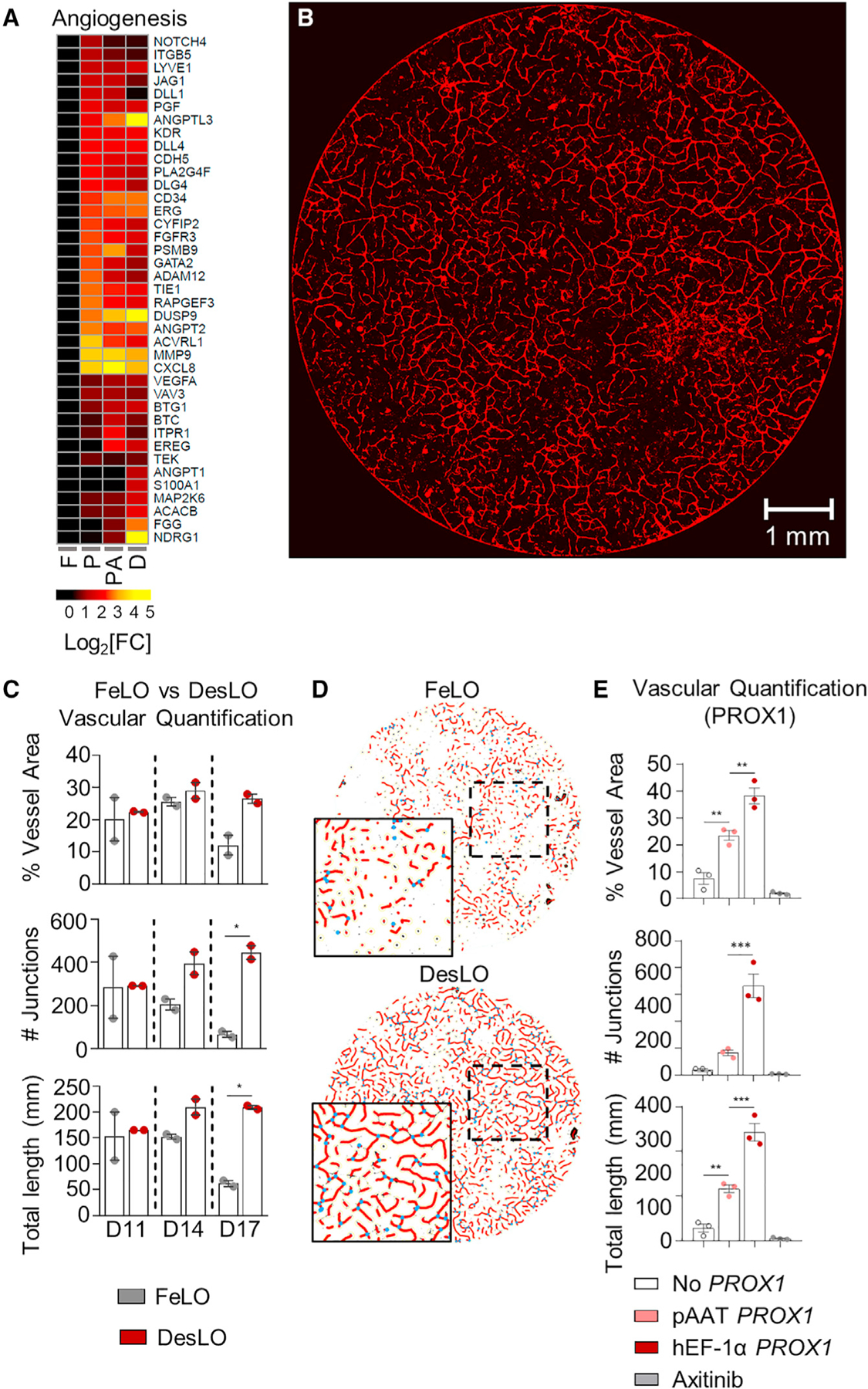
(A) Heatmap showing the expression relative to FeLO of angiogenesis-related genes in FeLO (F), PROX1 (P), PROX1+ATF5 (PA), and DesLO (D) transduced at day 5 (except FeLO) in day 17 tissues. (n = 2, mean shown).
(B) Immunofluorescence staining for CD31 showing vascular network on day 17 DesLO tissue. Image of 8-mm diameter culture is shown. Scale bar, 1 mm.
(C) AngioTool analysis of vessel area, vascular junctions, and total vessel length based on CD31 immunofluorescence staining of DesLO and control FeLO. Significance relative to control FeLO for each time point. D11, D14, D14: day 11, 14, 17, respectively (n = 2). *p < 0.05 determined by one-way ANOVA with Tukey’s multiple comparison test.
(D) Image interpretation of immunofluorescence staining of day 17 control FeLO and DesLO for CD31 generated by AngioTool analysis shows a visual increase in vascular networks in DesLO.
(E) AngioTool analysis of vessel area, vascular junctions, and total vessel length based on CD31 immunofluorescence staining of DesLO without PROX1, with PROX1 driven by pAAT, and PROX1 driven by hEF-1α with or without axitinib (n = 3). **p < 0.01, ***p < 0.001 determined by one-way ANOVA with Tukey’s multiple comparison test.
Data are represented as mean ± SEM for (C and E).
See also Figure S4.
To investigate the roles of PROX1 and ATF5 in the development of endothelial versus hepatic cell fates, we isolated ELCs and hepatobiliary cells. FACS analysis of the populations confirmed that transduction efficiency is equal among cell types (Figure S4C). Using qPCR to assay the genomic integration of transgene DNA, we determined that the endothelial population had a significantly lower integration of PROX1 (Figure S4D). FACS further showed that higher transgene levels increase the proportion of hepatic cells, corroborating the notion that higher PROX1 copy number can favor hepatic cell fate selection (Figure S4E).
To better understand the advancement of the vascular network in DesLO, we asked whether PROX1 expression in hepatocytes contribute to the development of vascular networks. We confined PROX1 gene circuit to the hepatocyte population by spatially restricting in hepatocytes using hepatocyte-specific promoter (pAAT). As a control, we used the ubiquitously active PROX1 circuit (hEF-1α-PROX1) or did not include a PROX1 circuit at all. pAAT-PROX1 DesLO showed a subtle but not significant decrease in PROX1 expression relative to hEF-1α-PROX1, possibly due to fewer PROX1-expressing cells (Figure S4F). Compared to tissue without PROX1, pAAT-PROX1 DesLO exhibited significantly improved vessel length, vessel area, and expression of CD34 and CD31 (Figures 4E and S4F). However, the number of vessel junctions, vessel length, and vessel area were significantly compromised relative to DesLO with hEF-1α-PROX1 (Figures 4E, S4F, and S4G). We also treated hEF-1α PROX1 DesLO cultures after day 7 with the small molecule axitinib, which selectively inhibits receptor tyrosine kinases (VEGFR1–3, PDGFRα, β, and c-kit). Axitinib completely ablated CD34 and CD31 expression and formation of vasculature in the presence of PROX1 expressing circuit (Figures 4E, S4F, and S4G).
Comparison of DesLO with Previously Developed Cells and Tissues
To examine DesLO relative to other available organoids, we performed a meta-analysis on available RNA-seq data of previously reported PSC-derived human liver organoids (Akbari et al., 2019; Asai et al., 2017; Ouchi et al., 2019), using CellNet. We also included data for monoculture of induced hepatocytes produced from fibroblasts via a cocktail of transcription factors (Du et al., 2014), along with primary hepatocytes and liver tissue from two independent studies as positive controls (Figures 5A and 5B). The positive controls showed high GRN and classification scores, confirming the robust network identification by CellNet. Notably, DesLO demonstrated the highest liver GRN and classification scores among all organoid samples, with only cryopreserved PHH, fresh PHH, and whole liver tissue scoring higher. DesLO achieved an approximately 50% higher GRN score than the next highest scoring organoid (Figure 5A). In the classification heatmap, the native human liver showed no apparent signatures from other organs. DesLO also had minimal aberrant signatures. However, we detected residual signatures associated with tissues, such as intestine/colon, fibroblasts, or kidney in other samples (Figure 5B). Liver bud samples (days 2 and 6) from Asai et al. showed low liver classification scores but they are prior to implantation, and in vivo maturation would be expected to occur when implanted (Asai et al., 2017; Takebe et al., 2013).
Figure 5. Comparison with Previously Developed Organoids and scRNA-Seq Analysis of DesLO.
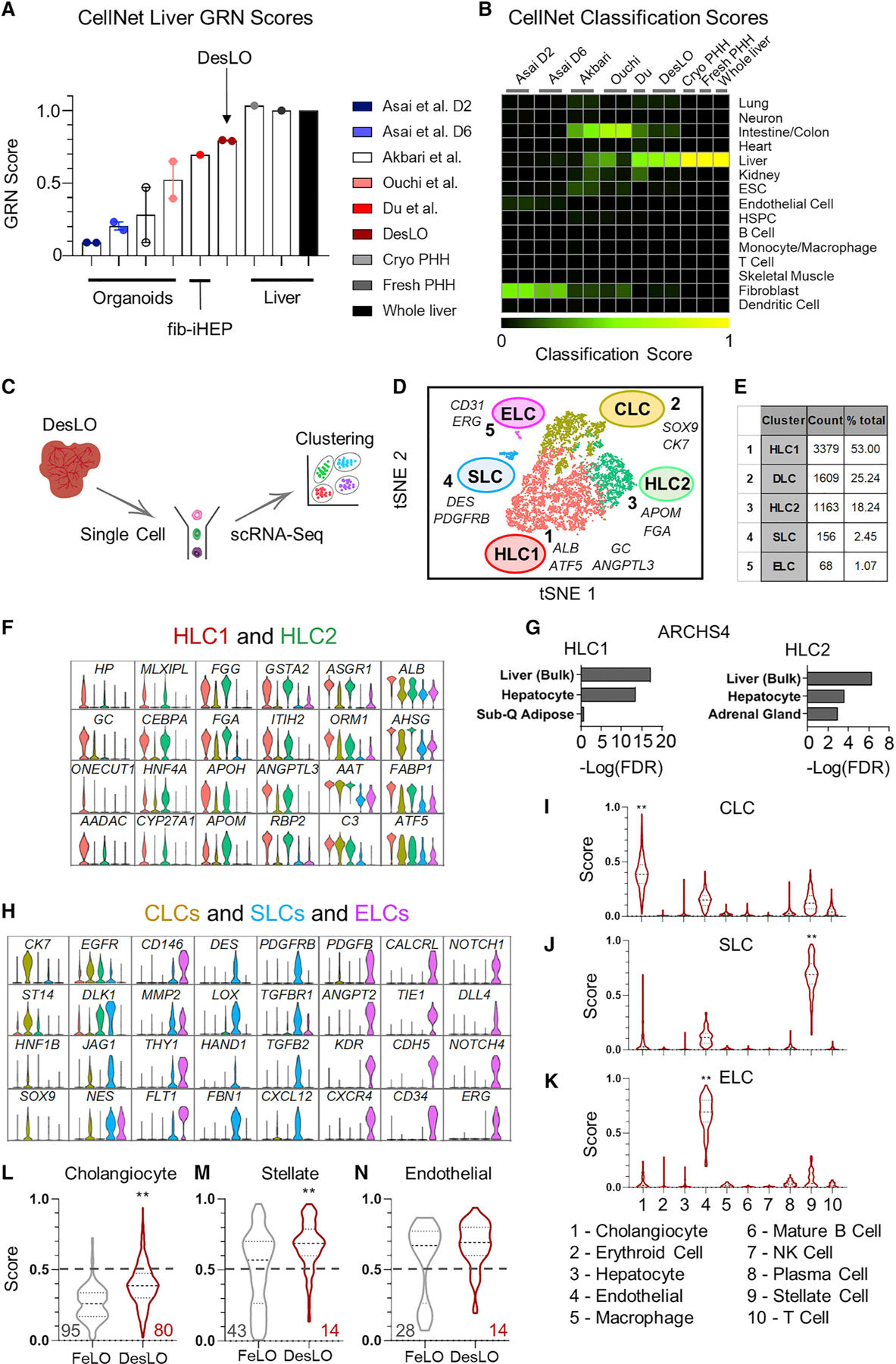
(A) CellNet meta-analysis of organoids showing GRN scores for liver organoids from hiPSCs, fibroblast-derived induced hepatocytes (fib-iHEP), cryopreserved primary human hepatocytes (Cryo PHH), freshly isolated primary human hepatocytes (Fresh PHH), and whole liver tissue. All samples are n = 2 except for Du et al, Fresh PHH, Cryo PHH, and Whole liver samples, which are n = 1.
(B) CellNet meta-analysis of organoids showing classification scores determined for selected previously reported liver organoids from hiPSCs, fib-iHEP, Cryo PHH, Fresh PHH, and whole liver tissue.
(C) Schematic of single-cell analysis for the organoids.
(D) t-distributed Stochastic Neighbor Embedding (tSNE) plot of scRNA-seq data analyzed using Seurat. Cluster numbers, names, and examples of enriched genes are displayed. HLC, hepatocyte-like cells; SLC, stellate-like cells; ELC, endothelial-like cells; CLC, cholangiocyte-like cells.
(E) Table showing the number of cells and percentage of the total population in each DesLO cluster.
(F) Violin plots showing hepatocyte associated genes upregulated in HLC clusters
(G) Enrichr analysis genes upregulated in HLC1 and HLC2 clusters against the ARCHS4 tissue library.
(H) Cholangiocyte-enriched genes upregulated in CLC, stellate-specific genes upregulated in SLC, and endothelial-specific genes upregulated in ELC clusters.
(I–K) Single-Cell net analysis of CLC (I), SLC (J), and ELC (K) clusters against a training set generated from human liver cells. **p < 0.0002 by KruskalWallis multiple comparison test.
(L-N) Comparison of classification scores between FeLO and DesLO for cholangiocyte (L), stellate cell (M), and endothelial cell (N) identities. The percentage of cells with classification scores below 0.5 are shown in gray for FeLO and red for DesLO. **p < 0.0001 by Mann-Whitney test
Data are represented as mean ± SEM for (A).
See also Figure S5.
Passaging and cryopreservation of PSC-derived liver organoids have not been shown before and have hampered their use across laboratories following generation. Hence, we tested whether DesLO can be passaged and cryopreserved. We show that after passaging and cryopreservation DesLO retains hepatic metrics such as the production of AAT and fibrinogen on day 17 (Figure S5A). Additionally, tissues re-establish their vascular structures (Figure S5B). The tissue can also be induced to grow in different conformations based on seeding substrate, providing another dimension of control for engineering applications. A time-lapse of DesLO grown on a 3D Matrigel surface demonstrates contraction into a condensed tissue (Figure S5C) that retains its vascular network (Figure S5D).
Single-Cell Analyses Show Heterotypic Cell Types and Improved Classification Scores in DesLO
The scRNA-seq analysis (Butler et al., 2018) identified five distinct clusters (Figures 5C–5E). Clusters 1 and 3 both show enrichment not only in well-known hepatic genes, such as CEBPA, HNF4A, MLXIPL, and ASGR1, but also differ in a subset of other genes. For instance, cluster 3 expresses higher levels of Glutathione S-Transferase Alpha 2 (GSTA2) and FGG, whereas cluster 1 was enriched in genes such as ALB, vitamin D binding protein (GC), and ANGPTL3 (Figure 5F). These two clusters, which align strongly with bulk liver tissue (Figure 5G), represent the pool of hepatocyte-like cells (HLC1 and HLC2) in DesLO. When we compared DesLO with FeLO, there was a visible increase in cells positive for liver transcription factors (e.g., MLXIPL, CREB3L3, and AHR); signaling molecules VEGFA and INHBE; lipid, and bile acid metabolism genes (e.g., ANGPTL3, APOH, CYP7A1, and FXR); acute phase response (e.g., HP, ORM1, ALB); and gluconeogenesis regulators (G6PC and PCK1) (Figure S5E). Additionally, expression of DLK1, which is known to mark fetal hepatocytes during development (Huang et al., 2019), decreases significantly from FeLO to DesLO (Figure S5E).
Cluster 2 was enriched for biliary markers CK7 and SOX9 and express higher ST14, a marker for a clonogenic subset of cholangiocytes (Figures 5H and S5F) (Li et al., 2017). Additionally, expression of several hepatic genes were lacking, most notably CEBPA, which is known to repress differentiation of hepatoblasts toward a biliary fate (Si-Tayeb et al., 2010a) (Figures 5H and S5F). Cluster 4 was enriched in DES, PDGFRβ and structural genes, including collagen type I alpha 1 chain (COL1A1), transgelin (TAGLN), and smooth muscle actin (ACTA2) (Figure 5H), and cluster 5 showed endothelial genes, including vascular endothelial growth factor receptor 1 (VEGFR1 or FLT1), VE-cadherin (CDH5), CD34, ERG, and CD31 (Figure 5H).
To further confirm and analyze the identity of non-hepatocyte clusters, we adapted SingleCellNet (SCN) as described in Tan et al. (Tan and Cahan, 2019) to utilize adult human liver scRNA-seq data (MacParland et al., 2018) as a training set to enable classification of non-parenchymal liver cell types. Using SingleCellNet, we quantitatively scored and confirmed the presence of cholangiocyte-like cells (CLCs, cluster 2), SLCs (cluster 4), and ELCs (cluster 5) (Figures 5I–5K). Alignment of DesLO ELC and SLC-upregulated genes with scRNA-seq data from primary liver stellate and endothelial cells also grouped DesLO ELCs closest to zone 1 liver sinusoidal endothelial cells (LSECs) and SLCs closest to stellate cells (Figure S5G) (MacParland et al., 2018).
In comparing DesLO versus FeLO, we detected higher classification scores in SLCs and CLCs following the formation of DesLO (Figures 5L and 5M). CLCs, ELCs, and SLCs in FeLO display a greater portion of cells exhibiting lower classification scores. However, the presence of these cells is decreased in DesLO (Figures 5L–5N).
We also examined GATA6, PROX1, ATF5, CYP3A4 levels in the engineered cell clusters. As confirmation that GATA6–2A-EGFP expression circuit is not active after the removal of doxycycline (a.k.a. leaky expression) we showed no GFP expression on day 17 (Figure S5H). We found higher endogenous GATA6 expression in the DesLO biliary and stellate clusters, but not in endothelial cells (Figure S5I). Additionally, scRNA-seq analysis shows that PROX1 and ATF5 are relatively enriched in hepatobiliary populations (Figure S5I). Levels of these genes further vary among the hepatobiliary clusters and require future investigation. CYP3A4 was not expressed in ELCs and SLCs based on confinement of Cas9 to AAT expressing cells in our experiments (Figure S5I).
Implantation in Mouse Models Demonstrates In Vivo Functions
To test the in vivo functionality of DesLO and its ability to engraft and provide therapeutic benefit, we implanted DesLO in mouse models of liver injury. We utilized FRGN mice (Wilson et al., 2014) in which liver injury is prevented with supplementation of 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC) in the drinking water (Vogel et al., 2004). Hepatocyte death and liver failure can be induced by the removal of NTBC (Figure 6A). Multiple DesLO were placed over the mesentery vascular bed or packed beneath the renal capsule (Figures 6B and S6A).
Figure 6. In Vivo Function of DesLO After Implantation in Mice.
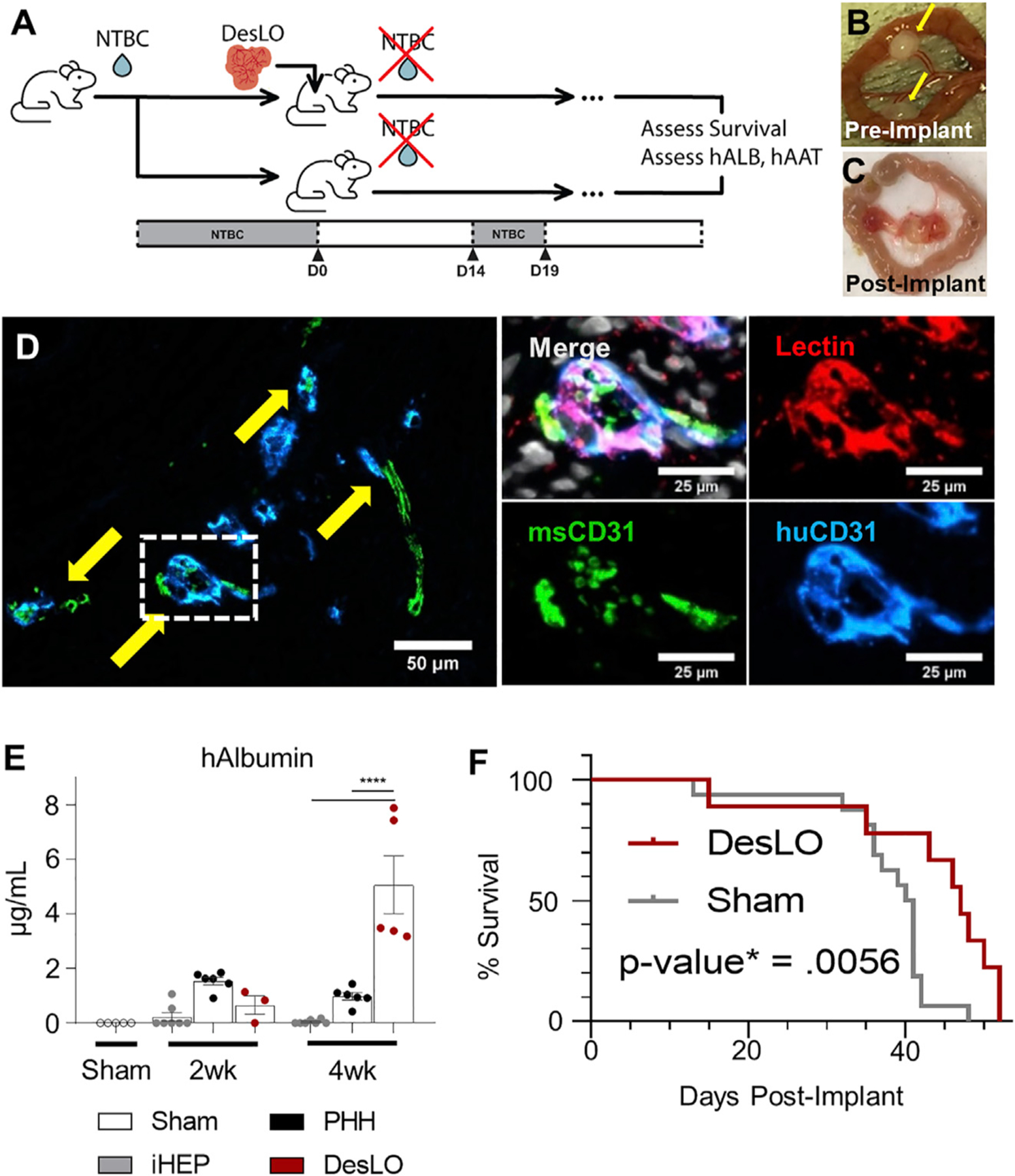
(A) Schematic of implantation into FRGN liver injury mouse model in vivo experiments. (NTBC: nitisinone; hALB: human albumin; hAAT: human alpha-1 antitrypsin).
(B) Fibrin-gelled DesLO tissue adhered to the mesentery. Implants indicated by yellow arrows.
(C) Mesentery-implanted DesLO harvested tissue after >4 weeks implanted shows tissue growth and vascularization.
(D) Integration of human and mouse CD31 shown via immunofluorescence staining of paraffin section. (Left) Arrows indicate sites of overlap. (Right) Increased zoom of indicated selection denoted in the left image by a dotted rectangle. Lectin denotes vasculature from both species. Scale bars, full image, 50 µm; zoom 25 µm.
(E) ELISA quantification of human albumin protein in mouse serum shows an increase of human albumin between 2 and 4 weeks post-implantation of DesLO (n = 5) at mesentery site, while detected human albumin decreases over this period in iHEP (n = 7, from 3 lots) and PHH (n = 6, from 3 lots). Sham (n = 5) used as control. ****p < 0.0001 determined by ANOVA with Tukey’s multiple comparison test.
(F) Kaplan Meier survival curve of FRGN mice with and without DesLO subrenal implantations. Log-rank p value = 0.0056 by Mantel-Cox test. Implant n = 9, Sham n = 11.
Data are represented as mean ± SEM for (E).
See also Figure S6.
Harvested DesLO showed vascularization at the site of implantation (Figures 6C and S6B). Upon harvest of the implants, histology revealed human and mouse vascular integration via species-specific markers for CD31, with lectin staining the endothelial cells from both species (Figure 6D). Patches of hepatic cells producing human albumin were also identified from stained paraffin-embedded sections of the recovered implant (Figure S6C). Secretion of human Albumin and AAT into host serum at two weeks post-implant was highest in the PHH control, but by four weeks post-implant PHH secretion had decreased while DesLO secretion increased to significantly higher levels (Figures 6E and S6D). Implantation of tissues transduced with each factor alone revealed that human Albumin secretion was largely driven by PROX1 (Figure S6E). When we examined the survival of mice receiving DesLO following complete removal of NTBC we found a moderate but statistically significant advantage over controls that were most evident after 40 days post liver injury (Figure 6F). To extend the applicability of these findings we also used the TK-NOG mouse model and found similarly high levels of human Albumin in the serum (Figure S6F).
DISCUSSION
Through this study, we demonstrate that the integration of GRN assessment and engineering will offer a tool to steer in vitro organogenesis and improve the products of iPSC-derived tissue engineering. Reverse engineering of GRNs in FeLO and comparison with adult human organs using CellNet offered a path to rationally select gene candidates for fate programing toward the human liver. To this end, we genetically instructed FeLO to advance to DesLO following GRN analysis and later assessed the tissue at the level of chromatin, evolved GRNs in single cells and tissue, as well as liver function in vitro and in vivo. The improvement in liver-specific GRN in DesLO was mirrored in part via physiological secretion of hepatocyte-specific proteins such as Albumin in vitro as well as in vivo and improved in CYP3A4 enzymatic function as one proxy for drug metabolism. Notably, established GRNs were responsive to perturbation and feedback regulation, as shown via FXR-mediated CYP7A1 repression and antifibrotic responses. These characteristics could closely mimic primary cells and were superior to immature FeLO and tested iHeps. The epigenetic trends reported via ATAC-Seq corroborated RNA-seq analysis and supported the existence of a robust liver identity program at the level of chromatin states in FeLO and DesLO. The high similarity between FeLO and DesLO ATAC-Seq results suggests that the establishment of chromatin profiles may largely occur during the early stages of organoid development. Such chromatin states can be exploited through changes in transcriptional machinery, including the availability of cell type and stage-specific transcription factors to direct tissue development.
Rigorous, quantitative assessment of cell identities in organoids is not frequently practiced in the field, making it difficult to perform side-by-side comparisons of synthetic tissues or the fidelity of cells generated through alternate protocols (Tan and Cahan, 2019). We performed GRN meta-analysis across a set of PSC-derived liver organoids with PHH and liver as positive controls. We showed DesLO achieved superior alignment to the human liver with minimal aberrant signatures as compared with FeLO or other liver organoids tested. Using the SingleCellNet platform we could interrogate the cell identity of non-hepatocyte populations in relation to their in vivo counterparts. We showed the presence of stellate, endothelial, and biliary cells. These cells exhibit superior classification scores in DesLO as compared with FeLO, suggesting an improved commitment to the target fates and co-maturation of clusters (Figures 5L–5N). This might be due to the direct function of PROX1 and ATF5 gene circuits or non-cell-autonomous effects via intercellular communications across lineages. These findings are also in line with observed phenotypic improvements for vascular formation (Figure 4) and stellate cell response in DesLO (Figures S3H and S3I).
Engineering a vascular network in organoids is an important objective in the field (Grebenyuk and Ranga, 2019; Yin et al., 2016) and allows for nutrient and oxygen supply and engraftment in vivo. DesLO implantation under the renal capsule resulted in sustained levels of human albumin in the mouse blood, which was not the case for primary hepatocytes without a vascular network (Figure 6E). Further optimization of the ectopic implantation in terms of tissue mass, encapsulation methods, or implant site can facilitate improvements in therapeutic survival outcome after implantation of DesLO. These data show the utility of GRN engineering to genetically program vasculogenesis within multicellular systems.
Spatial control of cues can be accomplished in cultures using inhomogeneous stimuli such as light or patterned biomaterials (Brassard and Lutolf, 2019). However, these techniques necessitate a predetermined knowledge of target locations and fall behind in linking cell state with their actuation. Engineering synthetic gene circuits instead enable a type of spatial control that can be achieved via linking the cell state to circuit function as part of its design process (Ebrahimkhani and Ebisuya, 2019; Velazquez et al., 2018). This capacity was exploited in our study via a hepatic progenitor specific promoter (pAAT) to confine and study SynTF(CYP3A4) or PROX1 function. pAAT-PROX1 circuits could partially promote DesLO vascularization. These data suggest a new role for non-cell-autonomous PROX1 expression in the hepatocyte population that regulates vascular formation in the developing liver, possibly via cell-cell communication (e.g., VEGF signaling). PROX1 or ATF5 was previously used in combination with several other transcription factors to convert fibroblasts to mono-cultures of human hepatocytes (Du et al., 2014; Nakamori et al., 2017). While our data are in line with those reports, using a multicellular system enabled the identification of new GRNs and pathways triggered by PROX1 or ATF5 expression in the course of human liver development.
Engineering CRISPR-based synthetic transcription factors has not been practiced in multicellular settings such as organoids. It can offer a possibility to turn on or off endogenous loci on demand usually without a need to probe upstream GRNs. Our CRISPR-mediated transcriptional activation of CYP3A4 in AAT expressing cells triggered a global change in hepatic identity and GRNs. It was shown before that CYP3A4 can metabolize bile acids, which then activates transcription of nuclear receptors (Bodin et al., 2005). Additionally, bile acid and derivatives promote hepatic differentiation (Sawitza et al., 2015). Thus, it is possible that CYP3A4-produced metabolites induce hepatocyte maturation via autocrine feedback loops that warrant future investigations. Future work can take advantage of fully assembled CRISPR-based layered genetic circuits (Kiani et al., 2014; Nissim et al., 2014) embedded in hiPSCs that can be activated based on cell state, decreasing the need for delivery of genetic cargo during differentiation in organoids. Such approaches can also avoid any potential supraphysiological levels of overexpressed transcription factors, taking advantage of endogenous transcriptional programs and multiplexity (e.g. different gRNAs). Human iPSC lines with genetically embedded programs can also facilitate expansion and scaling of the derived organoids. This strategy reduces the need for finding a complex media formulation that can support heterotypic cells (e.g., endothelial, pericyte, and epithelial) while alleviates the cost to produce and mature multicellular tissues. However, careful testing and selection of gRNAs will be necessary to avoid any off-target effects. Additionally, potential immunological responses against components of genetic circuits or their delivery vehicles warrant future studies.
Taken together, the ability to direct tissue development and vascularity solely through the application of genetic circuits provides a proof of concept model for the feasibility of synthetic genetic control to advance multicellular systems. DesLO developed here could not reflect every facet of human liver function such as a high level of urea production or CYP2C19 activity. Further maturation of tissue characteristics can be possible through iterative cycles of GRN assessment and engineering or combination with other techniques. Other tissue engineering approaches offer ways for cell-extrinsic control during in vitro morphogenesis (Brassard and Lutolf, 2019). Application of tissue scaffolding and microfluidics (Homan et al., 2019; Nikolaev et al., 2020) as well as bioprinting (Brassard et al., 2020) can direct cell state, tissue shape, as well as vascular development (Brassard and Lutolf, 2019; Yin et al., 2016). Interfacing cell-intrinsic genetic programming and computational systems biology with those other strategies can further advance the development of novel human-based multicellular platforms.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mo R. Ebrahimkhani (mo.ebr@pitt.edu)
Materials Availability
Plasmids used in this work were all purchased or generated directly from the sources indicated in the Key Resources Table. Any plasmids cloned from these sources reported in this manuscript are available upon request from the Lead Contact.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AAT | R&D | Cat# AF1268; RRID:AB_354707 |
| Albumin | Bethyl | Cat# A80–229A, RRID:AB_67018 |
| CD146 | R&D | Cat# AF932, RRID:AB_355721 |
| CD31 | Cell Signaling Technologies | Cat# 3528, RRID:AB_2160882 |
| CD31 | Cell Signaling Technologies | Cat# 77699, RRID:AB_2722705 |
| CD34 | Abcam | Cat# ab81289, RRID:AB_1640331 |
| CDX2 | BioGenex | Cat# AM392, RRID:AB_2650531 |
| CEBPA | R&D | Cat# AF7094, RRID:AB_10973004 |
| CK18 | Santa Cruz Biotechonology | Cat# sc-32329, RRID:AB_627849 |
| Desmin | Santa Cruz Biotechonology | Cat# sc-7559, RRID:AB_639081 |
| Desmin | Santa Cruz Biotechonology | Cat# sc-23879, RRID:AB_627416 |
| E-Cadherin | Cell Signaling Technologies | Cat# 3195, RRID:AB_2291471 |
| EPCAM | Cell Signaling Technologies | Cat# 36746, RRID:AB_2799105 |
| ERG | Abcam | Cat# ab92513, RRID:AB_2630401 |
| FOXA2 | R&D | Cat# AF2400, RRID:AB_2294104 |
| GFP | Abcam | Cat# ab13970; RRID:AB_300798 |
| HNF4A | Cell Signaling Technologies | Cat# 3113; RRID:AB_2295208 |
| NANOG | R&D | Cat# AF1997; RRID:AB_355097 |
| NANOG | Cell Signaling Technologies | Cat# 3580; RRID:AB_2150399 |
| Nestin | Santa Cruz Biotechonology | Cat# sc-23927; RRID:AB_627994 |
| SOX17 | R&D | Cat# AF1924; RRID:AB_355060 |
| T/Brachyury | R&D | Cat# AF2085; RRID:AB_2200235 |
| AF-488 anti-chicken | Jackson Immunoresearch Laboratories | Cat# 703–545-155; RRID:AB_2340375 |
| AF-594 anti-chicken | Jackson Immunoresearch Laboratories | Cat# 703–585-155; RRID:AB_2340377 |
| AF-488 anti-goat | Thermo Fisher | Cat# A-11055; RRID:AB_2534102 |
| AF-594 anti-goat | Thermo Fisher | Cat# A-11058; RRID:AB_2534105 |
| AF-647 anti-goat | Thermo Fisher | Cat# A-21447; RRID:AB_141844 |
| AF-488 anti-rabbit | Thermo Fisher | Cat# A-21206; RRID:AB_2535792 |
| AF-594 anti-rabbit | Jackson Immunoresearch Laboratories | Cat# 711–585-152; RRID:AB_2340621 |
| AF-647 anti-rabbit | Thermo Fisher | Cat# A-31573; RRID:AB_2536183 |
| AF-488 anti-mouse | Thermo Fisher | Cat# A-21202; RRID:AB_141607 |
| AF-594 anti-mouse | Jackson Immunoresearch Laboratories | Cat# 715–585-151; RRID:AB_2340855 |
| AF-647 anti-mouse | Thermo Fisher | Cat# A-31571; RRID:AB_162542 |
| AF-594 anti-Sheep | Thermo Fisher | Cat# A-11016; RRID:AB_2534083 |
| AF-647 anti-Sheep | Thermo Fisher | Cat# A21448; RRID:AB_1500712 |
| APC-CD34 | Miltenyi | Cat# 130–090-954, RRID:AB_244349 |
| PE-CD146 | Miltenyi | Cat# 130–097-939, RRID:AB_2660768 |
|
Bacterial and Virus Strains | ||
| NEB 5 alpha competent E. coli | New England Biolabs | Cat# C2987H |
|
Biological Samples | ||
| Human Adult Liver Total RNA | Cell Applications, Inc | Cat# 1H21–50 |
|
Chemicals; Peptides; and Recombinant Proteins | ||
| Axitinib | Cell Signaling Technology | Cat# 12961S |
| Phosphate buffered saline | Corning | Cal# 21–040-CV |
| Polybrene | Millipore-Sigma | Cat# TR-1003-G |
| Thawing/Plating Cocktail A | Thermo Fisher Scientific | Cat# CM3000 |
| Cell Maintenance Cocktail B | Thermo Fisher Scientific | Cat# CM4000 |
| Rat tail collagen 1 | Thermo Fisher Scientific | Cat# A1048301 |
| GW4064 | Sigma Aldrich | Cat# G5172–5MG |
| Chenodeoxycholic acid | Cayman Chemical | Cat# 10011286 |
| Human recombinant FGF19 | Peprotech | Cat# 100–32 |
| human recombinant TGFβ1 | Peprotech | Cat# 100–21 |
| Human recombinant HGF | Peprotech | Cat# 100–39H |
| Human recombinant VEGF-165 | Peprotech | Cat# 100–20 |
| Dimethylsulfoxide | Sigma Aldrich | Cal# D2650–100ML |
| TRIzol Reagent | Thermo Fisher Scientific | Cat# 15596018 |
| Y-27632 Dihydrochloride | Stem Cell Technologies | Cat# 72305 |
| Puromycin Dihydrochloride | Sigma Aldrich | Cat# P8833 |
| Doxycycline hyclate | Sigma Aldrich | Cat# D9891 |
| Chloroform | Sigma Aldrich | Cat# 288306 |
| DMEM | Gibco | Cat# 11960069 |
| DMEM/F12 | Gibco | Cat# 11320082 |
| Williams E Medium | Thermo Fisher Scientific | Cat# A1217601 |
| mTeSR 1 | Stem Cell Technologies | Cat# 85850 |
| mFresR | Stem Cell Technologies | Cat# 05855 |
| STEMdiff APEL | Stem Cell Technologies | Cat# 28995 |
| Accutase | Stem Cell Technologies | Cat# 07922 |
| hESC qualified matrigel | Corning | Cat# 354277 |
| GFR, LDEV free matrigel | Corning | Cat# 354230 |
| Lipofectamine 3000 Transfection Reagent | Thermo Fisher Scientific | Cat# L3000001 |
| Super PiggyBac Transposase Expression Vector | System Biosciences | Cat# PB210PA-1 |
| RNeasy Plus Mini Kit | QIAGEN | Cat# 74134 |
| AllPrep DNA/RNA Mini Kit | QIAGEN | Cat# 80204 |
| SYBR green power up | Thermo Fisher Scientific | Cat# A25742 |
| Normal donkey serum | Jackson Immunoresearch Laboratories | Cat# 017–000-001 |
| Prolong Diamond Antifade | Thermo Fisher Scientific | Cat# P36970 |
| Xylenes | Fisher Chemical | Cat# X3S-4 |
| Anhydrous Ethanol | Fisher Chemical | Cat# A405P-4 |
| DAPI | Thermo Fisher Scientific | Cat# 62248 |
| Heocsht 33342 | Thermo Fisher Scientific | Cat# H3570 |
| Indocyanine Green | Sigma | Cat# 21980–100MG-F |
| Oil Red O | Sigma | Cat# O1391–250ML |
| Cytodex3 Microcarrier Beads | Sigma | Cat# C3275–10G |
| Anti-TRA-1–60 MicroBeads, human | Miltenyi Biotec | Cat# 130–100-832 |
| Anti-CD34 MicroBeads, human | Miltenyi Biotec | Cat# 130–046-702 |
| Anti-CD146 MicroBeads, human | Miltenyi Biotec | Cat# 130–093-596 |
| Nitisinone (NTBC) | Sigma | Cat# PHR1731–1G |
| Ganciclovir Sodium Salt | Santa Cruz Biotechnology | Cat# sc-394139B |
|
Critical Commercial Assays | ||
| Bethyl Albumin ELISA KIT | Bethyl | Cat# E80–129 |
| SERPINA1 (AAT) ELISA | Genway Biotech | Cat# GWB-1F2730 |
| Fibrinogen ELISA | Genway Biotech | Cat# GWB-C5E724 |
| C3 ELISA | Immunology Consultants Laboratory | Cat# E-80C3 |
| ANGPTL3 ELISA | RayBiotech | Cat# ELH-ANGPTL3–1 |
| Periodic Acid-Schiff (PAS) Kit | Millipore-Sigma | Cat# 395B-1KT |
| Total bile acid | Cell Biolabs | Cat# STA-631 |
| Urea assay | Bioassay Systems | Cat# DIUR-100 |
| qPCR Lentivirus titration kit | Applied Biological Materials | Cat# LV900 |
| CYP2C19 P450-Glo™ assay | Promega | Cat# V8881 |
| CYP3A4 P450-Glo™ Assay | Promega | Cat# V9001 |
|
Deposited Data | ||
| Bulk RNA sequencing of FeLO, engineered FeLO conditions, DesLO, and Liver | This paper | GEO Accession GSE159491 |
| Day 2, 6 liver bud, cryopreserved human hepatocytes, and whole liver RNA sequencing data | Asai et al., 2017 | GEO Accession GSE85223: GSM2262397, GSM2262398, GSM2262403, GSM2262404, GSM2262407, and GSM2262406 |
| FeLO and DesLO day 17 single cell RNA sequencing | This paper | GEO Accession GSE159491 |
| iPSC, FeLO, and DesLO ATACseq data | This paper | GEO Accession GSE159491 |
| Human liver organoids in differentiation medium RNA sequencing data | Akbari et al., 2019 | GEO Accession GSE130075: GSM3731529 and GSM3731530 |
| Human liver organoids D25 | Ouchi et al., 2019 | GEO Accession GSE130075: GSM3731529 and GSM3731530 |
| Fibroblast, transdifferentiated hepatocytes, and freshly isolated hepatocytes | Du et al., 2014 | GEO Accession GSE54066: GSM1306654 and GSM1306653 |
| Adult human liver single cell RNA sequencing | MacParland et al., 2018 | GEO Accession GSE115469 |
|
Experimental Models: Cell Lines | ||
| HEK293FT | Thermo Fisher Scientific | Catalog# R70007 |
| PGP1 rttA3 TRE GATA6–2A-EGFP | This paper | N/A |
| Primary Human Hepatocytes | MGH Cell Resource Core | lot# HH-083 |
| Primary Human Hepatocytes | Lonza | Catalog# HUCPG; lot# HUM17299A, lot# HUM180851 |
| Cellartis Enhanced hiPS-HEP v2 | Takara Bio Inc | Catalog# Y10133, Y10134 |
| iCell Hepatocytes | Stem Cell Technologies | Catalog# R1104 |
|
Experimental Models: Organisms/Strains | ||
| Mouse: TK-NOG, NOD.Cg-Prkdcscid Il2rgtm1Sug Tg(Alb-TK) 7–2/ShiJic | Taconic Biosciences | Cat# 12907-M |
| Mouse: FRG® KO on NOD | Yecuris Corporation | Cat# 10–0008 |
|
Oligonucleotides | ||
| CYP3A4 sgRNA Target Sequence 1: ACTCAAAGGAGGTCAGTGAG | This paper | N/A |
| CYP3A4 sgRNA Target Sequence 2: TGATTCTTTGCCAACTTCCA | This paper | N/A |
| Primers for qPCR | This paper | See Table S1 |
|
Recombinant DNA | ||
| psPax2 | Trono Lab Packaging and Envelope Plasmids (Unpublished) | Addgene Plasmid# 12260 |
| pCMV-VSV-G | Stewart et al., 2003 | Addgene Plasmid# 8454 |
| MS2-P65-HSF1-GFP | Konermann et al., 2015 | Addgene Plasmid# 61423 |
| U6-sgRNA-MS2 | Konermann et al., 2015 | Addgene Plasmid# 61424 |
| Cas9M4-VP64 | Mali et al., 2013 | Addgene Plasmid# 47319 |
| pENTR_L1_hGATA6–2A-EGFP_L2 | Guye et al., 2016 | N/A |
| PB-TAG-ERP2 | Kim et al., 2016 | Addgene Plasmid# 80479 |
| PROX1 transcript variant 2 cDNA | Origene | Cat# RC200081 |
| ATF5 transcript variant 1 cDNA | Genecopoeia | Cat# F0925 |
| CREB3L3 complete CDS cDNA | DNASU Plasmid Repository | Clone ID# HsCD00080068 |
| MLXIPL transcript variant 1 cDNA | DNASU Plasmid Repository | Clone ID# HsCD00820703 |
| Promoter: AAT Sequence: AGGTATCTTGC TACCAGTGGAACAGCCACTAAGGATTC TGCAGTGAGAGCAGAGGGCCAGCTAAG TGGTACTCTCCCAGAGACTGTCTGAC TCACGCCACCCCCTCCACCTTGG ACACAGGACGCTGTGGTTTCTGAGC CAGGTACAATGACTCCTTTCGGTAAG TGCAGTGGAAGCTGTACACTGCCCAG GCAAAGCGTCCGGGCAGCGTAGGC GGGCGACTCAGATCCCAGCCAGTGGA CTTAGCCCCTGTTTGCTCCTCCGATAA CTGGGGTGACCTTGGTTAATATTCA CCAGCAGCCTCCCCCGTTGCCCCTCT GGATCCACTGCTTAAATACGGACG AGGACAGGGCCCTGTCTCCTCAGC TTCAGGCACCACCACTGACCTGG GACAGTGAATCGTAAGTGCTT |
This paper | N/A |
|
Software and Algorithms | ||
| CellNet | P. Cahan Lab | https://github.com/pcahan1/CellNet |
| SingleCellNet | P. Cahan Lab | https://github.com/pcahan1/singleCellNet/ |
| R version 3.6.2 | The Comprehensive R Archive Network | https://cran.r-project.org/ |
| R Studio version 1.2.5033 | RStudio Inc. | https://rstudio.com/ |
| Cell Ranger | 10x Genomics | https://www.10xgenomics.com/ |
| Graphpad Prism 8 | Graphpad Software Inc. | https://www.graphpad.com/ |
| FlowJo | Becton, Dickson and Co. | https://www.flowjo.com/ |
| IGV (version 2.7.2) | Broad Institute | https://software.broadinstitute.org/software/igv/ |
| Angiotool (version 0.6a) | NIH National Cancer Institute Center for Cancer Research | https://ccrod.cancer.gov/confluence/display/ROB2/Downloads |
| ImageJ (version 1.48v) | NIH | https://imagej.nih.gov/ij/ |
| Enrichr | A. Ma’ayan Lab | https://amp.pharm.mssm.edu/Enrichr/ |
| Seurat 3 (version 3.6.2) | R. Satija Lab | https://satijalab.org/seurat/ |
Data and Code Availability
RNA-Seq, scRNA-Seq, and ATAC-Seq source data have been deposited at GEO, accession number GSE159491. This paper also analyzes existing, publicly available data. These datasets’ accession numbers are provided in the Key Resource Table. All other data is available upon request.
The Seurat pipeline for the analysis of single cell RNA sequencing data is available for download in R via Bioconductor and through the Satija Lab website (https://satijalab.org/seurat/). The specific pipeline used is described in the STAR Methods. CellNet and SingleCellNet code is available from github (https://github.com/pcahan1/CellNet and https://github.com/pcahan1/singleCellNet/) respectively.
Additional scripts used to generate the figures reported in this paper are available in the Seurat (https://satijalab.org/seurat/) and pheatmap (https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf) documentation.
Any additional information required to reproduce this work is available from the Lead Contact
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animal husbandry and experiments were performed after ethical committee review and in accordance to The Institutional Animal Care and Use Committee at Arizona State University. The Fah−/− / Rag1−/− / Il2rgnull mouse strain on NOD background (FRGN) was purchased from Yecuris. The knockout of Fah necessitates the constant supplementation of 2-[2-nitro-4-(trifluoromethyl) benzoyl] cyclohexane-1,3-dione (NTBC, or nitisinone, 16 mg/L) in the feeding water in order to prevent accumulation of toxin in the liver. Both and female FRGN mice age were used for this study (with studies starting at 8–12 weeks of age). The NOD.Cg-PrkdcscidIl2rgtm1Sug Tg(Alb-TK)7–2/ShiJic (TK-NOG) strain was purchased from Taconic. TK-NOG mice have a hepatocyte specific expression of a Herpes Simplex Virus-1 Thymidine Kinase (TK) downstream of the albumin promoter which, upon systemic delivery of Ganciclovir (GCV), exhibits hepatocyte-specific ablation. Mice were bred (FRGN mice only, TK-NOGs from Taconic cannot be bred) housed, fed, and monitored in accordance with the protocols approved by the Institutional Animal Care and Use Committee at Arizona State University and all animals were carefully monitored for signs of morbidity and discomfort by research and veterinary staff. Experiments utilized Male only for TK-NOG and both male and female for FRGN. There were no differences noticed in results based on sex.
Primary Human Hepatocytes
Cryopreserved primary human hepatocytes were purchased from from Massachusetts General Hospital Cell Resource Core and Lonza. The use of samples for research was approved by ethical committees and informed consent was obtained from donors when appropriate. No personally identifying information has been released.
METHOD DETAILS
Lentiviral Production and Titration
HEK293FT cells (Life Technologies) were grown according to the manufacturer’s instructions in a humidified incubator at 37 °C with 5% CO2. The day before transfection 8 million HEK293FT cells were seeded on a collagen I-coated (Gibco A10483–01) 15 cm2 tissue culture treated cell culture dish. On the day of transfection cells were co-transfected with 15 µg psPAX2 (Addgene Plasmid 12260), 3.75 µg pCMV-VSV-G (Addgene Plasmid 8454), and 11.25 µg of the plasmid to be packaged using 90 µg of linear polyethylenimine (Polysciences, Inc 23966–1). Medium was changed the next morning and the supernatant was collected after 48 and 72 hours. Pooled supernatant was filtered through a 0.45 µm low protein binding filter (Corning) and concentrated in an Amicon Ultra 15 filter columns (100 kDa cutoff, Millipore) at 4,000g for 23 minutes. The concentrated virus was then aliquoted, snap frozen, and stored at —80 °C. Lentiviral concentrate was diluted 2000-fold in phosphate buffered saline (PBS) and titered via qRT-PCR using a commercially available kit (ABM LV900). Titers were calculated according to manufacturer’s instructions.
Vector Design and Construction
sgRNA, MPH, and dCas9 Lentiviral Vectors
The MS2-P65-HSF1-GFP (Addgene plasmid ID: 61423) MCP-fused transcriptional activator (MPH) was amplified and sub-cloned into a gateway entry vector via golden gate-based reaction for further cloning into a lentiviral gateway destination vector. To generate U6-sgRNA-MS2 plasmids, 20bp guide sequences were inserted into the sgRNA-MS2 cloning backbone (Addgene plasmid ID: 61424) at the BbsI site via golden gate-based reaction. After screening, U6-sgRNA-MS2 for two target sites were amplified and sub-cloned into a gateway entry vector via golden gate-based reaction. The U6-sgRNA-MS2 (x2) and MPH entry vectors were cloned into a lentiviral destination vector via gateway cloning. dCas9 (Addgene plasmid ID: 47319) was amplified and sub-cloned into a gateway entry vector via golden gate-based reaction and then cloned with the AAT promoter into a lentiviral destination vector using gateway cloning.
Transcription Factor Vectors
ATF5 transcript variant 1 cDNA was purchased from Origene (Cat. #RC200081), PROX1 transcript variant 2 cDNA was purchased from GeneCopoeia (Cat. #F0925), CREB3L3 complete CDS cDNA was purchased from DNASU (Clone ID# HsCD00080068), and MLXIPL transcript variant 1 cDNA was purchased from DNASU (Clone ID# HsCD00820703). These plasmids were individually amplified from their respective vectors and sub-cloned into gateway entry vectors via golden gate reaction. The PROX1 entry vector was cloned with gateway entry vectors for either the hEF-1α or AAT promoter into a lentiviral gateway destination vector via gateway cloning. The ATF5, CREB3L3, and MLXIPL entry vectors were cloned with a gateway entry vector for the hEF-1α promoter into a lentiviral gateway destination vector via gateway cloning. For constructing the dox-inducible expressing GATA6 vector, pENTR_L1_hGATA6–2A-EGFP_L2 previously published (Guye et al., 2016) was cloned into an All-in-One PiggyBac transposon destination vector from Addgene (Addgene plasmid ID: 80479) via gateway reaction.
Guide RNA Design
Guide RNA (gRNA) was designed to target the proximal promoter region of CYP3A4. Target loci were selected using the SAM gRNA design tool http://sam.genome-engineering.org/database/(Konermann et al., 2015) or custom designed.
Magnetic Depletion of TRA-1–60 Expressing Cells
TRA-1–60-expressing cells were depleted from cultures using the Miltenyi MACS separation system after 5 days of doxycycline induction. Cells were dissociated with accutase and depletion was performed using TRA-1–60 antibody-conjugated magnetic beads (Miltenyi Biotech) according to the manufacturer’s instructions.
Lentiviral Transduction
Pre-aliquoted lentiviral concentrate was thawed quickly and maintained on ice before use. hiPSCs were transduced as single-cell suspensions in Matrigel-coated 48-well cell culture plates in mTeSR-1 containing 8µg/mL polybrene,10 µM Y-27632, and 1µg/mL doxycycline. The medium was changed the following day. Multiplicity of infection (MOI) was determined by dividing the number of lentiviral infectious units (calculated from the lentivirus titration step) by the number of cells being seeded with the virus. Where indicated, the lentiviral MOI was 9 for low MOI and 150 for high MOI.
Cell Culture
All cells and tissues were cultured in a humidified incubator at 37 °C and 5% CO2. Our hiPSC lines were cultivated under sterile conditions in mTeSR-1 (Stem Cell Technologies, Vancouver) changed daily. Tissue culture plates were coated for 1 hour at room temperature with BD ES-qualified Matrigel (BD Biosciences) diluted according to the manufacturer’s instructions in ice cold DMEM/F-12 with 15 mM HEPES medium (Thermo Scientific). Routine passaging was performed by incubating hiPSC colonies for 5 minutes in Accutase (Sigma) at 37 °C, collecting the suspension and adding 5mL DMEM/F-12 medium containing 10 µM Y-27632, centrifuging at 300g for 5 minutes, and resuspending in DMEM/F-12 supplemented with 10 µM Y-27632 for counting. Cells were seeded at a cell density of 25,000 cells per cm2.
Primary cryopreserved human hepatocytes were obtained from Lonza and the MGH Cell Resource Core (refer to Key Resources Table for catalog and lot information). Hepatocytes were thawed, seeded, and cultured according to distributors’ instructions with the following held constant between both distributors: hepatocytes were seeded in rat tail collagen I (Thermo scientific)-coated 48-well plates at a seeding density of 175,000 viable hepatocytes per cm2, human cryopreserved hepatocyte thawing medium (Lonza) was used for recovery following thaw, Thawing/Plating Cocktail A + 5% fetal bovine serum was used for seeding, and Cell Maintenance Cocktail B was used for maintenance of hepatocytes with William’s Medium E (Thermo Scientific) as basal medium. Function was assessed via staining as well as ELISA, CYP activity, and urea production by day 2 of in vitro culture using at least 2 unique donors with age and sex matching (male ages 40–57). Results were normalized by number of seeded cells. For albumin assay we additionally included samples from day 3 and showed in a separate group. Human stem cell-derived hepatocytes (iHEP) were purchased from Takara Bio Inc. and Stem Cell Technologies, and were thawed and cultured according to the manufacturer’s protocols, and assessed at 8 days. HEK293FT cells (Thermo Scientific) were cultured under sterile conditions according to the user guide instructions.
GATA6-Engineered Cell Line Generation
rtTA expressing PGP1 hiPSCs previously generated (Guye et al., 2016) were transfected using Lipofectamine 3000 (Thermo Fisher Scientific) with Super PiggyBac Transposase (System Biosciences) and the PiggyBac transposon vector with hGATA6–2A-EGFP under control of the tetracycline responsive element promoter. Transfected cells were selected by adding 0.5µg/mL puromycin to the mTeSR1 maintenance medium.
Generation of FeLO and DesLO
FeLO is produced as explained before (Guye et al, 2016) except we used the engineered cells developed by GATA6 transposition. The GATA6-engineered hiPSCs were seeded at a density of 25,000 cells per cm2 in mTeSR-1 supplemented with 10 µM Y-27632. The next day, the medium was changed to mTeSR-1 with 1 µg/mL doxycycline to induce expression of the GATA6 transgene and replaced daily for 5 days. On day 5 the medium was changed to APEL, fully defined, serum-free and animal component-free medium (Stem Cell Technologies, Vancouver), and replaced daily until the day of harvest. For DesLO generation, cells were dissociated with accutase, depleted of TRA-1–60+ undifferentiated cells, transduced as described above with lentiviruses (PROX1/ATF5 transgenes and SynTF(CYP3A4)) on day 5, and seeded at a density of 300,000 cells per cm2 in mTeSR-1 supplemented with 1µg/mL doxycycline and 10 µM Y-27632. The next day, medium was replaced with mTeSR-1 with 1 µg/mL doxycycline. The medium was switched the following day to APEL and replaced daily. FeLO controls were generated through the same protocol as DesLO described above but were either not transduced or transduced with dCas9 (without gRNA) and mKate2 lentivirus. Samples were harvested at day 17 of culture unless otherwise indicated. For experiments in which axitinib (Cell Signaling Technology Cat. #12961S) was used, it was administered in the culture medium upon the switch to APEL on day 7 at 50 nM. For FXR regulation assessment studies day 14 DesLO media was supplemented with either 1:1000 dimethylsulfoxide (DMSO) vehicle, 5 µM GW4064, 50 µM Chenodeoxycholic acid (CDCA), or 25 µM FGF19 with media refreshed daily until harvest at day 17. For fibrosis induction and mitigation experiment, day 14 DesLO media was supplemented with either 1:1000 dimethylsulfoxide (DMSO) vehicle, 5 ng/mL TGFβ1, or 5 ng/mL TGFβ1 and 5 µM GW4064 together with media refreshed daily until harvest at day 17.
DesLO Passaging
DesLO was generated as described, but rather than switching to APEL on day 7 of culture, tissue was maintained on mTeSR-1 supplemented with with 1µg/mL doxycycline until day 10. On day 10 tissue was dissociated with accutase and reseeded at 300,000 cells per cm2 in mTeSR-1 supplemented with 1µg/mL doxycycline and 10 µM Y-27632. From this point the DesLO were cultured as previously described post-transduction for twelve days with APEL.
DesLO Freezing and Thawing
DesLO was generated as described, but rather than switching to APEL on day 7 of culture, tissue was dissociated using accutase and frozen in mFresR cryopreservation medium supplemented with 10 µM Y-27632. Cells were thawed and seeded into the same size well from which the vial was frozen. The next day, culture medium was switched to APEL and tissue was cultured for nine more days until day 17 of total culture.
MACS Bead Isolation of FeLO and DesLO
On day 15 of culture FeLO and DesLO were dissociated using trypsin. MACS bead isolation for CD34+ cells was performed according to manufacturer’s instructions (Miltenyi 130–100-453). MACS bead isolation for CD146+ cells was performed on the CD34− population according to manufacturer’s instructions (Miltenyi 130–093-596). DNA for the CD34+ the CD34−/CD146− populations was extracted using the Allprep DNA/RNA Plus Micro Kit (QIAGEN, Cat. #80284). Quantitative PCR was performed on genomic DNA with expression normalized to genomic Albumin and relative gene expression calculated using 2−ΔΔCT method.
Flow Cytometry
To test transduction efficiency DesLO was generated as described, with the exception that also included was an expression vector for mKate2 under control of the hEF-1α promoter. Transduced (DesLO) and untransduced (FeLO) tissues were dissociated at day 15 of culture using trypsin (Gibco 15400054) and analyzed using a Thermo Fisher Attune NxT Cell Analyzer flow cytometer. Analysis were performed using FlowJo 2.
Tissue Harvest, RNA Extraction, qRT-PCR
Tissues were harvested and lysed by adding 500µL Trizol (Life Technologies) directly to the tissue culture well and storing at −80°C. For extraction, lysate was thawed on ice, 100µL chloroform was added, vortexed for 30 seconds, and centrifuged at 12,000g for 15 minutes at 4°C. After centrifugation the aqueous phase was transferred to a QIAGEN gDNA eliminator column and centrifuged at 10,000g for 1 minute at room temperature. The flow-through was mixed with an equal volume of 70% EtOH and transferred to an RNEasy mini spin column. The manufacturer protocol for the RNEasy Plus Mini Kit (QIAGEN) was followed for the rest of the procedure. cDNA was synthesized using the high Capacity cDNA reverse transcription (Applied Biosystems). qRT-PCR was performed using the SYBR Green intercalating dye (ThermoFisher Scientific). Expression was normalized to 18S ribosomal RNA and relative gene expression was calculated using 2−ΔΔCT method. Primers used for qRT-PCR are listed in Table S1.
Immunofluorescence Staining
Staining on glass coverslips
Cells were grown on Matrigel-coated 8mm or 12mm diameter circular glass coverslips. Cultures were fixed for 20 minutes in 4% paraformaldehyde (Electron Microscopy Sciences) at room temperature. Coverslips were then washed three times with PBS followed by 15 minutes permeabilization with 0.2% Triton X-100 in PBS. Subsequently the coverslips were washed three times in washing buffer (0.05% Tween-20 in PBS) for 5 minutes and blocked for 20 minutes in 200 µl wash buffer plus 5% normal donkey serum (Jackson ImmunoResearch Laboratories). The primary antibodies were diluted in 5% normal donkey serum in PBS and incubated with the tissues 1 hour at room temperature followed by three washes in wash buffer for 5 minutes each. The secondary antibodies were diluted in 5% normal donkey serum in PBS and incubated with the tissues 1 hour at room temperature followed by three washes in wash buffer for 5 minutes each. Afterwards, the coverslips were mounted on microscopy glass slides using ProLong Diamond Antifade (Life Technologies), cured overnight at room temperature and then sealed with nail polish. Antibody list is in the STAR Methods.
Paraffin Embedded Sectioning and Staining
Tissue samples harvested from DesLO implanted mice washed in PBS and incubated in 4% paraformaldehyde at 4°C overnight. The samples were washed 3 times with PBS, transferred to a 70% ethanol solution, and delivered to Mayo Clinic Histology Core Laboratory for paraffin embedding and sectioning (4–5um slices). The returned slides were submerged 3 times with fresh xylene for 5 minutes then washed twice for 10 minutes in 100% ethanol, followed by twice for 10 minutes in 95% ethanol, then twice more in deionized (DI) water. Antigen retrieval was performed by submerging the slides in 1x citrate buffer (Abcam) and kept at the brink of boiling for 15 minutes in a microwave oven. The slides were then washed 3 times in DI water for 5 minutes, then once in PBS. Samples were blocked and permeablized in PBS + 8% donkey serum + 0.2% TritonX-100 for 2 hours at room temperature. Primary antibodies were incubated with the samples overnight at 4°C, rinsed 3 times in DI water, then washed 15 minutes in PBS. The secondary antibodies were added in PBS + 2% donkey serum for 2 hours at room temperature. Tissues were then counterstained with either DAPI or Hoechst, rinsed 3 times in DI water, washed for 15 minutes in PBS, and sealed with a glass coverslip for imaging using either Diamond Antifade (Thermo Scientific) or Vectorshield (Vector Labs).
Indocyanine Green, Oil Red O, and Periodic acid-Schiff Staining
Indocyanine green (Cat. #21980–100MG-F) was added to cultures at 1mg/mL for 30 minutes at 37°C, washed 3x in culture medium, and imaged. For Oil Red O (Cat. #O1391–250ML), cultures were fixed in 4% paraformaldehyde for 20 minutes at room temperature, rinsed 3x with DI water, treated with 60% isopropanol for 5 minutes, incubated with Oil Red O for 15 minutes, rinsed with DI water until there was no detectable red solution in the wash water, and then imaged. For Periodic acid-Schiff (Cat. #395B-1KT) cultures were fixed in 4% paraformaldehyde for 20 minutes, rinsed 3x with 1X PBS, incubated in periodic acid for 5 minutes, washed 3x with DI water, incubated for 15 minutes Schiff’s reagent, rinsed 3x with DI water, and imaged.
Image Acquisition and Processing
Images were acquired using the Leica DMi8 automated scanning microscope or Leica TCS SP5 confocal microscope and processed using ImageJ software (NIH). Any contrast adjustments were made in individual channels and applied evenly across the whole image in that channel. Contrast and color balance for color images was applied evenly across the whole image. AngioTool software (NIH) was used to quantify the vascular formation metrics from single channel grayscale images of vascular stains (CD31 staining). 3D reconstructions were generated using the Leica TCS SP5 confocal microscope to generate z-stacks spanning ~200um deep into the tissues and using ImageJ to construct a 3D volume from the stacks.
HNF4A+ Normalization
DesLO were stained for HNF4A, imaged using a DMi8 scanning fluorescent microscope, and quantified using ImageJ with the following pipeline: Threshold > Make Binary > Fill Holes > Watershed > Analyze Particles. Averaged results were used for normalization of DesLO in ELISA assays, CYP3A4 activity, and urea.
Enzyme-linked Immunosorbent Assays (ELISA)
Samples were assayed for AAT (Genway Biotech), albumin (Bethyl Labs), C3 (Immunology Consultants Laboratory), and ANGPTL3 (RayBiotech) using commercially available ELISA kits. Cultures being assayed for human albumin were switched to daily feeding with William’s medium E prior to sampling for measurement with ELISA due to the presence of human albumin in APEL medium. Sample dilutions were optimized to attain detection in the linear range of the standard curves for each individual assay.
Cytochrome P450 Activity Assays
The CYP3A4 and CYP2C19 activity assays were performed according to the manufacturer’s instructions for Cell-based assays (P450-Glo Assays, Promega) and analyzed with a luminometer. Activity was calculated using a beetle luciferin standard curve measured with the samples.
Total Bile Acid and Urea Assays
The total bile acid assay (Cell Biolabs) and Urea assay (Cell Biolabs) were used to measure the bile acid and urea concentrations in cultures. The cell supernatant was assessed at multiple dilutions to optimize detection, and the assays were performed according to the manufacturer’s protocol.
Mouse Studies and Implants
For implantation, engineered hiPSCs were seeded on 50% Matrigel 3-D gels (1:1 RGF Matrigel and DMEM/F12) in 48-well plates to allow contraction of cell layer into a dense tissue or adhered to Matrigel-coated Cytodex3 microcarrier beads (Sigma, 1000:1 cell to bead ratio) in 96-well round bottom plates instead of Matrigel monolayer coated tissue culture polystyrene. DesLO tissues were implanted either beneath the renal capsule or on the mesentery, and secured in place using a fibrin gel supplemented with 50 ng/mL hHGF and 20 ng/mL hVEGF-165 to promote survival and angiogenesis of the tissue for integration with host vasculature. For survival studies, FRGN (Yecuris) mice between 8 and 12 weeks old were used. For the TK-NOG studies male mice between 8 and 12 week old were used, and ganciclovir (50 mg/kg, IP), which is innocuous to normal human and mouse cells, was administered to induce damage of the mouse liver parenchymal cells at day 7, 10, and 20 after implantation of 10–12 DesLOs (~3 million cells) onto the mesentery. For FRGN studies, nitisinone (NTBC) was removed from the drinking water directly after implantation of up to 5 DesLOs under the renal capsule, or 10–12 DesLOs onto the mesentery. The NTBC treatment was cycled once, being added to the drinking water at 16mg/L after 14 days for 5 days, and then removed permanently. For control experiments, sham surgeries were performed identical to the implants but with no implanted tissues. Primary human hepatocyte (3 lots) and iHEP (3 lots) controls were generated by embedding 3 million cells in Matrigel, securing them to the mesentery with the same fibrin gel and growth factor recipe as DesLO, and maintaining under the same conditions as the DesLO-implanted mice. Blood was drawn from mice using cheek puncture. Kaplan-Meier survival analysis was performed with Prism 8 (GraphPad Software Inc.).
Bulk RNA sequencing of liver, FeLO, and DesLO tissues
RNA was extracted from samples as described above and sent to the UCLA Technology Center for Genomics and Bioinformatics for library preparation and sequencing. Libraries for RNA-Seq were prepared with KAPA Hyper Stranded RNA-Seq Kit. The workflow consists of mRNA enrichment, cDNA generation, and end repair to generate blunt ends, A-tailing, adaptor ligation and PCR amplification. Different adaptors were used for multiplexing samples in one lane. Sequencing was performed on Illumina NextSeq500 for a single read 75bp run. Data quality check was done on Illumina SAV. Demultiplexing was performed with Illumina Bcl2fastq2 v 2.17 program. A raw FASTQ quality check was performed using FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were then mapped to the latest UCSC transcript set using Bowtie2 version 2.1.0 (Langmead and Salzberg, 2012) and the gene expression level was estimated using RSEM v1.2.15 (Li and Dewey, 2011). EdgeR’s (Robinson et al., 2010) TMM (trimmed mean of M-values) algorithm was used to normalize the gene expression data. Reads were then mapped to the latest UCSC genome set using Bowtie2 and Tophat (Trapnell et al., 2009). The resulting BAM file allowed for collection of information on the alignment via PicardTools (http://broadinstitute.github.io/picard/) CollectRNASeqMetrics program. A Genebody analysis was performed using the ngsplot (Shen et al., 2014) toolkit.
Enrichr Analysis and Heatmaps for Pathways
Differential expression data was generated from the TMM normalized reads (counts per million, CPM) by dividing the average counts of each sample over the average counts for the reference condition for each gene. The resulting fold change list was used to generate a list of genes for each condition with at least two reads and ≥ 2 fold change over the reference condition. The genes from each resulting list were imported into the EnrichR web browser application, and the results used to generate alignment scoring and significance data for cell, pathway, function, and ontology analyses. For scRNA-Seq data gene lists were generated using Seurat for each cluster including genes with adj. p-values <.05 and fold change expression of at least 1.6-fold over the average gene expression of the other clusters. The lists were then submitted to EnrichR for enrichment analysis and results displayed using Graphpad Prism 8.
Genes from liver pathways illustrated using heatmaps were defined using available pathway lists from Enrichr (Chen et al., 2013; Kuleshov et al., 2016) and from additional publications. Genes were binned according to enrichment at each stage of DesLO engineering (F = untransduced FeLO, P = PROX1, PA = PROX1 + ATF5, D (DesLO) = PROX1 + ATF5 + SynTF(CYP3A4)) by fold change relative to untransduced FeLO for genes with at least two reads. Bins are defined as genes that have over two-fold enrichment for P, genes that have over two-fold enrichment for PA and not P, and genes that have over two-fold enrichment for D and not P or PA. Up to 15 genes for P enrichment, 5 genes for PA enrichment, and 5 genes for D enrichment were shown per pathway in heatmaps (depicted as log2 fold change expression over FeLO), with occasional addition of select genes of interest from the pathway in order to show the expression of key genes of interest. Due to significant overlap of gene lists included, the Fat metabolism and Cholesterol and lipid metabolism pathway results were combined after binning and displayed together as ‘‘Fat and lipid metabolism’’. The same was done for the combination of the Bile Acid Secretion and FXR Signaling pathways. In the case of no expression in genes of interest, fold change of expression was calculated using the lowest detectable expression value of the sample for estimated comparison if no reads were detected. For assessment of differentially expressed genes across the entire genome, the counts/million were again divided by the average FeLO expression and genes with greater than 2 fold-change increase or decrease were selected. The resulting gene list was used to generate a heamap divided into 9 k-means clusters. Gene set enrichment analysis was done using Enrichr. For Clusters 8 and 9, genes upregulated in DesLO greater than 2 fold-change over the FeLO reference were selected for Enrichr analysis. The positive control of human liver tissue RNA (Cell Applications 1H21–50) was added to analysis by request. While consistent preparation and analysis parameters were used, there is still a possibility of batch effects differences for this sample.
Assay for Transposase-Accessible Chromatin Using Sequencing (ATAC-Seq)
Day 17 DesLO were in acquired in single cell suspension by incubation with trypsin for 10 minutes at 37°C, followed by gentle pipetting using a serological pipette to dislodge and dissociate aggregates. Cells were frozen in mFresR cryopreservation medium. ATAC-Seq library preparation and sequencing reactions were conducted at GENEWIZ, LLC. (South Plainfield, NJ, USA). Live cell samples were thawed, washed, and treated with DNAse I (Life Tech, Cat. #EN0521) to remove genomic DNA contamination. Live cell samples were quantified and assessed for viability using a Countess Automated Cell Counter (ThermoFisher Scientific, Waltham, MA, USA). After cell lysis and cytosol removal, nuclei were treated with Tn5 enzyme (Illumina, Cat. #20034197) for 30 minutes at 37°C and purified with Minelute PCR Purification Kit (QIAGEN, Cat. #28004) to produce tagmented DNA samples. Tagmented DNA was barcoded with Nextera Index Kit v2 (Illumina, Cat. #FC-131–2001) and amplified via PCR prior to a SPRI Bead cleanup to yield purified DNA libraries. The sequencing libraries were clustered on a single lane of a flow cell. After clustering, the flow cell was loaded on the Illumina HiSeq instrument (4000 or equivalent) according to manufacturer’s instructions. The samples were sequenced using a 2x150bp Paired End (PE) configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS). Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into FASTQ files and de-multiplexed using Illumina’s bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification.
ATAC-seq Data Pre-processing
Raw ATAC-seq reads first went through the pipeline of FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) for quality control. Reads with low sequencing quality and adapter sequences were trimmed out by software Trimmomatic (Bolger et al., 2014). Then the surviving reads were aligned to reference genome hg19 by Burrows-Wheeler Aligner (Li and Durbin, 2009). Reads with low mapping quality were filtered out and only primary alignments were kept by SAMtools (Li et al., 2009). Duplicate reads were marked by Picard tool http://broadinstitute.github.io/picard/. Once the alignment was done, MACS (Zhang et al., 2008) was applied for peak calling.
Differential Binding Site Analysis
In order to compare the differential binding sites between pairwise conditions across hiPSC, FeLO and DesLO, peaks called from each individual sample were pooled together will common peak regions. Read count for each pooled peak region within each sample were quantified by R package ‘DiffBind’ http://bioconductor.org/packages/release/bioc/html/DiffBind.html. Based on peak enrichment, correlation plot and PCA plot were generated to visualize the correlation/similarity across the samples. Differential binding sites test were performed by R package ‘DESeq2’ (Love et al., 2014). By cutoff of FDR=5% and absolute log2(fold change) greater than 1, top peaks were selected, representing genomic regions that are more open in one condition compared with the other one. Selected peak regions were visualized by tool IGV (Robinson et al., 2011).
Single Nucleotide Polymorphism (SNP) Analysis
Genomic data for the hiPSC line used in this study was obtained from the personal genome project (https://my.pgp-hms.org). SNPs were extracted for CYP3A4 region (chr7:99354583–99381811) by reference genome hg19.
10x Genomics Sample Preparation for Next-generation Sequencing
Samples were prepared as described by the 10x Genomics Single Cell 3′ v2 Reagent Kit user guide. Day 17 DesLO were acquired in single cell suspension by incubation with trypsin for 10 minutes at 37°C, followed by gentle pipetting using a serological pipette to dislodge and dissociate aggregates. Two washes in PBS −/− + 0.04% BSA were performed with the cells re-suspended at a final concentration of 1000 cells/µL in PBS −/− + 0.04% BSA. A live cell count was performed with a hemocytometer using Trypan Blue to identify dead cells. Following counting, cells and 10x Genomics reagents were loaded into the single cell cassette, with a target of 6000 single cells for analysis, accounting for predicted cell loss and doublets as laid out in the user guide for the Chromium Single-Cell 3’ Reagent Kit (10x Genomics). After generation of GEMs, the cDNA library was prepared by ASU Genomics Core staff following the appropriate steps determined by the 10x Genomics user guide. Libraries were sent to Novogene for sequencing by an Illumina HiSeq X for an intended read depth of 100,000 reads per cell with 150 bp paired end reads. Our downstream analysis from the sequencing data estimates the actual number of cells sequenced was between 5000 and 7000 which would yield >80k reads per cell. The 10x Genomics CellRanger pipeline was used to align reads to the reference genome (GRCh38.84) appended with transgene sequences, to assign reads to individual cells, and to estimate gene expression based on UMI counts (Zheng et al., 2017).
Single cell data was excluded based on high mitochondrial genome transcript ratio and either high or low feature or UMI counts. Genes with UMI counts in fewer than 5 cells were removed from consideration. For scRNA-Seq data processing and cluster analysis using Seurat (Butler et al., 2018; Satija et al., 2015), we used the following general standardized pipeline for processing of the Cell Ranger output: Normalization, feature selection, scaling (including cell cycle regression), principal component analysis (PCA), and clustering. Elbow plots and permuted p-values were used to assist in determining the optimal number of principal components (PCs) needed to summarize the datasets without losing a significant amount of variation. The quality of a range of clustering resolution values were assessed using enrichment of cluster marker genes (genes differentially upregulated in a given cluster relative to all other clusters) with liver cell type-specific genes. As a quality check, PC and resolution metrics were modulated to yield fewer or additional clusters to confirm that chosen parameters resulted in the most biologically relevant clustering. Visualization was achieved by the use of tSNE plots identifying cells, clusters, and selected gene expression in each cell, as well as heatmaps and violin plots showing the expression level of genes by cluster. Expression per cell by cluster was also extracted and visualized using Graphpad Prism 8.
For assessment of hepatocyte clusters we used Enrichr to assess the list of genes with greater than 1.5 fold change expression (over the average gene expression) for the HLC1 and HLC2 clusters separately using the ARCHS4 library to determine similarity to gene expression of cell and tissue types. To further assess endothelial and stellate-like cells, the top 25 differentially expressed genes in LSEC zone1, LSEC zone 2, periportal endothelial, and stellate cell clusters from the available primary liver scRNA-Seq data of MacParland et al. (2018), were cross referenced against ≥1.5-fold upregulated (compared to average expression of the genes across all clusters) genes from the ELC and SLC clusters from the DesLO scRNA-Seq. The top 25 genes from MacParland et al. were displayed in a binary heatmap for LSEC zone 1, LSEC zone 2, periportal endothelial, and stellate cell clusters along with the DesLO ELC and SLC clusters.
CellNet Analysis of FeLO Time Lapse and DesLO Bulk RNA
We used the bulk RNA-Seq CellNet (Radley et al., 2017) pipeline to quantify gene expression estimates as previously described, aligning to reference genome GRCh38. Classification and network analysis were performed using the human cnProc cnProc_HS_R-S_Apr_05_2017, which is trained on 15 cell and tissue types from mostly adult sources. Source code are available through GitHub (https://github.com/pcahan1/CellNet).
For meta-analysis, we used CellNet to quantify gene expression, and performed classification and gene regulatory network analysis for bulk RNA sequencing samples from past studies (Akbari et al., 2019; Asai et al., 2017; Du et al., 2014; Ouchi et al., 2019) in the same manner as we did with our FeLO and DesLO bulk RNA seq samples. We compared the classification scores and liver gene regulatory network status across all studies using classification heatmaps and bar plots.
KeyGenes Analysis
Raw gene pseudocounts produced by SALMON (Patro et al., 2017) were run through the KeyGenes algorithm (https://github.com/DavyCats/KeyGenes). Human fetal (excluding the maternal endometrium) (Roost et al., 2015) and adult data (Fagerberg et al., 2014); Illumina Body Map 2.0) were used as training data.
SingleCellNet Analysis
We used single cell dataset from human livers (MacParland et al., 2018) as training sets for SingleCellNet platform (Tan and Cahan, 2019). We then analyzed non-parenchymal liver cells in DesLO and FeLO using SingleCellNet.
QUANTIFICATION AND STATISTICAL ANALYSIS
For studies in which statistical analyses were performed, at least three biologically independent replicates were used unless otherwise indicated. Statistical comparisons of three or more conditions were performed using one-way ANOVA and multiple comparisons testing on means using Tukey’s method with a p-value < 0.05 as the threshold for significance. This method was used where significance has been reported without indicating method. Otherwise, for comparisons between only two conditions, one or two tailed t-tests was used with a p-value < 0.05 as the threshold for significance and are indicated as such. We used the non-parametric Mann-Whitney U test to determine assess the significance of Single CellNet classification scores. Log-rank p-values for the FRGN survival study were determined using the Mantel-Cox test.
Supplementary Material
Highlights.
Analysis of gene regulatory network identifies maturation transcription factors
Advancement of maturation and vascularity via synthetic gene circuits and CRISPRa
Computational analyses and benchmarking against human organoids and livers
Modeling of hepatic functions in vitro and in vivo
ACKNOWLEDGMENTS
We thank all members of Ebrahimkhani and Kiani laboratories for discussions and assistance in this study. We thank the Mayo Clinic Histology Core Laboratory for assistance with histology as well as the ASU Department of Animal Care and Technologies for mouse husbandry. We also thank the UCLA Technology Center for Genomics and Bioinformatics, Novogene, and Genewiz for RNA sequencing. This work was supported by an R01 from the National Institute of Biomedical Imaging and Bioengineering (EB028532), an R01 from the National Heart Lung and Blood Institute (HL141805), and the New Investigator Award from Arizona Biomedical Research Council (ADHS16–162402) to M.R.E. as well as support from the Pittsburgh Liver Research Center (NIH-NIDDK P30DK120531). The research reported in this publication was also partly supported by an R01 from the National Institute of Biomedical Imaging and Bioengineering (EB024562) to S.K. It was also partly supported by the National Institute of General Medical Sciences Award Number T32GM008208. P.C. was supported by R35GM124725. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cels.2020.11.002.
DECLARATION OF INTERESTS
M.R.E, S.K., P.C., J.J.V., and R.L. have submitted a patent (WO2019237124) for the work included in this publication.
REFERENCES
- Akbari S, Sevinç GG, Ersoy N, Basak O, Kaplan K, Sevinç K, Ozel E, Sengun B, Enustun E, Ozcimen B, et al. (2019). Robust, long-term culture of endoderm-derived hepatic organoids for disease modeling. Stem Cell Rep 13, 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang LT, Tan AKY, Autio MI, Goh SH, Choo SH, Lee KL, Tan J, Pan B, Lee JJH, Lum JJ, et al. (2018). A roadmap for human liver differentiation from pluripotent stem cells. Cell Rep 22, 2190–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai A, Aihara E, Watson C, Mourya R, Mizuochi T, Shivakumar P, Phelan K, Mayhew C, Helmrath M, Takebe T, et al. (2017). Paracrine signals regulate human liver organoid maturation from induced pluripotent stem cells. Development 144, 1056–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodin K, Lindbom U, and Diczfalusy U (2005). Novel pathways of bile acid metabolism involving CYP3A4. Biochim. Biophys. Acta 1687, 84–93. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri H, and Davidson EH (2002). Modeling transcriptional regulatory networks. BioEssays 24, 1118–1129. [DOI] [PubMed] [Google Scholar]
- Brassard JA, and Lutolf MP (2019). Engineering stem cell self-organization to build better organoids. Cell Stem Cell 24, 860–876. [DOI] [PubMed] [Google Scholar]
- Brassard JA, Nikolaev M, Hübscher T, Hofer M, and Lutolf MP (2020). Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat. Mater [DOI] [PubMed]
- Busskamp V, Lewis NE, Guye P, Ng AH, Shipman SL, Byrne SM, Sanjana NE, Murn J, Li Y, Li S, et al. (2014). Rapid neurogenesis through transcriptional activation in human stem cells. Mol. Syst. Biol 10, 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, and Collins JJ (2014). CellNet: network biology applied to stem cell engineering. Cell 158, 903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H (2016). Modeling development and disease with organoids. Cell 165, 1586–1597. [DOI] [PubMed] [Google Scholar]
- Coll M, Perea L, Boon R, Leite SB, Vallverdú J, Mannaerts I, Smout A, El Taghdouini A, Blaya D, Rodrigo-Torres D, et al. (2018). Generation of hepatic stellate cells from human pluripotent stem cells enables in vitro modeling of liver fibrosis. Cell Stem Cell 23, 101–113.e7. [DOI] [PubMed] [Google Scholar]
- Davidson MD, Ware BR, and Khetani SR (2015). Stem cell-derived liver cells for drug testing and disease modeling. Discov. Med 19, 349–358. [PMC free article] [PubMed] [Google Scholar]
- Del Dosso A, Urenda JP, Nguyen T, and Quadrato G (2020). Upgrading the physiological relevance of human brain organoids. Neuron 107, 1014–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Wang J, Jia J, Song N, Xiang C, Xu J, Hou Z, Su X, Liu B, Jiang T, et al. (2014). Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell 14, 394–403. [DOI] [PubMed] [Google Scholar]
- Ebrahimkhani MR, and Ebisuya M (2019). Synthetic developmental biology: build and control multicellular systems. Curr. Opin. Chem. Biol 52, 9–15. [DOI] [PubMed] [Google Scholar]
- Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebenyuk S, and Ranga A (2019). Engineering organoid vascularization. Front. Bioeng. Biotechnol 7, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guye P, Ebrahimkhani MR, Kipniss N, Velazquez JJ, Schoenfeld E, Kiani S, Griffith LG, and Weiss R (2016). Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat. Commun 7, 10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han CY (2018). Update on FXR biology: promising therapeutic target? Int. J. Mol. Sci 19, 2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway EM, Capeling MM, and Spence JR (2019). Biologically inspired approaches to enhance human organoid complexity. Development 146, dev166173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, Donahee M, Wang DY, Mansfield TA, Kliewer SA, et al. (2003). Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev 17, 1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre JV, et al. (2019). Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods 16, 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhao X, Wang J, Cheng Y, Wu Q, Wang B, Zhao F, Meng L, Zhang Y, Jin M, and Xu H (2019). Distinct roles of Dlk1 isoforms in bi-potential differentiation of hepatic stem cells. Stem Cell Res. Ther 10, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, Ellis E, van Wenum M, Fuchs SA, de Ligt J, et al. (2015). Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, et al. (2005). Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab 2, 217–225. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Kakinuma S, Onodera M, Miyajima A, and Nakauchi H (2008). Prospero-related homeobox 1 and liver receptor homolog 1 coordinately regulate long-term proliferation of murine fetal hepatoblasts. Hepatology 48, 252–264. [DOI] [PubMed] [Google Scholar]
- Kiani S, Beal J, Ebrahimkhani MR, Huh J, Hall RN, Xie Z, Li Y, and Weiss R (2014). CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat. Methods 11, 723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Oceguera-Yanez F, Sakurai C, Nakagawa M, Yamanaka S, and Woltjen K (2016). Inducible Transgene Expression in Human iPS Cells Using Versatile All-in-One piggyBac Transposons. Methods Mol. Biol 1357, 111–131. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dorrell C, Canaday PS, Pelz C, Haft A, Finegold M, and Grompe M (2017). Adult mouse liver contains two distinct populations of cholangiocytes. Stem Cell Rep 9, 478–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares I, et al. (2018). Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun 9, 4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, and Church GM (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol 31, 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamori D, Akamine H, Takayama K, Sakurai F, and Mizuguchi H (2017). Direct conversion of human fibroblasts into hepatocyte-like cells by ATF5, PROX1, FOXA2, FOXA3, and HNF4A transduction. Sci. Rep 7, 16675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamori D, Takayama K, Nagamoto Y, Mitani S, Sakurai F, Tachibana M, and Mizuguchi H (2016). Hepatic maturation of human iPS cell-derived hepatocyte-like cells by ATF5, c/EBPα, and PROX1 transduction. Biochem. Biophys. Res. Commun 469, 424–429. [DOI] [PubMed] [Google Scholar]
- Nikolaev M, Mitrofanova O, Broguiere N, Geraldo S, Dutta D, Tabata Y, Elci B, Brandenberg N, Kolotuev I, Gjorevski N, et al. (2020). Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 585, 574–578. [DOI] [PubMed] [Google Scholar]
- Nissim L, Perli SD, Fridkin A, Perez-Pinera P, and Lu TK (2014). Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell 54, 698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi R, Togo S, Kimura M, Shinozawa T, Koido M, Koike H, Thompson W, Karns RA, Mayhew CN, McGrath PS, et al. (2019). Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab 30, 374–384.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Gómez-Lechón MJ, Castell JV, and Jover R (2008). ATF5 is a highly abundant liver-enriched transcription factor that cooperates with constitutive androstane receptor in the transactivation of CYP2B6: implications in hepatic stress responses. Drug Metab. Dispos 36, 1063–1072. [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, and Davidson EH (2011). A gene regulatory network controlling the embryonic specification of endoderm. Nature 474, 635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radley AH, Schwab RM, Tan Y, Kim J, Lo EKW, and Cahan P (2017). Assessment of engineered cells using CellNet and RNA-seq. Nat. Protoc 12, 1089–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo G, Renga B, Mencarelli A, Pellicciari R, and Fiorucci S (2005). Role of FXR in regulating bile acid homeostasis and relevance for human diseases. Curr. Drug Targets Immune Endocr. Metab. Disord 5, 289–303. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roost MS, van Iperen L, Ariyurek Y, Buermans HP, Arindrarto W, Devalla HD, Passier R, Mummery CL, Carlotti F, de Koning EJ, et al. (2015). KeyGenes, a tool to probe tissue differentiation using a human fetal transcriptional atlas. Stem Cell Rep 4, 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma JV, and Ward PA (2011). The complement system. Cell Tissue Res 343, 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawitza I, Kordes C, Götze S, Herebian D, and Häussinger D (2015). Bile acids induce hepatic differentiation of mesenchymal stem cells. Sci. Rep 5, 13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A, Ye J, Yu N, Guez F, Bedford DC, Neale GA, Cordi S, Brindle PK, Lemaigre FP, Kaestner KH, and Sosa-Pineda B (2014). Prox1 ablation in hepatic progenitors causes defective hepatocyte specification and increases biliary cell commitment. Development 141, 538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, and Nestler E (2014). ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Lemaigre FP, and Duncan SA (2010a). Organogenesis and development of the liver. Dev. Cell 18, 175–189. [DOI] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, and Duncan SA (2010b). Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 51, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen ISY, Hahn WC, Sharp PA, et al. (2003). Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 9, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura R, Jha DK, Han A, Soria-Valles C, da Rocha EL, Lu YF, Goettel JA, Serrao E, Rowe RG, Malleshaiah M, et al. (2017). Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, et al. (2013). Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484. [DOI] [PubMed] [Google Scholar]
- Takebe T, and Wells JM (2019). Organoids by design. Science 364, 956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, and Cahan P (2019). SingleCellNet: a computational tool to classify single cell RNA-seq data across platforms and Across species. Cell Syst 9, 207–213.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez JJ, Su E, Cahan P, and Ebrahimkhani MR (2018). Programming morphogenesis through systems and synthetic biology. Trends Biotechnol 36, 415–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A, van Den Berg IE, Al-Dhalimy M, Groopman J, Ou CN, Ryabinina O, Iordanov MS, Finegold M, and Grompe M (2004). Chronic liver disease in murine hereditary tyrosinemia type 1 induces resistance to cell death. Hepatology 39, 433–443. [DOI] [PubMed] [Google Scholar]
- Wang W, Du Z, Yan J, Ma D, Shi M, Zhang M, Peng C, and Li H (2014). Mesenchymal stem cells promote liver regeneration and prolong survival in small-for-size liver grafts: involvement of c-Jun N-terminal kinase, cyclin D1, and NF-κB. PLoS One 9, e112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EM, Bial J, Tarlow B, Bial G, Jensen B, Greiner DL, Brehm MA, and Grompe M (2014). Extensive double humanization of both liver and hematopoiesis in FRGN mice. Stem Cell Res 13, 404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus AM, and Löser P (2011). Present state and future perspectives of using pluripotent stem cells in toxicology research. Arch. Toxicol 85, 79–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Mead BE, Safaee H, Langer R, Karp JM, and Levy O (2016). Engineering stem cell organoids. Cell Stem Cell 18, 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, and Schwab M (2013). Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther 138, 103–141. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Rezvani M, Harbell J, Mattis AN, Wolfe AR, Benet LZ, Willenbring H, and Ding S (2014). Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature 508, 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq, scRNA-Seq, and ATAC-Seq source data have been deposited at GEO, accession number GSE159491. This paper also analyzes existing, publicly available data. These datasets’ accession numbers are provided in the Key Resource Table. All other data is available upon request.
The Seurat pipeline for the analysis of single cell RNA sequencing data is available for download in R via Bioconductor and through the Satija Lab website (https://satijalab.org/seurat/). The specific pipeline used is described in the STAR Methods. CellNet and SingleCellNet code is available from github (https://github.com/pcahan1/CellNet and https://github.com/pcahan1/singleCellNet/) respectively.
Additional scripts used to generate the figures reported in this paper are available in the Seurat (https://satijalab.org/seurat/) and pheatmap (https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf) documentation.
Any additional information required to reproduce this work is available from the Lead Contact


