Abstract
Background
The discovery of selection signatures has enabled the identification of genomics regions under selective pressure, enhancing knowledge of evolutionary genotype-phenotypes. Sex chromosomes play an important role in species formation and evolution. Therefore, the exploration of selection signatures on sex chromosomes has important biological significance.
Results
In this study, we used the Cross Population Extend Haplotype Homozygosity Test (XPEHH), F-statistics (FST) and EigenGWAS to assess selection signatures on the Z chromosome in 474 broiler chickens via Illumina chicken 60 K SNP chips. SNP genotype data were downloaded from publicly available resources. We identified 17 selection regions, amongst which 1, 11 and 12 were identified by XPEHH, FST, and EigenGWAS, respectively. Each end of the Z chromosome appeared to undergo the highest levels of selection pressure. A total of 215 candidate genes were located in 17 selection regions, some of which mediated lipogenesis, fatty acid production, fat metabolism, and fat decomposition, including FGF10, ELOVL7, and IL6ST. Using abdominal adipose tissue expression data of the chickens, 187 candidate genes were expressed with 15 differentially expressed genes (DEGs) in fat vs. lean lines identified. Amongst the DEGs, VCAN was related to fat metabolism. GO pathway enrichment analysis and QTL annotations were performed to fully characterize the selection mechanism(s) of chicken abdominal fat content.
Conclusions
We have found some selection regions and candidate genes involving in fat metabolism on the Z chromosome. These findings enhance our understanding of sex chromosome selection signatures.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12863-021-00971-6.
Keywords: Z chromosome, SNP, Selection signature, Population genetics, Gene expression
Background
The domestication of chickens in Asia (Gallus gallus) occurred around 5400 BC with Darwin suggesting their evolution from red jungle fowl [1, 2]. Chickens hold value from an evolutionary perspective as they provide information that bridges knowledge between mammals and other vertebrates [3]. Domestic chickens have genetically adapted to unique habitats through strong genetic and phenotypic alterations. To date, an array of specialized commercial populations and inbred chicken lines have subsequently been developed.
Selection has many effects on the genome. Allele frequencies and polymorphism underlying selection are expected to change. With the availability of high-quality draft sequences of the chicken genome, high-density single nucleotide polymorphism (SNP) genotyping chips, and whole-genome re-sequencing technologies, the detection of selection signatures on the chicken genome have been reported. Rubin et al. [4] identified the TSHR gene (thyroid stimulating hormone receptor) as a prominent selection signature in all domestic chickens. Guo et al. [5] identified 413 candidate genes in Xishuangbanna fighting chickens that were related to aggressive behavior, including BDNF, NTS and GNAO1. Boschiero et al. [6] revealed more than 300 regions of selection with many important genes, including AKAP6, IGFBP2 and IGF1R, associated with fat deposition and muscle development.
Sex chromosomes play an important role in species formation and evolution. Mcvicker et al. [7] analyzed the selective forces that shape hominid evolution and found that under natural selection, the selection pressure of sex chromosomes (12–40%) exceeded those of the autosome (19–26%). The selection pressure of autosomes and sex chromosomes is different, and when considering sex-specific dosage compensation, genes on the sex chromosomes are more directly and efficiently selected than those on autosomes [8, 9]. The size of the chicken Z chromosome is approximately 83 Mb, accounting for 7.9% of the chicken genome. The Z chromosome contains 1345 genes, and some genes, including FGF10 (fibroblast growth factor 10), ELOVL7 (ELOVL fatty acid elongase 7) and ACO1 (aconitase 1, soluble), regulated fat deposition and development. Previous studies have focused on the selection signatures of chicken autosomes, but the selection signals of the chicken Z chromosome less well studied. Zhang et al. [10] only identified PC1/PCSK1 gene, located on the Z chromosome, related to abdominal fat traits used selection signals and genome-wide association analysis based on NEAUHLF (Northeast Agricultural University broiler lines divergently selected for abdominal fat content) population. It is therefore necessary to identify as many selection signatures as possible on the Z chromosome in chickens.
In this study, we used the XPEHH, FST and EigenGWAS methods to identify the selection signatures associated with abdominal fat in the Z chromosomes of NEAUHLF populations. Through the integration of gene microarrays in the adipose tissue of NEAUHLF populations, we investigated the expression profiles of the candidate genes on selection regions at 7 weeks of age. Gene annotations and functional enrichment were implemented to elucidate the significance of the identified selection signatures to fat containing traits.
Results
Population structure
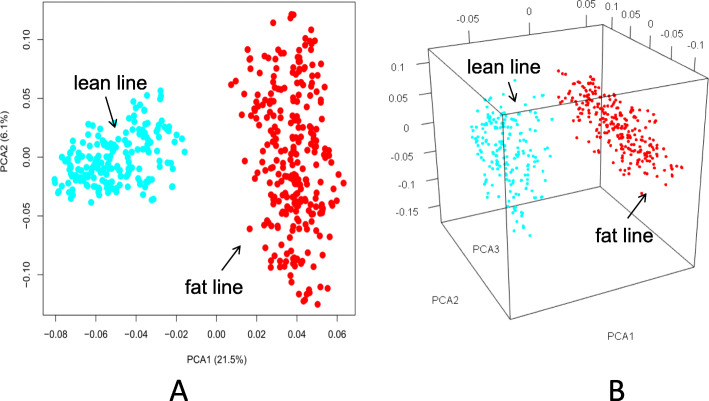
We performed principal component analysis (PCA) on 1937 SNPs on the Z chromosome to identify individual patterns. The first principal component (21.5% of the total variance) could separate the two lines (Fig. 1a). The second principal component (6.1% of the total variance) primarily revealed genetic differences in the fat lines, whilst the third principal component (5.3% of the total variance) primarily revealed differences in the lean lines (Fig. 1b).
Fig. 1.
Population structure based on Z chromosome SNPs using principal component analysis. The subgraph (a) and (b) represent two-dimensional and three-dimensional PCA images
Selection signatures on the Z chromosome
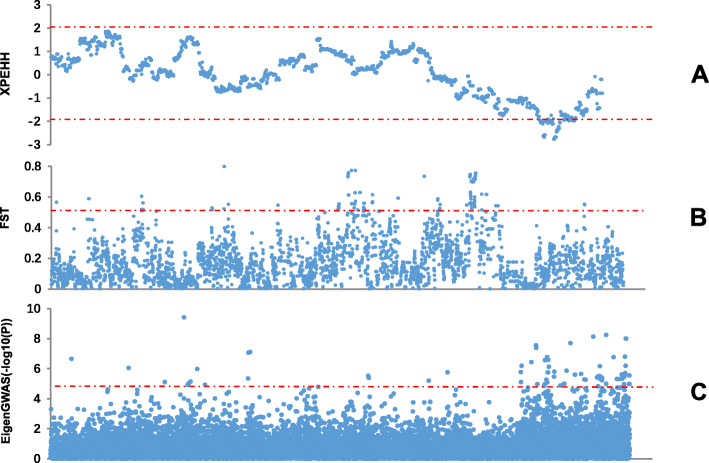
Chicken Z chromosome selection signatures were identified between fat and lean line populations. Table 1 summarizes the selection signatures obtained using XPEHH, FST and EigenGWAS. For the fat-lean line pair, 1, 11 and 12 selection regions were identified using XPEHH, FST and EigenGWAS methods, respectively (Table 2 and Fig. 2). A total of 17 candidate regions were identified by merging these regions. The majority of the identified selection regions were present on both ends of the Z chromosome, accounting for about 53%. Amongst them, one candidate region (61.68–73.63 Mb) was identified by all methods, and five candidate regions were identified by FST and EigenGWAS. There were 5 and 6 candidate regions only identified by FST and EigenGWAS method, respectively.
Table 1.
Selection signatures in the two chicken lines
| Items | Fat - Lean | ||
|---|---|---|---|
| XPEHH | FST | EigenGWAS | |
| Number of significant SNPs | 50 | 98 | 83 |
| Number of regions | 1 | 11 | 12 |
| Average length (Mb) | 2.24 | 0.63 | 1.71 |
| Total length (Mb) | 2.24 | 6.88 | 20.53 |
Table 2.
Selection regions on the Z chromosome and candidate genes detected in the regions
| Candidate Region (Mb) | FST | EigenGWAS | XPEHH | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region (Mb) | Top snp | FST | Major Genes | Region (Mb) | Top snp | P-value | Major Genes | Region (Mb) | Top snp | XP EHH | Major Genes | |
| 0.55–0.75 | 0.55–0.75 | rs14688003 | 0.56 | WDR7 | ||||||||
| 1.91–2.31 | 1.91–2.31 | rs312273884 | 2.24E-07 | SLC14A2 | ||||||||
| 4.53–4.66 | 4.53–4.66 | rs16704177 | 0.59 | |||||||||
| 6.95–7.35 | 6.95–7.35 | rs14689552 | 2.40E-05 | KIAA1328 | ||||||||
| 9.25–9.65 | 9.25–9.65 | rs14784526 | 9.10E-07 | ARHGEF39 | ||||||||
| 11.23–11.57 | 11.23–11.57 | rs16781096 | 0.57 | SLC1A3, | ||||||||
| 13.95–14.35 | 13.95–14.35 | rs16103326 | 7.97E-06 | FGF10 | ||||||||
| 16.75–19.95 | 19.81–19.95 | rs14753903 | 0.52 | 16.75–17.42 | rs16760225 | 7.23E-06 | IL6ST | |||||
| 17.80–18.20 | rs16099146 | 1.05E-06 | PLK2 | |||||||||
| 18.71–19.11 | rs736458094 | 1.18E-05 | ELOVL7 | |||||||||
| 21.22–23.50 | 21.22–21.84 | rs14755170 | 0.66 | MAST4 | 22.90–23.50 | rs313728591 | 7.91E-08 | IQGAP2 | ||||
| 28.08–28.19 | 28.08–28.19 | rs16106682 | 0.55 | KDM4C | ||||||||
| 32.13–33.63 | 32.13–32.53 | rs16108416 | 2.11E-05 | BNC2 | ||||||||
| 33.03–33.63 | rs314666735 | 1.66E-05 | ADAMTSL1 | |||||||||
| 36.03–40.49 | 36.03–36.25 | rs16081040 | 0.54 | ANXA1 | 39.07–39.87 | rs16781643 | 3.03E-06 | FRMD3 | ||||
| 37.29–37.85 | rs16768474 | 0.68 | VPS13A | |||||||||
| 38.04–38.89 | rs13817231 | 0.59 | TLE4 | |||||||||
| 39.22–39.45 | rs14787078 | 0.54 | ||||||||||
| 40.33–40.49 | rs14787751 | 0.57 | NTRK2 | |||||||||
| 47.72–51.04 | 47.72–47.58 | rs15597824 | 0.59 | FER | 48.33–51.04 | rs314662658 | 6.33E-06 | EFNA5 | ||||
| 50.89–51.02 | rs14766378 | 0.73 | FAM174A | |||||||||
| 52.48–53.77 | 52.50–52.89 | rs16770232 | 0.53 | SLC49A3 | 52.48–52.88 | rs14768956 | 1.78E-06 | PCGF3 | ||||
| 53.37–53.77 | rs14769714 | 2.52E-05 | ATP5I | |||||||||
| 55.98–60.28 | 55.98–57.39 | rs14751311 | 0.66 | PCSK1 | ||||||||
| 58.12–58.85 | rs16758604 | 0.60 | NR2F1 | |||||||||
| 59.90–60.28 | rs14748688 | 0.54 | CETN3 | |||||||||
| 61.68–73.63 | 71.43–71.58 | rs14781326 | 0.55 | PRR16 | 61.68–62.12 | rs14774149 | 6.47E-07 | 65.73–67.97 | rs14776247 | −2.78 | DNAJC25 | |
| 62.78–63.58 | rs14773275 | 3.68E-07 | VCAN | |||||||||
| 63.88–64.28 | rs16118182 | 2.81E-08 | CKMT2 | |||||||||
| 64.82–66.08 | rs14774834 | 1.72E-07 | SLC46A2 | |||||||||
| 66.24–66.67 | rs13788226 | 2.66E-07 | DNAJC25 | |||||||||
| 66.84–67.88 | rs16775262 | 1.59E-06 | ELAVL2 | |||||||||
| 68.59–68.99 | rs16776912 | 2.02E-08 | MOB3B | |||||||||
| 69.41–70.09 | rs14780063 | 2.38E-05 | ||||||||||
| 70.35–71.36 | rs16125209 | 2.28E-06 | ACO1 | |||||||||
| 72.16–73.63 | rs14781762 | 7.41E-09 | KCNN2 | |||||||||
| 78.84–82.42 | 78.84–79.27 | rs15992604 | 5.72E-09 | CDKN2A | ||||||||
| 79.67–80.45 | rs14685714 | 1.75E-07 | ZNF608 | |||||||||
| 80.62–82.42 | rs16683478 | 9.91E-09 | PAX5 | |||||||||
Fig. 2.
Selection signatures on the Z chromosome. The subgraphs (a), (b) and (c) are the selection signatures detected between the populations using the XPEHH, FST and eigenGWAS methods, respectively
Candidate gene annotations for functional analysis
According to the chicken gene annotation data (Gallus gallus 6.0) in the ENSEMBL database, we detected 215 candidate genes within 17 selection regions. Supplementary Table 1 summarizes the genes in each selection region on the Z chromosome. A number of genes were found to regulate lipogenesis, fatty acid production, fat metabolism, or fat decomposition, including FGF10, ELOVL7 and IL6ST.
To reveal the biological functions of the genes within the identified regions, gene Ontology (GO) pathway enrichment analyses were performed using DAVID (v6.7) [11]. Significant GO functional terms (P < 0.05) are listed in Table 3, but these terms were not significant upon Benjamini correction.
Table 3.
Functional enrichment analysis of the selected genes
| Category | GO ID | Term | P value |
|---|---|---|---|
| Biological Progresses | GO:0043171 | peptide catabolic process | 0.0195 |
| Cellular Components | GO:0005892 | acetylcholine-gated channel complex | 0.0231 |
| GO:0045211 | postsynaptic membrane | 0.0204 | |
| Molecular Function | GO:0070006 | Metalloaminopeptidase activity | 0.0154 |
| GO:0042166 | acetylcholine binding | 0.0230 | |
| GO:0004889 | acetylcholine-activated cation-selective channel activity | 0.0230 |
A total of 1229 QTLs were found on the chicken Z chromosome in the QTLdb database (https://www.animalgenome.org/cgi-bin/QTLdb/GG/index). The selection signatures overlapped on 132 QTLs of health, physiology, exterior and production categories. Interestingly, 45 of the candidate genes that overlapped with the QTL region were related to abdominal fat weight, 22 were related to liver weight, 12 were related to food intake, 11 to residual food intake, and 18 to food conversion ratios.
Expression profiles of the candidate genes in the selection regions
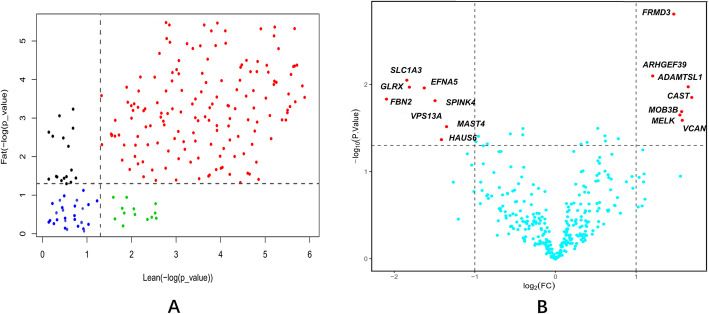
We extracted probe sets of the Gene Chip Chicken Genome Array to represent all candidate genes located on the selection regions. A total of 427 probes representing 187 of the 215 candidate genes were identified. As shown in Fig. 3, 168 and 174 genes (p < 0.05) were expressed in lean and fat chicken lines at 7 weeks, respectively (Fig. 3a), whilst 15 DEGs (P < 0.05, fold change > 2) between fat and lean lines were identified (Fig. 3b). Seven genes showed significantly higher expression in the fat line (e.g. ADAMTSL1, FRMD3, MOB3B, ARHGEF39, VCAN, CAST and MELK) and eight genes in the lean line (e.g. HAUS6, MAST4, VPS13A, SPINK4, SLC1A3, GLRX, FBN2 and EFNA5).
Fig. 3.
Candidate gene expression profiles in the adipose tissue of the NEAUHLF. The subgraph (a) is the gene expression profile of two chicken strains. The x-axis and y-axis is - log (P_value) of lean and fat line respectively, and the threshold value was p < 0.05. The red points indicate 155 genes that are expressed in the abdominal fat of both thin and fat lines, while the blue points indicate 28 genes that are not expressed in either line. The 13 green points indicate genes expressed in the lean line but not in the fat line and vice versa for the 19 black points.. The subgraph (b) shows the differential expression of genes in adipose tissues in fat and lean lines at the 7th week of age. The threshold is P < 0.05, fold change > 2. The red points indicate genes that are differentially expressed in the two chicken lines
Discussion
High-density SNPs chips permit the identification of genome-wide selection signatures using site frequent spectrums, population differentiation, and linkage disequilibrium, with known strengths and weaknesses. In this study, we used three complementary statistical approaches (FST, XPEHH, EigenGWAS) to explore the selection signatures on the Z chromosome to minimize bias and false positives in the broiler chickens. The FST method is best suited for the detection in events occurring in the more distant past [12]. The FST method is a powerful tool to detect signatures based on group differentiation. The XPEHH test compares extended haplotype homozygosity between populations to detect selection signatures, which are segregated in populations and represent points of ongoing selection. XPEHH is therefore useful for the detection of entirely or approximately fixed loci [13]. The XPEHH test is an LD-based method, and LD is expected to extend over longer distances in regions under recent selection. So, selection regions detected by XPEHH were much wider [14]. Ma et al. [15] pointed out that the FST method may bring a higher false positive rate compared with XPEHH. The EigenGWAS algorithm combines the statistical framework of GWAS with eigenvector decomposition to identify selection signatures in the genomes of the underlying population. The EigenGWAS method uses multi-point information to identify core SNPs and grid windows, and can identify potential loci during selection, and a larger number of selection regions than FST and XPEHH [16]. Due to the similarities and differences principles between FST, XPEHH and EigenGWAS, there are differently selection regions can be obtained using the different statistical approaches.
The sex (X) chromosome undergoes more drift than autosomes, as its effective population size (Ne) is three-quarters that of autosomes [17]. McVicker et al. [7] found that X chromosome has suffered higher selection pressure than autosomes. The NEAUHLF broiler population came from bi-directional divergently selected for abdominal fat content. Zhang et al. [10] found that four candidate regions of chromosome Z were identified as selection signature using long-range heterozygosity changes or allele frequency differences methods, and the 0.73 Mb PC1/PCSK1 region of the Z chromosome was the most heavily selected region based on genome-wide using the NEAUHLF populations. The Z chromosome contains some genes involving fat metabolism, such as FGF10, ELOVL7 and ACO1. However, Zhang et al. [10] did not identify selection signatures overlapped these genes. Selection signatures determined by multiple methods are deemed more credible [15, 18]. So, in this study, we used more methods to independently identify potential selection regions on the Z chromosome related on abdominal fat development to verify and supplement the previous findings. We detected three regions overlapped Zhang’s results using the FST method based on population differentiation (lean vs fat lines) or the EigenGWAS method in this study. Furthermore, we identified 14 other selection regions. These novel selection regions will provide specific gene targets for the control of chicken fatness traits or other traits genetically correlated with fatness. For example, we identified 61.68–73.63 Mb regions detected by three methods, and 69 genes that overlapped with the region, including DNAJC25, GNG10 and AKAP2. Interestingly, DNAJC25 is a member of DNAJ gene family identified by Liu et al. [19] as highly expressed in chicken liver tissue using transcriptome sequencing analysis. The DNAJB6 gene, located on gga2, is a member of the DNAJ gene family and has a similar sequence to the DNAJC25 gene. Jin et al. [20] previously found that the DNAJB6 gene was expressed in the abdominal fat and liver tissues of the 14th generation NEAUHLF population, and was differentially expressed between the fat line and the lean line. Moreover, the expression level of DNAJB6 in abdominal adipose tissue was significantly negatively correlated with abdomen fat weight and abdomen fat percentage [20].
In this study, there are 215 candidate genes overlapped 17 selection regions on chromosome Z. Amongst the candidate genes, IL6ST, ELOVL7, CKMT2 and FGF10 genes were also identified by Gholami et al. [21] in three commercial layer breeds and 14 non-commercial breeds. The VCAN, ST8SIA4, FBN2, ERAP1 and CAST were also identified by Fu et al. [14], showing 10 regions of high confidence for selection on the Z chromosome, detected in male Cornish lines (a meat type breed), and female lines from White Rock (a dual-purpose breed). We also found that the majority of candidate genes expressed in the adipose tissue of G8 NEAUHLF fat and lean lines, and 15 genes (including MAST4, SPINK4, VCAN, EFNA5, ARHGEF39, etc) in the adipose tissue significantly differed between fat vs. lean birds using microarray gene expression data. Among them, MAST4 encodes a microtubule-associated serine/threonine kinase; SPINK4 is a serine peptidase inhibitor; and ARHGEF39 encodes a rho guanine nucleotide exchange factor key to Rho mediated signal transduction.
According to known gene functions, some candidate genes were associated with the fat content of chickens, such as the FGF10 gene. FGF10 is a mesenchymal factor affecting epithelial cells. Matsubara et al. [22] reported that FGF10 when secreted in chicken adipose tissue contributes to adipogenesis, and is down regulated during the early stages of chicken adipocyte differentiation. Konishi et al. [23] showed that FGF10 stimulates proliferation in the white adipose tissue of mice. In addition, Yamasaki et al. [24] highlighted FGF10 as an important intercellular signaling molecule during lipogenesis that is abundantly expressed in the adipose tissue of adult rats.
Due to high species conservation, the identified genes related to human or mice obesity traits may hold importance for adipose deposition in chickens, such as ELOVL7, IL6ST, IQGAP2, PAX5 and CKMT2 (Table 4). ELOVL7 shows altered affinity for the elongation of precursor fatty acids and mediates the extension of saturated fatty acids of up to 24 carbon atoms [26]. IL6ST is an IL-6 transducer and a potent modulator of fat metabolism in humans, known to increase fat oxidation and fatty acid re-esterification [25]. IQGAP2 deficiency influences hepatic free fatty acid uptake, fatty acid synthesis, and lipogenesis, suggesting its importance in obesity [27]. PAX5 is a paired box 5 gene for which Melka et al. [29] performed GWAS in human adolescents from the French-Canadian founder population, revealing the association of its locus with total fat mass (TFM) and body mass index (BMI) in 6.4 and 3.7% of TFM and BMI heritability estimates, respectively. These results imply that PAX5 plays a key role in obesity regulation. CKMT2 (creatine kinase, mitochondrial 2) is a creatine kinase isoenzyme. Müller et al. [28] showed that CKMT2 is an effective modulator of ATP synthase coupled respiration and is exclusively expressed in human brown adipose tissue. CKMT2 also regulates energy metabolism.
Table 4.
Candidate genes in the selection regions and their functions
| Gene symbol | Location (Mb) | Full name | Function of association |
|---|---|---|---|
| FGF10 | 13.97–14.03 | Fibroblast growth factor 10 | Promotes fat formation [22] |
| IL6ST | 17.02–17.06 | Interleukin 6 signal transducer | Increases fat oxidation and fatty acid re-esterification [25]. |
| ELOVL7 | 18.87–18.91 | ELOVL fatty acid elongase 7 | Extension of saturated fatty acids [26] |
| IQGAP2 | 23.46–23.58 | IQ motif containing GTPase activating protein 2 | Influences the liver uptake of free fatty acids, fatty acid synthesis and adipogenesis [27] |
| CKMT2 | 63.95–63.98 | Creatine kinase, mitochondrial 2 | Regulation of energy metabolism, expression in brown adipose tissue [28] |
| PAX5 | 82.01–82.12 | Paired box 5 | Regulates obesity [29] |
Conclusion
In this study, 17 selection regions were screened through the analysis of selection signatures in the chicken Z chromosome, including 215 candidate genes, some of which are involved in lipogenesis, fatty acid production, fat metabolism and fat decomposition, such as FGF10, ELOVL7, IL6ST. Moreover, in the candidate region, using abdominal fat expression data from chickens, 187 candidate genes were identified as expressed in the fat and lean lines, with 15 genes identified as differentially expressed. GO pathways enrichment and QTL annotations provided additional information on the selection mechanism(s) of chicken abdominal fat content. The culmination of these data enhances our understanding of sex chromosome selection signatures and their role in fat deposition in chickens.
Methods
Genotype data and population
SNP genotype data were downloaded from GEO Datasets on the NCBI website (GEO accession: GSE58551) [30]. Based on Illumina chicken 60 K SNP chips, 48,035 SNPs from 28 autosomes and Z chromosomes in 475 male broilers of the 11th generation (G11) (203 lean lines and 272 fat lines) were identified from NEAUHLF [10]. We mapped the SNP loci on the Z chromosome of all birds to the chicken reference genome (Gallus gallus 6.0), resulting in 1973 SNP loci. We applied QC measurements on the SNP loci on the Z chromosome of all birds using PLINK (v1.90) software: (1) SNP loci call rates of 0.95; (2) Sample call rates of 0.95; and (3) Minor allele frequencies (MAF) of 0.01 were discounted. In total, 1937 SNPs and 474 birds were investigated to detect selective sweeps in the chicken sex chromosomes. Specific details of broiler breeding strategy have been described by Zhang et al. [10].
Principal component analysis
We performed PCA to distinguish population structures using EigenGWAS software [16] based on 1937 SNPs on the Z chromosome. The first ten eigenvalues and their corresponding eigenvectors were then calculated.
Detection of selection signatures
Extended haplotype homozygosity (EHH) scores measure the probability that two randomly selected chromosomes carry a tested core haplotype that is homozygous at all SNPs [31]. XPEHH scores can detect selective sweeps in which a selected allele has achieved fixation in one population but remains polymorphic in another [32]. FST can identify genomic regions with strongly differing or differentially fixed variants in alleles frequency between different populations, which is the conventional measure of population genetic differentiation. FST is defined as follows:
where MSP is the mean square error within the populations, MSG represents the mean square error between the two populations, and nc represents the average sample size of the entire population after correction [33, 34]. The EigenGWAS algorithm combines the statistical framework of genome-wide association studies with eigenvector decomposition to identify selection signatures on the underlying genome [16]. The EigenGWAS method uses the single marker regression model for association tests. However, its phenotype is different from the phenotype of typical GWASs and it is an individual-level eigenvector derived from genotype data. The model can be described as the following equation:
where yki is the k th eigenvector value of individual j; xij is the value of the j th SNP for individual j; bi is the regression coefficient for the i th SNP. EigenGWAS can be used as a method to find the selection signatures among the population or across a gradient of ancestry. In this study, XPEHH, FST, and EigenGWAS were used to detect selective footprints on the chicken Z chromosome. We used SHAPEIT (v2.12) software to generate haplotype data based on the SNPs data. We used the LD package of R (v3.6.1) to compute the XPEHH values. The threshold of the XPEHH at a significance level of 0.05 was ±2. Using VCFtools [35], the average FST value of all SNPs in each sliding window (window size: 100 Kb, step size: 10 Kb) was calculated. We determined the threshold for the outlier FST sliding windows average based on the following formula:
Among them, Q1 is the lower quartile (first quartile); Q3 is the upper quartile (third quartile). In this study, the FST value of the sliding window greater than threshold (0.51) is defined as a selection signature. For EigenGWAS, EMMAX software [36] was used for single-marker regression. Threshold P-values of 0.05/1937 = 2.58 × 10−5 were used to confirm statistically significant differences. In this study, the candidate regions were determined within 1 Mb of each other regions identified by different methods.
To reveal the biological functions of the selection signatures, additional analyses were performed. We identified candidate genes within the selection regions using chicken gene annotation data from the Ensembl database. We then used the online software DAVID (v6.7) [11] to perform GO analysis based on the candidate genes obtained. Thirdly, selection regions were mapped onto QTL obtained from the chicken QTL database (https://www.animalgenome.org/cgi-bin/QTLdb/GG/index).
Gene expression profiles
Gene expressions in the abdominal adipose tissue of seven-week-old NEAUHLF broilers were evaluated in chicken genome arrays. According to Wang et al. [37], the raw data set has been standardized using Affymetrix Microarray Suite 4.0 software and uploaded to the GEO database (GEO accession number: GSE8010). We downloaded the GSE8010 data set for subsequent gene differential expression analysis [37]. The ten birds were selected based on the percentage of abdominal fat (AFP) at 7-weeks for the G 8 of NEAUHLF broilers: the 5 chickens with the highest AFP in fat line and the 5 chickens with the lowest AFP in lean line. A one-way ANOVA was used to statistically compare the DEGs between fat and lean line chickens.
Supplementary Information
Additional file 1: Supplementary Table 1.
Acknowledgements
The authors would like to thank the Poultry Breeding Group of the College of Animal Science and Technology and Key Laboratory of Chicken Genetics and Breeding of Ministry of Agriculture and Rural Affairs at the Northeast Agricultural University for uploading SNP genotype data and gene microarray data.
Abbreviations
- XPEHH
Cross Population Extend Haplotype Homozygosity Test
- FST
F-statistics
- DEG
Differentially expressed genes
- SNP
Single nucleotide polymorphism
- NEAUHLF
Northeast Agricultural University broiler lines divergently selected for abdominal fat content
- PCA
Principal component analysis
- GO
Gene Ontology
- QTL
Quantitative trait loci
- TFM
Total fat mass
- BMI
Body mass index
- MAF
Minor allele frequencies
- EHH
Extended haplotype homozygosity
- AFP
Percentage of abdominal fat
Authors’ contributions
ZW and TW conceived the project. TW, MZ, JG and YG performed the bioinformatics and data analysis. TW and ZW wrote the manuscript. KD and PW collected the samples and data. All authors read and approved the final manuscript.
Funding
This research was supported by the Academic Backbone Project of Northeast Agricultural University (No.15XG14), NEAU Research Founding for Excellent Young Teachers (2010RCB29), Natural Science Foundation of China (No.31101709) and Open Projects of Key Laboratory of Chicken Genetics and Breeding Ministry of Agriculture and Rural Affairs (No.CGB-201706). Funding bodies played no role in the design of the study or analysis or interpretation of data or in writing the manuscript.
Availability of data and materials
The SNP genotype data and the 7-week-old abdominal fat expression data of broilers used in this study were obtained from the Gene Expression Omnibus (GEO) database at the National Center for Biotechnology Information (NCBI) under accession numbers GSE58551 and GSE8010.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tao Wang and Meng Zhou contributed equally to this work and share first authorship.
References
- 1.Xiang H, Gao J, Yu B, Zhou H, Cai D, Zhang Y, Chen X, Wang X, Hofreiter M, Zhao X. Early Holocene chicken domestication in northern China. Proc Natl Acad Sci U S A. 2014;111(49):17564–17569. doi: 10.1073/pnas.1411882111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu YP, Wu GS, Yao YG, Miao YW, Luikart G, Baig M, Beja-Pereira A, Ding ZL, Palanichamy MG, Zhang YP. Multiple maternal origins of chickens: out of the Asian jungles. Mol Phylogenet Evol. 2006;38(1):12–19. doi: 10.1016/j.ympev.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 3.International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432(7018):695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 4.Rubin CJ, Zody MC, Eriksson J, Meadows JR, Sherwood E, Webster MT, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464(7288):587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- 5.Guo X, Fang Q, Ma C, Zhou B, Wan Y, Jiang R. Whole-genome resequencing of Xishuangbanna fighting chicken to identify signatures of selection. Genet Sel Evol. 2016;48(1):62. doi: 10.1186/s12711-016-0239-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boschiero C, Moreira GCM, Gheyas AA, Godoy TF, Gasparin G, Mariani PDSC, Paduan M, Cesar ASM, Ledur MC, Coutinho LL. Genome-wide characterization of genetic variants and putative regions under selection in meat and egg-type chicken lines. BMC Genomics. 2018;19(1):83. doi: 10.1186/s12864-018-4444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McVicker G, Gordon D, Davis C, Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5(5):e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006;38(1):47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 9.Graves JA. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124(5):901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Hu X, Wang Z, Zhang Y, Wang S, Wang N, Ma L, Leng L, Wang S, Wang Q, Wang Y, Tang Z, Li N, da Y, Li H. Selection signature analysis implicates the PC1/PCSK1 region for chicken abdominal fat content. PLoS One. 2012;7(7):e40736. doi: 10.1371/journal.pone.0040736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cadzow M, Boocock J, Nguyen HT, Wilcox P, Merriman TR, Black MA. A bioinformatics workflow for detecting signatures of selection in genomic data. Front Genet. 2014;5:293. doi: 10.3389/fgene.2014.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll S, Gaudet R, Schaffner SF, Lander ES, International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallée C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe'er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Altshuler D, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Tsunoda T, Johnson TA, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Zeng C, Zhao H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Gibbs RA, Belmont JW, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Wheeler DA, Yakub I, Gabriel SB, Onofrio RC, Richter DJ, Ziaugra L, Birren BW, Daly MJ, Altshuler D, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L'Archevêque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen J, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449(7164):913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu W, Lee WR, Abasht B. Detection of genomic signatures of recent selection in commercial broiler chickens. BMC Genet. 2016;17(1):122. doi: 10.1186/s12863-016-0430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Y, Zhang H, Zhang Q, Ding X. Identification of selection footprints on the X chromosome in pig. PLoS One. 2014;9(4):e94911. doi: 10.1371/journal.pone.0094911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen GB, Lee SH, Zhu ZX, Benyamin B, Robinson MR. EigenGWAS: finding loci under selection through genome-wide association studies of eigenvectors in structured populations. Heredity (Edinb) 2016;117(1):51–61. doi: 10.1038/hdy.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heyer E, Segurel L. Looking for signatures of sex-specific demography and local adaptation on the X chromosome. Genome Biol. 2010;11(1):203. doi: 10.1186/gb-2010-11-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu XX, Pan JF, Zhao QJ, He XH, Pu YB, Han JL, Ma YH, Jiang L. Detecting selection signatures on the X chromosome of the Chinese Debao pony. J Anim Breed Genet. 2018;135(1):84–92. doi: 10.1111/jbg.12314. [DOI] [PubMed] [Google Scholar]
- 19.Liu T, Jiang W, Han D, Yu L. DNAJC25 is downregulated in hepatocellular carcinoma and is a novel tumor suppressor gene. Oncol Lett. 2012;4(6):1274–1280. doi: 10.3892/ol.2012.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin P, Wu X, Xu S, Zhang H, Li Y, Cao Z, Li H, Wang S. Differential expression of six genes and correlation with fatness traits in a unique broiler population. Saudi J Biol Sci. 2017;24(4):945–949. doi: 10.1016/j.sjbs.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gholami M, Erbe M, Gärke C, Preisinger R, Weigend A, Weigend S, Simianer H. Population genomic analyses based on 1 million SNPs in commercial egg layers. PLoS One. 2014;9(4):e94509. doi: 10.1371/journal.pone.0094509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsubara Y, Aoki M, Endo T, Sato K. Characterization of the expression profiles of adipogenesis-related factors, ZNF423, KLFs and FGF10, during preadipocyte differentiation and abdominal adipose tissue development in chickens. Comp Biochem Physiol B Biochem Mol Biol. 2013;165(3):189–195. doi: 10.1016/j.cbpb.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Konishi M, Asaki T, Koike N, Miwa H, Miyake A, Itoh N. Role of Fgf10 in cell proliferation in white adipose tissue. Mol Cell Endocrinol. 2006;249(1–2):71–77. doi: 10.1016/j.mce.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Yamasaki M, Emoto H, Konishi M, Mikami T, Ohuchi H, Nakao K, Itoh N. FGF-10 is a growth factor for preadipocytes in white adipose tissue. Biochem Biophys Res Commun. 1999;258(1):109–112. doi: 10.1006/bbrc.1999.0594. [DOI] [PubMed] [Google Scholar]
- 25.Van Hall G, Steensberg A, Sacchetti M, Fischer C, Keller C, Schjerling P, et al. Interleukin-6 stimulates lipolysis and fat oxidation in humans. J Clin Endocrinol Metab. 2003;88(7):3005–3010. doi: 10.1210/jc.2002-021687. [DOI] [PubMed] [Google Scholar]
- 26.Guillou H, Zadravec D, Martin PG, Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: insights from transgenic mice. Prog Lipid Res. 2010;49(2):186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Chiariello CS, LaComb JF, Bahou WF, Schmidt VA. Ablation of Iqgap2 protects from diet-induced hepatic steatosis due to impaired fatty acid uptake. Regul Pept. 2012;173(1–3):36–46. doi: 10.1016/j.regpep.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller S, Balaz M, Stefanicka P, Varga L, Amri EZ, Ukropec J, Wollscheid B, Wolfrum C. Proteomic analysis of human Brown adipose tissue reveals utilization of coupled and uncoupled energy expenditure pathways. Sci Rep. 2016;6(1):30030. doi: 10.1038/srep30030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melka MG, Bernard M, Mahboubi A, Abrahamowicz M, Paterson AD, Syme C, Lourdusamy A, Schumann G, Leonard GT, Perron M, Richer L, Veillette S, Gaudet D, Paus T, Pausova Z. Genome-wide scan for loci of adolescent obesity and their relationship with blood pressure. J Clin Endocrinol Metab. 2012;97(1):E145–E150. doi: 10.1210/jc.2011-1801. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Du ZQ, Dong JQ, Wang HX, Shi HY, Wang N, et al. Detection of genome-wide copy number variations in two chicken lines divergently selected for abdominal fat content. BMC Genomics. 2014;15(1):517. doi: 10.1186/1471-2164-15-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419(6909):832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 33.Weir BS, Hill WG. Estimating F-statistics. Annu Rev Genet. 2002;36(1):721–750. doi: 10.1146/annurev.genet.36.050802.093940. [DOI] [PubMed] [Google Scholar]
- 34.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12(12):1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang HM, Sul JH, Service SK. Zaitlen NA, Kong SY, Freimer NB, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42(4):348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang HB, Li H, Wang QG, Zhang XY, Wang SZ, Wang YX, Wang XP. Profiling of chicken adipose tissue gene expression by genome array. BMC Genomics. 2007;8(1):193. doi: 10.1186/1471-2164-8-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1.
Data Availability Statement
The SNP genotype data and the 7-week-old abdominal fat expression data of broilers used in this study were obtained from the Gene Expression Omnibus (GEO) database at the National Center for Biotechnology Information (NCBI) under accession numbers GSE58551 and GSE8010.