Abstract
Abstract
COVID-19 is characterized by an unprecedented abrupt increase in the viral transmission rate (SARS-CoV-2) relative to its pandemic evolutionary ancestor, SARS-CoV (2003). The complex molecular cascade of events related to the viral pathogenicity is triggered by the Spike protein upon interacting with the ACE2 receptor on human lung cells through its receptor binding domain (RBDSpike). One potential therapeutic strategy to combat COVID-19 could thus be limiting the infection by blocking this key interaction. In this current study, we adopt a protein design approach to predict and propose non-virulent structural mimics of the RBDSpike which can potentially serve as its competitive inhibitors in binding to ACE2. The RBDSpike is an independently foldable protein domain, resilient to conformational changes upon mutations and therefore an attractive target for strategic re-design. Interestingly, in spite of displaying an optimal shape fit between their interacting surfaces (attributed to a consequently high mutual affinity), the RBDSpike–ACE2 interaction appears to have a quasi-stable character due to a poor electrostatic match at their interface. Structural analyses of homologous protein complexes reveal that the ACE2 binding site of RBDSpike has an unusually high degree of solvent-exposed hydrophobic residues, attributed to key evolutionary changes, making it inherently “reaction-prone.” The designed mimics aimed to block the viral entry by occupying the available binding sites on ACE2, are tested to have signatures of stable high-affinity binding with ACE2 (cross-validated by appropriate free energy estimates), overriding the native quasi-stable feature. The results show the apt of directly adapting natural examples in rational protein design, wherein, homology-based threading coupled with strategic “hydrophobic ↔ polar” mutations serve as a potential breakthrough.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s00894-021-04779-0.
Keywords: COVID-19, SARS-CoV-2, Protein design, Complementarity, Competitive inhibitor, Homology-based threading in rational protein design
Introduction
The world is currently facing an unprecedented global health crisis due to the sudden pandemic outbreak of a “naturally evolving” [1] virus known as Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) [2, 3]. The disease condition associated with SARS-CoV-2 known as COVID-19 was first reported in human subjects in the city of Wuhan, China, in December 2019 [4]. In a span of 13 months, more than 110.75 million people got infected with a death toll rising to 24,55,131 (WHO report, as of 4:21 pm CET, February 21st, 2021). The situation has challenged the very foundation of our existing global health management system, threatening with economic crisis that has never been faced before. The SARS-CoV-2 is a positive stranded RNA virus and a β-coronavirus. It shares a significant amount of genomic identity (79.5%) with its related previous strain SARS-CoV which got outspread in 2003 as an endemic, affecting more than 8000 individuals. However, the fatal impact and current nature of the SARS-CoV-2 pandemic is indicative of an altogether different functional nature of the virus from that of SARS-CoV [5]. Both SARS-CoV and SARS-CoV-2 were originated from bat but the immediate host from which SARS-CoV-2 got transmitted to human remains to be unclear [5]. The high person-to-person transmission rate of SARS-CoV-2 due to an efficient immune evasion and infectivity are of great concerns from the human intervention perspective [6–14]. So, taking into consideration of the potential pre- and post-symptomatic transmissibility of SARS-CoV-2, it is an urgent biomedical need to contain the spreading of this virus either by designing antiviral drugs or by vaccine development.
The host receptor recognition by SARS-CoV-2 and its entry mechanisms are important determinants of viral infectivity, tissue tropism, and pathogenesis. Alongside, these are also the key targets to modulate host immune surveillance and intervene the viral entry into host cells. Mature SARS-CoV-2 expresses envelop anchored trans-membrane Spike (S) glycoproteins that mediate the host cell entry. Distinct pre- and post-fusion conformational states of the S protein have very recently been structurally identified by cryo-electron microscopic (EM) studies [15] with the proposition of a “surprisingly low kinetic barrier” for the conformational transition. Primed by a conformation dependent proteolytic cleavage, the membrane fusion thus not only acts as the necessary mechanism for the host cell entry of the viral genetic material but also leads to two kinetically-related yet distinct conformations of the S protein. The pre-fusion conformation represents the full-length S protein, while the post-fusion form is a cleaved fragment left embedded on the viral membrane after the cleavage [15]. The post-fusion form is presumed to have subsequent functions, not only limited to the membrane fusion alone, for being strategically decorated with N-linked glycans [15]. Being the initial mediator of the essential host –pathogen interaction cascade, the pre-fusion form appears to be the more vulnerable [15] of the two forms. Both the forms are found to be biologically expressed and assembled as trimers; the post-fusion form is an elongated coiled coil and is thus more stable and rigid. The full-length pre-fusion S protein consist of two domains, the S1 receptor binding domain (henceforth referred to as RBDSpike) and the S2 membrane fusion domain, wherein, the three S1 receptor binding heads are situated on the top of the trimeric membrane fusion S2 stalk [16]. The S1 domain is further consisted of two subdomains—the N-terminal subdomain (NTD) and the C-terminal subdomain (CTD) [17]. The pre-fusion conformation has been resolved structurally at 2.8 to 3.3 Å resolution by several recent cryo-EM studies capturing minor variations between its different (closed/stabilized) states (PDB ID: 6VXX [18], 6CRZ [19], 6XR8 [15])—in all of which the RBDSpike remains structurally unaltered (see Supplementary Fig. S1). In addition to the RBDSpike, the pre-fusion form also contains a receptor binding motif (RBMSpike), both of which reside in the CTD in S1 unit [17].
The host cell entry of the SARS-CoV-2 involves a cascade of molecular interactions which has been revealed to be triggered by the binding of the RBDSpike to human angiotensin-converting enzyme-2 (ACE2) embedded on the membranes of human lung cells [20–22]. The experimental structure of SARS-CoV-2 RBDSpike conjugated with the human ACE2 receptor has also been resolved by X-ray crystallography at 2.68 Å (PDB ID: 6VW1) [21]. Upon this RBDSpike–ACE2 interaction, the Spike protein requires a proteolytic cleavage at its S1/S2 junction for S2 to gain an irreversible conformational change which leads to a successful host cell entry. A furin1 cleavage site has exclusively been found in the S1/S2 boundary of SARS-CoV-2 Spike protein recently [16]. Interestingly, RBDSpike has a stronger affinity for ACE2 than that of the whole Spike protein. This implies a more complex mechanism behind the molecular access of SARS-CoV-2 into the host cell [21]. Moreover, the S1 trimer continuously switches between a “lying down” and a “standing up” position onto the S2 subunits [15, 16, 23]. When S1 (composed of three monomeric RBDSpike units) is at a “lying down” position (or “down” state), it remains hidden and unexposed enabling the SARS-CoV-2 to escape the host immune surveillance [16]. It is only the “standing up” position (or, “up” state) of S1 that enables it to bind with the ACE2 receptor with a higher affinity compared to that of other related SARS-CoV. Taking into consideration these intricate complex features, SARS-CoV-2 stands out to be one of the most challenging pathogens ever to be contained. In addition, there are significant evolutionary differences in the antigenic properties of SARS-CoV and SARS-CoV-2 [18] in spite of sharing 70% sequence similarity in their RBDs and docking to an identical site in the ACE2 receptor. These strategic critical differences potentially lead to the ineffectiveness of a panel of monoclonal antibodies raised against SARS-CoV towards the neutralization of SARS-CoV-2 [24]. So, developing an effective vaccine targeting the S protein of SARS-CoV-2 remains complex and might take more time than can be afforded in this emergency. So, it is of high value to explore alternative means to design effective antivirals/bio-therapeutics that can successfully target the SARS-CoV-2 host cell entry thereby curbing down its infectivity.
Since the onset of the current pandemic, enormous efforts are continuously being made for repurposing already approved drugs [25–27], unfortunately with very limited success. Developing strategically designed small molecules and screening them against the viral infectivity is another approach to find a potential inhibitor to block key interactions of SARS-CoV-2 with host cells. Despite some initial promising outcomes, in most of the cases, these drugs are unable to stop the spread of COVID-19. Thus, developing strategic molecules to block the guest-host binding remains a clinically unmet goal. To that end, peptide-based approaches to design antiviral bio-therapeutics might be a fruitful alternative strategy [28]. The availability of experimental atomic structures of the SARS-CoV-2 RBDSpike complexed with the ACE2 receptor [19, 29] serves as a great resource for this purpose, helping in the detailed understanding of the binding mechanism, and thereby, facilitating the design. The binding affinity of SARS-CoV-2 with the “ACE2 peptidase domain α-helix” is much stronger than SARS-CoV. Designing a peptide disruptor would therefore be an ideal choice over screening of small molecule inhibitors because of its higher efficacy in covering the extended protein contact interface, potentially acting as a compelling competitive inhibitor [28, 30].
In this present study, we aim to design non-reactive structural mimics of SARS-CoV-2 RBDSpike which can serve as potential competitive inhibitors for its binding to the host ACE2 receptor. These polypeptide-based mimics have been designed to bind stably with high affinity to the interacting surface of ACE2 containing multiple contact hotspots. They would thus potentially interfere with the binding of the native SARS-CoV-2 RBDSpike to ACE2 by already occupying the binding sites. To that end, we adapted a protein design approach with iterative cycles of screening followed by Molecular Dynamics (MD) simulations of the finally selected structural mimics. The objective of the exercise was to examine the dynamic stability of the prescribed binary protein-protein interaction (PPI) complexes formed with ACE2. We followed two alternative sampling strategies for the design, based on (i) alteration of hydrophobic character of the mutable amino acids at the RBDSpike–ACE2 interface and (ii) homology-based threading followed by performing strategic “gain-of-function” mutations. Scoring of the designed binary PPI complexes were based on shape and electrostatic complementarities {Sc, EC} [31, 32] which are essential prerequisites of binding affinity and stability and may thus be envisaged as coordinate driven representative measures of the same, as reasoned in the paper. The SARS-CoV-2 RBDSpike is an independently foldable protein domain and remains resilient to conformational changes yet after acquiring a series of mutations along evolution. The prescribed designed protein binary complexes are therefore expected to fold naturally as self-sustaining protein units.
Interestingly, the RBDSpike–ACE2 interaction in SARS-CoV-2 appears to have a quasi-stable character in spite of having a high affinity for the interaction. The effect is more pronounced if compared to its evolutionary ancestor, SARS-CoV. This can be further envisaged as having a bouncing nature of the ligand upon receptor binding. This enables the molecule to quickly get released from its receptor-site to be able to bind to a greater number of amenable receptors in nearby cells. It therefore appears that the key molecular player of the most determining interaction in COVID-19 has an inherent structural potential to have a high interaction cross-section with its cognate receptor. This has been vividly surveyed and discussed in the light of molecular evolution of the RBDSpike from SARS-CoV to SARS-CoV-2. Taken together, the current study has both a basic and an applied content and provides a novel approach to design polypeptide-based inhibitors against SARS-CoV-2 RBDSpike–ACE2 binding. Subsequent wet lab experiments and testing of the prescribed designed sequences on biological subjects may potentially offer an alternative powerful therapeutic strategy to combat SARS-CoV-2, due to be carried out in the next phase.
Materials and methods
Details of experimental structures used in the study
For the all-important pre-fusion form of the viral Spike protein, we used the cryo-EM structures in its “closed state” (PDB ID: 6VXX; solved at 2.8 Å, 22812 protein atoms), that of a “stabilized variant” (PDB ID: 6CRZ; solved at 3.30 Å, 25024 protein atoms) and that of the recent most full-length S protein (PDB ID: 6XR8; solved at 2.9 Å, 25995 protein atoms) from the Protein Data Bank [33]. As a single representative structure, 6VXX was preferred among the three for having the best resolution. The three structures had variation in minor details (missing loops, glycans, etc.) which was reflected in their all-atom RMS deviation upon pairwise structural superposition (average, 3.75 Å for an average length of 23,255 aligned non-hydrogen protein atoms). The same average RMS deviation for the RBDSpike in the three structures was even lower (2.2 Å) for a stretch of ~190 aligned residues. Visual structural investigation confirmed that this small deviation was due to the conformational variation of the disordered loop regions while the relative orientation of the secondary structural elements (helices and sheets) was virtually identical in all structures (see the “Results and discussion” section). When the three structures were further superposed (in turn) onto the same (ligand) domain in the ACE2-bound binary complex (PDB ID: 6VW1), the average all atom RMS deviation reduced even further to 1.36 Å.
The other coordinate files used in the core-study correspond to ligand-receptor protein complexes, pertaining to most if not all representative structures of the RBDSpike (Receptor Binding Domain, UNP Residues: 323–502)-Angiotensin-Converting-Enzyme (ACE2) receptor available at the Protein Data Bank [33] till date (28/02/2021). The ones that were of prime importance among these are RBDSpike of SARS-CoV complexed with human ACE2 (PDB ID: 2AJF; solved at 2.9 Å) and RBDSpike of SARS-CoV-2 complexed with human ACE2 (PDB ID: 6VW1; solved at 2.68 Å). Among the rest of the structures used, there were human strains of the ancestral viral RBDSpike (i.e., the 2002–2003 SARS-CoV) complexed with human-civet chimeric receptors (two of them, PDB ID: 3D0G; solved at 2.8 Å and PDB ID: 3D0H; solved at 3.1 Å). There was also civet strain of the viral RBDSpike complexed with human ACE2 (PDB ID: 3SCJ; solved at 3.0 Å) and RBDSpike from SARS-CoV epidemic strain complexed with human-civet chimeric receptor ACE2: (PDB ID: 3SCL, solved at 3.0 Å). Importantly, the only binary PPI complex representative of CoV-2 (PDB ID: 6VW1) had a human ACE2 receptor in it.
Additionally, equivalent/similar binary PPI complexes from MERS (PDB ID: 4L72; MERS-CoV complexed with human DPP4, solved at 2 Å) and Ebola (PDB ID: 5F18; viral glycoprotein bound to its endosomal receptor Niemann-pick C1, solved at 3 Å) were also assembled as a means to compare the receptor-ligand binding in terms of affinity and stability from complementarity estimates. Patches of residues missing due to poor electron densities were modeled using MODELLER [34], wherever applicable. These missing patches essentially mapped to an equivalent stretch in all the RBDSpike which was a disordered loop far from the ACE2 binding site. All sequence alignments, pairwise and multiple, were performed by Muscle [35]. Sequence similarities wherever calculated used the EMBOSS stretcher web-tool implementing its global alignment module (https://www.ebi.ac.uk/Tools/psa/emboss_stretcher/).
Protein design: side-chain threading and shaking the designed binary PPI complexes
Our approach was to target and build inhibitors of the SARS-CoV-2 RBDSpike to block its binding sites in the ACE2 receptor. Hence, all mutations were performed on the native ligand molecule alone while retaining the native sequence of the receptor. To fit and thread the mutated side-chains on the native template, Scwrl4.0 [36] was used which samples the side-chain conformations from the Dunbrack’s Rotamer library and has its unique fast way of optimally removing steric clash. Subsequent to fitting the mutated side-chains on the native main-chain coordinates, side-chain coordinates of the unaltered amino acid residues were retained from the original native structure (6VW1, chain E). Existing polar hydrogen atoms generated by Scwrl4.0 was subsequently trimmed and all hydrogen atoms were rebuilt afresh by the program REDUCE (v.3.3) [37]. REDUCE geometrically builds hydrogen atoms on the existing heavy atom coordinates by analyzing the local hydrogen bond network, flips -CO and -NH2 groups in amidino groups of asparagine, glutamine and takes care of resonating states of histidine as appropriate to the given context. The rebuilt structures were then energy minimized by 500,000 steps of steepest dissent and 50,000 steps of conjugate gradient method in Gromacs and were subsequently undertaken for short (10 ns) all atom molecular dynamic simulations (refer to the “Molecular dynamic simulation (short and long)” section) as a means to consider vibrational perturbation (or shake) due to the performed multi-mutations on the native RBD. The short simulations ensure necessary structural relaxation of the designed binary PPI complexes (upon multi-mutations) by allowing sufficient main- and side-chain flexibility. The designed structures are hence released from being trapped in local energy minima. The post-run time-evolved snapshots (after 10 ns) were taken as the final designed structures.
Scrambled sequences as negative control
To serve as negative controls, a pool of scrambled sequences was constructed having an identical composition to that of the presumably potential solutions obtained from the protein design results (i.e., its different variant protocols adapted). For these sequences (hits), amino acid compositions were computed and grouped into six classes: C1. hydrophobic and branched-chain (Ala, Val, Leu, Ile, Met), C2. hydrophobic and aromatic (Phe, Tyr, Trp), C3. polar (Ser, Thr, Asn, Gln), C4. positively charged (Lys, Arg, His), C5. negatively charged (Asp, Glu), and C6. helix breaker and disulfide forming (Gly, Pro, Cys). Compositions (in terms of percentage of each class) were averaged over the “hits” which served as a compositional consensus. Randomly reshuffled sequences were then generated (hundreds of them) with identical compositions implementing the Fisher-Yates Shuffle algorithm (http://www.programming-algorithms.net/article/43676/Fisher-Yates-shuffle). These “scrambled” sequences together served as potential negative controls to the computational prediction (see the “Results and discussion” section), as a means to physically verify and cross-validate the importance of crucial and/or conserved amino acid positions in the native sequence (as in 6VW1, chain E) over and above merely meeting the compositional criteria.
Contact map at the interface
Amino acid residues buried upon association/complexation (i.e., interfacial residues) were identified by a net (non-zero) change in their atomic solvent Accessible Surface Areas (ASA’s) between their bound and free forms. In other words, an interfacial residue is preconditioned by ∆ASAresidue ≠ 0, where, ∆ASAresidue = ∑∆ASAatoms_of_the_residue. ∆ASA for each ith atom in the residue was computed in the following way:
| 1 |
where ASAbound(i) and ASAfree(i) refer to the ASA’s of each ith atom of the same residue in its bound and free forms. The interfacial atomic contacts were identified when any two heavy atoms coming from two amino acid residues residing at each molecular interfacial surface were found within 4 Å of each-other. A slight relaxation (4.5 Å) of this very stringent cutoff was also attempted. This collection of residue-wise atomic contacts served as the contact map at the receptor-ligand interface—which were vividly and explicitly used as one of the indicators to choose the mutations for the protein design experiment. The same standard cutoff was also used to identify salt-bridges [38, 39] at the receptor-ligand interface.
Shape and electrostatic complementarity
The semi-empirical function of shape correlation statistic (Sc) as formulated by Lawrence and Colman [31] was adopted as a mean to evaluate the Shape Complementarity of the binary PPI complexes at their interface. The program Sc (version 2.0, © Michael Lawrence) attributed to the original paper was used to serve the purpose. Implicit to this program, first, the molecular (Connoly) surfaces [40] were constructed, sampled at 15 dots/Å2 for both interacting molecular partners separately. The nearest neighboring dot surface points were identified within a maximum distance of 3.5 Å and the following measure (Sdp) computed for each pair of nearest neighboring dot points:
| 2 |
where and refer to the unit normal vectors, one outwardly and the other inwardly oriented, corresponding to the two dot points A and B coming from the two interfacial molecular surfaces; d is their distance and w is a scaling constant set to 0.5. Median of this distribution is taken as Sc.
Electrostatic Complementarity (EC) at the protein-protein interfaces was adopted as originally prescribed by McCoy et al., [32] wherein, the surface electrostatic potential was computed for each interfacial protein surface twice, one time each for the contribution of each partner molecule (taken as “target” and “neighbor”). The surface electrostatic potentials were computed by numerically solving the Poisson-Boltzmann equation (using Delphi v8.4 [41]) implementing its finite difference method, wherein, the protein dielectric was modeled as a smooth Gaussian function of distance from its center of mass [42]). This returns two troughs of potential values for each interfacial surface and the negative of the Pearson’s correlation is defined as the EC at each interfacial surface (see Eq. 3). The average of the two ECs obtained for the two interfacial surfaces (EC1, EC2) is taken as EC at the interface:
| 3 |
In the above equation (Eq. 3), if an interacting molecular surface consisting of N dot surface points is taken as the “target” molecule (or object), Φ(i) represents the electrostatic potential on its ith point realized due to its own atoms and Φ’(i), due to the charged atoms of its molecular partner, taken as “neighbor.” and are the mean potentials of Φ(i) and Φ’(i), i = 1..N, respectively. EC1,2 may interchangeably represent both EC1,2 and EC2,1 with the necessary swapping of “target” and “neighbor” and the corresponding potential terms (Φ ↔ Φ’).
Force-field parameters (atomic partial charges and Van der Waals radii) for the surface-bound carbohydrates (as in 6XR8) were generated following the methodology reported in a recent study on glycan shielding of the SARS-CoV-2 spike protein [43] using the glycoprotein builder available at GLYCAM-Web (www.glycam.org).
Both Sc and EC are essentially correlation measures ranging from −1 (perfect anti-complementarity) to 1 (perfect complementarity) having the same sense of directions (higher the better). The non-rigid optimal ranges for Sc and EC can be reasonably approximated as (0.55, 0.75) and (0.45, 0.65), respectively, as has been found in protein (binary) complexes coming from a wide variety of biological origins [31, 32]. Such a zone comprising of these ranges (optimal zone) can be viewed analogous to the basin of attraction of optimal states (attractors).
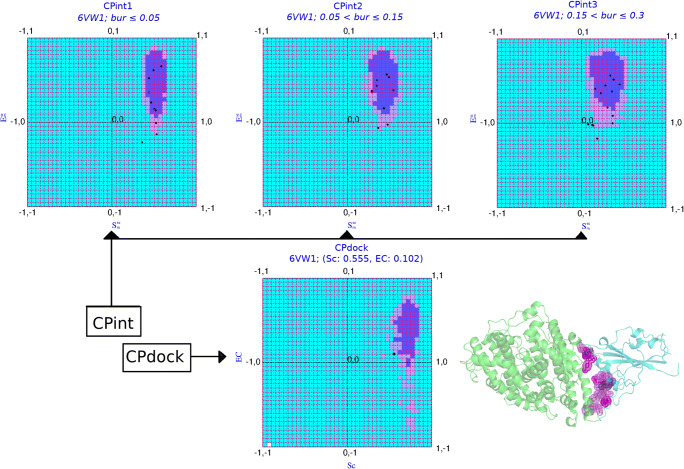
Complementarity Plot (CPint and CPdock)
Complementarity Plot [44–48] refers to a two-dimensional plot of the ordered-pair values of shape and electrostatic complementarities along its X and Y axes. The plot estimates the probabilistic correctness of an experimentally solved or a computationally built atomic model of a globular protein or a protein-protein complex, based on harmony of the embedded side-chains at their respective protein environments with their local and non-local neighborhood. The harmony with respect to the local and non-local neighborhood is estimated in terms of shape and electrostatic complementarity of buried and partially buried amino acid residues. The complementarity plot has three variants. The first two of them, namely, CP and CPint, are residue-wise plots plotting the ordered-pair complementarity values computed for buried or partially buried amino acid residues at the protein interior and interface, respectively. The third variant, CPdock, was originally proposed as a protein-protein docking scoring function [48] and is based on the aforementioned single {Sc, EC} values obtained for the whole protein-protein interface (see Fig. 1). In all the three variant plots, the resultant points may be found located to either of the three regions in the plots: “probable,” “less probable,” or “improbable” based on their probabilistic feasibility to fit into a folded protein or a protein (binary) complex model. Furthermore, as can be revealed from Fig. 1, the “probable” and “less probable” regions in CPdock (and those in the other variant CP’s) are primarily covered (>85% area) by the first (+, +) quadrant of the plot with Sc, EC both attaining positive values. Such “both positive” points would thus render a higher probabilistic feasibility of two proteins to interact and this probability would increase with the closeness of the point from the “probable” and/or “less probable” regions. Depending on the requirement, both CPint and CPdock were used in the study. CPdock was used for screening and scoring of the protein complexes, while CPint was utilized for shortlisting and identifying the amino acid residues at the interface to be attempted for mutations in the protein-design pipeline.
Fig. 1.
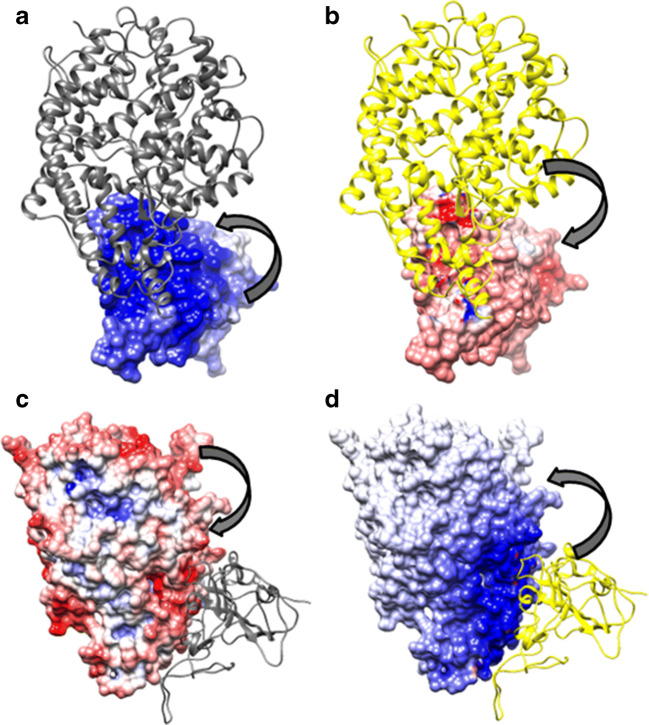
The Complementarity Plots (CPint and CPdock). The composite figure represents the two variants of the Complementarity Plot CPint and CPdock. CPint (upper panel) is the residue-wise plot, plotting the residue-wise complementarity estimates, Sm vs. Em [43] for interfacial residues—which is further distributed into three sub-plots (CPint1, CPint2, CPint3) based on their burial of solvent exposure (bur) of the plotted residues. CPdock (lower panel) is for the whole interface {Sc, EC}. The inner island colored in “purple,” the outer rim in “mauve,” and the rest in “sky blue” corresponds to the “probable,” “less probable,” and “improbable” regions of the plots. The pictorial demonstration is made on the very structure of 6VW1 (i.e., the RBDSpike–ACE2 complex in CoV-2) displayed at the right-bottom of the composite diagram. The interfacial residues of the ligand (RBDSpike: cyan cartoon) which are in physical contact with the receptor (ACE2: orange-yellow) are presented as their van der Waal’s dot surfaces colored according to their corresponding residences in CPint (“probable”: violet, “less probable”: magenta, “improbable”: violet-purple)
Accessibility score
The accessibility score (rGb) compares the hydrophobic burial profile (i.e., the distribution of amino acids as a function of solvent exposure) of a globular protein or a protein-protein complex with respect to corresponding native distributions, enumerated from standard databases. The score is also applicable to peptide fragments or protein domains. The accessibility score is an integral part of the structure validation protocol prescribed in the Complementarity Plot [45, 46]. Mathematically, the score is based on normalized conditional probability (or propensity) estimates of residue types given their burial (and hence the name: rGb) and can be formulated as follows:
| 4 |
where Nres is the sequence length of the input polypeptide chain and Pri is the propensity of a particular amino acid (Val, Asn, His, etc.) to acquire a particular degree of solvent exposure.
A value of rGb > 0.011 [45] (and higher the better) renders the input atomic model affirmative with regard to the “native-like” distribution of amino acids as a function of solvent exposure while a value less than that means hydrophobic residues are exposed to the solvent causing the molecule stay in an unfavorable/frustrated disordered (high entropy) state. A negative value emphasizes this instability which may be extended to explain the reaction-prone nature of the said fragment.
Fold recognition
Complementarity-based fold recognition measures (CSgl, CScp) [44] were implemented to test the compatibility of the designed sequences to the fold (i.e., main-chain trajectory) of the RBDSpike. A (μ - 3σ)2 baseline on the complementarity scores (CSgl:2.4, CScp:0.01) was set as a threshold value to determine the compatibility of a designed sequence to the given fold (μ, σ taken from the original reference).
Molecular dynamic simulation (short and long)
Molecular dynamic simulations were used in the study to serve a twofold purpose. As described previously (refer to the “Protein design: side-chain threading and shaking the designed binary PPI complexes” section), the short simulations were run implicit to the design protocol as a mean to incorporate vibrational perturbation to the in silico designed binary PPI complex. In contrast, long simulations were conducted to study the dynamics and stability of the binding of the finally selected binary PPI complexes (i.e., the proposed “optimal solutions”). In addition, the native binary PPI complex (6VW1) was also undertaken in the long MD simulation, as a mean to set baselines. In either case, the same protocol was followed but for changing the duration of the production phase. Explicit-water Molecular Dynamics (MD) simulation was performed in GROMACS v.2018.1 [49, 50] using the AMBER99SB-ILDN protein force-field [51], TIP3P water model and “solvent” as the ion replacing system associated with the MD package. Periodic boundary conditions were used; solvation and charge neutralization of the proteins were subsequently followed by two rounds of energy minimization (500,000 steps of steepest descent followed by 50,000 steps of conjugate gradient) using the in-built PROMD module [50] within GROMOS96. The energy minimized protein–solvent system was then equilibrated in an NVT ensemble followed by an NPT ensemble for 100 ps and 5 ns, respectively. The initial temperature set for the NVT ensemble was 100 K which was gradually raised to 300 K at constant volume and was kept the same for the entire NPT equilibration while the pressure being maintained at 1 bar. The simulation systems were large, consisting of a total number of 246,148 (±10) atoms at an average. The production runs were done in an NPT mode for 200 ns for the long MD simulation runs (10 ns for the short ones) with a time-step of 2 fs for each equilibrated protein–solvent system. The “cubic” simulation boxes were built by considering an initial length of at least 13 Å from the surface of the protein binary complex (placed at the center of the box) to each cubic face. This led to an average box-dimension of ~135 Å × 135 Å × 135 Å of the simulated solvated systems. To maintain constant temperature, Berendsen’s temperature bath was used with a coupling constant of 2 ps, while barostat with a coupling constant of 1 ps was used to regulate the constant pressure. The “LINCS” algorithm was used to restrain bond-lengths for all bonds. For the short simulation runs, the final snapshots (at 10 ns) were stored and used as the final designed structures for scoring. For the long simulation runs, trajectories were written at an interval of 2 ps, resulting in 100,000 frames (or time-stamps). Binding stability and other related dynamical analyses were all performed on the post-equilibrium 200 ns long trajectories (for the finally selected designed protein binary PPI complexes). For each simulated protein-complex trajectory, all post-simulation analyses were done 2000 snapshots collected at 100 ps interval. This sampling may be considered of sufficient resolution to capture the molecular events under investigation.
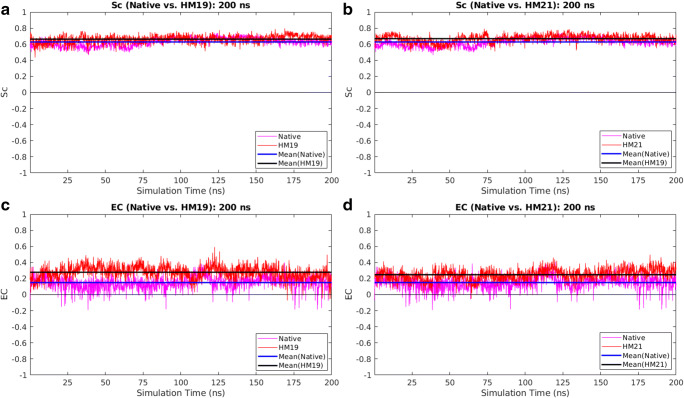
Measuring the dynamic stability of the proposed “optimal” solutions
To quantitatively assess the dynamic stability of the proposed “optimal” solutions, CPdock was run on their whole dynamic trajectory (i.e., on the selected collection of snapshots representing the trajectory). Alongside analyzing the dynamic persistence of the attained Sc, EC values individually, the ordered pair treatment of {Sc, EC} was also invoked by estimating the distance of the corresponding points in the plot from the “probable” region. To that end, a 2D Euclidean distance measure (E2d) was formulated based on a binary logic. If the {Sc, EC} point in the plot was found to be located on a “probable” grid, then E2d was set to zero. Otherwise, E2d was computed as the 2D Euclidean grid distance from the mid-point of the nearest edge belonging to the “probable” grid nearest to that point in the plot. It can be formally proven that E2d is a metric in an R2 vector-space (proof not given).
Estimating changes in binding/interaction energies for the proposed “optimal” solutions
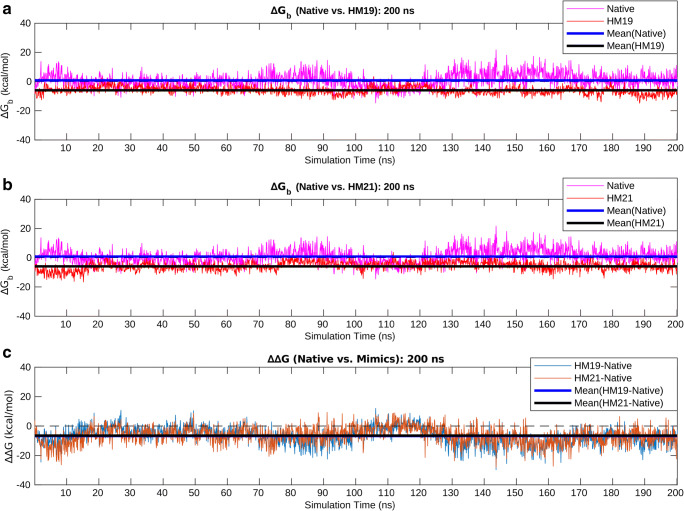
As a mean to cross-validate the directed designs performed based on the complementarity measures (Sc, EC), binding/interaction energies (ΔGbinding) of the native and the selected designed protein complexes (i.e., the proposed “optimal” solutions) were estimated along their 200 ns trajectories using the standalone (C++ with boost library) version (v.4) of FoldX (http://foldxsuite.crg.eu/) [52, 53]. FoldX performs fast computation of ΔGbinding/folding for proteins and PPI complexes directly from their high-resolution 3D coordinates (using full atomic description) and can efficiently be used to probe the cumulative effect of multiple mutations in stability of protein folds and/or protein-complexes from the corresponding ΔΔG estimates [54, 55]. Its advanced empirical force field includes van der Waals terms, solvent interaction terms (both polar and hydrophobic), hydrogen bonds, electrostatic contribution to free energies, atomic steric overlaps as well as the entropy cost for backbone and side-chain conformational changes. It is particularly impressive for protein engineering and stability analysis for its careful parameterization of the energy terms using empirical data from actual protein engineering experiments [52, 53].
To estimate the binding/interaction energies (ΔGbinding) between the receptor and the ligand chains in the native and a selected designed binary PPI complex (mimic), structural snapshots were sampled at 100 ps interval from their corresponding 200 ns MD simulation trajectories (resulting in 2000 time-stamps for each). Then, for each snapshot, FoldX was run using the command AnalyseComplex with the complexWithDNA parameter set to “false.” The resultant ΔGbinding values obtained for the native were then subtracted from the corresponding values of the selected designed mimic along their time evolution profiles to yield an equivalent time-evolved profile of their changes due to the performed directed design (as formulated in the following equation):
| 5 |
where i denotes the time-stamp in the corresponding time evolution profiles.
Time-series averages and standard deviations were further computed for all three parameters coupled with analyses of the corresponding time-series plots.
Discriminating two population-distributions by accounting for the deviations from an expected distribution
Wherever applicable, χ2 tests following an N-bin model (df3=N-1) were conducted to discriminate between two population-distributions (say, native and non-native) with the χ2-statistic being computed by the following equation:
| 6 |
where E(i) represents the frequency “under the null hypothesis” expected for the ith bin, while, O(i) denotes the actually observed frequency for that same (ith) bin.
Buried surface area
Buried surface area (BSA) of a “target” molecular object is the surface area of the object that gets buried on the neighboring surface upon a binary association/complexation. This was computed by taking the difference between the Accessible Surface Areas (ASA’s) of the target in its free and bound forms in the following way:
| 7 |
where ASAbound and ASAfree refer to the ASA’s of the “target” when free (i.e., separated from the rest of the molecule/complex), and when united/bound to the neighbor. Atoms at the association/interaction-interface conditioned by ∆ASA ≠ 0 were computed as in Eq. 1 of the “Contact map at the interface” section.
Results and discussion
Molecular evolution of the SARS-CoV-2 RBDSpike: reviewing key residues
SARS-CoV-2 has a high rate of transmission in human [56–59] (though the fatality rate is low) while transmitting only nominally within other close species (civet, rodents, ferrets, other primates, etc.). Evolutionary genomic studies have revealed that the RBDSpike is the most variable part of the corona virus genome [20, 60]. Furthermore, recent literature on the proximal origin of SARS-CoV-2 [1] has highlighted the essential effective difference between RBDSpike of CoV and CoV-2 to be localized within a 51 amino acid stretch (residues: 442–491 in CoV; 455–505 in CoV-2) on their (evolutionarily mapped) ACE2 binding sites. Let this stretch be henceforth referred to as the “Spike-RBD-hotspot.” A visual structural examination revealed that the stretch primarily mapped to a long partially folded disordered loop with a small anti-parallel β-strand embedded in it (see Supplementary Fig. S2). The hotspot region includes six “critical” amino acid positions that physically bind to the receptor out of which five are mutated in CoV-2 with respect to CoV (Y442 → L455, L472 → F486, N479 → Q493, D480 → S494, T487 → N501) [1]. The overall composition or physicochemical consensus (in terms of hydrophobicity, charge, polarity, aromaticity, amino acid volume, etc.) upon these evolutionary changes remains almost unaltered in the two viral species. The only noticeable effective difference is in the mutation of one negatively charged amino acid to a polar residue (D480: CoV → S494: CoV-2). In a sense, the mutations collectively appear to be a reshuffling of the overall discrete sequence space (consisting of the aforementioned crucial positions). So, based on the above hypothesis [1], it is quite surprising that how this small, localized change could alone lead to such an incredibly high increase in transmission rate in CoV-2 with respect to that in CoV. To portray a more comprehensive picture of the evolutionary event, the observation window was broadened to the aligned full-length sequences of the two homologous protein domains (RBDSpike). As a matter of fact, the total number of point mutations between RBDSpike of CoV and CoV-2 are found to be 17, 12 out of which have an alternating hydrophobic character (i.e., polar/charged ↔ hydrophobic). Interestingly, all these mutations are situated within the “Spike-RBD-hotspot” defined above.
Affinity and stability of binding from local and non-local measures of complementarity
The coupling between the dual attributes of complementarity is well known in biomolecular recognition, concerning shape and electrostatic matching of the interacting molecular surfaces [44, 61–64]. It was also realized subsequently that shape complementarity (Sc) is a necessary criterion for macro-molecular binding while electrostatic complementarity (EC) is sufficient [61, 65, 66]. For oligomer formation in proteins, where large surface area (~1600 Å2) [67] are required to get buried upon complexation, surfaces have to be carefully tailored for the complementary interlocking of side-chains at the interface. This close association between the interacting molecular partners enhances the effective match between their protrusions and crevices so that extended areas can move into close contact [31, 44, 68, 69]. A poor complementarity in shape between two macro-molecular surfaces, therefore, stands out to be a strong forbidding factor for their close association. For example, two purely convex surfaces (say, two spheroids or ellipsoids) lack the steric fit to bind.
On the other hand, complementarity in surface electrostatic potential serves as a secondary criterion in macro-molecular interactions, especially for proteins. The inter-relation of electrostatic forces and protein stability is well known [62]. For example, optimizing Coulomb interactions through charge substitution on the protein surface leads to increased stability [70–73]. However, the same may not be achieved by a mere non-strategic increase in the net charge (positive or negative) as electrostatic repulsion may interfere within the folded state [70, 74, 75]. Along the same line, complementarity in surface charge and/or net charge were ruled out as the representative complementarity term in protein binary complexes [32] and was corrected by redefining EC as the correlation in surface electrostatic potentials. Sub-optimal EC values (even negative values) have been found to result occasionally from unfavorable or repulsive interactions in protein complexes, also in protein-ligand interactions [76], often compensated by strong counterbalancing geometric fit [63]. Such instances have been found in statistically considerable proportion (in ~20% of the cases) in native protein-protein complexes [65], wherein, compensatory elevated Sc values have frequently been recorded [65]. Such obligate interactions4 are generally found to be transient in nature, often linked with signaling pathways [77–80].
The long- and short-range nature of the forces giving rise to EC and Sc, respectively, leads to their corresponding stringent and relaxed criteria. Accordingly, the height and width of the “probable” regions vary in the complementarity plots (see Fig. 1). From this conceptual platform, it is quite logical to envisage shape complementarity (Sc) as an attractant factor in macro-molecular interaction representing the mutual affinity of the two molecular partners to engage into physical binding. On the other hand, since adequate electrostatic matching at the interaction-surface works favorably to stabilize the bound protein-complex, EC may plausibly be treated as the analogous structural parameter representing binding stability.
Evolution of the CoV-2 RBDSpike–ACE2 interaction dynamics
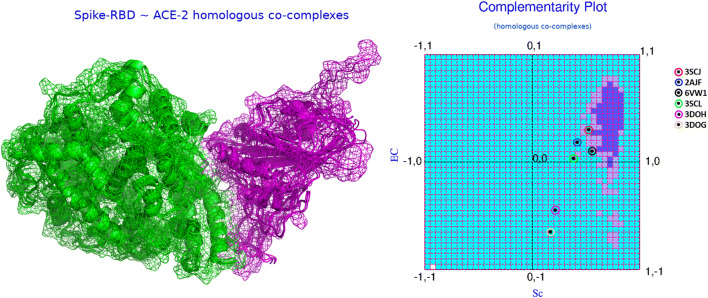
Based on the conceptual foundations discussed in the “Affinity and stability of binding from local and non-local measures of complementarity” section, the relative Sc and EC values (see Table 1) computed for SARS-CoV (2AJF) and SARS-CoV-2 (6VW1) were insightful. 2AJF has an Sc of 0.417 with an EC of 0.185. Together, these values rationalize the binding, both numbers are appreciably positive, falling in the “both-positive” (+, +) first quadrant of CPdock (refer to the “Complementarity plot (CPint and CPdock)” section). However, the ordered pair {Sc, EC} values also indicate that the binding is sub-optimal with respect to their corresponding reference ranges—which is clearly reflected from the location of the corresponding point in CPdock (see Fig. 2). In more elaborate terms, the point falls outside the optimal or near-optimal zones, i.e., outside the “probable” and “less probable” regions in the plot (refer to the “Complementarity plot (CPint and CPdock)” section). In contrast, in 6VW1, Sc is found to be 0.555 (14% increase w.r.t. CoV) while EC is as low as 0.102 (~5% drop w.r.t. CoV). Again, both values are positive, the resultant {Sc, EC} point in CPdock hits the first (+, +) quadrant of the plot (see Fig. 2), thereby, rationalizing the binding (refer to the “Complementarity plot (CPint and CPdock)” section). Visual investigation of the two {Sc, EC} points from 2AJF (CoV), 6VW1 (CoV-2) side-by-side on CPdock further revealed their comparative interaction dynamics which is evolutionarily insightful. Biochemical solution studies elsewhere [21] had already confirmed that the RBDSpike has a significantly greater affinity towards ACE2 relative to that in CoV. The same is also reflected in their corresponding Sc values. The 14% increase in Sc in CoV-2 relative to that in CoV actually makes the Sc value hit its non-rigid optimal range (refer to the “Shape and electrostatic complementarity” section). As a result of this appreciably increased shape matching, the RBDSpike in CoV-2 would have a much higher affinity for ACE2 than that of CoV and would therefore be attracted much faster to its cognate receptor. However, at the same time, it renders a sub-optimal EC value (0.102) upon interacting with ACE2. In elaborate terms, the receptor and the ligand contact-surfaces share just 10% match between their surface electrostatic potentials coming from the electric fields of their own and that of their partner’s (see Fig. 3). By definition (refer to the “Shape and electrostatic complementarity” section), this means weak anti-correlation in surface potentials at the interface, as the close association of two perfectly anti-correlated electrostatic surfaces would ideally return a value of EC = 1 [32]. Hence, yet being attracted to ACE2 faster than that in CoV, the RBDSpike in CoV-2 would also get released from the receptor faster as the unfavorable electrostatic interactions would act against a stable binding. The lower stability in the ACE2-bound binary PPI complex in CoV-2 relative to that in CoV can also be cross-validated by comparing the “dG_separated” values for both, computed by structure driven thermodynamic calculations using Rosetta [23]. Interestingly, in spite of the sub-optimal EC, the increase in Sc in CoV-2 relative to CoV results in a right-shift along the horizontal axis of the corresponding resultant point (CoV-2) in CPdock making the point map to the near optimal zone (~ “less probable” region). Overall, the RBDSpike–ACE2 interaction in CoV-2 does appear to have a quasi-stable character in spite of having a high affinity. At the same time, it is also interesting to reveal that a disease with such a high rate of transmission is actually triggered by a quasi-stable interaction—which may potentially instigate parallel research endeavors to further explore the phenomenon at more complex molecular hierarchies.
Table 1.
Comparison of the complementary estimates of the homologous RBDSpike bound binary PPI complexes
| PDB ID | SPIKE RBD source | Strain | ACE2 | Sc | EC | CSl | Pcount | Nlsp | Nimp |
|---|---|---|---|---|---|---|---|---|---|
| 3D0G | 2002–2003 SARS-CoV | Human | Human-civet chimeric | 0.168 | −0.648 | −6.56 | 58.3 | 0 | 4 |
| 3D0H | 2002–2003 SARS-CoV | Civet* | Human-civet chimeric | 0.211 | −0.445 | −1.92 | 47.1 | 2 | 5 |
| 3SCL | SARS-CoV | Epidemic | Human-civet chimeric | 0.382 | 0.034 | 1.18 | 22.9 | 6 | 5 |
| 3SCJ | Predicted SARS-CoV | Civet | Human | 0.523 | 0.301 | 1.45 | 20.0 | 7 | 0 |
| 2AJF | SARS-CoV | Human | Human | 0.417 | 0.185 | 1.09 | 23.5 | 5 | 3 |
| 6VW1 |
SARS-COV-2 COVID-19 |
Chimeric | Human | 0.555 | 0.102 | 1.15 | 14.7 | 5 | 3 |
In the table, Pcount refers to the percentage of interfacial residues falling in the “improbable” regions of the plot. Nlsp and Nimp refer to numbers of interfacial residues falling, respectively, in the “less probable” and “improbable” regions of the residue-wise Complementarity Plot (CPint; see the “Complementarity Plot (CPint and CPdock)” section)
*Paguma larvata
Fig. 2.
The dynamics of RBDSpike–ACE2 binding from complementarity estimates. The left panel of the figure shows the superposed RBDSpike–ACE2 binary complexes from the homologs (see the “Evolution of the CoV-2 RBDSpike–ACE2 interaction dynamics” section) in cartoon covered with mesh representation. The receptors and the ligands are colored in green and magenta, respectively. The right panel shows the mapping of their corresponding {Sc, EC} points in CPdock as per mentioned in the embedded legend
Fig. 3.
Electrostatic surface of the native RBDSpike–ACE2 binary complex. a and b Map the electrostatic potential surface of the ligand due to the electric fields coming from the ligand itself (self) and the receptor (partner), respectively. Likewise, c and d map the electrostatic surface of the receptor due to the electric fields coming from the receptor (self) and the ligand (partner), respectively. Atomic coordinates of the RBDSpike–ACE2 binary complex are taken from PDB ID: 6VW1. In each panel, the thick arrows indicate whether the surface potentials are due to “self” (a, c) or “partner” (b, d). Further, in each panel, the molecular partner represented as “cartoon” is colored “yellow,” if it is contributing to the potential (i.e., in case of partner-potentials), and, “dim gray” otherwise (self-potentials). The electrostatic surface coloring was done in Chimera [81] using Delphi [41] electrostatic focusing files (.cube) with a color scale set to −10 kT/e for “pure blue” to +10 kT/e for “pure red.” As can be seen, there is very little match of counter-colors (red and blue’s) between corresponding patches on both “contact surfaces” (ligand and receptor) due their respective self- and partner-potentials—which means weak anti-correlation due to unfavorable electrostatic interactions. The potential values portrayed in a and b yields EC1,2 = 0.055 while those portrayed in c and d yields EC2,1 = 0.149 (where, 1 and 2 in the subscripts of EC refer to the ligand and the receptor, respectively) together leading to EC = 0.102 (see Eq. 2 and the “Comparative stability of the RBDSpike conformers influencing their switch” section)
In order to carry out a comparison among the available homologs, Sc, EC were computed for all six RBDSpike–ACE2 binary complexes (refer to the “Details of experimental structures used in the study” section) and were plotted together in CPdock. Both Sc and EC hit values in their corresponding sub-optimal to near-optimal ranges (see the “Shape and electrostatic complementarity” section) making the corresponding points scattered around the “improbable” and “less probable” regions of CPdock. Noticeably, the civet strain, 3SCJ has the closest approach (see Fig. 2) to optimality (see the “Shape and electrostatic complementarity” section) in terms of the combined {Sc, EC} ordered pair, corresponding to its relative closeness from the “probable” region of CPdock (compared to the other candidates in the set). Interestingly, the {Sc, EC} points corresponding to all the homologs was found to cluster around the left-bottom (south west) of the “probable” (optimal) region in CPdock (see Fig. 2b). Such a distribution of points in CPdock is indicative of sub-optimal quasi-stable binding of the two molecular partners along evolution. This was also prominent from a structural display of the molecular interface (see Fig. 2a). For instance, there were no deep grooves or any binding pockets on the receptor where the ligand may stably fit with high affinity. Neither there were signs of any conformation-induced knotting upon binding nor other known/intuitive structural models that might map to “high affinity stable binding.” Rather, the binding appears to be reminiscent of a “molecular handshake” [82] rather than a molecular hug or cling, both from CPdock and from the corresponding structural displays. It is also noteworthy that the part of the “ACE2 peptidase domain (PD)” that physically binds to RBDSpike is actually a single α-helix, known as the “ACE2 PD α1 helix.” The same relative trends among the homologous structures (see Table 1) are also naturally reflected from CP-based global (Complementarity score, CSl) and local measures [45, 46].
Comparison with equivalent protein complexes from MERS and Ebola
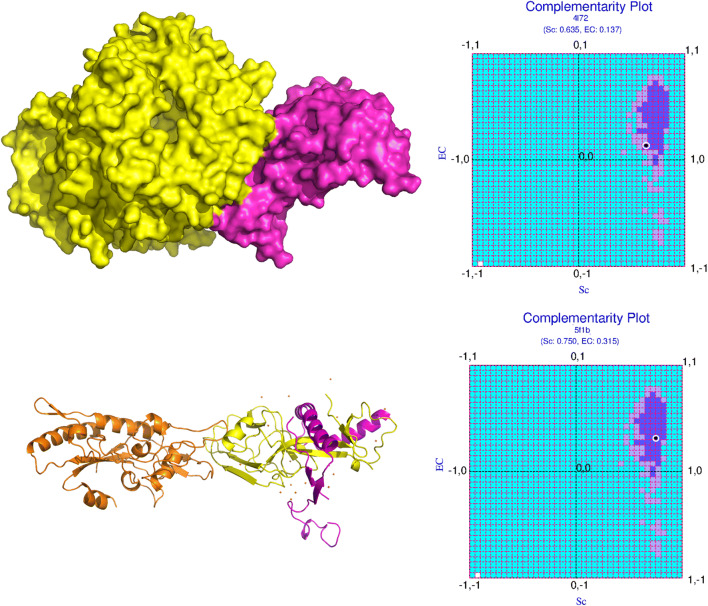
As a point of reference, equivalent protein (binary) complexes from other deadly viral diseases in human were surveyed in a likewise manner. MERS (PDB ID: 4L72) CoV RBD, when bound to its human-receptor Dipeptyl transferase (DPP4) had substantially better shape fit and electrostatic matching along extended mutually compatible surfaces (see Fig. 4, upper panels). On the other hand, the Ebola Viral Glycoprotein, bound to its endosomal receptor Niemann-Pick C1, displayed signatures of knotting upon binding induced conformational changes naturally having far greater surface fit coupled with optimal electrostatic matching (see Fig. 4, lower panels).
Fig. 4.
Analogous binary PPI complexes of SARS-CoV-2-RBDSpike–ACE2 in MERS and Ebola: dynamics of binding from complementarity. The upper and lower panels of the figure represent the binary complexes in MERS (PDB ID: 4L72) and Ebola (PDB ID: 5F1B), respectively, their structures on the left and the corresponding mapping of their {Sc, EC} points in CPdock on the right
Comparative stability of the RBDSpike conformers influencing their switch
As discussed in the “Introduction” section, pioneering EM studies [15] have revealed a “surprisingly low kinetic barrier” for the conformational transition between the pre- and post-fusion forms of the Spike protein. The key mediator of this conformational transition is the RBDSpike domain which, when proximal to the ACE2 expressing lung cells, switches from its native “down” (RBDdown) to active “up” (RBDup) forms primed by a conformation dependent proteolytic cleavage. This cleavage along with the conformational switch, together, set the RBDSpike free and enable it to bind to ACE2 concomitantly. Intuitively, the RBDdown is structurally preferred over RBDup as the “down” state is also known to be functionally coupled to its ability to escape the host immune surveillance. To that end, we carried out a comparison based on the proposed complementarity measures (Sc, EC) computed independently on RBDdown and RBDup (as “target” objects) with respect to their respective (local, global) neighborhoods in order to reveal if the said preference (RBDdown over RBDup) can indeed be portrayed from the relative numbers. In addition, surface area buried upon association (BSA, see the “Buried surface area” section) for both forms (RBDdown, RBDup) was also considered as a third measure of comparison. Thus, essentially, we surveyed to which of its two surrounding neighborhoods ((i) as embedded within the native Spike or (ii) as in complex with ACE2) does the RBDSpike (as the “target” molecular object) feel more harmonious. Notably, binding and folding in proteins can be treated equivalently based on the concept of complementarity [44], wherein, folding can be envisaged as the self-docking of the interior components of a protein-chain/domain onto their respective native environments, consistent with short- and long-range forces sustaining the native fold. To that end, the trimeric RBDdown was contemplated to have self-docked onto the rest of the (native) Spike protein.
The full-length Spike protein in its native pre-fusion form is a biological trimer (PDB ID: 6XR8, bio-assembly-1). Thus, structurally, RBDdown is actually an assemblage of three symmetry-related RBDSpike (down) units while they remain integral to the Spike protein, serving as its limbs. On the other hand, RBDup refers to the post-cleavage S1 fragment(s) entrapped as the ligand chain(s) in the RBDSpike–ACE2 binary complex, which again is a biological monomer (6VW1, two bio-assemblies, both monomeric). The proposed mechanism for the viral host cell entry [15] also clearly portrays this “trimeric → monomeric” switch of the RBDSpike (RBDdown → RBDup) upon binding to ACE2. Thus, as would be appropriate, RBDdown was taken as the trimeric association of the RBDSpike (down) units embedded in the full-length Spike protein (6XR8) while its neighborhood consisted of the “rest of the Spike protein” (barring the RBDdown). On the other hand, RBDup was retained (as throughout the paper) as the ligand (E) chain in 6VW1 with the receptor (A chain) ACE2 serving as its neighborhood. The three following calculations were then performed:
-
(i)
EC for RBDdown in native Spike (ECRBD_down) was computed (from 6XR8), and compared with the equivalent measure (ECRBD_up, referred to as EC1,2 in Fig. 3) already computed for RBDup (referred to as the “ligand” in 6VW1: see the “Evolution of the CoV-2 RBDSpike–ACE2 interaction dynamics” section). For ECRBD_down, RBDdown served as the “target” (refer to the “Shape and electrostatic complementarity” section) while the “rest of the Spike protein” served as its global neighborhood.
-
(ii)
Likewise, Sc for RBDdown (target) in native Spike (ScRBD_down) was computed (from 6XR8), and compared with the equivalent measure (ScRBD_up) already computed for RBDup (refer to the “Evolution of the CoV-2 RBDSpike–ACE2 interaction dynamics” section). Likewise to that of ECRBD_up, RBDdown also served as the “target” for computing ScRBD_down while its local neighborhood was sampled from the “rest of the Spike protein”. To that end, the local neighborhood of RBDdown was delineated by collecting those residues (from the “rest of the Spike protein”) which were found within a relaxed Cα-Cα cut-off of 12 Å from any residue in RBDdown (see Supplementary Fig. S3). The calculation was also repeated at a 15-Å cut-off which returned the same ScRBD_down. The over-relaxed cut-offs ensured not to miss out any potential neighboring atoms, while, at the same time, helped to speed up the calculations.
-
(iii)
BSA was computed (see the “Buried surface area” section) for RBDdown (target) in native Spike (BSARBD_down), and compared with that of RBDup (target) in complex with ACE2 (BSARBD_up).
The expected preference for “down” over “up” forms in RBDSpike was reflected from all three measures (EC, Sc, BSA) (see Supplementary Table S1). Although the ECRBD_Down (referred to as EC1,2 in Supplementary Fig. S4) was fairly low (0.254), the correlation is over 16,847 points (p value <0.00001; significant at p < 0.01) and the value is 4.5 times more than that of ECRBD_Up (0.055) computed over 762 points (p value, 0.129293; not significant even at p < 0.1). ECRBD_Up is the same measure referred to as EC1,2 in Fig. 3 (where RBDUp is referred to as the “ligand”). The corresponding shape complementarities also followed a similar trend (ScRBD_Down = 0.617; ScRBD_Up = 0.566), though, as expected (refer to the “Affinity and stability of binding from local and non-local measures of complementarity” and “Evolution of the CoV-2 RBDSpike–ACE2 interaction dynamics” sections), the difference was nominal. The preference is perhaps most pronounced and direct from the corresponding BSA values. While, in the native Spike (6XR8), BSARBD_down amounts to 6306.1 Å2 over 1538 atoms at the interface (∆ASA ≠ 0, see the “Buried surface area” section), BSARBD_up reduced to 875.3 Å2 over 189 interfacial atoms in the ACE2-bound complex (6VW1). Thus, both the relative BSA and the relative number of atoms buried upon association/complexation are more than 7 to 8 times higher in RBDdown (in “Spike native”) to that of RBDup in complex with ACE2. So, it is clear and unmistakable from all three measures that RBDSpike indeed prefers to stay in the passive “down” state till it reaches the primary site of infection, while, switching over to its more active “up” state only when proximal and exposed to the ACE2 receptors. This structural preference of RBDSpike (for “down” over “up”), in effect, serves to aid as a “transient” molecular switch to trigger the membrane fusion and host cell entry of the virus. The fact that such transitions are energetically costly and are therefore expected to be kinetically driven perfectly aligns with the finding of the dissociated cleaved S1/S2 complex in absence of ACE2 and the adopted post-fusion conformer of the S2 fragment under (membrane mimicking) mild detergent conditions, which together reveals the “surprisingly low kinetic barrier” for the conformational transition [15].
Reaction-prone nature of the ACE2 binding site in SARS-CoV-2 RBDSpike
As elaborated in the above sections, when compared with analogous ligand-receptor binary PPI complexes from related viral strains in the human host, the RBDSpike–ACE2 interface in SARS-CoV-2 does appear to be different and rare. All analyses unequivocally indicate that the interface maps to protein binary complexes involving transient interactions [79] which is likely to be causally linked to its presumably unique modus operandi. To cross-validate this observation, other independent approaches were also adopted concerning the study of the interface. This included (i) calculation of the accessibility score (rGb) of the binary PPI complex and different relevant molecular fragments, and (ii) a detailed analysis of the contact map at the interface. As a matter of interest, 6VW1 (i.e., the only representative interface structure from CoV-2) alone was chosen for the analyses.
As detailed in the “Materials and methods” section (see the “Accessibility score” section), a value of rGb greater than 0.011 (and higher the better) qualifies a globular protein/protein complex/peptide fragment/protein domain to be considered native-like in terms of hydrophobic burial or the distribution of amino acid residues with respect to solvent exposure. Any value less than this empirical threshold renders the input protein molecule non-native like which physically means that hydrophobic residues are exposed to the solvent. This would cause the molecule to stay in an unfavorable/frustrated disordered (high entropy) state. A negative value virtually guarantees this instability which may be extended to depict a reaction-prone nature of the said protein fragment.
With this understanding, rGb was computed for the (i) whole native protein binary complex (referred to as 6VW1_AE in Table 2) and its different relevant molecular fragments, namely, (ii) the ligand chain (chain E of 6VW1) or the RBDSpike alone (6VW1_E in Table 2), (iii) the “Spike-RBD-hotspot” (residues 455–505, refer to the “Molecular evolution of the SARS-CoV-2 RBDSpike: reviewing key residues” section) where all key mutations are localized (6VW1_E_hotspot in Table 2), and (iv) the actual ACE2 binding site or the collection of mapped interfacial residues on chain E as found in the contact map (6VW1_E_bs in Table 2). Interestingly, the rGb scores were found to be decreasing in large fractions from (i) 6VW1_AE to (iv) 6VW1_AE_bs following the descending order of size of the input protein fragment. The relative numbers clearly indicate that the binary PPI complex has the most optimum (or native-like) distribution of hydrophobic burial (rGb 0.052, see Table 2) in the whole set which is substantially better than the ligand chain alone (rGb 0.028). The high negative value (rGb −0.055) obtained for 6VW1_E_bs speaks for its high reaction-proneness [83]. In other words, the high degree of unfavorable hydrophobic exposure makes the ACE2 binding site in RBDSpike critically scurried or strained in its free state. Thus, it is always in a crisis need to embed itself within a befitting complementary surface of an appropriate binding partner.
Table 2.
Reaction proneness of the ACE2 binding site on RBDSpike surveyed by the accessibility (rGb) score
| Input protein fragment | rGb |
|---|---|
| 6VW1_AE | 0.052 |
| 6VW1_E | 0.028 |
| 6VW1_E_hotspot | 0.019 |
| 6VW1_E_bs | −0.055 |
| Threshold for native-like features | 0.011 |
Description of the input protein fragments are as detailed in the “Reaction-prone nature of the ACE2 binding site in SARS-CoV-2 RBDSpike” section
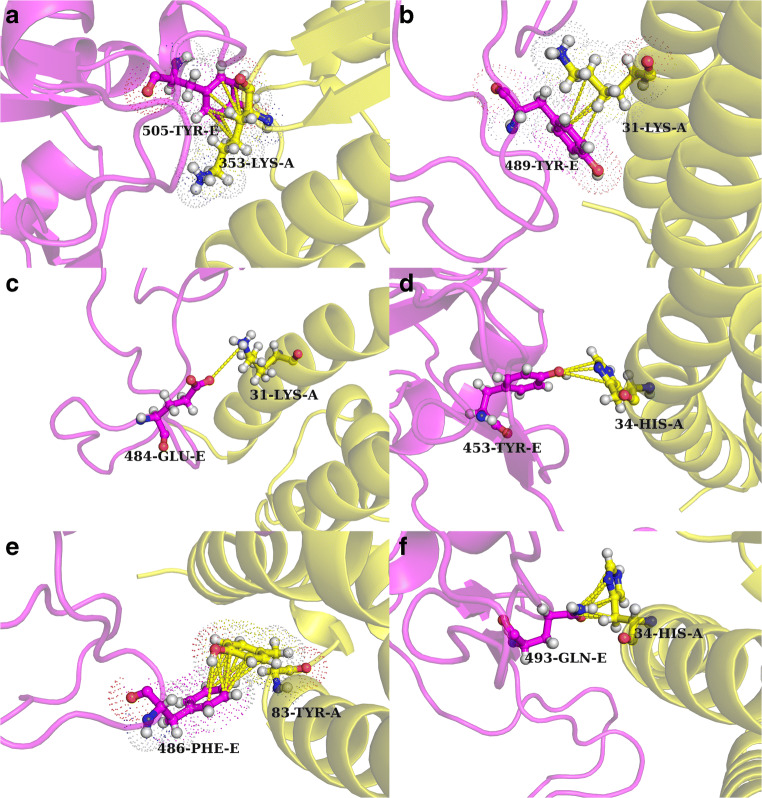
For another level of cross-checking, the contact map at the interface (see the “Contact map at the interface” section) was also rigorously scrutinized. The interface was large with an accessible surface area buried upon complexation (∆ASA) of 1644.4 Å2 considering both molecular partners. It involved 23 inter-residue contacts between the residues coming from the two molecular partners totaling 96 pairwise atomic contacts between their side-chain atoms. The interface appears to have many rare interesting features. From the rGb calculations stated above, it was already clear that the RBDSpike interfacial surface had several exposed hydrophobic residues; hence, it is perhaps of no surprise that the contact map consisted of several hydrophobic residues coming from the ligand (RBDSpike). Interestingly enough, most of these hydrophobic residues were found to be in contact with hydrophilic residues coming from the receptor. Furthermore, a large majority of these hydrophobic residues were in fact bulky aromatic amino acids (see Supplementary Table S2). They were mostly found to be in contact with either “elongated positively charged” (Lys) or “aromatic yet polar amino acids” (His) coming from the receptor. The corresponding interactions mapped to close hydrophobic packing between extended chains of successive mythelene groups (-(-CH2)4) of the lysine(s) and the aromatic ring (31-Lys-A–489-Tyr-E, 353-Lys-A–505-Tyr-E) (see Fig. 5a, b). There were also instances of polar interactions involving aromatic components (34-His-A–453-Tyr-E) (see Fig. 5d), although, there were no clear signatures of any cation–Π or Π-Π stacking between the charged residues and the aromatic rings. However, there were instances of regular aromatic stacking with a slide and an open angle separating the otherwise-parallel aromatic rings (83-Tyr-A–486-Phe-E) (see Fig. 5e). Also, there were hydrophobic packing (79-Leu-A–486-Phe-E, 34-His-A–455-Leu-E) and electrostatic interactions involving polar atoms (24-Gln-A–487-Asn-E, 42-Gln-A–498-Gln-E, 34-His-A–493-Gln-E) (see Fig. 5f). Interestingly, there was a salt-bridge (31-Lys-A–484-Glu-E) as well at the interface (see Fig. 5c) whose presence may be further destabilizing due to desolvation effects—as has been found for salt-bridges in general at protein-interfaces [32, 38, 84]. Overall, it genuinely appears that the interface high potential to harbor and withstand unfavorable electrostatic interactions—which may be causal to the resultant sub-optimal electrostatic complementarity (EC = 0.102).
Fig. 5.
The RBDSpike–ACE2 interface in SARS-CoV-2: non-trivial interactions. a and b Extended packing between aromatic rings and consecutively connected mythelene groups of elongated charged amino acids. c The only salt-bridge found at the interface. d and f Instances of polar atom-mediated interactions involving an aromatic ring while e presents aromatic stacking with a slide and an open angle. Atomic coordinates of the RBDSpike–ACE2 binary complex are taken from PDB ID: 6VW1
Inherent evolutionary features of RBDSpike naturally aiding the design of its structural mimics
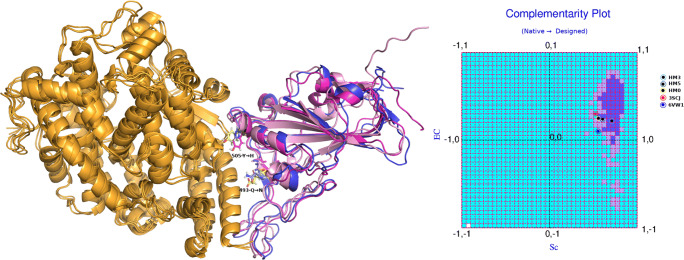
The primary objective of the current study was to develop non-virulent structural mimics of the RBDSpike that could bind to the ACE2 receptor stably with high affinity. For convenience, let these binary PPI complexes be henceforth referred to as “ACE2-complexes” pertaining to the corresponding RBDSpike-ligands (native and designed). These designed mimics would thus serve as potential competitive inhibitors of the viral RBDSpike by occupying the binding sites on the ACE2 receptors. To that end, a protein design approach was adopted aiming to raise the EC of the designed ACE2-complexes (from their sub-optimal native reference value: EC6VW1 = 0.102) while retaining or raising Sc at or from its already near-optimal range (Sc6VW1 = 0.555). The conceptual foundations of the “plausibility of the design strategy” relied on a twofold fact. Firstly, the RBDSpike is an independently foldable domain which is self-sustained as a protein unit and can undergo folding independent to that of the rest of the Spike protein [21]. Secondly, the RBDSpike is resilient to conformational changes upon multi mutations, as has been evident from structural analyses (refer to the “Evolution of the CoV-2 RBDSpike–ACE2 interaction dynamics” section) of the homologs. This means that the basic fold in RBDSpike remains unaltered in spite of the evolutionary sequence variations. The pairwise sequence similarity of the CoV RBDSpike sequences with respect to 6VW1 (CoV-2) was found to be ~69%. RMS deviations (Cα) upon superposing the CoV RBDSpike–ACE2 structures (refer to the “Details of experimental structures used in the study” section) onto 6VW1 were found varying from 1.29 Å (for 3SCL) to 3.18 Å (for 3D0G) (see Supplementary Fig. S5). Furthermore, there were virtually no conformational changes of the RBDSpike upon binding to the ACE2 receptor with respect to its structure in free form (6VXX). RMS deviation (Cα) upon superposing the RBDSpike from 6VW1 onto the free and full structure of the Spike protein (6VXX) was 0.893 Å. Together this means that one may simply administer the finally selected designed mimics without having to bother about their folding (ab-initio) as long as their sequences fit the fold. Test of this fitness with the given fold (i.e., fold compatibility) of the designed sequences was made by state-of-the-art scoring functions for fold recognition (refer to the “Fold recognition” section).
The protein design strategy: sampling and scoring
As mentioned in several earlier sections, a protein-design approach was adopted aiming to develop non-reactive structural mimics of the RBDSpike which may serve as potential competitive inhibitors of the native viral Spike protein to act against the viral pathogenicity. As was found out, the interacting surfaces of CoV-2 RBDSpike and ACE2 has a high shape fit (Sc: 0.555) mapping to its optimal range (refer to the “Shape and electrostatic complementarity” section) coupled with a sub-optimal electrostatic matching at the interface (EC: 0.102). Together, these may be interpreted in terms of having a high affinity yet with a low stability upon binding. Aligned observations have also been proclaimed by biochemical solution assays [21] and calculation of structure based thermodynamic parameters [23] carried out in other studies. This quasi-stable nature of the binding potentially triggers a fast-release of the ligand from the receptor, making them amenable to interact with a greater number of cells having surface-exposed ACE2 receptors. So, the primary objective in the designed RBDSpike mimics was to increase the EC at the interface which would make the interaction more stable. Combining the shape affinity factor, the design problem aimed to improve EC while retaining Sc at least native-like in that “near-optimal to optimal” range. Experimental structural studies in an aligned direction have already demonstrated the favorable effect of key residue substitutions performed across the whole C-terminal domain of the CoV-2 Spike protein harboring the RBDSpike (see Supplementary Fig. 1). Such key-substitutions have been found to strengthen the RBDSpike–ACE2 interaction leading to a 4-fold increased affinity for receptor binding than that of the native ACE2-complex (see the “Inherent evolutionary features of RBDSpike naturally aiding the design of its structural mimics” section) [24]. For our purpose, we had chosen to operate on the RBDSpike itself. When the native binary PPI complexes from the homologs (refer to the “Details of experimental structures used in the study” section) were superposed onto 6VW1, the average pairwise Cα-RMS deviation was found to be 2.05 Å. This evolutionary structural conservation meant that mutations at the ligand (RBDSpike) interface can directly be performed on the native ACE2-complex (6VW1) itself. In a sense, the bound binary PPI complexes were treated like unified globular proteins, wherein, the design protocol may be considered analogous to performing a “hydrophobic core design” or a “full sequence design” in globular proteins. Any protein design protocol has two essential steps: (i) sampling and (ii) scoring. For the current study, sampling (i.e., incorporating strategic mutations) was attempted by essentially two approaches, consistent with the main objective of raising the EC while retaining an at least native-like Sc. In the first of the two approaches, attempts were made to alter the hydrophobic character of the amino acid residues at the interface while keeping their shape and size as similar as possible. Intuitively, this could alter and possibly raise the EC while keeping Sc similar. An equivalent strategy, earlier, was found fruitful in incorporating unbalanced partial charges into native globular protein interiors and detecting the local “electrostatic” errors in-turn [45]. In the second approach, homologous sequences (i.e., direct examples from nature) that were already found to hit appreciably higher EC values were threaded on the native RBDSpike template in 6VW1. Strategic mutations were performed on this threaded homologous sequence based on the contact map at the interface. All mutations in the aforementioned two approaches were performed on the ligand molecule alone retaining the receptor as it is. Scoring and raking of the binary PPI complexes were primarily based on the complementarity measures (refer to the “Shape and electrostatic complementarity” and “Complementarity plot (CPint and CPdock)” sections). Fitness or compatibility of each designed sequence with respect to the native fold was tested by fold recognition measures also based on complementarity (refer to the “Accessibility score” section).
Design strategy-1: altering the hydrophobic character of the amino acids
First, from the distribution of interfacial amino acid residues of the ligand chain (6VW1_E) in the residue-wise Complementarity Plots (CPint), residues falling in the “less-probable” and “improbable” regions (see Fig. 1) were accumulated. They were then united with critical residue positions on the ACE2 binding site (the “Spike-RBD-hotspot,” residues 455–505: see the “Molecular evolution of the SARS-CoV-2 RBDSpike: reviewing key residues” section) said to be harboring determining evolutionary mutations [1]. The full set (S1) consisted of 11 amino acids in total (see Supplementary Table S2) and but for the case of 417-Val the rest of the residues were covered within the aforementioned “hotspot” region. Out of the 11 amino acids selected, four were bulky aromatics, three branched chain hydrophobic, and the rest polar. As a first trial (strategy-1a), mutations were made in this set of 11 residues alone. The raw combinatorial space considering all possible amino-acid mutations is of the astronomical order. To curtail it down to the limits of finite sampling, ad-hoc filters involving semi-empirical rules of thumb (detailed as follows) were judiciously incorporated. Each designed sequence was unique as the sampling involved random seeds. Coupled with the random seeds a weighting scheme was further adopted. For 50% of cases, the amino acids were mutated to (i) residues with alternating hydrophobic character and/or structural properties (S↔S, A↔S, V↔T, L↔N, F↔Y, L↔D, I↔M, M↔R, E↔R, E↔Q, D↔N, R↔M, R↔E, etc.: antonymous changes) and for the other 50%, to (ii) amino acids with similar properties (G↔P, V↔L, F↔W, K↔R, E↔D, Q↔N, H↔Y, S↔T, etc.: synonymous changes). Care was taken to retain their size and/or shape as much as possible. This 1:1 ratio of weights was further varied from 2:1 to 1:1. The intent was to raise the residue-wise electrostatic complementarity (Em) of amino acids falling into the “improbable”/“less probable” regions of CPint in such extents that they can make it to the “probable” regions. It was subsequently realized that electrostatic matching is essentially a global effect and need not necessarily affect the mutated residue itself. Hence, in an alternative approach (strategy-1b), the contact map of the interface was surveyed (refer to the “Contact map at the interface” section) and the ligand residues involved in this set (S2: 13 of them) were chosen as the target positions (see Supplementary Table S2) to perform the mutations keeping the same sampling strategy. There was appreciable overlap (~46%) between the two sets, S1 and S2.
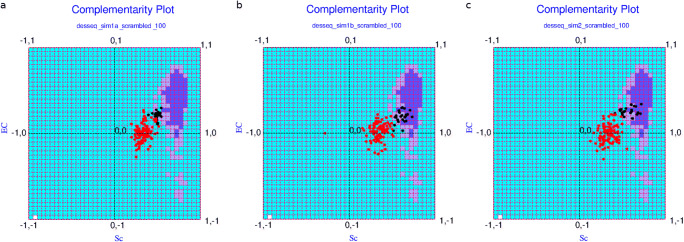
For each of the two aforementioned strategies (1a and 1b) 50 redesigned sequences were constructed and tested in CPdock. Each individual case was carefully scrutinized with visual intervention at all stages of the design protocol. When plotted in CPdock, they were fairly closely spaced creating a south-west island (see Fig. 6a) relative to the center of the optimal zone in CPdock (i.e., the “probable” region). The points were more closely clustered for the first set (strategy-1a) relative to the second (strategy-1b) in terms of both complementarity measures: Sc, EC as reflected in their corresponding range of obtained values (Set-1a: [0.394, 0.544] in Sc, [0.113, 0.298] in EC; Set-1b: [0.514, 0.733] in Sc, [0.042, 0.314] in EC for strategies 1a and 1b, respectively).
Fig. 6.
The solution space: from alteration of hydrophobic character to homology-based design. a, b, and c The solution space for strategies 1a, 1b, and 2, respectively (as referred in sections “Design strategy-1: altering the hydrophobic character of the amino acids” and “Design strategy-2: homology-based protein design: taking templates from nature itself”). The red dots represent the {Sc, EC} points obtained for the corresponding scrambled sequences
In spite of being more closely clustered, Set-1a mapped to values further away from the optimal zone relative to Set-1b. On the other hand, Set-1b appeared to have a greater chance of returning false positive points falling in the “improbable” regions (sub-optimal zones) of the plot (see Fig. 6b). The top 25 sequences from each set were then filtered based on their residence in CPdock (relative to the optimal zone). All filtered sequences successfully passed the test for fold-compatibility (averages 2.76 ± 0.17 in CSgl; 0.016 ± 0.0001 in CScp). These sequences were more closely spaced in CPdock relative to the corresponding original sets. Set-1b mapped more into the “probable”/“less probable” regions (i.e., optimal/near-optimal zone) relative to Set-1a, though, with a greater number of false-positives (see Fig. 6a, b). To serve as negative controls, “scrambled” sequences (refer to the “Scrambled sequences as negative control” section) were generated for each set by random reshuffling of the designed sequences and plotted alongside the “hits” in the two sets (1a and 1b). Clear discriminatory clusters were obtained for the “hits” and the “scrambled” sequences (refer to the “Scrambled sequences as negative control” section) with virtually no overlap (see Fig. 6). All points in the corresponding “random” clusters (the “red dots” in Fig. 6) representing the scrambled sequences were found at the “improbable” regions of the plot, indicating that they were unambiguously sub-optimal.
Design strategy-2: homology-based protein design: taking templates from nature itself
In several well-posed hard-to-solve bioinformatics problems, direct adoption of empirical natural strategies [85–88] coupled with trial-and-error modulations has found much scope and penetration. This includes the very problem of protein structure prediction (considered to be the “holy grail of structural biology”) or other related sub-problems emerging from the core of the protein folding problem (e.g., fold recognition [44], protein design [89], etc.). The “fragment assembly simulated annealing” strategy [87, 90] as in Rosetta is based on natural examples—which is arguably the best structure prediction methodology till date. With the same intuition, we also attempted the direct use of empirical natural examples in our design pipeline, as an alternative to changing the hydrophobic character of amino acids at the interface (strategy-1, a and b). In that line, we picked up the RBDSpike sequence from 3SCJ (i.e., the civet strain from predicted SARS-CoV; see Table 1) motivated by its complementarity estimates (Sc: 0.523, EC: 0.301)—together which stood out to be the best among the homologous. Consequently, 3SCJ also had the closest approach to the “probable” region of CPdock (see Fig. 2) relative to the other homologous, which is to say the closest to being an optimal solution. The sequence of 3SCJ and 6VW1 were aligned, and the aligned 3SCJ sequence (target) was directly threaded onto the main-chain trajectory of the ligand in 6VW1 (template). The threading protocol followed three simple rules of thumb. (R1) For a deletion in the target sequence with respect to the template, the template amino acid was incorporated to fill the gap. (R2) In case of substitution(s), the obvious choice was the target amino acid. (R3) For identical amino acids in the corresponding positions in the template and the target, choosing either of the two meant the same. As a matter of fact, there were no insertions in the target with respect to the template (i.e., no gaps in the template).
Subsequent to threading, dynamic perturbations were introduced to the designed binary PPI complexes (refer to the “Molecular dynamic simulation (short and long)” section) and the final atomic models were surveyed for their contact maps at the receptor–ligand interface. Absurdities in atomic contacts (design artifacts) such as those between two positively or two negatively charged amino acids (Lys-Lys, Glu-Asp, etc.) were obviated, wherever found, by mutating the corresponding amino acid in the originally threaded sequence (e.g., Lys → Glu, Glu → Arg, etc.). Such “artifact cleaning mutations” were chosen based on overall knowledge of atomic interactions in proteins. Such mutations often involved alteration in the hydrophobic character of the amino acids as well. This process gave rise to an iterative (threading → mutation → contact-map)n cycle in the protein design pipeline. Each resultant contact map was rigorously and manually scrutinized wherein other mutable positions were jotted down that could intuitively raise the EC while retaining the Sc. At instances, drastic changes like deleting a bulky side-chain (e.g., Phe → Ala) were also attempted. Charged amino acids were introduced as well as eliminated to favor and forbid the formation of salt-bridges. To eliminate the negative charge in Glu, Asp, they were mutated to corresponding polar variants (Gln, Asn). Attempts were also made to deliberately incorporate extended hydrophobic packing (i.e., introducing Ile, Met at strategic places, etc.) as well as aromatic stacking (introducing Tyr, His, etc.) at the interface. The final evaluation of the binary PPI complexes was made by the complementarity measures and their mapping in CPdock. Again, a total of 50 redesigned alternatives were constructed and tested in CPdock. Among the given alternatives, this set could fairly cover all non-redundant “presumably sensitive” point mutations and their combinations. Each individual case was carefully scrutinized with visual structural intervention of their redesigned interfaces to remove design artifacts. When plotted in CPdock, their population distribution in a close cluster ensured empirical thresholds in both measures to be naturally satisfied (Scmin: 0.402, ECmin: 0.173). In other words, the range of values obtained in the whole set were tight in both complementarity measures (Sc: [0.421, 0.723], EC: [0.178, 0.342]). Obtaining such tightly spaced numbers does not seem to be possible by random design or a mere reshuffling of sequence. To test this, scrambled sequences (refer to the “Scrambled sequences as negative control” section) were generated and undertaken in the same analysis. Just as the cases for strategies 1a and 1b, clear discriminatory clusters were obtained for the hits and the scrambled sequences (see Fig. 6) with practically no overlap. The disjointedness of the two clusters was clearer and more convincing than the earlier two sets (strategies 1a and 1b).
An apparent saturation was ensured in terms of covering arguably the whole spectrum of “sensitive” mutations attempted on the plausible mutational hot-spots. The analyses were greatly helped by the rigorous repeated use of visual structural examination. Interestingly, shape complementarity of the “hits” in this third set (strategy-2) has a much wider range (~ 1.5 to 2 times) than that of electrostatic complementarity, compared to the other two sets (strategies 1a, 1b). More interestingly, there was not a single case with the EC raised to 40%. The difference in geometric fit among the designed sequences may cause from mutations either resulting in undue holes being created at the interface or leading to short contacts. The two events involve truncation and forced incorporation of bulky groups (e.g., Gly → Trp and Tyr → Val, respectively) at the designed interface. At the same time, there appears to be natural evolutionary constraints on the upper limit of EC at this interface, which does not seem possible to be oversteped by different levels of protein engineering using the pull of 20 naturally occurring amino acids. The resultant EC values (natural as well as designed) physically mean quasi-stable to stable binding. The ones that are stable (i.e., optimal in terms of CPdock) were the ones of interest to be considered further. Overall, there appears to be strong natural and evolutionary control over the dynamics of RBDSpike–ACE2 binding. The top 25 sequences were filtered based on their residence relative to the optimal zone in CPdock, and considered further. The filtering also accompanied careful individual visual re-scrutiny of their interface. It is but trivial that these sequences were more closely spaced in CPdock and mapped to the “probable”/“less probable” regions (i.e., optimal/near-optimal zones). Again, all filtered sequences were successfully validated for fold-compatibility (averages 2.84 ± 0.16 in CSgl; 0.017 ± 0.0002 in CScp).
It was unambiguous from the comparison of the three plots pertaining to the three different design-sets (see Fig. 6) that the predicted solutions gradually improved from Set-1a, Set-1b to Set-2 reflected in the gradual north-eastern shift of the clusters (black dots in the plots). In other words, the homology-based design performed the best among the three. It was also evident from these results that the “scrambled” sequences may indeed serve as negative controls in the future experimental validation of the current hypothesis.
A demonstrative example is cited in Fig. 7, wherein, a case consisting of three designed sequences (HM0, HM3, HM5) selected from the pool (Set-2) collectively portrays the impact of strategic point mutations. For HM5, the designed sequence contains a single point mutation (493-Q → N) with respect to the initially threaded sequence (3SCJ_E on 6VW1_E, referred to as HM0 in Fig. 7). In the third case (HM5), the designed sequence further contains a second strategic point mutation (505-Y → H) over and above the earlier mutation. Here in this particular triad, the one with the single point mutation (HM3) gives somewhat better numbers (Sc: 0.710, EC: 0.224) than the one (HM5) with the additional aromatic mutation (Sc: 0.605, EC: 0.243), both better than the threaded sequence alone (HM0; Sc: 0.563, EC: 0.248). This demonstrates the scope and benefit of strategic point mutations to be invoked on the threaded homologous sequence to further improve the solution. Taken together with the natives (6VW1_E, 3SCJ_E), the results show a gradual shift towards a more balanced optimal solution upon threading (HM0) followed by subsequent strategic point mutations (HM3, HM5). The full-length sequences of these designed RBDSpike mimics are provided in Supplementary Dataset S1.
Fig. 7.
Homology-based design of the CoV-2 RBDSpike: signatures of stable high affinity binding. The top panel displays the superposed ACE2-complexes (see the “Inherent evolutionary features of RBDSpike naturally aiding the design of its structural mimics” section) of HM0, HM3, and HM5 with their designed RBDSpike chains colored in light pink, magenta, and tv_blue, respectively. The mutations are highlighted in the form of sticks. The bottom panel shows the mapping of their corresponding {Sc, EC} points in CPdock as per mentioned in the embedded legend
Dynamic persistence of the binding of the selected designed structural mimics
Two best predicted solutions (HM19, HM21) designed from strategy-2 were undertaken for long MD simulations (refer to the “Molecular dynamic simulation (short and long)” section) to study the dynamic persistence of the binding parameters. As a mean to set the baseline, the native ACE2-complex (6VW1) was also included in the calculation. HM19 and HM21 had originally attained {Sc, EC} values of {0.614, 0.276} and {0.687, 0.310}, respectively. To that end, all atom explicit-water MD simulation production runs were performed for 200 ns each, wherein, the simulated coordinates were accumulated at an interval of 100 ps resulting in 2000 snapshots (or time-stamps) for each simulated protein-complex. The post-simulation analyses commenced with collecting all snapshots pertaining to each trajectory and superposing them (using TM-align [91]) onto their respective templates (i.e., the starting structures of their respective MD simulations). The time-averaged Cα-RMS deviations of these superposed coordinates were found to be 2.50 (±0.38) Å, 2.66 (±0.39) Å for the designed ACE2-complexes (see the “Inherent evolutionary features of RBDSpike naturally aiding the design of its structural mimics” section) pertaining to HM19 and HM21, respectively (see Supplementary Fig. S6). In contrast, the native-average was ~1.5 times more with ~1.8 times the fluctuations (3.82 ± 0.66 Å) than both mimics. The dynamic persistence of the complementarity measures was analyzed by running CPdock on each sampled snapshot along the trajectory for each of the three subjects (HM19, HM21, native) followed by drawing their time-series plots individually for Sc, EC (see Fig. 8), and their statistical analysis.
Fig. 8.
Time-series plots of Sc and EC for the selected designed structural mimics in comparison to the native. a and b Plot the time-evolved Sc profiles for HM19 and HM21, respectively, alongside that of the native using different colors (magenta: native, red: mimics; as given in the legend-box), while, c and d plot their corresponding time-evolved EC profiles. The thicker lines drawn parallel to the X-axis plotted in different colors (blue: native, black: mimics, as also given in the legend-box) represent their corresponding time-series averages. Both Sc and EC are correlation measures, defined in the range of [−1, 1]. The X-axis represents the simulation time (in units of ns)
A direct comparison of the original and time-evolved values (averages and standard deviations) for the complementarity measures (Sc, EC) can be made from the corresponding time-series plots (see Fig. 8) as well as from Table 3. For HM19 and HM21, the time-series averages (and standard deviations) were, respectively, found to be 0.664 (±0.048), 0.669 (±0.049) for Sc, and 0.278 (±0.082), 0.248 (±0.074) for EC while the same for the native was found to be 0.628 (±0.050) for Sc and 0.149 (±0.080) for EC. Thus, by and large, both complementarity measures fairly retain their original trends and nuances as revealed from their respective initial values (see Table 3) in all three subjects. The numbers further suggest that the primary differentiating descriptor between the native and the designed mimics is indeed EC, while, the shape descriptor (Sc) serves as a (threshold-dependent) necessary criterion for the complexation, as it does generally for macro-molecular binding per se (refer to the “Affinity and stability of binding from local and non-local measures of complementarity” section). In more elaborate terms, Sc, once into its optimal range (refer to the “Shape and electrostatic complementarity” section), converges further to a more optimized narrower range (dependent on the particular protein co-complex system) with time, irrespective of their fine-grained structural difference brought about by the strategic design(s) (see Table 3). The difference between the corresponding ECs (designed vs. native) however persists throughout the entire 200 ns simulated trajectories. Notably, the native EC originally falling into the sub-optimal range (EC6vw1 = 0.102), largely remains in the same (sub-optimal) range throughout the course of the entire simulation run. On the other hand, the improvement brought about by the strategic design is fairly retained with time in both selected designed mimics. Equally notable is the fact that EC values for the designed mimics (original as well as time-evolved) regularly and consistently hit the crucial “near-optimal to optimal” range (refer to the “The protein design strategy: sampling and scoring” section) indicating stable electrostatic matching at the designed interfaces. These observations are consistent with the original proposition that the native ACE2-complex (6VW1) forms with high affinity, but lacks stability over time due to sub-optimal electrostatic matching at its interface. On the other hand, the directed design enables the mimics (HM19, HM21) with the potential to bind to ACE2 with equivalent high affinity, and also to remain bound stably over time.
Table 3.
Complementarity (Sc, EC) and its time-evolution for the selected designed binary complexes compared to the native
| ACE2-complexes | Sc | EC | ΔGbinding (kcal/mol) | ||
|---|---|---|---|---|---|
| Original | Time-evolved | Original | Time-evolved | Time-evolved | |
| HM19 | 0.614 |
0.664 (±0.048) |
0.276 |
0.278 (±0.082) |
−5.939 (±2.581) |
| HM21 | 0.687 |
0.669 (±0.049) |
0.310 |
0.248 (±0.074) |
−5.634 (±3.011) |
| Native | 0.555 |
0.628 (±0.050) |
0.102 |
0.149 (±0.080) |
0.854 (±4.981) |
The original Sc, EC values (as obtained before the corresponding long MD simulations) alongside their time-evolved averages (and standard deviations) are tabulated in a row-wise tabular format for the native and designed binary complexes. In addition, the corresponding binding free-energies (ΔGbinding) for each subject (ACE2-complex: see the “Inherent evolutionary features of RBDSpike naturally aiding the design of its structural mimics” section) are also tabulated
Within the entire 200 ns trajectories, Sc could maximally be raised to 0.797 and 0.793 for HM19 and HM21 while their corresponding highest EC values attained were 0.592 and 0.497, respectively. All numbers unequivocally indicate that the binding is dynamically stable and of high affinity. The directed improvement in the matching of electrostatic surface potentials for HM19 and HM21 are portrayed in Fig. 9 and Supplementary Fig. S7, respectively, citing the MD-snapshot(s) with their highest attained EC values. A comparison with Fig. 3 reveals the improvement in EC from the sub-optimal to the optimal range.
Fig. 9.
Electrostatic surface representation of one of the best predicted designed binary complexes (for HM19). a–d The electrostatic surface map of the snapshot (picked up from its 200 ns MD simulation trajectory) with the highest attained EC value for HM19 (the “The protein design strategy: sampling and scoring” section). The rest of the figure may be described likewise to that of Fig. 3. Briefly, a and c represent “self-potentials” while b and d represent “partner-potentials” realized on the ligand and receptor surface, respectively, for HM19. Self- and partner-potentials are as defined in the legend of Fig. 3. Arrows indicate whether the surface potentials are due to “self” (a, c) or “partner” (b, d). Coloring of “cartoons” are as in Fig. 3. A direct comparison with Fig. 3 clearly shows that the match in counter-colors (red and blue) improves appreciably between corresponding patches on the contact surfaces (due to their respective self- and partner-potentials) with respect to that of the native ACE2-complex (see the “Inherent evolutionary features of RBDSpike naturally aiding the design of its structural mimics” section). This reflects that the native weak anti-correlation in electrostatic surface potential could be significantly strengthened by the strategic design
Similar dynamical trends are also reflected from the time-series plots for E2d (see Supplementary Fig. S8)—which estimates the 2D Euclidean distance of a plotted {Sc, EC} point in CPdock from the “probable” region of the plot (refer to the “Measuring the dynamic stability of the proposed ‘optimal’ solutions” section). To note, E2d renders a value of “zero” if the point falls into the “probable” region. For E2d, the native has substantially greater fluctuations (see Supplementary Fig. S8) compared to both HM19 and HM21 at different patches of the simulation trajectories. Overall, this leads to a standard deviation of ~2.5 times higher in the native than in both of the designed ACE2-complexes (see the “Inherent evolutionary features of RBDSpike naturally aiding the design of its structural mimics” section). Also, notably, the time-series average for the native E2d is more than 4 times to that of the designed ACE2-complexes. In contrast, the same time-series averages for both HM19 and HM21 are almost identical to each-other and close to zero. All the numbers unambiguously indicate the dynamic stability of the designed ACE2-complexes relative to that of the native.
Implicit to the E2d analysis, distribution of {Sc, EC} points (coming from each snapshot in a given trajectory) across the three defined regions in CPdock (refer to the “Fold recognition” section) was also surveyed for each ACE2-complex (pertaining to HM19, HM21, native). While, for HM19, the fraction of snapshots falling into the “probable,” “less probable,” and “improbable” regions of CPdock were 78.05, 20.05, and 14.45%, respectively, the same fractional counts for HM21 were found to be 77.8, 20.9, and 1.3%. In great contrast, the “less probable” and “improbable” regions together populated 56% of the native trajectory (“probable” 43.5%, “less probable” 42.55%, “improbable” 13.95%). Overall, the numbers collectively suggest clear improvements from native instability to stable binding in the designed ACE2-complexes over time. As a formal test of significance (of the obtained deviations), we performed a χ2 test between the native and each of the designed sets from their respective raw counts using a 3-bin model (i.e., df = 3–1: “probable,” “less probable,” “improbable”; χ20.05 = 5.991). The χ2 method is traditionally associated with the Complementarity Plot(s) through several earlier applications using the plot(s) as discerning discriminatory metric(s) between different population-distributions [44, 45, 48]). For the current cause, the “null hypothesis” assumed “no significant improvement in stability over time upon the directed design” and that “the deviations from the native distribution were simply obtained by chance”. In reality, however, the resultant χ2 values (see Eq. 6 defined in the “Buried surface area” section) were computed to be 1001.375 and 990.654 for HM19 and HM21, respectively, both more than 160 times to that of the (above-quoted) χ20.05 for a 3-bin model. This literally rules out even the slightest of chances to accept the proposed “null hypothesis” and concludes instead that the deviations from the frequencies distributed under the “null hypothesis” are indeed significant and must not have occurred by chance. The fact that the selected designed ACE2-complexes (for both HM19 and HM21) are largely contained within the “near-optimal to optimal” regions of the CPdock over time is also reflected from their three-dimensional population density plots (see Supplementary Fig. S9).
Further, as a mean to cross-validate the predicted improvement in binding stability reflected from the complementarity measures (Sc, EC), binding/interaction energies (ΔGbinding) of the native () and the selected designed ACE2-complexes () were computed using FoldX (refer to the “Estimating changes in binding/interaction energies for the proposed ‘optimal’ solutions” section) along their corresponding (200 ns) simulated trajectories. This was followed by computing their directed difference () following Eq. 5 (defined in the “Estimating changes in binding/interaction energies for the proposed ‘optimal’ solutions” section) and drawing time-series plots individually for all three free-energy-difference terms (see Fig. 10). Time-series averages (and standard deviations) of the corresponding ΔGbinding terms were found to be −5.939 (± 2.581) kcal/mol and −5.634 (± 3.011) kcal/mol for the ACE2-complexes in HM19 and HM21, while, only amounting to 0.854 (± 4.981) kcal/mol for the native ACE2-complex. The obtained native average seems to be of potential physical significance, since it hits a near-zero value in meaning that the dynamic persistence of the native ACE2-complex is only mildly favored thermodynamically. The associated standard deviation of ~ ± 5 kcal/mol reflecting high dynamic fluctuations (μ = 6σ5) in the native ΔGbinding further suggests that the native ACE2-complex (6VW1) is indeed energetically unstable over time. Together, this favorably speaks for a model of quasi-stable binding/interaction. Given that the purpose of the complexation here is to switch on the membrane fusion and viral entry to the host cell [15], a transient (quasi-stable) nature in the interaction of the native RBDSpike and ACE2 is indeed intuitively expected, perhaps also reflected from the appreciably low (and sub-optimal) native-EC values all-throughout. Also, the “surprisingly low kinetic barrier” revealed for the preceding event (see the “Comparative stability of the RBDSpike conformers influencing their switch” section) does seem to add to the proposition. Notably, the proposition of the “low kinetic barrier” for the conformational switching of the Spike protein (“pre” to “post”-fusion forms) is purely based on experimental biophysical and structural data, wherein, they have found the dissociated “cleaved S1/S2 complex” in absence of ACE2 as well as the adopted “post-fusion conformer of the S2 fragment” under mild detergent conditions mimicking a membrane environment [15].
Fig. 10.
Time-series plots of binding/interaction energies for the selected designed structural mimics and their changes with respect to the native. Panels a and b Plot the time-evolved profiles (as defined in the “Estimating changes in binding/interaction energies for the proposed ‘optimal’ solutions” section) for HM19 and HM21, respectively, alongside that of the native (: the “Estimating changes in binding/interaction energies for the proposed ‘optimal’ solutions” section) using different colors (∆Gb in Fig.10: magenta: native, red: mimics; as given in the embedded legend-boxes). c The corresponding difference plots ( :: see Eq. 4 defined in the “Estimating changes in binding/interaction energies for the proposed ‘optimal’ solutions” section) for the mimics (HM19: magenta; HM21: red; as also given in the legend-boxes). The thicker lines drawn (in all three panels) parallel to the X-axes represent the corresponding time-series averages of the plotted parameters with their colors and descriptions given in the legend-box (blue: native, black: mimics, for a and b; blue: HM19, black: HM21 for c). The X-axis represents the simulation time (in units of ns)
The relative improvement in binding stability over time brought about by the strategic design is also reflected from the high negative time-averaged values (Table 3) and their appreciably low standard deviations (roughly scaling to μ = 2σ for both HM19 and HM21). As a result, the corresponding values are also equally negative (HM19 −6.793 ± 5.990 kcal/mol; HM21 −6.487 ± 5.781 kcal/mol)—which further confirms the predicted improvement in their thermodynamic stability over time. Thus the improvement in binding stability predicted from complementarity (EC in particular) is also clearly reflected in the corresponding free energy estimates of the binding events, over time.
Nullifying the feasibility of the proposed designed therapeutics to compete with the ACE2–angiotensin II binding
Angiotensin Converting Enzyme 2 (ACE2), a vital counter-regulatory component of the Renin-Angiotensin System (RAS), has recently got great attention in COVID-19 research for acting as a doorway to SARS-CoV-2 into the host cells [92–96]. Upon low blood flow, kidney cells convert the circulating pro-renins into renins which further take part in catalyzing the angiotensinogen secreted by liver cells into angiotensin I [95]. The membrane-bound Angiotensin Converting Enzyme (ACE) present on vascular endothelial cell surface in the lungs, thereafter, converts angiotensin I into angiotensin II which is an amphipathic linear octa-peptide that serves as a vasoconstrictor [95]. As a result, angiotensin II causes blood vessels to be constricted to increase blood pressure through engaging type 1 angiotensin receptor (AT1R) [96, 97]. Angiotensin II also increases blood pressure by stimulating adrenal cortex cells to secrete the aldosterone hormone. So, under normal physiological condition, a fine balance between ACE2–angiotensin II and ACE2–Ang-(1–7) has to be maintained in order to control the blood pressure and inflammation. As because SARS-CoV-2 utilizes the membrane bound ACE2 receptor to gain entry into host cells, so this is a condition where the viral Spike protein bound ACE2 receptors will be less available to angiotensin II. As a result, an equilibrium shift towards the increased activity of ACE2–angiotensin II might drive acute lung injury. Furthermore, according to the current hypothesis, SARS-CoV-2–ACE2 binding causes increased internalization and shedding off of the ACE2 receptor making it further unavailable to angiotensin II and thereby causing less production of Ang-(1–7). This can induce blood pressure along with direct parenchymal injury [98].
Our current work has considered the possibility of whether or not our designed plausible therapeutics can compete with the binding site of angiotensin II on ACE2 and may thereby disrupt the balance in RAS. In this regard, the NMR structure of angiotensin II (PDB ID: 1N9V) was surveyed which has little conformational deviations among its 21 models (average RMS deviation: 0.187 ± 0.09 Å upon aligning to MODEL-1 in PyMol). When, 1N9V (MODEL-1 taken as the representative structure) was superposed onto the ligand (E) chain of 6VW1, the peptide is found distant from the ACE2 binding site (see Supplementary Fig. S10) having an RMS deviation of 4.28 Å. Based on a pairwise sequence alignment (in CLUSTAL-OMEGA [99]), the angiotensin II sequence was then threaded onto “6VW1_E_bs,” the ACE2 binding site on RBDSpike (refer to the “Reaction-prone nature of the ACE2 binding site in SARS-CoV-2 RBDSpike” section). The corresponding atomic model was subsequently built which resulted in an RMS deviation of 3.46 Å considering a stretch of just eight mapped amino acids. Thus, the two molecular objects does not seem to have any appreciable structural resemblance. Furthermore, when this built atomic model is placed onto the RBDSpike–ACE2 complex (6VW1), it has no proximity with the ACE2 receptor (displayed as solid surface in Supplementary Fig. S10, bottom panel). No atoms were found at the native RBDSpike–ACE2 interface. Naturally, a small bent linear octa-peptide like angiotensin II (see Supplementary Fig. S10, top panel) finds little chance to fit into a plausible binding model with the Spike protein binding site in ACE2—which is no more than a single α-helix (refer to the “Evolution of the CoV-2 RBDSpike–ACE2 interaction dynamics” section). Rather, a deep groove or a pocket is generally required to engulf such small molecules without having the necessity to have a proper shape and/or electrostatic match at the interface [100–102]. Thus, the two ligands (angiotensin II and RBDSpike) have no good reason to compete for an identical binding site on ACE2. Also, it is well-known that unlike protein–protein binding, where large interacting surfaces (~1600 Å2 on average) [103] need to be carefully tailored to fit into each-other over extended areas, a small-molecule ligand (or co-factor) can present far greater conformational variation upon binding to different binding pockets—which, in-turn, exhibit more variability in shape and physicochemical attributes than can be accounted for by the adopted conformational multiplicity of the ligand [44, 100–102]. This further nullifies the possibility of a binding conflict with angiotensin II at the Spike binding site of ACE2.
Having said that, the actual binding site of angiotensin II on ACE2 is not yet known experimentally. To that end, further computational structural investigation of the two available individual partner molecules were carried out to gain some more intuitive insights into their plausible binding mode, followed by performing a molecular docking of the two.
The membrane bound ACE2 receptor represents the extra-membrane domain of the corresponding integral membrane protein. A closer look into its structure reveals that it is an all-α protein-domain (helical bundle) resembling the shape of an elongated spheroid and thereby forming a percolative channel fairly open to the aqueous solvent at either pole. It should thus mostly be facing an aqueous environment supported by having accordingly a bulk majority of hydrophilic regions. This was confirmed by the BRANEart webserver (http://babylone.3bio.ulb.ac.be/BRANEart/index.php) which analyzes strength, stability, and weaknesses of different regions of membrane proteins [104] and colors them accordingly (blue: hydrophilic, white: neutral, red: hydrophobic). BRANEart further lists a residue-wise “Membrane Propensity” score, defined in the range of −1 (red: hydrophobic) to +1 (blue: hydrophilic) computed by a linear regression machine trained on a collection of statistical potentials. From numerically as well as from the visual outputs (see Supplementary Fig. S11A), it was evident that indeed most part of the ACE2 structure (~85.5%) prefers to stay in polar (aqueous) environments. These hydrophilic regions are interspersed with neutral/mildly hydrophobic patches coming from some of the component helices, thereby forming an amphipathic6 open inner-groove, partially exposed to the solvent at either poles. A small molecule thus has a great chance to pervade and slip through the long axis of the open-inner groove and be sustained there stably—which appears to be genuinely plausible for an open-ended amphipathic linear octa-peptide like that of angiotensin II (see Supplementary Fig. S11B). To test this structural hypothesis, two docking studies were performed using the popular protein-docking webserver Cluspro (v.2) [105, 106]: (a) docking of angiotensin II vs. ACE2 and (b) docking of angiotensin II vs. the RBDSpike–ACE2 binary PPI complex.
As was anticipated from the structural hypothesis, the results of the first docking test (a) indeed revealed that angiotensin II prefers to diffuse through the open inner-groove of ACE2 and be contained stably at the protein core. The top 10 docked poses (as ranked and returned by Cluspro) upon superposition onto the ACE2 global frame of reference (as in 6VW1) were invariably found to hit the inner groove/core of the protein (see Fig. 11a) which has no structural conflict with the binding of RBDSpike (displayed alongside the docked poses in the same image). As can be expected, the same results were virtually reproduced in the second docking test (b) even within the larger structural context of the RBDSpike–ACE2 binary PPI complex, fed in as the receptor (see Supplementary Fig. S12). The top ranked docked binary complex (from (a)) was further surveyed in BRANEart which resulted in compatible hydrophilicity/hydrophobicity profiles for the two binding partners (angiotensin II and ACE2) in their bound form (see Fig. 11b). Thus the docking results are very much in accordance with the structural hypothesis stated and reasoned above—which practically nullifies all realistic chances of a potential conflict between the two bindings. Taken together, there does not seem to be any convincing structural rationale to favor a plausible interference caused by the proposed therapeutic intervention to the RAS via ACE2.
Fig. 11.
Docking and structural analysis in view of angiotensin II - binding to ACE2 with reference to the RBDSpike–ACE2 complexation in COVID-19. a The Cluspro docking results of Angiotensin II (PDB ID: 1N9V, MODEL 1) docked onto ACE2 (6VW1, chain A). The ligand chain of 6VW1 (chain E) is also displayed alongside the docked poses (for clarity). The 10 top-ranked docked poses of the ligand (angiotensin II) are displayed both as cartoon and dots (surface points) for better focus. b The BRANEart visual output of the top ranked angiotensin II ACE2 docked binary complex. The figure in b is regenerated in PyMol from the .pml file provided in the BRANEart output. Coloring of structural regions follow the coloring scheme specified in the color bar: blue: hydrophilic, white: neutral, red: hydrophobic (see the “Dynamic persistence of the binding of the selected designed structural mimics” section)
Comparing the proposed therapeutic intervention with the current state-of-the-art
One of the prime focuses of the recent research advances on anti-viral therapeutics for SARS-CoV-2 has been on utilizing the already available knowledge on the host cell entry mechanisms of SARS-CoV, MERS, and other coronaviruses. Three general pathways that could lead to the development of potential antiviral therapeutics are (i) repurposing through the testing of pre-existing antiviral drugs, (ii) by high throughput screening of small molecules, and (iii) through the redevelopment of new drugs or neutralizing antibodies or vaccines. Our current study proposes a non-trivial protein design approach to develop antiviral therapeutics that might act as potential competitive inhibitors of the SARS-CoV-2 RBDSpike. After gaining insight into host cell entry mechanisms, importantly through the revelation of X-ray crystallographic structure of SARS-CoV-2 Spike protein binding to its cognate receptor, ACE2, on human cells [21, 24, 107], the drug-designing methods are primarily revolving around the S protein subdomain blockers for obvious reasons.
There are also peptide-based approaches involving strategic contextual design of hybrid and fusion peptides. Such a hybrid peptide has been computationally constructed by linking two discontinuous fragments of ACE2 (residues 22–44 and 351–357) by a linker glycine [108]. In addition to designing of small peptides from ACE2 sequence, clinical-grade soluble hACE2 has proven to be a promising therapeutic candidate molecule which has shown to block the entry and growth of SARS-CoV-2 in the blood vessel and kidney organoids system [109]. In order to develop potential therapeutics against SARS-CoV-2, researchers have also targeted the HR1 (heptad repeat 1) and HR2 domains in the S2 subunit besides targeting RBDSpike (S1) [110]. Lipo-peptide such as EK1C4 has been demonstrated to be the most potent fusion inhibitor [110, 111]. Further, evidences have been put forward in support of significant efficacies of peptide inhibitors derived from the HR2 domain which can block the fusion of the viral and the host cell membranes [112].
Alternatively, it has been shown by wet-lab experiments in hACE2-expressing cells that the recombinant RBDSpike could block the entry of both the SARS-CoV and SARS-CoV-2 into the host cells [113]. A recent MD simulation study coupled with bio-layer interferometry [114] has targeted the “ACE2 PD α1 helix” (refer to the “Evolution of the CoV-2 RBDSpike–ACE2 interaction dynamics” section) where the SARS-CoV-2 RBDSpike binding actually occurs. This 23-mer peptide fragment (residues 21–43) can effectively bind to SARS-CoV-2 RBDSpike at a very low nano-molar affinity (Kd = 47 nM) thereby posing a high possibility to interfere with the viral entry into host [114]. Importantly, although their peptide-based drug designing approach means to bypass the alteration in ACE2 physiological functions, the actual effect of their RBDSpike blocker still remains to be checked in terms of titters in human system. Such approaches are essentially aiming for an “antigen arrest” before the pathogen reaches the host pulmonary system. A similar approach has also been adapted using nanobodies for directed delivery of neutralizing antibodies of RBDSpike [115]. In complete contrast, our approach takes the alternative route to develop therapeutics which may potentially block the RBDSpike binding site on the cognate receptor, ACE2. We take advantage of the quasi-stable native binding of RBDSpike to ACE2 in SARS-CoV-2 and aim to appreciably increase the binding stability while retaining near-native high affinity. The mutations were directly performed on the native experimental RBDSpike–ACE2 complex. The proposed designed variants are the end-products of cycles of rigorous computational screening through high-level structural descriptors, and the predicted improvement in binding stability in their corresponding ACE2-complexes (see the “Inherent evolutionary features of RBDSpike naturally aiding the design of its structural mimics” section) over time is also cross-validated by appropriate free energy estimates. The proposed “high affinity stable binding” in the predicted ACE2-complexes pertaining to the designed structural mimics should therefore serve as the basis of their potential usage as blockers of the native Spike protein for its cognate receptor. Aligned approaches have shown the effect of key residue substitutions in SARS-CoV-2-CTD (see Supplementary Fig. 1) leading to a fourfold increased affinity for receptor binding than that of the native binary PPI complex [24]. We further structurally cross-checked that the designed RBDSpike mimics do not seem to have a realistic chance to cause a potential conflict with the binding of angiotensin II to ACE2, and therefore presents only a thin feasibility to interfere with the native physiological function of ACE2 (refer to the “Nullifying the feasibility of the proposed designed therapeutics to compete with the ACE2–angiotensin II binding” section). Furthermore, the prescribed RBDSpike mimics being substantially smaller in size (of the order of 1/100th) than those of the full virus particles should be able to reach the binding sites at a much faster time-scale.
Although other groups have followed a more direct approach (“antigen arrests” as well as “immunization”) to prevent RBDSpike binding to ACE2 through designing mini-proteins [116], peptide blockers [114], nanobodies [115], and vaccines [117–119], we have chosen a more indirect and unconventional (reverse) approach in our proposed bio-therapeutic design. The reasons for our choice are as follows:
Firstly, in the absence of the viral infection, the ACE2–angiotensin II binding is not known to transmit any molecular signal leading to transcription of downstream genes [120, 121]. So, from that end, the proposed therapeutics do not appear to not cause any further impact on the intra-cellular downstream signaling. The second benefit is related to the “systemic clearance” of the therapeutics after their course of action—which is a common concern to all administered competitive inhibitors. It is well known that SARS-CoV-2 infection is associated with ACE2 downregulation [121, 122] mostly by endocytic internalization of ACE2, and also influenced by some other unknown mechanisms. The proposed RBDSpike mimics will likewise be internalized in the form of their ACE2-complexes (see the “Inherent evolutionary features of RBDSpike naturally aiding the design of its structural mimics” section), however, with the definite advantage of not carrying with them the rest of the viral particle. Additionally, the designed mimics being significantly smaller in size than the viral particle would likely have a faster approach to ACE2. By virtue of potentially having a greater stability (as all the results unequivocally indicate), they would thus occupy the viral attachment sites on the host cell membrane, eventually out-competing the viral binding (and infection). So, that way, the designed mimics would actually act against the endocytic internalization of the native RBDSpike, and at the same time, inhibit the host cell entry of the viral particle, by the proposed membrane fusion mechanisms [15]. The suggested downregulation of ACE2 will thus (in all probability) be only short-termed followed by a fast restoration of the physiological homeostasis both in terms of ACE2 and angiotensin II. Moreover, the internalization of ACE2-complexes (see the “Inherent evolutionary features of RBDSpike naturally aiding the design of its structural mimics” section) pertaining to the proposed designed mimics will naturally ensure the metabolism of the therapeutics and their systemic clearance. Thirdly, SARS-CoV-2 being extremely pleiotropic in nature, its titer(s) in individuals of different age groups, gender, and with different medical conditions might be challenging to evaluate. Since our reverse approach is aimed to block the ACE2 receptor which is native to the individual (rather than a foreign body), the precise doses of the therapeutics will likely be easier to determine. Considering these salient advantageous features, we preferred the reverse approach.
The proposed method, however, comes with certain potential caveats. Firstly, the predictions are purely computational (however, based on available experimental structures), yet to be validated in the wet lab. Secondly, important part of the structural hypothesis is based on available knowledge and current understanding of the viral entry mechanisms, part of which are also currently at a hypothesis level. Thirdly, the mode of administration (oral/intravenous/inhalation) is yet to be determined through wet lab experiments. Fourthly, cytokine storms (as immune responses) [123, 124] are found to be triggered upon binding of coronavirus with ACE2 and the consequences of the proposed therapeutic(s) to that end is yet to be tested again by wet lab experiments.
Conclusions
Quasi-stable binding appears to be one of the essential features of SARS-CoV-2 RBDSpike–ACE2 interaction. Having said that, the ligand possesses a high affinity towards its cognate receptor in the human host. This “affinity-stability trade-off” seems to be fine-tuned during evolution in the corresponding protein family and fold—as revealed from the study of homologous binary PPI complexes (refer to the “Evolution of the CoV-2 RBDSpike–ACE2 interaction dynamics” section). The current study unravels this fine-tuning from coordinate-driven local and non-local complementarity measures, {Sc, EC}, and validates the findings by appropriate free energy estimates. While being counterbalanced by compensatory shape constraints (attributed to high affinity), attainment of sub-optimal electrostatic matching at the interface certainly appears to be a characteristic feature of this binary (RBDSpike–ACE2) association, conserved through evolution. Even in the designed binary PPI complexes, other than a low fraction of snapshots (varying from 1.5 to 5.5%) in the long MD simulations, the maximally elevated EC value was found to be not more than 40% (refer to the “Design strategy-1: altering the hydrophobic character of the amino acids” and “Design strategy-2: homology-based protein design: taking templates from nature itself” sections). That too, given the design-protocol being directed to raise the EC (refer to the “The protein design strategy: sampling and scoring” section). The amino acid composition of the ACE2 binding site of the extracellular RBDSpike does seem to be non-trivial (compared to those in native globular proteins) involving solvent-exposed hydrophobic residues. This appears to be causally related to the conformational switch between RBDdown and RBDup states (the “Comparative stability of the RBDSpike conformers influencing their switch” section) and the corresponding change in the membrane environment it encounters. This has profound impact on the transitioning residues, effectively comprising the ACE2 binding site in RBDSpike. As a consequence, the RBDSpike–ACE2 interface consists of a bunch of hydrophobic–polar interactions coupled with weak aromatic stabilization. The relative stability of the native RBDdown over its more proactive RBDup state (in the ACE2-complex) is clear and unambiguous from all comparative structural measures (the “Comparative stability of the RBDSpike conformers influencing their switch” section) which is well known to help the SARS-CoV-2 to remain in the native “down” state until host cell proximity enabling them to escape the host immune surveillance. This effectively renders their conformational switch (RBDdown → RBDup) to be kinetically driven, and similar concussions have also been drawn from biochemical wet-lab experiments, collectively unraveling a “transient conformational switch” (the “Comparative stability of the RBDSpike conformers influencing their switch” section). Furthermore, the structure of the native RBDSpike–ACE2 complex is reminiscent of a molecular handshake rather than a hug or a cling (refer to the “Evolution of the CoV-2 RBDSpike–ACE2 interaction dynamics” section) like those found in analogous binary PPI complexes in other related respiratory viral disorders (refer to the “Comparison with equivalent protein complexes from MERS and Ebola” section). Together, this leads to the high reaction-prone nature of the RBDSpike. Also, the RBDSpike–ACE2 interaction is intricately coupled with the host-protease mediated peptide-cleavage (as detailed in the “Introduction” section), which, being an enzymatic reaction, involves covalent bond-breaking and bond-making. The interaction is thus concomitantly linked to the ephemeral formation of a transition state (TS) involving a saddle point, as is natural to enzyme kinetics. This also strongly speaks in favor of the quasi-stable nature of the RBDSpike–ACE2 interaction. Quantum chemical calculations (in combination with molecular mechanics) may be invoked to reveal the plausible mechanism of the associated enzymatic “cleavage” reaction (outside the scope of the current study). The improvement in binding stability predicted from complementarity (EC in particular) is also clearly reflected in the corresponding free energy estimates of the binding events, over time (the “Dynamic persistence of the binding of the selected designed structural mimics” section). The time-evolved native ΔGbinding values are further suggestive of the quasi-stable nature of interaction, concomitantly coupled to the transient (“pre” to “post”-fusion) conformational switch of the Spike protein (the “Comparative stability of the RBDSpike conformers influencing their switch” and “Dynamic persistence of the binding of the selected designed structural mimics” sections). This quasi-stable nature of the interaction has been utilized beneficially towards the directed design of the structural mimics aimed to serve as plausible blockers of the RBDSpike–ACE2 interaction. The objective of the exercise was to improve the interaction-stability while keeping intact the already attained high affinity so that the designed mimics can actually block the host-pathogen interaction by stably occupying the binding sites on the receptor. Accordingly, EC was directed to be raised in an iterative protein design cycle while retaining the Sc at least native-like. As a matter of fact, both complementarity measures could be raised substantially in the best predicted designed ACE2-complexes with respect to their native estimates. Starting from a native {Sc, EC} value of {0.555, 0.102}, we could statistically hit the (0.6, 0.7) and (0.3, 0.4) ranges in Sc and EC, respectively (refer to the “Design strategy-2: homology-based protein design: taking templates from nature itself” section), for the designed interfaces, in spite of the evolutionary constraints in EC. Together, these ranges in {Sc, EC} fall in the optimal zone for high affinity stable binding in protein binary complexes—as revealed from the mapping of the corresponding points in CPdock. This was possible by means of realizing the benefits of directly adapting natural examples in the design process. Thus, the use of homology-based design coupled with strategic mutations altering the hydrophobic characters of key amino acids appeared to be judicious in achieving the desired goal. Hence, the paper may also be viewed to have presented a design methodology per se, the applicability and robustness of which are to be tested across related host-pathogen systems.
Supplementary information
(PDF 1854 kb)
Acknowledgments
HKP would like to acknowledge the Wolfson College at the University of Cambridge (UK) for fellowship (Jr, B1), MIIC at Linkoping University (Sweden) and University College London (UK) for their activity support. We thank Mr. Souparno Adhikary, Department of Biochemistry, University of Calcutta, for his initial initiative for simulation study. We also sincerely thank Dr. Abhirup Bandyopadhyay, Institute De Neurosciences Des Systems, Aix-Marseille University, France; Dr. Sumit Mondal, Vidyasagar College, Kolkata, India; and Mr. Dhruba Ghosh, Department of Chemistry, University of Manitoba, Canada, for their help, motivations, and valuable discussions during the revision. SB conveys his sincere gratitude to Sri Gautam Basu, renowned literary figure and language expert, for the valuable cross-disciplinary discourses on the use and evolution of analytical language and logic during the work.
Authors’ contributions
The initial idea about a competitive inhibitor approach was conceived by HKP in a discussion with SB. Based on this idea, SB conceptualized the problem, designed the complete study, and performed all the calculations. DC figured out the optimal methodology for the free energy calculations, followed by executing them alongside SB. DC also substantially contributed to other necessary discussions during the revision. DB helped with the molecular dynamic simulation part, contributed with results, and participated with SB in many fruitful discussions, especially during the revision. PS contributed with background survey, developed and drafted most of the basic structure of the introduction, and actively helped in revising the manuscript. HKP intervened at different phases with crucial updates, technical inputs, terminological corrections, and language- and style-edits. SB wrote the main draft of the paper with active help primarily from PS and rigorously revised the paper until satisfactory. All authors have read and approved the final manuscript.
Availability of data and material
Included as electronic supplementary material.
Code availability
As detailed for each software application in the “Materials and methods” section.
Declarations
Conflicts of interest
N/A
Ethics approval
N/A
Consent to participate
N/A
Consent for publication
N/A
Footnotes
Proprotein convertase of the host
μ: mean; σ: standard deviation
df: degree of freedom
interactions required instantaneous/short-termed, e.g., those involved in signal transduction
μ: mean; σ: standard deviation
Having both hydrophobic and polar regions
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Andersen KG, Rambaut A, Lipkin WI, et al. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu H, Chan JF-W, Wang Y et al (2020) Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 10.1093/cid/ciaa410 [DOI] [PMC free article] [PubMed]
- 7.Dong L, Tian J, He S, et al. Possible vertical transmission of SARS-CoV-2 from an infected mother to her newborn. JAMA. 2020;323:1846–1848. doi: 10.1001/jama.2020.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferretti L, Wymant C, Kendall M et al (2020) Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science 368. 10.1126/science.abb6936 [DOI] [PMC free article] [PubMed]
- 9.Kim Y-I, Kim S-G, Kim S-M et al (2020) Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 10.1016/j.chom.2020.03.023 [DOI] [PMC free article] [PubMed]
- 10.Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morawska L, Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020;139:105730. doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanche S, Lin YT, Xu C et al (2020) High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerging Infect Dis 26. 10.3201/eid2607.200282 [DOI] [PMC free article] [PubMed]
- 13.Tong Z-D, Tang A, Li K-F, et al Potential presymptomatic transmission of SARS-CoV-2, Zhejiang Province, China, 2020—Volume 26, Number 5—May 2020—Emerging Infectious Diseases Journal—CDC. 10.3201/eid2605.200198 [DOI] [PMC free article] [PubMed]
- 14.Wei WE (2020) Presymptomatic transmission of SARS-CoV-2—Singapore, January 23–March 16, 2020. MMWR Morb Mortal Wkly Rep 69:. 10.15585/mmwr.mm6914e1 [DOI] [PMC free article] [PubMed]
- 15.Cai Y, Zhang J, Xiao T et al (2020) Distinct conformational states of SARS-CoV-2 spike protein. Science. 10.1126/science.abd4251 [DOI] [PMC free article] [PubMed]
- 16.Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS-CoV-2. PNAS. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaimes JA, André NM, Chappie JS, et al. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walls AC, Park Y-J, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercurio I, Tragni V, Busto F et al (2020) Protein structure analysis of the interactions between SARS-CoV-2 spike protein and the human ACE2 receptor: from conformational changes to novel neutralizing antibodies. Cell. Mol. Life Sci. 10.1007/s00018-020-03580-1 [DOI] [PMC free article] [PubMed]
- 24.Wang Q, Zhang Y, Wu L, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKee DL, Sternberg A, Stange U, et al. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res. 2020;157:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu C, Liu Y, Yang Y et al (2020) Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed]
- 27.Zhou Y, Hou Y, Shen J, et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery. 2020;6:1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Josephson K, Ricardo A, Szostak JW. mRNA display: from basic principles to macrocycle drug discovery. Drug Discov. Today. 2014;19:388–399. doi: 10.1016/j.drudis.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Wan Y, Shang J, Graham R et al (2020) Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. Journal of Virology:94. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed]
- 30.Smith MC, Gestwicki JE. Features of protein-protein interactions that translate into potent inhibitors: topology, surface area and affinity. Expert Rev. Mol. Med. 2012;14:e16. doi: 10.1017/erm.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, Chandana Epa V, Colman PM. Electrostatic complementarity at protein/protein interfaces. J. Mol. Biol. 1997;268:570–584. doi: 10.1006/jmbi.1997.0987. [DOI] [PubMed] [Google Scholar]
- 33.Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucl Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eswar N, Webb B, Marti-Renom MA et al (2006) Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics 05:–Unit-5.6. 10.1002/0471250953.bi0506s15 [DOI] [PMC free article] [PubMed]
- 35.MUSCLE: multiple sequence alignment with high accuracy and high throughput. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC390337/. [DOI] [PMC free article] [PubMed]
- 36.Krivov GG, Shapovalov MV, Dunbrack RL. Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 38.Basu S, Mukharjee D. Salt-bridge networks within globular and disordered proteins: characterizing trends for designable interactions. J. Mol. Model. 2017;23:206. doi: 10.1007/s00894-017-3376-y. [DOI] [PubMed] [Google Scholar]
- 39.Basu S, Biswas P. Salt-bridge dynamics in intrinsically disordered proteins: a trade-off between electrostatic interactions and structural flexibility. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2018;1866:624–641. doi: 10.1016/j.bbapap.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Connolly ML. Analytical molecular surface calculation. J. Appl. Crystallogr. 1983;16:548–558. doi: 10.1107/S0021889883010985. [DOI] [Google Scholar]
- 41.DelPhi Suite: New developments and review of functionalities - Li - 2019 - Journal of Computational Chemistry - Wiley Online Library. https://onlinelibrary.wiley.com/doi/full/10.1002/jcc.26006. [DOI] [PMC free article] [PubMed]
- 42.Li L, Li C, Zhang Z, Alexov E. On the dielectric “constant” of proteins: smooth dielectric function for macromolecular modeling and its implementation in DelPhi. J. Chem. Theory Comput. 2013;9:2126–2136. doi: 10.1021/ct400065j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grant OC, Montgomery D, Ito K, Woods RJ (2020) Analysis of the SARS-CoV-2 spike protein glycan shield: implications for immune recognition. bioRxiv 2020.04.07.030445. 10.1101/2020.04.07.030445 [DOI] [PMC free article] [PubMed]
- 44.Basu S, Bhattacharyya D, Banerjee R. Self-complementarity within proteins: bridging the gap between binding and folding. Biophys. J. 2012;102:2605–2614. doi: 10.1016/j.bpj.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basu S, Bhattacharyya D, Banerjee R. Applications of complementarity plot in error detection and structure validation of proteins. Indian J. Biochem. Biophys. 2014;51:188–200. [PubMed] [Google Scholar]
- 46.Basu S, Bhattacharyya D, Wallner B. SARAMAint: the complementarity plot for protein–protein interface. Journal of Bioinformatics and Intelligent Control. 2014;3:309–314. doi: 10.1166/jbic.2014.1103. [DOI] [Google Scholar]
- 47.Basu S, Bhattacharyya D, Banerjee R. SARAMA: a standalone suite of programs for the complementarity plot—a graphical structure validation tool for proteins. Journal of Bioinformatics and Intelligent Control. 2013;2:321–323. doi: 10.1166/jbic.2013.1059. [DOI] [Google Scholar]
- 48.Basu S. CPdock: the complementarity plot for docking of proteins: implementing multi-dielectric continuum electrostatics. J. Mol. Model. 2017;24:8. doi: 10.1007/s00894-017-3546-y. [DOI] [PubMed] [Google Scholar]
- 49.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- 50.Scott WRP, Hünenberger PH, Tironi IG, et al. The GROMOS biomolecular simulation program package. J. Phys. Chem. A. 1999;103:3596–3607. doi: 10.1021/jp984217f. [DOI] [Google Scholar]
- 51.K L-L, S P, K P, et al (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. In: Proteins. https://pubmed.ncbi.nlm.nih.gov/20408171/. [DOI] [PMC free article] [PubMed]
- 52.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 53.Schymkowitz J, Borg J, Stricher F, et al. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rasafar N, Barzegar A, Mehdizadeh Aghdam E. Design and development of high affinity dual anticancer peptide-inhibitors against p53-MDM2/X interaction. Life Sci. 2020;245:117358. doi: 10.1016/j.lfs.2020.117358. [DOI] [PubMed] [Google Scholar]
- 55.Insights from engineering the Affibody-Fc interaction with a computational-experimental method - PubMed. https://pubmed.ncbi.nlm.nih.gov/28472513/. [DOI] [PubMed]
- 56.Shereen MA, Khan S, Kazmi A, et al. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shim E, Tariq A, Choi W, et al. Transmission potential and severity of COVID-19 in South Korea. Int. J. Infect. Dis. 2020;93:339–344. doi: 10.1016/j.ijid.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bi Q, Wu Y, Mei S, et al (2020) Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. The Lancet Infectious Diseases 0: 10.1016/S1473-3099(20)30287-5 [DOI] [PMC free article] [PubMed]
- 59.Yuen K-S, Ye Z-W, Fung S-Y et al (2020) SARS-CoV-2 and COVID-19: the most important research questions. Cell Biosci 10. 10.1186/s13578-020-00404-4 [DOI] [PMC free article] [PubMed]
- 60.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, Friesner RA, Berne BJ. Competition of electrostatic and hydrophobic interactions between small hydrophobes and model enclosures. J. Phys. Chem. B. 2010;114:7294–7301. doi: 10.1021/jp100772w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou H-X, Pang X. Electrostatic interactions in protein structure, folding, binding, and condensation. Chem. Rev. 2018;118:1691–1741. doi: 10.1021/acs.chemrev.7b00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gabb HA, Jackson RM, Sternberg MJ. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J. Mol. Biol. 1997;272:106–120. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 64.Bk S, Id K. Protein docking and complementarity. J. Mol. Biol. 1991;221:327–346. doi: 10.1016/0022-2836(91)80222-g. [DOI] [PubMed] [Google Scholar]
- 65.Basu S, Wallner B. Finding correct protein-protein docking models using ProQDock. Bioinformatics. 2016;32:i262–i270. doi: 10.1093/bioinformatics/btw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsuchiya Y, Kinoshita K, Nakamura H. Analyses of homo-oligomer interfaces of proteins from the complementarity of molecular surface, electrostatic potential and hydrophobicity. Protein Eng Des Sel. 2006;19:421–429. doi: 10.1093/protein/gzl026. [DOI] [PubMed] [Google Scholar]
- 67.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 68.Banerjee R, Sen M, Bhattacharya D, Saha P. The jigsaw puzzle model: search for conformational specificity in protein interiors. J. Mol. Biol. 2003;333:211–226. doi: 10.1016/j.jmb.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 69.Yan Y, Huang S-Y. Pushing the accuracy limit of shape complementarity for protein-protein docking. BMC Bioinformatics. 2019;20:1–10. doi: 10.1186/s12859-019-3270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dell’Orco D, Xue W-F, Thulin E, Linse S. Electrostatic contributions to the kinetics and thermodynamics of protein assembly. Biophys. J. 2005;88:1991–2002. doi: 10.1529/biophysj.104.049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Makhatadze GI, Loladze VV, Gribenko AV, Lopez MM. Mechanism of thermostabilization in a designed cold shock protein with optimized surface electrostatic interactions. J. Mol. Biol. 2004;336:929–942. doi: 10.1016/j.jmb.2003.12.058. [DOI] [PubMed] [Google Scholar]
- 72.Marti DN, Rudolf Bosshard H. Electrostatic interactions in leucine zippers: thermodynamic analysis of the contributions of Glu and his residues and the effect of mutating salt bridges. J. Mol. Biol. 2003;330:621–637. doi: 10.1016/S0022-2836(03)00623-5. [DOI] [PubMed] [Google Scholar]
- 73.Ojennus DD, Lehto SE, Wuttke DS. Electrostatic interactions in the reconstitution of an SH2 domain from constituent peptide fragments. Protein Sci. 2003;12:44–55. doi: 10.1110/ps.0227903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Loladze VV, Makhatadze GI. Removal of surface charge-charge interactions from ubiquitin leaves the protein folded and very stable. Protein Sci. 2002;11:174–177. doi: 10.1110/ps.29902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schwehm JM, Fitch CA, Dang BN, et al. Changes in stability upon charge reversal and neutralization substitution in staphylococcal nuclease are dominated by favorable electrostatic effects. Biochemistry. 2003;42:1118–1128. doi: 10.1021/bi0266434. [DOI] [PubMed] [Google Scholar]
- 76.Kahraman A The geometry and physicochemistry of protein binding sites and ligands and their detection in electron density maps. 221
- 77.Maleki M, Vasudev G, Rueda L. The role of electrostatic energy in prediction of obligate protein-protein interactions. Proteome Sci. 2013;11:S11. doi: 10.1186/1477-5956-11-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Z, Witham S, Alexov E. On the role of electrostatics on protein-protein interactions. Phys. Biol. 2011;8:035001. doi: 10.1088/1478-3975/8/3/035001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kundrotas PJ, Alexov E. Predicting 3D structures of transient protein-protein complexes by homology. Biochim. Biophys. Acta. 2006;1764:1498–1511. doi: 10.1016/j.bbapap.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 80.Kundrotas PJ, Alexov E. Electrostatic properties of protein-protein complexes. Biophys. J. 2006;91:1724–1736. doi: 10.1529/biophysj.106.086025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera–a visualization system for exploratory research andanalysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 82.Patel DJ. A molecular handshake. Nature. 1994;367:688–689. doi: 10.1038/367688a0. [DOI] [PubMed] [Google Scholar]
- 83.Levine L, Edsall G. Tetanus toxoid: what determines reaction proneness? J. Infect. Dis. 1981;144:376–376. doi: 10.1093/infdis/144.4.376. [DOI] [PubMed] [Google Scholar]
- 84.Honig B, Yang AS. Free energy balance in protein folding. Adv. Protein Chem. 1995;46:27–58. doi: 10.1016/S0065-3233(08)60331-9. [DOI] [PubMed] [Google Scholar]
- 85.Chaudhury S, Berrondo M, Weitzner BD, et al. Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS One. 2011;6:e22477. doi: 10.1371/journal.pone.0022477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Simoncini D, Zhang KYJ. Efficient sampling in fragment-based protein structure prediction using an estimation of distribution algorithm. PLoS One. 2013;8:e68954. doi: 10.1371/journal.pone.0068954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rohl CA, Strauss CEM, Misura KMS, Baker D (2004) Protein structure prediction using Rosetta. In: Methods in Enzymology. Academic Press, pp. 66–93 [DOI] [PubMed]
- 88.Lyskov S, Gray JJ. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008;36:W233–W238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leaver-Fay A, Tyka M, Lewis SM, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Meth Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 91.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aleksova A, Ferro F, Gagno G et al COVID-19 and renin-angiotensin system inhibition: role of angiotensin converting enzyme 2 (ACE2)—is there any scientific evidence for controversy? Journal of Internal Medicine n/a. 10.1111/joim.13101 [DOI] [PMC free article] [PubMed]
- 93.Ingraham NE, Barakat AG, Reilkoff R, et al (2020) Understanding the renin-angiotensin-aldosterone-SARS-CoV-Axis: A Comprehensive Review. European Respiratory Journal. 10.1183/13993003.00912-2020 [DOI] [PMC free article] [PubMed]
- 94.South AM, Tomlinson L, Edmonston D, et al. Controversies of renin–angiotensin system inhibition during the COVID-19 pandemic. Nat. Rev. Nephrol. 2020;16:305–307. doi: 10.1038/s41581-020-0279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peach MJ. Renin-angiotensin system: biochemistry and mechanisms of action. Physiol. Rev. 1977;57:313–370. doi: 10.1152/physrev.1977.57.2.313. [DOI] [PubMed] [Google Scholar]
- 96.Paz Ocaranza M, Riquelme JA, García L, et al. Counter-regulatory renin-angiotensin system in cardiovascular disease. Nat. Rev. Cardiol. 2020;17:116–129. doi: 10.1038/s41569-019-0244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sparks MA, Crowley SD, Gurley SB, et al. Classical renin-angiotensin system in kidney physiology. Compr Physiol. 2014;4:1201–1228. doi: 10.1002/cphy.c130040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kuba K, Imai Y, Rao S, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences: Clustal Omega for many protein sequences. Protein Sci. 2018;27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stockwell GR, Thornton JM. Conformational diversity of ligands bound to proteins. J. Mol. Biol. 2006;356:928–944. doi: 10.1016/j.jmb.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 101.Kahraman A, Morris RJ, Laskowski RA, Thornton JM. Shape variation in protein binding pockets and their ligands. J. Mol. Biol. 2007;368:283–301. doi: 10.1016/j.jmb.2007.01.086. [DOI] [PubMed] [Google Scholar]
- 102.Kahraman A, Morris RJ, Laskowski RA, et al. On the diversity of physicochemical environments experienced by identical ligands in binding pockets of unrelated proteins. Proteins. 2010;78:1120–1136. doi: 10.1002/prot.22633. [DOI] [PubMed] [Google Scholar]
- 103.Janin J, Chothia C. The structure of protein-protein recognition sites. J. Biol. Chem. 1990;265:16027–16030. doi: 10.1016/S0021-9258(17)46181-3. [DOI] [PubMed] [Google Scholar]
- 104.Mbaye MN, Hou Q, Basu S, et al. A comprehensive computational study of amino acid interactions in membrane proteins. Sci. Rep. 2019;9:1–14. doi: 10.1038/s41598-019-48541-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Desta IT, Porter KA, Xia B, et al. Performance and its limits in rigid body protein-protein docking. Structure. 2020;28:1071–1081.e3. doi: 10.1016/j.str.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kozakov D, Hall DR, Xia B, et al. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 108.Huang X, Pearce R, Zhang Y. De novo design of protein peptides to block association of the SARS-CoV-2 spike protein with human ACE2. Aging (Albany NY) 2020;12:11263–11276. doi: 10.18632/aging.103416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Monteil V, Kwon H, Prado P, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xia S, Liu M, Wang C, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xia S, Yan L, Xu W, et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Science Advances. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xia S, Zhu Y, Liu M, et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cellular & Molecular Immunology. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tai W, He L, Zhang X, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cellular & Molecular Immunology. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang G, Pomplun S, Loftis AR, et al (2020) Investigation of ACE2 N-terminal fragments binding to SARS-CoV-2 spike RBD. bioRxiv 2020.03.19.999318. 10.1101/2020.03.19.999318
- 115.J H, A LB, Rr R, et al (00:00:00.0) Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat Struct Molec Biol. 10.1038/s41594-020-0469-6 [DOI] [PubMed]
- 116.Cao L, Goreshnik I, Coventry B, et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020;370:426–431. doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pollet J, Chen W-H, Strych U. Recombinant protein vaccines, a proven approach against coronavirus pandemics. Adv Drug Deliv Rev. 2021;170:71–82. doi: 10.1016/j.addr.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Malladi SK, Singh R, Pandey S et al (2020) Design of a highly thermotolerant, immunogenic SARS-CoV-2 spike fragment. J Biol Chem. 10.1074/jbc.RA120.016284 [DOI] [PMC free article] [PubMed]
- 119.Voysey M, Clemens SAC, Madhi SA, et al. Safety and efficacy of the ChAdOx1nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. The Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mahmoud G, Kaiming W, Anissa V, et al. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System. Circ Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Samavati L, Uhal BD (2020) ACE2, Much More Than Just a Receptor for SARS-COV-2. Front Cell Infect Microbiol 10. 10.3389/fcimb.2020.00317 [DOI] [PMC free article] [PubMed]
- 122.Ciulla MM (2020) SARS-CoV-2 downregulation of ACE2 and pleiotropic effects of ACEIs/ARBs. Hypertens Res 1–2. 10.1038/s41440-020-0488-z [DOI] [PMC free article] [PubMed]
- 123.Chen I-Y, Chang SC, Wu H-Y, et al. Upregulation of the chemokine (C-C motif) ligand 2 via a severe acute respiratory syndrome coronavirus spike-ACE2 signaling pathway. J Virol. 2010;84:7703–7712. doi: 10.1128/JVI.02560-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chang Y-J, Liu CY-Y, Chiang B-L, et al. Induction of IL-8 release in lung cells via activator protein-1 by recombinant baculovirus displaying severe acute respiratory syndrome-coronavirus spike proteins: identification of two functional regions. J Immunol. 2004;173:7602–7614. doi: 10.4049/jimmunol.173.12.7602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1854 kb)
Data Availability Statement
Included as electronic supplementary material.
As detailed for each software application in the “Materials and methods” section.