Abstract
The organization and dynamics of human genome govern all cellular processes – directly impacting the central dogma of biology – yet are poorly understood, especially at large length scales. Chromatin, the functional form of DNA in cells, undergoes frequent local remodeling and rearrangements to accommodate processes such as transcription, replication and DNA repair. How these local activities contribute to nucleus-wide coherent chromatin motion, where micron-scale regions of chromatin move together over several seconds, remains unclear. Activity of nuclear enzymes was found to drive the coherent chromatin dynamics, however, its biological nature and physical mechanism remain to be revealed. The coherent dynamics leads to a perpetual stirring of the genome, leading to collective gene dynamics over microns and seconds, thus likely contributing to local and global gene-expression patterns. Hence, a possible biological role of chromatin coherence may involve gene regulation.
Keywords: chromatin dynamics, genome self-organization, chromatin coherent motions, active nucleoplasm, genome biophysics, gene regulation
Introduction
The human genome consists of about 2 m of DNA organized into chromosomes, tightly packed inside a cell nucleus barely 10 μm in diameter [1]. Chromosomes are comprised of linear DNA molecules wrapped around nucleosomes, made of core histone proteins, forming a chromatin fiber [1, 2]. Twenty years ago, the Human Genome Project revealed the sequence of the human genome [3], while in the past decade chromosome conformation capture techniques uncovered its 3D static folded state [4–7]. Such a picture is extraordinarily rich and intricate, elucidating the static structure, architecture, topology and organization of the genome inside the nucleus at many length scales: The chromatin fiber is spatially organized into a hierarchy of loops, leading to formation of topologically associated domains, further grouped into A and B compartments, corresponding to transcriptionally active and inactive regions, respectively, and finally, at larger length scales, chromosome territories [4–7].
Although the static folded state of the genome has been described in detail, it remains unclear how to reconcile this picture with its dynamic nature, both biochemical and biophysical. Chromatin serves as a template for number of biological processes such as transcription, replication and DNA repair. Therefore, when needed, DNA has to be accessible to the molecular machinery of these processes, inevitably leading to local chromatin rearrangements and spatial displacements of both DNA and its molecular machinery. Figure 1 illustrates the close interplay between local changes in chromatin structure due to dynamical and ATP-dependent DNA transactions, such as transcription and replication, and chromatin dynamics at different timescales and length scales. The biochemistry of such DNA transactions has been extensively studied over several decades [1, 2], but the mechanistic picture behind their contributions to the physical motion of chromatin remains elusive [6–10]. Recent advances in the microscopy and image processing techniques open new avenues for elucidating the genome’s positional fluctuations in vivo, illuminating its physical behavior in space and time. These efforts are revealing unusual motion of chromatin, different from free diffusion of small molecules or motor-driven motion of cytoskeletal filaments. In this review, we survey the current knowledge about the large-scale physical movements of the genome in live cells and their possible impact on chromatin physiology.
Figure 1:
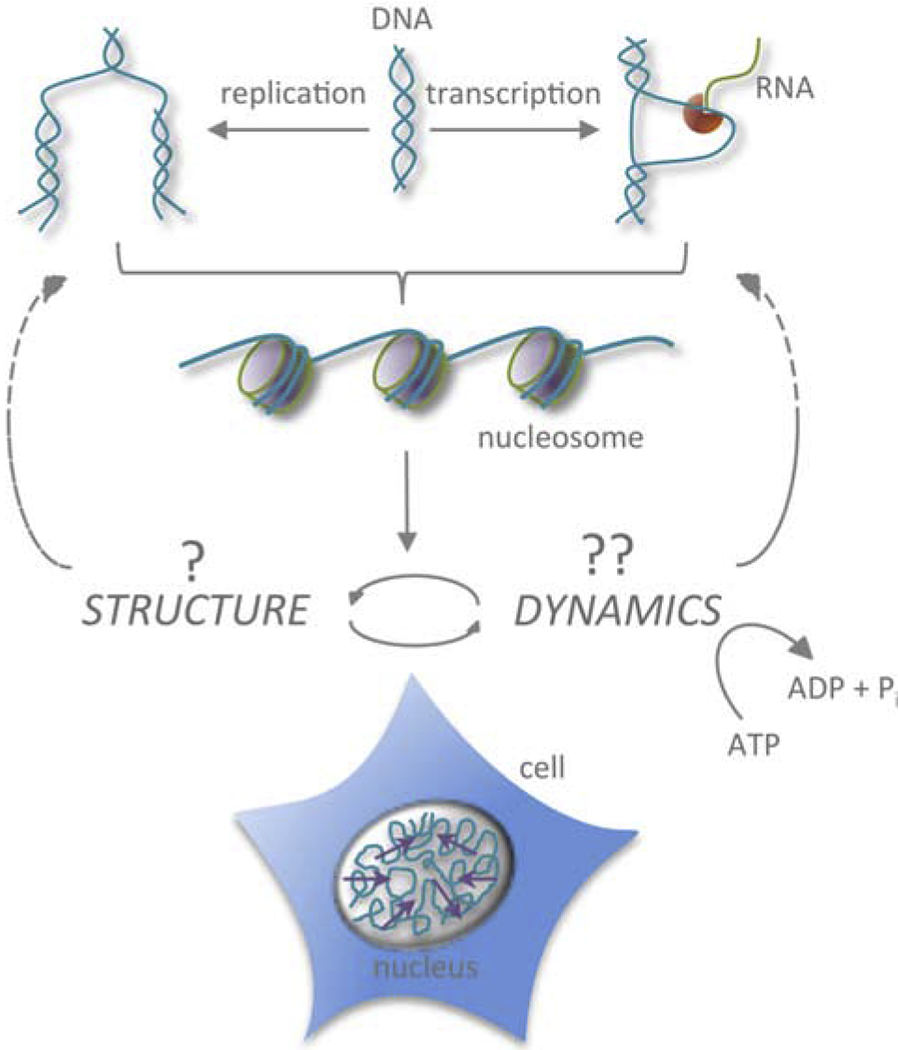
Chromatin structure versus dynamics. Cartoon illustrating a close interplay between local changes in chromatin structure due to dynamical and ATP-dependent DNA transactions, such as transcription and replication, and chromatin dynamics at different length scales and timescales.
Chromatin dynamics: from nucleosomes to genome-wide motions
The human genome undergoes large-scale reorganizations during interphase, the time between two cell divisions: First, highly compact mitotic chromosomes decondense into loosely packed interphase chromosomes, each of which presents a long linear polymer and occupies a well defined region, chromosome territory, in the cell nucleus [1, 11]. Next, the genome is duplicated, followed by chromosome condensation, preparing genome for segregation during mitosis [1]. Notably, nuclear size monotonously increases during interphase [12]. In addition, numerous local chromosome transactions occur at specific DNA sites, such as transcription, DNA replication and DNA repair, contributing to local chromatin dynamics during the cell cycle [Fig. 1].
Historically, chromatin dynamics has been studied by tracking motions of fluorescently labeled nuclear proteins highlighting chromatin structures of interest; from nucleosomes [13–15], single genes [16–25], nuclear proteins, enzymes and machineries [26–33], subchromosomal foci [34, 35] to entire chromosome territories [36, 37]. In turn, measuring fluorescence recovery after photobleaching [38–41] and photoactivation [42, 43] of nuclear proteins, has illuminated their kinetics. All of these approaches provide an invaluable insight into the timescales and length scales of dynamic processes in the cell nucleus. These studies found chromatin dynamics largely subdiffusive to diffusive, with an occasional directed motion. The dynamical information obtained by single particle tracking reports on local chromatin dynamics of a tracked entity. How these local motions amount or contribute to the global genome-wide dynamics remains an open question.
Recent progress shedding light on the global genome-wide chromatin dynamics in vivo came from a new spectroscopy-based method Displacement Correlation Spectroscopy (DCS) [44]. DCS is a microscopy-based image correlation method, which introduced spatiotemporal spectroscopy analysis into the dynamic image correlation processing. It maps chromatin dynamics over time intervals, while concurrently sampling all time intervals accessible by the experiment. Using transgenic histones H2B-GFP as markers of chromatin position and high-resolution spinning disc confocal microscopy, this noninvasive technique enables measurement of chromatin dynamics in real time across the entire nucleus in live cells [Fig. 2A], while simultaneously probing different timescales and length scales [44]. Using DCS chromatin dynamics was found to be subdiffusive [44]. Strikingly, DCS revealed two distinct timescales and length scales in nucleus-wide chromatin dynamics: (i) local, fast movement consistent with single particle tracking studies [Fig. 2B], and (ii) a slower, coherent movement – which is new – where chromatin in large regions (~ 3-5 μm) moved in same direction for several seconds [Fig. 2C & D] [44]. Both types of motion occur in the nucleus concurrently and superposed. The discovery of coherent chromatin motion was later corroborated by high-resolution imaging of local motion of single nucleosomes and replication domains [45, 46]. Recently, DCS-like correlative imaging and spectroscopy analysis has reproduced nucleus-wide measurements of the coherent motion in U2OS cells over the same time intervals and length scales as DCS revealed in HeLa cells [44, 47]. This speaks to the reliability and robustness of the correlative spectroscopy analysis introduced in DCS [44] as well as generality of the phenomenon of coherent chromatin motion in the interphase nucleus.
Figure 2:
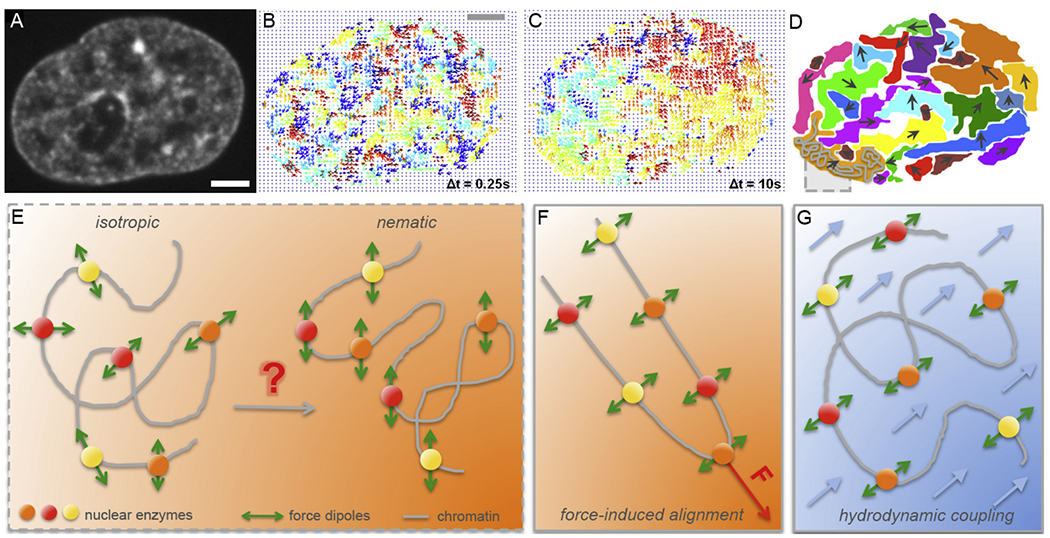
Micron-scale coherence of interphase chromatin. (A-D) Mapping of interphase chromatin dynamics using Displacement Correlation Spectroscopy (DCS) [44]: (A) A fluorescent micrograph of chromatin visualized by histones H2B-GFP in a live HeLa cell nucleus. (B) DCS map of chromatin displacements displayed as vectors (arrows) for Δt = 0.25 s. The displacement vectors are color-coded by their direction, i.e., the vectors of the same color correspond to a motion in the same direction. Note, chromatin motion at timescale of Δt = 0.25 s is uncorrelated. (C) DCS map of chromatin displacements for Δt = 10 s. Chromatin motion shows regions, where chromatin displacement vectors are correlated, i.e., chromatin motion is coherent in those regions at timescale of Δt = 10 s. (D) Cartoon highlighting the observed regions of coherency in (C). A grey box highlighting a chromatin fiber (grey) inside a coherent region (orange). (E) Schematics illustrating nuclear enzymes acting on chromatin fiber as force dipoles. In an isotropic state force dipoles are pointing into random directions, whereas in a nematic state force dipoles are ordered in parallel and can lead to coherent chromatin motion over large length scales. (F-G) Two hypotheses of inducing a local nematic order of force dipoles: (F) force-induced alignment by a local action of a transient force, and (G) hydrodynamic coupling, i.e. fluid-mediated interactions between force dipoles (fluid in blue, fluid flows indicated by blue arrows). (A-C) Adapted from [44]. Scale bar, 2 μm.
Micron-scale coherence of chromatin motion
The fact that interphase chromatin moves coherently over microns and seconds, has major implications for organization of nuclei on micron-second scales and collective gene behavior [Fig. 2A–D]. Strikingly, regions of coherent motion were found to extend beyond the boundaries of single chromosome territories suggesting elastic coupling of motion over length scales much larger than genes [44]. It is unclear if the coherence has a biological role or is an epiphenomenon occurring as a result of other biological processes. However, even as an epiphenomenon, coherence allows groups of genes to travel together for several seconds. Such collective gene dynamics might have important implications for the local and global gene expression patterns in both space and time, impacting the spatiotemporal evolution of gene regulation.
To elucidate the biological role of coherent motion, it is critical to understand the biophysical mechanism behind it. Active (ATP-dependent) processes were shown to drive coherent motion without the involvement of cytoplasmic cytoskeletal forces, thus pointing to the activity of nuclear AT-Pases [44, 48]. The list of possible candidates is long. Interestingly, upon inhibiting transcription, replication or topoisomerase activity, coherent motion becomes eliminated [44, 47]. Surprisingly, the local chromatin displacements become uncoupled, yet their amplitude significantly increases [44]. Inducing direct DNA damage in the form of DNA double stranded breaks, and thus initiating the DNA damage response, led to the same effect [44]. This suggests that DNA damage response activated upon perturbing of DNA-related processes (e.g. transcription, replication) is responsible for the elimination of the chromatin coherence by relaxing the chromatin and preventing the long-distance communication of forces. In other words, a loss of function of a nuclear ATPase leads to a gain in function by activating the DNA damage response [44]. This connection requires that we understand the chromatin dynamics associated with the DNA damage response in order to elucidate the dynamical contribution of nuclear enzymes to the emergence of coherent motion. Moreover, it implies that coherent chromatin motion is a signature of a healthy cell.
In parallel to the experimental efforts, new theories have been developed to illuminate the physical origin of coherent chromatin motion. Such approaches conceptually and mechanistically account for the non-equilibrium sources of activity without knowing their molecular identity. Motivated by the DCS measurements, a first chromatin hydrodynamics theory was developed [44, 49]. In this theory, hydrodynamic (fluid-mediated) interactions between the chromatin and nucleoplasm were considered, while simultaneously accounting for two types of sources of non-equilibrium activity: scalar and vector events [49]. Scalar events represent the local condensation and decondensation of chromatin that is mostly caused by the chromatin remodelers [49, 50]. In contrast, vector events are generated by nuclear enzymes such as polymerase II, helicase and topoisomerase II, which can be represented by a force dipole, thus possessing both a magnitude and direction [49]. A force dipole consists of two equally large, but opposite forces, corresponding to a force that an enzyme applies on the chromatin fiber and an opposite force applied on the surrounding fluid by virtue of Newton’s third law [Fig. 2E]. Hence force dipoles induce flows that in turn transmit forces on the chromatin fiber. Such flow-mediated interaction is referred to as hydrodynamic coupling. This theory predicts that the chromatin concentration fluctuations (condensation/decondensation) dominate chromatin dynamics at short length scales, while the force-dipole activity dominates the long length-scales [49].
Notably, the physical size of active nuclear enzymes such as RNA polymerase II, helicase, or topoisomerase is ~ 5 nm and the coherent regions span 3–5 μm, thus a single force dipole is unlikely to drive the coherent motion. Instead, this suggests that the nucleus-wide coherent motion might be caused by a collective effort of ATP-dependent force-generating nuclear enzymes, as supported by experiments [44]. It could be hypothesized that such an effect can be achieved by a nematic (parallel) alignment of force dipoles [Fig. 2E] in the following ways: (i) a local transient force applied on the chromatin fiber might align the force dipoles along the chromatin fiber [Fig. 2F], or (ii) through hydrodynamic coupling, i.e., fluid-mediated interactions, between force dipoles [Fig. 2G]. In fact, both effects might occur in the nucleus at the same time, with force-induced alignment nucleating a region of coherent motion and hydrodynamic coupling growing the size of the coherent region.
Numerical simulations of hydrodynamic self-interactions of the chromatin fiber with dipolar activity identified the presence of extensile force dipoles (outward forces) as required for generation of the coherent chromatin motion [48]. The extensile force dipoles were also shown to generate organized nucleoplasmic flows and lead to a local nematic alignment of the chromatin fiber. In contrast, contractile force dipoles (inward forces) lead to an accelerated Brownian-like chromatin dynamics and no fiber alignment [48].
An alternative approach omits the presence of nucleoplasm and fluid-mediated interactions, instead using a 3D-conformational space of the chromatin fiber emerging from a quasi-equilibrium energy landscape [51]. To generate such an energy landscape, Langevin dynamics at an effective temperature was assumed, which implicitly captures the chromatin activity and its non-equilibrium nature. This model was also able to capture the large-scale coherency of chromatin motion [51]. Another hydrodynamics-free model explores the spatiotemporal dynamics of interphase chromosomes describing chromatin as a heteropolymer directly informed by the HiC data of human chromosomes [52]. To mimic chromatin activity, an isotropic noise was used and the large-scale spatial correlation of local chromatin displacements was again successfully recapitulated [52]. Moreover, chromatin dynamics was found to be glassy with a wide range of subdiffusive behavior [53].
In all of these models, activity of nuclear enzymes acting on the chromatin fiber is required to generate coherent chromatin motion, as corroborated by experiments. However, while one group of models finds the chromatin fluid-mediated interactions to be critical for generating the spatial coherence in chromatin dynamics, the other group of models suggests that specific conformations of the fiber, as revealed by HiC or determined by proposed energy landscapes, might be of importance. The former suggests that nucleoplasm plays a major role in large-scale chromatin dynamics, whereas the latter might indirectly imply nucleoplasm in facilitating formation of the preferred chromatin fiber conformations.
Role of nucleoplasm in chromatin dynamics
To date, the observed active motion of interphase chromatin has been attributed purely to the enzymatic activity occurring directly at the chromatin fiber. The presence of nucleoplasm was either neglected or considered to be a passive solvent, in which the chromatin is immersed. However, the nucleoplasm is a complex fluid, and possibly active itself [Fig. 3]. It contains not only RNA and proteins, but also a plethora of subnuclear bodies such as nucleoli, speckles and Cajal bodies [Fig. 3A] [1]. Thus, a next-order description of the nucleoplasm would be a colloidal suspension containing polydisperse colloidal particles (50nm – 3μm) [Fig. 3B]. These particles, e.g. nucleoli or nucleoplasmic enzymes, might not be only advected by the flows generated by the active chromatin fiber, but also directly contribute to chromatin dynamics by interacting hydrodynamically and/or sterically with the chromatin fiber [Fig. 3C–D]. Moreover, recent observations of nucleolar coalescence in human cells in vivo suggest that such a dynamical event in the nucleoplasm might lead to a large-scale chromatin reorganization and dynamics in the nucleus [Fig. 3F] [54, 55]. Intriguingly, velocities measured for the growth of radius of nucleolar neck between two coalescing nucleoli were similar to velocities measured for interphase chromatin [Fig. 3F] [44, 54]. Furthermore, many subnuclear bodies are known to be sites of active processes themselves, e.g., ribosomal synthesis in nucleoli, RNA splicing in speckles [1]. Such internal activity might also in some cases contribute to nucleoplasmic and chromatin motions.
Figure 3:
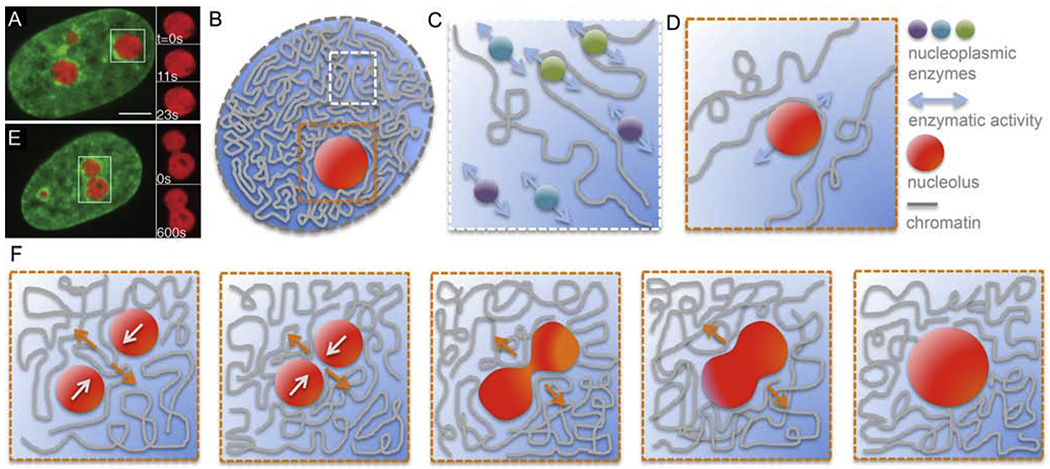
Role of nucleoplasm in chromatin dynamics. (A) Micrograph of a HeLa cell nucleus with fluorescently labeled chromatin (green, H2B-GFP) and nucleoli (red, NPM-mApple). Inset shows an enlarged boxed nucleolus at times t = 0, 11 and 23 s [54]. (B) Cartoon of a cell nucleus containing chromatin (grey), subnucleolar bodies such as nucleolus (red), and nucleoplasm (blue). White box highlights a region containing chromatin and nucleoplasm, orange box shows a region around a nucleolus. (C) Enlarged view of the white boxed region from (B) showing a schematics of free nucleoplasmic enzymes interacting hydrodynamically with each other leading to a nematic (parallel) alignment of their force dipoles. This could lead to a generation of local nucleoplasmic flows contributing to local chromatin dynamics. (D) Enlarged view of the orange boxed region from (B) showing a schematics of a moving nucleolus sterically interacting with chromatin fiber and thus leading to local rearrangement and motion of chromatin. This might occur also for nucleoplasmic enzymes or other subnuclear bodies. (E) Micrograph of a HeLa cell nucleus with fluorescently labeled chromatin (green, H2B-GFP) and two coalescing nucleoli (red, NPM-mApple). Inset shows an enlarged view of the boxed nucleolar coalescenc at times t = 0 and 11 s [54]. (F) Schematics of nucleolar coalescence leading to large-scale chromatin reorganization and dynamics inside the nucleus. Orange and white arrows indicate the local motion of chromatin and nucleoli, respectively. (A) and (E) adapted from [54]. Scale bar, 5 μm.
Recently, liquid-liquid phase separation (LLPS) of nucleoplasmic proteins has been implicated in genome organization and function. For example, nucleoli, the largest nuclear organelles and sites of ribosomal biogenesis, were shown to form via LLPS and behave like liquid droplets [54–57]. Similarly, transcription machinery at actively expressed genes and DNA-repair machinery at double-stranded DNA breaks were found to form liquid-like condensates [58–61]. Moreover, LLPS of heterochromatin HP1 proteins was uncovered to drive the formation of heterochromatin in genome [62, 63]. Indeed, HP1 association with parts of chromatin fiber was found to lead to a microphase separation of heterochromatin and euchromatin [64, 65]. In addition, chromatin fiber itself was suggested to have an intrinsic capacity to undergo local LLPS in the nucleoplasm [66]. It is conceivable that kinetics of such processes could directly impact genome organization as well as its local and large-scale dynamics.
Remarkably, both nucleoplasm and the chromatin fiber are comprised of active (i.e., ATP-driven) and passive (i.e., thermally driven) components. Interestingly, previous studies of colloidal mixtures found that a presence of actively driven particles leads to phase separation of active and passive colloids [67]. Similar behavior has been shown for polymer mixtures containing active and passive polymers, and was in fact proposed to facilitate positioning of chromosomes and formation of transcriptionally active and inactive genomic regions, euchromatin and heterochromatin, respectively [53, 68, 69]. Thus, activity-driven phase separations, where active entities phase separate from passive entities, might also need to be considered in our efforts to elucidate the nucleus-wide motion of genome. Indeed, in non-equilibrium systems such as chromatin and nucleoplasm, combined effects of microphase, activity-driven and liquid-liquid phase separations could occur, leading to new physical phenomena.
From genome biophysics to gene regulation
The physical characteristics of chromatin and nucleoplasm are inseparable from their biological functions. Their material properties are critical for all cellular processes as they directly impact the central dogma of biology [70, 71]. For example, the viscoelastic nature of the chromatin and the rheological behavior of the nucleoplasm affect the timescales and length scales of local and nucleus-wide chromatin rearrangements, as well as molecular and organelle transport inside the nucleus. Thus, an insight into the physical properties of the cell nucleus and its constituents might prove integral in understanding the genome self-organization and dynamics. This is not trivial considering its non-equilibrium nature. Recent microrheology studies employing artificial as well as naturally occurring cellular probes (e.g., nucleolus) found that nucleoplasm viscosity ranges from 25-3000 Pa·s [54, 72–74]. Such viscosity was proposed to be apparent, i.e. effective, containing viscous effects from chromatin-related and nucleoplasmic enzymatic activity in addition to the viscosity of a passive fluid [54, 55]. In turn, we might speculate that by altering such activity, effective material properties of nuclear constituents could be manipulated locally and globally across the nucleus.
A spatiotemporal modulation of genomic material properties could indeed influence nucleus-wide behavior of many processes such as gene expression or DNA replication. A recent study revealed that kinetics of biomolecular crowding in the cell nucleus directly influences the kinetics and efficiency of the transcriptional machinery [75]. Moreover, altering the supra-nucleosomal physical structure of chromatin was proposed to allow for the controlled modulation of global patterns in gene expression [76, 77]. Similarly, collective gene dynamics corresponding to the micron-scale coherent motion of chromatin might lead to concerted gene expression, implying that local conformation changes at short timescales could lead to temporal fluctuations in global gene-expression patterns. Hence, a possible biological role of the coherent chromatin motion may involve gene regulation, contributing to transcriptional heterogeneity through local chromatin rearrangements.
Conclusions and perspective
The genome is a prominent example illustrating the critical interplay between biology, physics and chemistry in a living system. Physical properties of the genome and its nuclear environment are inseparable from chromatin physiology. Hence, insights from biophysics, polymer physics and statistical mechanics are critical to elucidate genome self-organization and non-equilibrium dynamics.
Specifically, elucidating the biophysical mechanism underlying the micron-scale coherent motion of chromatin might provide insight into spatiotemporal organization of the genome at large scales. Moreover, the coherency facilitates collective gene motion, causing groups of genes to travel together for several seconds. This phenomenon might have numerous implications, especially for gene regulation. For example, the constant local stirring of the genome through coherent motion might lead to local changes in the chromatin fiber conformation, accessibility, as well as intragenomic interactions, all of which might play a key role in gene regulation. Furthermore, chromatin motion is closely linked to the motion of nucleoplasm. Thus, nucleoplasmic flows accompanying coherent chromatin motion could contribute to gene regulation by facilitating the distribution of transcription machinery in the cell nucleus. Such local advective flows might accelerate the otherwise diffusion-limited transport. In addition, considering the complex and active nature of the nucleoplasm, we might need to rethink its role in both genome organization and dynamics.
Future studies are needed to connect the detailed picture of static genome folding with its statistics of folding conformations, large-scale dynamics and gene-expression patterns. The biomedical implications of such approaches are far reaching, as they might lead to therapies able to treat complex multi-gene diseases such as cancer.
Acknowledgements
AZ would like to thank members of the Zidovska lab for fruitful discussions. This work was supported by the National Institutes of Health Grant R00-GM104152, the National Science Foundation Grants CAREER PHY-1554880, CMMI-1762506 and New York University MRSEC DMR-1420073, and NYU Whitehead Fellowship for Junior Faculty in Biomedical and Biological Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- [1].Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, Walter P, Molecular Biology of the Cell., Garland Science, New York, 2014. [Google Scholar]
- [2].Van Holde KE, Chromatin, Springer Science & Business Media, 2012. [Google Scholar]
- [3].Collins FS, Morgan M, Patrinos A, The human genome project: lessons from large-scale biology, Science 300 (2003) 286–290. [DOI] [PubMed] [Google Scholar]
- [4].Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. , Comprehensive mapping of long-range interactions reveals folding principles of the human genome, Science 326 (2009) 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bonev B, Cavalli G, Organization and function of the 3D genome, Nat. Rev. Genet 17 (2016) 661–678. [DOI] [PubMed] [Google Scholar]
- [6].Dekker J, Marti-Renom MA, Mirny LA, Exploring the three-dimensional organization of genomes: Interpreting chromatin interaction data, Nat. Rev. Genet 14 (2013) 390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gibcus JH, Dekker J, The hierarchy of the 3D genome, Mol. Cell 49 (2013) 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hübner MR, Spector DL, Chromatin dynamics, Annu. Rev. Biophys 39 (2010) 471–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bickmore WA, van Steensel B, Genome architecture: Domain organiztion of interphase chromosomes, Cell 152 (2013) 1270–1284. [DOI] [PubMed] [Google Scholar]
- [10].Sazer S, Schiessel H, The biology and polymer physics underlying large-scale chromosome organization, Traffic 19 (2018) 87–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cremer T, Cremer M, Chromosome territories, Cold Spring Harbor Perspectives in Biology 2 (2010) a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chu F-Y, Haley SC, Zidovska A, On the origin of shape fluctuations of the cell nucleus, Proc. Natl. Acad. Sci. U.S.A 114 (2017) 10338–10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Xu J, Ma H, Jin J, Uttam S, Fu R, Huang Y, Liu Y, Super-resolution imaging of higher-order chromatin structures at different epigenomic states in single mammalian cells, Cell Rep. 24 (2018) 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nagashima R, Hibino K, Ashwin S, Babokhov M, Fujishiro S, Imai R, Nozaki T, Tamura S, Tani T, Kimura H, et al. , Single nucleosome imaging reveals loose genome chromatin networks via active RNA polymerase II, J. Cell Biol 218 (2019) 1511–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]; • In this study, authors use single-nucleosome imaging to investigate chromatin dynamics, revealing the constraints posed by active RNA polymerase II on the local chromatin movements.
- [15].Ashwin S, Nozaki T, Maeshima K, Sasai M, Organization of fast and slow chromatin revealed by single-nucleosome dynamics, Proc. Natl. Acad. Sci. U.S.A (2019) 201907342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Marshall W, Straight A, Marko J, Swedlow J, Dernburg A, Belmont A, Murray A, Agard D, Sedat J, Interphase chromosomes undergo constrained diffusional motion in living cells, Curr. Biol 7 (1997) 930–939. [DOI] [PubMed] [Google Scholar]
- [17].Belmont AS, Straight AF, In vivo visualization of chromosomes using lac operator-repressor binding., Trends Cell Biol. 8 (1998) 121–4. [DOI] [PubMed] [Google Scholar]
- [18].Levi V, Ruan Q, Plutz M, Belmont AS, Gratton E, Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope, Biophys. J 89 (2005) 4275–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chuang C-H, Carpenter AE, Fuchsova D, Hohnson T, de Lanerolle P, Belmont AS, Long-range directional movement of an interphase chromosome site., Curr. Biol 16 (2006) 825–831. [DOI] [PubMed] [Google Scholar]
- [20].Weber SC, Spakowitz AJ, Theriot JA, Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci, Proc. Natl. Acad. Sci. U.S.A 109 (2012) 7338–7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B, Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system, Cell 155 (2013) 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lampo TJ, Kennard AS, Spakowitz AJ, Physical modeling of dynamic coupling between chromosomal loci, Biophys. J 110 (2016) 338–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Germier T, Kocanova S, Walther N, Bancaud A, Shaban HA, Sellou H, Politi AZ, Ellenberg J, Gallardo F, Bystricky K, Real-time imaging of a single gene reveals transcription-initiated local confinement, Biophys. J 113 (2017) 1383–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Amitai A, Holcman D, Encounter times of chromatin loci influenced by polymer decondensation, Phys. Rev. E 97 (2018) 032417. [DOI] [PubMed] [Google Scholar]
- [25].Khanna N, Zhang Y, Dudko JSLOK, Murre C, Chromosome dynamics near the sol-gel phase transition dictate the timing of remote genomic interactions., Nat. Commun 10 (2019) 2771. [DOI] [PMC free article] [PubMed] [Google Scholar]; • In this paper, authors show how the local conformation and organization of the chromatin fiber influences the timing of local genomic interactions.
- [26].Misteli T, Protein dynamics: implications for nuclear architecture and gene expression, Science 291 (2001) 843–847. [DOI] [PubMed] [Google Scholar]
- [27].Carmo-Fonseca M, Platani M, Swedlow JR, Macromolecular mobility inside the cell nucleus, Trends Cell Biol. 12 (2002) 491–495. [DOI] [PubMed] [Google Scholar]
- [28].Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH, In vivo dynamics of RNA polymerase II transcription, Nat. Struct. Mol. Biol 14 (2007) 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bronstein I, Israel Y, Kepten E, Mai S, Shav-Tal Y, Barkai E, Garini Y, Transient anomalous diffusion of telomeres in the nucleus of mammalian cells, Phys. Rev. Lett 103 (2009). [DOI] [PubMed] [Google Scholar]
- [30].Stixová L, Bártová E, Matula P, Daněk O, Legartová S, Kozubek S, Heterogeneity in the kinetics of nuclear proteins and trajectories of substructures associated with heterochromatin, Epigenetics Chromatin 4 (2011) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, Darzacq X, Real-time dynamics of RNA polymerase II clustering in live human cells, Science 341 (2013) 664–667. [DOI] [PubMed] [Google Scholar]
- [32].Hinde E, Kong X, Yokomori K, Gratton E, Chromatin dynamics during DNA repair revealed by pair correlation analysis of molecular flow in the nucleus, Biophys. J 107 (2014) 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Eaton JA, Zidovska A, Structural and dynamical signatures of local DNA damage in live cells, Biophysical Journal (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bornfleth H, Edelmann P, Zink D, Cremer T, Cremer C, Quantitative motion analysis of subchromosomal foci in living cells using four-dimensional microscopy., Biophys. J 77 (1999) 2871–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Küpper K, Joffe B, Thormeyer T, von Hase J, et al. , Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks, Chrom. Res 14 (2006) 707–733. [DOI] [PubMed] [Google Scholar]
- [36].Zink D, Cremer T, Saffrich R, Fischer R, Trendelenburg MF, Ansorge W, Stelzer EH, Structure and dynamics of human interphase chromosome territories in vivo, Hum. Genet 102 (1998) 241–251. [DOI] [PubMed] [Google Scholar]
- [37].Edelmann P, Bornfleth H, Zink D, Cremer T, Cremer C, Morphology and dynamics chromosome territories in living cells., Biochim. Biophys. Acta 1551 (2001) M29–39. [DOI] [PubMed] [Google Scholar]
- [38].Abney JR, Cutler B, Fillbach ML, Axelrod D, Scalettar BA, Chromatin dynamics in interphase nuclei and its implications for nuclear structure, J. Cell Biol 137 (1997) 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Misteli T, Gunjan A, Hock R, Bustin M, Brown DT, Dynamic binding of histone H1 to chromatin in living cells, Nature 408 (2000) 877. [DOI] [PubMed] [Google Scholar]
- [40].Phair RD, Misteli T, High mobility of proteins in the mammalian cell nucleus, Nature 404 (2000) 604. [DOI] [PubMed] [Google Scholar]
- [41].Kimura H, Cook PR, Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B, J. Cell Biol 153 (2001) 1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mora-Bermúdez F, Gerlich D, Ellenberg J, Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase, Nature Cell Biol. 9 (2007) 822. [DOI] [PubMed] [Google Scholar]
- [43].Wiesmeijer K, Krouwels IM, Tanke HJ, Dirks RW, Chromatin movement visualized with photoactivable GFP-labeled histone H4, Differentiation 76 (2008) 83–90. [DOI] [PubMed] [Google Scholar]
- [44].Zidovska A, Weitz DA, Mitchison TJ, Micron-scale coherence in interphase chromatin dynamics, Proc. Natl. Acad. Sci. U.S.A 110 (2013) 15555–15560. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• In this study, authors present the discovery of micron-scale coherent chromatin motion in live cells using a new method Displacement Correlation Spectroscopy.
- [45].Nozaki T, Imai R, Tanbo M, Nagashima R, Tamura S, Tani T, Joti Y, Tomita M, Hibino K, Kanemaki MT, et al. , Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging, Mol. Cell 67 (2017) 282–293. [DOI] [PubMed] [Google Scholar]
- [46].Xiang W, Roberti MJ, Hériché J-K, Huet S, Alexander S, Ellenberg J, Correlative live and super-resolution imaging reveals the dynamic structure of replication domains, J. Cell Biol 217 (2018) 1973–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shaban HA, Barth R, Bystricky K, Formation of correlated chromatin domains at nanoscale dynamic resolution during transcription, Nuc. Ac. Res 46 (2018) e77–e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Saintillan D, Shelley MJ, Zidovska A, Extensile motor activity drives coherent motions in a model of interphase chromatin, Proc. Natl. Acad. Sci. U.S.A 115 (2018) 11442–11447. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The authors investigate hydrodynamic self-interactions of the chromatin fiber and implicate extensile motor activity (outward forces) in driving the chromatin coherent motion.
- [49].Bruinsma R, Grosberg AY, Zidovska A, Chromatin hydrodynamics, Biophys. J 106 (2014) 1871–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Racki LR, Narlikar GJ, Atp-dependent chromatin remodeling enzymes: two heads are not better, just different, Curr. Op. Gen. Dev 18 (2008) 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Di Pierro M, Potoyan DA, Wolynes PG, Onuchic JN, Anomalous diffusion, spatial coherence, and viscoelasticity from the energy landscape of human chromosomes, Proc. Natl. Acad. Sci. U.S.A 115 (2018) 7753–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The authors study chromatin dynamics by exploring a 3D-conformational space of the chromatin fiber emerging from a quasi-equilibrium energy landscape and reproduce the coherent chromatin motion.
- [52].Liu L, Thirumalai GSD, Hyeon C, Chain organization of human interphase chromosome determines the spatiotemporal dynamics of chromatin loci, PLoS Comput. Biol 14 (2018) e1006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shi G, Liu L, Hyeon C, Thirumalai D, Interphase human chromosome exhibits out of equilibrium glassy dynamics, Nat. Commun 9 (2018) 3161. [DOI] [PMC free article] [PubMed] [Google Scholar]; • In this paper, authors present a model accounting for both euchromatin and heterochromatin, showing that chromatin exhibits glassy dynamics with coherent motion on micron scale.
- [54].Caragine CM, Haley SC, Zidovska A, Surface fluctuations and coalescence of nucleolar droplets in the human cell nucleus, Phys. Rev. Lett 121 (2018) 148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Caragine CM, Haley SC, Zidovska A, Nucleolar dynamics and interactions with nucleoplasm in living cells, eLife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study reveals the non-equilibrium nature of the nucleoplasm through observation of nucleolar shape and dynamics.
- [56].Brangwynne CP, Mitchison TJ, Hyman AA, Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes, Proc. Natl. Acad. Sci. U.S.A 108 (2011) 4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP, Coexisting liquid phases underlie nucleolar subcompartments, Cell 165 (2016) 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, Cisse II, Mediator and RNA polymerase II clusters associate in transcription-dependent condensates, Science 361 (2018) 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall’Agnese A, Hannett NM, Spille J-H, Afeyan LK, Zamudio AV, Shrinivas K, et al. , Pol II phosphorylation regulates a switch between transcriptional and splicing condensates, Nature 572 (2019) 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, Altmeyer M, Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments, EMBO J. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pessina F, Giavazzi F, Yin Y, Gioia U, Vitelli V, Galbiati A, Barozzi S, Garre M, Oldani A, Flaus A, et al. , Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors, Nat. Cell Biol (2019) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH, Phase separation drives heterochromatin domain formation, Nature 547 (2017) 241. [DOI] [PMC free article] [PubMed] [Google Scholar]; • In this study, authors propose that heterochromatin formation is driven by liquid-liquid phase separation of HP1α in the nucleoplasm.
- [63].Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ, Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin, Nature 547 (2017) 236. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The authors propose that liquid-liquid phase separation of HP1α in the nucleoplasm plays a key role in heterochromatin formation.
- [64].MacPherson Q, Beltran B, Spakowitz AJ, Bottom–up modeling of chromatin segregation due to epigenetic modifications, Proc. Natl. Acad. Sci. USA 115 (2018) 12739–12744. [DOI] [PMC free article] [PubMed] [Google Scholar]; • In this study, authors present evidence that heterochromatin epigenetic marks lead to a microphase separation of heterochromatin and euchromatin.
- [65].Falk M, Feodorova Y, Naumova N, Imakaev M, Lajoie BR, Leonhardt H, Joffe B, Dekker J, Fudenberg G, Solovei I, Mirny L, Heterochromatin drives compartmentalization of inverted and conventional nuclei, Nature 570 (2019) 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The authors show heterochromatic and heterochromatin-lamina interactions to be necessary for heterochromatin segregation in inverted and conventional nuclei, respectively.
- [66].Gibson BA, Doolittle LK, Schneider MW, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK, Organization of chromatin by intrinsic and regulated phase separation, Cell 179 (2019) 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Stenhammar J, Wittkowski R, Marenduzzo D, Cates ME, Activity-induced phase separation and self-assembly in mixtures of active and passive particles, Phys. Rev. Lett 114 (2015) 018301. [DOI] [PubMed] [Google Scholar]
- [68].Ganai N, Sengupta S, Menon GI, Chromosome positioning from activity-based segregation, Nuc. Ac. Res 42 (2014) 4145–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Smrek J, Kremer K, Small activity differences drive phase separation in active-passive polymer mixtures, Phys. Rev. Lett 118 (2017) 098002. [DOI] [PubMed] [Google Scholar]
- [70].Crick FH, On protein synthesis 12 (1958) 138. [PubMed] [Google Scholar]
- [71].Crick F, Central dogma of molecular biology, Nature (London) 227 (1970) 561. [DOI] [PubMed] [Google Scholar]
- [72].Tseng Y, Lee JS, Kole TP, Jiang I, Wirtz D, Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking, J. Cell Sci 117 (2004) 2159–2167. [DOI] [PubMed] [Google Scholar]
- [73].de Vries AH, Krenn BE, van Driel R, Subramaniam V, Kanger JS, Direct observation of nanomechanical properties of chromatin in living cells, Nano Lett. 7 (2007) 1424–1427. [DOI] [PubMed] [Google Scholar]
- [74].Celedon A, Hale CM, Wirtz D, Magnetic manipulation of nanorods in the nucleus of living cells, Biophys. J 101 (2011) 1880–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Shim AR, Nap RJ, Huang K, Almassalha LM, Matusda H, Backman V, Szleifer I, Dynamic crowding regulates transcription, Biophys. J (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; • In this study, authors demonstrate that kinetics of biomolecular crowding in the cell nucleus directly influences the efficiency of transcriptional machinery.
- [76].Almassalha LM, Tiwari A, Ruhoff P, Stypula-Cyrus Y, Cherkezyan L, Matsuda H, Cruz MD, Chandler JE, White C, Maneval C, et al. , The global relationship between chromatin physical topology, fractal structure, and gene expression, Sci. Rep 7 (2017) 41061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Almassalha LM, Bauer GM, Wu W, Cherkezyan L, Zhang D, Kendra A, Gladstein S, Chandler JE, VanDerway D, Seagle B-LL, et al. , Macrogenomic engineering via modulation of the scaling of chromatin packing density, Nat. Biomed. Eng 1 (2017) 902. [DOI] [PMC free article] [PubMed] [Google Scholar]


