Abstract
Background
Coagulase-negative staphylococci (CNS) survive as commensals of skin, anterior nares and external canals of human and were regarded as non-infectious pathogens. However, they are emerging as a major cause of nosocomial infectious due to their ability to form biofilms and high resistance to several classes of antibiotics. This study examines the biofilm forming abilities of 214 clinical CNS isolates using phenotypic and genotypic methods, and determines their antibiotic susceptibility patterns.
Methods
A total of 214 clinical isolates collected from different clinical samples were identified as CNS and their antibiotic susceptibility determined by CLSI guidelines. The biofilm forming ability of all isolates was determined by three phenotypic methods; Congo red agar (CRA) method, tube adherence method (TM) and tissue culture plate (TCP) method and by genotypic method for the detection of icaAD genes.
Results
Among all the isolates, S. epidermidis (57.5%) was found the most frequently, followed by S. saprophyticus (18.7%), S. haemolyticus (11.2%), S. hominis (7%), and S. capitis (5.6%). Antibiotic susceptibility pattern demonstrated 91.6% isolates were resistant to penicillin and 66.8% to cefoxitin while 91.1% isolates were susceptible to chloramphenicol. Constitutive and inducible clindamycin resistant phenotype as measured by D-test was seen among 28% and 14.5% of isolates respectively. Tissue culture plate method detected biofilm production in 42.1% isolate followed by 31.8% through tube method while 20.1% isolates were found to produce slime in Congo red agar method. The genotypic assay revealed presence of icaA and icaD genes in 19.2% isolates.
Conclusion
The study shows a high prevalence of biofilm formation and inducible clindamycin resistance in CNS isolates, indicating the importance of in-vitro biofilm production test and D-test in routine laboratory diagnostics. Implementation of efficient diagnostic techniques for detection of biofilm production in clinical samples can help manage staphylococcal infections and minimize risks of treatment failures in hospitals.
Keywords: Coagulase-negative staphylococci, Biofilm production, Antibiotic susceptibility
Background
Presently, the wide-spread use of implants in modern healthcare facilities has substantially increased the risk of device associated infections, which in turn has drastically increased the rate of mortality and morbidity. Non-pathogenic indigenous microbiota of skin, nares and other mucosal surfaces of human, coagulase-negative staphylococci (CNS) are opportunistic pathogens accounting as one of the most common etiologic agents of device related infections [1, 2]. Over the past few decades, CNS, especially Staphylococcus epidermidis has emerged as a major cause of nosocomial infections [3–5]. The pathogen, now regarded as a life threatening, causes septicemia, meningitis, endocarditis and other serious invasive infections. Immunocompromised individuals or patients undergoing treatment with indwelling devices such as catheters and tips are more vulnerable to contract the CNS infections. Therefore, paradoxically, medical devices aimed to improve the treatment outcome, have often contributed to the prevalence of nosocomial infections [1, 5, 6], increasing the clinical significance of CNS.
Biofilm production is one of the major arsenals of saprophytic microbiota to become an opportunistic pathogen [7, 8]. Through biofilms, the CNS can adhere to and colonize on biotic as well as abiotic surfaces. Likewise, the damaged host tissue caused by prolonged antibiotic use and the implanted devices facilitate the survival, proliferation, and virulence of these pathogens [4, 6]. The biofilm of CNS is composed of the layer of extracellular polymeric substance called polysaccharide intercellular adhesion (PIA) matrix, which is encoded by ica operon (icaADBC genes) [9]. Beneath the biofilm matrix, bacteria are protected from physical, chemical, and biological stresses imposed by the antibiotics and the host immune cells [4]. Indeed, increased evidences show that the bacteria embedded within biofilms are difficult to kill by the antibiotics that are effective against planktonic cells, leading to the treatment failures of biofilm infections [1, 10, 11]. Moreover, the dispersal phenomenon of biofilms also serves as a source to metastatic infections [1, 4, 10]. There are mounting evidences of antibiotic resistance among biofilm forming clinical staphylococcal isolates. Furthermore, polymicrobial proximity within the biofilm might facilitate horizontal exchange of genetic information leading to development of antibiotic resistant clonal population [7]. Therefore, considering the clinical significance of biofilm associated infections, prevention and management of CNS infections remain a priority for the betterment of public health. This warrants the implementation of efficient methods to detect biofilm production among clinical samples in routine laboratory diagnosis.
The antimicrobial susceptibility testing (AST) of a clinical isolate is crucial for the optimal antimicrobial treatment of infected patients. This practice has been even more critical considering the single or multidrug resistant microorganisms [12]. Studies have reported clinical antibiotic resistance including that against erythromycin, macrolides, lincosamides and streptogramins type B (MLSB) in clinical CNS isolates [13, 14]. Many studies have reported the prevalence of antibiotic resistance including MLSB resistance S. aureus isolates from different regions of Nepal [15–22]. Recent studies although have reported antibiotic resistance and biofilm in clinical CNS isolates [22, 23], the prevalence of MLSB resistance in the CNS isolates are lacking in Nepal. Herein, we report the prevalence of antibiotic resistance and biofilm production among clinical CNS isolates collected from two tertiary care hospitals of Nepal.
Methods
Isolation and identification of coagulase-negative staphylococci (CNS)
A descriptive cross sectional study was carried out at two tertiary care hospitals of Nepal; B & B hospital and Kathmandu Institute of Technology (KIST) Medical College & Hospital. A total of 214 isolates were collected from various clinical samples including central venous catheter (CVC), catheter tip, suction tip, drain tip, Double J (DJ) stent, tracheal tip, endotracheal tip, blood, wound/pus and urine.
The elution of the bacterial cells from CVC or catheters was done by following standard microbiological protocol routinely followed in the hospital. The catheter tips were collected in sterile container and then mixed with 2 ml of nutrient broth (NB). After mixing by vortexing, loop-full of the suspension was streaked on MacConkey Agar (MA), Blood Agar (BA) plate and further processed for bacterial identification. The isolates were identified as staphylococci according to the standard microbiological procedure which included colony morphology on BA and mannitol salt agar (MSA), Gram staining and biochemical tests including catalase, slide and tube coagulase tests and oxidative/fermentative (O/F) test [25]. Once the isolates were identified as CNS, they were classified into species following simplified scheme proposed by Cunha et al., [26] using several biochemical tests. After identification of species, the isolates were stored in tryptic soy broth (TSB) with glycerol in freezer at −20 °C for further use.
Phenotypic and genotypic characterization of biofilm formation
Both phenotypic and genotypic methods were used for the detection of biofilm production in all the isolates. ATCC 35983 S. epidermidis strain was used as positive control for biofilm production in all assays performed.
Congo red agar method (CRA)
The screening of biofilm production was performed using CRA media; a qualitative method as described by Freeman et al., [27]. The medium composed of Brain Heart Infusion (BHI) agar supplemented with sucrose and a dye; Congo red. These plates were inoculated with CNS isolates, incubated aerobically at 37 ºC for 24 h and were interpreted based on the qualitative observation of colored colonies formation on the CRA plates. The black colonies with dark consistency were regarded as strong biofilm producers while the pink colonies were regarded as biofilm non-producers. The experiments were performed in triplicates and repeated three times.
Tube adherence method (TM)
This qualitative method for the detection of biofilm formation was performed as described by Christensen et al. [28]. A loop-full of microorganism was inoculated in trypticase soy broth (TSB) supplemented with 1% glucose. The tubes were incubated at 37 ºC for 24 h. The tubes were decanted, washed with PBS (pH 7.3) for 4 times and dried. Tubes were then stained with 0.1% crystal violet for 15 min. Excess stain was removed by washing with deionized water for 3 times. The tubes were then dried in inverted position and observed for biofilm formation. In this assay, biofilm formation was considered positive when a visible film was observed along the inner wall and bottom of tube. Depending on this, isolates were scored as 0 for absence, + for weak, + + for moderate, and + + + for strong biofilm formation. The experiments were performed in triplicates and repeated three times.
Tissue culture plate (TCP) method
All the isolates were screened for their ability to produce biofilm by this quantitative method as previously described by Christensen et al. [28] with slight modification [22]. In this assay, a loop-full of organism was inoculated in 5 mL TSB supplemented with 1% glucose and incubated at 37º C for 24 h. The overnight culture was diluted 1:100 with fresh media and 0.2 mL of this diluted culture was inoculated into individual wells of sterile polystyrene 96 well flat bottom tissue culture plates and incubated at 37 ºC for 24 h. After incubation, the content of tissue culture plate was removed by gentle tapping, and washed with PBS (pH 7.2) 4 times to remove free flowing planktonic bacteria. Biofilms formed by adherent sessile bacteria in the plate were fixed with 2% sodium acetate. It was then stained with 0.1% crystal violet for 15 min at room temperature. Excess stain was rinsed off by washing with deionized water for 4 times and plates were dried. Optical density (OD) of stained adherent bacteria was measured with micro ELISA auto reader at OD 630 nm. OD values from sterile medium, fixative and dye were averaged and subtracted from all test values. The experiments were performed in triplicates and repeated three times. Bacterial adherence was classified based on OD values of the individual isolates. Mean OD value < 0.120, 0.120–0.240 and > 0.240 were classified as non/weak, moderate and strong biofilm adherence respectively.
Detection of ica genes
The genomic DNA was extracted as previously described [22] using the DNA extraction Kit (Thermo Fischer). The sequences of icaA and icaD (accession number U43366) were taken from the GenBank sequence of the National Center for Biotechnology Information (NCBI) database. Primers specific for icaA and icaD were forward 5′-TCTCTTGCAGGAGCAATCAA, reverse 5′-TCAGGCACTAACATCCAGCA generating a product size of 188 bp and forward 5′-ATGGTCAAGCCCAGACAGAG, reverse 5′-CGTGTTTTCAACATTTAATGCAA with a product size of 198 bp respectively [22]. A 25 μl of reaction mixture consisted of MgCl2 (2.5 mM), Taq DNA polymerase (1U), each dNTPs (100 μM), each primer (1 μM) and DNA extract (200 ng). DNA amplification was carried out with following parameters: preheating at 95 °C for 5 min followed by 35 cycles of amplification (denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s and elongation at 72 °C for 30 s) and termination at 72 °C for 2 min. The PCR product was analyzed in 2% agarose gel stained with SYBR safe dye (Invitrogen).
Antimicrobial susceptibility testing (AST)
AST of the isolates was performed on Mueller Hinton Agar (MHA) by modified Kirby-Bauer disk diffusion method recommended by clinical laboratory standard institution (CLSI) guidelines [29]. The antimicrobial discs (HiMedia Laboratories) used in the study were: penicillin (10 units), ciprofloxacin (30 µg/disc), tetracycline (30 µg/disc), clindamycin (2 µg/disc), chloramphenicol (30 µg/disc), cefoxitin (30 µg/disc), erythromycin (15 µg/disc), cotrimoxazole (1.25/23.75 µg/disc) and gentamicin (10 µg/disc). The cefoxitin disc was used to detect methicillin resistance. The Staphylococcus aureus ATCC 25923 was used as reference strain for analyzing AST results.
Screening of inducible clindamycin resistance
The double disc diffusion test or D zone test outlined in CLSI document M100-S25 [29] was performed to determine if the erythromycin resistant isolates expressed inducible clindamycin resistance. Erythromycin (15 µg) disc was placed at a distance of 15 mm (edge to edge) from clindamycin (2 µg) on Mueller Hinton agar plates previously inoculated with 0.5 McFarland bacterial suspensions. Plates were analyzed after 18 h of incubation at 37 °C. Interpretation of the inhibition zone diameters was as follows: If an isolate was erythromycin resistant and clindamycin susceptible, with a D-shaped inhibition zone around the clindamycin disc, it was considered positive for inducible resistance (D-test positive, iMLSB phenotype). If the isolate was erythromycin resistant and clindamycin susceptible, with both zones of inhibition showing a circular shape, the isolate was considered to be negative for inducible resistance (D test negative, MS phenotype), but to have an active efflux pump. If the isolate was resistant to both drugs, it was considered to have the macrolide–lincosamide–Streptogramin B constitutive (cMLSB phenotype) [30].
Statistical analysis
The statistical analysis was performed using SPSS 17.0 (SPSS Inc., Chicago, USA) software. Chi-square test was used to compare between groups and P values < 0.05 were considered statistically significant.
Results
Isolation and identification of CNS
A total of 214 isolates were identified as CNS from various clinical samples following standard microbiological procedure using different biochemical tests [25]. Five species were identified among all CNS isolates including S. epidermidis (57.5%); the most frequently isolated species followed by S. saprophyticus (18.7%), S. haemolyticus (11.2%), S. homonis (7%) and S. captis (5.6%) (Fig. 1).
Fig. 1.
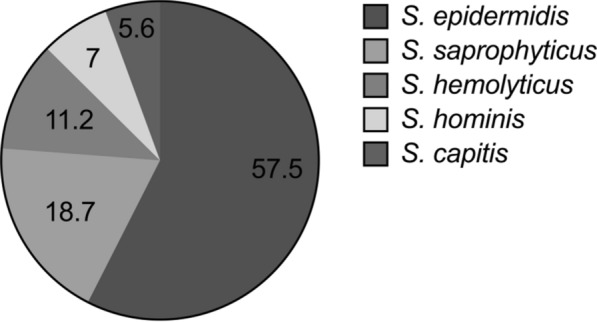
Percentage of CNS isolated from various clinical samples
Frequency of CNS isolated from different clinical specimens
S. epidermidis was isolated from all specimen types received in the laboratory. It was most frequently isolated from blood (52, 42.3%) followed by wound/pus (34, 27.6%). Similarly, S. saprophyticus was also mostly isolated from blood (12, 30%) and wound/pus (12, 30%). Different implant devices were also found to harbor CNS. All CNS were isolated from catheter tip except S. hominis. However, S. hominis were isolated from CVC, blood, wound/pus and urine. On the other hand, S. saprophyticus and S. haemolyticus were not isolated from suction tip, drain tip, tracheal tip and endotracheal tip. Similarly, S. capitis were isolated only from CVC, catheter tip, blood, and wound/pus (Table 1).
Table 1.
Frequency of CNS isolated from different clinical specimens
| CNS | Clinical specimen | Total | ||||||
|---|---|---|---|---|---|---|---|---|
| CVC | Catheter tip | Blood | Wound/pus | Urine | Others | |||
| S. epidermidis | 8 (6.5%) | 14 (11.4%) | 52 (42.3%) | 34 (27.6%) | 11(8.9%) | 4 (3.3%) | 123 (57.5%) | |
| S. saprophyticus | 2 (5.0%) | 2 (5.0%) | 12 (30.0%) | 12 (30.0%) | 11(27.5%) | 1(2.5%) | 40 (18.7%) | |
| S. hemolyticus | 6 (25.0%) | 6 (25.0%) | 5 (20.8%) | 5 (20.8%) | 1(4.2%) | 1(4.2%) | 24 (11.2%) | |
| S. hominis | 1 (6.7%) | – | 10 (66.7%) | 3 (20.0%) | 1 (6.7%) | – | 15 (7.0%) | |
| S. capitis | 3 (25.0%) | 3 (25.0%) | 3 (25.0%) | 2 (16.7%) | – | 1(8.3%) | 12 (5.6%) | |
CVC central venous catheter; others (Suction tip, Drain tip, Double J stent, Tracheal tip, Endotracheal tube)
Antibiotic susceptibility pattern among CNS isolates
Antibiotic susceptibility testing of 9 clinically relevant antibiotics was performed against all the collected isolates. These antibiotics were selected because of their common use in clinical practice in hospitals of Nepal. Among all CNS, 91.6% isolates were found to be resistant to penicillin. Mostly, S. epidermidis (114, 92.7%), S. saprophyticus (35, 87.5%) and S. haemolyticus (23, 95.8%) were susceptible to chloramphenicol. Similarly, S. capitis (12, 100%) and S. hominis (14, 93.3%) were susceptible to tetracycline (Table 2). Cefoxitin disc was used to detect methicillin resistance, which was observed in 66.8% CNS isolates. All five CNS species were found to be methicillin resistant with 73 (59.3%) S. epidermidis, 29 (72.5%) S. saprophyticus, 21 (87.5%) S. haemolyticus, 10 (66.7%) S. hominis and 10 (83.3%) S. capitis (Table 2).
Table 2.
Antimicrobial susceptibility pattern of CNS isolates
| Antibiotics | Total (n = 214) | S. epidermidis (n = 123) | S. saprophyticus (n = 40) | S. hemolyticus (n = 24) | S. hominis (n = 15) | S. capitis (n = 12) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | S | R | S | R | S | R | S | R | S | R | S | |
| CNS | ||||||||||||
| Penicillin | 196 (91.6%) | 18 (8.4%) | 111 (90.2%) | 12 (9.8%) | 39 (97.5%) | 1 (2.5%) | 23 (95.8%) | 1 (4.2%) | 12 (80%) | 3 (20%) | 11 (91.7%) | 1 (8.3%) |
| Ciprofloxacin | 76 (35.5%) | 138 (64.5%) | 39 (31.7%) | 84 (68.35) | 7 (17.5%) | 33 (82.5%) | 17 (70.8%) | 7 (29.2%) | 5 (33.3%) | 10 (66.75) | 8 (66.7%) | 4 (33.3%) |
| Tetracycline | 27 (12.6%) | 187 (87.4%) | 11 (8.9%) | 112 (91.1%) | 8 (20%) | 32 (80%) | 7 (29.2%) | 17 (70.8%) | 1 (6.7%) | 14 (93.3%) | - | 12 (100%) |
| Clindamycin | 63 (29.4%) | 151 (70.6%) | 30 (24.4%) | 93 (75.6%) | 13 (32.5%) | 27 (67.5%) | 9 (37.5%) | 15 (62.5%) | 5 (33.3%) | 10 (66.7%) | 6 (50%) | 6 (50%) |
| Chloramphenicol | 19 (8.9%) | 195 (91.1%) | 9 (7.3%) | 114 (92.7%) | 5 (12.5%) | 35 (87.5%) | 1 (4.2%) | 23 (95.8%) | 2 (13.3%) | 13 (86.7%) | 2 (16.7%) | 10 (83.3%) |
| Cefoxitin | 143 (6.5%) | 71 (33.2%) | 73 (59.3%) | 50 (40.7%) | 29 (72.5%) | 11 (27.5%) | 21 (87.5%) | 3 (12.5%) | 10 (66.7%) | 5 (33.3%) | 10 (83.3%) | 2 (16.7%) |
| Erythromycin | 155 (72.4%) | 59 (27.6%) | 80 (65%) | 43 (35%) | 34 (85%) | 6 (15%) | 21 (87.5%) | 3 (12.5%) | 10 (66.7%) | 5 (33.3%) | 10 (83.3%) | 2 (16.7%) |
| Cotrimoxazole | 80 (37.4%) | 134 (62.6%) | 46 (37.4%) | 77 (62.6%) | 14 (35%) | 26 (65%) | 10 (41.7%) | 14 (58.3%) | 6 (40%) | 9 (60%) | 4 (33.3%) | 8 (66.7%) |
| Gentamicin | 45 (21%) | 169 (79%) | 17 (13.8%) | 106 (86.2%) | 8 (20%) | 32 (80%) | 14 (58.3%) | 10 (41.7%) | – | 15 (100%) | 6 (50%) | 6 (50%) |
R Resistant, S Sensitive
Inducible clindamycin resistance
Clindamycin is a useful drug in the treatment of serious infections caused by Staphylococci due to its excellent tissue penetration, good oral absorption, and is an alternative to penicillin allergic patients. However, it has been indicated that approximately 45% of erythromycin resistant S. aureus isolates have inducible MLSB resistance that would go unrecognized if erythromycin and clindamycin disc are not placed appropriately during routine antibiotic susceptibility test [29, 31]. Among 214 CNS, 155 (72.4%) were resistant to erythromycin. MS phenotype (D test) was performed for these isolates and showed that 60 (28%) isolates were resistant to both erythromycin and clindamycin indicating constitutive MLSB phenotype. Out of 151 clindamycin sensitive isolates, positive D test was observed among 31(14.5%) isolates, indicating inducible MLSB phenotype. Negative D-test results were obtained among 70 (32.7%) isolates, whereas, the remaining 53 (24.8%) isolates were susceptible to both erythromycin and clindamycin. Constitutive and inducible MLSB phenotype was 51 (23.8%) and 25 (11.7%) among methicillin resistant CNS and 9 (4.2%) and 6 (2.8%) in methicillin sensitive CNS respectively. Both constitutive and inducible MLSB phenotype was predominant among MRCNS as compared to MSCNS (Table 3).
Table 3.
Erythromycin and clindamycin susceptibility testing
| Phenotypes | MRCNS | MSCNS | Total | |||
|---|---|---|---|---|---|---|
| (n) | % | (n) | % | (n) | % | |
| E-S, CD-S | 18 | 8.4 | 35 | 16.4 | 53 | 24.8 |
| E-R, CD-R (constitutive MLSB) | 51 | 23.8 | 9 | 4.2 | 60 | 28 |
| E-R, CD-S (inducible MLSB, D-positive) | 25 | 11.7 | 6 | 2.8 | 31 | 14.5 |
| E-R, CD-S (MS, D-negative) | 49 | 22.9 | 21 | 9.8 | 70 | 32.7 |
| Total | 143 | 66.8 | 71 | 33.2 | 214 | 100 |
MRCNS Methicillin resistant CNS, MSCNS Methicillin sensitive CNS, E Erythromycin, CD Clindamycin, R Resistant, S Sensitive, MLSB Macrolides, Lincosamides and Streptogramin B
Biofilm formation among CNS isolates
Biofilm production was assessed by both phenotypic and genotypic methods. The slime production ability of identified clinical CNS isolates was screened by CRA method. Among 214 CNS, 20 (9.3%) isolates produced black colonies indicating strong positive for polysaccharide production. The qualitative assessment of the amount of biofilm production was done by tube adherence method (TM) which showed strong production (+ + +) among 44 (20.6%) isolates and moderate biofilm production among 24 (11.2%) isolates. Biofilm production was determined quantitatively by tissue culture plate (TCP) method that demonstrated strong and moderate biofilm producers in 35 (16.4%) and 55 (25.7%) CNS isolates. Among all CNS species, S. epidermidis was the most frequent species to produce biofilm in all phenotypic methods. In addition, genotypic assay also revealed this species harboring both icaA and icaD genes more frequently. The ica genes were detected in 41 (19.2%) of all CNS isolates. Among all methods, TCP detected biofilm production in a greater number of isolates in all species (Table 4).
Table 4.
Determination of biofilm formation in CNS by genotypic and phenotypic methods
| Biofilm | CNS isolates | Total (n = 214) | ||||
|---|---|---|---|---|---|---|
| S. epidermidis | S. saprophyticus | S. haemolyticus | S. hominis | S. capitis | ||
| (n = 123) | (n = 40) | (n = 24) | (n = 15) | (n = 12) | ||
| Congo red agar (CRA) | ||||||
| Strong | 12 (9.8%) | 2 (5.0%) | 3 (12.5%) | 3 (20.0%) | - | 20 (9.3%) |
| Moderate | 12 (9.8%) | 2 (5.0%) | 5 (20.8%) | 2 (13.3%) | 2 (16.7%) | 23 (10.7%) |
| Weak/none | 99 (80.5%) | 36 (90.0%) | 16 (66.7%) | 10 (66.7%) | 10 (83.3%) | 171 (79.9%) |
| Tube adherence method (TM) | ||||||
| Strong | 33 (26.8%) | 4 (10.0%) | 2 (8.33%) | 3 (20.0%) | 2 (16.7%) | 44 (20.6%) |
| Moderate | 9 (7.3%) | 6 (15.0%) | 6 (25.0%) | 3 (20.0%) | - | 24 (11.2%) |
| Weak/none | 81 (65.9%) | 30 (75.0%) | 16 (66.7%) | 9 (60.0%) | 10 (83.3%) | 146 (68.2%) |
| Tissue culture plate (TCP) | ||||||
| Strong | 23 (18.7%) | 8 (20.0%) | 3 (12.5%) | 1 (6.7%) | - | 35 (16.4%) |
| Moderate | 32 (26.0%) | 9 (22.5%) | 4 (16.7%) | 7 (46.7%) | 3 (25.0%) | 55 (25.7%) |
| Weak/none | 68 (55.3%) | 23 (57.5%) | 17 (70.8%) | 7 (46.7%) | 9 (75.0%) | 124 (57.9%) |
| ica gene | ||||||
| Present | 29 (23.6%) | 5 (12.5%) | 2 (8.33%) | 3 (20.0%) | 2 (16.7%) | 41 (19.2%) |
| Absent | 94 (76.4%) | 35 (87.5%) | 22 (91.7%) | 12 (80.0%) | 10 (83.3%) | 173 (80.8%) |
Determination of biofilm formation among methicillin resistant CNS isolates
The biofilm production as detected by different phenotypic methods were higher in methicillin resistant than methicillin sensitive isolates but was not found to be statistically significant. Similarly, the ica genes were also found in higher number among methicillin resistant CNS than methicillin sensitive CNS but it was not found to be statistically significant (Table 5).
Table 5.
Biofilm formation in CNS in relation to methicillin susceptibility
| Biofilm | MRCNS | MSCNS | p-value |
|---|---|---|---|
| Congo red agar (CRA) | 0.322 | ||
| Present | 26 (18.2%) | 17 (23.9%) | |
| Absent | 117 (81.8%) | 54 (76.1%) | |
| Tube adherence method (TM) | 0.447 | ||
| Present | 43 (30.1%) | 25 (35.2%) | |
| Absent | 100 (69.9%) | 46 (64.8%) | |
| Tissue culture plate (TCP) | 0.529 | ||
| Present | 58 (40.6%) | 32 (45.1%) | |
| Absent | 85 (59.4%) | 39 (54.9%) | |
| ica genes | 0.824 | ||
| Present | 28 (19.6%) | 13 (18.3%) | |
| Absent | 115 (80.4%) | 58 (81.7%) |
MRCNS Methicillin resistant CNS, MSCNS Methicillin sensitive CNS
The icaAD genes are present in all clinical sample type
The study revealed that 14 (17.07%) CNS isolates from blood samples (n = 82) and 14 (25%) from wound/pus samples (n = 56) harbored ica genes. Likewise, ica genes were also present in six (30%) biomaterial isolates collected from CVC (n = 20), four (16%) from catheters (n = 23), and one each from DJ stenting (n = 2), ET tip (n = 1), and urine samples (n = 24) (Fig. 2). The identification of icaAD genes was done by PCR of DNA extracted from the CNS isolates using the primer listed in method section (Fig. 3).
Fig. 2.
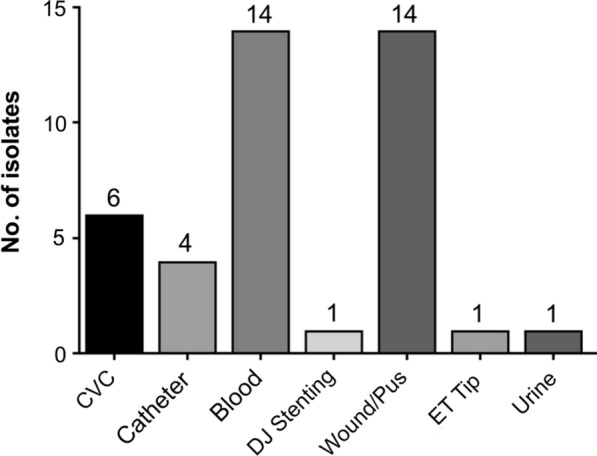
Distribution of CNS isolates possessing ica genes in different clinical specimens
Fig. 3.
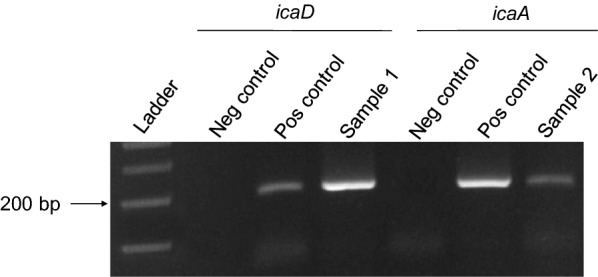
Representative picture of PCR amplification of icaA (188 bp) and icaD (198 bp) genes in CNS isolates
Antibiotic susceptibility pattern of isolates that harbor ica genes
The antibiotic susceptibility patterns of the isolates harboring ica genes were determined. The study showed no significant difference in antibiotic susceptibility between icaAD positive and icaAD negative isolates. Methicillin resistance was observed more among the isolates harboring ica genes than those lacking it. Altogether, the result shows that not only biofilm formation but other factors are responsible for antibiotic resistance among CNS (Table 6).
Table 6.
Antibiotic susceptibility pattern of ica positive isolates
| Antibiotics | ica genes | p-value | |||
|---|---|---|---|---|---|
| Present | Absent | ||||
| Susceptible | Resistant | Susceptible | Resistant | ||
| Penicillin | 4 (1.9%) | 37 (17.3%) | 14 (6.5%) | 159 (74.3%) | 0.119 |
| Ciprofloxacin | 28 (13.1%) | 13 (6.1%) | 110 (51.4%) | 63 (29.4%) | 0.334 |
| Tetracycline | 38 (17.8%) | 3 (1.4%) | 149 (69.6%) | 24 (11.2%) | 6.590 |
| Clindamycin | 32 (15.0%) | 9 (4.2%) | 119 (55.6%) | 54 (25.2%) | 3.332 |
| Chloramphenicol | 38 (17.8%) | 3 (1.4%) | 157 (73.4%) | 16 (7.5%) | 0.231 |
| Cefoxitin | 13 (6.1%) | 28 (13.1%) | 58 (27.1%) | 115 (53.7%) | 0.049 |
| Erythromycin | 15 (7.0%) | 26 (12.1%) | 44 (20.6%) | 129 (60.3%) | 2.256 |
| Cotrimoxazole | 29 (13.6%) | 12 (5.6%) | 105 (49.1%) | 68 (31.8%) | 1.460 |
| Gentamicin | 35 (16.4%) | 6 (2.8%) | 134 (62.6%) | 39 (18.2%) | 1.464 |
Discussion
The coagulase-negative staphylococci (CNS) including S. epidermidis are ubiquitous in nature; reside on the skin of healthy individuals as normal flora. In fact, due to this phenomenon, CNS has been emerged as common nosocomial pathogens. In addition, the ability to form biofilms on biotic as well as abiotic surfaces have made them successful human pathogens causing persistent infections leading to serious health problems. Given that biofilm is an important virulence factor that is mostly associated with antibiotic resistance for these pathogens, early detection in clinical specimen would impose significant impact in management of staphylococcal nosocomial infections. Taking this into consideration, the study was carried out to investigate the prevalence of biofilm producing CNS and antibiotic resistant strains in different clinical samples collected from two tertiary care hospitals of Nepal.
Among 214 CNS, S. epidermidis was the most common isolate accounting for more than half (57.4%) of total numbers (n = 123) followed by S. saprophyticus (n = 40, 18.6%), S. haemolyticus (n = 24, 11.2%), S. hominis (n = 15, 7%) and S. capitis (n = 12, 5.6%) (Table 1). Previous studies have also reported S. epidermidis as the most common isolates among CNS [24, 32–34]. While S. epidermidis was isolated form all clinical samples, it was more commonly isolated from blood, wound/pus. S. saprophyticus along with blood and wound/pus was also isolated from urine. The high number of S. saprophyticus in urine is consistent with the previous studies [24, 35] may be due to its adhering capacity to wall of urinary tract. Importantly, all CNS species were isolated from different medical devices indicating their ability to cause device associated infections.
Antibiotic resistance is a major human health problem. The present result also demonstrated that majority of isolates were 91.6% and 72.4% resistant to penicillin and erythromycin respectively. However, the study also demonstrated majority of isolates being susceptible to chloramphenicol (91.1%) and tetracycline (87.4%). Such high frequency of susceptibility pattern of chloramphenicol is consistent with the previous study by [24, 33, 36, 37], but their study showed lower susceptibility in case of erythromycin and penicillin. Indeed, similar susceptibility profile for all CNS strains was found as that of S. epidermidis strains. This indicates CNS are still susceptible to the first line drug and being cheap, these antibiotics could be used for the treatment of CNS infections caused by CNS in a resource limiting country like Nepal.
Among all CNS isolates, constitutive MLSB were present among 51 (23.8%) isolates and inducible MLSB among 25 (11.7%) isolates. While similar frequency of inducible MLSB was demonstrated by Perez et al., [38] as high as 50% CNS were found to be positive in study by Schreckenberger et al., [14]. These results indicate that constitutive and inducible MLSB resistance is dependent on the hospitals and the geographical area. Nonetheless, chances of inducible MLSB can be reduced by implementing reliable diagnostics of MLSB in the clinical samples.
Biofilm formation remains the most important mechanism of pathogenicity among staphylococci [10]. Although CRA and TM methods detected the biofilm production in a smaller number of isolates, TCP method was able to detect biofilm production in 90 (42.1%) isolates. Similar frequency of biofilm production in clinical CNS isolates was reported previously [39–41]. However, the higher frequency (65.38%) of biofilm production was reported by Shrestha et al., [23] in a study, conducted in tertiary care hospital of eastern Nepal as well as the study by Oliveira et al., [42], who reported 75% of CNS producing biofilm. Likewise, PCR amplification in our study revealed only 41 (19.2%) isolates possessing both icaA and icaD genes. This rate is lower as compared to the previous studies [33, 40]. The possible reasons are the various factors such as environment, nutrition, sub-inhibitory concentration of certain antibiotics, and stress (temperature, osmolarity) might play a significant role in biofilm formation resulting in varied frequency of biofilm producers among clinical isolates [43–47].
Among 41 icaAD positive isolates, only 7 (17%) showed the positive result in CRA method. This is in contrast to the previous studies by Zhou et al., [48] who reported all icaAD positive isolates also produced black colonies on CRA method. Similarly, good correspondence between possession of icaAD genes and CRA positivity was reported by de Silva et al., [2]. Our result shows that not all biofilm producers as per phenotypic assays possessed ica genes. Several previous studies have reported this phenomenon [49–52]. We speculated that while most of the staphylococcal isolates which form biofilm are dependent on the icaAD genes, other factors such as teichoic acids also contribute to form biofilm [33]. It may also be possible that inactivation of ica operon occurs due to the insertion of the IS256 insertion sequence. Similarly, the ica-negative, non-slime-producing isolates likely represent strains with alternate means of adhesion, such as microbial surface components recognizing adhesive matrix molecules (MSCRAMMs). These findings reinforce the opinion that several mechanisms besides slime production are responsible for bacterial adhesion and hence biofilm production [43].
Our study revealed ica genes were present mostly among blood and wound/pus samples (14 isolates each). Our results are consistent with the study previously performed [40] where quarter of the isolates from blood cultures and catheter tips produced biofilm. However, in contrast, we detected lesser frequency of biofilm production in the biomaterial sample like CVC, catheter, DJ stenting and ET tip. Interestingly, we did not observe significant difference of antibiotic resistance among biofilm producers and non-producers. Similar results were also reported previously [53] with no difference in antimicrobial resistance between biofilm-producing and non-producing S. epidermidis. Nonetheless, altogether, the percentage of biofilm production was higher among S. epidermidis than in other CNS isolates indicating that biofilm production is an important virulence factor for the pathogenicity of S. epidermidis. However, the results also support the idea that neither biofilm nor the icaAD genes could alone be used as biomarker of clinical significance, as suggested previously [2].
Conclusion
This study identified S. epidermidis as the most frequent species (57.5%) with the highest rate of biofilm production in all CNS examined. Among all phenotypic methods, TCP method detected biofilm production in higher percentage of CNS. We observed high prevalence of methicillin resistance as well as the presence of both constitutive and inducible MLSB phenotype in these clinical CNS isolates. However, the result showed no significant difference in the prevalence of antibiotic resistance between biofilm producers and non-producers CNS.
Acknowledgements
Authors would like to thank B & B hospital and KIST teaching hospital for allowing collecting the samples. We also highly appreciate National College for providing laboratory facilities to perform the experiments.
Abbreviations
- AST
Antibiotic susceptibility test
- BA
Blood agar
- BHI
Brain heart infusion
- CLSI
Clinical and Laboratory Standards Institute
- cMLSB
Constitutive macrolide, lincosamide, streptogramin B
- CNS
Coagulase-negative staphylococci
- CRA
Congo red agar
- CVC
Central venous catheter
- DJ stent
Double J stent
- D-test
Constitutive and inducible clindamycin resistance test
- iMLSB
Inducible macrolide, lincosamide, streptogramin B
- MHA
Mueller Hinton agar
- MLSB
Macrolide, lincosamide, streptogramin B
- MRCNS
Methicillin resistant coagulase-negative staphylococci
- MSA
Mannitol salt agar
- MSCNS
Methicillin sensitive coagulase-negative staphylococci
- MSCRAMMs
Microbial surface components recognizing adhesive matrix molecules
- PBS
Phosphate buffer saline
- PCR
Polymerase chain reaction
- PIA
Polysaccharide intercellular adhesion
- TCP
Tissue culture plate
- TM
Tube adherence method
- TSB
Tryptic soy broth
Authors' contributions
SM and NS conceived the study. SM carried out experiments. AS, AV and SM analyzed data. SM, NS, AS and SP drafted the manuscript and all authors contributed to preparing the final version of the manuscript. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
The ethical clearance and consent to participate was approved by Nepal Health Research Council (Reg. no. 213/2015) and Institutional Review Committee (IRC) of respective hospitals. The consent to participate was not required as all the investigated isolates were taken as part of standard routine diagnostic purposes.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sarita Manandhar, Email: sarita.manandhar@trc.tu.edu.np.
Neeraj Shrivastava, Email: nsrivastava1@amity.edu.
References
- 1.de Araujo GL, Coelho LR, de Carvalho CB, Maciel RM, Coronado AZ, Rozenbaum R, Ferreira-Carvalho BT, Figueiredo AM, Teixeira LA. Commensal isolates of methicillin-resistant Staphylococcus epidermidis are also well equipped to produce biofilm on polystyrene surfaces. J Antimicrob Chemother. 2006;57(5):855–864. doi: 10.1093/jac/dkl071. [DOI] [PubMed] [Google Scholar]
- 2.de Silva GD, Kantzanou M, Justice A, Massey RC, Wilkinson AR, Day NP, Peacock SJ. The ica operon and biofilm production in coagulase-negative staphylococci associated with carriage and disease in a neonatal intensive care unit. J Clin Microbiol. 2002;40(2):382–388. doi: 10.1128/JCM.40.02.382-388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arciola CR, Gamberini S, Campoccia D, Visai L, Speziale P, Baldassarri L, Montanaro L. A multiplex PCR method for the detection of all five individual genes of ica locus in Staphylococcus epidermidis. A survey on 400 clinical isolates from prosthesis-associated infections. J Biomed Mater Res A. 2005;75(2):408–413. doi: 10.1002/jbm.a.30445. [DOI] [PubMed] [Google Scholar]
- 4.von Eiff C, Peters G, Heilmann C. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect Dis. 2002;2(11):677–685. doi: 10.1016/S1473-3099(02)00438-3. [DOI] [PubMed] [Google Scholar]
- 5.Otto M. Staphylococcus epidermidis–the 'accidental' pathogen. Nat Rev Microbiol. 2009;7(8):555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziebuhr W, Hennig S, Eckart M, Kranzler H, Batzilla C, Kozitskaya S. Nosocomial infections by Staphylococcus epidermidis: how a commensal bacterium turns into a pathogen. Int J Antimicrob Agents. 2006;28(Suppl 1):S14–20. doi: 10.1016/j.ijantimicag.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 7.Mertens A, Ghebremedhin B. Genetic determinants and biofilm formation of clinical Staphylococcus epidermidis isolates from blood cultures and indwelling devises. Eur J Microbiol Immunol. 2013;3(2):111–119. doi: 10.1556/EuJMI.3.2013.2.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kloos WE, Bannerman TL. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7(1):117–140. doi: 10.1128/CMR.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mack D, Riedewald J, Rohde H, Magnus T, Feucht HH, Elsner HA, Laufs R, Rupp ME. Essential functional role of the polysaccharide intercellular adhesin of Staphylococcus epidermidis in hemagglutination. Infect Immun. 1999;67(2):1004–1008. doi: 10.1128/IAI.67.2.1004-1008.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15(2):167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arciola CR, Baldassarri L, Montanaro L. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J Clin Microbiol. 2001;39(6):2151–2156. doi: 10.1128/JCM.39.6.2151-2156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Eiff C, Jansen B, Kohnen W, Becker K. Infections associated with medical devices: pathogenesis, management and prophylaxis. Drugs. 2005;65(2):179–214. doi: 10.2165/00003495-200565020-00003. [DOI] [PubMed] [Google Scholar]
- 13.Sasirekha B, Usha MS, Amruta JA, Ankit S, Brinda N, Divya R. Incidence of constitutive and inducible clindamycin resistance among hospital-associated Staphylococcus aureus. Biotech. 2014;4(1):85–89. doi: 10.1007/s13205-013-0133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreckenberger PC, Ilendo E, Ristow KL. Incidence of constitutive and inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci in a community and a tertiary care hospital. J Clin Microbiol. 2004;42(6):2777–2779. doi: 10.1128/JCM.42.6.2777-2779.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belbase A, Pant ND, Nepal K, Neupane B, Baidhya R, Baidya R, Lekhak B. Antibiotic resistance and biofilm production among the strains of Staphylococcus aureus isolated from pus/wound swab samples in a tertiary care hospital in Nepal. Ann Clin Microbiol Antimicrob. 2017;16(1):15. doi: 10.1186/s12941-017-0194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adhikari RP, Shrestha S, Barakoti A, Amatya R. Inducible clindamycin and methicillin resistant Staphylococcus aureus in a tertiary care hospital, Kathmandu Nepal. BMC Infect Dis. 2017;17(1):483. doi: 10.1186/s12879-017-2584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ansari S, Nepal HP, Gautam R, Rayamajhi N, Shrestha S, Upadhyay G, Acharya A, Chapagain ML. Threat of drug resistant Staphylococcus aureus to health in Nepal. BMC Infect Dis. 2014;14:157. doi: 10.1186/1471-2334-14-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhatta DR, Cavaco LM, Nath G, Kumar K, Gaur A, Gokhale S, Bhatta DR. Association of Panton Valentine Leukocidin (PVL) genes with methicillin resistant Staphylococcus aureus (MRSA) in Western Nepal: a matter of concern for community infections (a hospital based prospective study) BMC Infect Dis. 2016;16:199. doi: 10.1186/s12879-016-1531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khatri S, Pant ND, Bhandari R, Shrestha KL, Shrestha CD, Adhikari N, Poudel A. Nasal carriage rate of methicillin resistant Staphylococcus aureus among health care workers at a tertiary care hospital in Kathmandu Nepal. J Nepal Health Res Council. 2017;15(35):26–30. doi: 10.3126/jnhrc.v15i1.18009. [DOI] [PubMed] [Google Scholar]
- 20.Khanal A. Khanal A, Gaire A, Khanal A, Estrada R, Ghimire R, Panthee S: Methicillin-resistant Staphylococcus aureus in Nepal: a systematic review and meta-analysis. Int J Infect Dis. 2020;2020(103):48–55. doi: 10.1016/j.ijid.2020.11.152. [DOI] [PubMed] [Google Scholar]
- 21.Manandhar S, Singh A, Varma A, Pandey S, Shrivastava N. Evaluation of methods to detect in vitro biofilm formation by staphylococcal clinical isolates. BMC Res Notes. 2018;11(1):714. doi: 10.1186/s13104-018-3820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manandhar S, Singh A, Varma A, Pandey S, Shrivastava N. Biofilm producing clinical Staphylococcus aureus isolates augmented prevalence of antibiotic resistant cases in tertiary care hospitals of Nepal. Front Microbiol. 2018;9:2749. doi: 10.3389/fmicb.2018.02749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shrestha LB, Bhattarai NR, Khanal B. Antibiotic resistance and biofilm formation among coagulase-negative staphylococci isolated from clinical samples at a tertiary care hospital of eastern Nepal. Antimicrob Resist Infect Control. 2017;6:89. doi: 10.1186/s13756-017-0251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shrestha LB, Bhattarai NR, Rai K, Khanal B. Antibiotic resistance and mecA gene characterization of coagulase-negative staphylococci isolated from clinical samples in Nepal. Infect Drug Resist. 2020;13:3163–3169. doi: 10.2147/IDR.S274163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheesebrough M: District laboratory practice in tropical countries. Cambridge: Cambridge University Press; 2012, 2nd Edition. pp 116–123.
- 26.Cunha Mde L, Sinzato YK, Silveira LV. Comparison of methods for the identification of coagulase-negative staphylococci. Mem Inst Oswaldo Cruz. 2004;99(8):855–860. doi: 10.1590/S0074-02762004000800012. [DOI] [PubMed] [Google Scholar]
- 27.Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol. 1989;42(8):872–874. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22(6):996–1006. doi: 10.1128/JCM.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clinical laboratory and Standard Institutions: Performance standards for Antimicrobial Susceptibility Testing. CLSI Document M100-S25 2015, Twenty-Fifth International Supplement.
- 30.Steward CD, Raney PM, Morrell AK, Williams PP, McDougal LK, Jevitt L, McGowan JE, Jr, Tenover FC. Testing for induction of clindamycin resistance in erythromycin-resistant isolates of Staphylococcus aureus. J Clin Microbiol. 2005;43(4):1716–1721. doi: 10.1128/JCM.43.4.1716-1721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiebelkorn KR, Crawford SA, McElmeel ML, Jorgensen JH. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J Clin Microbiol. 2003;41(10):4740–4744. doi: 10.1128/JCM.41.10.4740-4744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreira RB, Iorio NL, Malvar KL, Nunes AP, Fonseca LS, Bastos CC, Santos KR. Coagulase-negative staphylococci: comparison of phenotypic and genotypic oxacillin susceptibility tests and evaluation of the agar screening test by using different concentrations of oxacillin. J Clin Microbiol. 2003;41(8):3609–3614. doi: 10.1128/JCM.41.8.3609-3614.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seng R, Kitti T, Thummeepak R, Kongthai P, Leungtongkam U, Wannalerdsakun S, Sitthisak S. Biofilm formation of methicillin-resistant coagulase negative staphylococci (MR-CoNS) isolated from community and hospital environments. PLoS ONE. 2017;12(8):e0184172. doi: 10.1371/journal.pone.0184172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El MF, MOufti MB, Reem Abdel Hammed Harfoush, and Hoda Mohamed Aly Owais Characterization of some genotypic and phenotypic traits of biofilm producing clinical isolates of methicillin resistant Staphylococcus Epidermidis. Am J Infect Dis Microbiol. 2015;3(3):7. [Google Scholar]
- 35.Barbier F, Ruppe E, Hernandez D, Lebeaux D, Francois P, Felix B, Desprez A, Maiga A, Woerther PL, Gaillard K, et al. Methicillin-resistant coagulase-negative staphylococci in the community: high homology of SCCmec IVa between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2010;202(2):270–281. doi: 10.1086/653483. [DOI] [PubMed] [Google Scholar]
- 36.Wojtyczka RD, Orlewska K, Kepa M, Idzik D, Dziedzic A, Mularz T, Krawczyk M, Miklasinska M, Wasik TJ. Biofilm formation and antimicrobial susceptibility of Staphylococcus epidermidis strains from a hospital environment. Int J Environ Res Public Health. 2014;11(5):4619–4633. doi: 10.3390/ijerph110504619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Z, Mkrtchyan HV, Cutler RR. Antibiotic resistance and mecA characterization of coagulase-negative staphylococci isolated from three hotels in London UK. Front Microbiol. 2015;6:947. doi: 10.3389/fmicb.2015.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez J, Limansky A, Toresani I, Ebner G, Di Bartolomeo S, de Inocenti I, Pretto G, Salazar N, Laferrara M, Bottiglieri M, et al. Distribution of capsular types and antimicrobial susceptibility of Streptococcus agalactiae causing infections in Argentina] Rev Argent Microbiol. 2004;36(2):63–67. [PubMed] [Google Scholar]
- 39.Martini R, Horner R, Rampelotto RF, Garzon LR, Nunes MS, Teixeira MD, Graichen DA. Investigation of Biofilm Formation in Coagulase-Negative Staphylococci Isolated from Platelet Concentrate Bags. Rev Inst Med Trop Sao Paulo. 2016;58:1. doi: 10.1590/S1678-9946201658001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prasad S, Nayak N, Satpathy G, Nag HL, Venkatesh P, Ramakrishnan S, Ghose S, Nag TC. Molecular & phenotypic characterization of Staphylococcus epidermidis in implant related infections. Indian J Med Res. 2012;136(3):483–490. [PMC free article] [PubMed] [Google Scholar]
- 41.Arciola CR, Campoccia D, Baldassarri L, Donati ME, Pirini V, Gamberini S, Montanaro L. Detection of biofilm formation in Staphylococcus epidermidis from implant infections. Comparison of a PCR-method that recognizes the presence of ica genes with two classic phenotypic methods. J Biomed Mater Res A. 2006;76(2):425–430. doi: 10.1002/jbm.a.30552. [DOI] [PubMed] [Google Scholar]
- 42.Oliveira A, Cunha Mde L. Comparison of methods for the detection of biofilm production in coagulase-negative staphylococci. BMC Res Notes. 2010;3:260. doi: 10.1186/1756-0500-3-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arciola CR, Campoccia D, Ravaioli S, Montanaro L. Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Front Cell Infect Microbiol. 2015;5:7. doi: 10.3389/fcimb.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirani ZA, Aziz M, Khan MN, Lal I, Hassan NU, Khan SI. Biofilm formation and dispersal of Staphylococcus aureus under the influence of oxacillin. Microb Pathog. 2013;61–62:66–72. doi: 10.1016/j.micpath.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Dobinsky S, Kiel K, Rohde H, Bartscht K, Knobloch JK, Horstkotte MA, Mack D. Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J Bacteriol. 2003;185(9):2879–2886. doi: 10.1128/JB.185.9.2879-2886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foulston L, Elsholz AK, DeFrancesco AS, Losick R. The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. mBio. 2014;5(5):e01667–01614. doi: 10.1128/mBio.01667-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moormeier DE, Bose JL, Horswill AR, Bayles KW. Temporal and stochastic control of Staphylococcus aureus biofilm development. mBio. 2014;5(5):e01341–01314. doi: 10.1128/mBio.01341-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou S, Chao X, Fei M, Dai Y, Liu B. Analysis of Staphylococcus Epidermidis icaA and icaD genes by polymerase chain reaction and slime production: a case control study. BMC Infect Dis. 2013;13:242. doi: 10.1186/1471-2334-13-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin Z, Yang X, Yang L, Jiang J, Ou Y, Molin S, Qu D. Formation and properties of in vitro biofilms of ica-negative Staphylococcus epidermidis clinical isolates. J Med Microbiol. 2007;56(Pt 1):83–93. doi: 10.1099/jmm.0.46799-0. [DOI] [PubMed] [Google Scholar]
- 50.Nasr RA. Biofilm formation and presence of icaAD gene in clinical isolates of staphylococci. Egypt J Med Human Genet. 2012;13(3):6. doi: 10.1016/j.ejmhg.2012.04.007. [DOI] [Google Scholar]
- 51.Chaieb K, Mahdouani K, Bakhrouf A. Detection of icaA and icaD loci by polymerase chain reaction and biofilm formation by Staphylococcus epidermidis isolated from dialysate and needles in a dialysis unit. J Hosp Infect. 2005;61(3):225–230. doi: 10.1016/j.jhin.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 52.Kogan G, Sadovskaya I, Chaignon P, Chokr A, Jabbouri S. Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol Lett. 2006;255(1):11–16. doi: 10.1111/j.1574-6968.2005.00043.x. [DOI] [PubMed] [Google Scholar]
- 53.Klingenberg C, Aarag E, Ronnestad A, Sollid JE, Abrahamsen TG, Kjeldsen G, Flaegstad T. Coagulase-negative staphylococcal sepsis in neonates. Association between antibiotic resistance, biofilm formation and the host inflammatory response. Pediatr Infect Dis J. 2005;24(9):817–822. doi: 10.1097/01.inf.0000176735.20008.cd. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


