Abstract
Colorectal cancer with a Kirsten rat sarcoma 2 viral oncogene homolog (KRAS) gene mutation is considered to be resistant to anti-EGFR agents. G12D is the most common KRAS mutation in colorectal cancer, followed by G12V and G13D. According to clinical and basic research data, patients with colorectal cancer exhibiting G12D and G12V KRAS mutations are resistant to anti-EGFR agents; however, this is not true of G13D and other minor mutations, which are still not well understood. The current study focused on minor KRAS mutations (G12A, G12C, G12S, Q61H and A146T) and evaluated whether these were resistant to anti-EGFR antibodies using a mix culture assay. The results demonstrated that all KRAS mutations, including minor mutations, were resistant to two anti-EGFR agents: Cetuximab and panitumumab. The combined effect of MEK and BCL-XL inhibition on colorectal cancer cells with KRAS minor mutations were subsequently evaluated. The combined effect of MEK and BCL-XL inhibitors was confirmed in all KRAS minor mutations. The sensitivity of AMG510, a novel KRAS G12C selective inhibitor, was also assessed. The mix culture assay revealed that AMG510 selectively exerted an antitumor effect on colon cancer cells with a G12C KRAS mutation. The combination of MEK and BCL-XL inhibition markedly enhanced the effect of AMG510 in colon cancer cells. The current study suggested that AMG510 may have potential clinical use in combination with MEK and BCL-XL inhibitors in the treatment of patients with colorectal cancer exhibiting the G12C KRAS mutation.
Keywords: colorectal cancer, Kirsten rat sarcoma 2 viral oncogene homolog, G12C, MEK, BCL-XL, ABT263, trametinib, AMG510
Introduction
KRAS is a small 21 kDa GTP-binding protein that plays a role in transmitting growth signals downstream of EGFR. KRAS mutations are found in 45-50% of colorectal cancers, of which ~90% are found at codon 12 and codon 13 (1-3). It is thought that the presence of KRAS mutation maintains GTP-binding activity and enhances downstream growth signals (4). Subgroup analysis of large-scale Randomized Controlled Trials showed that cetuximab and panitumumab were effective in wild-type patients with no mutations in exon 2 (codons 12 and 13) of the KRAS gene, whereas anti-EGFR antibody treatment was not effective in the KRAS mutant (1,2,5). Currently, administration of anti-EGFR antibody is not indicated for colorectal cancer patients with any KRAS mutation. However, it has been reported that treatment with cetuximab prolonged progression-free survival (PFS) and overall survival (OS) in patients with KRAS G13D mutation colorectal cancer (6). In addition, there are a few reports that KRAS G13D mutation colorectal cancer may be sensitive to anti-EGFR antibodies in vitro (6,7). As other KRAS mutations, codon 61 and 146 mutations (with frequencies of ~2%) are known. Imamura et al reported that colorectal cancer with codon 61 and 146 mutations have similar clinicopathological features to exon 2 (codons 12, 13) mutations (3). In the report, anti-EGFR antibody treatment was ineffective in all colorectal cancers with codon 61 mutations, whereas it was effective in some codon 146 mutation cases. KRAS mutations are more frequent in the order of G12D, G12V and G13D, three of which account for approximately 75% (1-3). In our report, these three mutations are referred to as major mutations. Otherwise, the next most frequent G12A, G12C, G12S, Q61H and A146T were described as minor KRAS mutations. To assess the sensitivity of molecularly targeted drugs for KRAS mutations, Mix Culture Assays (8,9) were performed. First, we evaluated the resistance of EGFR drugs to minor KRAS mutations in colorectal cancer cells, the sensitivity of MEK and BCL-XL inhibitors, and their combined effects. Furthermore, we evaluated the effect of a novel KRAS-G12C selective inhibitor, AMG510, and its combination effects with MEK and BCL-XL inhibitors in colorectal cancer cells.
Materials and methods
Cell culture
CACO-2 cells, a human colorectal cancer cell line, were purchased from RIKEN Cell Bank and maintained in DMEM (Gibco; Thermo Fisher Scientific, Inc.). Cells were incubated with 10% fetal bovine serum and penicillin/streptomycin at 37˚C and 5% CO2.
Antibodies and reagents
The following antibodies were used: Monoclonal mouse FLAG (cat. no. 014-22383; 1:1,000 for western blotting; FUJIFILM Wako Pure Chemicals Corporation); monoclonal rabbit ERK; cat. no. 4695; 1:1,000), monoclonal rabbit p-ERK (cat. no. 4376; 1:1,000), monoclonal mouse MEK1/2 (cat. no. 4694; 1:1,000) and monoclonal rabbit p-MEK1/2 (cat. no. 9121; 1:1,000) all purchased from Cell Signaling Technology, Inc.; monoclonal mouse β-actin (cat. no. sc-47787; 1:2,000) purchased from Santa Cruz Biotechnology, Inc. The secondary antibodies polyclonal goat anti-mouse (cat. no. P0447; 1:5,000) IgG and polyclonal goat anti-rabbit (cat. no. P0448; 1:5,000) IgG conjugated with HRP were obtained from Dako; Agilent Technologies, Inc. Cetuximab and panitumumab were purchased from Merck and Takeda Pharmaceutical Company and 7-aminoactinomycin D (7-AAD) was purchased from BioLegend. Trametinib, ABT263 and AMG510 were purchased from Cayman Chemical, LC Laboratories and Selleck Chemicals.
Construction and sequencing of vectors
Total mRNA of CACO-2 cells was extracted using NucleoSpin RNAplus (Takara Bio, Inc.) and cDNA was synthesized by using PrimeScript™ RT reagent Kit and PrimeScript RT Master Mix (Takara Bio, Inc.). KRAS-4B carrying a C-terminal FLAG was amplified using PCR and KRAS mutants of G12D, G12V, G13D, G12A, G12C, G12S, Q61H and A146T were created using In-Fusion® HD Cloning Kit (Takara Bio, Inc.). DNA sequences of all the constructs were confirmed using ABI 3130xl Genetic Analyzer using BigDye® Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Inc.).
The method of creating these vectors is shown in the paper by Koyama et al (9).
Retroviral transduction of the KRAS mutations
KRAS wild and mutated genes, G12D, G12V, G13D, G12A, G12C, G12S, Q61H, and A146T, were inserted into the multiple cloning site of pMXs-IRES-GFP vector (Cell Bio-Lab, Inc.). For retroviral transduction, these vectors were transfected into the amphotropic packaging cells, Phoenix, using PEI MAX (Polysciences Inc.). The virus-containing supernatants were harvested 24 and 48 h after gene transduction, and CACO-2 cells were infected with the retroviral particles on RetroNectin (Takara Bio, Inc.) coated plates. We confirmed transduction efficiency of pMXs-IRES-GFP vector as a GFP-positive ratio measured using a flow cytometer (BD FACSCanto II) and analyzed with Kaluza 2.1 software (Beckman Coulter, Inc.), and cells from the 10th passage were used for the Mix Culture Assay, as shown below.
Protein sample preparation and western blotting
After introducing the KRAS wild and mutant genes into CACO-2, western blotting was performed to confirm the gene transfer. CACO-2 cells were lysed in RIPA lysis buffer (cat. no. sc-24948; Santa Cruz Biotechnology, Inc.) containing 1 mM PMSF on ice for 30 min. The lysates were separated by centrifugation at 10,000 x g for 10 min at 4˚C and the resultant supernatant was collected as the total cell lysate. Protein was quantified using a Pierce BCA Protein assay kit (Thermo Fisher Scientific, Inc.) and 10 µg protein was separated using SDS-PAGE gel and then electroblotted onto a PVDF membrane. The membrane was blocked with Tris-buffered saline containing 5% non-fat dry milk and 1% Tween-20 for 1 h at room temperature and then probed using the primary antibodies at 4˚C overnight. The membrane was then incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at 4˚C, which was detected by enhanced chemiluminescence using Immobilon Western HRP (Amersham ECL Prime Western Blotting Detection Reagent; Cytiva). We used Bolt™ 4-12% Bis-Tris Plus Gels, 12 wells (Invitrogen), and laded 10 µg of protein into one lane in the order of ladder, Mock, KRAS wild, G12V, G12C, ladder, Mock, KRAS wild, G12V, G12C in one gel. After transfer to the PVDF membrane, the membrane was cleaved in the center and reacted separately with the primary antibodies MEK and pMEK. Similarly, we cut the same membrane in half and perform Western blotting of ERK and pERK, FLAG and β-actin. This experiment has been conducted three times separately.
Mix culture assay
We have developed and reported a mixed culture assay for stable and reliable screening of effective therapeutic targets, transduced with the pMXs-IRES-GFP vector, and analyzed using a flow cytometer (8,9). For this assay, we retrovirally transduced wild-type KRAS, major mutant KRAS genes (G12D, G12V, G13D) and minor mutant KRAS genes (G12A, G12C, G12S, Q61H, and A146) into CACO-2 cells using the pMX-IRES-GFP vector. A high gene-transduction efficiency of ≥90%, which was determined using the GFP-positive rate (%) measured using a flow cytometer, was obtained. After gene transfer, GFP expression in gene transduced cells stabilizes at ~7 passages, so cells with 7-10 passages are used in the experiment. Parental cells (GFP negative) and gene-transduced cells (GFP positive) were mixed ideally at a 1:1 ratio. It is impossible to keep the GFP positive rate constant at 50%. Approximately plus or minus 10% is considered to be an acceptable range (8,9). On the first day, the mixed cells were seeded at a 20% confluency on a 12-well plate and were cultured for 12 days with molecular targeting agents. They were then passaged at a 5:1 ratio before reaching confluence. On day 12, the cells were harvested and stained with 7-AAD. The population that was 7-AAD-negative, which represents viable cells, was gated and the GFP-positive ratio of these populations was determined using a flow cytometer. We calculated the relative proliferation ratio (RPR) using the following formula, the day 0 GFP-positive rate (%) (A), and the day 12 GFP-positive rate (%) (B). The outline of this experimental system, the calculation method for RPR, and the experimental example are shown in our previous report (9).
 |
Here, a low RPR indicates that the GFP-positive cell population was sensitive to the drug, while a high RPR indicates drug resistance. Using this system, we evaluated the drug sensitivities of KRAS-transduced cells to several molecular targeting drugs.
Statistical analysis
The data are expressed as mean ± standard deviation of 4 experiments for each assay. Statistical significance of the results was evaluated using Student's t-test and one-way ANOVA, followed by Bonferroni correction. Differences were considered significant at a P<0.05.
Results
Sequence analysis confirmed the correct gene sequence of the KRAS mutations inserted in the pMXs-IRES vector
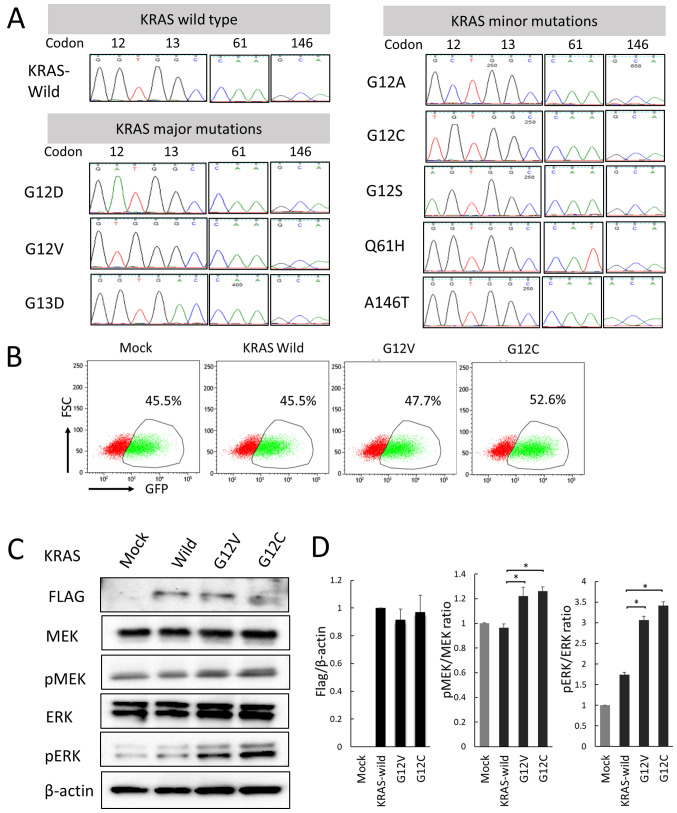
G12D, G12V, and G13D KRAS mutations, which are frequently found in colorectal cancer, are referred to as KRAS major mutations, and other mutations (G12A, G12C, G12S, Q61H, and A146T) are referred to as KRAS minor mutations in this study. It was confirmed that correctly sequenced wild KRAS, major KRAS mutations (G12D, G12V, and G13D) and minor KRAS mutations (G12A, G12C, G12S, Q61H, and A146T) had been inserted into pMX-IRES-GFP using an ABI 3130xl Genetic Analyzer (Fig. 1A). GFP-positive rates in a mixed state in Mock and KRAS mutations were confirmed by flow cytometer (Fig. 1B). Transfection efficiency of KRAS wild and mutant genes was confirmed by Western blotting with FLAG-specific antibodies. Furthermore, it was confirmed by western blotting that MEK and ERK were activated by the introduction of the mutant KRAS gene (Fig. 1C).
Figure 1.
Sequence analysis of genes inserted into the pMXs-IRES-GFP vector. Flow cytometery and western blotting demonstrated successful ectopic KRAS gene transduction. (A) The KRAS mutant genes inserted into the vector were amplified using the BigDye™ Terminator v3.1 Cycle Sequencing kit with a vector primer (5'-GACGGCATCGCAGCTTGGATACAC-3') and analyzed using an ABI 3130 Genetic Analyzer. (B) Flow cytometery analysis of GFP-positive rates in a mixed state with GFP-positive transgenic cells (mock, KRAS wild, G12V and G12C) and GFP-negative parental cells. (C) Transduction of KRAS wild and mutant (G12V and G12C) genes were confirmed via western blotting using a FLAG-specific antibody. MEK and ERK phosphorylation in CACO-2 cells was also examined via western blotting. (D) Normalized band intensity of FLAG relative to actin, pMEK relative to MEK and pERK relative to ERK. *P<0.05 vs. KRAS-wild type. KRAS, Kirsten rat sarcoma 2 viral oncogene homolog; GFP, green fluorescent protein; p, phosphorylated.
Flow cytometers and western blots showed successful ectopic KRAS gene transduction
To confirm successful transduction with pMX-IRES GFP, Mutant KRAS promotes cell proliferation via the upregulation of ERK phosphorylation., wild-type and mutant KRAS genes carrying a C-terminal FLAG were inserted into pDON-5Neo DNA vectors, which were retrovirally transduced into CACO-2 and SW48 human CRC cells expressing wild-type KRAS. Transduction was confirmed using an anti-FLAG antibody (Fig. 1C). The effect of KRAS gene mutation on cell proliferation were then examined. Mutant KRAS-transduced CACO-2 cells exhibited a significantly higher rate of proliferation compared with cells transduced with wild-type KRAS. The RAS/MEK/ERK signaling pathway is known to act downstream of the KRAS gene and regulate key cellular activities including differentiation, proliferation and survival. Therefore, p-MEK and p-ERK expression was examined using western blotting, and the ratios of p-protein/total protein were significantly higher in all mutant KRAS cells compared with in wild-type cells (Fig. 1C and D). Subsequently, other pathways associated with the RAS/MEK/ERK pathway were examined using the Mix Culture assay system.
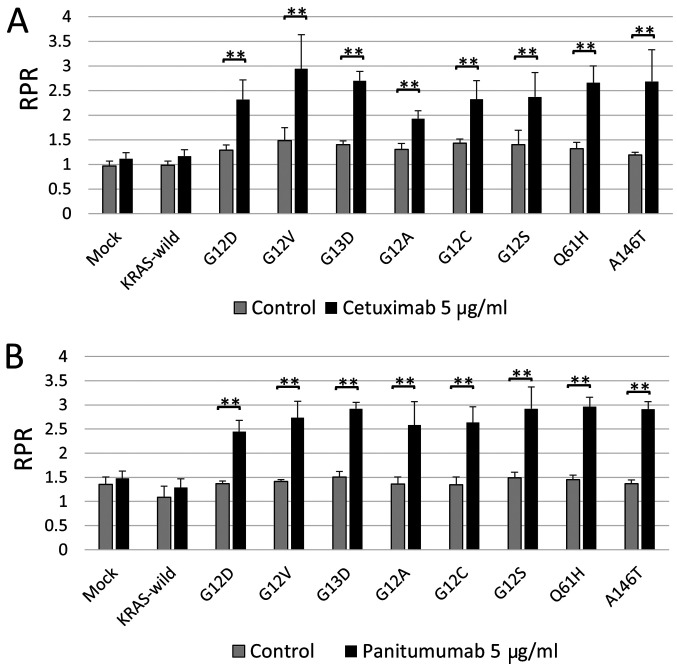
All major and minor KRAS mutations in CACO-2 cells contributed to resistance to anti-EGFR agents, cetuximab, and panitumumab
Do G13D and minor KRAS mutations really contribute to resistance to anti-EGFR agents? The Mix Culture Assay was used to answer this clinical question. As a result, RPR did not change significantly in KRAS wild type, but in all KRAS mutations, RPR increased significantly in the anti-EGFR-administered group, compared to the non-treated group. That is, it is suggested that all KRAS mutations, including G13D and minor mutations, contribute to anti-EGFR antibody resistance (Fig. 2).
Figure 2.
KRAS mutations contributed to anti-EGFR antibody drug resistance in CACO-2 cells. (A) The RPR with cetuximab administration was significantly elevated in all KRAS G12D, G12V, G13D, G12A, G12C, G12S, Q61H and A146T mutants. (B) Similar results were obtained following the administration of panitumumab. **P<0.01 vs. drug-free control. KRAS, Kirsten rat sarcoma 2 viral oncogene homolog; RPR, relative proliferation ratio.
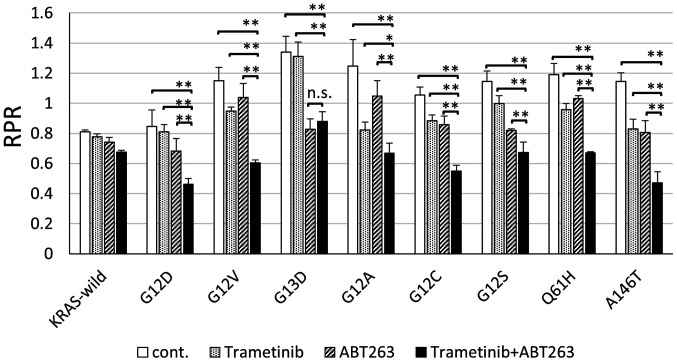
The Mix Culture Assay showed that the combination of MEK and BCL-XL inhibition was effective as a treatment target for all KRAS mutations
We reported that the combination of MEK and BCL-XL inhibition was effective as a targeting treatment for KRAS major mutations of colorectal cancer cells (9). We also analyzed whether simultaneous inhibition of MEK and BCL-XL had potent tumor suppressor effects on KRAS minor mutant colorectal cancer cells. In all minor KRAS mutations, the combination of MEK and BCL-XL inhibitors significantly reduced RPR, compared to drug-free controls. It was suggested that MEK and BCL-XL inhibition could be effective treatments, even for minor KRAS mutations (Fig. 3).
Figure 3.
Mix culture assay results revealed that the combination of MEK and BCL-XL inhibitors effectively targeted all KRAS mutations. In all KRAS mutations, the combination of trametinib and ABT263 significantly reduced RPR when compared with drug-free controls. In addition, the RPR of trametinib in combination with ABT263 was significantly reduced in all KRAS mutation groups compared with trametinib administered alone. In all KRAS mutants, except for G13D, the RPR of trametinib combined with ABT263 was significantly reduced compared with ABT263 alone. *P<0.05 and **P<0.01. KRAS, Kirsten rat sarcoma 2 viral oncogene homolog; RPR, relative proliferation ratio; cont., control; n.s., not significant.
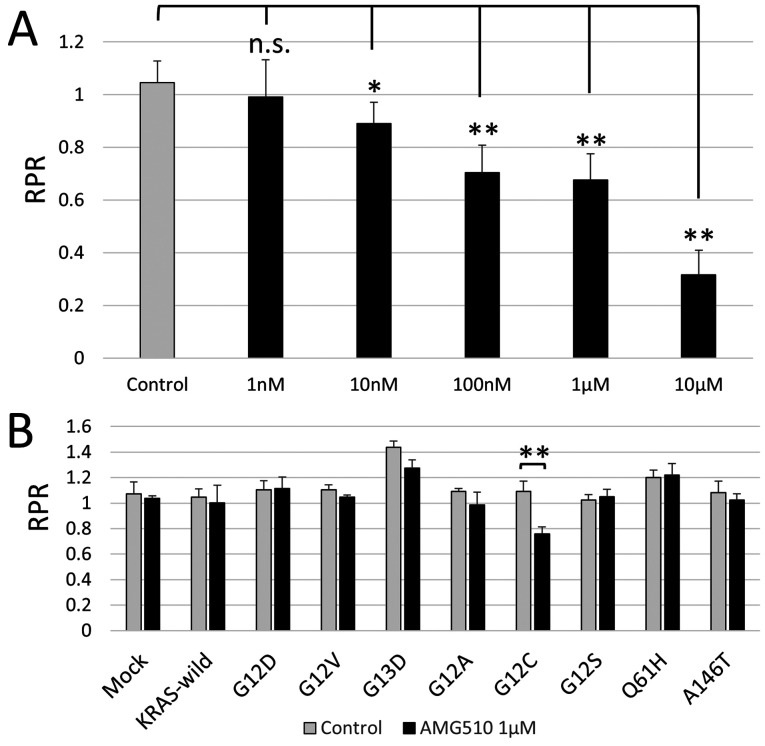
AMG510 selectively suppresses the growth of colorectal cancer cells with G12C KRAS mutation in vitro
The results of the phase I clinical trial for the solid tumor of AMG510, a novel KRAS G12C inhibitor, was reported at the 2019 American Oncology Society. AMG510 showed an antitumor effect on KRAS G12C mutation in non-small cell lung cancer (NSCLC), but unfortunately it did not have sufficient clinical effects on colorectal cancer (Trial ID: NCT03600883). The frequency of G12C in colorectal cancer is ~3-4% of KRAS mutations (3,10), the fourth highest, and finding an effective treatment for it is very important, from a clinical standpoint. First, using Mix Culture Assay, it was examined whether AMG510 has an antitumor effect on the CACO-2 cells into which the KRAS G12C gene was introduced. The results showed that there was a clear concentration dependent RPR decrease and that AMG 510 selectively inhibited KRAS G12C in colon cancer cells in vitro (Fig. 4A). Next, in order to evaluate the selectivity of AMG510 for KRAS G12C, Mix Culture Assays of KRAS-wild, G12D, G12V, G13D, G12A, G12C, G12S, Q61H and A146T were performed. The results showed that RPR was only significantly reduced in G12C (n=4, P<0.01). It was confirmed that the effect of AMG510 is selective for G12C and does not cross-react with other KRAS mutations. This result also demonstrates that the Mix Culture Assay is a powerful experimental tool for screening effective drugs that target gene mutations (Fig. 4B).
Figure 4.
Mix culture assay results demonstrated a selective AMG510 inhibitory effect when targeting KRAS G12C in colon cancer cells. (A) The RPR of G12C transgenic cells decreased in an AMG510 concentration-dependent manner. (B) Following AMG510 administration, the RPR of the G12C was greatly reduced. There was a significant difference in G13D; however, there was a slight decrease in RPR. No changes in RPR were observed in KRAS wild type or other KRAS mutations. *P<0.05 and **P<0.01 vs. drug-free control. KRAS, Kirsten rat sarcoma 2 viral oncogene homolog; RPR, relative proliferation ratio; n.s., not significant.
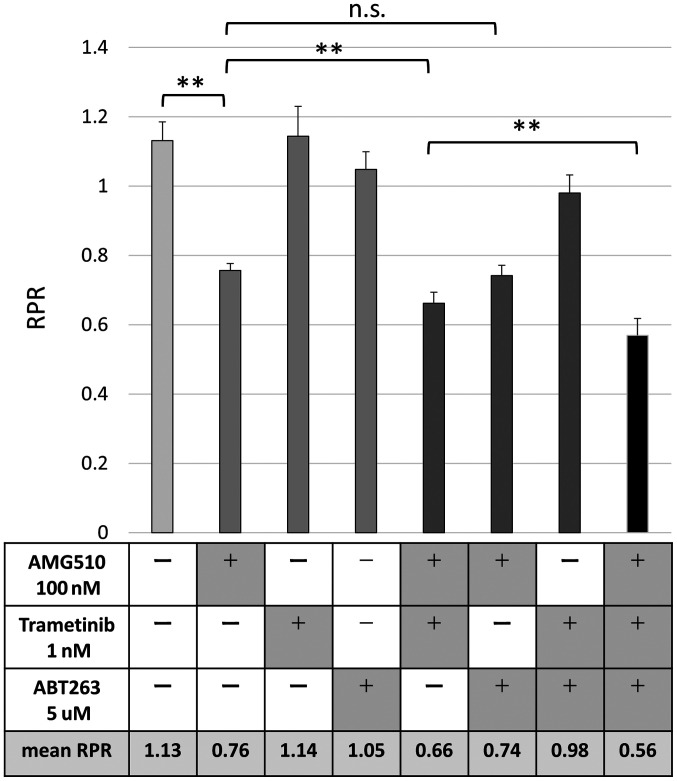
The combination of AMG510, MEK inhibitor, and BCL-XL inhibitor has potent anti-tumor effects on colorectal cancer cells with G12C KRAS mutation
The results of the Phase Ⅰ trial were presented at American Society of Clinical Oncology (ASCO) in 2019, and AMG510 proved to be effective for NSCLC, but it is not expected to be as effective in colon cancer. Since the effects of AMG510 are observed in CACO-2 cells in vitro, we hypothesized that clinical effects could be expected if there is a drug that enhances the effects of AMG510. In our previous study, we demonstrated that inhibition of MEK and BCL-XL was an effective targeting therapy for colorectal cancer cells with KRAS mutation (9). We evaluated the combined effect of AMG510, MEK inhibitor, and BCL-XL inhibitor in colorectal cancer cells using a Mix Culture Assay. Finally, G12C-transfected CACO-2 cells were used to evaluate the changes in RPR due to the combination of AMG510, MEK inhibitor, and BCL-XL inhibitor. The combination of MEK inhibitor, trametinib, significantly enhanced the RPR-lowering effect of AMG510, but BCL-XL inhibitor, ABT263, did not show such combination effect. Surprisingly, the combined use of the three drugs showed a further RPR-lowering effect, suggesting that simultaneous inhibition of KRAS, MEK, and BCL-XL may contribute to a further antitumor effect (Fig. 5).
Figure 5.
AMG510 is enhanced by MEK inhibition, but not ABT263. The combined use of the three inhibitors revealed a strong antitumor effect that targeted Kirsten rat sarcoma 2 viral oncogene homolog G12C. The MEK inhibitor, trametinib, significantly enhanced the RPR-lowering effect of AMG510, whereas the BCL-XL inhibitor, ABT263, did not. The combination of the three drugs revealed an additional RPR-lowering effect. **P<0.01. RPR, relative proliferation ratio; n.s., not significant.
Discussion
Our study focused on minor KRAS mutations and evaluated drug sensitivity using a unique method called Mix Culture Assay (9). There are three important points in our research. The first is the selection of KRAS mutations G12D, G12V, G13D, G12A, G12C, G12S, Q61H, and A146T. The sum of these eight mutations accounts for over 90% of KRAS mutations and can be considered the most clinically encountered KRAS mutations. The second point is the selection of a cell line without mismatch repair (MMR) deficiency. Many colorectal cancer studies use MMR-deficient colorectal cancer cell lines. Since MMR-deficient colorectal cancer has the characteristic of being clinically resistant to anticancer drugs (11), we believe that it may have a completely different phenotype from the usual colorectal cancer. In fact, the studies above mentioned (6,7) use MMR-deficient colon cancer cell lines, such as SW48, HT-116, and Lovo, which suggests that colorectal cancer cells with KRAS G13D mutation were sensitive to anti-EGFR antibody. In our study, CACO-2 cells were selected because the use of a cell line with the common genetic alterations of colorectal cancer was thought to have clinically significant consequences. The genetic status of CACO-2 cells was considered a usual colorectal cancer with MMR-proficiency, TP53 mutation, and APC mutation. Although we needed to study using multiple cell lines, only CACO-2 was available for human colorectal cell lines that meet the two conditions of RAS wild-type and MSS.A third important point is the use of a unique experimental procedure, called Mix Culture Assay, to evaluate the therapeutic targets for KRAS mutations. It may seem complicated at a first glance, but it is a simple research method to evaluate whether the GFP-positive rate increases or decreases by co-culturing KRAS-mutant-gene transfected cells (GFP-positive) and parental cells (GFP-negative) for a certain period under drug administration. RPR is a mathematical formula created to quantify the experimental results. In this assay, the number of cells is measured using a flow cytometer, and stable results are obtained. Details of this method can be found in the reported study by Koyama (9). The usual proliferation assay is carried out in different cell dishes or different mice under the same conditions, and the evaluation method is performed by counting the total cell number or measuring tumor size. In the Mix Culture Assay, KRAS mutant cells are cultured with internal control cells in the same cell culture dish with the same drug concentration, and the results are calculated as relative ratios using a flow cytometer. We believe that the Mix Culture Assay is an extremely useful tool to screen effective drugs targeting a certain gene mutation. Furthermore, we believe that the Mix Culture Assay is a very useful experimental tool for verifying the combined effect of multiple antitumor agents. On the other hand, the limitation of this assay is only in vitro validation, which only reveals the relative difference in drug susceptibility between transgenic GFP-positive cells and control GFP-negative cells. In other words, even if the RPR is low and the agent is expected to be highly effective, it needs to be further verified in vitro and further confirmed in vivo. On the contrary, even if the RPR is high and resistance can be expected, it is necessary to further verify whether the drug is really ineffective in vitro and in vivo (8,9).
The majority of KRAS mutations in colorectal cancer includes G12D, G12V and G13D, which are reported to account for approximately 75% of the total mutations (1-3). Based on clinical and basic research data, there is no doubt that G12D and G12V mutations contribute to anti-EGFR antibody resistance. On the other hand, it has been reported that anti-EGFR antibody may be effective for colorectal cancers with KRAS G13D mutation (6,7). For the 61 and 146 codon mutations, clinical data suggested that 61 codon-mutated colorectal cancers may be resistant to anti-EGFR; on the other hand, 146 codon-mutated colorectal cancers may be susceptible to anti-EGFR (3). However, this report is a very limited analysis of the few patient data, and this susceptibility cannot be further clarified. Clinically, all colorectal cancers with KRAS mutations are considered resistant to anti-EGFR antibodies, and cetuximab and panitumumab have not been used for them. Are all KRAS mutations contributing to anti-EGFR antibody resistance? Our study gave a clear answer to that question. All KRAS mutations at codons 12, 13, 61, and 146 have clearly been shown to contribute to resistance to the anti-EGFR drugs, cetuximab and panitumumab. (Fig. 2).
Previously, we reported that the combined inhibition of MEK and BCL-XL was effective against major mutations in KRAS: G12D and G12V (9). This study showed that MEK inhibitors and BCL-XL had similar effects on all other KRAS minor mutations.
Furthermore, we also evaluated the effect of a novel G12C-specific inhibitor, AMG510, by using the Mix Culture Assay. G12C is the most common KRAS mutation in NSCLC. Why are the treatment effects of colorectal cancer and lung cancer different? A similar phenomenon is observed in BRAF-mutant colorectal cancer. BRAF inhibitors are effective in treating BRAF-mutant lung cancer and melanoma, but not in treating colorectal cancer. This happens due to the previously reported reason that inhibition of BRAF activates CRAF and enhances downstream proliferation and anti-apoptotic signals (12,13). Since KRAS mutation activates downstream MEK, it has been thought that it could be an effective therapeutic target for KRAS mutant cancer; however, at present, MEK inhibition alone has not been established as an effective therapeutic method. MEK inhibition has been reported to activate other molecules, such as PI3K (14,15), ERBB2(16), FGFR (17), YAP (18), BCL-XL (9,19), and CDK4/6 (20-22) to amplify proliferative and anti-apoptotic signals. This phenomenon is called feedback/paradoxical activation and it is expected that this signal may also be induced by AMG510(23). Blocking this signal under administration of AMG510 enhances the effect of AMG510 and could be an effective clinical treatment. We developed this Mix Culture Assay to stably evaluate the combined effect of multiple drugs on KRAS mutations. Koyama et al screened for effective drugs in combination with MEK inhibitors in KRAS mutant colorectal cancer cells (9). As a result, it was reported that BCL-XL inhibitors potentiated the effects of MEK inhibitors, while BRAF inhibitor, PI3K inhibitor, and CDK4/6 inhibitor had no effects on MEK actuation. In our study, it was confirmed that the combination of MEK and BCL-XL inhibitors was effective for minor KRAS mutations. Furthermore, it was clarified that the anti-tumor effect of AMG510 on KRAS G12C mutant colorectal cancer was enhanced by MEK inhibitor, but not by BCL-XL. Surprisingly, it was shown that the three-drug combination has a dramatic antitumor effect on G12C KRAS colorectal cancer.
Mix Culture Assays demonstrated that all KRAS mutations, including minor KRAS mutations, contributed to anti-EGFR antibody drug resistance. It was also demonstrated that the combination of MEK and BCL-XL inhibitors has antitumor effects on all minor KRAS mutations. Furthermore, it was revealed in vitro that MEK inhibitor enhanced the selective antitumor effect of AMG510, a novel KRAS G12C selective inhibitor, in colorectal cancer cells. Furthermore, the combination of AMG510, MEK inhibitor, and BCL-XL inhibitor showed a strong antitumor effect on G12C colon cancer in vitro.
Acknowledgements
Not applicable.
Funding Statement
Funding: This work was supported by the Japan Society for the Promotion of Science KAKENHI (grant no. JP18K15238).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MKi, DK, MT and YS designed the current study. MKi, MKo, SN, SM, NH and TE performed the experiments, analyzed the data and wrote the manuscript. YM, FM, ST, MKu, HT and YY designed the experiments, and interpreted the data. All authors read and approved the final manuscript. YM and MKo confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, et al. K-ras Mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 2.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 3.Imamura Y, Lochhead P, Yamauchi M, Kuchiba A, Qian ZR, Liao X, Nishihara R, Jung S, Wu K, Nosho K, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: Cohort study and literature review. Mol Cancer. 2014;13(135) doi: 10.1186/1476-4598-13-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 5.Dahabreh IJ, Terasawa T, Castaldi PJ, Trikalinos TA. Systematic review: Anti-epidermal growth factor receptor treatment effect modification by KRAS mutations in advanced colorectal cancer. Ann Intern Med. 2011;154:37–49. doi: 10.7326/0003-4819-154-1-201101040-00006. [DOI] [PubMed] [Google Scholar]
- 6.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 7.Kumar SS, Price TJ, Mohyieldin O, Borg M, Townsend A, Hardingham JE. KRAS G13D mutation and sensitivity to cetuximab or panitumumab in a colorectal cancer cell line model. Gastrointest Cancer Res. 2014;7:23–26. [PMC free article] [PubMed] [Google Scholar]
- 8.Kitazawa M, Hida S, Fujii C, Taniguchi S, Ito K, Matsumura T, Okada N, Sakaizawa T, Kobayashi A, Takeoka M, Miyagawa SI. ASC induces apoptosis via activation of caspase-9 by enhancing Gap junction-mediated intercellular communication. PLoS One. 2017;12(e0169340) doi: 10.1371/journal.pone.0169340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koyama M, Kitazawa M, Nakamura S, Matsumura T, Miyazaki S, Miyagawa Y, Muranaka F, Tokumaru S, Okumura M, Yamamoto Y, et al. Low-dose trametinib and Bcl-xl antagonist have a specific antitumor effect in KRAS-mutated colorectal cancer cells. Int J Oncol. 2020;57:1179–1191. doi: 10.3892/ijo.2020.5117. [DOI] [PubMed] [Google Scholar]
- 10.AACR Project GENIE: Powering precision medicine through an International Consortium. Cancer Discov. 2017;7:818–831. doi: 10.1158/2159-8290.CD-17-0151. AACR Project GENIE Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Des Guetz G, Schischmanoff O, Nicolas P, Perret GY, Morere JF, Uzzan B. Does microsatellite instability predict the efficacy of adjuvant chemotherapy in colorectal cancer? A systematic review with meta-analysis. Eur J Cancer. 2009;45:1890–1896. doi: 10.1016/j.ejca.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 12.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 14.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, Ryan DP, Meyerhardt JA, Benes C, Settleman J, et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J Clin Invest. 2011;121:4311–4321. doi: 10.1172/JCI57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turke AB, Song Y, Costa C, Cook R, Arteaga CL, Asara JM, Engelman JA. MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer Res. 2012;72:3228–3237. doi: 10.1158/0008-5472.CAN-11-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manchado E, Weissmueller S, Morris JP IV, Chen CC, Wullenkord R, Lujambio A, de Stanchina E, Poirier JT, Gainor JF, Corcoran RB, et al. A Combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534:647–651. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin L, Sabnis AJ, Chan E, Olivas V, Cade L, Pazarentzos E, Asthana S, Neel D, Yan JJ, Lu X, et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet. 2015;47:250–256. doi: 10.1038/ng.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM, Greninger P, Brown RD, Godfrey JT, Cohoon TJ, et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell. 2013;23:121–128. doi: 10.1016/j.ccr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziemke EK, Dosch JS, Maust JD, Shettigar A, Sen A, Welling TH, Hardiman KM, Sebolt-Leopold JS. Sensitivity of KRAS-mutant colorectal cancers to combination therapy that cotargets MEK and CDK4/6. Clin Cancer Res. 2016;22:405–414. doi: 10.1158/1078-0432.CCR-15-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao Z, Le Blanc JM, Wang C, Zhan T, Zhuang H, Wang P, Yuan Z, Lu B. Coadministration of Trametinib and Palbociclib radiosensitizes KRAS-mutant non-small cell lung cancers in vitro and in vivo. Clin Cancer Res. 2016;22:122–133. doi: 10.1158/1078-0432.CCR-15-0589. [DOI] [PubMed] [Google Scholar]
- 22.Lee MS, Helms TL, Feng N, Gay J, Chang QE, Tian F, Wu JY, Toniatti C, Heffernan TP, Powis G, et al. Efficacy of the combination of MEK and CDK4/6 inhibitors in vitro and in vivo in KRAS mutant colorectal cancer models. Oncotarget. 2016;7:39595–39608. doi: 10.18632/oncotarget.9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan MB, Fece de la Cruz F, Phat S, Myers DT, Wong E, Shahzade HA, Hong CB, Corcoran RB. Vertical pathway inhibition overcomes adaptive feedback resistance to KRAS G12C inhibition. Clin Cancer Res. 2020;26:1633–1643. doi: 10.1158/1078-0432.CCR-19-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.