Abstract
Local translation can provide a rapid, spatially targeted supply of new proteins in distal dendrites to support synaptic changes that underlie learning. Learning and memory are especially sensitive to manipulations of translational control mechanisms, particularly those that target the initiation step, and translation initiation at synapses could be a means of maintaining synapse specificity during plasticity. Initiation predominantly occurs via recruitment of ribosomes to the 5’ mRNA cap by complexes of eukaryotic initiation factors (eIFs), and the interaction between eIF4E and eIF4G1 is a particularly important target of translational control pathways. Pharmacological inhibition of eIF4E-eIF4G1 binding impairs formation of memory for aversive Pavlovian conditioning as well as the accompanying increase in polyribosomes in the heads of dendritic spines in the lateral amygdala (LA). This is consistent with a role for initiation at synapses in memory formation, but whether eIFs are even present near synapses is unknown. To determine whether dendritic spines contain eIFs and whether eIF distribution is affected by learning, we combined immunolabeling with serial section transmission electron microscopy (ssTEM) volume reconstructions of LA dendrites after Pavlovian conditioning. Labeling for eIF4E, eIF4G1, and eIF2α – another key target of regulation – occurred in roughly half of dendritic spines, but learning effects were only found for eIF4E, which was upregulated in the heads of dendritic spines. Our results support the possibility of regulated translation initiation as a means of synapse-specific protein targeting during learning and are consistent with the model of eIF4E availability as a central point of control.
Keywords: eukaryotic initiation factor, electron microscopy, polyribosome, dendritic spine, learning and memory, amygdala, fear conditioning
Graphical Abstract

Memory formation requires initiation of new protein synthesis as well as protein synthesis in dendrites, but it is unknown whether initiation can happen within dendritic spines. We combined immunolabeling with serial section 3D electron microscopy to examine the distribution of three initiation factors, eIF4E, eIF4G1, and eIF2α in dendritic spines of the lateral amygdala during formation of a Pavlovian conditioning memory. Top row: Electron micrographs showing labeled spines (arrows) with synapses (arrowheads). Bottom left: Segment of dendrite reconstructed from serial sections with synapses shown in red. Bottom right: The percentage of spines containing labeling for eIF4E was higher after training, but there were no differences for eIF4G1 or eIF2α labeling.
Introduction
Protein synthesis in the time period immediately after learning is necessary for supporting memory-related synapse remodeling (Davis and Squire, 1984; Mayford et al., 2012; Rosenberg et al., 2014; Segal, 2017). Translation is heavily regulated, and it is now clear that learning does not simply require new proteins, but instead relies on an orchestrated balance of translational control mechanisms (Kelleher and Bear, 2008; Groppo and Richter, 2009; Darnell, 2011; Buffington et al., 2014; Santini et al., 2014). Translation occurs in neuronal processes, where it may serve as a means of spatiotemporal protein targeting at synaptic sites (Wang et al., 2010; Liu-Yesucevitz et al., 2011; Holt and Schuman, 2013; Rangaraju et al., 2017). The presence of polyribosomes and mRNA in dendritic spines suggests that local translation could be regulated at the level of individual synapses, potentially helping to maintain synapse specificity during learning-related synaptic plasticity (Steward and Levy, 1982; Tiruchinapalli et al., 2003; Kao et al., 2010; Ostroff et al., 2010; Dynes and Steward, 2012). There is evidence that local translation plays a role in learning and memory, but this comes mainly from studies of mRNA trafficking (Miller et al., 2002; Nakayama et al., 2017; Roy et al., 2020). In addition, the spatial scale of translational control in neurons is unknown (Rangaraju et al., 2017) and whether translation regulation mechanisms could mediate synapse-specific protein targeting – perhaps explaining their particular importance in learning and memory – is an open question.
The most complex regulation of translation is at the initiation step, in which ribosomes and the initiator tRNA are recruited to the target mRNA strand. Initiation is coordinated by a family of eukaryotic initiation factors (eIFs) and begins with the formation of a complex between eIF4E and eIF4G, which then binds the 5’ cap of the target mRNA and recruits a complex of the 40S ribosome and Met-tRNA bound to eIF2. The two major targets of regulation are eIF4E, whose availability to bind to eIF4G is controlled by repressor proteins, and the activity of eIF2, which is controlled by phosphorylation of its α subunit (Groppo and Richter, 2009; Sonenberg and Hinnebusch, 2009; Borden and Volpon, 2020). Dysfunction of eIF4E or eIF2α leads to impairment of various forms of learning and learning-related synaptic plasticity (Gkogkas et al., 2010; Buffington et al., 2014; Santini et al., 2014) as well as intellectual functions in humans (Kelleher and Bear, 2008; Darnell, 2011; Kapur et al., 2017). A variety of mechanisms are known to transport mRNA into dendrites and maintain it in a dormant state (Hutten et al., 2014; Buxbaum et al., 2015), and a single synapse could be the locus of translational control if initiation occurs on dormant mRNA in dendritic spines. Consistent with this possibility, we have found by serial section transmission electron microscopy (ssTEM) reconstruction that polyribosomes are upregulated in dendritic spines of the rat lateral amygdala (LA) during consolidation of aversive Pavlovian conditioning (Ostroff et al., 2010), and that the drug 4EGI-1, which inhibits initiation by interfering with the eIF4E-eIF4G interaction, impairs both formation of the memory and the associated polyribosome upregulation (Hoeffer et al., 2011; Ostroff et al., 2017).
Initiation at individual synapses is not the only explanation for this, however. Ribosomes can be stalled on mRNAs after initiation, and most dendritic polyribosomes in cultured neurons in fact appear to be stalled (Richter and Coller, 2015; Langille et al., 2019). Meanwhile, there is also evidence that actively translating mRNAs are mobile in cultured dendrites (Wu et al., 2016). Polyribosomes, either stalled or active, could be transported into spines by mRNA trafficking mechanisms, making mRNA transport, not initiation, the key regulator of synapse-specific protein targeting. Dendritic spines in cultured neurons and cortex have been reported to contain immunolabeling for eIF4E (Asaki et al., 2003; Smart et al., 2003), but whether other eIFs are present and how they are distributed among spines is unknown. To determine whether eIFs are present in LA spines and whether they are redistributed during memory formation, we combined ssTEM with immunohistochemistry for eIFs in LA dendrites following Pavlovian conditioning. We found that eIFs are indeed present in dendritic spines, and that their distribution after learning is consistent with cap-dependent initiation in spines.
Materials and Methods
Subjects and behavior:
Adult male Sprague-Dawley rats (Hilltop) weighing approximately 300g were housed singly on a 12-hour light cycle with food and water ad libitum. Experiments were conducted during the animals’ light cycle. Behavior experiments were conducted as previously described (Ostroff et al. 2010). Briefly, animals were habituated on two consecutive days to a lit conditioning chamber equipped with a speaker and a grid floor. On the third day, one group of animals were trained with five auditory tones (30 s, 5 kHz, 80 dB) co-terminating in a mild footshock (0.7 mA, 1 s), while a control group was placed in the chamber but not presented with tones or shocks. All procedures were approved by the Institutional Animal Care and Use Committee of New York University.
Antibodies:
The following antibodies and dilutions were used: Bethyl Labs rabbit polyclonal anti-eIF4E (A301-154A; lot# A301-154A-1; RRID:AB_2097708) at 1:500, Abnova mouse polyclonal anti-eIF4G1 (H00001981-A01; lot# 08213-2A9; RRID:AB_462490) at 1:500; Cell Signaling mouse monoclonal anti-eIF2α (L57A5; lot# 3; RRID:AB_836874) at 1:500; Vector Laboratories biotinylated polyclonal goat anti-rabbit (BA-1000; RRID:AB_2313606) or goat anti-mouse (BA-9200; RRID:AB_2336171) secondary antibodies at 1:200. Preadsorption controls were run for the two polyclonal primary antibodies by incubating them for 1 hour in a 10:1 (by weight) excess of immunizing peptide before use. Preadsorption eliminated 98% of labeled structures, as described previously (Ostroff et al., 2019).
Immunolabeling and electron microscopy:
For eIF4E and eIF4G1 labeling, rats were deeply anesthetized with chloral hydrate and perfused transcardially with ~50 ml of heparinized saline (154mM NaCl/2mM CaCl2/4mM MgCl2 in 0.01M PIPES buffer at pH 7.4) followed by 500ml of perfusion fixative (0.5% gluteraldehyde/4% paraformaldehyde/2mM CaCl2/4mM MgCl2 in 0.1M PIPES buffer, pH7.4). For eIF2α labeling, the heparinized saline was 0.01M phosphate buffer with 154 mM NaCl, and the perfusion fixative was 0.25% glutaraldehyde/4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4. All fixation and labeling procedures were carried out at room temperature. Brains were rinsed in buffered saline (0.01 M fixation buffer with 154 mM NaCl) and sectioned coronally at 40um. Sections from the left hemisphere containing the caudal LA were reacted for 30 minutes with 1% sodium borohydride then blocked in 1% BSA in buffered saline and incubated overnight in primary antibody in 1% BSA. Labeling for eIF4E and eIF4G1 was performed on alternating sections from the same rats. Sections were incubated for 30min in secondary antibody followed by 30 min in Vector Laboratories ABC-peroxidase (PK-6100; RRID:AB_2336819), then reacted with 1mM 3,3’-diaminobenzidine in 0.003% H2O2 for 5 min. The LA was dissected out of each section and processed for serial EM as previously described (Ostroff et al., 2010). Briefly, sections were postfixed in reduced osmium (1.5% potassium ferrocyanide/1% osmium tetroxide) followed by 1% osmium tetroxide, dehydrated in ethanol containing 1.5% uranyl acetate, and embedded in epon resin (LX-112, Ladd Industries). Serial 45 nm sections were cut on an ultramicrotome (Leica) in the ventral-to-dorsal direction, perpendicular to the plane of vibratome sectioning. Sections were picked up on pioloform-coated slot grids and imaged at 100kV on a JEOL 1200EX II electron microscope with an AMT digital camera. Grids were not counterstained to avoid the possibility of staining artifacts obscuring immunolabel.
Reconstruction and analysis:
Image alignment, reconstruction, and measurements were done using Reconstruct software (Fiala, 2005; RRID:SCR_002716) with experimenters blind to experimental condition. Series consisted of an average of 99 ± 4 sections. All dendrites on the central section of each series were examined and segments of spiny dendrites with a diameter less than 1μm (average 0.68 ± 0.02 μm) and length between 2 and 7 μm (average 3.87 ± 0.12) were reconstructed. There were no significant differences between antibodies or groups in either measure. Frequencies were calculated using inclusion lengths measured from the first complete protrusion at the ventral end of the series to the final protrusion at the dorsal end. Synapse morphology was designated as asymmetric (putatively excitatory) or symmetric (inhibitory or modulatory) according to established morphological criteria (Peters et al., 1991). Approximately 11% of dendritic protrusion origins gave rise to more than one protrusion. These branched protrusions did not differ in any measures from protrusions arising singly, thus branches were treated as single protrusions for analysis. Approximately 2% of dendritic protrusions carried more than one asymmetric synapse, 4% carried both a symmetric and an asymmetric synapse, and 4% lacked a synapse. Spines were defined as protrusions carrying at least one synapse, and protrusions lacking synapses were excluded from spine analyses. For analysis of sub-regions of spines, the base was defined as the plane where the spine neck arises from the dendrite and a boundary of 150nm on either side was used to assign labeling to the shaft, base, or neck.
Statistics:
Analyses were conducted with STATISTICA software (Tibco Inc.). For all comparisons, means were compared using hierarchical ANOVAs with subjects nested into group to test for significant interactions. The breakdown of dendrite and spine numbers by rat, group, and antibody are given in Table 1. Bar graphs show means and standard errors, and exact p, F, and η2 values are given in Table 2.
Table 1.
Composition of dataset
| Group | # of Subjects | Antibody | # of Dendrites | # of Spines | spines/μm (mean ± std. err.) |
|---|---|---|---|---|---|
| control | 3 | eIF4E eIF4G1 |
17 (7, 6, 4) 13 (4, 5, 4) |
225 (85, 60, 80) 185 (59, 40, 86) |
2.70 ± 0.26 2.66 ± 0.34 |
| trained | 3 | eIF4E eIF4G1 |
23 (9, 6, 8) 17 (6, 6, 5) |
257 (101, 59, 97) 193 (72, 74, 47) |
2.83 ± 0.22 2.89 ± 0.26 |
| control | 2 | eIF2α | 7 (4, 3) | 82 (50, 32) | 2.78 ± 0.44 |
| trained | 2 | eIF2α | 9 (6, 3) | 117 (78, 39) | 2.06 ± 0.44 |
Table 2.
Effects of training on eIF4E labeling
| Figure | Variable (spines / μm) | Training effect | Subject (training) effect | ||||
|---|---|---|---|---|---|---|---|
| F | p | Partial η2 | F | p | Partial η2 | ||
| 2E | labeled | 12.53 | 0.0012 | 0.269 | 0.79 | 0.5416 | 0.085 |
| 2E | unlabeled | 7.52 | 0.0097 | 0.181 | 2.19 | 0.0906 | 0.205 |
| 2H | labeled heads | 14.11 | 0.0006 | 0.293 | 1.71 | 0.1706 | 0.167 |
| 2H | labeled necks | 8.45 | 0.0064 | 0.199 | 3.15 | 0.0263 | 0.271 |
| 2I | labeled in one location | 12.77 | 0.0011 | 0.273 | 0.69 | 0.6033 | 0.075 |
| 2I | labeled in >1 location | 6.76 | 0.0137 | 0.166 | 3.04 | 0.0304 | 0.263 |
| 2J | labeled head only | 6.02 | 0.0194 | 0.151 | 0.53 | 0.7162 | 0.058 |
| 2K | labeled head and neck | 7.28 | 0.0108 | 0.176 | 2.62 | 0.0520 | 0.236 |
| 3C | percent spines labeled | 18.53 | 0.0001 | 0.353 | 0.80 | 0.5306 | 0.087 |
| 3D | percent spine heads labeled | 16.85 | 0.0002 | 0.319 | 1.44 | 0.2408 | 0.138 |
| 3D | percent spine necks labeled | 8.37 | 0.0064 | 0.189 | 2.09 | 0.1018 | 0.189 |
| 4B | labeled, PSD < 0.1 μm2 | 8.41 | 0.0065 | 0.198 | 0.22 | 0.9228 | 0.026 |
| 4B | labeled, PSD > 0.2 μm2 | 8.54 | 0.0061 | 0.201 | 1.43 | 0.2437 | 0.144 |
| 4D | unlabeled, PSD < 0.15 μm2 | 4.47 | 0.0420 | 0.116 | 1.71 | 0.1703 | 0.168 |
| 4F | labeled head, PSD < 0.1 μm2 | 5.37 | 0.0266 | 0.136 | 0.49 | 0.7438 | 0.054 |
| 4F | labeled head, PSD > 0.2 μm2 | 8.54 | 0.0061 | 0.201 | 1.43 | 0.2437 | 0.144 |
| 5D | labeled, with spine apparatus | 10.93 | 0.0022 | 0.243 | 1.60 | 0.1979 | 0.158 |
| 5D | unlabeled, with spine apparatus | 5.37 | 0.0266 | 0.137 | 2.38 | 0.0710 | 0.219 |
| 5D | labeled, without spine apparatus | 4.78 | 0.0357 | 0.123 | 0.55 | 0.6997 | 0.061 |
| 5D | unlabeled, without spine apparatus | 4.19 | 0.0485 | 0.110 | 1.27 | 0.3026 | 0.130 |
| 5F | head labeled, with spine apparatus | 10.93 | 0.0022 | 0.243 | 2.06 | 0.1083 | 0.195 |
| 5F | neck labeled, with spine apparatus | 13.17 | 0.0009 | 0.279 | 6.32 | 0.0006 | 0.427 |
| 5F | base labeled, with spine apparatus | 5.39 | 0.0264 | 0.137 | 2.08 | 0.1056 | 0.196 |
| 5F | head labeled, without spine apparatus | 4.37 | 0.0442 | 0.114 | 1.61 | 0.1954 | 0.159 |
| 5I | labeled, with spine apparatus, PSD < 0.15 μm2 | 4.40 | 0.0435 | 0.114 | 0.94 | 0.4548 | 0.099 |
| 5F | labeled, with spine apparatus, PSD > 0.2 μm2 | 8.54 | 0.0061 | 0.201 | 1.43 | 0.2437 | 0.144 |
Results
To investigate dendritic eIF distribution during memory consolidation, we used aversive Pavlovian conditioning (also known as threat or fear conditioning), an extensively studied behavioral paradigm which produces robust learning in a single training session (LeDoux, 2000; Maren, 2001). Formation of the long-term memory is impaired by intra-LA infusion of general protein synthesis inhibitors (Schafe and LeDoux, 2000; Maren et al., 2003) or 4EGI-1 (Hoeffer et al., 2011) immediately after learning. The memory is resistant to a protein synthesis inhibitor infused after 6 hours, confirming the importance of translation in the first few hours (Schafe and LeDoux, 2000). The temporal dynamics of learning-induced translation in the LA have not been worked out in detail, but studies of memory formation in hippocampus consistently report a protein synthesis-dependent phase lasting less than three hours after learning, followed sometimes by a second phase whose timing varies between learning paradigms (Bourtchouladze et al., 1998; Quevedo et al., 1999). A variety of signaling pathways induced by a single trial of avoidance learning peak within 30 to 90 minutes across several brain areas but diverge thereafter, further supporting the importance of molecular changes during the first hours (Izquierdo et al., 2006).
To capture eIF distribution associated with this period of active memory formation, we trained animals with five tone-shock pairings and collected tissue 1 hour after the first pairing, which was 32.5 minutes after the last pairing. In previous work, we found an increase in polyribosomes in spines after paired training relative to training with unpaired shocks and tones or control exposure to the conditioning box (Ostroff et al., 2010, 2017). Polyribosome distribution did not differ between unpaired training and box exposure, but unpaired training did produce aversive conditioning to the chamber and inhibitory conditioning to the tone, alongside reduced synapse size relative to the box control group. It is possible that unpaired tones and shocks would result in eIF redistribution in the absence of polyribosome redistribution, and a non-learning group would be necessary to detect this. Because ssTEM entails a large investment in each image set, there are practical limits on the number of animals that can be included in a study. To get a fuller picture of eIF distribution, we chose to prioritize additional antibodies over additional behavioral groups. Conditioning box exposure was used as the behavioral control for paired training, as exposure to a familiar chamber does not induce learning. The experimental workflow is shown in Figure 1a. Freezing during the tone was scored to confirm acquisition of the tone-shock association in the trained group (Figure 1b).
Figure 1.
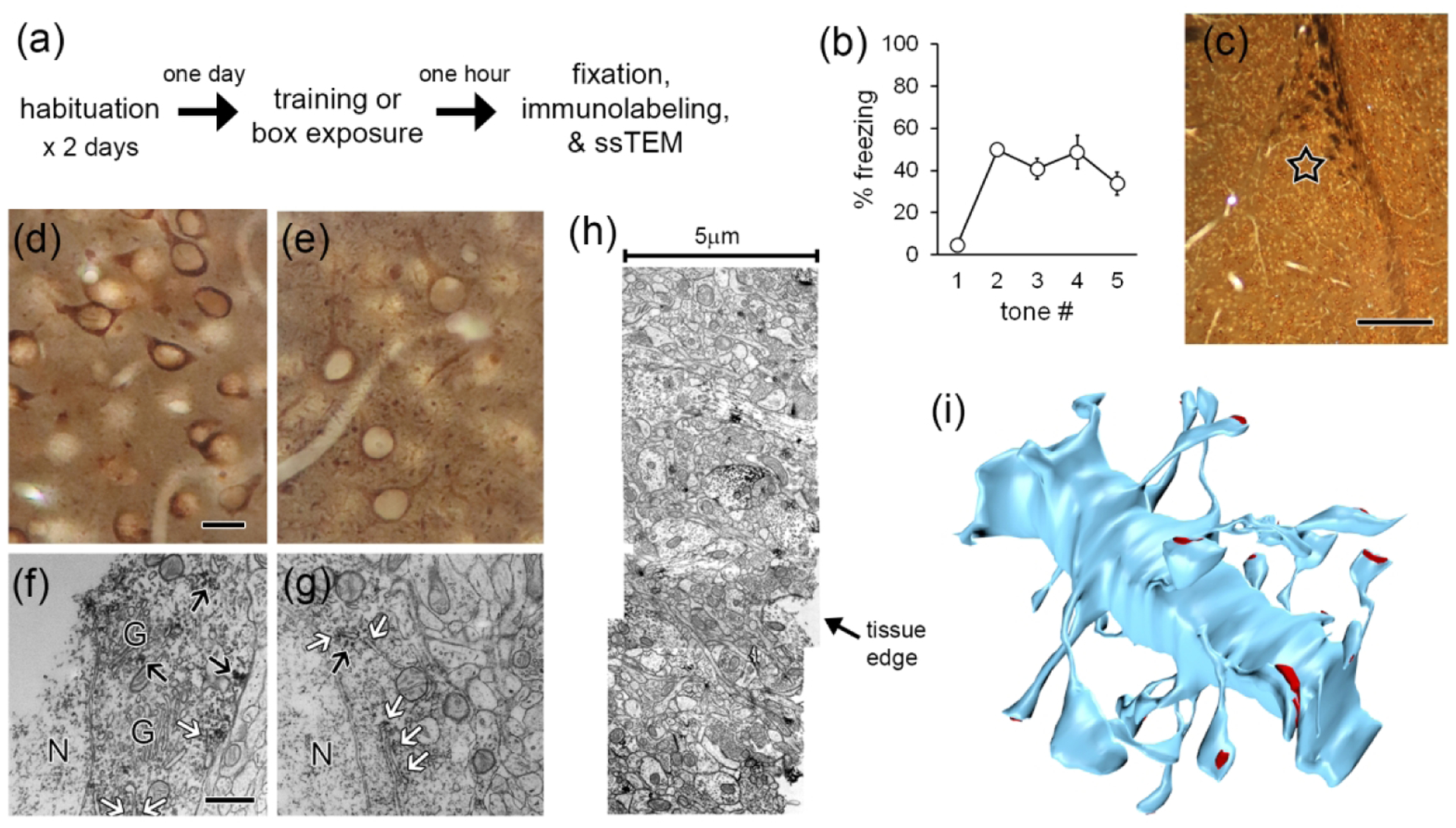
Methods. a) Experimental workflow. b) Freezing to tones during the training session by the rats used for ssTEM. c) Lateral amygdala section immunolabeled for eIF4E and embedded for EM, showing location of sampling for ssTEM (star). d,e) Brightfield image of LA cells labeled for eIF4E (d) and eIF4G1 (e) in EM sample blocks. f,g) Electron micrographs of cell bodies showing labeling for eIF4E (f) and eIF4G1 (g) on rough endoplasmic reticulum (white arrows) and on cytoplasmic polyribosomes (black arrows). h) Montage of three sections along edge of tissue labeled for eIF4E. i) Dendrite from eIF4E labeled tissue reconstructed by ssTEM. Synapses shown in red. Scale = 250 mm in (c); 20 mm in (d,e); 500 nm in (f,g).
Excitatory projection neurons in the LA have spiny dendrites while aspinous dendrites belong to inhibitory interneurons (McDonald, 1982, 1992; McDonald and Pearson, 1989), so the presence of spines ensured that only excitatory neuron dendrites were analyzed. To determine whether eIF4E and eIF4G are present in excitatory LA dendrites and spines, we performed pre-embedding immunohistochemistry for these proteins on alternating sections before processing the tissue for ssTEM. A region of the dorsolateral subdivision of the LA at a mid-caudal level was chosen for ssTEM (Figure 1c). For optimal sensitivity of immunolabeling, we used a chromogenic detection method and only used one antibody on each tissue section. Although this approach does not reveal colocalization, it maximizes the likelihood of detecting labeled spines, as the antibodies and labeling reagents do not interfere with each other. DAB was chosen as the chromogen because it produces a diffuse reaction product that tends to spread across serial sections, in contrast to the more compact crystals formed by VIP or tetramethylbenzidine. It is therefore easier to detect DAB in small spines, as a single crystal of another label could be ambiguous. DAB labeling cannot be directly quantified, but our goal was to assess the presence and distribution of eIFs in spines, so we prioritized sensitivity over ease of quantification.
For both eIF4E and eIF4G1, polyclonal antibodies raised against the C-terminal region were used. The site of interaction between eIF4E and eIF4G1 is not near the C-terminal of either molecule (Mader et al., 1995; Morino et al., 2000; Grüner et al., 2016), and it is therefore possible that the antibodies recognize both the unbound proteins and the eIF4F complex. The 4E binding proteins (4E-BPs), which are major repressors of eIF4E activity, bind at the eIF4G binding site and similarly should not directly block the C-terminus (Marcotrigiano et al., 1999; Tomoo et al., 2005). The C-terminus of eIF4E is part of the cap-binding pocket and is deflected, but not occluded, by binding to mRNA (Marcotrigiano et al., 1997; Tomoo et al., 2002). The C-terminus of eIF4G, meanwhile, does not bind the protein’s other major binding partner, eIF4A, and appears to be modulatory (Morino et al., 2000).
At the light level, labeling for both initiation factors was seen in the cytoplasm of the cell bodies, proximal dendrites, and processes throughout the neuropil, but not in the nuclei (Figure 1d,e). At the EM level, immunolabel in cell bodies was found associated with rough endoplasmic reticulum and with free polyribosomes as well as sporadically in the cytoplasm, but was conspicuously absent from the Golgi apparatus (Figure 1f,g). This staining pattern is consistent with expected subcellular sites of translation. Because the antibodies only penetrated a few microns into fixed tissue, analysis was restricted to the region closest to the vibratome edge. To maximize our usable image volumes, three adjacent fields along the tissue edge were imaged and montaged (Figure 1h). Spiny dendrites and their synapses were reconstructed in 3D (Figure 1i) and the distribution of immunolabel was quantified.
Learning-induced upregulation of eIF4E in dendritic spines
Immunolabeling for both eIF4E and eIF4G1 was visible in dendritic spine heads (Figure 2a,b), necks (Figure 2c), and bases (Figure 2d). Quantification of the frequency of labeled and unlabeled spines along dendritic segments in the control group revealed that a majority (57%) of spines were labeled for eIF4G1, but not for eIF4E (29%). The frequency of spines with eIF4E labeling was higher in the trained group and the frequency of unlabeled spines was lower, but there were no changes in eIF4G1 labeling (Figure 2e). The mean frequencies of spines with eIF4G1 labeling in both groups were nearly identical to the frequency of spines with eIF4E labeling in the trained group. Although this is intriguing to note, direct comparison between the two patterns could be misleading because the detection sensitivity of the antibodies may differ. The frequency of labeled and unlabeled spines was not correlated for either antibody in either condition (Figure 2f,g), suggesting that the presence of labeling is not directly regulated at the level of the dendritic segment. When the presence of labeling was broken down by location in spines, more spines with eIF4E labeling in the head and neck were found in the trained group, but there were no effects on eIF4G1 labeling (Figure 2h). In the trained group the most frequent location for eIF4E label was the spine head, while spine bases were more frequently labeled for eIF4G1.
Figure 2.
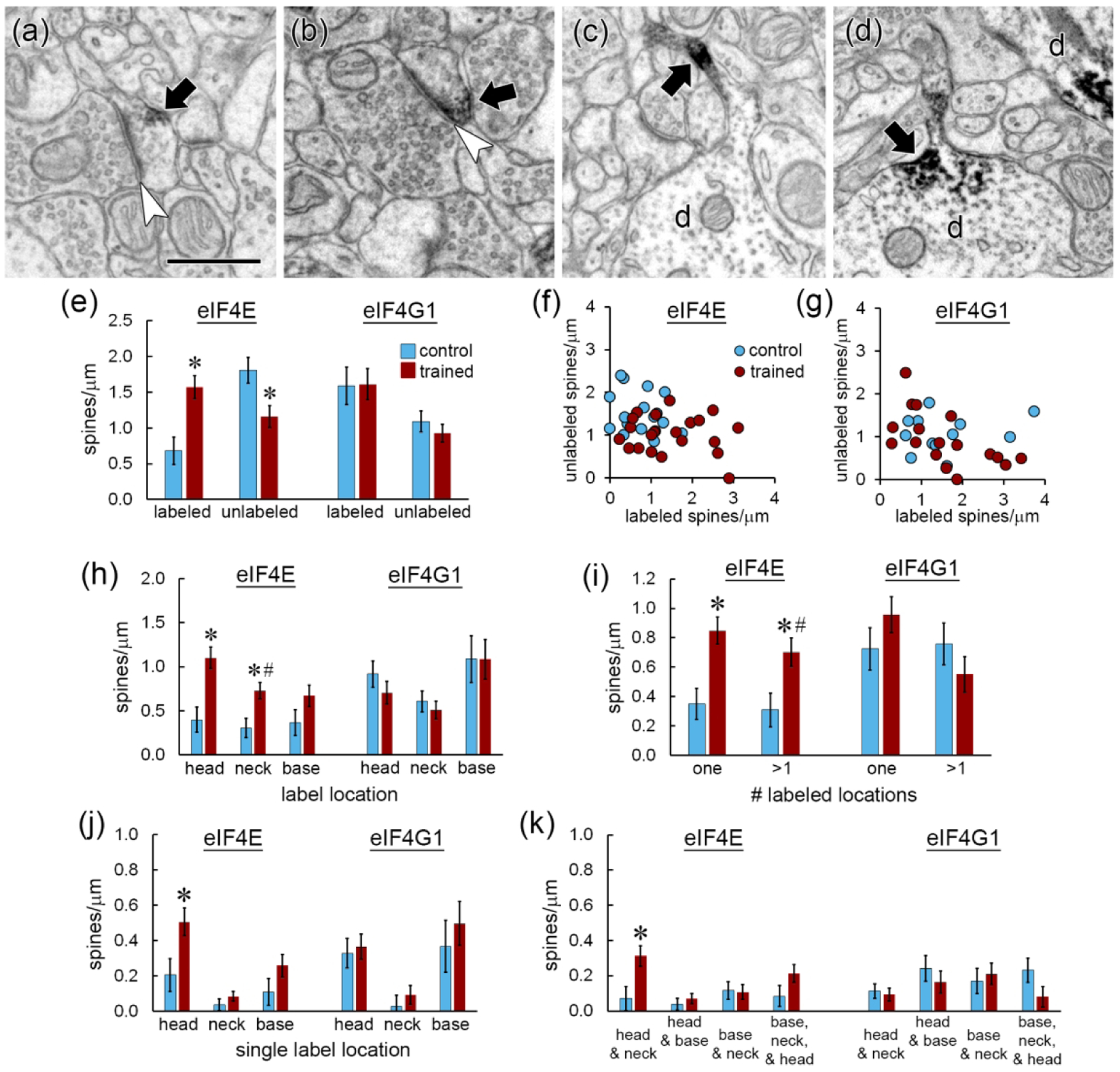
Distribution of immunolabel for eIF4E and eIF4G1. a,b) Immunolabel (arrows) for eIF4E (a) and eIF4G1 (b) in the heads of dendritic spines forming asymmetric synapses (arrowheads). c,d) Dendrites (d) with immunolabel (arrows) for eIF4E in a spine neck (c) and a spine base (d). Another dendrite (d) containing label in its shaft is visible in the upper right corner of (d). Scale = 500nm. e) The frequency of spines labeled for eIF4E, but not eIF4G, increased after learning, with a corresponding decrease in unlabeled spines. f,g) There were no correlations between the frequency of labeled and unlabeled spines for eIF4E (f) or eIF4G1 (g). h) The frequency of spine heads and necks labeled for eIF4E, but not eIF4G1, increased after learning. i) The frequency of spines with eIF4E label in one or multiple locations increased with learning. j) Among spines with only one labeled location, only eIF4E-labeled spine heads increased. k) Among spines with multiple labeled locations, spines with eIF4E in the head and neck were increased. * p < 0.05
To examine how labeling was distributed within spines, we classified labeled spines by the number of labeled locations. Labeling for eIF4E and eIF4G1 was commonly seen in multiple locations within a spine, with similar numbers of spines having label in single and multiple locations. More spines were found with both labeling patterns for eIF4E in the trained group (Figure 2i). Of the spines with label in a single location, the increase in eIF4E-labeled spines was specific to the head location (Figure 2j). Very few spines had labeling for either eIF4E or eIF4G1 in the neck only, suggesting that labeling in the neck reflects diffusion towards or away from accumulation in the base or head. Of spines with multiple labeled locations, only those with both head and neck label were significantly higher in the trained group (Figure 2k). Overall, these analyses show upregulation of spines with eIF4E labeling in the head, and sometimes also in the neck, during memory formation. In contrast, there were no group differences in spine labeling for eIF4G1.
Distribution of eIF2α in dendritic spines
In addition to eIF4E interaction, the second major target of translational control mechanisms is eIF2α, whose regulation has been implicated in amygdala-based memory (Trinh and Klann, 2013; Jian et al., 2014). To investigate whether the distribution of eIF2α in spines resembles that of eIF4E, we repeated the learning experiment on a new cohort of animals and immunolabeled the tissue for eIF2α. The antibody we used was raised against the full-length protein, so in theory it could recognize eIF2α whether free or in the preinitiation complex. Qualitatively, labeling for eIF2α resembled that of eIF4E and eIF4G1, with reaction product distributed throughout the cell bodies, proximal dendrites, and neuropil processes, but largely sparing the nucleus (Figure 3a). Like the other initiation factors, eIF2α labeling was found in dendritic spine heads (Figure 3b), as well as necks and bases. Because of the tissue preparation conditions required for sensitive labeling of eIF2α, the morphology of this material was relatively poorer and the yield of dendrites suitable for reconstruction was lower than in the eIF4E/eIF4G1 experiment. Many spines could not be unequivocally reconstructed along the dendrites thereby rendering absolute frequency measurements inaccurate, so we limited analysis of this material to percentages of labeled spines. Whereas the percentage of spines labeled for eIF4E was higher in the trained group, the percentage of spines labeled for eIF2α, like eIF4G1, was unchanged (Figure 3c). At the sub-spine level, the percentage of spine heads and necks with eIF4E label was higher in the trained group (Figure 3d), but there were no differences in any location for eIF4G1 (Figure 3e) or eIF2α (Figure 3f) labeling.
Figure 3.
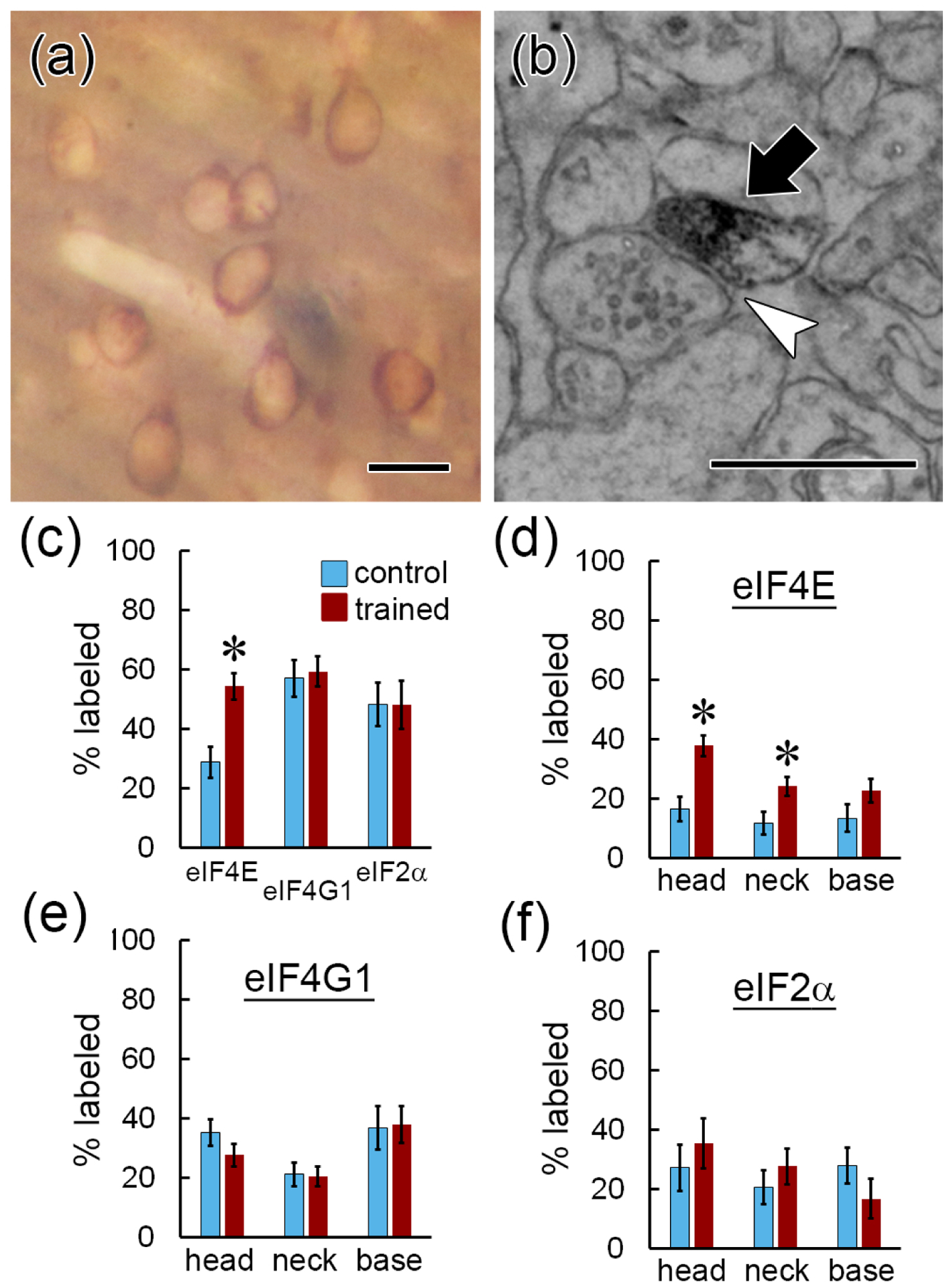
Comparison between eIF4E, eIF4G1, and eIF2α distribution. a) Brightfield image of eIF2α immunolabel in LA tissue prepared for EM. b) EM image of eIF2α immunolabeling in a dendritic spine head (arrow) forming an asymmetric synapse (arrowhead). Scale = 500nm. c) A higher percentage of dendritic spines are labeled for eIF4E, but not eIF4G1 or eIF2α, after learning. d–f) A higher percentage of dendritic spines heads and necks contain eIF4E (d), but not eIF4G1 (e) or eIF2α (f), after learning. * p < 0.05 Scale = 20mm in (a); 500 nm in (b).
Accumulation of eIF4E label in large and small spine heads
The spine enlargement that occurs with synapse strengthening is believed to be a substrate for long-term memories (Bourne and Harris, 2007; Kasai and Fukuda, 2010), and translation is necessary for plasticity-associated spine enlargement in the rat hippocampus (Fifkova et al., 1982; Tanaka et al., 2008). Proliferation of small spines is associated with eIF4E manipulation; small spine outgrowth was observed in the LA when 4EGI-1 was infused after training (Ostroff et al., 2017), but was also seen in the prefrontal cortex of mice overexpressing eIF4E (Santini et al., 2013). To determine whether eIF distribution differed between spines of different sizes, we measured the area of the post-synaptic density (PSD), which is correlated with spine volume and surface area in the LA (Ostroff et al., 2010). PSD area was quantified in two dimensions for all spines by directly measuring the area of PSDs cut en face or by measuring the length of cross-sectioned PSDs across serial sections as shown in Figure 4a. Synapse size ranged from 0.003 – 0.849 μm2 and did not follow a normal distribution, with nearly 90% of the synapses in the dataset smaller than 0.1 μm2. To determine how labeling for eIF4E and eIF4G1 were distributed across the range of spine sizes, spine frequency was binned. There were more eIF4E-labeled spines in the trained group at both the smaller (0.05 – 0.1 μm2) and largest (> 0.2 μm2) ends of the range (Figure 4b), and fewer unlabeled spines in the intermediate (0.1 – 0.15 μm2) range (Figure 4c). Spines with eIF4G1 labeling spanned the full range of synapse sizes, but there were no differences between the groups (Figure 4d). As seen for eIF4E, large spines lacking eIF4G1 label were very scarce (Figure 4e). When eIF4E-labeled spines were analyzed by size and label location, the trained group had more labeled spine heads in the 0.05 – 0.1 μm2 and >0.2 μm2 bins (Figure 4f), but no size-specific effects were seen in the neck (Figure 4g) or base (Figure 4h) locations.
Figure 4.
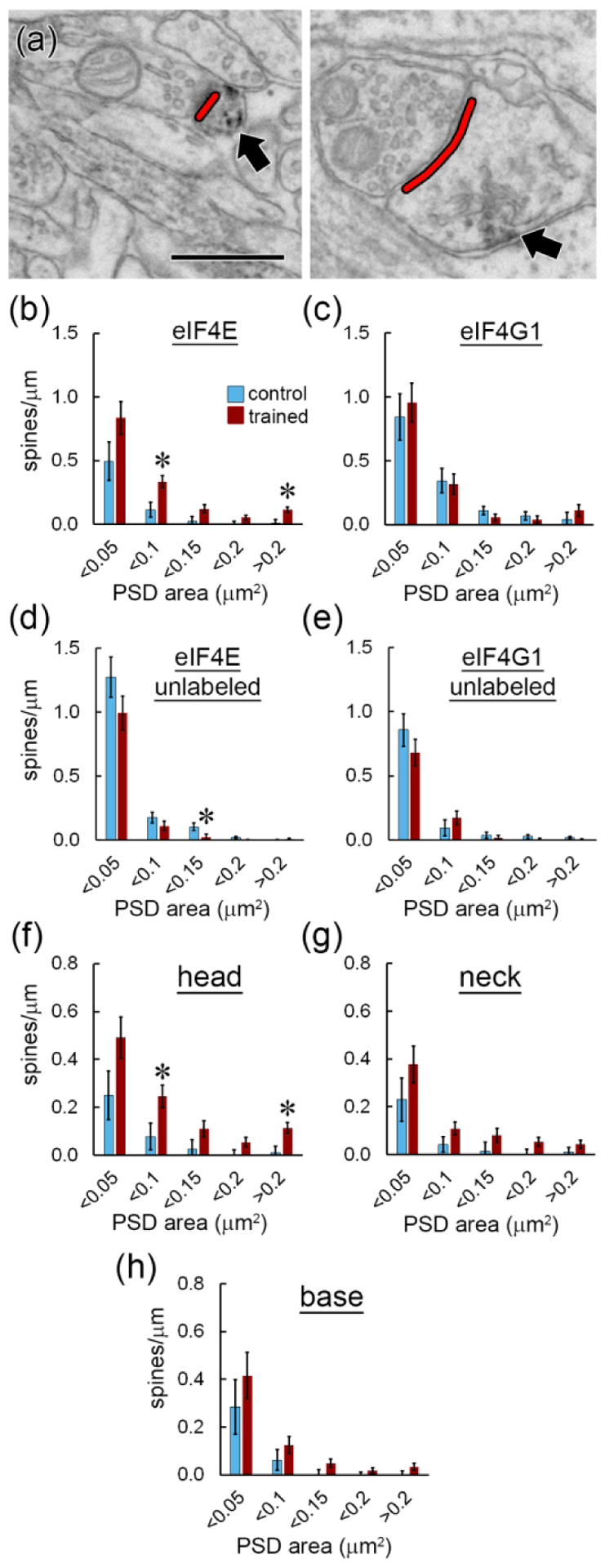
Synapse size. a) EMs showing measurement of postsynaptic density length (red) on spine heads containing label for eIF4E (arrows). Scale = 500 nm. b–e) Spine frequency binned by synapse size for spines with (b) or without (c) eIF4E labeling and with (d) or without (e) eIF4G1 labeling. f–h) Frequency of spines binned by synapse size with eIF4E labeling in the head (f), neck (g), or base (h). * p < 0.05
Differential distribution of labeling in spines with and without a spine apparatus
The spine apparatus is a membranous organelle found in the largest dendritic spines (Westrum et al., 1980; Spacek, 1985; Ostroff et al., 2010). It is a specialization of the continuous smooth endoplasmic reticulum (sER) network that extends throughout the dendritic arbor, and consists of membranous calcium-containing cisterns interleaved with dense actin plates (Fifkova et al., 1983; Deller et al., 2000; Capani et al., 2001). The function of the spine apparatus has not been determined, but it contains Golgi markers (Pierce et al., 2001) and thus could have a role in post-translational processing as well as calcium signaling. Immunolabeling for eIFs was found in spines with and without a spine apparatus (Figure 5a–c), and there were more spines of both types labeled for eIF4E in the trained group (Figure 5d). There were no group differences in eIF4G1 labeling distribution with respect to the spine apparatus (Figure 5e). Within spines, the trained group had more labeling for eIF4E in the heads, necks, and bases of spines with a spine apparatus, but only in the heads of spines without a spine apparatus (Figure 5f). Again, there were no group differences in eIF4G1 labeling (Figure 5g).
Figure 5.
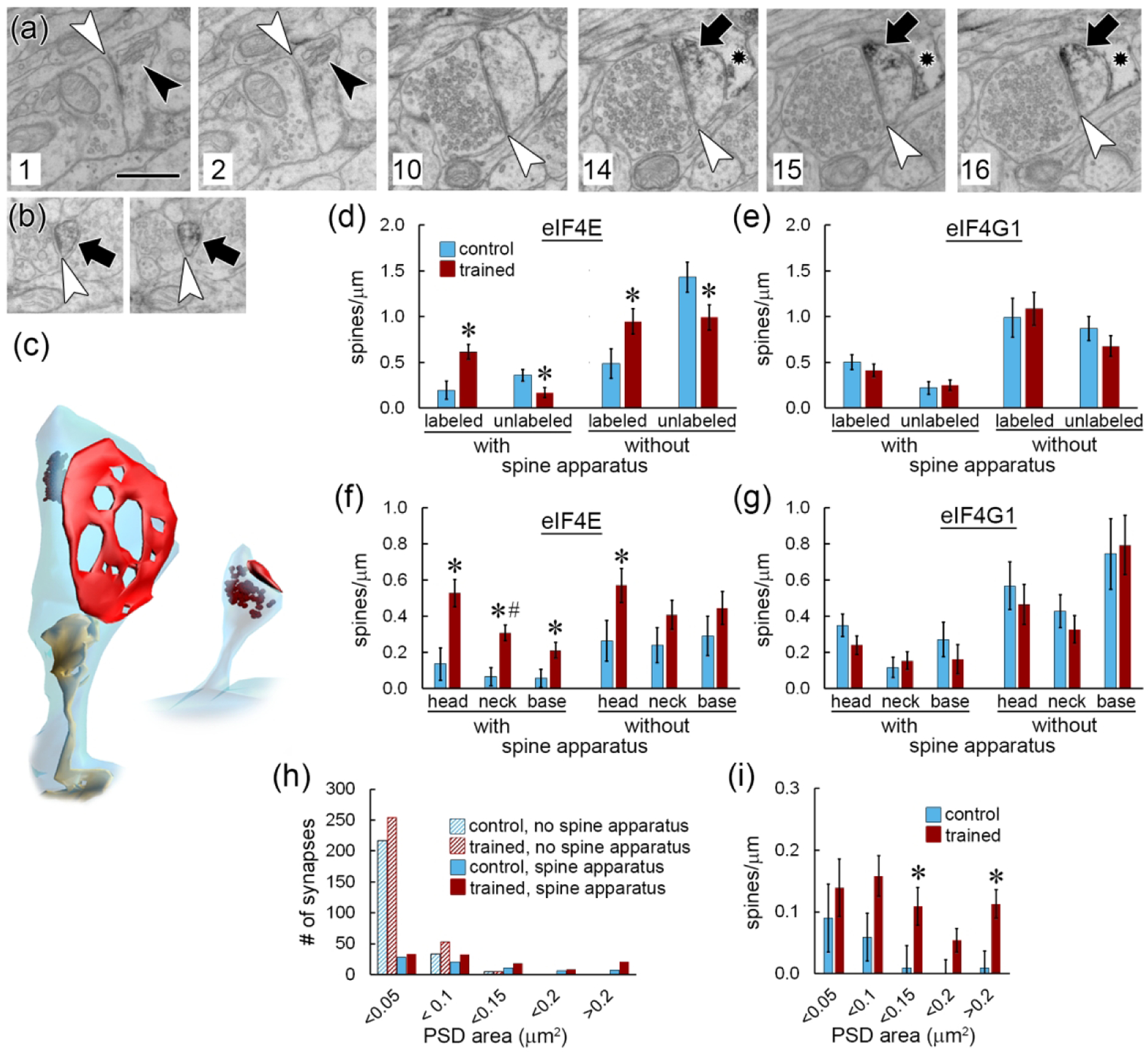
Spine apparatus. a) Six non-consecutive serial EM images through a large spine forming an asymmetric synapse (white arrowheads) and containing a spine apparatus (black arrowheads) and immunolabel for eIF4E (arrows). Section numbers are shown at lower left. An astrocytic process (star) containing immunolabel is visible in sections 14–16. Scale = 500nm. b) Two consecutive serial EM images of a spine without a spine apparatus labeled for eIF4E. c) 3D reconstructions of the spines shown in (a) and (b) with spine apparatus in yellow, synapse in red, and label in brown. d,e) Frequency of spines with and without eIF4E (d) or eIF4G1 (e) labeling broken down by the presence of a spine apparatus. f,g) Spine frequency by label location and presence of a spine apparatus for eIF4E (f) and eIF4G1 (g). h) Histogram showing the synapse size distribution for all spines in the dataset by training group and presence of a spine apparatus. i) Frequency of spines with a spine apparatus and eIF4E immunolabel, binned by synapse size. * p < 0.05; # significant interaction with subject.
Spines that contain a spine apparatus are several-fold larger on average than other spines (Ostroff et al., 2010); across all spines in the analyzed dataset, spines without a spine apparatus had a mean synapse size of 0.034 ± 0.003 μm2, compared to 0.128 ± 0.006 μm2 for spines with a spine apparatus. We find that nearly all spines with synapses >0.1 μm2 contain a spine apparatus, making it a universal feature of the largest spines. The spine apparatus is not exclusively found in large spines, however, but is present across the entire range of synapse sizes (Figure 5h). When spines with a spine apparatus were binned by synapse size, the trained group had more eIF4E-labeled spines only in the largest (>0.2 μm2) and intermediate (0.1 – 0.15 μm2) bins (Figure 5i). Among small spines, therefore, the training only affected eIF4E labeling in those without a spine apparatus.
eIF labeling in dendritic shafts
Immunolabel for eIFs was present in the majority of dendritic shafts. As in dendritic spines, the DAB reaction product in shafts was not found in the form of isolated puncta, but typically consisted of irregular groupings of dark puncta and patches of speckling spread through swaths of cytoplasm (Figure 6a). The diffuse, amorphous nature of the label does not allow quantification of the number of antibody molecules. We instead performed a semi-quantitative evaluation of label distribution in the shafts by measuring the volume of cytoplasm that contained labeling as a percentage of each dendrite’s cylindrical shaft volume. Labeling for eIF4E was present throughout an average of 7.1% of the shaft cytoplasm (range: 0 – 32.4%), and eIF4G1 labeling was in 30.7% (range: 0 – 100%). There were no differences between groups (Figure 6b). Although this is not a quantitative analysis of protein levels, the range of values suggests that eIFs are non-uniformly distributed in dendritic shafts and that eIF4G1 has a more extensive presence than eIF4E. Plotting the labeled shaft volume against dendrite diameter showed no relationship (6c,d). The frequency of unlabeled spines likewise appears unrelated to the extent of shaft labeling (Figure 6e,f), but the frequency of labeled spines looks to be higher in the most heavily labeled dendrites relative to the least (Figure 6g,h).
Figure 6.
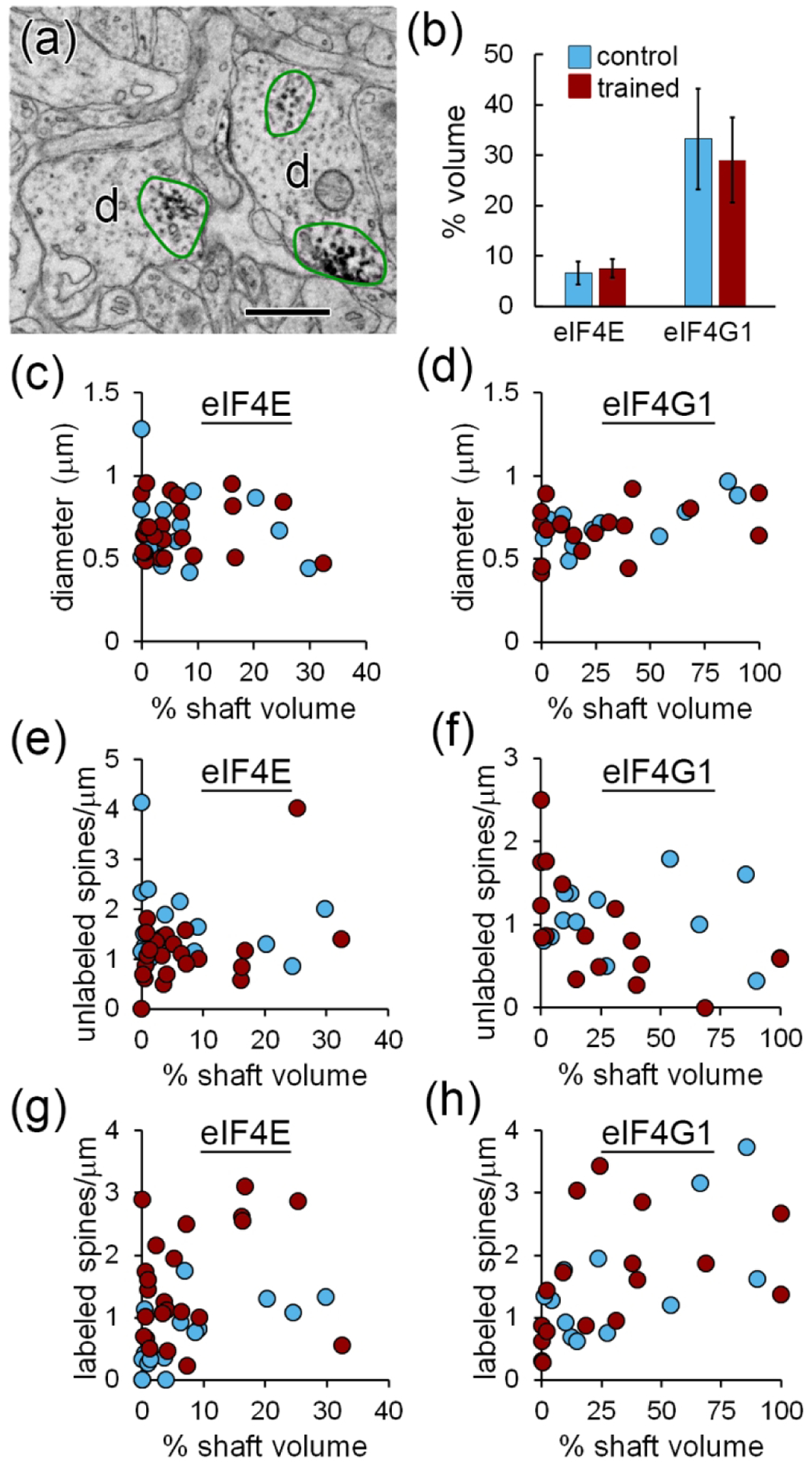
Labeling in dendritic shafts. a) Two dendrites (d) with patches of cytoplasm containing immunolabeling for eIF4E outlined in green. b) Average percent of dendritic shaft volume containing immunolabel for eIF4E or eIF4G1. c,d) Plots of percent shaft volume with label for eIF4E (d) and eIF4G1 (d) versus diameter for each dendrite. e–h) Plots of percent shaft volume with label for eIF4E (e,g) and eIF4G1 (f,h) versus frequency of unlabeled spines (e–f) and labeled spines (g–h).
Comparison of eIF labeling with polyribosome distribution in spines
If polyribosomes in spines are locally assembled via cap-dependent initiation, we would expect some correspondence between the spine distribution of eIFs and polyribosomes. Due to the nature of our labeling procedure, we were unable to reliably identify polyribosomes in dendritic spines in this study. To preserve contrast between the label and the tissue, we omitted the usual counterstaining that enhances polyribosomes. A few polyribosomes were visible amid deposits of immunolabel in spines (Figures 1a, 5a16, and 5b1 for example), as well as in dendrites and especially in cell bodies, where they are highly abundant, but none were observed in unlabeled dendritic spines. Because of the lack of counterstaining, we cannot conclude that these spines lack polyribosomes. Likewise, the absence of polyribosomes in labeled spines could not be determined because the immunolabeling was often dense enough to occlude structures in the cytoplasm.
The Pavlovian training protocol, timing of tissue collection, and analysis methods used here were identical to those used in our ssTEM study of the effects of 4EGI-1 on polyribosomes (Ostroff et al., 2017), so we compared the frequencies of spine polyribosomes from that study (Figure 7a) with eIF labeling to put the datasets in context. Polyribosomes were found in approximately 30% of dendritic spines in the vehicle-infused control group and approximately half of spines in the vehicle-infused trained group, similar to the percentages of spines labeled for eIF4E (Figure 7b). In both studies, effects differed by sub-spine location and the presence of a spine apparatus, so we compared these measures as well. In the heads of spines with a spine apparatus, both eIF4E labeling and polyribosomes were more frequent in the trained group versus the control, and 4EGI-1 blocked the effect of training but did not reduce the control level of polyribosomes (Figure 7c). Both eIF4E and polyribosomes were also upregulated in the heads of spines without a spine apparatus, but in this case both drug groups did have fewer polyribosomes (Figure 7d). The spine base and neck locations did not differ, so spines with labeling or polyribosomes in the base, neck, or both were pooled. There was an effect of training on eIF4E labeling, but not polyribosomes, in the base/neck location of spines with a spine apparatus (Figure 7e). In the bases and necks of spines without a spine apparatus, there were more polyribosomes in the trained groups with no effect of 4EGI-1, and no significant effect on eIF4E (Figure 7f).
Figure 7.
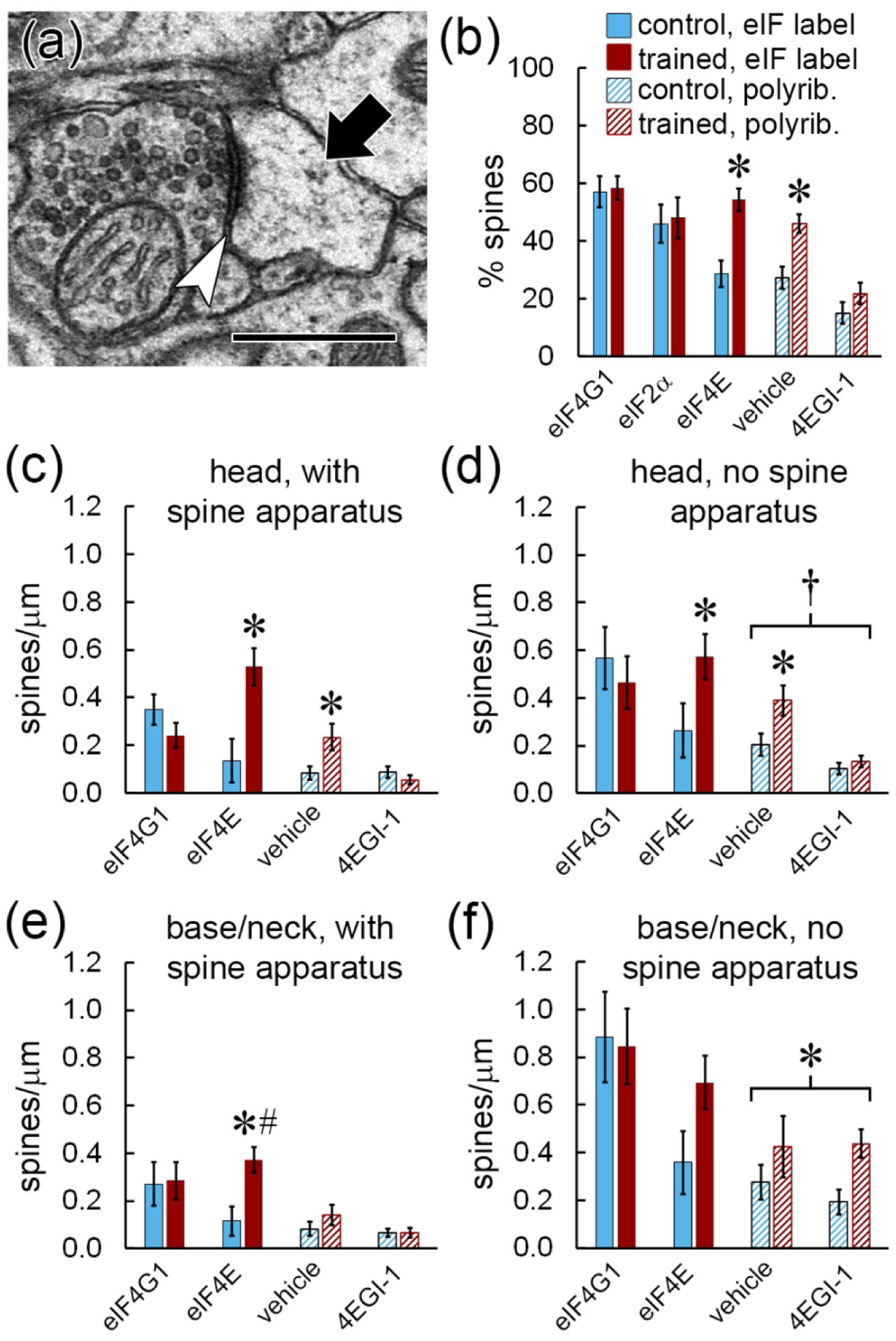
Comparison with polyribosome distribution. a) EM of a polyribosome (arrow) in the head of a dendritic spine forming a synapse (arrowhead). Scale = 500 nm. B) Percentage of all spines labeled for eIFs or containing polyribosomes. c–f) Effects of learning on the frequency of spines with immunolabel for eIF4G1 or eIF4E, or with polyribosomes after infusion of vehicle or 4EGI-1. Data are broken down by location of label in the spine head (c,d) or base/neck (e,f) and the presence (c,e) or absence (d,f) of a spine apparatus. Labeling data in (c) and (d) replotted from Figure 5f,g, and all polyribosome data replotted from Ostroff et al. (2017). * p < 0.05; # significant interaction with subject; † effect of 4EGI-1
The data shown in Figure 7 are summarized in Figure 8. The effect of learning on eIF4E distribution was similar to the effect on cap-dependent (4EGI-1-sensitive) but not cap-independent (4EGI-1-insensitive) polyribosomes: eIF4E and cap-dependent polyribosomes accumulated in the heads of dendritic spines, while cap-independent polyribosomes, but not eIF4E, accumulated in the bases and necks of spines lacking a spine apparatus. Although the other eIFs were distributed throughout dendrites and spines, the selective upregulation of eIF4E in locations where cap-dependent polyribosomes accumulate is consistent with eIF4E serving as the rate limiting component of initiation. Overall, our data indicate that the machinery for translation initiation is localized near individual synapses, and that initiation may be regulated in a highly spatially-specific manner during memory consolidation via availability of eIF4E.
Figure 8.
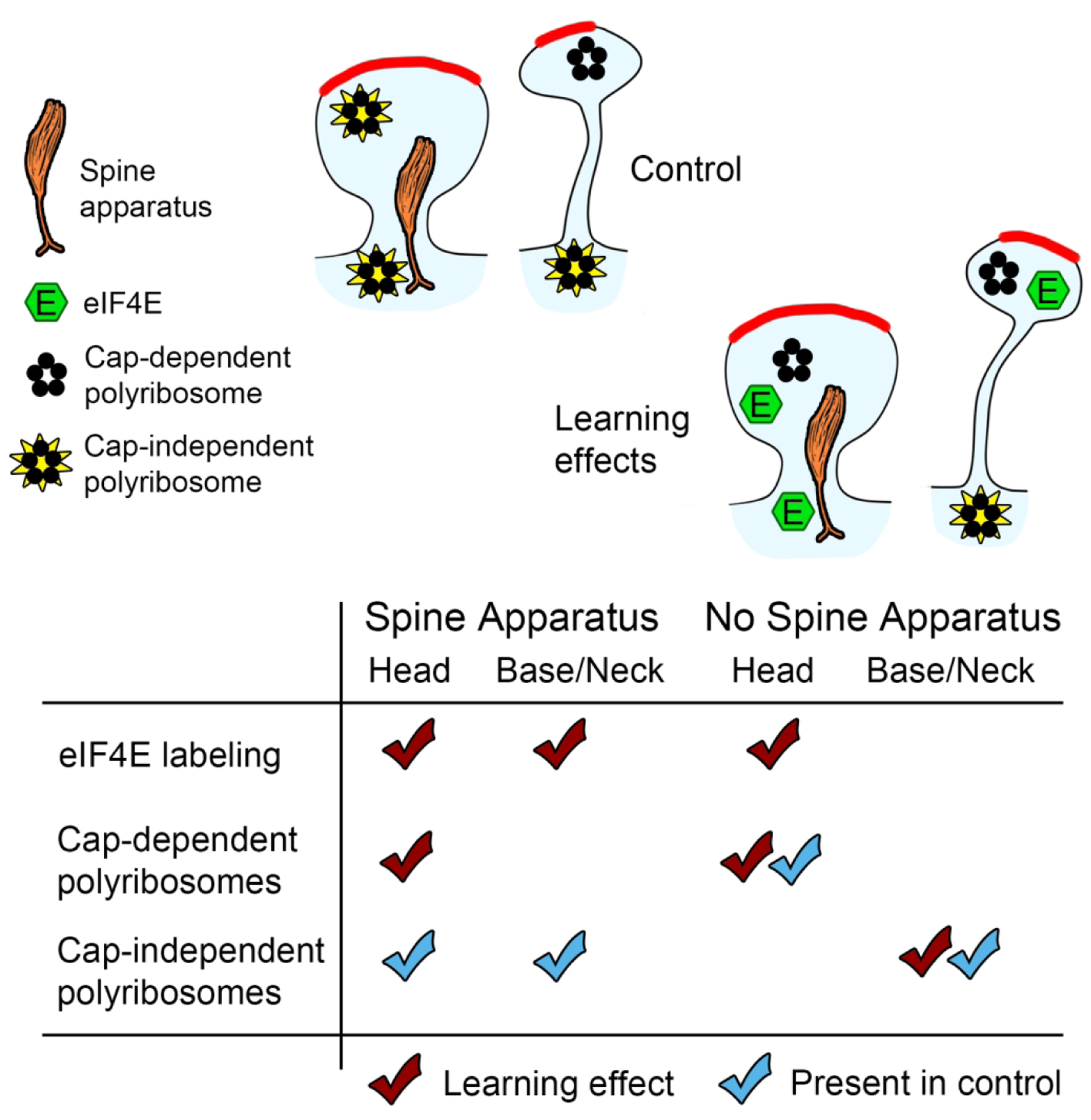
Summary of learning effects on eIF4E labeling, cap-dependent (4EGI-1 sensitive) and cap-independent (4EGI-1 insensitive) polyribosomes. Effects on eIF4E from this study, effects on polyribosomes from Ostroff et al. (2017).
Discussion
Our results indicate that three different eIFs localize to dendritic spines, suggesting that de novo protein synthesis not only occurs within dendritic spines, but is initiated there as well. Translation initiation within a spine could be a component of autonomous regulation of the synaptic proteome; a spine with its own complement of mRNA and translation machinery could remodel in response to synaptic activity without affecting its neighbors, potentially preserving the fidelity of a memory network. Proteins do not seem to be restricted to single spines, however. Many synaptic proteins are highly motile within dendrites and move into and out of spines on a timescale of minutes to hours (Hanus and Schuman, 2013; Bourke et al., 2018). If proteins come and go from synapses via diffusion and transport mechanisms, local translation may not necessarily be a spine-specific targeting mechanism. Instead, pools of locally-translated proteins could be available to all synapses within a dendritic region, with local synaptic activity determining the composition of the protein pool. Indeed, the synaptic tagging model of heterosynaptic plasticity posits just such spatiotemporally-regulated sharing of new proteins among synapses, with local synthesis a likely protein source (Sajikumar et al., 2007; Doyle and Kiebler, 2011; Redondo and Morris, 2011). If proteins are shared among synapses, why not initiate all translation in the dendritic shaft? One possibility is that initiation in a spine would bias protein availability towards that spine and its immediate neighbors.
A variety of dendritic mRNA transport mechanisms have been identified, and different mechanisms appear to be selective for specific transcripts (Miller et al., 2002; Buxbaum et al., 2015; Nakayama et al., 2017; Roy et al., 2020). Once transported into dendrites, mRNA can remain dormant in dendrites until it is unmasked by synaptic activity (Hutten et al., 2014), so localization of mRNA may be a means of priming certain locations for translation of specific proteins. There is evidence that some mRNAs are targeted at a fine spatial scale; in cultured neurons Arc and dendrin mRNA are restricted to the base and proximal neck of spines, while CaMKIIα and β-actin mRNA localize to spine heads (Tiruchinapalli et al., 2003; Kao et al., 2010; Dynes and Steward, 2012). Translation initiation in spines could create localized hotspots of specific proteins determined by the distribution of mRNA. It is notable that we did not find eIFs in all spines, and that only eIF4E, considered the key control point in translation (Amorim et al., 2018), appeared in more spines during memory formation. Translational control mechanisms are complex, and because components of multiple signaling pathways, mRNA, and translation machinery must be present and active in a single location, translation effectively serves as a coincidence detector. If eIFs and mRNAs are selectively localized to particular spines, either at baseline or in response to activity, the local protein composition in a dendritic region could serve as a readout of recent synaptic activity patterns. As opposed to restricting protein availability, translation initiation within spines could instead restrict translational control such that translation-competent spines can influence the plasticity properties of neighboring synapses via protein production. Exploitation of translational control pathways to achieve synapse-specific regulation of the local proteome could be a reason for the central role of translational control in plasticity and perhaps explain why intellectual functions such as learning are uniquely sensitive to disruption of translation regulation (Kelleher and Bear, 2008; Darnell, 2011; Kapur et al., 2017).
Besides selective localization, eIFs offer ample opportunities for nuanced control of translation via biochemical regulation. Multiple different signaling pathways and effector molecules separately target eIF4E and eIF2α (Trinh and Klann, 2013; Amorim et al., 2018), for example, so cap-dependent translation in a spine could require concurrent activation of potentially orthogonal triggers. Two key regulators of eIF4E, the mechanistic target of rapamycin complex 1 (mTORC1) and mitogen-activated kinase (MAPK) pathways, are downstream of synaptic activity and have well-established roles in multiple forms of synaptic plasticity, memory, and neurological disease are key regulators of eIF4E (Kelleher et al., 2004; Hoeffer and Klann, 2010). The fragile X mental retardation protein (FMRP) and its binding partner, the cytoplasmic FMRP interacting protein (CYFIP1), are also direct repressors of eIF4E (Genheden et al., 2015). Mice that overexpress eIF4E or lack its repressors FMRP or CYFIP1 have deficits in synaptic plasticity and learning, as well as abnormal spine morphology similar to that seen in Fragile X and other intellectual disorders (Fiala et al., 2002; Grossman et al., 2006; De Rubeis et al., 2013; Santini et al., 2013). Regulation of eIF2α is less studied than that of eIF4E, but its activity is modulated by kinases whose activity is necessary for normal learning and cognitive function, suggesting that it too could have a role in local translation (Costa-Mattioli et al., 2007; Trinh and Klann, 2013). The experimental evidence is highly suggestive of eIFs, and eIF4E in particular, serving as substrates for biochemical signal integration during memory formation.
Because we used eIF antibodies that were not specific to phosphorylation or complexing with major regulators, we were able to observe the dynamics of eIF localization separately from biochemical regulation. Fewer than 60% of spines contained labeling for any eIF, and only a third of spines were labeled for eIF4E in the control condition. Similar proportions of spines were labeled for eIF4G1 and eIF2α in both groups, but fewer were labeled for eIF4E in the control group. The proportion of spines with eIF4E labeling was higher in the trained group, suggesting a scenario in which some spines are primed for translation by the presence of eIF4G1 and eIF2α, but initiation does not occur until synaptic activity triggers delivery of eIF4E. This is consistent with the canonical view that eIF4E is the rate limiting factor in initiation, but without multiplexed labeling we do not know how often all three eIFs are present in the same spines. The four-fold difference between the spatial extent of eIF4G1 versus eIF4E labeling in the dendritic shafts also argues for limited eIF4E availability as a regulatory mechanism. The possibility of differences in sensitivity between the antibodies makes direct comparisons difficult, however. In addition, it is possible that our antibody was less efficient at binding to eIF4E in complex with its repressor proteins, and the increased labeling reflects activation of pre-existing eIF4E. Interestingly, a quantitative proteomic analysis of HeLa cells found similar copy numbers of eIF4G1 and eIF2α protein, which were almost double that of eIF4E (Kulak et al., 2014). Copy number data are not available for neurons or their compartments, but could shed light on the relative role of local copy number versus biochemical mechanisms in translation regulation.
The source of eIFs in spines is another open question. A substantial amount of immunolabeling for eIFs was present in dendritic shafts, and transport from the shaft is an obvious possibility. In an earlier study of LA dendrites we found a positive correlation between polyribosome density in dendritic shafts and spines regardless of learning condition (Ostroff et al., 2010), indicating that translation sites in shafts and spines are not zero-sum and are regulated in parallel. The spatial extent of eIF labeling in shafts was likewise not depleted relative to spine labeling in the current study. Whether this reflects relative copy numbers is unknown, but a widespread upregulation of translation machinery appears more likely than a net shift from shafts into spines. Dendritic eIFs could be transported from the soma, but there is evidence that they are locally translated themselves. RNA transport granules in cultured neurons do not contain eIF4E, eIF4G1, or eIF2α (Krichevsky and Kosik, 2001), but mRNA for all three has been found in hippocampal neuropil (Cajigas et al., 2012; Nakayama et al., 2017). An alternative explanation for the correspondence between eIF4E and polyribosome distribution is that the polyribosomes are translating eIF4E, perhaps as part of a synaptically-triggered translation cascade.
A particularly intriguing aspect of our data is the correspondence between eIF4E distribution and the distribution of polyribosomes in our earlier study (Ostroff et al., 2017). Although this supports the general model of eIF4E as rate-limiting, it is not a given that all polyribosomes represent recent eIF4E-mediated initiation or that all initiation creates polyribosomes. Polyribosomes may not be actively translating; stalled polyribosomes may be transported in dendrites for later reactivation, and live imaging of cultured neurons found that most dendritic polyribosomes are stalled (Richter and Coller, 2015; Langille et al., 2019). There is also growing evidence that a significant amount of translation occurs independently of eIF4E by a variety of alternative cap-dependent or cap-independent mechanisms (de la Parra et al., 2018; Borden and Volpon, 2020). Initiation via internal ribosome entry sites, which bypasses the cap, occurs in dendrites and is biased towards specific transcripts (Pinkstaff et al., 2001). Although polyribosomes are traditionally assumed to represent the default mode of translation, two studies have reported that a substantial amount of translation occurs on monosomes, with one showing that translation in dendrites is biased towards monosomes (Heyer and Moore, 2016; Biever et al., 2020). Polyribosomes are abundant in dendrites but extremely scarce in axons, yet in a previous study we found immunolabeling for eIF4E, eIF4G1, eIF2α, and ribosomal protein s6 in presynaptic boutons and axons in the LA (Ostroff et al., 2019), consistent with reports of translation machinery despite the absence of polyribosomes in peripheral axons (Zheng et al., 2001; Merianda and Twiss, 2013). Since eIF4E and polyribosomes apparently represent subsets of translation sites, their correspondence suggests that memory formation triggers specific translation mechanisms in specific sites near synapses. Overall, our results are consistent with a high degree of spatial precision in the regulation of local translation, and with eIF localization as a mechanism of translational control.
Acknowledgements:
We are grateful to Nikita Gupta for expert technical assistance.
Funding statement:
This work was supported by NIH MH083583, MH094965, and MH119517 to LEO, and NS034007 and NS047384 to EK.
Footnotes
Data availability statement: Full image sets are available from the corresponding author upon reasonable request.
Conflict of interest statement: The authors declare no competing financial interests.
References
- Amorim IS, Lach G, Gkogkas CG (2018) The Role of the Eukaryotic Translation Initiation Factor 4E (eIF4E) in Neuropsychiatric Disorders. Front Genet 9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaki C, Usuda N, Nakazawa A, Kametani K, Suzuki T (2003) Localization of translational components at the ultramicroscopic level at postsynaptic sites of the rat brain. Brain Res 972:168–176. [DOI] [PubMed] [Google Scholar]
- Biever A, Glock C, Tushev G, Ciirdaeva E, Dalmay T, Langer JD, Schuman EM (2020) Monosomes actively translate synaptic mRNAs in neuronal processes. Science 367:eaay4991. [DOI] [PubMed] [Google Scholar]
- Borden KLB, Volpon L (2020) The diversity, plasticity, and adaptability of cap-dependent translation initiation and the associated machinery. RNA Biol 17:1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke AM, Bowen AB, Kennedy MJ (2018) New approaches for solving old problems in neuronal protein trafficking. Mol Cell Neurosci 91:48–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne J, Harris KM (2007) Do thin spines learn to be mushroom spines that remember? Curr Opin Neurobiol 17:381–386. [DOI] [PubMed] [Google Scholar]
- Bourtchouladze R, Abel T, Berman N, Gordon R, Lapidus K, Kandel ER (1998) Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn Mem 5:365–374. [PMC free article] [PubMed] [Google Scholar]
- Buffington SA, Huang W, Costa-Mattioli M (2014) Translational control in synaptic plasticity and cognitive dysfunction. Annu Rev Neurosci 37:17–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum AR, Yoon YJ, Singer RH, Park HY (2015) Single-molecule insights into mRNA dynamics in neurons. Trends Cell Biol 25:468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM (2012) The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74:453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capani F, Martone ME, Deerinck TJ, Ellisman MH (2001) Selective localization of high concentrations of F-actin in subpopulations of dendritic spines in rat central nervous system: a three-dimensional electron microscopic study. J Comp Neurol 435:156–170. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Stern E, Gamache K, Colina R, Cuello C, Sossin W, Kaufman R, Pelletier J, Rosenblum K, Krnjević K, Lacaille J-C, Nader K, Sonenberg N (2007) eIF2alpha phosphorylation bidirectionally regulates the switch from short-to long-term synaptic plasticity and memory. Cell 129:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC (2011) Defects in translational regulation contributing to human cognitive and behavioral disease. Curr Opin Genet Dev 21:465–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis HP, Squire LR (1984) Protein synthesis and memory: a review. Psychol Bull 96:518–559. [PubMed] [Google Scholar]
- de la Parra C, Ernlund A, Alard A, Ruggles K, Ueberheide B, Schneider RJ (2018) A widespread alternate form of cap-dependent mRNA translation initiation. Nat Commun 9:3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, Pasciuto E, Li KW, Fernández E, Di Marino D, Buzzi A, Ostroff LE, Klann E, Zwartkruis FJT, Komiyama NH, Grant SGN, Poujol C, Choquet D, Achsel T, Posthuma D, Smit AB, Bagni C (2013) CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron 79:1169–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deller T, Merten T, Roth SU, Mundel P, Frotscher M (2000) Actin-associated protein synaptopodin in the rat hippocampal formation: Localization in the spine neck and close association with the spine apparatus of principal neurons. J Comp Neurol 418:164–181. [DOI] [PubMed] [Google Scholar]
- Doyle M, Kiebler MA (2011) Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J 30:3540–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynes JL, Steward O (2012) Arc mRNA docks precisely at the base of individual dendritic spines indicating the existence of a specialized microdomain for synapse-specific mRNA translation. J Comp Neurol 520:3105–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC (2005) Reconstruct: a free editor for serial section microscopy. J Microsc 218:52–61. [DOI] [PubMed] [Google Scholar]
- Fiala JC, Spacek J, Harris KM (2002) Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res Brain Res Rev 39:29–54. [DOI] [PubMed] [Google Scholar]
- Fifkova E, Anderson CL, Young SJ, Van Harreveld A (1982) Effect of anisomycin on stimulation-induced changes in dendritic spines of the dentate granule cells. J Neurocytol 11:183–210. [DOI] [PubMed] [Google Scholar]
- Fifkova E, Markham JA, Delay RJ (1983) Calcium in the spine apparatus of dendritic spines in the dentate molecular layer. Brain Res 266:163–168. [DOI] [PubMed] [Google Scholar]
- Genheden M, Kenney JW, Johnston HE, Manousopoulou A, Garbis SD, Proud CG (2015) BDNF stimulation of protein synthesis in cortical neurons requires the MAP kinase-interacting kinase MNK1. J Neurosci 35:972–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkogkas C, Sonenberg N, Costa-Mattioli M (2010) Translational control mechanisms in long-lasting synaptic plasticity and memory. J Biol Chem 285:31913–31917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppo R, Richter JD (2009) Translational control from head to tail. Curr Opin Cell Biol 21:444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AW, Elisseou NM, McKinney BC, Greenough WT (2006) Hippocampal pyramidal cells in adult Fmr1 knockout mice exhibit an immature-appearing profile of dendritic spines. Brain Res 1084:158–164. [DOI] [PubMed] [Google Scholar]
- Grüner S, Peter D, Weber R, Wohlbold L, Chung MY, Weichenrieder O, Valkov E, Igreja C, Izaurralde E (2016) The structures of eIF4E-eIF4G complexes reveal an extended interface to regulate translation initiation. Mol Cell 64:467–479. [DOI] [PubMed] [Google Scholar]
- Hanus C, Schuman EM (2013) Proteostasis in complex dendrites. Nat Rev Neurosci 14:638–648. [DOI] [PubMed] [Google Scholar]
- Heyer EE, Moore MJ (2016) Redefining the Translational Status of 80S Monosomes. Cell 164:757–769. [DOI] [PubMed] [Google Scholar]
- Hoeffer CA, Cowansage KK, Arnold EC, Banko JL, Moerke NJ, Rodriguez R, Schmidt EK, Klosi E, Chorev M, Lloyd RE, Pierre P, Wagner G, LeDoux JE, Klann E (2011) Inhibition of the interactions between eukaryotic initiation factors 4E and 4G impairs long-term associative memory consolidation but not reconsolidation. Proc Natl Acad Sci U S A 108:3383–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Klann E (2010) mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci 33:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Schuman EM (2013) The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80:648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S, Sharangdhar T, Kiebler M (2014) Unmasking the messenger. RNA Biol 11:992–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo I, Bevilaqua LRM, Rossato JI, Bonini JS, Medina JH, Cammarota M (2006) Different molecular cascades in different sites of the brain control memory consolidation. Trends Neurosci 29:496–505. [DOI] [PubMed] [Google Scholar]
- Jian M, Luo YX, Xue YX, Han Y, Shi HS, Liu JF, Yan W, Wu P, Meng SQ, Deng JH, Shen HW, Shi J, Lu L (2014) eIF2α dephosphorylation in basolateral amygdala mediates reconsolidation of drug memory. J Neurosci 34:10010–10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao D-I, Aldridge GM, Weiler IJ, Greenough WT (2010) Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proc Natl Acad Sci U S A 107:15601–15606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur M, Monaghan CE, Ackerman SL (2017) Regulation of mRNA Translation in Neurons—A Matter of Life and Death. Neuron 96:616–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Fukuda M (2010) Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci 33:121–129. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, Bear MF (2008) The autistic neuron: troubled translation? Cell 135:401–406. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, Govindarajan A, Jung HY, Kang H, Tonegawa S (2004) Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell 116:467–479. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS (2001) Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32:683–696. [DOI] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11:319–324. [DOI] [PubMed] [Google Scholar]
- Langille JJ, Ginzberg K, Sossin WS (2019) Polysomes identified by live imaging of nascent peptides are stalled in hippocampal and cortical neurites. Learn Mem 26:351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux JE (2000) Emotion Circuits in the Brain. Annu Rev Neurosci 23:155–184. [DOI] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B (2011) Local RNA translation at the synapse and in disease. J Neurosci 31:16086–16093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader S, Lee H, Pause A, Sonenberg N (1995) The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol 15:4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK (1997) Cocrystal structure of the messenger RNA 5’ cap-binding protein (elF4E) bound to 7-methyl-GDP. Cell 89:951–961. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK (1999) Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of elF4G. Mol Cell 3:707–716. [DOI] [PubMed] [Google Scholar]
- Maren S (2001) Neurobiology of Pavlovian fear conditioning. Annu Rev Neurosci 24:897–931. [DOI] [PubMed] [Google Scholar]
- Maren S, Ferrario CR, Corcoran KA, Desmond TJ, Frey KA (2003) Protein synthesis in the amygdala, but not the auditory thalamus, is required for consolidation of Pavlovian fear conditioning in rats. Eur J Neurosci 18:3080–3088. [DOI] [PubMed] [Google Scholar]
- Mayford M, Siegelbaum SA, Kandel ER (2012) Synapses and memory storage. Cold Spring Harb Perspect Biol 4:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald AJ (1982) Neurons of the lateral and basolateral amygdaloid nuclei: a Golgi study in the rat. J Comp Neurol 212:293–312. [DOI] [PubMed] [Google Scholar]
- McDonald AJ (1992) Projection neurons of the basolateral amygdala: a correlative Golgi and retrograde tract tracing study. Brain Res Bull 28:179–185. [DOI] [PubMed] [Google Scholar]
- McDonald AJ, Pearson JC (1989) Coexistence of GABA and peptide immunoreactivity in non-pyramidal neurons of the basolateral amygdala. Neurosci Lett 100:53–58. [DOI] [PubMed] [Google Scholar]
- Merianda T, Twiss J (2013) Peripheral nerve axons contain machinery for co-translational secretion of axonally-generated proteins. Neurosci Bull 29:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M (2002) Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron 36:507–519. [DOI] [PubMed] [Google Scholar]
- Morino S, Imataka H, Svitkin YV., Pestova TV, Sonenberg N (2000) Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol 20:468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Ohashi R, Shinoda Y, Yamazaki M, Abe M, Fujikawa A, Shigenobu S, Futatsugi A, Noda M, Mikoshiba K, Furuichi T, Sakimura K, Shiina N (2017) RNG105/caprin1, an RNA granule protein for dendritic mRNA localization, is essential for long-term memory formation. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroff LE, Botsford B, Gindina S, Cowansage KK, Ledoux JE, Klann E, Hoeffer C (2017) Accumulation of polyribosomes in dendritic spine heads, but not bases and necks, during memory consolidation depends on cap-dependent translation initiation. J Neurosci 37:1862–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroff LE, Cain CK, Bedont J, Monfils MH, LeDoux JE (2010) Fear and safety learning differentially affect synapse size and dendritic translation in the lateral amygdala. Proc Natl Acad Sci U S A 107:9418–9423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroff LE, Santini E, Sears R, Deane Z, Kanadia RN, Ledoux JE, Lhakhang T, Tsirigos A, Heguy A, Klann E (2019) Axon TRAP reveals learning-associated alterations in cortical axonal mRNAs in the lateral amgydala. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Palay S, Webster H (1991) The fine structure of the nervous system, 3rd ed. New York: Oxford University Press. [Google Scholar]
- Pierce JP, Mayer T, McCarthy JB (2001) Evidence for a satellite secretory pathway in neuronal dendritic spines. Curr Biol 11:351–355. [DOI] [PubMed] [Google Scholar]
- Pinkstaff JK, Chappell SA, Mauro VP, Edelman GM, Krushel LA (2001) Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc Natl Acad Sci U S A 98:2770–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevedo J, Vianna MRM, Roesler R, De-Paris F, Izquierdo I, Rose SPR (1999) Two time windows of anisomycin-induced amnesia for inhibitory avoidance training in rats: Protection from amnesia by pretraining but not pre-exposure to the task apparatus. Learn Mem 6:600–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju V, tom Dieck S, Schuman EM (2017) Local translation in neuronal compartments: how local is local? EMBO Rep 18:693–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo RL, Morris RGM (2011) Making memories last: the synaptic tagging and capture hypothesis. Nat Rev Neurosci 12:17–30. [DOI] [PubMed] [Google Scholar]
- Richter JD, Coller J (2015) Pausing on Polyribosomes: Make Way for Elongation in Translational Control. Cell 163:292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg T, Gal-Ben-Ari S, Dieterich DC, Kreutz MR, Ziv NE, Gundelfinger ED, Rosenblum K (2014) The roles of protein expression in synaptic plasticity and memory consolidation. Front Mol Neurosci 7:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Shiina N, Wang DO (2020) More dynamic, more quantitative, unexpectedly intricate: Advanced understanding on synaptic RNA localization in learning and memory. Neurobiol Learn Mem 168:107149. [DOI] [PubMed] [Google Scholar]
- Sajikumar S, Navakkode S, Frey JU (2007) Identification of compartment- and process-specific molecules required for “synaptic tagging” during long-term potentiation and long-term depression in hippocampal CA1. J Neurosci 27:5068–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Huynh TN, Klann E (2014) Mechanisms of translation control underlying long-lasting synaptic plasticity and the consolidation of long-term memory. Prog Mol Biol Transl Sci 122:131–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, Kaphzan H, Klann E (2013) Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature 493:411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafe GE, LeDoux JE (2000) Memory Consolidation of Auditory Pavlovian Fear Conditioning Requires Protein Synthesis and Protein Kinase A in the Amygdala. J Neurosci 20:RC96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M (2017) Dendritic spines: Morphological building blocks of memory. Neurobiol Learn Mem 138:3–9. [DOI] [PubMed] [Google Scholar]
- Smart FM, Edelman GM, Vanderklish PW (2003) BDNF induces translocation of initiation factor 4E to mRNA granules: Evidence for a role of synaptic microfilaments and integrins. Proc Natl Acad Sci U S A 100:14403–14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spacek J (1985) Three-dimensional analysis of dendritic spines. II. Spine apparatus and other cytoplasmic components. Anat Embryol 171:235–243. [DOI] [PubMed] [Google Scholar]
- Steward O, Levy W (1982) Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci 2:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka J, Horiike Y, Matsuzaki M, Miyazaki T, Ellis-Davies GC, Kasai H (2008) Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science 319:1683–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ (2003) Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci 23:3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoo K, Matsushita Y, Fujisaki H, Abiko F, Shen X, Taniguchi T, Miyagawa H, Kitamura K, Miura KI, Ishida T (2005) Structural basis for mRNA Cap-Binding regulation of eukaryotic initiation factor 4E by 4E-binding protein, studied by spectroscopic, X-ray crystal structural, and molecular dynamics simulation methods. Biochim Biophys Acta - Proteins Proteomics 1753:191–208. [DOI] [PubMed] [Google Scholar]
- Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, Ishida T, Taniguchi T, Hasegawa H, Terashima A, Sasaki M, Katsuya Y, Kitamura K, Miyoshi H, Ishikawa M, Miura KI (2002) Crystal structures of 7-methylguanosine 5′-triphosphate (m7GTP)- and P1–7-methylguanosine-P3-adenosine-5′, 5′-triphosphate (m7GpppA)-bound human full-length eukaryotic initiation factor 4E: Biological importance of the C-terminal flexible region. Biochem J 362:539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh MA, Klann E (2013) Translational control by eIF2α kinases in long-lasting synaptic plasticity and long-term memory. Neurobiol Learn Mem 105:93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DO, Martin KC, Zukin RS (2010) Spatially restricting gene expression by local translation at synapses. Trends Neurosci 33:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrum LE, Jones DH, Gray EG, Barron J (1980) Microtubules, dendritic spines and spine appratuses. Cell Tissue Res 208:171–181. [DOI] [PubMed] [Google Scholar]
- Wu B, Eliscovich C, Yoon YJ, Singer RH (2016) Translation dynamics of single mRNAs in live cells and neurons. Science 352:1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JQ, Kelly TK, Chang B, Ryazantsev S, Rajasekaran AK, Martin KC, Twiss JL (2001) A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J Neurosci 21:9291–9303. [DOI] [PMC free article] [PubMed] [Google Scholar]


