Abstract
The prior existence of human ACE2 protein-expressing mice used to study SARS-CoV and the rapid development of mouse-adapted virus strains, has allowed the study of SARS-CoV-2 in mice, even as we are still learning about its natural pathology in humans. With myriad genetically altered strains on the C57BL/6 background and the abundance of immunological reagents available to interrogate its immune responses, the C57BL/6 mice may provide useful insight into the immunology of SARS-CoV-2 infection and vaccination. In order to conduct more detailed studies on their T cell responses to vaccines and infection, the epitopes eliciting those responses must be characterized in further detail. Here, we mapped CD8 T cell epitopes within the receptor binding domain of the SARS-CoV-2 spike protein in C57BL/6 mice. Our study identified five major CD8 T cell epitopes in immunized C57BL/6 mice, including one, VVLSFELL, presented by H-2Kb and common between SARS-CoV and SARS-CoV-2.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has emerged as one of the most devastating pandemics in a century. The global response to this threat has been swift, leading to the development of multiple safe and efficacious vaccines(1–3).
The primary target for SARS-CoV-2 vaccine studies in humans is the spike (S) protein(4), the surface protein on coronaviruses essential for antibody-mediated neutralization of viral particles. Although the two mRNA-based vaccines now approved for emergency use authorization by the United States FDA elicit strong antibody responses, they also elicit CD8 T cell responses to the S protein(5, 6), as do S-encoding adenoviral vectors of other leading vaccine candidates(7, 8). Future studies may define the protective effect of CD8 T cell responses, especially in the latter. Indeed, in a recent study of COVID-19 patients, CD4 and CD8 T cell responses were independently associated with less severe disease(9).
Despite the availability of multiple mouse models of disease, mechanistic studies into the roles for T cells in vaccine-mediated protection and immunity derived from natural infection have been hampered by limited knowledge of the SARS-CoV-2 antigens targeted by CD8 T cells. To define the epitopes contained within the RBD of the S protein, we used a subunit vaccine platform composed of recombinant RBD protein antigen in combination with an adjuvant containing agonistic anti-CD40 antibody and the TLR3 agonist poly(I:C)(10). With peptide stimulation and subsequent cytokine staining, we identified five major and two minor CD8 T cell epitopes in immunized C57BL/6 mice. Furthermore, we defined the MHC class I-restriction as H-2Kb for a peptide epitope that is shared between both SARS-CoV and SARS-CoV-2.
Materials and Methods
Mice and immunizations
All experiments involving mice were conducted following protocols approved by the University of Colorado Institutional Animal Care and Use Committee (IACUC) according to guidelines provided by the Association for Assessment and Accreditation of Laboratory Animal Care. C57BL/6 mice were obtained from the Jackson Laboratory and were subsequently bred in specific-pathogen-free facilities at the University of Colorado Anschutz Medical Campus. Experiments were performed in 6–12-week-old female mice. Mice were immunized via tail-vein injection with 100 μg or 200 μg, of SARS-CoV-2 spike RBD protein plus adjuvant. SARS-CoV-2 RBD protein (Wuhan-Hu-1; GenBank: MT380724.1) was expressed by transfection of Expi293 cells with a His-tagged vector (a gift from F. Krammer, Icahn School of Medicine at Mount Sinai, New York, NY)(11) and subsequently purified from cell culture supernatants by the University of Colorado Cell Technologies Shared Resource. Immunizations were adjuvanted with 40 μg poly(I:C) (Invivogen), and 40 μg anti-CD40 (clone FGK4.5, BioXCell). Vaccines were made immediately prior to immunization and injected in a total volume of 200 μl.
RMA-S MHC class I-stabilization assay
To determine the MHC class I-restriction of SARS-CoV-2 peptide epitopes shown to induce RBD-specific CD8 T cell responses in immunized mice, we employed the murine TAP-deficient RMA-S lymphoma cell line, which is derived from C57BL/6 mice(12, 13). RMA-S cells were cultured overnight at 27°C to stabilize unloaded MHC class I H-2Db and H-2Kb on the cell surface. RMA-S cells containing peptide-empty H-2Db and H-2Kb were coincubated with indicated CD8 T cell peptides at 10 μM for 5 h at 37°C. We tested the 15-mer peptides representing the 5 major SARS-CoV-2 RBD epitopes revealed in these studies: S1–14,319, S337–351, S401–415, S477–491, S505–519/509–523, and the 8-mer S511–518. Peptides with known H-2Kb, H-2Db, and H-2Kb/H-2Db restriction, respectively, were included as controls: OVA257–264, LCMV NP396–404, and LCMV GP33–41. After 5 h at 37°C, MHC class I stabilization was quantified by flow cytometry using anti-mouse antibodies directed against H-2Kb (clone AF6-14-8) and H-2Db (clone 28-14-8).
Flow cytometry
Seven days after immunization, single cell suspensions generated from spleens were subjected to ACK red blood cell lysis and counted using a Vi-Cell automated cell counter (Beckman Coulter). For in vitro stimulation assays, 1 × 106 cells were incubated with 1 μg/ml peptide and 3 μg/ml brefeldin A for 5 h at 37°C in complete media (RPMI 1640 containing 10% FBS, 10 mM HEPES, 0.1 mM β-ME, 0.1 mM non-essential amino acids, 0.1 mM sodium pyruvate, 2 mM L-glutamine and penicillin-streptomycin). After stimulation, cells were surfaced-stained with CD8α-BV421 (clone 53.67, BioLegend), CD4-FITC (GK1.5, BioLegend), B220-PE-Cy7 (clone RA3-6B2, Tonbo), and a fixable viability dye (Ghost Dye Red 780, Tonbo) for 10 min at room temperature. After staining for surface antigens, cells were fixed and permeabilized with FoxP3 fixation/permeabilization buffers (Tonbo) for 15 min at room temperature. After fixation and permeabilization, cells were washed in perm/wash buffer and stained for intracellular cytokines using IFNγ-APC (XMG1.2, Tonbo) and TNFα-PE (MP6-XT22, BD Biosciences) diluted in perm/wash buffer for 30 min at room temperature. After a final wash, flow cytometry data were acquired on a four-laser (405, 488, 561, 638 nm) CytoFLEX S flow cytometer (Beckman Coulter) and analysis was performed using FlowJo (version 10.7.1; BD Biosciences).
Peptides
Crude preparations of 58 peptides covering the SARS-CoV-2 spike RBD protein (GenBank: MT380724.1), derived from isolate Wuhan-Hu-1, were generated (ChinaPeptides), comprising 15-mer peptides overlapping by 11 amino acids. Highly purified (>96% purity) VVSLFELL peptide was also prepared (ChinaPeptides).
Statistical Analysis
Prism (version 9.01, GraphPad) was used to plot data and perform one-way ANOVA tests with Dunnett’s multiple comparisons test to compare all values to stimulation with an irrelevant peptide (HSV glycoprotein B498–504).
Results
One week following vaccination via intravenous injection with 100 μg purified, recombinant SARS-CoV-2 RBD protein adjuvanted with poly(I:C) and anti-CD40, splenic CD4 and CD8 T cells from C57BL/6 mice were evaluated by ex vivo peptide restimulation and subsequent intracellular cytokine staining for IFNγ and TNFα and flow cytometric analysis. Cells were stimulated using a peptide library of 15-mers, overlapping by 11 amino acids, covering the entire RBD protein (Table I). No CD4 T cell responses to RBD peptides were revealed for C57BL/6 mice by this analysis, however, several major CD8 T cell epitopes were identified. Five peptides were determined to generate statistically significant IFNγ responses in a one-way ANOVA analysis, including S1–14,319, S337–351, S401–415, S477–491, and S505–519/509–523 (Fig. 1A). The latter sequences, spanning S505–523, aligned with a previously identified SARS-CoV CD8 T cell epitope, VVLSFELL(14). Using this same 8-mer sequence, S511–518 (511*) was determined to be the minimal epitope for SARS-CoV-2 (Fig. 1A). Two additional minor epitopes were confirmed in an experiment where antigen dose was increased to 200 μg (Fig. 1B). In this experiment, the three strongest epitopes each elicited IFNγ production in roughly 3% of CD8 T cells, each, whereas S529–343 and S389–403 elicited significant, but relatively modest CD8 T cell responses at about 0.3% of CD8 T cells. Representative flow cytometry plots show most of the CD8 T cells responding to peptide restimulation stain positive for both IFNγ and TNFα, with negligible background cytokine production in negative control wells (stimulated with HSVgB498–505) (Fig. 1C).
Table I.
Amino acid sequences for peptides used in in vitro cytokine stimulation assays.
| Spike protein aa# | Sequence |
|---|---|
| 1–14, 319 | MFVFLVLLPLVSSQR |
| 5–14, 319–323 | LVLLPLVSSQRVQPT |
| 9–14, 319–327 | PLVSSQRVQPTESIV |
| 14, 323–331 | SQRVQPTESIVRFPN |
| 321–335 | QPTESIVRFPNITNL |
| 325–339 | SIVRFPNITNLCPFG |
| 329–343 | FPNITNLCPFGEVFN |
| 333–347 | TNLCPFGEVFNATRF |
| 337–351 | PFGEVFNATRFASVY |
| 341–355 | VFNATRFASVYAWNR |
| 345–359 | TRFASVYAWNRKRIS |
| 349–363 | SVYAWNRKRISNCVA |
| 353–367 | WNRKRISNCVADYSV |
| 357–371 | RISNCVADYSVLYNS |
| 361–375 | CVADYSVLYNSASFS |
| 365–379 | YSVLYNSASFSTFKC |
| 369–383 | YNSASFSTFKCYGVS |
| 373–387 | SFSTFKCYGVSPTKL |
| 377–391 | FKCYGVSPTKLNDLC |
| 381–395 | GVSPTKLNDLCFTNV |
| 385–399 | TKLNDLCFTNVYADS |
| 389–403 | DLCFTNVYADSFVIR |
| 393–407 | TNVYADSFVIRGDEV |
| 397–411 | ADSFVIRGDEVRQIA |
| 401–415 | VIRGDEVRQIAPGQT |
| 405–419 | DEVRQIAPGQTGKIA |
| 409–423 | QIAPGQTGKIADYNY |
| 413–427 | GQTGKIADYNYKLPD |
| 417–431 | KIADYNYKLPDDFTG |
| 421–435 | YNYKLPDDFTGCVIA |
| 425–439 | LPDDFTGCVIAWNSN |
| 429–443 | FTGCVIAWNSNNLDS |
| 433–447 | VIAWNSNNLDSKVGG |
| 437–451 | NSNNLDSKVGGNYNY |
| 441–455 | LDSKVGGNYNYLYRL |
| 445–459 | VGGNYNYLYRLFRKS |
| 449–463 | YNYLYRLFRKSNLKP |
| 453–467 | YRLFRKSNLKPFERD |
| 457–471 | RKSNLKPFERDISTE |
| 461–475 | LKPFERDISTEIYQA |
| 465–479 | ERDISTEIYQAGSTP |
| 469–483 | STEIYQAGSTPCNGV |
| 473–487 | YQAGSTPCNGVEGFN |
| 477–491 | STPCNGVEGFNCYFP |
| 481–495 | NGVEGFNCYFPLQSY |
| 485–499 | GFNCYFPLQSYGFQP |
| 489–503 | YFPLQSYGFQPTNGV |
| 493–507 | QSYGFQPTNGVGYQP |
| 497–511 | FQPTNGVGYQPYRVV |
| 501–515 | NGVGYQPYRVVVLSF |
| 505–519 | YQPYRVVVLSFELLH |
| 509–523 | RVVVLSFELLHAPAT |
| 513–527 | LSFELLHAPATVCGP |
| 517–531 | LLHAPATVCGPKKST |
| 521–535 | PATVCGPKKSTNLVK |
| 525–539 | CGPKKSTNLVKNKCV |
| 529–541, 2xH | KSTNLVKNKCVNFHH |
| 533–541, 6xH | LVKNKCVNFHHHHHH |
| HSVgB 498–505 | SSIEFARL |
| 511*–518 | VVLSFELL |
Figure 1. Epitope mapping of CD8 T cell responses to SARS-CoV-2 RBD protein in C57BL/6 mice.
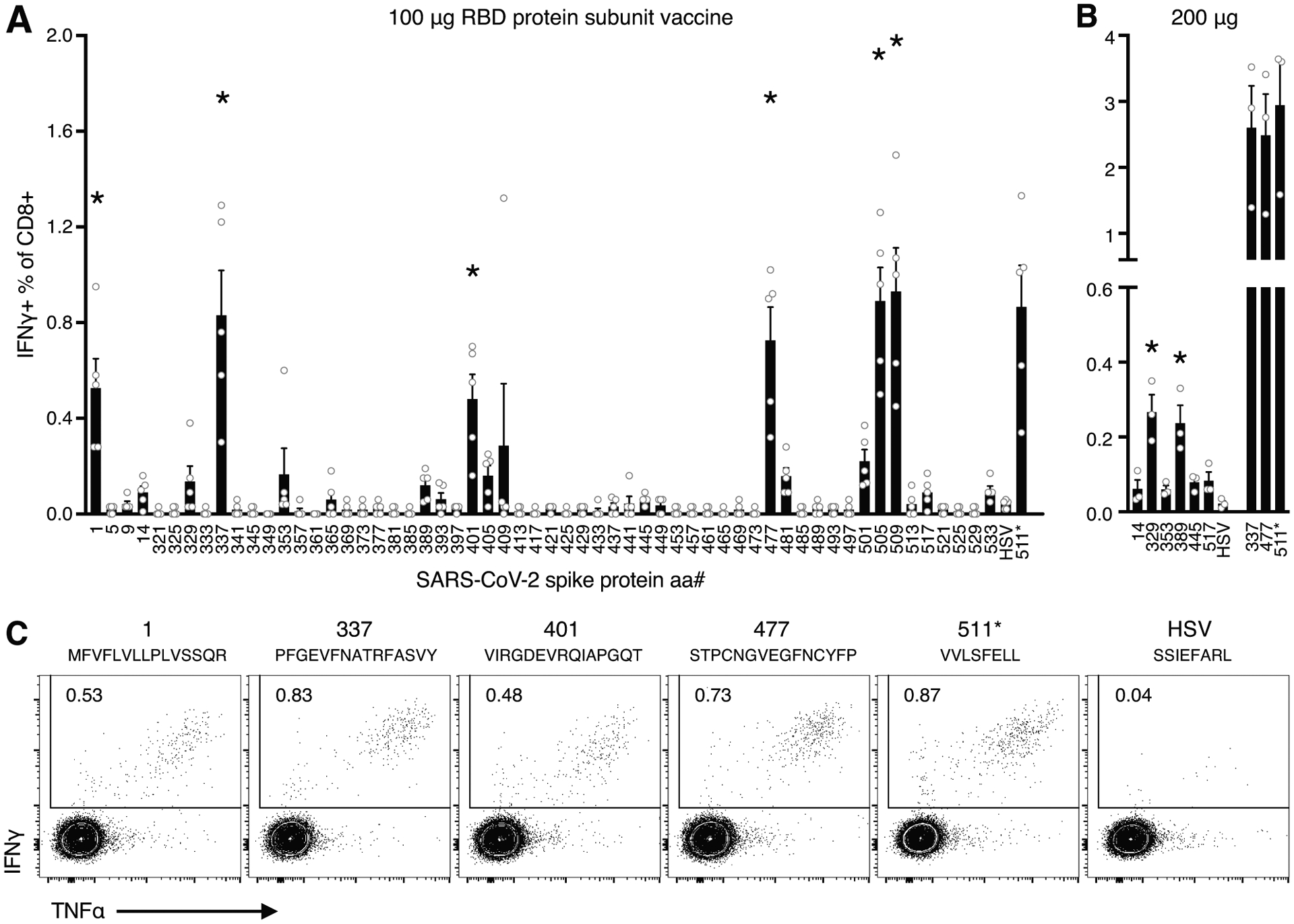
Five mice were immunized with RBD protein plus adjuvant and their spleens harvested one week later. A) The percentage of CD8 T cells staining for IFNγ after a 5 h incubation with individual 15-mer peptides spanning SARS-CoV-2 RBD. Responses that were significantly greater than those induced by an irrelevant peptide (HSVgB498–504), as determined by Dunnett’s multiple comparisons test (where p<0.01), were indicated by an asterisk. B) The percentage of CD8 T cells staining for IFNγ for the six potential minor epitopes and three of the major epitopes identified in A) in mice immunized 200 μg of RBD plus adjuvant. C) Representative intracellular IFNγ and TNFα staining. Cells were pre-gated on lymphocytes, singlets, live cells, and CD8+CD4−B220−.
These data suggest a promiscuity of the peptide VVLSFELL (S511–518) for MHC of multiple haplotypes, here eliciting responses in C57BL/6 mice and in another recent publication, S511–525 elicited responses in BALB/c mice immunized with a DNA-based vaccine encoding the S protein(15). Using the MHC-I peptide binding prediction tool NetH2pan(16), the only 8–14-mer peptides predicted to bind H-2Kd or H-2Dd within S511–525 are VVLSFELL and VVVLSFELL (S510–518), which are both predicted to strongly bind H-2Dd. Interestingly, the 9-mer VVVLSFELL is also predicted to bind to H-2Db. To determine whether this epitope was restricted to H-2Kb and/or H-2Db, we performed a cell-based MHC-I stabilization assay. RMA-S cells were interrogated with the 15-mers S1–14,319, S337–351, S401–415, S477–491, S505–519, and S509–523, as well as the minimal 8-mer S511–518. RMA-S cells are deficient in the expression of the TAP peptide transporter, critical for stabilizing MHC-I through peptide loading in the endoplasmic reticulum. This results in little to no MHC-I expression on the cell surface at 37°C(12). However, when RMA-S cells are cultured at 27°C, empty H-2Db and H-2Kb MHC-I molecules accumulate on the cell surface. The addition of peptides able to bind to either Kb or Db, followed by shifting the cells to 37°C, permits identification of the MHC-I molecules (i.e., Kb, Db, or both) stabilized on the cell surface. Staining with antibodies specific for H-2Kb and H-2Db indicated that the 8-mer VVLSFELL (S511–518) was clearly restricted to H-2Kb (Fig. 2). In contrast, S505–519, and S509–523, which contain the S511–518 8-mer as well as the 9-mer VVVLSFELL, appeared to stabilize H-2Db, as predicted, with S509–523 stabilizing both Kb and Db. Results for the 15-mer peptides covering the remaining major epitopes were less clear, with the exception of S477–491, which also stabilized H-2Db. It is not surprising that the RMA-S assay was unable to define the restriction for every 15-mer, as it is likely a less sensitive measure of peptide binding as the cytokine staining of activated T cells, known to react to picomolar quantities of peptide-bound MHC(17). However, use of the MHC-I peptide binding prediction tool NetH2pan(16) indicated a likely VFLVLLPL epitope binding H-2Kb within S1–14,319, a NATRFASV epitope binding H-2Kb in S337–351, and a STPCNGVEGF epitope binding H-2Db in S477–491.
Figure 2. Determination of peptide MHC class I-restriction.
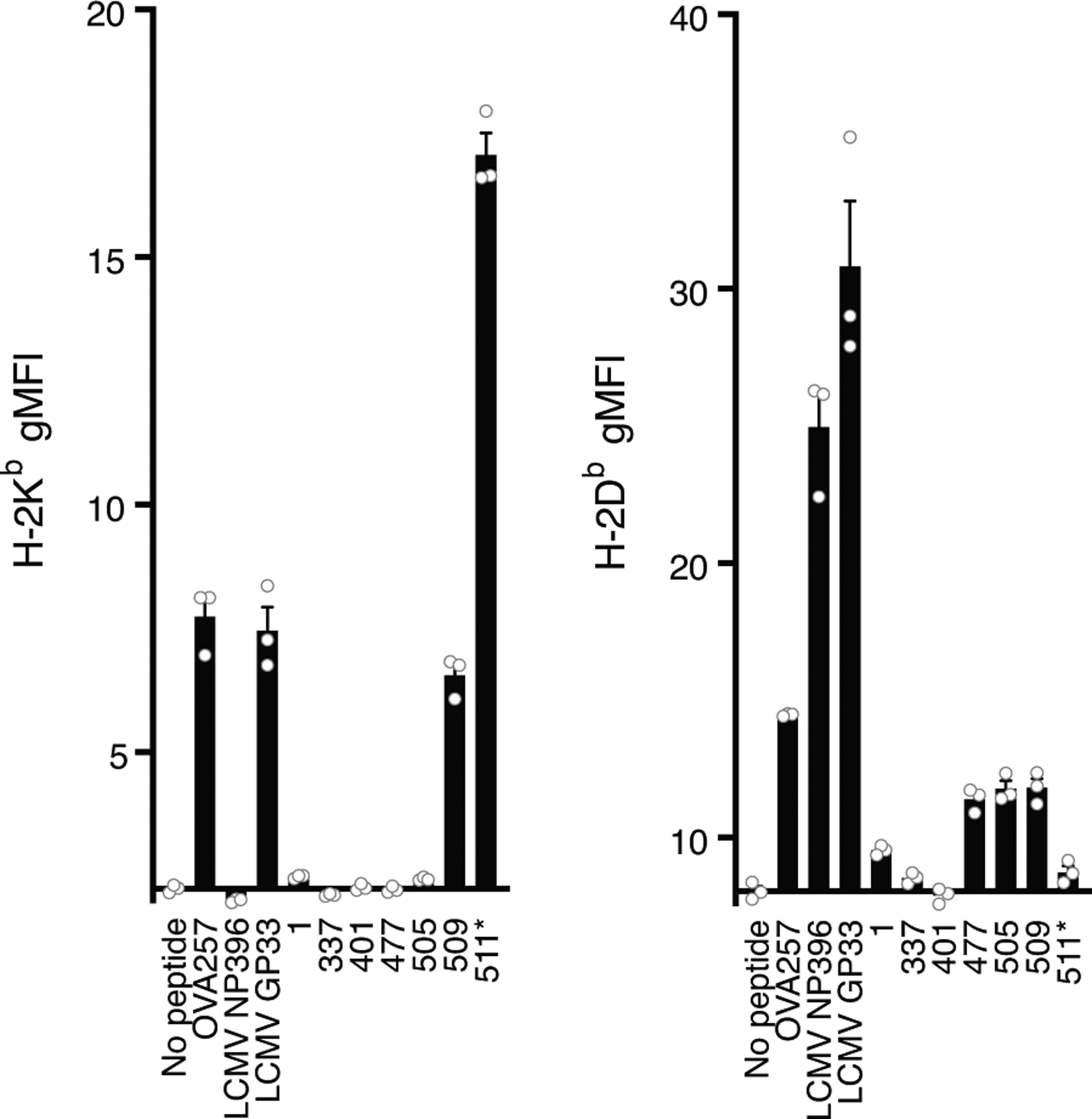
H-2Kb or H-2Kb staining of RMA-S cells 5 h after incubation with the indicated peptide. For both graphs, the x-axis intersects the y-axis at the average gMFI value for controls without peptide.
Discussion
The relative durability of the antibody responses to SARS-CoV-2 infection has been controversial, with initial studies reporting a dramatic early decline in titers that may leave patients susceptible to reinfection(18). More recent, much larger studies, however, indicate that neutralizing antibody titers persist for at least 5 months after infection(19). In line with these data, preliminary studies suggest the risk of reinfection remains very low, and is associated with asymptomatic disease(20). Yet, whether or not antibody responses ultimately demonstrate long-term durability, cellular immune responses are likely an important determinant of prolonged protection.
COVID-19 patients show T cell reactivity toward multiple proteins, including membrane (M), nucleocapsid (N) and non-structural proteins (NSPs)(21); in fact, one recent study identified an epitope within the nucleocapsid, N219–227, shared by both mouse (H-2Db) and human (HLA-A2) T cells(22). However, in serum isolated from PCR-confirmed SARS-CoV-2 positive patients, the primary target for neutralizing antibody is the S protein, with epitope specificity of neutralization directed against both the S protein RBD, and the S protein N-terminal domain (NTD)(23). As such, the S protein may experience greater pressure to mutate from one virus strain to another, and, thus, the T cell epitopes identified within S, are more likely to be unique to SARS-CoV-2 than those from other structural proteins. Indeed, the sequence identity between SARS-CoV and SARS-CoV-2 is 91% for both the membrane (M), nucleocapsid (N) proteins, whereas it is only 76% for S, and 73% for the RBD. In spite of this, we identified one epitope shared by the two viruses within the RBD, S511–518. Two of the five major epitopes (S337–351 and S401–415) had high sequence homology but were not known to the authors to be previously described epitopes for SARS-CoV. In addition, we identified two unique CD8 T cell epitopes – the sequence homology at S1–14,319 and S477–491 is only 50% and 40%, respectively, between SARS-CoV and SARS-CoV-2. Although the minimal epitope within S1–14,319, could comprise a hybrid peptide between the signal peptide and the RBD, not seen in natural infection, this is unlikely, given that NetH2pan predictions only predict MHCI binding for S1–8, and S3–10.
The combination of both conserved and unique epitopes within the RBD of the S protein may foster future investigations into serial infections using SARS-CoV and SARS-CoV-2 in either mouse-adapted coronavirus strains, or hACE2-expressing C57BL/6 mice. During infection, CD8 T cell responses to additional structural and non-structural proteins will undoubtedly also arise, as recently reported for the nucleocapsid protein(22), and each may contribute to viral control. Moreover, infection may elicit CD8 T cell responses to these epitopes to varying degrees compared to what we have reported here for vaccination, especially as CD8 T cells responding to immunogenic epitopes within other proteins compete for immunodominance. Nonetheless, we expect one or more of these epitopes to be involved in the infectious response, and we hope the data reported here will be a useful resource, reducing the financial and practical threshold for new studies of SARS-CoV-2 infection or vaccination in mice.
Acknowledgements
We would like to thank Lori Sherman and the CU Cancer Center Cell Technologies Shared Resource for producing the SARS-CoV-2 spike RBD protein. We would also like to thank Timothy Davis and the Peptide Core Facility at the University of Colorado Anschutz for technical assistance.
This work was supported by funds from the University of Colorado School of Medicine and the National Institutes of Health via National Institute of Allergy and Infectious Diseases grant AI148919 (R.M.K.).
References
- 1.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, and Zaks T. 2021. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med 384: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson EJ, Rouphael NG, Widge AT, Jackson LA, Roberts PC, Makhene M, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott AB, Flach B, Lin BC, Doria-Rose NA, O’Dell S, Schmidt SD, Corbett KS, Swanson PA, Padilla M, Neuzil KM, Bennett H, Leav B, Makowski M, Albert J, Cross K, Edara VV, Floyd K, Suthar MS, Martinez DR, Baric R, Buchanan W, Luke CJ, Phadke VK, Rostad CA, Ledgerwood JE, Graham BS, and Beigel JH. 2020. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med 383: 2427–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, and Gruber WC. 2020. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med 383: 2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krammer F 2020. SARS-CoV-2 vaccines in development. Nature 586: 516–527. [DOI] [PubMed] [Google Scholar]
- 5.Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott A, Flach B, Doria-Rose NA, Corbett KS, Morabito KM, O’Dell S, Schmidt SD, Swanson PA, Padilla M, Mascola JR, Neuzil KM, Bennett H, Sun W, Peters E, Makowski M, Albert J, Cross K, Buchanan W, Pikaart-Tautges R, Ledgerwood JE, Graham BS, and Beigel JH. 2020. An mRNA Vaccine against SARS-CoV-2 — Preliminary Report. N. Engl. J. Med 383: 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, Baum A, Pascal K, Quandt J, Maurus D, Brachtendorf S, Lörks V, Sikorski J, Hilker R, Becker D, Eller AK, Grützner J, Boesler C, Rosenbaum C, Kühnle MC, Luxemburger U, Kemmer-Brück A, Langer D, Bexon M, Bolte S, Karikó K, Palanche T, Fischer B, Schultz A, Shi PY, Fontes-Garfias C, Perez JL, Swanson KA, Loschko J, Scully IL, Cutler M, Kalina W, Kyratsous CA, Cooper D, Dormitzer PR, Jansen KU, and Türeci Ö. 2020. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 586: 594–599. [DOI] [PubMed] [Google Scholar]
- 7.Ewer KJ, Barrett JR, Belij-Rammerstorfer S, Sharpe H, Makinson R, Morter R, Flaxman A, Wright D, Bellamy D, Bittaye M, Dold C, Provine NM, Aboagye J, Fowler J, Silk SE, Alderson J, Aley PK, Angus B, Berrie E, Bibi S, Cicconi P, Clutterbuck EA, Chelysheva I, Folegatti PM, Fuskova M, Green CM, Jenkin D, Kerridge S, Lawrie A, Minassian AM, Moore M, Mujadidi Y, Plested E, Poulton I, Ramasamy MN, Robinson H, Song R, Snape MD, Tarrant R, Voysey M, Watson MEE, Douglas AD, Hill AVS, Gilbert SC, Pollard AJ, Lambe T, and Oxford COVID Vaccine Trial Group. 2021. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat. Med 27: 270–278. [DOI] [PubMed] [Google Scholar]
- 8.Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J, Liu J, Peter L, McMahan K, Tostanoski LH, He X, Martinez DR, Rutten L, Bos R, van Manen D, Vellinga J, Custers J, Langedijk JP, Kwaks T, Bakkers MJG, Zuijdgeest D, Rosendahl Huber SK, Atyeo C, Fischinger S, Burke JS, Feldman J, Hauser BM, Caradonna TM, Bondzie EA, Dagotto G, Gebre MS, Hoffman E, Jacob-Dolan C, Kirilova M, Li Z, Lin Z, Mahrokhian SH, Maxfield LF, Nampanya F, Nityanandam R, Nkolola JP, Patel S, Ventura JD, Verrington K, Wan H, Pessaint L, Van Ry A, Blade K, Strasbaugh A, Cabus M, Brown R, Cook A, Zouantchangadou S, Teow E, Andersen H, Lewis MG, Cai Y, Chen B, Schmidt AG, Reeves RK, Baric RS, Lauffenburger DA, Alter G, Stoffels P, Mammen M, Van Hoof J, Schuitemaker H, and Barouch DH. 2020. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 586: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, Belanger S, Abbott RK, Kim C, Choi J, Kato Y, Crotty EG, Kim C, Rawlings SA, Mateus J, Tse LPV, Frazier A, Baric R, Peters B, Greenbaum J, Ollmann Saphire E, Smith DM, Sette A, and Crotty S. 2020. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 183: 996–1012.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahonen CL, Doxsee CL, McGurran SM, Riter TR, Wade WF, Barth RJ, Vasilakos JP, Noelle RJ, and Kedl RM. 2004. Combined TLR and CD40 Triggering Induces Potent CD8 + T Cell Expansion with Variable Dependence on Type I IFN. J. Exp. Med 199: 775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Arunkumar GA, Jurczyszak D, Polanco J, Bermudez-Gonzalez M, Kleiner G, Aydillo T, Miorin L, Fierer DS, Lugo LA, Kojic EM, Stoever J, Liu STH, Cunningham-Rundles C, Felgner PL, Moran T, García-Sastre A, Caplivski D, Cheng AC, Kedzierska K, Vapalahti O, Hepojoki JM, Simon V, and Krammer F. 2020. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med 26: 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ljunggren H-G, Öhlén C, Höglund P, Franksson L, and Kärre K. 1991. The RMA-S lymphoma mutant; consequences of a peptide loading defect on immunological recognition and graft rejection. Int. J. Cancer 47: 38–44. [DOI] [PubMed] [Google Scholar]
- 13.Ross P, Holmes JC, Gojanovich GS, and Hess PR. 2012. A cell-based MHC stabilization assay for the detection of peptide binding to the canine classical class I molecule, DLA-88. Vet. Immunol. Immunopathol 150: 206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J, Zhao J, and Perlman S. 2010. T Cell Responses Are Required for Protection from Clinical Disease and for Virus Clearance in Severe Acute Respiratory Syndrome Coronavirus-Infected Mice. J. Virol 84: 9318–9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith TRF, Patel A, Ramos S, Elwood D, Zhu X, Yan J, Gary EN, Walker SN, Schultheis K, Purwar M, Xu Z, Walters J, Bhojnagarwala P, Yang M, Chokkalingam N, Pezzoli P, Parzych E, Reuschel EL, Doan A, Tursi N, Vasquez M, Choi J, Tello-Ruiz E, Maricic I, Bah MA, Wu Y, Amante D, Park DH, Dia Y, Ali AR, Zaidi FI, Generotti A, Kim KY, Herring TA, Reeder S, Andrade VM, Buttigieg K, Zhao G, Wu JM, Li D, Bao L, Liu J, Deng W, Qin C, Brown AS, Khoshnejad M, Wang N, Chu J, Wrapp D, McLellan JS, Muthumani K, Wang B, Carroll MW, Kim JJ, Boyer J, Kulp DW, Humeau LMPF, Weiner DB, and Broderick KE. 2020. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun 11: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeVette CI, Andreatta M, Bardet W, Cate SJ, Jurtz VI, Jackson KW, Welm AL, Nielsen M, and Hildebrand WH. 2018. NetH2pan: A computational tool to guide MHC peptide prediction on murine tumors. Cancer Immunol. Res 6: 636–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potter TA, Grebe K, Freiberg B, and Kupfer A. 2001. Formation of supramolecular activation clusters on fresh ex vivo CD8+ T cells after engagement of the T cell antigen receptor and CD8 by antigen-presenting cells. Proc. Natl. Acad. Sci. U. S. A 98: 12624–12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, Hemmings O, O’Byrne A, Kouphou N, Galao RP, Betancor G, Wilson HD, Signell AW, Winstone H, Kerridge C, Huettner I, Jimenez-Guardeño JM, Lista MJ, Temperton N, Snell LB, Bisnauthsing K, Moore A, Green A, Martinez L, Stokes B, Honey J, Izquierdo-Barras A, Arbane G, Patel A, Tan MKI, O’Connell L, O’Hara G, MacMahon E, Douthwaite S, Nebbia G, Batra R, Martinez-Nunez R, Shankar-Hari M, Edgeworth JD, Neil SJD, Malim MH, and Doores KJ. 2020. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol 5: 1598–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, McMahon M, Meade P, Mendu DR, Muellers K, Stadlbauer D, Stone K, Strohmeier S, Simon V, Aberg J, Reich DL, Krammer F, and Cordon-Cardo C. 2020. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 370: 1227–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lumley SF, O’Donnell D, Stoesser NE, Matthews PC, Howarth A, Hatch SB, Marsden BD, Cox S, James T, Warren F, Peck LJ, Ritter TG, de Toledo Z, Warren L, Axten D, Cornall RJ, Jones EY, Stuart DI, Screaton G, Ebner D, Hoosdally S, Chand M, Crook DW, O’Donnell A-M, Conlon CP, Pouwels KB, Walker AS, Peto TEA, Hopkins S, Walker TM, Jeffery K, and Eyre DW. 2021. Antibody Status and Incidence of SARS-CoV-2 Infection in Health Care Workers. N. Engl. J. Med 384: 533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, and Sette A. 2020. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 181: 1489–1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joag V, Wijeyesinghe S, Stolley JM, Quarnstrom CF, Dileepan T, Soerens AG, Sangala JA, O’Flanagan SD, V Gavil N, Hong S, Bhela S, Gangadhara S, Weyu E, Matchett WE, Thiede J, Krishna V, Cheeran MC-J, Bold TD, Amara R, Southern P, Hart GT, Schifanella L, Vezys V, Jenkins MK, Langlois RA, and Masopust D. 2021. Cutting Edge: Mouse SARS-CoV-2 Epitope Reveals Infection and Vaccine-Elicited CD8 T Cell Responses. J. Immunol 206: 931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang Q, Luo Y, Chan JFW, Sahi V, Figueroa A, Guo XV, Cerutti G, Bimela J, Gorman J, Zhou T, Chen Z, Yuen KY, Kwong PD, Sodroski JG, Yin MT, Sheng Z, Huang Y, Shapiro L, and Ho DD. 2020. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584: 450–456. [DOI] [PubMed] [Google Scholar]


