Abstract
Permanently frozen environments (glaciers, permafrost) are considered as natural reservoirs of huge amounts of microorganisms, mostly dormant, including human pathogens. Due to global warming, which increases the rate of ice-melting, approximately 4 × 1021 of these microorganisms are released annually from their frozen confinement and enter natural ecosystems, in close proximity to human settlements. Some years ago, the hypothesis was put forward that this massive release of potentially-pathogenic microbes—many of which disappeared from the face of the Earth thousands and even millions of years ago—could give rise to epidemics. The recent anthrax outbreaks that occurred in Siberia, and the presence of bacterial and viral pathogens in glaciers worldwide, seem to confirm this hypothesis. In that context, the present review summarizes the currently available scientific evidence that allows us to imagine a near future in which epidemic outbreaks, similar to the abovementioned, could occur as a consequence of the resurrection and release of microbes from glaciers and permafrost.
Supplementary Information
The online version of this article (10.1007/s42398-021-00184-8) contains supplementary material, which is available to authorized users.
Keywords: Microbial pathogens, Epidemics, Pandemics, Epidemiological surveillance, Global warming, Glaciers, Cryosphere, Permafrost, Global change
Introduction
More than one year has passed since the World Health Organization (WHO) announced COVID-19 outbreak a global pandemic whose etiologic agent, SARS-CoV-2, is a coronavirus of animal origin (Hu et al. 2020). This is a rather unprecedented circumstance in the history of humanity, if we consider that the closest historical antecedent (the so-called “Spanish flu”) affected societies whose level of development, economic interdependence and speed of communication were completely different from those that characterize today’s globalized world.
Despite the particular epidemiologic features of COVID-19, it is the third time that, in the last 20 years, an epidemic (or pandemic) originates from the rapid and massive dissemination of an emerging virus, whose circulation was limited for a long time to the natural (animal) reservoirs in which it multiplies. It seems that the virus jumped accidentally to humans which inadvertently entered in close contact with these animals (Hu et al. 2020).
It is a well-known fact that outbreaks caused by emerging pathogens originate most often from (1) the invasion of virgin territories by humans, or (2) close contact between humans and wildlife removed from its natural ecosystems. Nowadays, there is increasing concern about a third form of interaction between humans and potential emerging pathogens: the destruction or disappearance of natural barriers, many of which acted as ecological filters for millennia, isolating pristine and remote ecosystems from humans, animals and plants (Harding et al. 2011; Huss and Farinotti 2012). Such is the case of permanently frozen environments, like continental glaciers or permafrost, whose rapid and massive thawing releases an enormous amount of dormant-but-viable microorganisms entrapped therein from millennia.
In this review, we present and discuss some recent data that seems to support the hypothesis that, in a close future, this massive release of ice-entrapped modern and ancient microbes—including their genes and genomes—could lead to local epidemics and, perhaps, pandemics.
The cryosphere
The cryosphere is the portion of the Earth where water is permanently frozen, mainly in the form of glaciers and ice sheets (Vaughan et al. 2013). Thus, the term cryosphere is intimately related to the hydrosphere, but typically refers to poly-extreme environments characterized by extremely low temperatures, high UV radiation index, oligotrophy, and low water activity. This not only encompasses sea ice, lake ice, river ice, ice caps, ice sheets, frozen ground, glaciers and permafrost, but also seasonal snow.
According to the most recent report of the Intergovernmental Panel on Climate Change (IPCC 2019), the cryosphere is experiencing widespread, dramatic and pervasive changes, which can be observed and monitored from high mountains to the polar regions, to coasts and to the deep oceans. More recently, Slater et al. (2021) combined satellite observations and numerical models and quantified the global ice loss in about 28 trillion tons, from 1994 to 2017. In the last few years, several major events that occurred all around the world seem to confirm this trend (Table S1). The effects of such a gigantic ice loss on humans will be profound, and the societies will be forced to adapt to new and unexpected threats. As we will see, the consequences of the rapid melting of the cryosphere can go well beyond hydrological and climate changes.
Glacier thawing
Glaciers cover about one tenth of the land surface worldwide, accumulating approximately 170,000 km3 of ice (Huss and Farinotti 2012). Owing to their extreme conditions they were initially thought to be devoid of life. Nowadays, it is acknowledged that glaciers contain myriads of microorganisms, many of which are metabolically active and multiplying, although at very low rates; these microbes are confined to the small veins formed between ice crystals, where they benefit from the presence of liquid water and enough nutrients to sustain their basal metabolism for a long time (Price 2000). Glacier ice also harbor a huge diversity of dormant or anabiotic microorganisms (Anesio et al. 2017). This means that, even though these microorganisms perform basal metabolic functions and do not multiply, they may reactivate when the environment becomes more friendly. The existence of such a vast number of microorganisms in glaciers explains why many consider them as true repositories of microbial life, both ancient and modern (Miteva 2008).
On the other hand, glaciers are not homogeneous environments. In fact, at least three different ecosystems have been proposed to co-exist in a glacier: a supraglacial ecosystem, an englacial ecosystem and a subglacial ecosystem. All of them differ in several physicochemical parameters, including water content, solar radiation, nutrient abundance, pH and redox potential (Hodson et al. 2008; Anesio et al. 2017). Due to the technical and logistic difficulties to reach the subsurface layers of a glacier, the englacial and subglacial ecosystems have been much less studied than the supraglacial one. This is regrettable, because the great majority of ancient microorganisms are immured in the subsurface, from where they can be released when water flows downwards through moulins and crevasses, and reaches the bedrock (Wilhelm et al. 2013; Fegel et al. 2016). These microbe-loaded running waters quit the glacier as proglacial streams, and can travel long distances until they reach aquatic and terrestrial ecosystems, where they may cause a tremendous impact by disturbing their microbial diversity and community composition (Sommaruga 2015).
During the past 40 years, most glaciers have shrunk and retreated continuously (Pelto and Network 2018; IPCC 2019). In fact, based on the analysis of satellite imagery, Wouters et al. (2019) estimated that, in a 14-year period from 2002 to 2016, continental glaciers lost collectively 199 ± 32 Gt of their global mass per year, contributing to an 8 mm rise in the level of the sea. In the Tropical Andes, a massive decay of glacier ice has occurred so fast that some glaciers have already disappeared (they became “extinct”) and many other will vanish in the next few years (Rabatel et al. 2017). In this region of South America, glaciers lost 1.0 ± 05 Gt/year between 2000 and 2018 (Dussaillant et al. 2019). One of the most dramatic examples of the consequences of global warming on the cryosphere can be found in Venezuela, the first country in the world to have lost all of its glaciers (Braun and Bezada 2013; Ramírez et al. 2020) (Fig. 1).
Fig. 1.
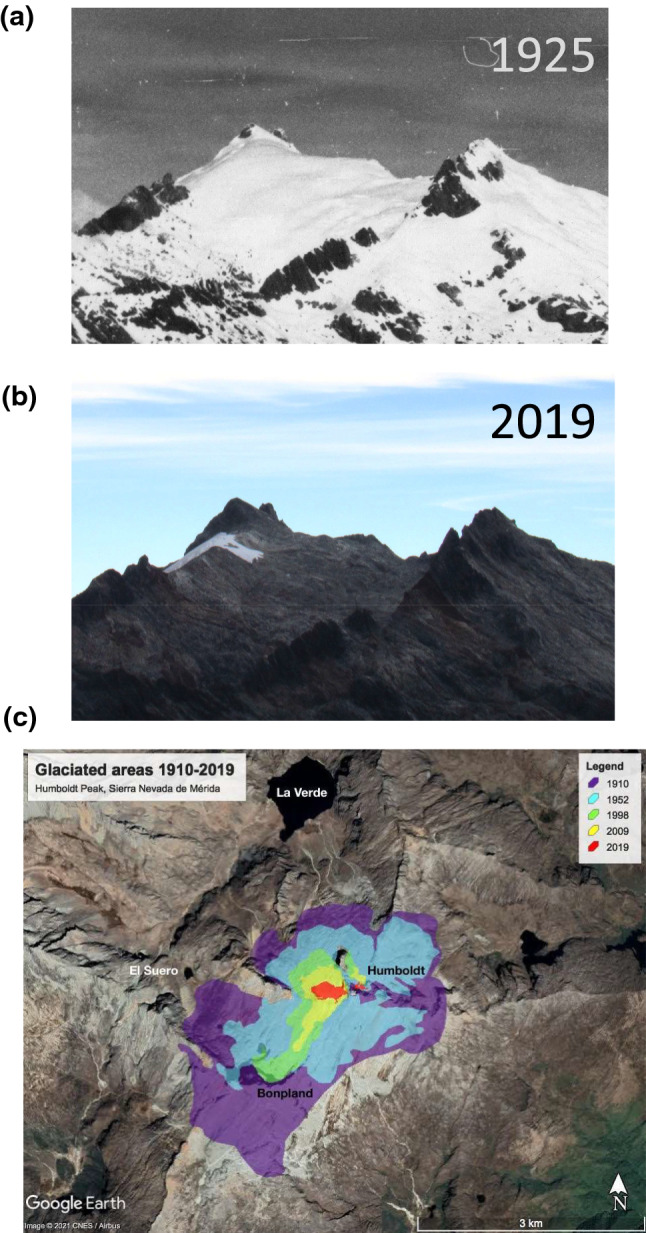
Glacier retreat in Venezuelan Andes. a La Corona glacier, then covering Humboldt and Bonpland peaks, seen from Pico Espejo in 1910 (Jahn 1925); b Same view in 2019 (
Copyright Luis Daniel Llambí, with permission). c Surface area for La Corona glacier (covering Humboltd and Bonpland peaks) between 1910 and 2019 (Copyright Alejandra Melfo, adapted from Ramírez et al. 2020)
If we consider that the average density of microorganisms in glacier ice varies between 102 and 107 cells/ml, approximately 4 × 1021 microbes are released each year, due to the melting of non-polar ice (Edwards 2015). From an epidemiological point of view this is disturbing, since many of these microbes—closely related to human pathogens—are considered “ancient” forms of life that became extinct from the surface of the Earth thousands and even millions of years ago (see below).
Permafrost thawing
On May 29th 2020 an oil tank in Siberia collapsed and released 21,000 tons of oil, polluting the Ambarnaya River and affecting an area of 180.000 m2. More recently, in September 2020, a giant hole of more than 50 m deep formed at the Yamal Peninsula and released tons of methane into the atmosphere. Both events were linked by a common phenomenon: the rapid thawing of permafrost.
Permafrost can be defined as a mixture of soil, sediment and gravel bound together by ice, whose temperature remains below 0 °C for at least two consecutive years (Brown et al. 1998). It covers approximately one fourth of the terrestrial surface on Earth (i.e. 27%) and is found at high latitudes in the Northern Hemisphere, particularly in Alaska, Siberia, Greenland, and the northern regions of Canada, Norway and Sweden (Schaefer et al. 2012). Permafrost can also be found in Antarctica and in alpine regions of Asia, South America and Northern Europe. Due to climate change, the temperature of permafrost has increased steadily since 1980, melting the superficial layers and shallowing the depth of frozen ground (Vaughan et al. 2013). It has been estimated that permafrost will melt more rapidly in the coming years, a phenomenon that will seriously modify its stability (Ding et al. 2019).
This thawing will certainly have many negative consequences. For instance, soils in the Arctic permafrost store about twice as much carbon as the atmosphere (Hugelius et al. 2011), and its uncontrolled release will contribute to global warming. Permafrost do also immure another important threat to humanity: tons of microbes, including many pathogens, ranging from 105 to 1010 cells/g (Altschuler et al. 2017).
Cryosphere as reservoir for past and present human pathogens
As we have already mentioned, even though permanently frozen environments represent one of the most poly-extreme environments for life, they immure an impressive amount of microorganisms mostly dormant, but viable (Anesio et al. 2017). And many of them have been identified as potential- or actual human pathogens.
For instance, Cryptococcus yeasts and bacterial coliforms have been detected in ice cores or meltwater runoff in different regions (Goodwin et al. 2012; Turchetti et al. 2015). Also, emerging pathogens like Aureobasidium melanogenum, Naganishisa albida, and Rhodotorula mucilaginosa, have been isolated from Arctic environments in Greenland and Svalbard (Perini et al. 2019). The yeasts exhibited several virulence-associated traits such as hemolytic ability, growth at 37 °C, resistance to antifungal agents, and production of siderophores. Recently, Mogrovejo-Arias et al. (2020) demonstrated the presence of hemolytic bacteria, potentially pathogenic, in environmental samples (including glacial meltwater) collected in the Svalbard archipelago, in the Norwegian Arctic. Further, a Brazilian group have also detected drug-resistant fungi, in Antarctic samples of rocks, soils, and seawater (Alves et al. 2019; de Menezes et al. 2020).
All of these pathogens are closely related to modern microbial strains. However, several studies also report on the presence of potentially pathogenic microorganisms in very old ice samples. While in many cases these ancient microbes have been detected only by microscopy (either optical or electronic) or by molecular methods, many other have been reactivated (“resurrected”) by culturing them in the laboratory under controlled conditions. In the case of bacteria, the ability to sporulate can account for this long-term persistence in a dormant state; this is also the case of cyst-forming protozoa or spore-forming fungi.
The existence of viable microbes in ancient ice is far from new. In 1982, a Russian group led by Sabit S. Abyzov reported the finding of numerous and highly-diverse viable bacteria in ancient ice cores collected from several kilometers deep above Lake Vostok (the largest subglacial lake in Antarctica) (Abyzov et al. 1982). A few years later, Brent Christner and his collaborators demonstrated that glaciers worldwide are indeed colonized by ancient bacteria, many of which can be easily reactivated in the laboratory (Christner et al. 2000, 2002). In 2013, Knowlton and his collaborators used culture-dependent and molecular methods (such as metagenomics and metatranscriptomics) to detect the presence of bacteria and fungi in Greenland’s and Antarctica’s glacial ice cores, ranging from 500 to 157,000 years old (Knowlton et al. 2013).
The presence of human pathogens in very old glacial ice has also been demonstrated. For example, Dancer et al. (1997) reactivated and isolated a wide variety of coliform strains from glacial-ice samples estimated to be 2,000 years old, collected in the Canadian Arctic.
Even though their viability is highly variable, other eukaryotic microorganisms have also been detected in ancient ice and resurrected with relative ease. For instance, in 2005 a group of Russian researchers found viable protozoa (including amoeboids, ciliates and flagellates) in permafrost samples collected from the Eastern sector of the Arctic tundra, with an estimated age ranging between 28,000 and 35,000 years old (Shatilovich et al. 2005). Since then, many more protozoa have been discovered in permanently frozen environments and further reactivated. These studies confirmed that microbes may survive for long periods of time (sometimes, up to one million years) at subzero temperatures, and in the absence of any oxygen and liquid water (Shatilovich et al. 2009, 2015; Stoupin et al. 2012; Shmakova et al. 2016, 2018; Malavin and Shmakova 2020). Very recently, Malavin and co-workers reported on the isolation of 35 amoeboid protists from frozen sediments, a discovery that motivated them to propose the existence of a so-called “frozen zoo” (Malavin et al. 2020). Following a different approach, García-Descalzo et al. (2013) detected the presence of 18S rDNA from protozoan species (Rhizaria) in Pyrenean glaciers.
Some of these amoeboids have been shown to be infected by giant double-stranded DNA viruses, identified as members of previously unknown families (Legendre et al. 2014, 2015) and potentially infectious to humans and animals (Tokarz-Deptula et al. 2019). Incidentally, as strange as it may seem, nematodes are also able to survive for thousand years in anabiosis, and have been resurrected from ancient glacier ice (Shatilovich et al. 2018).
Antibiotic-resistance genes in the cryosphere
Natural ecosystems –including frozen ones- are a vast source of genes, particularly resistance genes, some of which are supposed to be completely unknown (Wright 2010). In the case of glaciers, even the most pristine and remote have been shown to contain an astonishing amount of antibiotic-resistant microorganisms, able to thrive in vitro in the presence of high doses of antibiotics, once reactivated (Ushida et al. 2010; Segawa et al. 2013; Ball et al. 2014; Rondón et al. 2016; Tan et al. 2018; Rafiq et al. 2019; Ramírez et al. 2020).
Antibiotic resistant genes (ARGs) have been frequently detected in ancient bacteria isolated from permanently frozen environments. For instance, Mindlin et al. (2008) cultured bacterial strains from Eastern Siberian permafrost sediments, estimated to be up to 3-million-years-old; many of these strains harbored genes encoding resistance towards several antibiotics (e.g. chloramphenicol, gentamicin, kanamycin, streptomycin, tetracycline, and mercury compounds). Similarly, bacteria recovered and cultured from 3.5-million-years-old permafrost samples collected at Central Yakutia (Russia), were also shown to carry multiple ARGs (Kashuba et al. 2017). One of them, a Staphylococcus hominis MMP2 strain, harbored ARGs > 96% identical to genes conferring resistance to beta-lactams, aminoglycosides, phenicols and MLS (macrolide, lincosamide, and streptogramin B).
By following a metagenomics approach, D’Costa et al. (2011) also detected ARGs in permafrost samples from Alaska, aged 30,000 years-old, that encoded resistance to β-lactams, tetracycline and glycopeptide antibiotics. Noticeably, when cloned in Escherichia coli, a few of these genes turned out to be functional.
Particularly relevant (and worrying) is the occurrence of bacteria carrying integrons, in samples collected from distant and remote glaciers. These complex genetic elements are capable of capturing genes and gene cassettes, frequently encoding for antibiotic resistance; more importantly, integrons can be transferred horizontally within a microbial community as part of other mobile genetic elements, like plasmids and transposons (Gillings 2014). Further, integrons are at the origin of multidrug resistance in bacteria. In 2011, Petrova et al. detected the presence of a transposon in a Pseudomonas sp. strain isolated from a 15,000 to 40,000-year-old permafrost sample in Siberia. The transposon, conferring resistance to several antibiotics and chromate, included a class 1 integron, which was considered as the first example of its kind that originated in the “pre-antibiotic” era. More recently, Makowska et al. (2020) detected the occurrence of resistance integrons in the genomes of bacteria isolated and cultured from cryoconite holes and supraglacial ice collected in two Arctic- and two Caucasian glaciers.
Sequence-based metagenomics has been the preferred strategy to detect many of these ARGs or complex genetic elements, and to predict the resistant potential of microbial communities colonizing pristine ecosystems (Schmieder and Edwards 2012). Alternatively, functional metagenomics allowed identifying ARGs whose similarity to nucleotidic sequences in databases might not be obvious, by expressing them in heterologous hosts. This experimental approach, although more laborious, has also helped to apprehend the diversity of environmental genes, genetic elements and mechanisms behind resistance to antibiotics in ecosystems not subjected to anthropogenic disturbances. Both approaches had permitted to reveal that ancient ARGs predate the use of antibiotics for therapeutic purposes (Perry et al. 2016). There is also undisputable evidence that many ARGs, found today in pathogenic microorganisms, have arisen from the environmental resistome (i.e. the sum of all resistance genes present in diverse ecosystems throughout the world) (Wright 2010; Schmieder and Edwards 2012; Van Goethem et al. 2018).
On the other side, several studies have shown that many ancient ARGs retained their function when expressed in either homologous or heterologous hosts (D’Costa et al. 2011; Bhullar et al. 2012; Perron et al. 2015; Perry et al. 2016). However, the link between environmental ARGs and antibiotic-resistant human pathogens is not always clear. Hence, it has been proposed that more efforts should be made to test the hypothesis that some of these ancient ARGs might be transmitted to modern microbes by horizontal gene transfer (HGT); it has also been highlighted the importance to measure the rate of these HGT events, as well as to assess the environmental factors contributing to such genetic exchange (Wright 2010).
A landmark in the study of ancient ARGs was the isolation—in 2008—and subsequent sequencing—in 2014—of plasmids extracted from ancient permafrost bacteria, aged 15,000 to 40,000 years-old (Petrova et al. 2009, 2014). One of these plasmids contained streptomycin and tetracycline resistance determinants, which were transferred to “modern” plasmids, confirming their ability to spread into contemporary natural bacterial populations. More recently, another study confirmed the presence of ARG-containing plasmids in ancient permafrost bacteria (Kurakov et al. 2016).
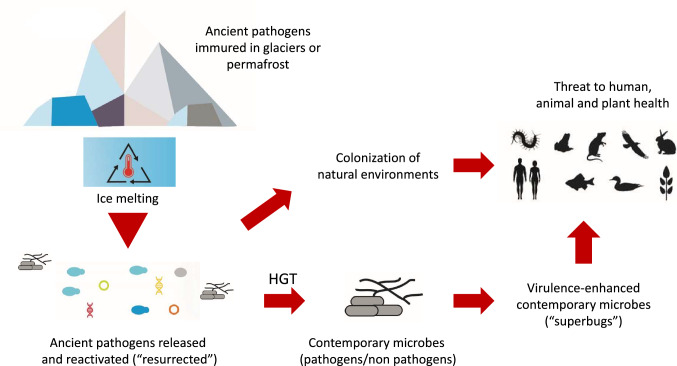
Thus, it is believed that, following ice thawing, some of these ARG-containing genetic elements might be transferred horizontally to modern microbes, giving rise to new strains of multi- and pan-resistant “superbugs” (Fig. 2).
Fig. 2.

Permanently-frozen environments as vast sources of antimicrobial resistance/virulence genes. Once released, these genetic elements can be transferred horizontally to contemporary microbes, likely transforming some of them into “superbugs”
Viruses in permanently frozen environments
Viral communities (= “viromes”) colonizing permanently frozen habitats exhibit astonishing taxonomic- and functional diversities; they also play paramount roles regulating the population dynamics of prokaryotic communities, and consequently, the biogeochemical cycles occurring within frozen environments (Rassner et al. 2016). The amount of virus-like-particles (VLP) in different glacial environments have been estimated to range between 0.9 × 106 ml−1 (Säwstrom et al. 2008; Rassner 2017) and 3.1 × 107 ml−1 (Wilson et al. 2000). Even though some of them can survive intracellularly within their natural hosts in frozen environments (e.g. the giant viruses of amoeboids mentioned earlier), others can be preserved abiotically in ice, as demonstrated by Shoham et al. (2012).
Pioneer studies about ice-immured viruses were published more than 20 years ago, and relied on visual detection of virus-like particles (VLP) by means of electron microscopy techniques (Wilson et al. 2000). For instance, Castello et al. (1999) detected tomato mosaic tobamovirus particles in 500 to approximately 140,000-year-old ice cores. A few years later, Priscu et al. (2006) discovered viral particles in ice cores collected from Antarctica, at more than 2.5 km depth above Lake Vostok, with an estimated age of several million years. Similarly, Castello et al. (2005) detected bacteriophages capable of infecting Bacillus subtilis strains in ice cores collected at great depth in Greenland, with an estimated age of 500–100,000 years. The same year, Zhang et al. (2006) reported on the presence of influenza-A virus particles in ice and water samples from three Siberian lakes. This study further demonstrated the ability of viruses to withstand the extreme conditions imposed by cold and/or permanently frozen environments.
In 2015, the presence of a wide variety of viruses in glacial environments was indisputably confirmed. In their work, Bellas et al. (2015) used massive parallel sequencing techniques to detect the presence of highly novel and diverse viral sequences in cryoconite holes, on the surface of Svalbard glaciers and also in the Greenland platform. A similar study, conducted by Paéz-Espino et al. (2016), allowed the identification of 125,000 partial viral genomes from 3,042 samples collected worldwide, including Antarctica. Further, Rassner et al. (2016) demonstrated that viruses carried by glacier streams are resilient to variations in temperature, and remain active during long periods of time, being able to infect bacteria in aquatic environments located downstream. Even more recently, Zhong et al. (2020) developed a method to concentrate viral particles from glacier ice samples, and subsequently to extract and purify viral DNA. This allowed them to characterize the viromes present in ice samples from the Guliya glacier, on the Tibetan plateau (with an estimated age of 500 and 15,000 years), by using massive parallel sequencing. Their results revealed the presence of 33 viral populations (including 28 new viral genera, unknown to date), half of which represent bacteriophages likely capable of infecting bacteria.
Giant viruses infecting amoeboids have also been detected and resurrected from frozen environments. Since their discovery in 1992, these viruses, able to infect protists and microalgae, have been isolated from diverse environments (La Scola et al. 2003; Aherfi et al. 2016). Noticeably, it is increasingly acknowledged that giant viruses could have an important impact on human health (Brandes and Linial 2019). For example, some works associate them with human diseases like adenitis, atopic pneumonia, and lymphoma, among others (Saadi et al. 2013a, b; Aherfi et al. 2016; Brandes and Linial 2019).
In 2014 and 2015, Legendre and co-workers isolated two giant viruses, namely Pithovirus sibericum and Mollivirus sibericum, from > 30,000-year-old ice samples collected in the Siberian permafrost. Both viruses were shown to be infective by following their propagation in axenic cultures of a modern strain of Acanthamoeba castellanii. Approximately at the same time, Ng et al. (2014) used metagenomics to detect DNA and RNA viruses, preserved in in a subarctic ice patch in frozen caribou feces. One of them, a DNA virus distantly related to plant- and fungi-infecting viruses, was able to infect the plant Nicotiana benthamiana when tested using a reverse genetics approach.
Limitations to assess the diversity of glacial viromes
As we can see, efforts have been made to try understanding the community composition and ecological niches of viruses in glacial ecosystems; unfortunately, we are still far from having a comprehensive knowledge concerning the role played by glacial viromes in frozen environments. Although it does not seem obvious at first glance, it is not an easy task to characterize viromes by culture-independent approaches (like metagenomics). This is because, contrary to what happens in the case of prokaryotes and eukaryotes, there are no universal molecular markers to perform a targeted search of viral genomes.
On the other hand, very limited information has been published about the acquisition of viral auxiliary metabolic genes (AMGs) by psychrophilic or psychrotolerant microbes. This is particularly relevant from a public health perspective because, in addition to improve resistance to frozen environments, AMGs could also provide microbes with new, potentially virulent genes. For instance, this is the case of the genes encoding the Shiga toxins in certain pathogenic strains of Escherichia coli (Boyd 2012). Giant viruses, in turn, do also represent vast reservoir of uncharacterized proteins encoded by orphan genes (ORFans), as shown by metagenomic approaches (Brandes and Linial 2019). However, these hypotheses remain to be properly tested.
In addition, efforts should be made to generate critical information to distinguish between the persistence and the infectivity of ice entrapped viruses. As stated before, an increasing number of studies report on the occurrence and abundance of viruses in permanently frozen environments. However, the information about the infectious potential of these viruses is still scarce. In fact, as stated by Rassner et al. (2016), frozen habitats are characterized by harsh environmental conditions that may strongly affect the ability of viruses to infect microbes. Also, viral genomes are susceptible to damage caused by high UV radiance and extremely low temperatures, two conditions present in glacial environments. Therefore, the infectivity of viruses entrapped in frozen environments should be demonstrated experimentally and without a doubt, by using rigorous and accurate laboratory methodologies, and proper biological models of infection, i.e. new extremophilic host systems to cultivate viruses isolated from extreme habitats (Holmes 2014; Dávila-Ramos et al. 2019). An example of such experimental approach is the study conducted in 2016 by Filippova and co-workers, to determine the occurrence of bacteriophages on the water column of a permanently ice-covered and oligotrophic lake in East Antarctica. The authors found a high diversity of bacteriophages with the ability to undergo a lysogenic cycle on modern bacteria, which indicated that the phage populations present in this frozen lake were, indeed, infective at extremely low temperatures.
Finally, in all the above mentioned examples and due to the methodology used, only double-stranded DNA viruses have been targeted, likely leaving aside much of the glacial viromes’ diversity. Therefore, new experimental approaches should be taken in order to better characterize the actual virome entrapped in permanently frozen environments (Fig. 3).
Fig. 3.
Harnessing the glacial virome by contemporary methods (Photography Copyright José Manuel Romero, with permission)
Are all these ancient microbes really so old?
Following publication of the discovery of influenza-A viruses in Siberian lakes’ ice (Zhang et al. 2006), some concerns were raised that disputed the validity of these results. Indeed, by performing a rigorous phylogenetic analysis, Worobey (2008) concluded that these viruses were contaminants derived from the same laboratory and argued against considering glaciers as virus reservoirs or repositories. Even though it was shown later that avian influenza viruses are markedly cryostable (Shoham et al. 2012), the debate highlighted the importance of implementing rigorous protocols to avoid false conclusions concerning isolation and reactivation of ancient microbes.
In fact, when dealing with permanently frozen environments, contamination of ice cores with modern DNA (or modern microorganisms) can be really hard to prevent. It is therefore essential to follow some strict rules in order to authenticate results from studies of ancient DNA, recovered from any kind of samples. In 2000, Cooper and Poinar summarized nine essential criteria to be followed by any researcher working with this type of genetic material. Even though others called for a more cognitive approach (Gilbert et al. 2005), nowadays it seems obvious to include appropriate controls aimed at detecting accidental contamination with modern DNA or modern microorganisms, and to look for independent replication of the results.
Processing very old ice samples is also a major issue that requires to follow stringent protocols that guarantee the absence of any undesirable contamination. Several decontamination procedures have been developed by Rogers et al. (2004a) and Christner et al. (2005) to remove all possible contaminants from the sample’ surfaces; these protocols have been further used by different groups (D’Elia et al. 2008; Knowlton et al. 2013; Miteva et al. 2016). However, no approach is meticulous enough and, recently, some modifications have been introduced to improve the performance of these protocols. For example, a new ultra-clean protocol for ancient glacial ice decontamination was developed by Zhong et al. (2020). The three-step procedure included band saw scrapping, ethanol washing and sterile water washing of the ice cores, before reaching non-contaminated-, “inner ice” layers. The same year, Saidi-Mehrabad et al. (2020) proposed a slightly different procedure to remove contaminants and extract DNA from permafrost samples with low biomass. Again, it combined bleaching, washing, and scraping of the samples, before conducting further experiments.
The future that awaits us …?
Continental glaciers and permafrost are vanishing. In some parts of the world, this process often occurs in front of our eyes, rapidly. As we have seen, microbes immured for millennia, are being released massively from their confinement, and run downhill with meltwater streams.
In 2004, Alvin Smith and his colleagues raised the worrying hypothesis that glacial environments could act as reservoirs of viruses pathogenic for humans. At the time it was formulated, the experimental evidence supporting this proposal was rather circumstantial, and based on the following arguments: (1) the detection of microorganisms in glacial ice cores; (2) the enormous density of viral particles in water and its close interaction with glaciers (particularly at the poles); and, (3) the constant release of micro-droplets containing viral particles, from the sea surface into the atmosphere, and their subsequent transport by the wind over long distances. Nowadays, there is almost no doubt that Smith et al. were right.
According to Rogers et al. (2004b) and Edwards (2015), the interaction which is currently taking place between ice-released ancient microorganisms and modern living beings may pose serious threats to other forms of life. Not only because pathogenic microbes are released from melting glaciers and permafrost, but also because their genes and genomes are also being disseminated in natural ecosystems; these genetic elements can be subsequently acquired by horizontal gene transfer by other contemporary species (Fig. 4). This exchange of genes and mobile genetic elements (plasmids, transposons, integrons, etc.) between ancient and modern microorganisms, has been termed “genome turnover” (Rogers et al. 2004a). This turnover normally occurs at a very high rate among microorganisms that co-exist in a given habitat, and may even trespass the barriers that exist between the three domains of life (Soucy et al. 2015). However, even though the scientific evidence is outstanding, the information concerning the potential threat posed by this “global resistome” is still scarce (Perry et al. 2016). Also limited is our knowledge about the role played by glacial environments as natural reservoirs of resistomes. Therefore, there is an urgent need to increase our efforts to better understand the possible consequences of the release of such massive amounts of ARGs (Surette and Wright 2017).
Fig. 4.
Potential threats posed by release of ancient pathogens from permanently-frozen environments. Ancient pathogens (or microbes carrying potentially harmful genetic elements, such as antibiotic-resistance plasmids or integrons), are released massively when ice melts, due to global warming. Once reactivated (or resurrected), these microbes enter natural environments, close to human settlements, posing serious threats to plant-, animal- and human health
A few outstanding examples, that clearly illustrate the threat posed to native communities by non-native pathogens, have been recently highlighted by Howenhyuse et al. (2018). In all of them, the effects on animal biodiversity (e.g. amphibian, crustacean), or else on human health were catastrophic. However, almost nothing is known concerning the consequences of biological invasions of native microbial communities by ice-released microbes. Thus, microcosm and mesocosm experiments—designed to mimic the natural conditions these microbes will encounter once liberated from their confinement—are crucial to characterize the infectivity and virulence of these extremotolerant microbes, as well as their potential to act as donors of genetic elements. Looking for past events of HGT by means of bioinformatics can also bring some important clues to understand genetic exchanges within natural communities colonizing glacial ice, as already suggested by Klassen and Foght (2011). In turn, this information would be fundamental to anticipate the fate of mobile genetic elements once the ice-entrapped microbes are released, reactivated and enter modern communities.
Not long ago, the concern about the potential threat posed to global public health by the glacial pathome (that is, all potentially pathogenic microorganisms trapped in glaciers worldwide) (Roossinck and García-Arenal 2015) received unexpected support from a fortuitous event. In July 2016, an outbreak of Anthrax took place in Yamal, Russia, that decimated a population of reindeers, killed a child and forced the hospitalization of more than a hundred pastors (Hueffer et al. 2020). The Bacillus anthracis strain at the origin of this outbreak was shown to be identical to other strains isolated from the tissues of dead animals immured for centuries in the Siberian permafrost, until it thawed due to global warming (Timofeev et al. 2019).
Since then, several authors have alerted about the resurrection of ancestral pathogens in documents ranging from prudence to alarm (Houldcroft and Underdown 2016; Barras 2017; Houwenhuyse et al. 2018; Miner et al. 2020). From a more rational and objective point of view, the occurrence of outbreaks caused by pathogens hitherto unknown, has highlighted the need to implement new surveillance strategies. In this regard, it is critical to complement the surveillance of “animal reservoirs” of pathogens (both natural and accidental) with the study of “ecosystem reservoirs”. Therefore, studies of microorganisms in glaciers and permafrost could help us to take a glimpse into the future, by understanding and anticipating the potential threats we will face when the confined pathomes, viromes and resistomes are released from their confinement.
Conclusion
Melting of the Earth’s cryosphere is occurring at an unprecedented rate, since the beginning of the 21st century. Despite repeated warnings by international organizations, the tendency of this global disaster seems unstoppable. Thus, tons of microbes immured for millennia in glacial ice or permafrost, many of which are either closely related to human and animal pathogens, are being reactivated and released into aquatic and terrestrial environments. Hence, as shown by recent outbreaks of diseases caused by supposed to be extinct microbial pathogens immured in glacial ice for centuries, there is a serious risk for future epidemics (or even pandemics) to happen more often.
In the past three decades, great scientific efforts have been made to better understand the diversity of the glacial-immured microbiome, as well as to predict the potential threats it might pose to humans (and other live beings), once resurrected and liberated from their glacial imprisonment.
Only four years ago, a scientific hypothesis related the extinction of Neanderthal populations with an infection of either viral or bacterial origin (Houldcroft and Underdown 2016). Although this hypothesis was received with skepticism at first, it is gaining an unexpected support at a time when humanity faces a pandemic of dramatic consequences. This further reinforces the concern that many feel about the possible re-emergence of infectious diseases, coming from the depths of thawing glaciers and permafrost, and thought to be eradicated from the Earth’s surface thousands or millions of years ago. Without reaching such extremes, it cannot be excluded that if we continue to disturb the stability of these ecosystems by modifying the global climate, the possibility of coming into contact with any of these ice-entrapped pathogens will increase substantially. This will also happen if we continue to go deeper, farther and for longer periods into the frozen territories for scientific, commercial or touristic reasons. Since this seems impossible to avoid, we shall be at least prepared for what might happen.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
RABG acknowledges support from Conacyt-DADC Project 311684 and Conacyt-Voc-Cientif Project 1004 grants. The authors would like to thank Dr. Eduardo Chica (University of Cuenca) for his comments and suggestions. We also thank Dr. Alejandra Melfo for providing a modified version of a figure showing the disappearance of La Corona glacier in time. Finally, we express our sincere gratitude to four anonymous reviewers, for their valuable comments, which helped to improve the quality of this manuscript.
Declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abyzov SS, Bobin NE, Koudryashov BB (1982) Quantitative assessment of microorganisms in microbiological studies of Antarctic glaciers. Biol Bull Acad Sci USSR 9:558–564 [Google Scholar]
- Aherfi S, Colson P, La Scola B, Raoult D (2016) Giant viruses of Amoebas: An Update. Front Microbiol 7:349. 10.3389/fmicb.2016.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler I, Goordial J, Whyte LG (2017) Microbial life in permafrost. In: Margesin R (ed) Psychrophiles: from biodiversity to biotechnology. Springer International Publishing, pp 153–179. 10.1007/978-3-319-57057-0_8
- Alves IM, Gonçalves VN, Oliveira FS, Schaefer CE, Rosa CA, Rosa LH (2019) The diversity, distribution, and pathogenic potential of cultivable fungi present in rocks from the South Shetlands archipelago, Maritime Antarctica. Extremophiles 23:327–336 [DOI] [PubMed] [Google Scholar]
- Anesio AM, Lutz S, Chrismas NAM, Benning LG (2017) The microbiome of glaciers and ice sheets. NPJ Biofilms Microbiomes 3:10. 10.1038/s41522-017-0019-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball MM, Gómez W, Magallanes X, Rosales R, Melfo A, Yarzábal LA (2014) Bacteria recovered from a high-altitude, tropical glacier in Venezuelan Andes. World J Microbiol Biotechnol 30(3):931–941 [DOI] [PubMed] [Google Scholar]
- Barras C (2017) Wakey, wakey. New Sci 234(3126):34–37. 10.1016/S0262-4079(17)30978-8 [Google Scholar]
- Bellas CM, Anesio AM, Barker G (2015) Analysis of virus genomes from glacial environments reveals novel virus groups with unusual host interactions. Front Microbiol 6:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhullar K, Waglechner N, Pawlowski A, Koteva K, Banks ED, Johnston MD, Barton HA, Wright GD (2012) Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS One 7(4):e34953 [DOI] [PMC free article] [PubMed]
- Boyd EF (2012) Bacteriophage-encoded bacterial virulence factors and phage-pathogenicity island interactions. Adv Virus Res 82:91–118. 10.1016/B978-0-12-394621-8.00014-5 [DOI] [PubMed] [Google Scholar]
- Brandes N, Linial M (2019) Giant Viruses—Big Surprises. Viruses 11:404. https://www.mdpi.com/1999-4915/11/5/404#cite [DOI] [PMC free article] [PubMed]
- Braun C, Bezada M (2013) The history and disappearance of glaciers in Venezuela. J Latin Am Geog 12(2):85–124. 10.1353/lag.2013.0016 [Google Scholar]
- Brown J, Ferrians OJ, Heginbottom JA, Melnikov ES (1998) Circum-Arctic map of permafrost and ground-ice conditions. National Snow and Ice Data Center/World Data Center for Glaciology. Digital Media, Boulder
- Castello JD, Rogers SO, Starmer WT, Catranis CM, Ma L, Bachand GD et al (1999) Detection of tomato mosaic tobamovirus RNA in ancient glacial ice. Polar Biol 22(3):207–212 [Google Scholar]
- Castello JD, Rogers SO, Smith JE, Starmer WT, Zhao Y (2005) Chapter 13. Plant and bacterial viruses in the Greenland Ice Sheet. In: Castello J, Rogers SO (eds) Life in Ancient Ice. Princenton University Press, Oxford , pp 196–207. 10.2307/j.ctt1dr350p.19
- Christner BC, Mosley-Thompson E, Thompson LG, Zagorodnov V, Sandman K, Reeve JN (2000) Recovery and identification of viable bacteria immured in glacial ice. Icarus 144:479–485 [Google Scholar]
- Christner BC, Mosley-Thompson E, Thompson LG, Zagorodnov V, Sandman K, Reeve JN (2002) Isolation and identification of bacteria from ancient and modern ice core archives. In: Casassa G, Sepulveda FV, Sinclair R (eds) Patagonian ice fields. A unique natural laboratory for environmental and climate change studies. Kluwer, New York, pp 9–16 [Google Scholar]
- Christner BC, Mikucki JA, Foreman CM, Denson J, Priscu JC (2005) Glacial ice cores: A model system for developing extraterrestrial decontamination protocols. Icarus 174:572–584 [Google Scholar]
- Cooper A, Poinar HN (2000) Ancient DNA: do it right or not at all. Science 289(5482):1139. 10.1126/science.289.5482.1139b [DOI] [PubMed] [Google Scholar]
- D’Costa V, King C, Kalan L et al (2011) Antibiotic resistance is ancient. Nature 477:457–461. 10.1038/nature10388 [DOI] [PubMed] [Google Scholar]
- D’Elia T, Veerapaneni R, Rogers SO (2008) Isolation of microbes from Lake Vostok accretion ice. Appl Environ Microbiol 74:4962–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancer SJ, Shears P, Platt DJ (1997) Isolation and characterization of coliforms from glacial ice and water in Canada’s High Arctic. J Appl Microbiol 82:597–609 [DOI] [PubMed] [Google Scholar]
- Dávila-Ramos S, Castelán-Sánchez HG, Martínez-Ávila L, Sánchez-Carbente MdR, Peralta R, Hernández-Mendoza A, Dobson ADW, Gonzalez RA, Pastor N, Batista-García RA (2019) A review on viral metagenomics in extreme environments. Front Microbiol 10:2403. 10.3389/fmicb.2019.02403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Menezes GCA, Porto BA, Amorim SS et al (2020) Fungi in glacial ice of Antarctica: diversity, distribution and bioprospecting of bioactive compounds. Extremophiles 24:367–376. 10.1007/s00792-020-01161-5 [DOI] [PubMed] [Google Scholar]
- Ding Y, Zhang S, Zhao L, Li Z, Kang S (2019) Global warming weakening the inherent stability of glaciers and permafrost. Sci Bull 64:245–253 [DOI] [PubMed] [Google Scholar]
- Dussaillant I, Berthier E, Brun F, Masiokas M, Hugonnet R, Favier V, Rabatel A, Pitte P, Ruiz L (2019) Two decades of glacier mass loss along the Andes. Nat Geosci 12:802–808. 10.1038/s41561-019-0432-5 [Google Scholar]
- Edwards A (2015) Coming in from the cold: Potential microbial threats from the terrestrial cryosphere. Front Earth Sci 3:10–13 [Google Scholar]
- Fegel TS, Baron JS, Fountain AG, Johnson GF, Hall EK (2016) The differing biogeochemical and microbial signatures of glaciers and rock glaciers. J Geophys Res Biogeosci 121:919–932 [Google Scholar]
- Filippova SN, Surgucheva NA, Sorokin VV et al (2016) Bacteriophages in Arctic and Antarctic low-temperature systems. Microbiology 85:359–366. 10.1134/S0026261716030048 [Google Scholar]
- García-Descalzo L, García-López E, Postigo M, Baquero F, Alcazar A, Cid C (2013) Eukaryotic microorganisms in cold environments: examples from Pyrenean glaciers. Front Microbiol 4:55. 10.3389/fmicb.2013.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MTP, Bandelt HJ, Hofreiter M, Barnes I (2005) Assessing ancient DNA studies. Trends Ecol Evol 20:541–544 [DOI] [PubMed] [Google Scholar]
- Gillings MR (2014) Integrons: past, present, and future. Microbiol Mol Biol Rev 78(2):257–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin K, Loso M, Braun M (2012) Glacial transport of human waste and survival of fecal bacteria on Mt. McKinley’s Kahiltna glacier, Denali National Park, Alaska. Arctic Antarct Alp Res 44(4):432–445 [Google Scholar]
- Harding T, Jungblut AD, Lovejoy C, Vincent WF (2011) Microbes in high arctic snow and implications for the cold biosphere. Appl Environ Microbiol 77(10):3234–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson A, Anesio A, Tranter M, Fountain A, Osborn M, Priscu J et al (2008) Glacial ecosystems. Ecol Monogr 78(1):41–67. http://www.jstor.org/stable/27646118. Accessed 23 Mar 2021
- Holmes EC (2014) Freezing viruses in time. PNAS 111(47):16643–16644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houldcroft CJ, Underdown SJ (2016) Neanderthal genomics suggests a pleistocene time frame for the first epidemiologic transition. Am J Phys Anthropol 160(3):379–388 [DOI] [PubMed] [Google Scholar]
- Houwenhuyse S, Macke E, Reyserhove L, Bulteel L, Decaestecker E (2018) Back to the future in a petri dish: origin and impact of resurrected microbes in natural populations. Evol Appl 11:29–41. 10.1111/eva.12538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Guo H, Zhou P et al (2020) Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 10.1038/s41579-020-00459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueffer K, Drown D, Romanovsky V, Hennessy T (2020) Factors contributing to Anthrax outbreaks in the Circumpolar North. EcoHealth 17(1):174–180. 10.1007/s10393-020-01474-z [DOI] [PubMed] [Google Scholar]
- Hugelius G, Virtanen T, Kaverin D, Pastukhov A, Rivkin F, Marchenko S, Romanovsky V, Kuhry P (2011) High-resolution mapping of ecosystem carbon storage and potential effects of permafrost thaw in periglacial terrain, European Russian Arctic. J Geophys Res Biogeosci 116:G03024. 10.1029/2010JG001606 [Google Scholar]
- Huss M, Farinotti D (2012) Distributed ice thickness and volume of all glaciers around the globe. J Geophys Res Earth Surf 117(4):1–10 [Google Scholar]
- IPCC (2019) Technical summary. In: Pörtner H-O, Roberts DC, Masson-Delmotte V, Zhai P, Poloczanska E, Mintenbeck K, Tignor M, Alegría A, Nicolai M, Okem A, Petzold J, Rama B, Weyer NM (eds) IPCC Special report on the ocean and cryosphere in a changing climate (in press)
- Kääb A, Leinss S, Gilbert A et al (2018) Massive collapse of two glaciers in western Tibet in 2016 after surge-like instability. Nat Geosci 11:114–120. 10.1038/s41561-017-0039-7 [Google Scholar]
- Kashuba E, Dmitriev AA, Kamal SM, Melefors O, Griva G, Römling U, Ernberg I, Kashuba V, Brouchkov A (2017) Ancient permafrost staphylococci carry antibiotic resistance genes. Microb Ecol Health Dis 28(1):1345574. 10.1080/16512235.2017.1345574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane JT (2018) Catastrophic glacier collapse. Nat Geosci 11:87. 10.1038/s41561-018-0063-2 [Google Scholar]
- Klassen JL, Foght JM (2011) Characterization of Hymenobacter isolates from Victoria Upper Glacier, Antarctica reveals five new species and substantial non-vertical evolution within this genus. Extremophiles 15:45–57. 10.1007/s00792-010-0336-1 [DOI] [PubMed] [Google Scholar]
- Knowlton C, Veerapaneni R, D’Elia T, Rogers SO (2013) Microbial Analyses of Ancient Ice Core Sections from Greenland and Antarctica. Biology 2:206–232. 10.3390/biology2010206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakov A, Mindlin S, Beletsky A, Shcherbatova N, Rakitin A, Ermakova A, Mardanov A, Petrova M (2016) The ancient small mobilizable plasmid pALWED1.8 harboring a new variant of the noncassette streptomycin/spectinomycin resistance gene aadA27. Plasmid 84–85:36–43. 10.1016/j.plasmid.2016.02.005 [DOI] [PubMed] [Google Scholar]
- La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D (2003) A giant virus in amoebae. Science 299:2033 [DOI] [PubMed] [Google Scholar]
- Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A, Lescot M, Poirot O, Bertaux L, Bruley C, Coute Y, Rivkina E, Abergel C, Claverie J (2014) Thirty-thousandyear-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci USA 111(11):4274–4279. 10.1073/pnas.1320670111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Lartigue A, Bertaux L, Jeudy S, Bartoli J, Lescot M, Alempic J, Ramus C, Bruley C, Labadie K, Shmakova L, Rivkina E, Couté Y, Abergel C, Claverie J (2015) In depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc Natl Acad Sci USA. 10.1073/pnas.1510795112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowska N, Zawierucha K, Nadobna P, Piątek-Bajan K, Krajewska A, Szwedyk J, Iwasieczko P, Mokracka J, Koczura R (2020) Occurrence of integrons and antibiotic resistance genes in cryoconite and ice of Svalbard, Greenland, and the Caucasus glaciers. Sci Total Environ. 10.1016/j.scitotenv.2020.137022 [DOI] [PubMed] [Google Scholar]
- Malavin S, Shmakova L (2020) Isolates from ancient permafrost help to elucidate species boundaries in Acanthamoeba castellanii complex (Amoebozoa: Discosea). Eur J Protistol. 10.1016/j.ejop.2020.125671 [DOI] [PubMed] [Google Scholar]
- Malavin S, Shmakova L, Claverie J-M, Rivkina E (2020) Frozen Zoo: a collection of permafrost samples containing viable protists and their viruses. Biodiv Data J. 10.3897/BDJ.8.e51586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindlin SZ, Soina VS, Petrova MA, Gorlenko ZhM (2008) Isolation of antibiotic resistance bacterial strains from Eastern Siberia permafrost sediments. Russ J Gen 44(1):27–34 [PubMed] [Google Scholar]
- Miner KR, Edwards A, Miller Ch (2020) Deep frozen Arctic microbes are waking up. Scientific American. https://www.scientificamerican.com/article/deep-frozen-arctic-microbes-are-waking-up/
- Miteva V (2008) Bacteria in snow and glacier ice. In: Margesin R, Schinner F, Marx JC, Gerday C (eds) Psychrophiles: from biodiversity to biotechnology. Springer, Berlin, Heidelberg, pp 31–50 [Google Scholar]
- Miteva V, Sowers T, Schupbach S, Fischer H, Brenchley J (2016) Geochemical and microbiological studies of nitrous oxide variations within the new NEEM Greenland ice core during the Last Glacial period. Geomicrobiol J 33:647–660 [Google Scholar]
- Mogrovejo-Arias DC, Brill FHH, Wagner D (2020) Potentially pathogenic bacteria isolated from diverse habitats in Spitsbergen. Svalbard Environ Earth Sci 79(5):1–9 [Google Scholar]
- Ng TF, Chen LF, Zhou Y, Shapiro B, Stiller M, Heintzman PD, Varsani A, Kondov NO, Wong W, Deng X, Andrews TD, Moorman BJ, Meulendyk T, MacKay G, Gilbertson RL, Delwart E (2014) Preservation of viral genomes in 700-y-old caribou feces from a subarctic ice patch. Proc Natl Acad Sci USA 111(47):16842–16847. 10.1073/pnas.1410429111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N et al (2016) Uncovering Earth’s virome. Nature 536(7617):425–430 [DOI] [PubMed] [Google Scholar]
- Pelto M, Network WGMS (2018) Alpine glaciers [in State of the Climate in 2017]. Bull Am Meteorol Soc 99(8):S23–S25 [Google Scholar]
- Perini L, Gostinčar C, Gunde-Cimerman N (2019) Fungal and bacterial diversity of Svalbard subglacial ice. Sci Rep 9:20230. 10.1038/s41598-019-56290-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron GG, Whyte L, Turnbaugh PJ, Goordial J, Hanage WP, Dantas G, Desai MM (2015) Functional characterization of bacteria isolated from ancient arctic soil exposes diverse resistance mechanisms to modern antibiotics. PLoS ONE 10(3):e0069533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Waglechner N, Wright G (2016) The prehistory of antibiotic resistance. Cold Spring Harb Perspect Med. 10.1101/cshperspect.a025197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova M, Gorlenko Z, Mindlin S (2009) Molecular structure and translocation of a multiple antibiotic resistance region of a Psychrobacter psychrophilus permafrost strain. FEMS Microbiol Lett 296(2):190–197. 10.1111/j.1574-6968.2009.01635.x [DOI] [PubMed] [Google Scholar]
- Petrova M, Gorlenko Z, Mindlin S (2011) Tn5045, a novel integron-containing antibiotic and chromate resistance transposon isolated from a permafrost bacterium. Res Microbiol 162:337–345 [DOI] [PubMed] [Google Scholar]
- Petrova M, Kurakov A, Shcherbatova N, Mindlin S (2014) Genetic structure and biological properties of the first ancient multiresistance plasmid pKLH80 isolated from a permafrost bacterium. Microbiology 160:2253–2263. 10.1099/mic.0.079335-0 [DOI] [PubMed] [Google Scholar]
- Price PB (2000) A habitat for psychrophiles in deep Antarctic ice. Proc Natl Acad Sci USA 97:1247–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priscu JC, Christner BC, Foreman CM, Royston-Bishop G (2006) Biological Material in Ice Cores. In: Elias SA (ed) Encyclopedia of quaternary science. Elsevier, Amsterdam, London
- Rabatel A, Ceballos JL, Micheletti N, Jordan E, Braitmeier M, González J, Mölg N, Ménégoz M, Huggel C, Zemp M (2017) Toward an imminent extinction of Colombian glaciers? Geogr Ann Ser A Phys Geogr. 10.1080/04353676.2017.1383015 [Google Scholar]
- Rafiq M, Hayat M, Zada S, Sajjad W, Hassan N, Hasan F (2019) Geochemistry and bacterial recovery from Hindu Kush Range glacier and their potential for metal resistance and antibiotic production. Geomicrobiol J 36:326–338. 10.1080/01490451.2018.1551947 [Google Scholar]
- Ramírez N, Melfo A, Resler LM, Llambí LD (2020) The end of the eternal snows: Integrative mapping of 100 years of glacier retreat in the Venezuelan Andes. Arctic Antarctic Alpine Res 52:563–581 [Google Scholar]
- Rassner SM (2017) Viruses in glacial environments. In: Margesin R (ed) Psychrophiles: from biodiversity to biotechnology, 2nd edn. Springer, Berlin, pp 111–131 [Google Scholar]
- Rassner SME, Anesio AM, Girdwood SE, Hell K, Gokul JK, Whitworth DE, Edwards A (2016) Can the bacterial community of a High Arctic glacier surface escape viral control? Front Microbiol 7:956. 10.3389/fmicb.2016.00956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SO, Starmer WT, Castello JD (2004a) Recycling of pathogenic microbes through survival in ice. Med Hypotheses 63(5):773–777 [DOI] [PubMed] [Google Scholar]
- Rogers SO, Theraisnathan V, Ma LJ, Zhao Y, Zhang G, Shin SG, Castello JD, Starmer WT (2004b) Comparisons of protocols for decontamination of environmental ice samples for biological and molecular examinations. Appl Environ Microbiol 70:2540–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondón J, Gómez W, Ball MM, Melfo A, Rengifo M, Balcázar W, Dávila-Vera D, Balza-Quintero A, Mendoza-Briceño RV, Yarzábal LA (2016) Diversity of culturable bacteria recovered from Pico Bolívar’s glacial and subglacial environments, at 4950 m Venezuelan Tropical Andes. Can J Microbiol 62(11):904–917. 10.1139/cjm-2016-0172 [DOI] [PubMed] [Google Scholar]
- Roossinck MJ, García-Arenal F (2015) Ecosystem simplification, biodiversity loss and plant virus emergence. Curr Opin Virol 10:56–62. 10.1016/j.coviro.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadi H, Pagnier I, Colson P, Cherif JK, Beji M, Boughalmi M et al (2013a) First isolation of Mimivirus in a patient with pneumonia. Clin Infect Dis 57:e127–e134. 10.1093/cid/cit354 [DOI] [PubMed] [Google Scholar]
- Saadi H, Reteno DG, Colson P, Aherfi S, Minodier P, Pagnier I et al (2013b) Shan virus: a new mimivirus isolated from the stool of a Tunisian patient with pneumonia. Intervirology 56:424–429. 10.1159/000354564 [DOI] [PubMed] [Google Scholar]
- Saidi-Mehrabad A, Neuberger P, Cavaco M et al (2020) Optimization of subsampling, decontamination, and DNA extraction of difficult peat and silt permafrost samples. Sci Rep 10:14295. 10.1038/s41598-020-71234-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Säwström C, Lisle J, Anesio AM et al (2008) Bacteriophage in polar inland waters. Extremophiles 12:167–175. 10.1007/s00792-007-0134-6 [DOI] [PubMed] [Google Scholar]
- Schaefer K, Lantuit H, Romanovsky V, Schuur EAG (2012) Policy implications of warming permafrost. United Nations Environment Programme Special Report, Nairobi, Kenya
- Schmieder R, Edwards R (2012) Insights into antibiotic resistance through metagenomic approaches. Future Microbiol 7(1):73–89. 10.2217/fmb.11.135 [DOI] [PubMed] [Google Scholar]
- Segawa T, Takeuchi N, Rivera A, Yamada A, Yoshimura Y, Barcaza G et al (2013) Distribution of antibiotic resistance genes in glacier environments. Environ Microbiol Rep 5(1):127–134 [DOI] [PubMed] [Google Scholar]
- Shatilovich AV, Shmakova LA, Gubin SV, Gudkov AV, Gilichinsky DA (2005) Viable protozoa in Late PIleistocene and Holocene permafrost sediments. Doklady Biol Sci 401:136–138 [DOI] [PubMed] [Google Scholar]
- Shatilovich AV, Shmakova LA, Mylnikov AP, Gilichinsky DA (2009) Ancient protozoa isolated from permafrost. In: Margesin R (ed) Permafrost soils. Springer, Berlin, Heidelberg, pp 97–115. 10.1007/978-3-540-69371-0_8
- Shatilovich A, Stoupin D, Rivkina E (2015) Ciliates from ancient permafrost: assessment of cold resistance of the resting cysts. Eur J Protistol 51:230–240. 10.1016/j.ejop.2015.04.001 [DOI] [PubMed] [Google Scholar]
- Shatilovich AV, Tchesunov AV, Neretina TV, Grabarnik IP, Gubin SV, Vishnivetskaya TA, Onstott TC, Rivkina EM (2018) Viable nematodes from Late Pleistocene permafrost of the Kolyma River lowland. Dokl Biol Sci 480:100–102 [DOI] [PubMed] [Google Scholar]
- Shmakova L, Bondarenko N, Smirnov A (2016) Viable species of Flamella (Amoebozoa: Variosea) isolated from ancient Arctic permafrost sediments. Protist 167:13–30. 10.1016/j.protis.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Shmakova LA, Karpov SA, Malavin SA, Smirnov AV (2018) Morphology, biology and phylogeny of Phalansterium arcticum sp. n. (Amoebozoa, Variosea), isolated from ancient Arctic permafrost. Eur J Protistol 63:117–129. 10.1016/j.ejop.2018.02.002 [DOI] [PubMed] [Google Scholar]
- Shoham D, Jahangir A, Ruenphet S, Takehara K (2012) Persistence of avian influenza viruses in various artificially frozen environmental water types. Influ Res Treat. 10.1155/2012/912326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater T, Lawrence IR, Otosaka IN, Shepherd A, Gourmelen N, Jakob L, Tepes P, Gilbert L, Nienow P (2021) Earth’s Ice Imbalance. Cryosphere 15:233–246. 10.5194/tc-15-233-2021 [Google Scholar]
- Smith AW, Skilling DE, Castello JD, Rogers SO (2004) Ice as a reservoir for pathogenic human viruses: specifically, caliciviruses, influenza viruses, and enteroviruses. Med Hypotheses 63(4):560–566 [DOI] [PubMed] [Google Scholar]
- Sommaruga R (2015) When glaciers and ice sheets melt: consequences for planktonic organisms. J Plankton Res 37(3):509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy SM, Huang J, Gogarten JP (2015) Horizontal gene transfer: building the web of life. Nat Rev Genet 16(8):472–482. 10.1038/nrg3962 [DOI] [PubMed] [Google Scholar]
- Stoupin D, Kiss AK, Arndt H, Shatilovich AV, Gilichinsky DA, Nitsche F (2012) Cryptic diversity within the choanoflagellate morphospecies complex Codosiga botrytis—phylogeny and morphology of ancient and modern isolates. Eur J Protistol 48:263–273. 10.1016/j.ejop.2012.01.004 [DOI] [PubMed] [Google Scholar]
- Surette M, Wright GD (2017) Lessons from the environmental antibiotic resistome. Annu Rev Microbiol 71:309–312 [DOI] [PubMed] [Google Scholar]
- Tan L, Li L, Ashbolt N, Wang X, Cui Y, Zhu X, Xu Y, Yang Y, Mao D, Luo Y (2018) Arctic antibiotic resistance gene contamination, a result of anthropogenic activities and natural origin. Sci Total Environ 621:1176–1184. 10.1016/j.scitotenv.2017.10.110 [DOI] [PubMed] [Google Scholar]
- Timofeev V, Bahtejeva I, Mironova R, Titareva G, Lev I, Christiany D et al (2019) Insights from Bacillus anthracis strains isolated from permafrost in the tundra zone of Russia. PLoS ONE 14(5):1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz-Deptuła B, Niedźwiedzka-Rystwej P, Czupryńska P, Deptuła W (2019) Protozoal giant viruses: agents potentially infectious to humans and animals. Virus Genes 55:574–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchetti B, Selbmann L, Blanchette RA, Di Mauro S, Marchegiani E, Zucconi L et al (2015) Cryptococcus vaughanmartiniae sp. Nov. and Cryptococcus onofrii sp. nov.: two new species isolated from worldwide cold environments. Extremophiles 19(1):149–159 [DOI] [PubMed] [Google Scholar]
- Ushida K, Segawa T, Kohshima S, Takeuchi N, Fukui K, Li Z, Kanda H (2010) Application of real-time PCR array to the multiple detection of antibiotic resistant genes in glacier ice samples. J Gen Appl Microbiol 56:43–52. 10.2323/jgam.56.43 [DOI] [PubMed] [Google Scholar]
- Van Goethem MW, Pierneef R, Bezuidt OKI, Van De Peer Y, Cowan DA, Makhalanyane TP (2018) A reservoir of “historical” antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 6(1):40. 10.1186/s40168-018-0424-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan DG, Comiso JC, Allison I et al (2013) Observations: cryosphere. In: Stocker TF, Qin D, Plattner GK (eds) Climate change 2013: the physical science basis. Cambridge University Press, Cambridge, pp 317–382 [Google Scholar]
- Wilhelm L, Singer GA, Fasching C, Battin TJ, Besemer K (2013) Microbial biodiversity in glacier-fed streams. ISME J 7:1651–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W, Lane D, Pearce D et al (2000) Transmission electron microscope analysis of virus-like particles in the freshwater lakes of Signy Island, Antarctica. Polar Biol 23:657–660. 10.1007/s003000000152 [Google Scholar]
- Worobey M (2008) Phylogenetic evidence against evolutionary stasis and natural abiotic reservoirs of influenza A virus. J Virol 82(7):3769–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters B, Gardner AS, Moholdt G (2019) Global glacier mass loss during the GRACE satellite mission (2002–2016). Front Earth Sci 7:96. 10.3389/feart.2019.00096 [Google Scholar]
- Wright GD (2010) Antibiotic resistance in the environment: a link to the clinic? Curr Opin Microbiol 13(5):589–594 [DOI] [PubMed] [Google Scholar]
- Zhang G, Shoham D, Gilichinsky D, Davydov S, Castello JD, Rogers SO (2006) Evidence of influenza A Virus RNA in Siberian Lake Ice. J Virol 80(24):12229–12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong ZP, Solonenko NE, Li YF, Gazitúa MC, Roux S, Davis ME, Van Etten JL, Mosley-Thompson E, Rich VI, Sullivan MB, Thompson LG (2020) Glacier ice archives fifteen-thousand-year-old viruses. BioRxiv. 10.1101/2020.01.03.894675 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.