Abstract
As a member of the ubiquitin-dependent proteasome degradation pathway, Ubiquitin carboxyl-terminal hydrolase-L1 (UCHL1) plays a key role in post-translational modification and protein degradation, and it is extensive and important for the regulation of various biological functions of the organism. However, its function remains unclear in goose growth performance. In this study, the full-length genomic DNA and coding region of UCHL1 gene was firstly cloned and characterized in Yangzhou goose. Tissue expression profile revealed that UCHL1 was exclusively expressed in brain and gonads. A novel single nucleotide polymorphisms c.-652C>T which is significantly related to 64-d body weight of Yangzhou goose was found in UCHL1 promoter region by comparative sequencing. Correlation analysis in a population of 405 geese showed that TT genotype individuals had higher body weight than CC individuals in male, but not in female geese. Dual-luciferase reporter assay indicated that the single nucleotide polymorphisms c.-652C>T is located at the core promoter region of UCHL1, and the promoter transcription activity was significantly increased (P < 0.01) when allele C changed to T. Geese with TT genotype had higher mRNA level of UCHL1 in brain tissue than those of CC genotype (P < 0.01). Compared with CC individuals, neuropeptide Y and AdipoR1 mRNA level was significantly higher in TT individuals (P < 0.05), while FAS mRNA level was lower in the TT individuals (P < 0.05). In summary, we identify a novel mutation in the promoter of UCHL1 gene, which can alter transcriptional activity of UCHL1 gene, and affect the growth performance of male goose.
Key words: UCHL1, SNP, promoter, Yangzhou goose, growth performance
INTRODUCTION
Yangzhou goose is one of Chinese indigenous goose breeds, mainly distributed in Jiangsu Province, China. Goose meat production has recently attracted increasing attention because of its high yield of breast meat, low caloric content, rapid growth, disease resistance, and easily fed with coarse fodder. The growth performance is a crucial trait of goose industry because it is closely related to the economic benefits. As the molecular mechanisms underlying the goose growth trait remains unclear, traditional phenotype selection is still the major method for improving growth performance in goose production. Molecular genetic markers are playing an increasingly important role in modern breeding of livestock because of their high selection accuracy and genetic stability. As a direct reflection of genetic variation at the DNA level, they have no relationship with gene expression, and are not affected by tissues, physiological development stages, and the environment (Grover and Sharma, 2016). Among them, molecular markers based on single nucleotide polymorphisms (SNPs) have been extensively studied in goose breeding work (Wu et al., 2014; Zhang et al., 2014; Yu et al., 2015; Zhang et al., 2015; Yu et al., 2017; Xia et al., 2018). Identification of growth-related functional genes and genetic markers are therefore of great significance for improving goose production.
Ubiquitin carboxyl-terminal hydrolase-L1 (UCHL1 / PGP 9.5) was first detected as a "brain-specific protein" in 1981. This protein is highly conserved and localized in neurons and neuroendocrine cells of vertebrates, accounting for 5% to10% of the total brain-soluble proteins, which is close to the levels of several glycolytic enzymes in human cells (Kandel, 1970; Rao, 2004; Day and Thompson, 2010). UCHL1 belongs to the deubiquitinase family and is involved in the ubiquitin-dependent proteolytic system. It is widely expressed in neuronal tissues and maintains ubiquitin balance in the body by binding and releasing ubiquitin (Osaka et al., 2003; Day and Thompson, 2010). Previous studies have shown that UCHL1 plays an important role in neurodegenerative diseases including Parkinson's disease and Alzheimer's disease (Setsuie and Wada, 2007), and also plays a role in tumorigenesis and tumor migration (Tezel et al., 2000; Mandelker et al., 2005; Wang et al., 2008; Aaron et al., 2010). Interestingly, there is increasing evidence that UCHL1 gene shows a unique role in muscle differentiation or lipid deposition, suggesting that its potential role in energy metabolism. Pedro et al. (2013) found that UCHL1 gene expression was up-regulated in subcutaneous abdominal fat tissue of obese men, which confirmed that UCHL1 gene was related to energy metabolism (Pedro et al., 2013). Piórkowska et al. (2018) also identified UCHL1 as a differentially expressed gene in the semi-membranous muscle and longissimus dorsal muscle of 2 Polish pig breeds differing in fat and meat qualities (Piórkowska et al., 2018). The UCHL1 protein level was continuously declined during cell differentiation. Knockdown of UCHL1 by siRNA resulted in a significant decrease in cell proliferation but marked acceleration of cell differentiation and myotube formation. These studies suggest that UCHL1 may play a role in myogenesis by promoting myoblast proliferation and inhibiting differentiation (Gao et al., 2017). In avian species, there have some research found that the transcriptional profile of UCHL1 changed in DF-1 cells infected with ALV-J compared to uninfected DF-1 cells (Fan et al., 2012). Replaceable neurons and neurodegenerative disease share depressed UCHL1 levels in male zebra finches and mouse brain (Lombardino et al., 2005), suggesting that UCHL1 levels may affect the expression of some neuropeptides. However, so far there has no report on whether UCHL1 can influence goose growth performance.
We propose that UCHL1 gene not only plays a vital role in tumors, nerve-related diseases, it may also participate in energy metabolism and muscle proliferation and differentiation. The present study aims to find out whether the UCHL1 gene affects weight gain in Yangzhou goose. The cDNA sequence of gene was firstly cloned, and the gene expression profiles were analyzed in different tissues. Comparative sequencing was used to find SNPs in UCHL1 promoter region which was related to body weight. The effect of the identified SNP on the expression of UCHL1 as well as hypothalamic appetite-related genes was evaluated. We hope that this study will provide candidate genes or molecular markers for the improvement of the goose growth performance.
MATERIALS AND METHODS
Ethics Statement
Animal experiments were conducted in accordance with the principles and specific guidelines presented in Guide for the Care and Use of Agricultural Animals in Research and Teaching, 3rd edition, 2010. All efforts were made to minimize any discomfort during goose slaughtering process.
Animals and Samples Preparation
One hundred and sixty-three male Yangzhou geese and2 hundred and forty-two female geese, provided by the breeding farm of Jiangsu Lihua Animal Husbandry CO. Ltd. (Jiangsu, China), were used in this study. During the experiments, geese were fed ad libitum with rice grain supplemented with green grass or water plants whenever possible. The feed was offered during daytime when the geese were released to an open area outside the house. The geese were exposed to natural lighting and temperature throughout this study. Individual record of 64-d body weight was obtained from Jiangsu Lihua Animal Husbandry CO. Ltd. Blood samples of all 405 geese were collected from wing vein for DNA extraction. Twelve geese (6 males and 6 females) were slaughter on D21 to collect the tissues including kidney, ovary, testis, small intestine, liver, abdominal fat, muscular stomach, breast muscle, hypothalamus, spleen, brain, cerebellum, and heart. The tissues were immediately frozen and stored in liquid nitrogen until total RNA extraction. Sixteen male geese with CC (n = 6) or TT (n = 10) genotype of UCHL1 gene were selected for brain sampling on D64.
UCHL1 Genomic DNA and CDS Region Cloning
According to the genomic sequence of duck UCHL1 gene (NC_040049.1), four pairs of primers (P1, P2, P3, and P4, Supplementary Table 1) were designed to clone the Yangzhou goose UCHL1 genomic sequence covering the full-length gene and upstream promoter region. PCR amplification was conducted in a final volume of 20 μL containing 1 μL DNA, 1 μL each primer (10 nM), 10 μL 2X Premix LA Taq (Takara, Dalian, China), and 7 μL dd H2O. The PCR reaction procedure were performed as follows: 98°C for 30 s, 32 cycles of amplification (98°C for 10 s, TM for 30 s and 72°C for 3 min) and a final extension at 72°C for 5 min. The PCR products were subjected to electrophoresis on 2% agarose gel and the target bands were excised under UV light. The target products were direct sequenced by a company (Tsingke, Nanjing, China). The obtained sequence was submitted to GeneBank database with accession number MT019969 and MN759312.
According to the chicken UCHL1 mRNA sequence (NM_001080212.1), one pair of primers (P5, Supplementary Table 1) were designed to clone the full-length CDS region of Yangzhou goose UCHL1 gene. PCR amplification was conducted in a final volume of 50 μL containing 2.5 μL first-strand cDNA, 2.5 μL each primer (10 nM), 25 μL 2X LA Taq Master Mix (Takara, Dalian, China), and 17.5 μL nuclease-free water. The PCR reaction procedure were performed as follows: 98°C for 30 s, 32 cycles of amplification (98°C for 10 s, 58°C for 30 s and 72°C for 1 min) and a final extension at 72°C for 2 min. The PCR products were subjected to electrophoresis on 2 % agarose gel, and the target bands were excised under UV light and purified using the E.Z.N.A. Gel Extraction Kit (Omega Bio-Tek, Doraville, GA) as recommended by the supplier. The purified products were cloned into the pClone007 vector (Tsingke, Nanjing, China), then transformed into the E. coli DH5α competent cells (Tsingke, Nanjing, China). PCR was used to identify the positive clones. The clones with goose UCHL1 cDNA fragments were sequenced by a company (Tsingke, Nanjing, China).
Identification of Body Weight Related Mutations
Twenty-six geese of high or low 64-d body weight (H 4725±144.45 g v.s L 3720±50.71 g, P< 0.001, n = 13 for each group) were chosen for comparative sequencing. The UCHL1 promoter region and full-length gene region were sequenced and compared between high and low individuals to identify candidate SNPs which are correlated with goose body weight.
Genotyping and Association Analysis
UCHL1 promoter region SNP c.-652T>C was genotyped for 405 Yangzhou geese by allele-specific PCR (AS-PCR). Two allele-specific primers and a universal primer were designed according to the protocol described previously (Bustos et al., 2000; Cosenza et al., 2008) (UCHL1-AS, UCHL1-C, UCHL1-T, Supplementary Table 1). The PCR reactions were performed in a total 20 μL system including 10 μL r-taq (Takara, Dalian, China), 1 μL forward primer, 1 μL reverse primer, 1 μL DNA template, and 7 μL ddH2O. The PCR reaction procedure were performed as follows: 94°C for 5 min, 32 cycles of amplification (94°C for 30 s, 57°C for 30 s, and 72°C for 30 s) and a final extension at 72°C for 7 min. The 262-bp PCR products were subjected to electrophoresis using 2 % agarose gel and the target bands were excised under UV light. Ten PCR products were randomly selected and sequenced (Tsingke, Nanjing, China) to confirm the veracity.
RNA Isolation and First Strand cDNA Synthesis
Total RNA was isolated from different tissues using a Trizol reagent (Invitrogen, Carlsbad, CA) according to the standard protocol, and subsequently treated with DNase I (Invitrogen). The quality of RNA samples was evaluated by electrophoresis on 1 % agarose gels. ProtoScript First Strand cDNA Synthesis kit (Takara, Dalian, China) was used to synthesize cDNA. The reverse transcription system contained 0.5 μg of total RNA, 2 μL of d (T) 23 VN (50 μM), and add nuclease-free water to 10 μL. The reaction procedure was performed as follows: 37°C for 15 min, then 85°C for 5 s. The reaction products were diluted to 50 μL with 40 μL nuclease-free water for PCR reaction and stored at -20°C.
Quantitative Real-Time PCR Analysis
Quantitative real-time PCR (qPCR) was used to determine the expression of UCHL1 (Supplementary Table 1, UCHL1-F, UCHL1-R) in various goose tissues including kidney, ovary, testis, small intestine, liver, abdominal fat, muscular stomach, breast muscle, hypothalamus, spleen, brain, cerebellum, as well as heart. The mRNA levels of UCHL1 and hypothalamic feeding-related neuropeptides gene (Primers in Supplementary Table 1) between c.-652T>C CC and TT genotype in the brain were also detect by qPCR, as well as in siRNA knockdown experiments in mouse hypothalamus cell line N38. SYBR® Green Master Mix (Vazyme, Nanjing, China) was used in a StepOne Plus Real-Time PCR system (Applied Biosystems). The PCR reaction (20 μL) consisted of 2 μL cDNA, 0.4 μL of each of primer (10 μmol), 0.4 μL ROX Reference Dye, 10 μL SYBR Green Master Mix, and 6.8 μL nuclease-free water. Amplification conditions were as followed: pre-denaturation at 95°C for 5 min, 40 cycles of amplification (95°C for 10 s and 60°C for 30 s). A melt curve analysis was performed from 60°C to 95°C by reading plate every 0.1°C. Each sample was detected for three times. Gene expression levels were calculated by 2−ΔΔCT method using GAPDH as an internal control (Adeola, 2018).
Preparation of Deleted Fragments of UCHL1 Gene Promoter
According to the sequence of goose UCHL1 gene we sequenced, 3 fragments of 1173, 767 and 421 bp upstream of the translation initiation site were amplified by using genomic DNA as a template with primer pairs including KpnI and HindIII restriction sites (underlined residues) (P6, P7, P8, Supplementary Table 1). The DNA was amplified in a 50-μL reaction mixture containing 25 μL of 2x Premix LA Taq, 2 μL (10 μmol/L) of each primer, 2 μL of DNA template, with sterilized distilled water to make up the final reaction volume of 50 μL. The PCR amplification protocol consisted of denaturation at 98°C for 30 s, 32 cycles of amplification (98°C for 10 s, Tm for 30S and 72°C for 1 Kb/min) and a final extension at 72°C for 7 min. The resulting PCR fragments and the pGL3-basic plasmid were digested using KpnI and HindIII. The 20 μL system of enzyme digestion consisted of 600 ng of PCR fragments or plasmid, 1 μL of KpnI or HindIII, 2 μL of 5 × Fast digest buffer, and sterilized distilled water to make up the final volume of 20 μL. PCR fragments or plasmid was digested for 1 h at 37°C using the KpnI or HindIII restriction enzyme. The target fragment was recycled through the column, and the pGL3-basic plasmid was recycled from agarose gel. The ligation mixtures of each PCR fragment and the plasmid consisted of 2.5 μL of 10 × ligation buffer, 1 μg of DNA fragments (KpnI/ HindIII), 1 μg of pGL3-basic plasmid (KpnI/ HindIII), and 1 μL of T4 DNA ligase (10 U/μL), then add water to 20 μL system. The ligation mixtures were then transformed into E. coli DH5α cells. The positive recombinant products were selected on Luria-Bertani agar plates containing 100 μg/mL ampicillin, and confirmed by PCR and DNA sequencing. The fragments of interest were recovered from the agarose gel, purified, and ligated by T4 DNA ligase, resulting in the recombinant plasmids pGL3-1173, pGL3-767, pGL3-421 (named by target fragment length).
Site-Directed Mutagenesis
Site-directed mutagenesis was performed using pGL3-767 (pGL3-T) as a template according to the Mut Express® II Fast Mutagenesis Kit*(Vazyme, Nanjing, China) with the -767 (T / C) site mutation primers (P9, Supplementary Table 1). The PCR amplification protocol consisted of denaturation at 95°C for 30 s, 30 cycles of amplification (95°C for 10 s, 62°C for 15S and 72°C for 1 min) and a final extension at 72°C for 5min. And the PCR products were stored at 4°C. Then the PCR product was digested with DpnI, ligated into pGL3 vector, placed in ice bath and cooled for 5 minutes, and then was transformed to obtain a mutated vector (pGL3-C).
Cell Transfection and Dual Luciferase Reporter Assay
293T cells were seeded on 12-well plates (at a density of 2 × 104 cells/well) 24 h before the start of transfection. Transfection using Lipofectamine 2000 was performed according to the manufacturer's protocol. The medium was replaced with antibiotic-free DMEM medium before transfection. 1500 ng of plasmid DNA (pGL3-basic, pGL3-1173, pGL3-767, pGL3-421, pGL3-T, pGL3-C) was transfected into the cells per well (plasmid pRL-TK was transfected simultaneously with the desired plasmid: internal control plasmid = 50: 1 ratio). 24 h after plasmid transfection, cell lysates were obtained and the luciferase activities were determined using the Dual-Luciferase Reporter Assay System according to the manufacturer's instructions (Harger and Dinman, 2003; Noguchi et al., 2012).
Bioinformatics Analysis
Sequence chromatograms were examined and edited by Chromas Version 2.23 (http://technelysium.com.au/). The sequences comparison was conducted by DNAMAN 8.0 (http://www.lynnon.com/). Related sequences were identified with ensembl (http://www.ensembl.org/index.html), Genbank (http://www.ncbi.nlm.nih.gov/genbank) and BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). The identification of ORFs were analyzed by ORF Finder tool of NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Protein Sequences were translated using DNAStar 5.02 (DNASTAR Inc.). Phylogenetic tree construction was performed by MEGA 7.0 (http://www.megasoftware.net/). The molecular weight and isoelectric point of this protein were analyzed using the ExPASy ProtParam tool (http://www.expasy.org/tools/protparam.html). The transcription factor was predicted by TRANSFAC®Professional(http://gene-regulation.com/).
RESULT
UCHL1 Gene Cloning and Mutation Detection
The goose UCHL1 genomic DNA of 7411 bp length was cloned by 4 pairs of primers designed according to duck UCHL1 genomic DNA, which covers 3071 bp upstream of ATG translation starting point. The 4340 bp starting from ATG translation starting point includes the full-length of goose UCHL1 gene. The obtained sequences were submitted to GenBank database with the accession number MT019969 and MN759312.
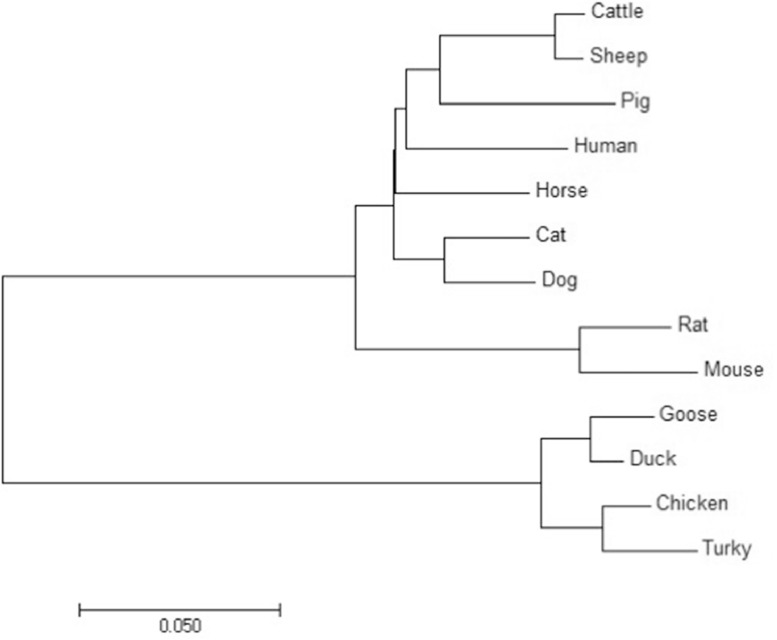
A 675 bp length of goose UCHL1 coding sequence was cloned from goose brain which included the translation initiation codon ATG and the stop codon TGA. Nine exons were identified in the CDS region when mapping UCHL1 cDNA sequence with genomic DNA sequence. The CDS sequence is predicted to encode a peptide of 224 amino acids with a molecular weight of 25038.43 Da, and its theoretical pI is 5.68. Its product is an unstable and hydrophilic protein, mainly distributed in the cytoplasm. According to the analyses with INTERPRO and PROSITE online software, UCHL1 contained protein domain of ubiquitin carboxyl-terminal hydrolase from amino acid residues 2-224. A conserved cysteine active-site (UCH_1) was located at amino acid residues 84–100. Gene ontology term prediction results indicated that UCHL1 is involved in ubiquitin-dependent protein catabolic process and protein deubiquitination. The nucleotide sequence of the coding region of the goose UCHL1 was highly similar to that of other avian species such as duck (97.6%), chicken (94.%) and turkey (92.9%), but was relatively less similar to human (75.5%), rat (73.4%), mouse (74.1%), horse (76.3%), pig (74.8%), cattle (75.9%), sheep (76.2%), dog (75.7%) and zebrafish (64.0%). The phylogenetic tree of UCHL1 CDS sequence was shown in Figure 1.
Figure 1.
The phylogenetic tree of UCHL1 CDS sequence was constructed with the neighbor-joining method by using MEGA7. A 1000 bootstrap replication was chosen to test the reliability of each branch. Bootstrap values are indicated as numbers at the branch nodes. CDS sequences of UCHL1 for these species were downloaded from nucleotide database of NCBI, those NCBI GenBank accession number is listed below: human (NM_004181.5), rat (NM_017237.3), mouse (NM_011670.2), cattle (NM_001046172.2), sheep (XM_004009789.3), horse (NM_001081820.1), pig (NM_213763.2), duck (XM_027457192.1), chicken (NM_001080212.1), cat (XM_011281913.2), dog (XM_022416363.1), zebrafish (NM_201477.1), turkey (XM_010710310.2).
Twenty-six geese of high or low 64-day body weight (n=13 for each group) were used to compare differences of UCHL1 genomic sequences by direct sequencing. Totally, 39 SNPs were revealed in 7411 bp length UCHL1 genomic sequences, 26 of which were distributed in intron region. A synonymous mutation c.195A>G was identified in exon 4. A mutation c.- 652C>T was found in the promoter region 652 bp upstream of ATG translation starting point. The frequency distribution of SNPs alleles in high and low groups was compared, which showed that allele frequencies of SNP c.- 652C>T in high and low groups were different (P < 0.01; Table 1).
Table 1.
The gene frequency between the high and low 64-day body weight groups.
| Genotype |
Allele |
|||||
|---|---|---|---|---|---|---|
| Groups | CC | CT | TT | C | T | Pearson Chi-square |
| Low group | 3 | 6 | 4 | 12 | 14 | 7.589 |
| High group | 0 | 3 | 10 | 3 | 23 | P < 0.01 |
SNP Genotyping and Association Analysis
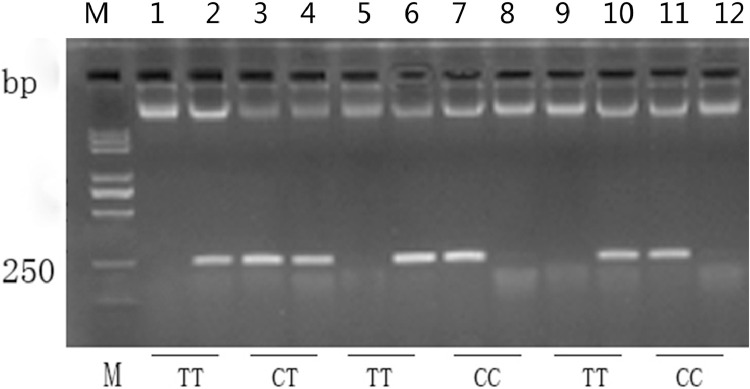
By allele specific PCR (AS-PCR), the mutation c.-652C>T in regulation region was genotyped in a total of 405 geese. Three different genotypes were detected: CC, CT and TT (Figure 2). In male geese, individuals with TT genotype showed a higher 64-day body weight compared with CC and CT ones (P < 0.01; Table 2). However, no difference was found among 3 genotypes in female geese, which indicated that the SNP c.-652C>T of UCHL1 was linked to growth performance in Yangzhou geese, and this effect might relate to gender.
Figure 2.
Genotyping of SNP c.-652C>T in UCHL1 gene by AS-PCR. M: Marker; Lane 1/2: sample 1, TT genotype; Lane 3/4: sample 2, CT genotype; Lane 5/6: sample 3, TT genotype; Lane 7/8: sample 4, CC genotype; Lane 9/10: sample 5, TT genotype; Lane 11/12: sample 6, CC genotype.
Table 2.
The association analysis between different genotypes of SNP c.-652C>T and body weight (BW) of 64-day geese in Yangzhou geese.
| Gender | Genotype | Number | BW |
|---|---|---|---|
| Male | CC | 15 | 3989 ± 51.13a |
| CT | 69 | 4063 ± 27.87a | |
| TT | 79 | 4204 ± 33.22b | |
| Female | CC | 52 | 3552 ± 44.61 |
| CT | 90 | 3531 ± 37.41 | |
| TT | 100 | 3526 ± 37.10 |
The BW is presented as mean ± SD. Multiple comparisons were performed using the Duncan multiple-range test.
a,bP < 0.05.
Identification of UCHL1 Core Promoter Region
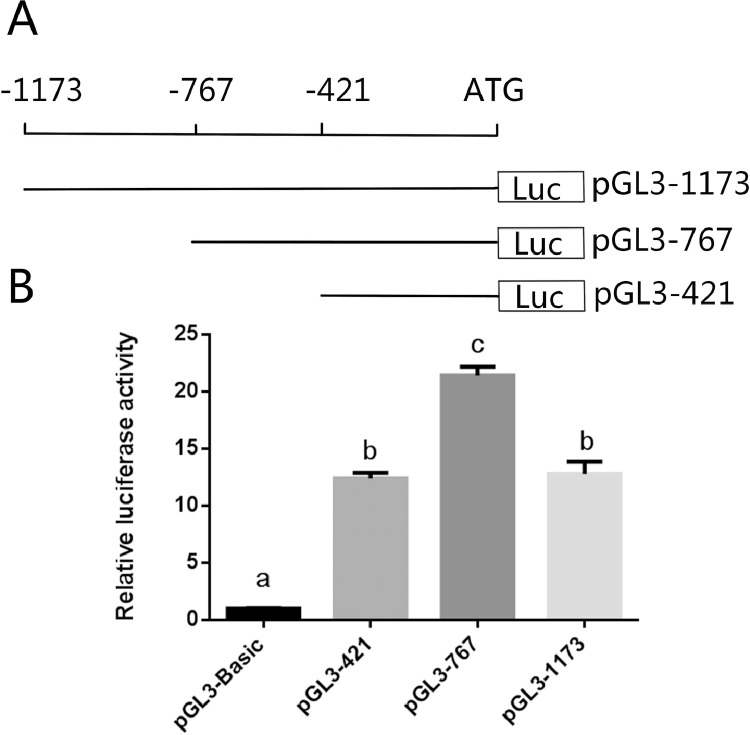
The core promoter region of the goose UCHL1 gene was predicted by the core promoter region prediction website (http://www.cbs.dtu.dk/services/Promoter/). The results showed that the transcription start site was located around c.-472bp of UCHL1 gene. Three overlapped fragments containing 1173 bp, 767 bp and 421 bp upstream of ATG were amplified and ligated in pGL3-basic vector to generate constructs pGL3-1173, pGL3-767 and pGL3-421, respectively. The exact location was shown in Figure 3A. The ligated plasmids were electrophoresed and the result of double enzyme digestion identification indicated luciferase reporter vector pGL3-1173, pGL3-767, pGL3-421 construction was successful. These positive constructs were sequenced and the results showed that the vector was successfully constructed. The transcription factor was predicted by TRANSFACProfessional (http://gene-regulation.com/). The results showed that there are 2 transcription factors (Oct-1, v-Myb) in the pGL3-1173 region (Figure 4A), and v-Myb also be predicted in the pGL3-767 region (Figure 4B), while there are no any transcription factors in the pGL3-421 region (Figure 4C) with the program of Match-1.0 Public. Interestingly, we found that TT genotype can bind transcription factor GR and AR, but CC genotype can't bind any transcription factor when we predicted it by the program of Patch (Figure 4D). The transcriptional activities of the different plasmids constructs were determined by dual-luciferase reporter assay in 293T cells. The results indicated that transcriptional activity of all the constructs was elevated at least twelve-fold greater than pGL3-basic without promoter (Figure 3B). The transcriptional activity of the construct pGL3-767 was highest of the 3 constructs. Specifically, pGL3-767 was significantly higher than that of pGL3-421 and pGL3-1173 (P < 0.001). The transcriptional activity of pGL3-421 was lower than those of pGL3-1173, but there was no significant difference in the relative fluorescence activity. These results indicated that the core promoter region of the UCHL1 gene is located between -421 bp and -767 bp.
Figure 3.
Identification of UCHL1 core promoter region. (A) The location of the 3 fragments upstream of the translation initiation site. (B) The relative fluorescence activity of the plasmids of pGL3-basic, pGL3-1173, pGL3-767 and pGL3-421. The different letters denote significant differences for mean comparisons (P < 0.001, n = 3).
Figure 4.
The predicted results of transcription factors in the truncated promoter regions. The transcription factor was predicted by TRANSFAC®Professional (http://gene-regulation.com/). (A) The predicted results of transcription factors in the pGL3-1173 region. (B) The predicted results of transcription factors in the pGL3-767 region. (C) The predicted results of transcription factors in the pGL3-421 region. (D) The predicted results of transcription factors between CC and TT genotype.
Effect of c.-652C>T on UCHL1 Promoter Transcriptional Activity
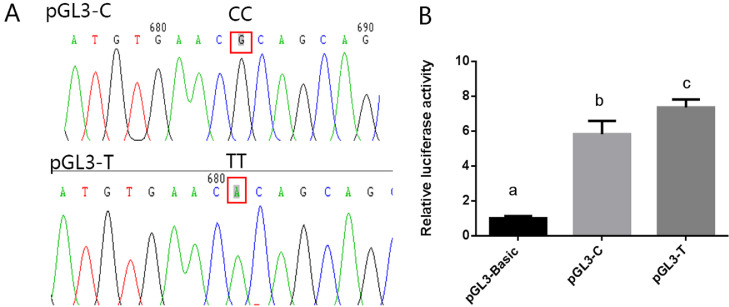
Because of the SNP c.-652C>T is exactly located in the region from -421 to -767 bp, we further investigated whether this mutation can influence UCHL1 promoter transcriptional activity. PCR was performed by c.-652 (C / T) site mutation primer using the pGL3-767 (pGL3-T) as template to obtain c.-652 C plasmid (pGL3-C). Double enzyme digestion and sequencing results showed that the mutant vector was successfully constructed (Figure 5A). The 293T cells were cotransfected with pRL-TK and pGL3-T, or pRL-TK and pGL3-C. pRL-TK and pGL3-Basic cotransfection group was used as a negative control. Twenty-four hours after transfection, cells were lysed and the luminescence values of the 2 luciferases were measured on the GloMax 20/20 instrument. The result showed that the promoter activity was significantly increased by 12.0% (P < 0.01) when the c.-652C allele was changed to c.-652T (Figure 5B).
Figure 5.
The transcriptional activities of the different genotype plasmids. (A) The sequencing result of the plasmids of pGL3-C and pGL3-T. (B) The relative fluorescence activity of the plasmids of pGL3-basic, pGL3-C and pGL3-T. Two-tailed student's t-tests were performed to compare relative fluorescence activity in different groups. The error bars represent the standard error of the mean. The different letters denote significant differences for mean comparisons (P < 0.05, n = 3).
UCHL1 mRNA Expression Profile in Yangzhou Geese Tissues
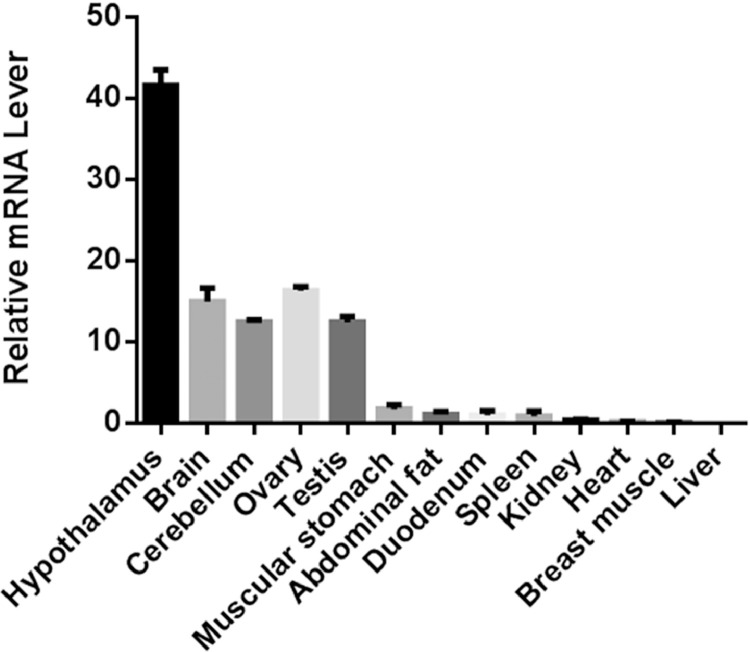
The expression levels of UCHL1 mRNA in 13 tissues (hypothalamus, brain, cerebellum, ovary, testis, abdominal fat, liver, kidney, spleen, breast muscle, muscular stomach, heart and duodenum) were evaluated by qPCR. The results showed that UCHL1 was specific expressed in the brain and gonads (Figure 6). Except for brain and gonads, the mRNA expression levels of UCHL1 gene in muscle, stomach, fat, duodenum, and spleen were relatively higher compared to other tissues.
Figure 6.
UCHL1 mRNA expression profile in Yangzhou geese tissues. qPCR was performed to evaluate the expression levels of UCHL1 mRNA in liver, ovary, testis, heart, breast muscle, hypothalamus, brain, abdominal fat, cerebellum, muscular stomach, duodenum, spleen and kidney.
UCHL1 mRNA Level Differs in Geese With Different Genotypes
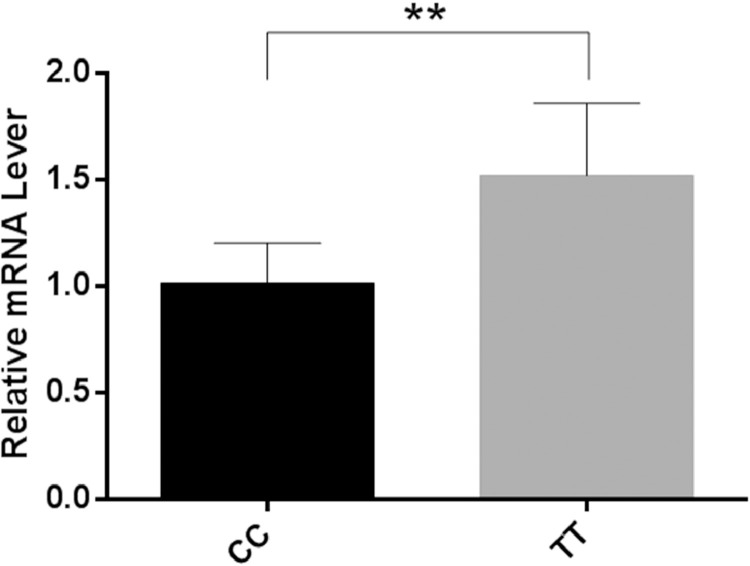
The relative expression level of UCHL1 in brain was compared between the 2 groups with CC and TT genotypes of SNP c.-652C>T. The results indicated that geese with TT genotype had higher mRNA expression level of UCHL1 in brain than those of CC genotype (P < 0.01) (Figure 7). Besides, TT genotype individuals showed higher 64-day body weight compared with CC individuals in our study (Table 1). The better body weight performance and higher mRNA levels observed in TT genotype individuals indicated that the mRNA expression level of UCHL1 was positively correlated with body weight performance in Yangzhou geese.
Figure 7.
The mRNA expression levels of UCHL1 gene in the brain of CC and TT genotype geese. Two-tailed student's t-tests were performed to compare gene expression in the 2 genotypes. The error bars represent the standard error of the mean. ** P < 0.01, n = 6 (CC genotype), n = 10 (TT genotype).
Effect of UCHL1 c.-652 C>T Mutation on Hypothalamic Feeding-Related Neuropeptides
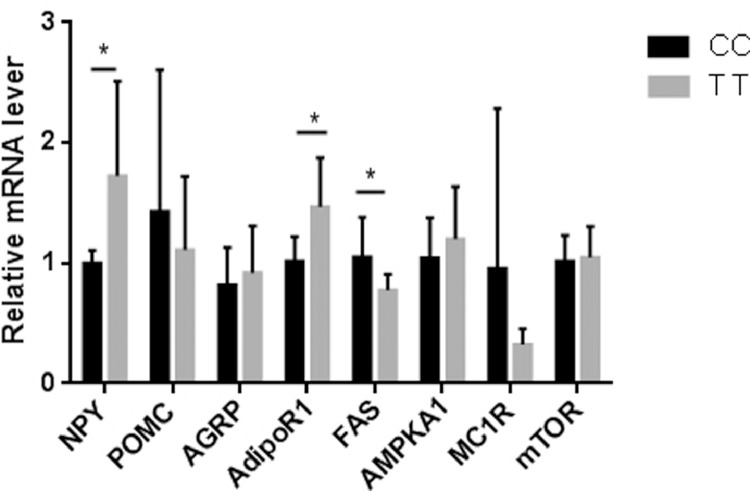
Because of the body weight is significantly different between 2 homozygotes of c.-625C>T, we investigated whether this mutation could cause changes of hypothalamic feeding-related neuropeptides. We examined the expression of feeding-related peptides in the brain of different genotypes, including neuropeptide Y (NPY), pro-opiomelanocortin AGRP, AdipoR1, FAS, AMPKA1, MC1R and mTOR. Compared with the CC groups, NPY and adipoR1 mRNA level was significantly increased in the TT groups (P < 0.05), while the FAS mRNA level was significantly decreased in the TT groups (P < 0.05) (Figure 8).
Figure 8.
The mRNA expression levels of Hypothalamic feeding-related neuropeptides gene in the brain of CC and TT genotype geese. The error bars represent the standard error of the mean. * P < 0.05, n = 6 (CC genotype), n = 10 (TT genotype). two-tailed student's t-test.
DISCUSSION
Body weight is an important trait for assessing the growth performance of goose, which is crucial for economic benefits and breeding. The 9-wk-old goose usually reaches the adult weight. In this study, the body weight of the 64-day Yangzhou goose were used to evaluate the growth performance. Recently, some studies have found that UCHL1 gene plays an important role in energy metabolism, obesity regulation, and myoblast proliferation and differentiation (Wang et al., 2011; Lee et al., 2015; Shruthi et al., 2016; Gao et al., 2017; Bollepalli et al., 2018). However, there is no report of UCHL1 gene related to goose growth performance.
As UCHL1 gene sequence was not available in goose, we firstly obtained the full-length genomic DNA and coding region sequence of goose UCHL1 gene by comparative cloning using duck and chicken sequences as references. Sequence homology alignment revealed that the CDS of goose UCHL1 is of high similarity to those of duck, chicken and other birds, which indicates that UCHL1 gene is relatively conserved among birds. Next, we scanned the promoter region of the goose UCHL1 gene by direct sequencing, and found a novel mutation c.-652C> T was significantly related to the body weight of the Yangzhou male goose but not female goose. It was further proved that this SNP was located in the core promoter region and could change the transcriptional activity of the UCHL1. According to the prediction results of transcription factors (Figure 4), TT genotype has higher transcription level than CC genotype, which may be due to the fact that TT genotype can bind GR and AR transcription factors, while CC genotype cannot bind any transcription factors, which leads to the difference of promoter transcription activity.
As a member of ubiquitin carboxy terminal hydrolase family of deubiquitinase, UCHL1 regulates protein degradation by catalyzing hydrolysis of C-terminal ubiquitin esters and amides. UCHL1 is exclusively expressed in neurons, neuroendocrine cells and gonads (Gao et al., 2019). Our result is consistent with the previous report, which shows that the highest level of UCHL1 is in hypothalamus, followed by brain, cerebellum, ovary and testis. Interestingly, we found that TT homozygote of c.-652C> T SNP had higher UCHL1 level in brain, and higher body weight as well. Given that hypothalamus is a key organ involved in neuroendocrine regulation of growth, we wondered if the effect of c.-652C> T on goose growth performance was mediated by hypothalamic feeding-related peptides.
The central nervous system controls food intake and energy expenditure via tight coordination between multiple neuronal populations. Specifically, 2 distinct neuronal populations exist in the arcuate nucleus of hypothalamus: the anorexigenic (appetite-suppressing) pro-opiomelanocortin neurons and the orexigenic (appetite-increasing) (NPY)/agouti-related peptide (AGRP) neurons (Sohn, 2015). Adiponectin (ADPN) and its related receptors (AdipoR1 and AdipoR2), fatty acid synthase (FAS), melanocortin system (MC1R), mechanistic target of rapamycin kinase(mTOR), and AMP-activated protein kinase alpha subunit (AMPKA1and AMPKA2) all play a critical role in the regulation of food intake and energy expenditure (Blankenship et al., 2016), and for this reason the mRNA level of these genes in goose brain were tested between CC and TT genotype. The result showed that compared to CC genotype, the mRNA level of NYP and adipoR1 in TT genotype are significant increased, while the mRNA level of FAS is significant decreased, and others have no significant change between the 2 genotypes. Then, we verified this result in mouse hypothalamus N38 cell lines by using siRNA interference experiments (primers showed in Supplementary Table 2). The results showed that siRNA could significantly reduce the amount of UCHL1 mRNA. Compared with the control groups, NPY, adipoR1 and ADPN mRNA level was significantly decreased, while the FAS and AMPKA1 mRNA level was significantly increased in the UCHL1 knockdown groups treated with siRNA (Supplementary Figure 1).
Thus, in geese, as the abundance of UCHL1 increases, the abundance of NPY and ADPN and their receptors (adipoR1 but not adipoR2) also increases, while the mRNA level of FAS decreases. NPY stimulates appetite and promotes lipid deposition in chicken (Liu et al., 2017). In mouse, ADPN stimulates AMP-activated protein kinase in the hypothalamus and increases food intake (Kubota et al., 2007), and ADPN has a remarkable effect on impairment of adipocyte differentiation, which contributes to the negative regulation of fat deposition in chicken (Yan et al., 2014). And the role of adiponectin in the regulation of feed intake in mammals is controversial (Coope et al., 2008; Huang et al., 2012). ADPN and its related receptors (AdipoR1 but not AdipoR2) were down regulated by UCHL1 gene knockdown experiment in N38 cells, as well as AdipoR1 was down regulated in TT genotype individuals. Downregulation of fatty acid synthesis via FAS depletion in white adipocytes of mature mice can stimulate neuronal signaling to control thermogenic programming in iWAT (Guilherme et al., 2017). Hypothalamic FAS is regulated by energy status in chickens and effect fatty acid synthesis on food intake (Dridi et al., 2006). Interestingly, ADPN has been shown to reduce FAS gene expression in 3T3-L1 cells (Lazra et al., 2015). Mitochondria play a key role in energy metabolism in many tissues such as skeletal and cardiac muscle, as well as in the liver, brain, and adipose tissue (Cedikova et al., 2016).
As one of mitochondrial biogenesis genes, AMPKA1 was up-regulated in UCHL1 knockdown group compared to control group in N38 cells, which probably indicated that higher expression of UCHL1 promotes less energy expenditure. In our study, we did not find any significant change in mTOR mRNA level. Consistently, in C2C12 cells, UCHL1 knockdown increased the myotube size, enhanced mTORC1 activity, and reduced mTORC2 activities as compared with control cells. UCHL1 knockdown did not change the major proteins of mTOR complex but decreased the protein turnover of PRAS40, an inhibitory factor of mTORC1 (Gao et al., 2019). Thus, UCHL1 may affect myoblast proliferation and differentiation by changing mTORC1 and mTORC2 activities, in spite of no changes in major proteins of mTOR complex.
Comprehensive expression level of hypothalamic feeding-related neuropeptides gene may affect appetite and growth performance of geese (Zhao et al., 2016; Clark et al., 2018; Rui et al., 2018). In our study, UCHL1 change the expression NPY, ADPN, FAS mRNA expression, resulting in changes in male goose growth performance. E2 affects the orexigenic NPY/ AGRP neurons in rapid but antagonistic manner (Stincic et al., 2018). This may explain why the change in the UCHL1 expression level did not affect the change in bodyweight of the female geese but in male geese.
In summary, we identify a novel mutation c.-652C>T in the promoter region of goose UCHL1 gene, which is linked with goose growth performance. The SNP can affect UCHL1 transcription activities, which may lead to changes in the expression level of hypothalamic feeding-related peptides. The SNP c.-652C>T therefore is a new promising genetic marker for goose growth performance selection.
ACKNOWLEDGMENTS
This work was supported by the Creation Project of Major New Varieties of Agriculture in Jiangsu [grant numbers PZCZ201738]. The funding source only provided financial support, and not played any role in study design, in the collection, analysis and interpretation of data, in the writing of the report, and in the decision to submit the article for publication. We thank Jiangsu Lihua animal Husbandry Co., Ltd. for providing animal samples for this study.
DISCLOSURES
The authors declared that they have no conflicts of interest to this work. We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2021.101089.
Appendix. Supplementary materials
REFERENCES
- Aaron L., Cross S.S., Catto J.W.F., Giancarlo P., Hamdy F.C., Ishtiaq R.J.P. Human prostate cancer cells express neuroendocrine cell markers PGP 9.5 and chromogranin A. Prostate. 2010;67:1761–1769. doi: 10.1002/pros.20654. [DOI] [PubMed] [Google Scholar]
- Adeola F. Normalization of gene expression by quantitative RT-PCR in human cell line: comparison of 12 endogenous reference genes. Ethiop. J. Health Sci. 2018;28:741–748. doi: 10.4314/ejhs.v28i6.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship K., Gilley A., Piekarski A., Orlowski S., Greene E., Bottje W., Anthony N., Dridi S. Differential expression of feeding-related hypothalamic neuropeptides in the first generation of quails divergently selected for low or high feed efficiency. Neuropeptides. 2016;58:31–40. doi: 10.1016/j.npep.2015.12.007. [DOI] [PubMed] [Google Scholar]
- Bollepalli S., Kaye S., Heinonen S., Kaprio J., Rissanen A., Virtanen K.A., Pietilainen K.H., Ollikainen M. Subcutaneous adipose tissue gene expression and DNA methylation respond to both short- and long-term weight loss. Int. J. Obes. (Lond) 2018;42:412–423. doi: 10.1038/ijo.2017.245. [DOI] [PubMed] [Google Scholar]
- Bustos A.D., Rubio P., Theoretical N.J.J., Genetics A. Molecular characterisation of the inactive allele of the gene Glu-A1 and the development of a set of AS-PCR markers for HMW glutenins of wheat. Theor. Appl. Genet. 2000;100:1085–1094. [Google Scholar]
- Cedikova M., Kripnerova M., Dvorakova J., Pitule P., Grundmanova M., Babuska V., Mullerova D., Kuncova J. Mitochondria in white, brown, and beige adipocytes. Stem Cells Int. 2016;2016 doi: 10.1155/2016/6067349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L.D., McCormick L.J., Velleman G.S. Effect of incubation temperature on neuropeptide Y and neuropeptide Y receptors in turkey and chicken satellite cells. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2018;219-220:58–66. doi: 10.1016/j.cbpa.2018.02.014. [DOI] [PubMed] [Google Scholar]
- Coope A., Milanski M., Araujo E.P., Tambascia M., Saad M.J., Geloneze B., Velloso L.A. AdipoR1 mediates the anorexigenic and insulin/leptin-like actions of adiponectin in the hypothalamus. FEBS Lett. 2008;582:1471–1476. doi: 10.1016/j.febslet.2008.03.037. [DOI] [PubMed] [Google Scholar]
- Cosenza G., Pauciullo A., Gallo D., Colimoro L., D'Avino A., Mancusi A., Research L.R.J.S.R. Genotyping at the CSN1TABLE S1 locus by PCR-RFLP and AS-PCR in a Neapolitan goat population. Small Rumin. Res. 2008;74:84–90. [Google Scholar]
- Day I.N., Thompson R.J. UCHL1 (PGP 9.5): neuronal biomarker and ubiquitin system protein. Prog. Neurobiol. 2010;90:327–362. doi: 10.1016/j.pneurobio.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Dridi S., Ververken C., Hillgartner F.B., Arckens L., Van der Gucht E., Cnops L., Decuypere E., Buyse J. FAS inhibitor cerulenin reduces food intake and melanocortin receptor gene expression without modulating the other (an)orexigenic neuropeptides in chickens. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006;291:R138–R147. doi: 10.1152/ajpregu.00899.2005. [DOI] [PubMed] [Google Scholar]
- Fan Z., Hu X., Zhang Y., Yu C., Qian K., Qin A. Proteomics of DF-1 cells infected with avian leukosis virus subgroup. J. Virus Res. 2012;167:314–321. doi: 10.1016/j.virusres.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Gao H., Freeling J., Wu P., Liang A.P., Wang X., Li Y. UCHL1 regulates muscle fibers and mTORC1 activity in skeletal muscle. Life Sci. 2019;233 doi: 10.1016/j.lfs.2019.116699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Hartnett S., Li Y. Ubiquitin C-Terminal Hydrolase L1 regulates myoblast proliferation and differentiation. Biochem. Biophys. Res. Commun. 2017;492:96–102. doi: 10.1016/j.bbrc.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover A., Sharma P.C. Development and use of molecular markers: past and present. Crit. Rev. Biotechnol. 2016;36:290–302. doi: 10.3109/07388551.2014.959891. [DOI] [PubMed] [Google Scholar]
- Guilherme A., Pedersen D.J., Henchey E., Henriques F.S., Danai L.V., Shen Y., Yenilmez B., Jung D., Kim J.K., Lodhi I.J., Semenkovich C.F., Czech M.P. Adipocyte lipid synthesis coupled to neuronal control of thermogenic programming. Mol. Metab. 2017;6:781–796. doi: 10.1016/j.molmet.2017.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger J.W., Dinman J.D. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.N., Qi J.H., Xiang L., Wang Y.Z. Construction of adiponectin-encoding plasmid DNA and overexpression in mice in vivo. Gene. 2012;502:87–93. doi: 10.1016/j.gene.2012.04.052. [DOI] [PubMed] [Google Scholar]
- Kandel E.R. Nerve cells and behavior. Sci. Am. 1970;223:57–67. doi: 10.1038/scientificamerican0770-57. passim. [DOI] [PubMed] [Google Scholar]
- Kubota N., Yano W., Kubota T., Yamauchi T., Itoh S., Kumagai H., Kozono H., Takamoto I., Okamoto S., Shiuchi T., Suzuki R., Satoh H., Tsuchida A., Moroi M., Sugi K., Noda T., Ebinuma H., Ueta Y., Kondo T., Araki E., Ezaki O., Nagai R., Tobe K., Terauchi Y., Ueki K., Minokoshi Y., Kadowaki T. Adiponectin stimulates AMP-activated protein kinase in the hypothalamus and increases food intake. Cell Metab. 2007;6:55–68. doi: 10.1016/j.cmet.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Lazra Y., Falach A., Frenkel L., Rozenberg K., Sampson S., Rosenzweig T. Autocrine/paracrine function of globular adiponectin: inhibition of lipid metabolism and inflammatory response in 3T3-L1 adipocytes. J. Cell Biochem. 2015;116:754–766. doi: 10.1002/jcb.25031. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Yoo J.Y., You Y.A., Kwon W.S., Lee S.M., Pang M.G., Kim Y.J. Proteomic analysis of fetal programming-related obesity markers. Proteomics. 2015;15:2669–2677. doi: 10.1002/pmic.201400359. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang G., Xiao Y., Shipp S.L., Siegel P.B., Cline M.A., Gilbert E.R. Peripheral neuropeptide Y differentially influences adipogenesis and lipolysis in chicks from lines selected for low or high body weight. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2017;213:1–10. doi: 10.1016/j.cbpa.2017.08.001. [DOI] [PubMed] [Google Scholar]
- Lombardino A.J., Li X.C., Hertel M., Nottebohm F. Replaceable neurons and neurodegenerative disease share depressed UCHL1 levels. Proc. Natl. Acad. Sci. U S A. 2005;102:8036–8041. doi: 10.1073/pnas.0503239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelker D.L., Yamashita K., Tokumaru Y., Mimori K., Howard D.L., Tanaka Y., Carvalho A.L., Jiang W.W., Park H.L., Kim M.S.J.C.R. PGP9.5 promoter methylation is an independent prognostic factor for esophageal squamous cell carcinoma. Cancer Res. 2005;65:4963. doi: 10.1158/0008-5472.CAN-04-3923. [DOI] [PubMed] [Google Scholar]
- Noguchi T., Ikeda M., Ohmiya Y., Nakajima Y. A dual-color luciferase assay system reveals circadian resetting of cultured fibroblasts by co-cultured adrenal glands. PLoS One. 2012;7:e37093. doi: 10.1371/journal.pone.0037093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka H., Wang Y.K., Takizawa S., Setsuie R., Li H., Sato Y., Nishikawa K., Sun Y.J., Sakurai M., Harada T.J.H.M.G. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum. Mol. Genet. 2003;12:1945–1958. doi: 10.1093/hmg/ddg211. [DOI] [PubMed] [Google Scholar]
- Pedro G.M., María Pilar M., José Alfredo M., o. M. S. María Jesús M.A.J.I.J. Differential proinflammatory and oxidative stress response and vulnerability to metabolic syndrome in habitual high-fat young male consumers putatively predisposed by their genetic background. Int. J. Mol. Sci. 2013;14:17238–17255. doi: 10.3390/ijms140917238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piórkowska K., Żukowski K., Ropka-Molik K., Tyra M., Gurgul A., Piórkowska K., Żukowski K., Ropka-Molik K., Tyra M., Gurgul A.J.G., Biology M. A comprehensive transcriptome analysis of skeletal muscles in two Polish pig breeds differing in fat and meat quality traits. Genet. Mol. Biol. 2018;41:125–136. doi: 10.1590/1678-4685-GMB-2016-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, M. 2004. Chapter 9 – neural cell specification during development. Pages 223–258 in Myelin Biology & Disorders. Academic Press, Cambridge, MA.
- Rui Wen, Xiang Gan, Shenqiang Hu, Shanyan Gao, Yan Deng. Evidence for the existence of de novo lipogenesis in goose granulosa cells. Poult. Sci. 2018;98:1023–1030. doi: 10.3382/ps/pey400. [DOI] [PubMed] [Google Scholar]
- Setsuie R., Wada K. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochem. Int. 2007;51:105–111. doi: 10.1016/j.neuint.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Shruthi K., Reddy S.S., Reddy P.Y., Shivalingam P., Harishankar N., Reddy G.B. Amelioration of neuronal cell death in a spontaneous obese rat model by dietary restriction through modulation of ubiquitin proteasome system. J. Nutr. Biochem. 2016;33:73–81. doi: 10.1016/j.jnutbio.2016.03.008. [DOI] [PubMed] [Google Scholar]
- Sohn J.W. Network of hypothalamic neurons that control appetite. BMB Rep. 2015;48:229–233. doi: 10.5483/BMBRep.2015.48.4.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stincic T.L., Ronnekleiv O.K., Kelly M.J. Diverse actions of estradiol on anorexigenic and orexigenic hypothalamic arcuate neurons. Horm. Behav. 2018;104:146–155. doi: 10.1016/j.yhbeh.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel E., Hibi K., Nagasaka T., Nakao A. PGP9.5 as a prognostic factor in pancreatic cancer. Int. J. Mol. Sci. 2000;6:4764–4767. [PubMed] [Google Scholar]
- Wang W., Li Q., Xu J., Cao X., Li H., Tang F., Chen Q., Yang J., Xu Z., Liu X. Over-expression of ubiquitin carboxy terminal hydrolase-L1 induces apoptosis in breast cancer cells. Int. J. Oncol. 2008;33:1037. doi: 10.3892/ijo_00000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.M., Yang H., Tian D.R., Cai Y., Wei Z.N., Wang F., Yu A., Han J.S. Proteomic analysis of rat hypothalamus revealed the role of ubiquitin-proteasome system in the genesis of DR or DIO. Neurochem. Res. 2011;36:939–946. doi: 10.1007/s11064-011-0423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Guo X., Zhang L., Hu D. Association between single nucleotide polymorphisms of fatty acid synthase and fat deposition in the liver of the overfed goose. Asian-Australas J. Anim. Sci. 2014;27:1244–1249. doi: 10.5713/ajas.2013.13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M., Wei W., Jiang Z., He D., Li Z., Yu S., Wang Q., Liu H., Chen J. A functional mutation in KIAA1462 promoter decreases glucocorticoid receptor affinity and affects egg-laying performance in Yangzhou geese. Int. J. Mol. Sci. 2018;19:1531. doi: 10.3390/ijms19051531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Yang H., Gan L., Sun C. Adiponectin-impaired adipocyte differentiation negatively regulates fat deposition in chicken. J. Anim. Physiol. Anim. Nutr. (Berl.) 2014;98:530–537. doi: 10.1111/jpn.12107. [DOI] [PubMed] [Google Scholar]
- Yu S., Chu W., Zhang L., Han H., Zhao R., Wu W., Zhu J., Dodson M.V., Wei W., Liu H., Chen J. Identification of laying-related SNP markers in geese using RAD sequencing. PLoS One. 2015;10 doi: 10.1371/journal.pone.0131572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Xia M., Alsiddig M.A., Liu H., Wei W., Chen J. Molecular cloning, alternative splicing and mRNA expression analysis of MAGI1 and its correlation with laying performance in geese. Br. Poult. Sci. 2017;58:158–165. doi: 10.1080/00071668.2016.1268251. [DOI] [PubMed] [Google Scholar]
- Zhang W.W., Xiao X., Gan J.K., Zhang X.Q., Kong L.N., Luo Q.B. Characterization of HSP70 and its expression in tissue: correlation with physiological and immune indices in goose (Anser cygnoides) serum. Genet. Mol. Res. 2015;14:12288–12298. doi: 10.4238/2015.October.9.17. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhu Z., Xu Q., Chen G. Association of polymorphisms of exon 2 of the growth hormone gene with production performance in Huoyan goose. Int. J. Mol. Sci. 2014;15:670–683. doi: 10.3390/ijms15010670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wang Q., Gong D., Yang B., Geng T. Identification of protective components that prevent the exacerbation of goose fatty liver: characterization, expression and regulation of adiponectin receptors. Comp Biochem Physiol B Biochem Mol Biol. 2016;194-195:32–38. doi: 10.1016/j.cbpb.2016.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.